Abstract
Visualization of signal transduction within live primary cilia constitutes a technical challenge due to its sub-micron dimensions and close proximity to the cell body. Using a genetically encoded calcium indicator targeted to primary cilia we visualized calcium signaling in cilia of mouse fibroblasts and kidney cells upon chemical or mechanical stimulation with high specificity, sensitivity and wide dynamic range.
The vertebrate primary cilium is a solitary hair-like structure that protrudes from the plasma membrane into the extracellular space, and functions as a sensory organelle for detecting diverse chemical1–3 and mechanical4,5 stimuli. Each cilium measures 0.2 μm in diameter and a few micrometers in length, and takes up only 1/10,000th of the total cell volume. Visualization of ciliary signal transduction evoked by extracellular stimuli has faced a technical challenge due to the organelle’s sub-micron size as well as the inability to dissect cilia specific signals from the vast activity volume in the main cell body. Emerging evidence supports the role of calcium ions (Ca2+) as a principal secondary messenger of ciliary signaling pathways. Ca2+ permeable channels of the transient receptor potential (TRP) channel family localize to primary cilia, and Ca2+ fluxes through these ion channels have been proposed to play major roles in mechanical, thermal and G protein-coupled receptor signal transduction6–9. We therefore set forth to monitor Ca2+ signaling activities within the primary cilium. An electrophysiological method to measure signals within primary cilia has been previously reported10, but the sub-micron size of this organelle makes patch-clamping the ciliary membrane difficult. Furthermore, this technique would not facilitate the simultaneous monitoring of multiple primary cilia. Synthetic Ca2+ indicator dyes offer the advantage of monitoring multiple cells, but often result in signal saturation of the entire cytosol that overwhelms local transient Ca2+ fluxes in specific subcellular compartments (Supplementary Fig. 1).
Here, we devise a strategy to specifically target genetically encoded Ca2+ indicators (GECIs) into primary cilia to distinguish cilia-specific Ca2+ signaling from that of the main cell body. We first evaluated a collection of ciliary targeting sequences (CTSs)11, which included two truncated peptides derived from the cytoplasmic tail of fibrocystin (CTS20 and CTS68), full-length 5-hydroxytriptamine (serotonin) receptor isoform 6 (5HT6), a fusion peptide consisting of the transmembrane domain of integrin β1 and the C terminus domain of Arl13b (Integrin-Arl13b; IA), as well as a 5HT6-CTS20 combination. Each CTS was tagged with GFP and evaluated for targeting efficiencies and effects on cilia morphology (Supplementary Fig. 2 and 3). 5HT6-GFP and IA-GFP demonstrated high cilia targeting efficiencies of 87% (131/150) and 85% (146/172), respectively (Supplementary Fig. 2a and 3e,g). None of the CTSs tested had an obvious effect on ciliation frequency, indicating that the overexpression of CTS sequences in cells did not adversely affect ciliogenesis (Supplementary Fig. 2b). Expression of 5HT6-GFP and IA-GFP however caused the average length of primary cilia to increase approximately two-fold, (Supplementary Fig. 2c). The elongated cilia length was in turn correlated with a higher frequency of morphological deformations (Supplementary Fig. 2d and 4), Nevertheless, the bulk of primary cilia expressing these two constructs exhibited regular morphology. This is further supported by transmission electron microscopy and immunofluorescence studies of 5HT6-expressing primary cilia, which demonstrated no obvious defects in its ultrastructure, as well as normal localization of key ciliary proteins (Supplementary Fig. 5,6).
We next constructed a collection of primary cilia-targeted GECI by fusing these CTS sequences with currently available GECIs, including intramolecular CFP and YFP fluorescence resonance energy transfer (FRET) indicators TNXXL12 and YC3.6013, as well as single fluorescence GFP indicators G-CaMP5G14 and G-GECO1.015 (see Supplementary Table 1 for a full list of generated constructs). We characterized the cilia targeting efficiency and signal dynamic range of each CTS-tagged GECI (Supplementary Fig. 7–9). 5HT6-G-GECO1.0 (Fig. 1a) demonstrated the greatest potential as a cilia-specific Ca2+ indicator in terms of functionality and targeting efficiency and is comparable with 5HT6-GFP in cilia targeting efficiency, ciliation efficiency and effects on ciliary structure (Fig. 1b, Supplementary Fig. 2–4). At basal state, 5HT6-G-GECO1.0 displayed weak GFP fluorescence within primary cilia, but exhibited an increase of 360.0% +/− 62.1% (SEM) in GFP fluorescence when stimulated with 2 μM ionomycin (Fig. 1c, Supplementary Fig. 9a, Supplementary Table 1, and Supplementary Video 1). All further experiments were conducted with 5HT6-G-GECO1.0.
Figure 1.
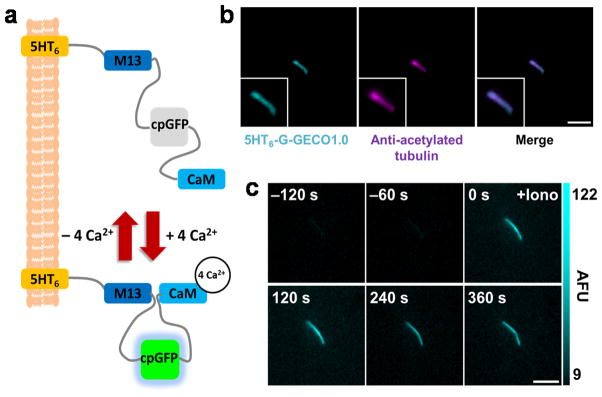
5HT6-G-GECO1.0 targets primary cilia and detects changes in ciliary Ca2+. (a) Schematic of 5HT6-G-GECO1.0. G-GECO1.0 contains M13 (a skeletal muscle light-chain kinase), a circularly permuted GFP (cpGFP), and Calmodulin (CaM). (b) A primary cilium from a NIH-3T3 cell expressing 5HT6-G-GECO1.0 stained with antibody against acetylated α-tubulin. Bar represents 3 μm. (c) Time-lapse imaging of a NIH-3T3 primary cilium expressing 5HT6-G-GECO1.0 treated with ionomycin (Iono). AFU stands for arbitrary fluorescence unit. Bar represents 5 μm.
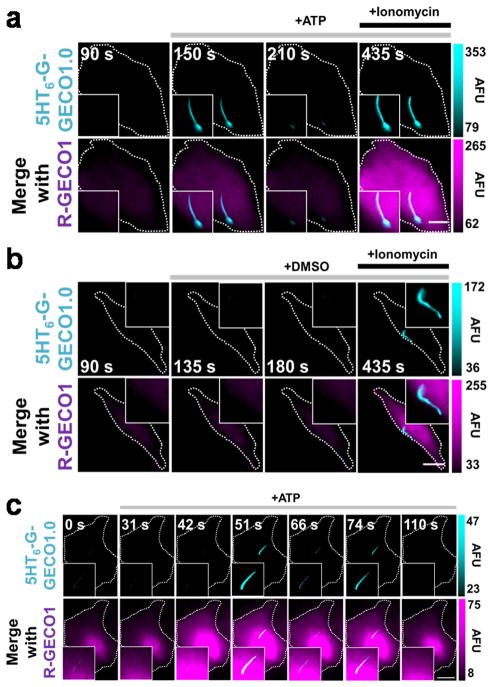
Adenosine triphosphate (ATP) triggers increases in cellular calcium levels via the activation of ATP-gated P2X calcium-permeable channels and/or G protein-coupled P2Y receptors which induce the mobilization of intracellular Ca2+ from inositol 1,4,5-trisphosphate-sensitive stores.. We therefore investigated whether ciliary Ca2+ changes could be detected in response to ATP stimulation. Through the co-expression of cytosolic R-GECO1 (a single red fluorescent GECI)15 and cilia-targeted 5HT6-G-GECO1.0 in NIH-3T3 cells, we detected a pronounced rise in cytosolic Ca2+ that was accompanied by a comparable increase in ciliary Ca2+ in 52.2% (12/23) of cells stimulated by 10 μM ATP (Fig. 2a). An average maximum increase of 53.9% +/− 20.4% and 54.3% +/− 10.0% in fluorescence intensity was observed in the cilia and the cytosol, respectively (Supplementary Fig. 10a). In contrast, vehicle control did not generate any Ca2+ response in the cytosol and primary cilia (Fig 2b and Supplementary Fig. 10b). To determine the source of observed ciliary Ca2+ fluxes, we increased our imaging frequency from 0.067 to 0.63 Hz. We observed that spikes in cytosolic Ca2+ clearly preceded ciliary Ca2+ spikes in 100% (14/14) of cells (Fig 2c, Supplementary Fig. 10c, and Supplementary Video 2). The average delay time between the cytosolic and ciliary Ca2+ elevation was 6.04 +/− 0.98 seconds (n = 14). Consistent with this, whenever Ca2+ oscillations are detected in the cytosol, correlated but delayed calcium spikes were also observed to occur within the cilia. (Fig. 2c, Supplementary Fig. 10c, and Supplementary Video 2). Furthermore, ciliary Ca2+ fluxes propagated in a base-to-tip direction within the ciliary lumen in 100% (14/14) of ATP-induced ciliary Ca2+ spikes detected (Supplementary Fig. 11 and Supplementary Videos3). Collectively, these observations suggest that the ciliary Ca2+ flux originates from the calcium stores in the cytosol. However, we cannot exclude other possibilities such as a contribution of ATP-gated P2X calcium-permeable channels localized at the base of primary cilia. By further increasing the imaging frequency to 1.5 Hz, we calculated the linear rate of Ca2+ propagation along the ciliary shaft to be 0.83 +/− 0.22 μm/s in cilia where the Ca2+ propagated the entire length of the lumen (n = 11). Of note, these numbers could be at least partially affected by buffering effect from overexpressed 5HT6-G-GECO1.0. The expression level of biosensors in general needs to be carefully controlled not to affect endogenous buffering. Because GFP-based GECIs are sensitive to changes in pH16, we further confirmed that ciliary pH did not change with ATP by using a newly developed ciliary pH biosensor, 5HT6-CFP-Venus (H148G) (Supplementary Fig. 12).
Figure 2.
5HT6-G-GECO1.0 detects ciliary Ca2+ influxes in response to ATP. (a,b) Fluorescence microscopy images of NIH-3T3 cell expressed with the indicated sensors showing response to ATP (a) or DMSO (b). Bar represents 5 μm. Time lapse images here were captured at 0.067 Hz. (c) High speed time lapse imaging (0.63Hz) reveals oscillations in cytosolic and ciliary Ca2+ levels in response to 10 μM ATP in NIH-3T3 cells expressing indicated sensors. Bar indicates 10 μm. AFU, arbitrary fluorescence unit.
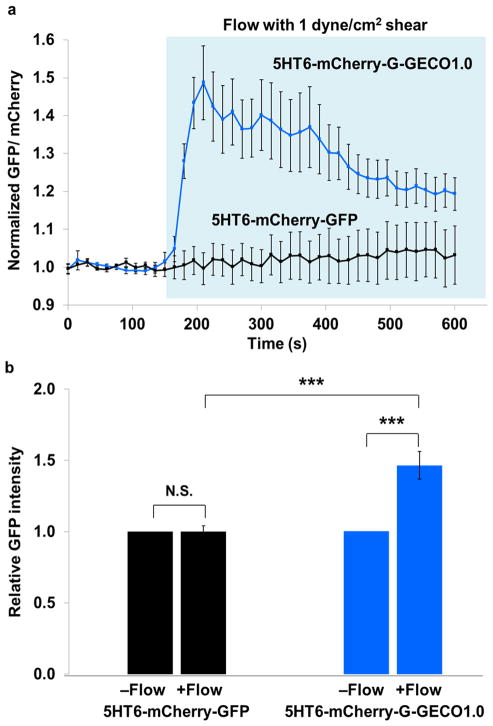
Finally, we asked whether we could detect Ca2+ dynamics within primary cilia when subjecting them to a mechanical stimulus. Fluid flow across the apical cell membrane induces the bending of the cilia, and the resultant shear force is commonly believed to activate cilia-localized Ca2+-permeable TRPP2 channels7. However, the critical step involving the entry of extracellular Ca2+ into the ciliary lumen has not been demonstrated. Therefore, we set up a fluid flow system to subject ciliated murine inner medullary collecting duct (mIMCD3) cells with defined laminar flow. In order to normalize for the anticipated flow-induced movements of cilia, cells were transiently expressed with 5HT6-mCherry-G-GECO1.0 such that cilia spatial movements could be visualized by the mCherry fluorescence marker (Supplementary Fig. 13a,b. Indeed, the initiation of fluid flow (with wall shear stress corresponding to a physiological value of 1 dyne/cm2) induced the immediate bending of cilia (Supplementary Fig. 13c and Supplementary Video 4), while specific bending behavior of each primary cilium was dependent on parameters such as spatial orientation and cilium length. Importantly, flow initiation also induced a pronounced increase in ciliary Ca2+ which initiated within 15 seconds of flow induction on average, and attained peak responses at 1-minute post flow induction (Fig. 3a, Supplementary Fig. 13c, and Supplementary Video 4). To validate that the observed changes in GFP fluorescence within primary cilia was indicative of genuine changes in GECI activity and not due to ciliary movement, we further compared results of 5HT6-mCherry-G-GECO1.0 with 5HT6-mCherry-GFP, which serves as a non-calcium responsive negative control. As expected, flow initiation did not induce a significant change in GFP fluorescence in 5HT6-mCherry-GFP expressing cilia (P = 0.9545), while cilia expressing 5HT6-mCherry-G-GECO1.0 exhibited an average 1.46-fold increase in GFP fluorescence 1-minute post flow induction (Fig. 3a,b and Supplementary Fig. 14). Further work is required to elucidate the source of Ca2+ signaling observed.
Figure 3.
Laminar fluid flow induces dynamic calcium signals in primary cilia (a) Fluorescence intensity of GFP divided by that of mCherry is indicated before and after flow administration. 5HT6-mCherry-G-GECO1.0 activity is shown in blue (18 primary cilia from 7 independent experiments) and 5HT6-mCherry-GFP (control) is in black (9 primary cilia from 3 independent experiments). Fluorescence signals have been normalized against baseline fluorescence before flow induction. Region highlighted in light blue indicates time points with flow. Error bars represent SEM. (b) Comparison of relative GFP intensities between 5HT6-mCherry-G-GECO1.0 and 5HT6-mCherry-GFP before and after one minute flow induction. In each case, the GFP intensity without flow has been normalized to a value of 1. Statistical analysis: 5HT6-mCherry-GFP −flow vs +flow (P = 0.9545); 5HT6-mCherry-G-GECO1.0 −flow vs +flow (P = 0.000153); 5HT6-mCherry-G-GECO1.0 +flow vs 5HT6-mCherry-GFP +flow (P = 0.000128).
The intermediate affinity of G-GECO1.0 for Ca2+ (Kd value of 749 nM)15 possesses sufficient sensitivity to detect changes in Ca2+ concentrations within primary cilia when cells are stimulated with ATP and mechanical flow. Nevertheless, other forms of signaling stimuli may induce lower concentration ranges of ciliary Ca2+ that may not be detectable by G-GECO1.0. For these applications, it may be necessary to target GECIs with lower Kd values to the primary cilia. In all, this visualization technique promises important insights into how sensory functions associated with primary cilia could be regulated by Ca2+ fluxes within the organelle. The successful application of the cilia-targeted GECI also serves as a proof-of-concept to extend this approach to visualize other signaling molecules within primary cilia.
Online Methods
DNA construction
Note: All DNA Plasmids are available through Addgene.
Construction of the β1Int-HaloTag-Arl13b/C-GFP (IA-GFP) expression vector
DNA encoding the human Arl13bC-terminal region (amino acids 355-428 of Arl13b)17 was amplified by PCR primers (5′-cccaagcttaggaaccaccgggtagaacc and 5′-aggtcgactgagatcacatcatgagcatca) and subcloned into pEGFP-C-CMV5 (a modified pCMV5 mammalian expression vector for C-terminal GFP-fusion protein). DNA encoding β1Int-HaloTag18 was then subcloned into pEGFP-C-CMV5/Arl13b[355-428] to make the β1Int-HaloTag-Arl13b/C-GFP expression vector. GenBank/EMBL/DDBJ accession numbers for Arl13b and β1 Integrin receptor are NP_001167621.1 and NM_002211, respectively.
Construction of the Lyn-GFP expression vector
GFP was subcloned into a Lyn-YFP (a gift from Marc Fivaz) in the Clontech pYFP-N1 vector using AgeI and BsrGI to replace YFP.
Construction of the 5HT6-GFP-CTS20 expression vector
DNA encoding 5HT6 flanked by AgeI was amplified by PCR primers (5′-ctactgaccggtcgccaccatggttccagagcccggccctgtcaacag and 5′-gctgacaccggtcctcctgcgctaccaccagcactgttcatgggggaaccaagtgg) from 5HT6-GFP19 (a gift from Akiko Seki and Tobias Meyer) in the Clontech pEGFP-N3 vector and then subcloned into a GFP-CTS2020 (a gift from Gregory Pazour) in the Clontech pEGFP-C2 vector at the AgeI site. CTS20 contains residues 1–20 of the N-terminal cytoplasmic tail of Fibrocystin. GenBank/EMBL/DDBJ accession numbers for 5HT6 and fibrocystinare NM_021358 and NM_153179.2, respectively.
Construction of the 5HT6-YC3.60 expression vector
DNA encoding 5HT6 flanked by HindIII was amplified by PCR primers (5′-catccgaagcttgccaccatggttccagagc and 5′-gcacctaagctttcctcctgcgcttcctcctgcgctctttgagattcgtcggaacacatgataatag) from 5HT6-GFP and then subcloned into a YC3.60 vector (a gift from Atsushi Miyawaki).
Construction of the 5HT6-G-GECO1.0 expression vector
DNA encoding 5HT6 flanked by BamHI was amplified by PCR primers (5′-cattcaggatccgccaccatggttccagagc and 5′-gcatctggatcctcctcctgcgctaccacca) from 5HT6-GFP and then subcloned into CMV-G-GECO1.0 vector (obtained from Addgene).
Construction of the 5HT6-G-CaMP expression vector
DNA encoding G-CaMP5G (a gift from Loren Looger) flanked with BamHI and HindIII was amplified by PCR primers (5′-ctactgggatccagtgctggtggtagcgcaggaggaatgggttctcatcatcatcatcatcatgg and 5′-gcaacatagttaagaataccagtcaatctttcac) and then subcloned into 5HT6-CFP-FKBP vector21 in replacement of CFP-FKBP.
Construction of the TNXXL-CTS20 expression vector
First, the stop codon was removed from the C terminus of the TNXXL vector (a gift from Oliver Griesbeck) by site directed mutagenesis (Stratagene) using PCR primers (5′-cgaggactacgaattctgcagatatccatcacactggcggcc and 5′-ggccgccagtgtgatggatatctgcagaattcgtagtcctcg). The resulting vector was digested with EcoRI and ligated to CTS20 digested with EcoRI from a GFP-CTS20 vector.
Construction of the TNXXL-CTS68 expression vector
The TNXXL vector with no stop codon was digested with EcoRI and then ligated to CTS68 digested with EcoRI from a GFP-CTS20 vector. CTS68 contains residues 1–68 of the N-terminal cytoplasmic tail of Fibrocystin.
Construction of the 5HT6-mCh-G-GECO1.0
G-GECO1.0 was digested from CMV-G-GECO1.0 vector using BamHI and EcoRI and subcloned into a 5HT6-mCherry (pmCherry-C1, Clontech) that had been digested with BglII and EcoRI.
Construction of the 5HT6-mCherry-GFP expression vector
GFP was digested from 5HT6-GFP (pEGFP-N3, Clontech) using Acc65I and BsrGI and subcloned into a 5HT6-mCherry (pmCherry-C1, Clontech) that had been digested with Acc65I.
Construction of the 5HT6-CFP-Venus(H148G) expression vector
5HT6-CFP was first constructed by subcloning 5HT6 into a CFP vector using NheI and AgeI. Venus(H148G) flanked with EcoRI and BamHI was then amplified by PCR primers (5′-catccggaattcgatggtgagcaagggcgagg and 5′-gcagtgggatccttacttgtacagctcgtccatgcc) and subcloned into the 5HT6-CFP vector.
Cell Culture and Transfection
NIH-3T3 cells and mIMCD3 cells containing an integrated FRT site in the genome (a kind gift from Randall Reed) were cultured in DMEM (Gibco) supplemented with 10% fetal bovine serum. For all transient transfections, cells were transfected with the respective DNA constructs by plating them directly in a transfection solution containing DNA plasmid and FuGENE HD (Roche). Cells are plated on poly-D-lysine-coated borosilicate glass Lab-Tek 8-well chambers (Thermo Scientific). Ciliogenesis was induced by serum starvation for 24 hours. For flow experiments, transfected cells were seeded into Microslide VI0.4 channels (ibidi) at a cell suspension density of approximately 1.3 x 106 cells/ml to achieve confluence.
Immunofluorescence
To mark primary cilia, NIH3T3 cells were fixed with 4% (w/v) paraformaldehyde, permeabilized with 0.1% (v/v) Triton X-100 and immunostained with mouse monoclonal anti-acetylated tubulin antibody (Sigma, T7451, 1:2000 dilution) and secondary anti-mouse antibody conjugated to Alexa Fluor 568 (Invitrogen, 1:1000 dilution). For immunostaining of IMCD3 cells, cells were grown on cover slips, transfected with 5HT6-YFP, cultured for 72h, washed with phosphate-buffered saline (PBS) and fixed with ice cold methanol at −20°C for 7 min. Fluorescent images were obtained using a LSM710 confocal microscope (Carl Zeiss) equipped with a Plan Apochromat ×100 oil immersion objective lens (NA 1.4) and processed using ImageJ software. The antibody list includes rabbit polyclonal antibodies against pericentrin (Babco, PRB-432C, 1:250 dilution), NPHP3 (Proteintech, 22026-1-AP, 1:1000 dilution), IFT88 (Proteintech, 13967-1-AP, 1:750 dilution), Arl13b (Proteintech, 17711-1-AP, 1:1000 dilution), Cep164 (Novus, 45330002, 1:4000 dilution), Cep290 (Bethyl Laboratories, A301-659A, 1:1000 dilution), and mouse monoclonal antibodies against acetylated α-tubulin (Sigma; 6-11B-1, T7451, 1:1000 dilution), γ-tubulin (Sigma; GTU-88, T-6557, 1:1000 dilution), and poly-Glu-tubulin (Enzo; GT335, ALX-804-885-C100, 1:1000 dilution). The secondary antibodies used in this study were Alexa Fluor 568-labelled anti-mouse IgG (Molecular Probes, 1:1500 dilution) and Alexa-Fluor 633-labelled anti-rabbit IgG (Molecular Probes, 1:2000 dilution).
Transmission electron microscopy (TEM)
For the ultrastructural analysis of cilia overexpressing 5HT6-GFP or GFP alone, these genes were introduced into NIH3T3 cells using a lentiviral expression system developed by Dr. Hiroyuki Miyoshi at the RIKEN BioResource Center22. For the expression of 5HT6-GFP, the cDNA was amplified with KOD DNA polymerase and ligated into the Eco47III site of CSII-CMV-MCS-IRES2-Bsd. For the expression of GFP alone, the CS-CDF-CG-PRE was used. These expression constructs were packaged into infectious viral particles22 and added to the NIH3T3 culture medium at the multiplicity of infection of >20. After the viral transduction, the cells expressing 5HT6-GFP were selected with 30 μM blasticidin. The expressions of 5HT6-GFP or GFP alone in the most if not all cells were confirmed by fluorescent microscopy. Preparations of the cells and observation of cilia by TEM were carried out principally according to the previous study with slight modifications23. Briefly, cultured cells were fixed with a half Karnovsky’s solution (2% paraformaldehyde, 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.5) supplemented with 1% tannic acid for 30 min at RT, followed by rinse with 10% sucrose in cacodylate buffer (pH 7.5) for three times. The cells were postfixed with 1% osmium tetroxide for 30 min on ice, followed by extensive irrigation with ice-cold distilled water. Subsequently, the cells were stained en bloc with 1% uranyl acetate in 50% ethanol for 2 hrs, dehydrated with a graded concentration of ethanol series, and embedded in epoxy resin. The cells in the epoxy block were cut by the LKB2088 ultramicrotome (Stockholm, Sweden), mounted onto formvar-reinforced single slot grids, and stained with uranyl acetate and lead citrate. The samples were observed under the Hitachi H-7500 transmission electron microscope (Tokyo, Japan). The images of ciliary cross sections were analyzed with ImageJ to measure the ciliary diameter.
Epi-fluorescence imaging
Most of the imaging experiments were performed on an Axiovert135TV epi-fluorescence microscope (Zeiss) with 63× oil objective (Zeiss), and images were collected by a QIClick charge-coupled device (CCD) camera (QImaging). For the dual-color epi-fluorescence imaging under flow conditions, IX-71 (Olympus) microscope was used together with a 40x oil objective (Olympus) and a CoolSNAP HQ CCD camera (Photometrics). Imaging was driven by Metamorph 7.5 imaging software (Molecular Devices). All calcium imaging experiments were performed in Dulbecco’s Phosphate-Buffered Saline (Gibco) containing 0.9 mM [Ca2+], except for the characterization of the cytosolic dynamic range of each CTS-GECI which was performed in DMEM with 25 mM HEPES (Gibco). All pH imaging experiments were performed in DMEM with 25 mM HEPES (Gibco). All imaging experiments were completed at room temperature (21–23 °C). FRET images were thresholded to remove background prior to any contrast adjustments.
Ciliary and cytoplasmic pH determination
NIH-3T3 cells were transfected with either 5HT6-CFP-Venus(H148G) for cilia measurements, or CFP plus Venus(H148G) for cytoplasmic measurements. For measurements of fluorescence ratios at known pH, cells were washed once with DMEM plus HEPES at the chosen pH, then allowed to sit in DMEM plus HEPES at the known pH containing 5 μM each of the H+ ionophores nigericin and monensin (both Sigma) for 5 minutes to equilibrate. Approximately 8–10 cilia were analyzed at each chosen pH point. All fluorescence ratios are normalized to an initial measurement at pH 7.4. For determination of pH in cilia and cytoplasm, cells were placed in DMEM plus HEPES at the standard pH of 7.4 with no H+ ionophores and imaged.
Flow system coupled with epi-fluorescence time-lapse imaging
Syringe pump (Model 230, KD Scientific) was used to provide uni-directional laminar flow when connected with cell-seeded micro-channel slides. DPBS (Gibco) was used as flow perfusate. Using the Poiseuille equation for rectangular channels, τ = 6μQ/bh2 (where τ = shear stress, μ = medium viscosity, Q = flow rate, b = channel width, h = channel height), a flow rate of 0.6 ml/min was provided by the syringe pump to provide a shear stress of approximately 1 dyne/cm2 within the micro-channel. In these experiments, only upright-positioned cilia were imaged. Each cilium was imaged at 0.067 Hz for 2 minutes before flow was initiated for a period of approximately 7.5 minutes. Imaging was continued for an additional 4 minutes after flow was stopped. At each time point, each primary cilium was imaged in the xy-plane with nine z-stacks (separated by 1 μm). Experiments were conducted at room temperature. A total of eighteen cells from seven independent experiments were imaged and quantified for 5HT6-mCherry-G-GECO1.0, while a total of nine cells from three independent experiments were imaged and quantified for 5HT6-mCherry-GFP. Notably, the flow-induced calcium response was found to be sensitive to environmental changes. Each imaging experiment was performed at room temperature (21–23 °C), and was completed within one hour after cells were taken out from 37 °C. Fluorescence images shown in Supplementary Figure 13c and Supplementary Video 4 are z-projections of nine consecutive xy-planes. GFP and corresponding mCherry cilia images have been normalized against background signal variation. GFP/mCherry cilia images were obtained by taking the ratio of background normalized GFP and background-normalized mCherry signal intensities, and represented in pseudo-color scale. These values have been further subjected to two other steps of normalization, (1) normalization against signal area variation and (2) normalization against basal signal intensities (before flow), and presented in graph plots in Figure 3a as normalized measurements of GFP/mCherryin response to flow stimulation.
Supplementary Material
Acknowledgments
We thank A. Seki and T. Meyer (Stanford University) for the 5HT6 construct, M. Fivaz (National University of Singapore) for Lyn-YFP, A. Miyawaki (RIKEN) for YC3.60, L. Looger (Janelia Farm) for GCaMP5G, G. Pazour (University of Massachusetts) for GFP-CTS20 and GFP-CTS68, O. Griesbeck (Max Planck Institute) for TNXXL and R. Reed (Johns Hopkins University) for mIMCD3 cells. We also thank Y. Okubo, K. Kanemaru (University of Tokyo) and H. Ishikawa (UCSF) for helpful comments on the manuscript. This study was supported in part by the US National Institute of Health (NIH) (GM092930, DK065655 and DK090868 -pilot funds provided by the Baltimore Polycystic Kidney Disease Research and Clinical Core Center-) to T.I., and other grants to S.C., K.N., S.T., K.K. and T.K. from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan and the Japan Society for the Promotion of Science (JSPS). S.C.P. is supported by the Agency for Science, Technology and Research in Singapore.
Footnotes
Accession Codes
GenBank/EMBL/DDBJ accession numbers for 5HT6, fibrocystin, Arl13b and β1 Integrin receptor are NM_021358, NM_153179.2, NP_001167621.1 and NM_002211, respectively.
Author Contributions
S.S., S.C.P., R.D., P.N.K. and T.I. generated constructs. K.K. developed the IA sequence under the supervision of T.K. The immunohistochemistry was performed by S.C., and K.N. and S.T. took the TEM images. S.S., S.C.P., R.D. and P.N.K. carried out cell biology experiments and microscopy under the supervision of T.I. S.C.P., S.S. and T.I. wrote the manuscript.
References
- 1.Christensen S, Clement C, Satir P, Pedersen L. J Pathol. 2012;226:172–184. doi: 10.1002/path.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singla V, Reiter JF. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 3.Berbari N, Johnson A, Lewis J, Askwith C, Mykytyn K. Mol Biol Cell. 2008;19:1540–1547. doi: 10.1091/mbc.E07-09-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Praetorius HA, Spring KR. J Membrane Biol. 2001;184:71–79. doi: 10.1007/s00232-001-0075-4. [DOI] [PubMed] [Google Scholar]
- 5.Whitfield JF. Cellular Signalling. 2008;20:1019–1024. doi: 10.1016/j.cellsig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Köttgen M, et al. The Journal of Cell Biology. 2008;182:437–447. doi: 10.1083/jcb.200805124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nauli SM, et al. Nat Genet. 2003;33:129–137. doi: 10.1038/ng1076. [DOI] [PubMed] [Google Scholar]
- 8.Belgacem YH, Borodinsky LN. Proceedings of the National Academy of Sciences. 2011;108:4482–4487. doi: 10.1073/pnas.1018217108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bai CX, et al. EMBO Rep. 2008;9:472–479. doi: 10.1038/embor.2008.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleene N, Kleene S. Cilia. 2012;1:17. doi: 10.1186/2046-2530-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nachury MV, Seeley ES, Jin H. Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mank M, et al. Nat Methods. 2008;5:805–811. doi: 10.1038/nmeth.1243. [DOI] [PubMed] [Google Scholar]
- 13.Horikawa K, et al. Nat Methods. 2010;7:729–732. doi: 10.1038/nmeth.1488. [DOI] [PubMed] [Google Scholar]
- 14.Akerboom J, et al. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, et al. Science. 2011;333:1888–1891. doi: 10.1126/science.1208592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mank M, Griesbeck O. Chem Rev. 2008;108:1550–1564. doi: 10.1021/cr078213v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.