Abstract
Diffusion barriers are universal solutions for cells to achieve distinct organizations, compositions, and activities within a limited space. The influence of diffusion barriers on the spatiotemporal dynamics of signaling molecules often determines cellular physiology and functions. Over the years, the passive permeability barriers in various subcellular locales have been characterized using elaborate analytical techniques. In this review, we will summarize the current state of knowledge on the various passive permeability barriers present in mammalian cells. We will conclude with a description of several conventional techniques and one new approach based on chemically-inducible diffusion trap (C-IDT) for probing permeable barriers.
Introduction
Diffusion is the random motion of molecules driven by thermal energy, resulting in molecular movement from areas of high concentration to areas of low concentration. As an energetically favorable process, diffusion occurs without energy expenditure and thus is a ubiquitous strategy used by cells for molecular transport. However, uncontrolled diffusion can be disadvantageous to achieving localized signaling, which often requires the spatial enrichment of signaling species and is critical to fundamental cellular functions such as cell polarity, growth, proliferation, and death [1–5]. To address this issue, cells have evolved diffusion barriers, which serve as gatekeepers in filtering molecules based on size, shape, charge, and other intrinsic properties. Diffusion barriers thus enable cellular compartmentalization and spatiotemporal control of signaling. Intracellular diffusion barriers exist at a variety of cellular structures, including the nuclear envelope, the annulus of spermatozoa, the leading edge of migrating cells, the cleavage furrow of dividing cells, and the budding neck of yeast as well as in cellular extensions such as primary cilia, dendritic spines, and the initial segment of the neuronal axon [1,6,7]. While some diffusion barriers exist constitutively in cells, others are highly dynamic. The importance of diffusion barriers is further underscored by the various human diseases which result from their dysfunction [3,6,8,9].
Diffusion barriers can be categorized into two major classes based on the substrates they affect: lateral diffusion barriers and permeability barriers. Lateral diffusion barriers localize in membranes and restrict the movement of molecules within the membrane plane such as transmembrane proteins and membrane lipids [1]. Conversely, permeability barriers embedded within membranes act as conduits regulating the movement of solutes through the membrane. Permeability barriers also localize within aqueous cellular compartments to hinder solute diffusion [2,10]. Generally speaking, lateral diffusion barriers are well characterized [1], primarily because membrane molecules move relatively slowly and are in a two-dimension environment, allowing easy observation of their dynamics. In contrast, it has been challenging to measure the dynamics of solutes in cells, owing to their generally fast diffusion as well as technical limitations in precisely determining the axial position of solute molecules inside living cells [10]. However, recent advances in microscopy techniques have enabled the refinement of our understanding of passive permeability barriers. In this review, we will provide an overview of the current knowledge of permeable diffusion barriers in various subcellular regions (summarized in Figure 1 and Table 1). Subsequently, we will describe six methods used to measure the dynamics of solutes in cellular aqueous compartments and their application to probing permeable diffusion barriers, with a particular focus on the strengths and weaknesses of these approaches (summarized in Table 2).
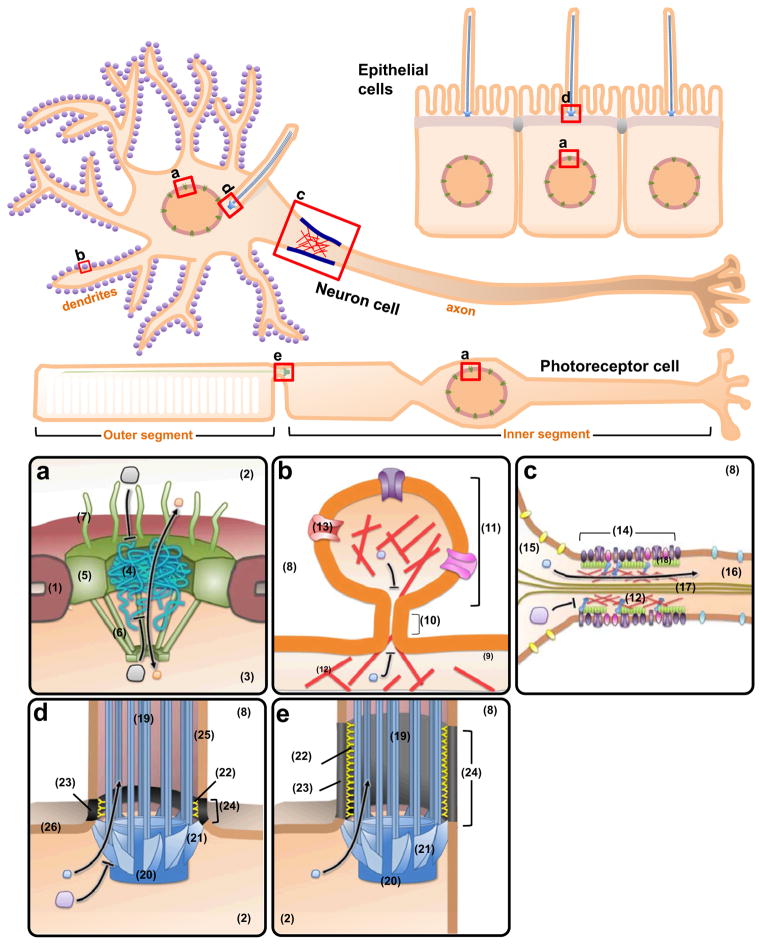
Figure 1. Passive permeability barriers in cells.
Cellular structures that harbor passive permeability barriers are represented by (a) nuclear pore complex, (b) dendritic spines, (c) axon initial segment, (d) primary cilium, and (e) connecting cilium. The detailed structures are denoted by Arabic numbers: (1) nuclear envelope (2) cytoplasm (3) nucleoplasm (4) central tube (5) scaffold ring (6) nuclear basket (7) cytoplasmic filament (8) extracellular space (9) dendrite (10) spine neck (11) spine head (12) actin filament (13) spine receptors (14) axon initial segment (15) somatodendritic compartment (16) axon (17) microtubules (18) ankyrin-G proteins (19) axoneme (20) basal body (21) transition fibers (22) Y-shaped linkers (23) ciliary necklace (24) transition zone (25) ciliary membrane (26) plasma membrane.
Table 1.
Summary of different permeable diffusion barriers in cells.
| Permeable diffusion barrier | Key components of diffusion barrier | Functional pore size for passive diffusion | Diffusion tracers | methods used to study | References |
|---|---|---|---|---|---|
| Nuclear Pore Complex | FG-rich nucleoporins | 9~10 nm | Dextran, gold particles, recombinant proteins | SPT, PAFP, FCS, FRAP, microinjection, CID | [12–15,40,44] |
| Dendritic spines neck | Unknown | <2.4 nm | GFP, Dextran, Ca2+ dye, | PAFP | [9] |
| Axon initial segment | Actin, ankyrin-G | >10 kDa, <70 kDa | Dextran, GFP | Microinjection, FRAP | [21] |
| Ciliary pore complex | Nucleoporin | ~8.43 nm (~650 kD) [31] or <67 kD [30] | Dextran, recombinant proteins, YFP-FKBP-POIs | CID, Microinjection, FRAP | [30,31] |
| Connecting cilium | Unkown | >81 kD | GFP, PAFP, arrestin, transducin, recoverin | PAFP | [26,28,29] |
| Permeable diffusion barrier in cytoplasm | Actin | 30~40 nm (average pore size) | Dextran, Ficoll, DNA, recombininat proteins, IgG | Microinjection, FRAP | [10,32,33] |
Table 2.
Summary of the approaches used to study the permeable diffusion barriers.
| Technique | Protein tracers | Non-protein tracers | Very fast tracers (ms) | Very slow tracers (hrs) | Tracking single molecule | Tracking population | Intact cells | In vitro system | Photo-damage |
|---|---|---|---|---|---|---|---|---|---|
| Single Particle Tracking (SPT) | ✓ | ✓ | ✓✓ | ✓✓ | ✓ | ✓ | ✓ | ✓✓ | |
| Photoactivatable Fluorescence Protein (PAFP) | ✓ | ✓✓✓ | ✓ | ✓ | ✓ | ✓✓✓ | |||
| Fluorescence Correlation Spectroscopy (FCS) | ✓ | ✓ | ✓✓✓ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| Fluorescence Recovery After Photobleaching (FRAP) | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓✓✓✓ | ||
| Microinjection | ✓ | ✓ | ✓✓✓ | ✓ | ✓ | ✓ | |||
| Chemically-inducible diffusion trapping (C-IDT) | ✓ | ✓ | ✓ | ✓ | ✓ |
A. Passive permeability barriers in cells
A1. Nuclear pore complex
The eukaryotic nucleus is surrounded by the nuclear envelope, a double layered membrane structure that functionally separates the nucleus from the cytosol. Communication between the cytosol and nucleus is regulated by specialized conduits, known as nuclear pore complexes (NPCs), which are anchored in the nuclear envelope at junctions between the inner and outer membrane (Figure 1a). Due to their importance in regulating nuclear function, NPCs are one of the most well-characterized diffusion barriers in cells [7]. Each NPC consists of several major domains: a central channel, a core scaffold that supports the central channel, a nuclear basket, and cytoplasmic filaments. Nuclear trafficking is regulated through the central channel created by the core scaffold, which is assembled by approximately 30 different nucleoporin proteins arranged in rotational symmetry to form a conduit-like structure. Two thirds of these nucleoporins constitute the core scaffold, while the remaining one third of nucleoporins extend out phenylalanine-glycine (FG)-rich repeats to fill the central channel of the NPC [7]. Importantly, these FG-rich proteins have been proposed to assemble into a molecular sieve that regulates nucleocytoplasmic shuttling [11]. This molecular sieve-like diffusion barrier, with a calculated diameter of ~9–10 nm, allows passive diffusion of molecules such as ions, metabolites, and cargo smaller than ~40 kDa [12]. Larger cargo, however, require active transport-mediated and signal-dependent mechanisms to pass through the NPC [2]. The largest cargo that can be actively transported across NPCs is ~39 nm, which approximates the narrowest region of the central channel (~50 nm) [7,13]. It is still unknown whether these passive and active transport systems occupy distinct or similar channels of the NPC [14,15]. In summary, NPCs enable cells to properly regulate the localization and mobility of molecules across the nuclear envelope.
A2. Dendritic spine neck
Dendritic spines are small membranous protrusions originating from neuronal dendrites, which serve as postsynaptic terminals of excitatory synapses. As postsynaptic compartments of chemical neurotransmission, dendritic spine structure and functional regulation can determine the strength with which a receiving neuron is stimulated by a given connection. Spines are dynamic structures, as they retract from the dendritic shaft when inactive and reform in response to stimulation [16]. Despite their morphological and temporal variability along dendrites, they typically have a relatively large head attached to the dendritic shaft by a narrow neck (Figure 1b) [17].
Postsynaptic induction requires NMDAR (NMDA-sensitive glutamate receptor)-mediated calcium influx and calcium-dependent signaling in the dendritic spine. During synaptic transmission, NMDAR-mediated calcium entry is tightly restricted to the spine head, with minimal calcium diffusion into the connected dendrite [18]. As a result, the spine neck has been suggested to restrict calcium diffusion and also appears to limit that of soluble GFP, indicating the existence of a passive permeability barrier at the neck of dendritic spines [9,16–18]. The strength of the cytosolic compartmentalization varies according to spine morphology and correlates inversely with the width of the neck, providing further evidence for a permeability barrier [18]. The neck of the dendritic spine thus effectively enables compartmentalized signaling in neurons.
A3. Axon initial segment
During development, axon and dendrite specification in neurons depends on the orchestration of positive and negative signals that regulate protein trafficking and cytoskeletal dynamics. Functional differentiation between axons and dendrites depends on the presence of different sets of proteins in their respective membranes. A lateral membrane diffusion barrier has been found at the initial segment of the axon, called the axon initial segment (AIS), which contributes to maintaining the asymmetric distribution of proteins in this compartment (Figure 1c) [19]. Actin, ankyrin-G, voltage-dependent sodium channels, neurofascin, and NrCAM have all been shown to concentrate within the AIS and directly contribute to its lateral diffusion barrier function [4,19,20]. A size-selective permeable diffusion barrier also exists in the cytoplasm of the AIS, with actin and ankyrin-G serving as key players in maintaining its functionality [21]. Taken together, the AIS barrier serves to maintain the polarity of axonal extensions from the neuronal soma [4,20].
A4. Primary cilium
The primary cilium is a hair-like membrane projection on the apical surface of many vertebrate cells [22]. The primary cilium acts as a sensory and signaling organelle which regulates several signaling systems such as phototransduction, olfaction, and developmental pathways [22]. Although the ciliary membrane is contiguous with the plasma membrane and the ciliary lumen is open to the cytoplasm (Figure 1d, e), the primary cilium is highly enriched with specific proteins that support ciliary functions. Disruption of canonical proteins that normally localize to and function within primary cilia leads to ciliary dysfunction and a suite of congenital conditions known as ciliopathies [23].
Each primary cilium contains several structurally distinct domains: the axoneme, basal body, transition fibers, Y-shaped linkers, ciliary necklace and transition zone (Figure 1d, e) [24]. The axoneme is composed of a radial array of nine doublet microtubules with no central pair of singlet microtubules (the 9+0 arrangement), while the basal body is a cytosolic microtubule-organizing center that is derived from the mother centriole. Transition fibers form a pinwheel-like structure radiating from the basal body and anchor to the most proximal region of the ciliary membrane. Y-shaped linkers connect the axoneme to a specialized membrane domain, known as the ciliary necklace. The transition zone describes a region between the axoneme and transition fibers where the Y-shaped linkers and the necklace can be observed (Figure 1d, e). The transition fibers cover the entrance to the ciliary lumen and the space between two consecutive transition fibers accommodates particles smaller than 60 nm in diameter [25]. It has been widely believed that transition fibers act as a diffusion barrier to contribute to the specialization of the privileged ciliary environment.
The diffusion of soluble molecules between the cytosol and the ciliary lumen has been well characterized in rod photoreceptor cells, which possess a connecting cilium (CC) that serves as a conduit between the cell body and the outer segment (OS)(Figure 1) [26]. The CC shares a similar structure with primary cilia but possesses an elongated transition zone (Figure 1e) [27]. Diffusion across the CC occurs during phototransduction when photons expose arrestin-binding sites on rhodopsin in the OS. The initial pool of arrestin in the OS is removed from the soluble environment by binding to rhodopsin and induces soluble arrestin from the cell body to diffuse across the CC into the OS due to the newly induced concentration gradient [28]. Investigation of the barrier properties of the CC using fluorescent proteins has demonstrated that monomers, dimers and trimers of green fluorescent protein (GFP) freely diffuse across the CC, suggesting that there is no fixed pore that limits the diffusion of soluble proteins up to 81 kDa [29].
The diffusion of soluble proteins into primary cilia of epithelial cells is beginning to be evaluated. Kee et al. found that molecules above 67 kDa are excluded from the primary cilia lumen by a fixed size NPC-like pore at the base of the primary cilia [30]. Particular nucleoporins were found to localize at the base of primary cilia, presumably as part of a permeable diffusion barrier in primary cilia [30]. In distinct contrast, we recently indicated that the interior of the cilium was accessible to proteins as large as 79 Å (650 kDa), and the kinetics of protein diffusion was exponentially limited by molecular size (personal communication) [31]. Together with computational modeling, they concluded that the permeable diffusion barrier of primary cilia behaves like a molecular sieve rather than a fixed pore. The discrepancy between these two studies may have resulted from the use of different techniques with various sensitivity and resolution (see below for thorough discussions on the techniques).
A5. Cytoplasm
The cytoplasm is a fairly crowded soluble environment filled with small solutes, soluble macromolecules, cytoskeleton, and membrane structures such as vesicles and organelles. Therefore solute diffusion is slower in cytoplasm than in saline, especially with increasing molecular size. For example, the diffusion of a 1000-kDa dextran molecule in the cytoplasm is fivefold slower than that in saline [10]. Cytoplasmic diffusion is primarily hindered by actin structures which serve as diffusion barriers with an average mesh size between 20 to 40 nm [10,32,33].
B. Experimental techniques to probe diffusion barriers in cells
The observation and quantification of molecular dynamics in living cells constitutes a general strategy to study functional properties of cellular diffusion barriers. Live-cell fluorescence microscopy represents a prevailing approach to such studies, primarily because it offers specificity and sensitivity on top of its inherently non-invasiveness nature. In this section, we will review five conventional imaging techniques commonly used to study diffusion properties. We will then describe a newly emerging technique that adapts chemically-inducible dimerization to investigating passive permeability barriers.
B1. Single-particle tracking
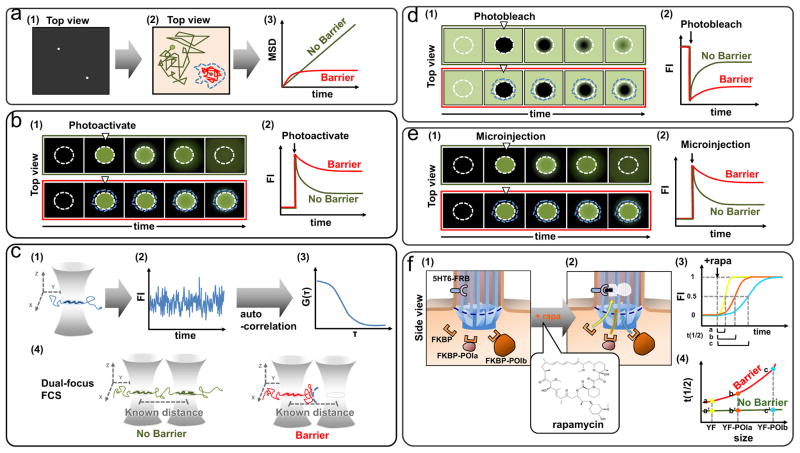
Single-particle tracking (SPT) consists of motion-tracking of individual molecules in cells by highly sensitive fluorescence microscopy techniques combined with live-cell imaging [34]. In the acquired time-lapse image sequences, individual fluorescent particles can be identified as bright spots on a dark background (Figure 2a1). The trajectory of the labeled fluorescent particles can be analyzed to identify and probe any diffusion barriers that affect their diffusion (Figure 2a2–3) [5]. Single-particle tracking (SPT) involves the selective labeling of specific proteins, chromatin sequences, or lipids with organic fluorophores, carbon nanotubes, fluorescent proteins, noble-metal nanoparticles, quantum dots or probes visible with transmitted light (gold or latex beads) [35]. The positions of these particles can be measured with as low as nanometer spatial and submillisecond temporal resolution using suitable camera detectors. At its inception, SPT was exclusively used to study the two-dimensional planar diffusion of membrane proteins rather than solute diffusion, which occurs in three dimensions, primarily because of limitations in determining particle z (axial) positions. Recently, the development of new fluorescence microscopy techniques such as spinning disk confocal microscopy, scanning confocal microscopy, and two-photon microscopy have allowed single molecules to be tracked by three-dimensional SPT with high temporal and spatial resolution in real time [35]. However, SPT remains limited when particles diffuse rapidly or are at saturating particle densities where individual particles often cross paths, making it challenging to track an individual particle.
Figure 2. Experimental approaches to probe passive permeability barriers in cells.
(a) Single particle tracking (SPT): (a1) Individual fluorescent particles can be identified as bright spots on a dark background by fluorescence microscopy. (a2) In the acquired image sequences, the trajectories of single particles (red line and green line) can be monitored and quantified. (a3) The mean-square-displacement (MSD) as a function of the lag time can be calculated from the trajectories of particles and provide information about the distance a particle has moved and also show the existence of diffusion barriers. (b) Photoactivatable fluorescence protein (PAFP): (b1) Photoactivation in the selected region (white circle with dash line) increases the fluorescence intensity of PAFPs. (b2) The spreading kinetics of PAFPs can be quantified and thus provide information about the diffusion rate of PAFPs and the existence of diffusion barriers. (c) Fluorescence correlation spectroscopy (FCS): (c1) Fluorescent molecules diffusing in and out of the confocal detection volume give rise to intensity fluctuations. (c2) Typical fluorescence intensities measured in time by FCS. (c3) The intensity trace can be calculated as the autocorrelation of the intensity fluctuations and measures the self-similarly of the signal by the equation: G(τ)= <δI(t)δI(t+τ)>/ <I(t)>2. The correlation curve provides information about the diffusion coefficients and average number of particles in the detection volume. (c4) Dual-focus FCS using two detection volumes separated by a known distance. The molecules in two detection volumes are cross-correlated and the precise time required for the same molecule to move between two detection volumes can be measured. This provides information about the diffusive behavior of molecules and can be used to probe diffusion barriers. (d) Fluorescence recovery after photobleaching (FRAP): (d1) Fluorescently labeled proteins in a region of interest (white circle with dash line) are photobleached using a laser. (d2) The recovery kinetics of fluorescently labeled proteins in the region of interest can be quantified to calculate diffusion coefficients and to identify potential diffusion barriers. (e) Microinjection: (e1) Diffusion tracers are microinjected into the region of interest (white circle with dash line). (e2) The spreading kinetics of injected tracers can be quantified to provide information on the diffusion coefficients and possible diffusion barriers.. (f) Chemically-inducible diffusion trapping (C-IDT): (f1) A primary cilia membrane marker, 5HT6, was used to tag FRB to primary cilia. Cytosolic tracers of various sizes such as FKBP, FKBP-POIa or FKBP-POIb are coexpressed with 5HT6-FRB in distinct compartments. (f2) After addition of rapamycin (rapa), cytosolic tracers are trapped in primary cilia by rapamycin-induced dimerization after diffusion through putative diffusion barriers. (f3) The concentration of tracers in primary cilia can be quantified and normalized to the range from 0 to 1. The time required for half-maximal (t1/2) accumulation in cilia for tracers were extracted from the kinetic curves (a, b, and c, are t1/2 of FKBP, FKBP-POIa, FKBP-POIb, and FKBP-POIc, respectively). (f4) The fitted curve for t1/2 as a function of the tracer size determines the nature of permeable barriers.
B2. Photoactivatable fluorescence protein
A photoactivatable fluorescence protein (PAFP) is capable of a dramatic increase in fluorescence intensity in response to irradiation with light of a specific wavelength, intensity, and duration [36]. A PAFP-tagged protein of interest (PAFP-POI) can be precisely photolabeled and tracked in a spatially and temporally controlled manner. Combined with live cell imaging, the diffusive properties of PAFP-POIs can be easily evaluated and thus used to probe diffusion barriers (Figure 2b). Although this method is specific and accurate with high spatiotemporal resolution, it cannot directly measure the dynamics of non-protein substrates. For instance, the dynamics of DNA or RNA in cells need to be measured by tracking the PAFP-tagged nucleotide-binding domain and binding of the protein may alter the diffusive properties of the nucleic acid substrates.
B3. Fluorescence correlation spectroscopy
Fluorescence correlation spectroscopy (FCS) measures fluorescence intensity fluctuations due to molecules diffusing in and out of a sub-femtoliter volume, usually defined by the focal volume of a confocal microscopy (Figure 2c1). The intensity fluctuations depend on the average number of fluorescent molecules in the detection volume and their diffusion coefficients. Increased diffusion results in more rapid fluctuations and reduces the probability that a particle initially found in the detection volume will be detected in the same area at a later time. This probability is quantified by the autocorrelation function (Figure 2c2–3) [37]. As a result, FCS derives the diffusion coefficients of molecules in cells.
Traditional FCS measurements using a single detection volume, however, cannot be used to study diffusion barriers due to a lack of spatial resolution. To overcome this limitation, dual-focus FCS using two confocal detection volumes separated by a well-defined distance of a few hundred nanometers has been developed (Figure 2c4) [38]. The molecules in two points are cross-correlated and the precise time required for the same molecule to be found in a given location away from the position at time zero can be measured (left panel, Figure 2c4). If there is a barrier to diffusion at any given location, it will require a longer time to find the particle at a position across the barrier (right panel, Figure 2c4) [39]. Indeed, this proof-of-concept has been demonstrated in the study of single molecule diffusion through NPCs. [40].
FCS has potential complications arising from photobleaching, due to its inability to distinguish between molecules bleaching and molecules diffusing out of the detection volume; both result in a drop in fluorescence intensity. Therefore, in very slow diffusion processes (requiring more laser excitation), molecular retention times are inevitably overestimated. Moreover, auto-fluorescence from other molecules present in the cytoplasm or membrane could also contribute to the constant background signal, causing molecular concentrations to be overestimated. These limitations can in principle be overcome by adjusting the excitation power and duration, while carefully calibrating optics and preparing experimental samples.
B4. Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching (FRAP) has been extensively used for diffusion measurements. Fluorescently labeled molecules in a region of interest are first photobleached with a high intensity laser, leaving a region devoid of fluorescent signal. The recovery of fluorescence intensity in this region occurs when non-bleached fluorescent molecules diffuse in from the surrounding area. Real-time measurements of the fluorescence recovery kinetics allow estimates of diffusion coefficients (Figure 2d).
FRAP is generally limited in measurement duration, such that long-tail recovering curves expected in anomalous diffusion are easily overestimated. Furthermore, diffusion rates of fast diffusing molecules, such as small soluble molecules, are always underestimated due to the relatively long photobleaching time required to overcome the rapid diffusion of surrounding non-photobleached molecules into the region of interest.
B5. Microinjection of diffusion tracers
Microinjection has been widely applied to studies of diffusion. Injection can be applied transiently and locally to increase the concentration of diffusion tracer molecules. In combination with live cell imaging, the dynamics of injected solutes can be monitored and the diffusion coefficient can be extracted from the spreading kinetics (Figure 2e). Microinjection has been used to study NPCs [15], primary cilia [30], AIS [21], and diffusion barriers in cytoplasm [32,33].
To determine the functional size of permeable diffusion barriers, the ideal injected diffusion tracers should meet the following conditions: (1) inert (unable to affect the cellular physiology and doesn’t have any affinity with unrelated endogenous proteins and diffusion barriers), (2) spherical shape, (3) stable, (4) appropriate fluorescence signal, (5) soluble in aqueous buffer (6) can form polymers with a continuous series of sizes; (7) neutral in electric charge. Dextran, ficoll, polyethylene glycol (PEG), gold particles and globular proteins have been frequently used as diffusion tracers [13,21,30,41]. Dextran, ficoll, and PEG can form polymers of various sizes and are essentially non-interacting macromolecules [41]. Ficoll and PEG, but not dextran, are relatively spherical macromolecules [41]. Gold particles are also spherical molecules with well-defined sizes and very low affinity with other endogenous components in cells. Globular proteins such as insulin, lysozyme, myoglobin and hemoglobin, although they are not neutral molecules and may have affinity to endogenous cellular components, are still frequently used to study permeable diffusion barriers more physiologically.
The delivery of microinjected diffusion tracers, can be precisely controlled by automated equipment Several current models of air-pressure-driven microinjectors allow fine-tuning of injection pressure, compensation pressure as well as injection duration, making the transferred dosage highly controlled in a reproducible manner. There is also a diverse array of diffusion tracers ranging from synthetic molecules such as pharmacological drugs to physiological molecules including nucleotides, peptides, recombinant proteins and macromolecules. The disadvantages of microinjection include potential damage to cells due to its invasive application and technical difficulties in injecting small structures such as dendritic spines.
B6. Chemically-inducible diffusion trap (C-IDT)
Chemically-inducible dimerization (CID) techniques have been widely used in cell biology for over 20 years since their inception [42]. The most common CID system relies on the rapamycin-inducible interaction of FK506-rapamycin-binding domain (FRB) and immunophilin FK506-binding protein-12 (FKBP) [42,43]. In a typical CID system, two chimeric proteins are coexpressed in cells [42,43]. One protein contains FRB and a plasma membrane anchor sequence that localizes it to the cytoplasmic leaflet of plasma membrane, while the other FKBP-tagged cytosolic protein of interest (FKBP-POI) freely diffuses throughout the cytoplasm. Addition of rapamycin rapidly traps FKBP-POIs at the membrane, where the FRB-tagged protein is localized (Figure 2f1–2) [42]. The kinetics of FKBP-POI motility from cytoplasm to the membrane can be quantified to provide information about its diffusion rate (Figure 2f3). Based on this methodology, two groups recently have improvised CID systems to probe the dynamics of soluble proteins across diffusion barriers in living cells. Raschbichler et al. generated a bipartite assay called NEX-TRAP (Nuclear EXport Trapped by RAPmycin) for the analysis of protein nuclear export in cells [44]. This method used the CID system to trap NLS-tagged proteins that have shuttled out of the nucleus to specific membrane compartments in the cytoplasm. In this manner, the kinetics of protein nuclear export can be conveniently represented by the relative amounts of NLS-tagged proteins trapped on the cytoplasmic membrane compartment, which in turn allows probing of the diffusion barrier properties of the NPC.
In contrast to the large-volume nucleus, the primary cilium is a tiny organelle, whose volume only comprises ~0.01% of the total cell volume. This has posed a great challenge to fluorescently visualizing the translocation of soluble proteins into this tiny organelle. However, Lin et al. recently overcame this technical difficulty with a newly developed CID-based methodology termed the chemically-inducible diffusion trap (C-IDT) at cilia to visualize diffusion of cytosolic proteins into primary cilia in real time in intact living cells (personal communication) [31]. In this system, a specific cilia-enriched membrane protein, the 5-hydroxytriptamine receptor 6 (5HT6), was tagged with FRB on its cytoplasmic tail, while cytosolic proteins to be tested for ciliary influx are fused with FKBP (Figure 2f1). Upon addition of a chemical dimerizer, cytosolic diffusion probe proteins that are able to diffuse between the cytosol and ciliary lumen are trapped inside primary cilia (Figure 2f2). The diffusion kinetics of FKBP-tagged cytosolic proteins into primary cilia can be visualized via wide-field fluorescence microscopy from the accumulating fluorescence signal (Figure 2f3). The advantages of C-IDT lie in the ability to rapidly trigger diffusion measurements without damaging the plasma membrane by microinjection or detergent-mediated permeabilization. Furthermore, analysis of the diffusion kinetics similarly provides estimates of diffusion coefficients by using Fick’s first law,
where [Fcy] and [Fci] are concentrations of the FKBP tagged proteins in the cytoplasm and cilia, respectively, D is the diffusion coefficient of the FKBP-tagged proteins, and x is the proposed length of a diffusion barrier. Estimates of the flux, J, can be obtained by quantifying the change in fluorescence intensity (proportional to FKBP tagged proteins) in cilia over time through an estimated cross sectional area provided by structural studies. Thus, diffusion coefficients of different sized FKBP-tagged proteins can be derived from C-IDT measurements and comparisons can be made between theoretical and experimentally-determined diffusion coefficients to shed light on the regulation of diffusion into cilia (Figure 2f4).
Despite its utility, the C-IDT technique presents a few limitations. First, the CID diffusion trap system is not suitable to study the diffusion of non-protein substrates. Second, to enable visualization and inducible dimerization of cytosolic proteins, each cytosolic diffusion probe has to be tagged with either FRB or FKBP fused with a fluorescent protein, placing a restriction on their minimal molecular size (e.g. YFP-FKBP, molecular weight, ~40 kDa; stokes radius, ~32 Å). Therefore, C-IDT cannot readily probe diffusion barriers with a pore size smaller than 40 kDa. The derivation of diffusion coefficients is also limited to early time points to minimize the effects of the lateral diffusion of FKBP-FRB pairs in the ciliary membrane.
Conclusions
Dysfunction of subcellular diffusion barriers severely impacts cellular functions, leading to various disease conditions such as cancers and brain disorders, triple A syndrome, and ciliopathies. Due to their importance in regulating cellular functions, several approaches have been established to evaluate the motion of solutes across diffusion barriers in cells. In this review, we have highlighted each approach while emphasizing their pros and cons. Measurements of solute motions in a given cellular aqueous compartment with present techniques requires specialized microscopy techniques and careful analysis. In contrast, a newly emerging approach such as C-IDT complements nicely with existing techniques, as it uses an ordinary wide-field fluorescence microscope. C-IDT is well suited to probing sub-micron compartments that have been otherwise challenging. Armed with these techniques, we continue our endeavor of elucidating elegant passive permeability architectures in living cells.
References
• of special interest
•• of outstanding interest
- 1•.Caudron F, Barral Y. Septins and the lateral compartmentalization of eukaryotic membranes. Dev Cell. 2009;16:493–506. doi: 10.1016/j.devcel.2009.04.003. A comprehensive review that covers the different lateral diffusion barrier in cells. [DOI] [PubMed] [Google Scholar]
- 2.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 3.Buffington SA, Rasband MN. The axon initial segment in nervous system disease and injury. Eur J Neurosci. 2011;34:1609–1619. doi: 10.1111/j.1460-9568.2011.07875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winckler B, Forscher P, Mellman I. A diffusion barrier maintains distribution of membrane proteins in polarized neurons. Nature. 1999;397:698–701. doi: 10.1038/17806. [DOI] [PubMed] [Google Scholar]
- 5••.Ruthardt N, Lamb DC, Brauchle C. Single-particle tracking as a quantitative microscopy-based approach to unravel cell entry mechanisms of viruses and pharmaceutical nanoparticles. Mol Ther. 2011;19:1199–1211. doi: 10.1038/mt.2011.102. A thorough review that covers the introduction of SPT and recent advances of SPT in three dimensions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6•.Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu Rev Cell Dev Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. A thorough review that covers the different mechniasms of ciliary influx of protein into cilia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–643. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 8.Cronshaw JM, Matunis MJ. The nuclear pore complex: disease associations and functional correlations. Trends Endocrinol Metab. 2004;15:34–39. doi: 10.1016/j.tem.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Bloodgood BL, Sabatini BL. Neuronal activity regulates diffusion across the neck of dendritic spines. Science. 2005;310:866–869. doi: 10.1126/science.1114816. [DOI] [PubMed] [Google Scholar]
- 10••.Dix JA, Verkman AS. Crowding effects on diffusion in solutions and cells. Annu Rev Biophys. 2008;37:247–263. doi: 10.1146/annurev.biophys.37.032807.125824. A thorough review that describes the molecular crowding of solutes in cellular aqueous comartments. It also introduces several experimental measurements of solute diffusion in cellular aqueous compartments. [DOI] [PubMed] [Google Scholar]
- 11.Grunwald D, Singer RH, Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475:333–341. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paine PL, Moore LC, Horowitz SB. Nuclear envelope permeability. Nature. 1975;254:109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- 13.Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mohr D, Frey S, Fischer T, Guttler T, Gorlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naim B, Brumfeld V, Kapon R, Kiss V, Nevo R, Reich Z. Passive and facilitated transport in nuclear pore complexes is largely uncoupled. J Biol Chem. 2007;282:3881–3888. doi: 10.1074/jbc.M608329200. [DOI] [PubMed] [Google Scholar]
- 16.Alvarez VA, Sabatini BL. Anatomical and physiological plasticity of dendritic spines. Annu Rev Neurosci. 2007;30:79–97. doi: 10.1146/annurev.neuro.30.051606.094222. [DOI] [PubMed] [Google Scholar]
- 17.Lee KF, Soares C, Beique JC. Examining form and function of dendritic spines. Neural Plast. 2012;2012:704103. doi: 10.1155/2012/704103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18•.Noguchi J, Matsuzaki M, Ellis–Davies GC, Kasai H. Spine-neck geometry determines NMDA receptor-dependent Ca2+ signaling in dendrites. Neuron. 2005;46:609–622. doi: 10.1016/j.neuron.2005.03.015. This work demonstrates that the geometry of the dendritic spine neck regulates the permeability of the diffusion barrier for Ca2+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bender KJ, Trussell LO. The physiology of the axon initial segment. Annu Rev Neurosci. 2012;35:249–265. doi: 10.1146/annurev-neuro-062111-150339. [DOI] [PubMed] [Google Scholar]
- 20.Hedstrom KL, Ogawa Y, Rasband MN. AnkyrinG is required for maintenance of the axon initial segment and neuronal polarity. J Cell Biol. 2008;183:635–640. doi: 10.1083/jcb.200806112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21••.Song AH, Wang D, Chen G, Li Y, Luo J, Duan S, Poo MM. A selective filter for cytoplasmic transport at the axon initial segment. Cell. 2009;136:1148–1160. doi: 10.1016/j.cell.2009.01.016. This work focuses on the diffusion barrier for solutes in the axon initial segment. The authors found that a size–selective permeable diffusion barrier exists in the cytoplasm of AIS. [DOI] [PubMed] [Google Scholar]
- 22.Singla V, Reiter JF. The primary cilium as the cell’s antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 23.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 25.Anderson RG. The three-dimensional structure of the basal body from the rhesus monkey oviduct. J Cell Biol. 1972;54:246–265. doi: 10.1083/jcb.54.2.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calvert PD, Strissel KJ, Schiesser WE, Pugh EN, Jr, Arshavsky VY. Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 2006;16:560–568. doi: 10.1016/j.tcb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 27.Horst CJ, Forestner DM, Besharse JC. Cytoskeletal-membrane interactions: a stable interaction between cell surface glycoconjugates and doublet microtubules of the photoreceptor connecting cilium. J Cell Biol. 1987;105:2973–2987. doi: 10.1083/jcb.105.6.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Vishnivetskiy SA, Chen J, Hurley JB, Gurevich VV, et al. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Najafi M, Maza NA, Calvert PD. Steric volume exclusion sets soluble protein concentrations in photoreceptor sensory cilia. Proc Natl Acad Sci U S A. 2012;109:203–208. doi: 10.1073/pnas.1115109109. This work focuses on the diffusion barrier for soluble proteins in the conneting cilium of photoreceptor cells. The authors found that monomers, dimers and trimers of green fluorescent protein (GFP) freely diffused across the CC, suggesting that there is no fixed pore that limits the diffusion of soluble proteins at least up to 81 kDa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Kee HL, Dishinger JF, Lynne Blasius T, Liu CJ, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nat Cell Biol. 2012;14:431–437. doi: 10.1038/ncb2450. This work focuses on the diffusion barrier for soluble proteins into primary cilia. The authors found that molecules above 67 kDa are excluded from the primary cilia lumen by a fixed size NPC-like pore at the base of the primary cilia. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Lin YC, Niewiadomski P, Lin B, Nakamura H, Phua SC, Jiao J, Levchenko A, Inoue T, Rohatgi R, Inoue T. Chemically-inducible diffusion trap at cilia (C-IDTc) reveals molecular sieve-like barrier. Nature Chemical Biology. 2013;9:437–443. doi: 10.1038/nchembio.1252. The first study to probe the diffuse barrier in primary cilia using chemical chemically-inducible dimerization technique. The authors found that a molecule-sieve like permeable barrier exists at the base of primary cilia which is accessible to proteins as large as 650 kDa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luby–Phelps K, Castle PE, Taylor DL, Lanni F. Hindered diffusion of inert tracer particles in the cytoplasm of mouse 3T3 cells. Proc Natl Acad Sci U S A. 1987;84:4910–4913. doi: 10.1073/pnas.84.14.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luby-Phelps K, Taylor DL, Lanni F. Probing the structure of cytoplasm. J Cell Biol. 1986;102:2015–2022. doi: 10.1083/jcb.102.6.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Michalet X, Berglund AJ. Optimal diffusion coefficient estimation in single-particle tracking. Phys Rev E Stat Nonlin Soft Matter Phys. 2012;85:061916. doi: 10.1103/PhysRevE.85.061916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hellriegel C, Gratton E. Real-time multi-parameter spectroscopy and localization in three-dimensional single-particle tracking. J R Soc Interface. 2009;6 (Suppl 1):S3–14. doi: 10.1098/rsif.2008.0313.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36•.Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Innovation: Photoactivatable fluorescent proteins. Nat Rev Mol Cell Biol. 2005;6:885–891. doi: 10.1038/nrm1741. A comprehensive review that covers the instructional use of photoactivatable proteins and the potential applications of photoactivatable proteins. [DOI] [PubMed] [Google Scholar]
- 37.Ries J, Schwille P. Fluorescence correlation spectroscopy. Bioessays. 2012;34:361–368. doi: 10.1002/bies.201100111. [DOI] [PubMed] [Google Scholar]
- 38.Dertinger T, Pacheco V, von der Hocht I, Hartmann R, Gregor I, Enderlein J. Two-focus fluorescence correlation spectroscopy: a new tool for accurate and absolute diffusion measurements. Chemphyschem. 2007;8:433–443. doi: 10.1002/cphc.200600638. [DOI] [PubMed] [Google Scholar]
- 39••.Digman MA, Gratton E. Imaging barriers to diffusion by pair correlation functions. Biophys J. 2009;97:665–673. doi: 10.1016/j.bpj.2009.04.048. This work develops a dual-FCS system to map diffusion barriers in cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cardarelli F, Gratton E. In vivo imaging of single-molecule translocation through nuclear pore complexes by pair correlation functions. PLoS One. 2010;5:e10475. doi: 10.1371/journal.pone.0010475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venturoli D, Rippe B. Ficoll and dextran vs. globular proteins as probes for testing glomerular permselectivity: effects of molecular size, shape, charge, and deformability. Am J Physiol Renal Physiol. 2005;288:F605–613. doi: 10.1152/ajprenal.00171.2004. [DOI] [PubMed] [Google Scholar]
- 42•.Derose R, Miyamoto T, Inoue T. Manipulating signaling at will: chemically-inducible dimerization (CID) techniques resolve problems in cell biology. Pflugers Arch. 2013 doi: 10.1007/s00424-012-1208-6. A comprehensive review that introduces the principle of CID systems and reviews the different applications of CID-based techniques. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43•.Putyrski M, Schultz C. Protein translocation as a tool: The current rapamycin story. FEBS Lett. 2012;586:2097–2105. doi: 10.1016/j.febslet.2012.04.061. A comprehensive review that covers the different CID-based techniques used to modulate molecular activities by translocation of specific effectors to their functional sites. [DOI] [PubMed] [Google Scholar]
- 44••.Raschbichler V, Lieber D, Bailer SM. NEX-TRAP, a novel method for in vivo analysis of nuclear export of proteins. Traffic. 2012;13:1326–1334. doi: 10.1111/j.1600-0854.2012.01389.x. This work uses chemically-inducible dimerization system to evaluate the nuclear export activity of protein. The method can also be utiltized to measure the kinetic of protein passing through the NPCs. [DOI] [PubMed] [Google Scholar]