Abstract
Because the colonic mucosa is in direct contact with digesta, luminal exposure to potentially carcinogenic or chemopreventive agents may be important in colorectal carcinogenesis, independently of the effects of systemic exposure through the circulation. To date, few biomarkers for luminal exposure have been identified, and isolation of reasonably good quality fecal human RNA remains difficult. In this study, we assessed the yield and quality of RNA extracted from 10 human stool samples after storage with several commercially available preservatives compared with stool samples immediately frozen in liquid nitrogen. This study shows that careful design of primer pairs which amplify a short length of DNA is key to obtaining interpretable and reproducible results. Moreover, the use of commercially available RNA preservation kits enables investigators to collect usable fecal samples from large populations. Of all the preservative methods tested, RNAlater had the best performance in terms of overall quality, quantity, and level of genomic DNA contamination, and thus deserves further investigation.
Introduction
Colorectal cancer is the second leading cause of cancer death in the United States (1). Because the colonic mucosa is in direct contact with digesta, luminal exposure to potentially carcinogenic or chemopreventive agents may be important in colorectal carcinogenesis, independently of the effects of systemic exposure through the circulation (2, 3). Although ∼55% of dry fecal weight is attributed to bacteria, ∼1.5 million colonic epithelial cells can nevertheless also be isolated per gram of stool (4, 5). Thus, exfoliated cells in stool may serve as an excellent source for examining the effects of the luminal environment on colorectal epithelial cells. In addition, fecal RNA analysis may expand the potential of RNA-based colorectal cancer screening (6, 7).
One of the major obstacles to developing fecal markers has been difficulty in collecting adequate samples for assays from a large number of subjects because standard fecal collection procedures require fresh or frozen samples. However, several commercial products have recently become available to preserve DNA and RNA for a period of time at room temperature. Whatman FTA cards have been designed to obtain both DNA and RNA. They are impregnated with a blend of chemicals that lyses cell membranes and denatures proteins on contact, leading to trapping, immobilization, and stabilization of nucleic acids (8, 9). RNAlater was originally developed for RNA preservation but has been used successfully to stabilize and store RNA and DNA (10,11). Silica gel is a drying agent that has been used for DNA/RNA isolation from field non–human primate fecal samples (10, 12). Paxgene is a clinically approved method for the preservation of RNA from blood (13) but has not been investigated previously for use with feces. In this pilot study, we assessed the yield and quality of RNA extracted from human stool samples processed with these products, in comparison with fresh frozen samples.
Materials and Methods
Sample Collection and Processing
The research protocol was approved by Wayne State University and the VA Medical Center Human Investigation Committees, and signed informed consent was obtained from each study participant. Fresh stool samples were collected in plastic containers from 10 patients being evaluated at the John D. Dingell VA Medical Center (Detroit, MI) who did not report any history of gastrointestinal surgery. Stool samples were immediately placed on ice for transfer to the laboratory for further processing.
Aliquots of 0.2 g of each stool specimen were processed with the following preservation methods: Whatman FTA card (W;Whatman), silica gel beads (S), 1.0 mL RNALater (R;Ambion), and 1.0 mL Paxgene (P;PreAnalytiX). The remainder of each sample was flash-frozen in liquid nitrogen and stored at −80°C (0.2 g was used for RNA extraction, and designated “N”). For W samples, feces were spread over two of the four quadrants of the FTA card, allowed to dry for ∼2 h at room temperature, and then placed in a protective barrier pouch with a silica gel desiccant packet. For S samples, feces were placed over silica gel beads (∼10 mL) and ∼1 cm of glass wool in a 50 mL tightly sealed sterile polypropylene tube. R and P samples were stored in 2 mL of sterile polypropylene tubes. After 5 days at ambient temperature, R and P samples were centrifuged, and pellets were transferred to −80°C together with W and S samples. Total RNA was extracted using the RNA PowerSoil (MO BIO Laboratories) method. Full aliquots were used for extraction, except W samples for which 20 FTA card-punches were used. The resultant RNA quantity and A260/280 ratio were assessed with a NanoDrop ND-1000 Spectrophotometer.
Reverse Transcription-PCR
Reverse transcription-PCR (RT-PCR) was used to amplify the housekeeping gene, β-actin. Reverse transcription was done with 150 ng of total RNA and 1.25 μmol/L of random hexamer in a 20 μL reaction containing 2.5 mmol/L of magnesium chloride, 250 μmol/L of each deoxynucleotide triphosphate, 10 units of RNase inhibitor, 10 mmol/L of DTT, and 15 units of MultiScribe Reverse Transcriptase (Applied Biosystems). The reactions were incubated at 25°C for 10 min and then held at 42°C for 15 min. Total cellular RNA isolated from human colon cancer HCT-116 cells was used as a positive control.
PCR was done using a GeneAmp PCR System 9600 (Perkin-Elmer). Each 50 μAL reaction contained 3 μL of reverse transcription reaction mix comprised of 200 μmol/L of each deoxynucleotide triphosphate, 0.15 μmol/L of each primer, and 2.5 units of AmpliTaq Gold DNA polymerase (Applied Biosystems). Because we chose a higher annealing temperature (>60°C), two temperatures (without a combined anneal-extend step) were employed with the following conditions: DNA polymerase activation and denaturing at 95°C for 10 min, 60 cycles of annealing at 94°C for 20 s and at 62°C for 1 min, and a final extension at 72°C for 10 min. Three primer pairs (Table 1) were tested to amplify the different sizes of β-actin cDNA. The RT-PCR products were evaluated by electrophoresis on 3% agarose gels stained with ethidium bromide. All experiments were done in duplicate to verify the results.
Table 1. PCR primer sets and expected amplicon sizes tested for the β-actin gene.
| Primer sets | Amplicon size | |
|---|---|---|
| (1) | Forward 5′-ACACTGTGCCCATCTACGAGG-3′ | 620 bp |
| Reverse 5′-AGGGGCCGGACTCGTCATACT-3′ | ||
| (2) | Forward 5′-GGACTTCGAGCAAGAGATGG-3′ | 234 bp |
| Reverse, 5′-AGCACTGTGTTGGCGTACAG-3′ | ||
| (3) | Forward 5′-CCCAGCACAATGAAGATCAA-3′ | 103 bp |
| Reverse 5′-ACATCTGCTGGAAGGTGGAC-3 |
Statistical Analysis
The distributional forms of all measurements were first assessed using both graphical and statistical testing procedures. As a result, all data were log-transformed to reduce the deviation from normality before descriptive analysis; however, anti-log values are presented in the Results. The correlation between the measurements with different preservation methods was assessed by Pearson correlation coefficients.
Results
RNA yield varied markedly among specimens. The mean yield was highest for P, followed by R and N, whereas that from W and S was low. Analysis of 260/280 ratios showed that samples frozen in liquid nitrogen and samples treated with R yielded the best quality RNA (1.6-2.0). A high degree of protein contamination (<1.6) was present in S and W samples, whereas higher RNA degradation (>2.0) was indicated (14) for samples treated with P (Table 2). There were negative correlations between P and N results (−0.54 for the 260/280 ratios and −0.25 for the concentrations), but positive correlations between the R and N results (0.44 and 0.23, respectively). The difference in the two correlation coefficients for the 260/280 ratios was statistically significant (P = 0.044).
Table 2. Means, 80% confidence intervals, coefficients of variation, and ranges of total RNA yields (ng/μL) and 260/280 ratios by preservation method tested for stool specimens.
| Preservation methods* | No. | Variables | Mean (80% confidence interval) | Coefficient of variation (range) |
|---|---|---|---|---|
| N | 10 | Concentration | 7.50 (4.36-12.88) | 61.4% (2.28-67.74) |
| 260/280 ratio | 1.65 (1.52-1.80) | 37.5% (1.20-2.20) | ||
| P | 10 | Concentration | 36.13 (20.52-63.61) | 36.1% (3.26-194.77) |
| 260/280 ratio | 2.04 (1.96-2.13) | 13.6% (1.63-2.19) | ||
| R | 10 | Concentration | 15.26 (7.89-29.50) | 55.3% (2.89-273.87) |
| 260/280 ratio | 1.73 (1.58-1.89) | 38.0% (1.17-2.18) | ||
| S | 10 | Concentration | 4.70 (3.98-5.55) | 24.5% (2.89-9.47) |
| 260/280 ratio | 1.55 (1.48-1.63) | 24.7% (1.35-1.98) | ||
| W | 10 | Concentration | 3.52 (2.82-4.40) | 40.6% (2.31-9.54) |
| 260/280 ratio | 1.48 (1.38-1.58) | 38.6% (1.16-1.90) |
N, liquid nitrogen; P, paxgene; R: RNAlater; S, silica gel; W, Whatman FTA card.
RNA electrophoresis showed some degradation in all samples (data not shown). A small amount of residual intact 28S RNA was identified in some R and N samples, but was only rarely identified in samples collected in the other media;the 28S RNA band was truncated to the lower molecular weight. DNA was clearly visible in most P samples and some N and R samples.
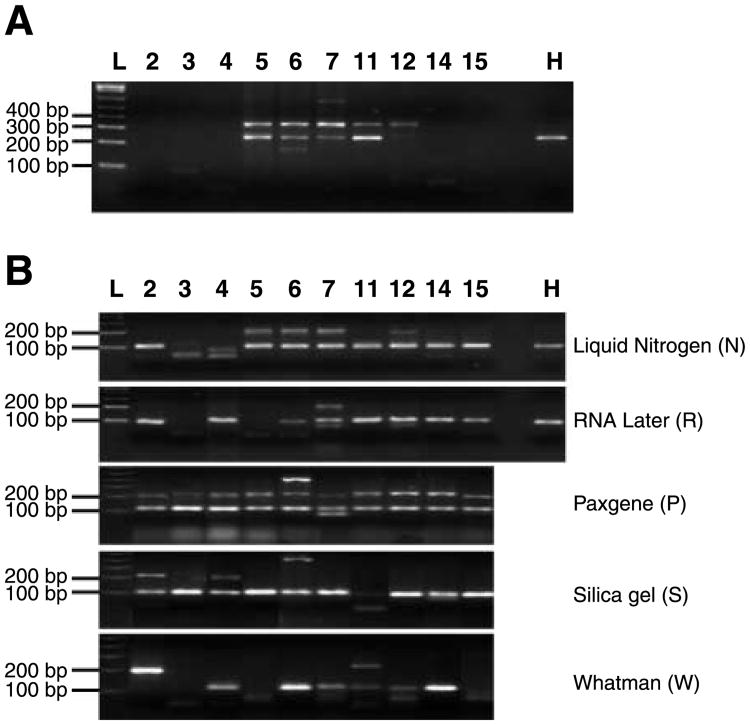
Successful PCR amplification of cDNA sequences was dependent on the primer sets tested. Using the first set of primers, the 620 bp amplicon was obtained from only 1 of the 10 samples tested (data not shown). The second primer yielded a positive band (234 bp) in 40% to 50% of the samples, regardless of preservation method. Fig. 1A shows examples of the 10 N samples. The most consistent positive results were obtained with the third primer pair with an expected product size of 103 bp (Fig. 1B). Among several preservation vehicles tested, P and S yielded the highest human RNA detection rate (10 of 10). N and R (8 of 10) and W (6 of 10) yielded the next higher detection rates for the 103 bp amplicon. The 215 bp PCR products in some samples (e.g., all P samples and several N samples) were thought to originate from contaminating genomic DNA, which includes a 112 bp intron in this part of the β-actin gene. By this criterion, only one R sample and two each of W and S samples seemed to have significant DNA contamination. A PCR product of unknown identity of ∼400 bp was observed in a sample from patient 7 (Fig. 1A) and in P and S samples from patient 6 (Fig. 1B), along with other unknown smaller bands, including a <200 bp fragment in patient 6 (Fig. 1A) and <100 bp fragments in several samples preserved by different methods (Fig. 1B).
Figure 1.
RT-PCR product analysis on agarose gel. Ten microliters of the PCR product was electrophoresed on 3% agarose gels containing ethidium bromide. L, a 1 kb plus molecular DNA ladder (Invitrogen). H, human colon cell line HCT-116. 2 to 15, patient ID numbers. A. Expression of the second primer pair products (234 bp) with the 10 liquid nitrogen preserved samples. B. Expression of the third primer pair products (103 bp).
Discussion
Human fecal RNA is an understudied type of biospecimen because it was believed that the isolation of human RNA with reasonably good quality is difficult. Even if stool specimens are processed immediately after defecation, RNA in exfoliated epithelial cells may have been subject to degradation in vivo over several hours or days depending on the stool transit time. In addition, bacterial enzymes may contribute to the degradation of human DNA/RNA. This feasibility study confirmed that degradation is common regardless of processing methods, as shown by the reduced size of the higher molecular weight ribosomal band and the presence of low molecular weight RNA. Furthermore, microbial RNA/DNA contamination may have contributed to unidentified extra PCR bands in some of the samples.
The present study shows that commercially available RNA preservation kits enable investigators to collect only a small aliquot of fecal samples, in place of whole stool specimens, for large populations. Although these kits have been tested for various samples including fecal non–human RNA (9–11, 13), to our knowledge, there are no other published studies reporting parallel comparisons of these methods for human fecal samples. Importantly, these methods are also useful in preserving human and microbial DNA, which we simultaneously evaluated (15).
In the present study, extraction of RNA with the MO BIO RNA PowerSoil method yielded human RNA from most samples. The lower yield with the W method was primarily due to the fact that the amount of stool used for extraction was only 5% of that used in the other methods. The final choice of method should depend on both overall quality and quantity of RNA/DNA, as well as the presence of PCR inhibitors, common in fecal specimens (7). The R method is potentially interesting because it gave the best quality RNA with acceptable human RNA detection rate.
There are some important technical considerations in the application of RT-PCR. RNA samples are often contaminated with genomic DNA. Relatively slight DNA contamination may lead to erroneous results in the analysis of gene expression. In this study, sense and antisense primers were chosen to reside in different exons (16). PCR products originating from contaminating DNA could be distinguished from the authentic cDNA product by their larger size, which includes intronic sequences and results in additional slower bands or no band on agarose gels. According to this criterion, preservation of fecal samples by the R method facilitated the preparation of DNA-free RNA for RT-PCR because only one of the R-preserved samples exhibited apparent genomic DNA contamination, in contrast to P-preserved samples, all of which showed genomic contamination, and N-preserved samples of which about half exhibited the genomic product. However, some protein contamination was present even in R and N samples because the average 260/280 ratio was <1.8 (17). However, the level of protein contamination we observed was considered acceptable for this type of complex sample (18, 19). Greater protein contamination may decrease the efficiency of the conversion of RNA to cDNA by blocking reverse transcriptase activity, as well as to inefficient PCR amplification by affecting DNA polymerase activity.
Another factor influencing the RT-PCR results is the amplicon length because templates for longer PCR products are less abundant in degraded samples. Our data and published results (20) suggest that amplicons should be designed as ∼100 bp in order to obtain evaluable results for this type of degraded RNA samples. RNA degradation is one of the most frequent reasons for sample variation. RNA isolation usually involves treatment with DNase, which is often not completely RNasefree and thus further degrade RNA. RNA degradation may be the reason for the loss of bands in samples from patient nos. 3 and 5 treated with R, and in the sample from patient no. 11 treated with S. Normalizing the expression of a specific gene to a “housekeeping” gene such as β-actin will allow relative mRNA quantification in degraded RNA samples correcting for variations in RNA concentrations. The present study suggests that such normalization is feasible even for stool specimens.
Acknowledgments
The authors thank Wayne State University Applied Genomics Technology Center and Director, Dr. Susan Land, for their technical expertise in RNA assessment.
Grant support: Research Enhancement Program of Wayne State University.
References
- 1.Ries LAG, Melbert D, Krapcho M, et al. SEER Cancer Statistics Review. National Cancer Institute; Bethesda, MD: 1975-2004. Available from: http://seer.cancer.gov/csr/1975_2004/, based on November 2006 SEER data submission, posted to the SEER web site, 2007. [Google Scholar]
- 2.Wurzelmann JI, Silver A, Schreinemachers DM, Sandler RS, Everson RB. Iron intake and the risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 1996;5:503–7. [PubMed] [Google Scholar]
- 3.Kato I, Dnistrian AM, Schwartz M, et al. Iron intake, body iron stores and colorectal cancer risk in women: a nested case-control study. Int J Cancer. 1999;80:693–8. doi: 10.1002/(sici)1097-0215(19990301)80:5<693::aid-ijc11>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 4.Iyengar V, Albaugh GP, Lohani A, Nair PP. Human stools as a source of viable colonic epithelial cells. FASEB J. 1991;5:2856–9. doi: 10.1096/fasebj.5.13.1655550. [DOI] [PubMed] [Google Scholar]
- 5.Desilets DJ, Davis KE, Nair PP, et al. Lectin binding to human colonocytes is predictive of colonic neoplasia. Am J Gastroenterol. 1999;94:744–50. doi: 10.1111/j.1572-0241.1999.00946.x. [DOI] [PubMed] [Google Scholar]
- 6.Davidson LA, Lupton JR, Miskovsky E, Fields AP, Chapkin RS. Quantification of human intestinal gene expression profiles using exfoliated colonocytes: a pilot study. Biomarkers. 2003;8:51–61. doi: 10.1080/1354750021000042268. [DOI] [PubMed] [Google Scholar]
- 7.Kanaoka S, Yoshida K, Miura N, Sugimura H, Kajimura M. Potential usefulness of detecting cyclooxygenase 2 messenger RNA in feces for colorectal cancer screening. Gastroenterology. 2004;127:422–7. doi: 10.1053/j.gastro.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 8.Subrungruang I, Mungthin M, Chavalitshewinkoon-Petmitrp, et al. Evaluation of DNA extraction and PCR methods for detection of Enterocytozoon bienuesi in stool specimens. J Clin Micrbiol. 2004;42:3490–4. doi: 10.1128/JCM.42.8.3490-3494.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogers CD, Burgoyne LA. Reverse transcription of an RNA genome from databasing paper (FTA®) Biotechnol Appl Biochem. 2000;31:219–24. doi: 10.1042/ba19990113. [DOI] [PubMed] [Google Scholar]
- 10.Whittier CA, Horne W, Slenning B, Loomis M, Stoskopf MK. Comparison of storage methods for reverse-transcriptase PCR amplification of rotavirus RNA from gorilla (Gorilla g. gorilla) fecal samples. J Virol Methods. 2004;116:11–7. doi: 10.1016/j.jviromet.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Santiago ML, Lukasik M, Kamenya S, et al. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pantroglodytes schweinfurthii) J Virol. 2003;77:7545–62. doi: 10.1128/JVI.77.13.7545-7562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wasser SK, Houston CS, Koehler GM, Cadd GG, Fain SR. Techniques for application of faecal DNA methods to field studies of Ursids. Mol Ecol. 1997;6:1091–7. doi: 10.1046/j.1365-294x.1997.00281.x. [DOI] [PubMed] [Google Scholar]
- 13.Habis AH, Vernon SD, Lee DR, Verma M, Unger ER. Molecular quality of exfoliated cervical cells: implications for molecular epidemiology and biomarker discovery. Cancer Epidemiol Biomarkers Prev. 2004;13:492–6. [PubMed] [Google Scholar]
- 14.Kim HS, Byun SH, Lee BM. Effects of chemical carcinogens and physicochemical factors on the UV spectrophotometric determination of DNA. J Toxicol Environ Health A. 2005;68:2081–95. doi: 10.1080/15287390500182503. [DOI] [PubMed] [Google Scholar]
- 15.Nechvatal J, Ram J, Marc Basson M, et al. Fecal collection, ambient preservation, and DNA extraction for PCR amplification of bacterial and human markers from human feces. J Microbiol Methods. doi: 10.1016/j.mimet.2007.11.007. In press. [DOI] [PubMed] [Google Scholar]
- 16.Nakajima-Iijima S, Hamada H, Reddy P, Kakunaga T. Molecular structure of the human cytoplasmic β-actin gene: interspecies homology of sequences in the introns. Proc Natl Acad Sci U S A. 1985;82:6133–7. doi: 10.1073/pnas.82.18.6133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsvik PA, Lie KK, Hevrøy EM. Do anesthetics and sampling strategies affect transcription analysis of fish tissues? BMC Mol Biol. 2007;8:48. doi: 10.1186/1471-2199-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar GN, Iyer S, Knowles NR. Extraction of RNA from fresh, frozen, and lyophilized tuber and root tissues. J Agric Food Chem. 2007;55:1674–8. doi: 10.1021/jf062941m. [DOI] [PubMed] [Google Scholar]
- 19.He LH, Zhao Y, Chen MJ, Pan YJ. An efficient method for DNA extraction from compost. Wei Sheng Wu Xue Bao. 2006;46:162–5. [PubMed] [Google Scholar]
- 20.Tong D, Schneeberger C, Leodolter S, Zeillinger R. Quantitative determination of gene expression by competitive reverse transcrip-tion-polymerase chain reaction in degraded RNA samples. Anal Biochem. 1997;251:173–7. doi: 10.1006/abio.1997.2280. [DOI] [PubMed] [Google Scholar]