Abstract
Objective
Systemic sclerosis (SSc) is a heterogeneous multifactorial disease dominated by progressive skin and internal organ fibrosis that is driven in part by Transforming Growth Factor-beta (TGF-β). An important downstream target of TGF-β is the Abelson (c-Abl) tyrosine kinase, and its inhibition by imatinib mesylate (Gleevec)attenuates fibrosis in mice. Here we examined the effect of c-Abl activation and blockade in explanted healthy control and SSc fibroblasts.
Methods
Skin biopsies and explanted fibroblasts from healthy subjects and patients with SSc were studied. Changes in genome-wide expression patterns in imatinib-treated control and SSc fibroblasts were analyzed by DNA microarray.
Results
Treatment of control fibroblasts with TGF-β resulted in activation of c-Abl and stimulation of fibrotic gene expression that was prevented by imatinib. Moreover, imatinib reduced basal collagen gene expression in SSc but not control fibroblasts. No significant differences in tissue levels of c-Abl and phospho-c-Abl were detected between SSc and control skin biopsies. In vitroimatinib induced dramatic changes in the expression of genes involved in fibrosis, cardiovascular disease, inflammation, and lipid and cholesterol metabolism. Remarkably, of the 587-imatinib-responsive genes, 91% showed significant change in SSc fibroblasts, but only 12% in control fibroblasts.
Conclusion
c-Abl plays a key role in fibrotic responses. Imatinib treatment results in dramatic changes in gene expression in SSc fibroblasts but has only modest effects in control fibroblasts. These data provide novel insights into the mechanisms underlying the antifibrotic effect of imatinib in SSc.
Systemic sclerosis (SSc) is a clinically heterogeneous disease of unknown etiology. In some patients, the disease course is dominated by progressive fibrosis in the skin and internal organs[1]. The pathogenesis of fibrosis in SSc is not well understood. Transforming Growth Factor-beta (TGF- β) causes fibroblast activation and differentiation and is implicated as a major factor[2]. The canonical Smad pathway propagates TGF-β-driven profibrotic signaling in fibroblasts, but non-canonical pathways have been shown to be important as well[3]. Abelson kinase (c-Abl) is a non-receptor tyrosine kinase that is best known for its role in chronic myelogenous leukemia[4]. Recent studies in fibroblasts suggest that TGF-β may play a role in c-Abl activation and consequent stimulation of extracellular matrix protein synthesis in vitro and in vivo[5–8]. The precise mechanism of c-Abl-mediated profibrotic signaling and its role in SSc remain incompletely understood.
The small molecule imatinib mesylate (Gleevec) is a tyrosine kinase inhibitor that binds to the c-Abl kinase domain thereby blocking substrate phosphorylation[9]. The drug is currently approved for the treatment of chronic myelogenous leukemia and gastrointestinal stromal tumors[10, 11]. Imatinib has been shown to modulate TGF-β signaling in vitro, and prevent fibrosis in mouse models of scleroderma and pulmonary fibrosis[5, 7, 8, 12, 13]. Recently, we showed that early growth response gene-1 (Egr-1) is a profibrotic downstream target of TGF-β and c-Abl that is important in the pathogenesis of SSc[6]. Skin biopsies from a subset of SSc patients treated with imatinib for one year revealed decreased Egr-1 expression compared to baseline levels[14]. Together these findings suggest a potential role for c-Abl in the pathogenesis of SSc, and imply that its pharmacological inhibition may be a useful approach to control fibrosis. However, recent clinical trials of imatinib therapy in fibrosis have shown variable results, with some studies demonstrating improvement in skin and lung fibrosis while others show no benefits [15–19].
Analysis of expression microarrays from explanted fibroblasts and skin biopsies has been used to identify signaling pathways potentially underlying the pathogenesis of SSc[20, 21]. Using DNA microarray, we showed that cultured fibroblasts from a subset of patients with the diffuse form of SSc displayed a TGF-β-responsive gene expression signature in vitro[22]. A recent study identified biologically relevant gene expression changes in skin biopsies from SSc patients treated with imatinib[23]. To better understand the role of c-Abl activation and its inhibition in SSc, we conducted a series of experiments utilizing explanted fibroblasts and skin biopsies. The purpose of this report is to define the anti-fibrotic effects of imatinib in explanted control and SSc fibroblasts in vitro, and to characterize the genome-wide changes in expression induced by imatinib in control and SSc fibroblasts.
Materials and Methods
Cell Culture and Reagents
Primary cultures of human dermal fibroblasts were established by explantation from neonatal foreskin or from biopsies of the affected dorsal forearm from patients with diffuse cutaneous SSc (dcSSc) as indicated. Abl-null/Arg-null (Abl−/−/Arg−/−) mouse embryonic fibroblasts stably expressing wild-type c-Abl or a kinase-inactive form of c-Abl were also used[6]. Skin biopsies were obtained in accordance with Northwestern University Institutional Review Board Guidelines, and propagated as previously described[24, 25]. When fibroblasts reached confluence, cultures were serum-starved or incubated with fresh media with 10% fetal calf serum (FCS) (Gibco BRL, Grand Island, NY). Media contained indicated concentrations of imatinib (gift from Novartis Pharmaceuticals (Basel, Switzerland)) dissolved in water and were adjusted to a final concentration of 10 µM (5.89 µg/ml), a concentration two times the peak plasma concentration at steady state in patients with gastrointestinal stromal tumors treated with imatinib[26]. In selected experiments, TGF-β1 (Pepro Tech, Rocky Hill, NJ) or PDGF-AB (Amgen, Thousand Oaks, CA) was added to the cultures for 24h. Experiments were performed in triplicate, and repeated two-three times with consistent results.
Western analysis
At the end of each experiment, whole cell lysates were prepared and subjected to immunoblot analysis, as previously described[25]. Briefly, equal amounts of proteins (50µg/lane) were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) in 4–15% gels, transferred to Immobilon-P membranes (Millipore, Billerica, MA) and immunoblotted overnight with primary antibodies specific for Type I collagen (1:400 dilution) (Southern Biotech, Birmingham, AL), or α-tubulin (1:1000 dilution) (Sigma-Aldrich, St. Louis, MO). Membranes were then incubated with appropriate secondary antibodies and subjected to enhanced chemiluminescence detection using ECL Reagent (Amersham-Pharmacia, Piscataway, NJ). Results were quantified using Image J (NIH software).
In some experiments, foreskin fibroblasts were transiently transfected with FLAG-tagged c-Abl expression vectors harboring a mutated nuclear translocalization signal (ΔNLS) (generous gift from Dr. Jean Y.J. Wang, University of California, San Diego, CA) as previously described[27]. After overnight serum deprivation, fibroblasts were incubated with TGF-β1 at indicated concentrations for 30–60 min with or without PDGF-AB [25 µM] for 20 min as a positive control. Whole cell lysates were isolated and immunoprecipitated with anti-FLAG (M2) antibody (Sigma-Aldrich, St. Louis, MO) and run in SDS-polyacrylamide gels, and transferred to membranes. After blocking with 10% nonfat dry milk, membranes were subjected to immunoblot analysis with primary antibody to phospho-serine/threonine/tyrosine followed by horseradish peroxidase-conjugated secondary antibodies (both Abcam, Cambridge, UK) as previously described[28]. Protein levels were quantified by scanning densitometry using Image J software.
Northern Analysis
For determination of mRNA levels, total RNA was isolated from fibroblasts using TRIZOL Reagent (Gibco BRL, GRAND Island, NY), and examined by Northern blot analysis as previously described [25]. Filters were sequentially hybridized with [32P]-labeled human cDNA probes for COL1A1 and COL1A2, TIMP-1, PAI-1, fibronectin, CTGF and 18S ribosomal RNA. Signal intensities were quantified by densitometry, and results in each sample were normalized for the 18S ribosomal RNA signal intensities.
c-Abl kinase assays
The regulation of cellular c-Abl kinase activity was determined by in vitro kinase assays[13]. Briefly, confluent foreskin fibroblasts were incubated with TGF-β1 at indicated concentrations in the presence or absence of imatinib [10µM] for 30 min. Cells were then harvested and lysed in kinase buffer[5]. Cellular c-Abl was immunoprecipitated using K12 antibody (Santa Cruz Biotechnology, Santa Cruz, CA), immune complexes were collected with protein A-Sepharose, washed twice and incubated in kinase buffer containing 0.5 µci [γ32P] ATP for 5 min as previously described[13]. Complexes were then subjected to SDS-PAGE followed by autoradiography. Total c-Abl levels were detected by Western blot analysis.
Cell proliferation assays
Early passage foreskin fibroblasts were seeded in quadruplet at 50% confluence and incubated in DMEM with 10% FCS. After 24 h, cells were counted to determine baseline cell numbers. Imatinib was added to the cultures at indicated concentrations, and cells were counted following 24 or 48 h further incubation.
Immunohistochemistry
Skin biopsies from the affected forearm of 13 patients with early diffuse cutaneous SSc or 6age-matched healthy adult control subjects were obtained. Paraffin-embedded tissue blocks were sectioned (5 µm thick) and immunostained using primary antibody to c-Abl (Cell Signaling; Cat# 2862; final concentration 1:100) or p-c-Abl (Abcam; Cat# ab62189; final concentration 1:100), followed by secondary antibodies (Dako Company, Carpinteria, CA). Sections from breast cancer tumors were stained in parallel as positive controls. A negative control consisted of substituting nonspecific rabbit IgG (Invitrogen, Carlsbad, CA) for primary antibody. Slides were counterstained with hematoxylin and eosin and visualized using a Zeiss Axioskop microscope. Positively stained fibroblastic cells were counted at 40X magnification by two independent observers. Photomicrographs of biopsies were taken with a CRi Nuance spectral (breast cancer controls) or DP71® Olympus camera.
Statistical analyses
A Student’s t-test and Fisher's exact test were used as appropriate to identify statistically significant differences between clinical variables and means of positively stained dermal fibroblasts. Two-sample t-tests were used to compare mRNA levels normalized for Col1A1 between control and each experimental group. Analyses were conducted using SPSS (Chicago, IL) and SAS (version9.2 Cary, NC) statistical software.
Microarray hybridization and data analysis
To examine changes in mRNA levels induced by imatinib at the genomewide level, fibroblasts explanted from skin biopsies of an age- and sex-matched healthy control subject and a patient with dcSSc were seeded in duplicate. At confluence, fibroblasts were placed in serum-free media and incubated with imatinib [10µM] for a further 24h. Total RNA was isolated at the beginning and at the end of the incubation period using Qiagen Mini Plus Kits (Qiagen, Valencia, CA). RNA integrity was determined using an Agilent Bioanalyzer (Santa Clara, CA). Fluorescently-labeled cDNA was prepared using labeling kits (TargetAmp 1-Round Aminoallyl-aRNA Ki; Epicentre, Madison, WI), followed by hybridization to Illumina Human HT-12Version 3 microarray chips (Illumina, San Diego, CA) containing 48,802 probes and expressed sequence tags. Raw signal intensities for each probe were obtained using Illumina Beadstudio data analysis software and imported to the Bioconductor lumi package for transformation and normalization [29–31]. The data were preprocessed using a variance stabilization transformation method [31] followed by quantile normalization. Data from probes that produced signals near or below background levels (estimated based on Illumina negative control probes) with all samples were discarded, leaving 20,746 probes that were further analyzed.
Statistical and Bioinformatics Analysis
To identify important changes in gene expression induced by imatinib, we calculated fold-change in expression comparing expression levels between treated and untreated healthy fibroblasts and treated and untreated SSc fibroblasts. Genes whose mean expression showed ≥2-fold change (post/pretreatment of the duplicate samples) were identified. Cluster analysis was used to identify major gene clusters. One-sample t-tests were used to determine if fold-change in genome-wide expression was significantly different from untreated healthy fibroblasts for each gene cluster. Two-sample t-tests were used to compare fold-change in gene expression in SSc fibroblasts between pre- and post-imatinib treatment for each gene cluster. The Bonferroni correction method was used to correct for multiple hypothesis testing. Ingenuity Pathway Analysis software and the Bioconductor package: GeneAnswers were used to interpret gene expression results in the context of relevant biochemical pathways and gene ontology (GO) categories. Changes in the expression of selected genes with biological relevance were validated by quantitative real-time PCR.
Quantitative Real-time Polymerase Chain Reaction
At the end of the incubation periods, total RNA was isolated from fibroblasts using Trizol reagent (Life Technologies, Grand Island, New York). For qPCR, 1µg of total RNA was reverse transcribed to cDNA in 20 µl reaction volume using cDNA Synthesis Supermix (Quanta Biosciences, Gaithersburg, MD). 8 µl cDNA, 2 µl primers (2 µM each) and 10 µl of 2× Power SYBR Mastermix (Applied Biosystems, Foster City, CA) were used for real-time qPCR that was performed in triplicate on an ABI 7300 Thermocycler (Applied Biosystems). Data were normalized to actin or 18S RNA, and fold change was represented as 2−ΔΔCt (2-((Ct target-Ct 18S)treatment - (Ct target-Ct 18S) non-treatment)). The primers used for real-time qPCR were: Col1A1 (forward 5’-GCTGGTGTGATGGGATTC-3’ and reverse 5’-GGGAACACCTCGCTCT-3’), Il-6 (forward 5’- AAATTCGGTACATCCTCGACGG-3’ and reverse 5’-GGAAGGTTCAGGTTGTTTTCTGC-3’), CTGF (forward 5’-AGCTGACCTGGAAGAGAACATTAAG-3’ and reverse 5’-GATAGGCTTGGAGATTTTGGGAGTA-3’) 18S (forward 5’-CATGAGAAGTATGACAACAGCCT-3’ and reverse 5’- AGTCCTTCCACGATACCAAAGT-3’), α-smooth muscle actin (forward 5’-CAGGGCTGTTTTCCCATCCAT-3′ and reverse 5’-GCCATGTTCTATCGGGTACTTC-3′)and actin (forward 5’-AATGTCGCGGAGGACTTTGAT-3’ and reverse 5’-AGGATGGCAAGGGACTTCCTG-3’).
Results
TGF-β induces c-Abl phosphorylation
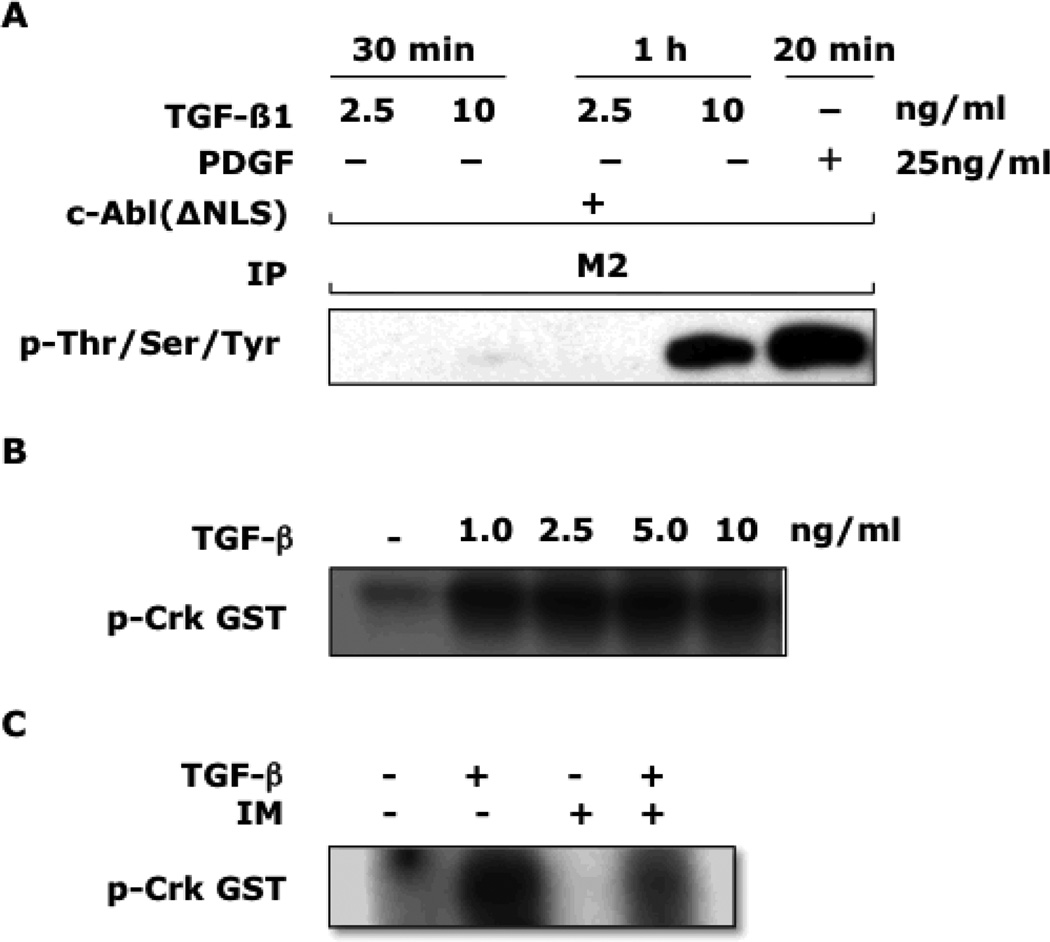
To evaluate the effect of TGF-β on c-Abl activity in fibroblasts, two complementary approaches were taken. First, confluent healthy skin fibroblasts transiently transfected with a FLAG-tagged c-Abl (ΔNLS) vector that encodes for a mutated c-Abl unable to undergo nuclear translocation to facilitate detection were incubated with TGF-β or PDGF, and whole cell lysates were subjected to immunoprecipitation with anti-FLAG, followed by Western analysis[32]. The results demonstrated that TGF-β stimulation resulted in potent phosphorylation of recombinant c-Abl that was comparable in magnitude to that induced by PDGF (Fig. 1A). Next, in vitro kinase assays were used to evaluate the effect of TGF- β on fibroblast c-Abl kinase activity. The results showed that low concentrations of TGF-β stimulated c-Abl kinase activity as assessed by substrate phosphorylation (Fig. 1B). As expected, preincubation of the fibroblasts with imatinib partially blocked this response (Fig. 1C). These results indicate that TGF-β induced both the phosphorylation and kinase activity of c-Abl in healthy dermal fibroblasts, and imatinib at pharmacologic concentrations abrogates the TGF-β-induced responses.
Figure 1. TGF-β stimulates c-Abl phosphorylation and Abl kinase activity.
A: Foreskin fibroblasts were transiently transfected with a FLAG-tagged c-Abl (ΔNLS) expression vector. 24 h later, TGF-β1 at indicated concentrations or PDGF [25 ng/ml] was added for a further 30 or 60 min (TGF-β) and 20 min (PDGF). Whole cell lysates were immunoprecipitated with anti-FLAG (M2) antibodies, and immunoprecipitates were subjected to Western analysis using antibodies to phospho-serine/threonine/tyrosine. Results are representative of two independent experiments. B and C:Fibroblasts were incubated with TGF-β at indicated concentrations in the presence (+) or absence (−) of imatinib [10µM] for 30 min. Whole cell lysates were immunoprecipitated with antibodies to c-Abl, and kinase activity assayed using (GST)-Crk as substrate.
Imatinib decreases the expression of fibrotic genes
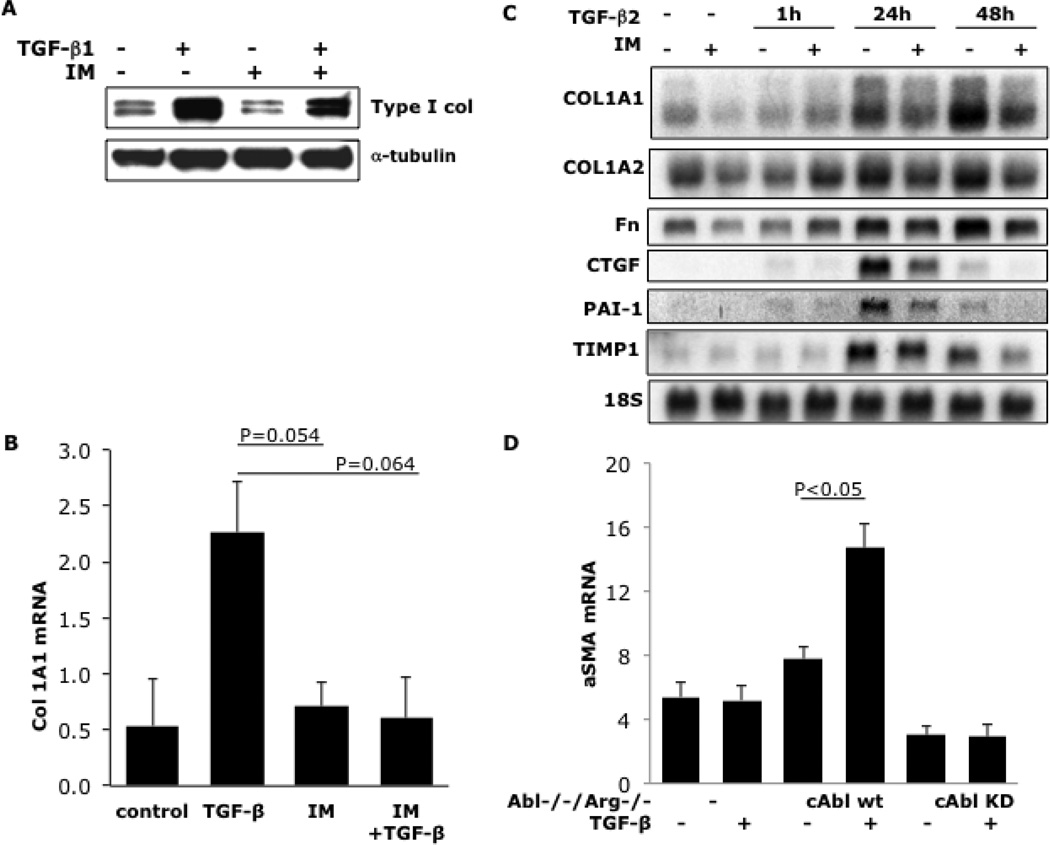
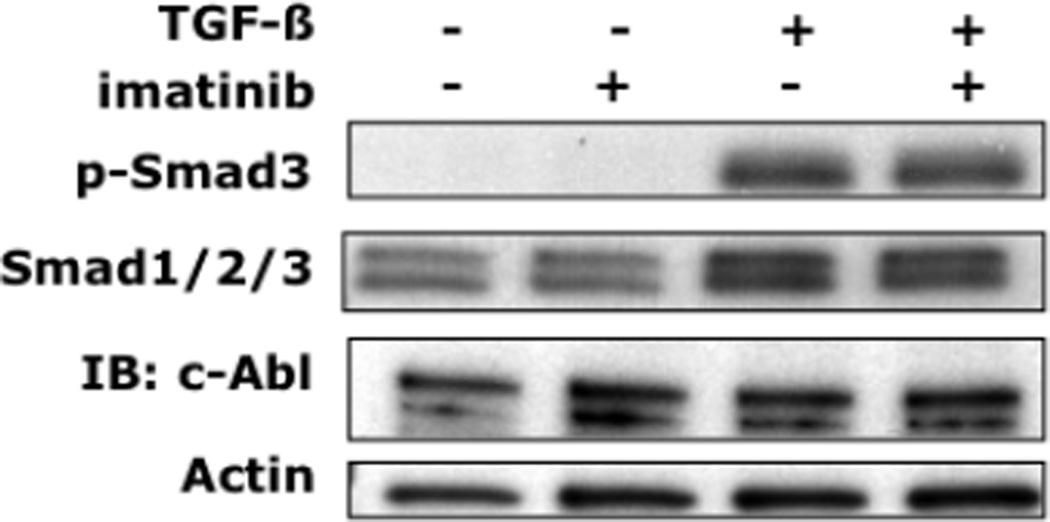
To determine the effect of c-Abl blockade on TGF-β-induced profibrotic responses, neonatal fibroblasts were incubated with TGF-β in the presence or absence of imatinib for 24h, and extracellular matrix gene expression was evaluated. The results showed that imatinib abrogated the stimulation of Type I collagen production induced by TGF-β (Fig. 2). The stimulation of other gene products important in fibrosis, including fibronectin, connective tissue growth factor, plasminogen activator inhibitor-1 and tissue inhibitor of metalloproteinases was also abrogated by imatinib (Fig. 2C). Importantly, at concentrations up to 10 µM, imatinib had no effect on fibroblast proliferation or survival as assessed by cell proliferation and MTT assays (data not shown). In order to further validate the role of c-Abl in TGF-β-induced fibrogenic responses, we used a complementary genetic approach. The results demonstrate that genetic loss of c-Abl completely abrogates TGF-β-induced stimulation of α-smooth muscle actin production (Fig. 2D). Smad 3 plays an important role in mediating the stimulation of collagen gene expression by TGF-β [33]. To examine the effect of imatinib on Smad3 activation, dermal fibroblasts were incubated with TGF-β for 30 min in the presence or absence of imatinib. As shown in Figure 3, TGF-β induced an increase in the levels of phosphorylated Smad3 that was not prevented by preincubation of the fibroblasts with imatinib.
Figure 2. Imatinib abrogates TGF-β stimulation of the expression of extracellular matrix genes, and c-Abl is important in mediating TGF-β-induced profibrotic responses.
Foreskin fibroblasts were pretreated with imatinib [10 µM] for 30 min prior to the addition of TGF-β [12.5 ng/ml]. Cells were harvested following a further incubation for 24h (A, B) or indicated periods (C). Whole cell lysates were subjected to Western analysis (A). Total RNA was subjected to real-time qPCR (B) or Northern analyses (C). The results of Western analyses quantitated by Image J software and normalized for α-tubulin, are representative of three independent experiments. D:Abl−/−/Arg−/− mouse embryonic fibroblasts stably expressing c-Abl (cAbl wt) or a kinase-inactive mutant form of c-Abl (cAbl KD) in parallel were incubated with TGF-β for 24h. Total RNA was subjected to real-time qPCR analysis. Results, expressed relative to GAPDH are means ± SD from triplicate determinations from a representative experiment.
Figure 3. Inhibition of c-Abl activity by imatinib is independent of Smad3.
Foreskin fibroblasts were pretreated with imatinib (+) [10µM] for 30 min prior to the addition of TGF-β 1 [10 ng/ml] (30 min). Whole cell lysates were subjected to Western analysis. Representative immunoblots. Identical responses were seen with mouse fibroblasts (data not shown).
Imatinib blocks TGF-β signaling in explanted SSc fibroblasts
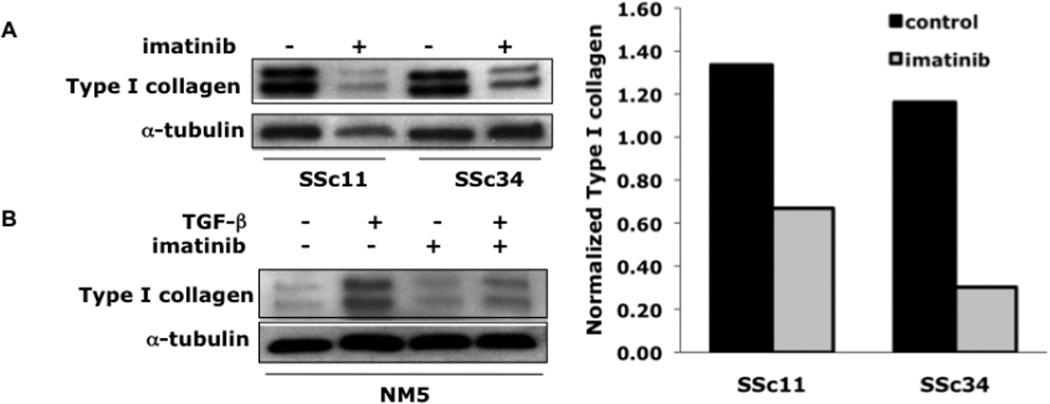
We next examined the effect of imatinib on collagen gene expression in fibroblasts explanted from two patients with dcSSc. Western analysis showed elevated expression of Type I collagen in unstimulated SSc fibroblasts that was comparable to levels in fibroblasts from an age-and sex-matched healthy control stimulated with TGF-β (Fig. 4). Incubation of SSc fibroblasts with imatinib caused a >80% reduction in Type I collagen gene expression (Fig. 4A). In contrast, imatinib had little effect on basal collagen gene expression in fibroblasts explanted from four healthy donors (Fig. 4B and data not shown).
Figure 4. Imatinib reduces Type I collagen levels in SSc fibroblasts.
(A) At early confluence, SSc fibroblasts were incubated in the absence (−) or presence (+) of imatinib [10µM] for 24h. Following 24h incubation, fibroblasts were harvested and subjected to Western analysis. (A, right panel) Results were quantified using Image J software. (B) Healthy adult dermal fibroblasts were pre-treated with imatinib [10µM] for 30min prior to the addition of TGF-β1 (12.5 ng/ml). NM5= healthy control.
p-c-Abl staining in SSc fibroblasts
To assess c-Abl expression and activation in SSc, skin biopsies from 13 patients with SSc and 6healthy controls were studied by immunohistochemistry. The clinical characteristics of the subjects are shown in Table 1. Six of the thirteen patients had early SSc disease duration defined as ≤1year between the onset of the first non-Raynauds symptom and the biopsy date. Three of 16 patients had interstitial lung disease defined as a forced vital capacity <70 % predicted on pulmonary function testing (PFT). Symptoms, abnormal echocardiography and/or PFT results raised suspicion for pulmonary artery hypertension in one subject who was referred for a right heart catheterization that revealed a mean pulmonary artery pressure <25 mm Hg.
Table 1.
Clinical characteristics of subjects
| Characteristic | SSc (n=13) | Controls (n=6) |
|---|---|---|
| Age, y mean (SD) |
48(12) | 36(15) |
| % Female (n)* | 100(13) | 0(0) |
| % White (n) | 69(9) | 50(3) |
| Disease duration, mo mean (SD) |
34(33) | N/A |
| Subtype (% dcSSc) | 85% | N/A |
| mRSS, median (range) | 18(4–36) | N/A |
| Pulmonary function tests, mean (SD) % predicted FVC DLCO |
77(18) 66(21) |
N/A |
| SSc complications: n(%) ILD PAH |
3(23) 0 |
N/A |
Skin biopsies from 13patients with scleroderma and 6 healthy controls were studied. mRSS:modified Rodnan skin score;ILD:interstitial lung disease;PAH:pulmonary arterial hypertension; dcSSc: diffuse cutaneous SSc; NA: Not applicable:
p<0.05.
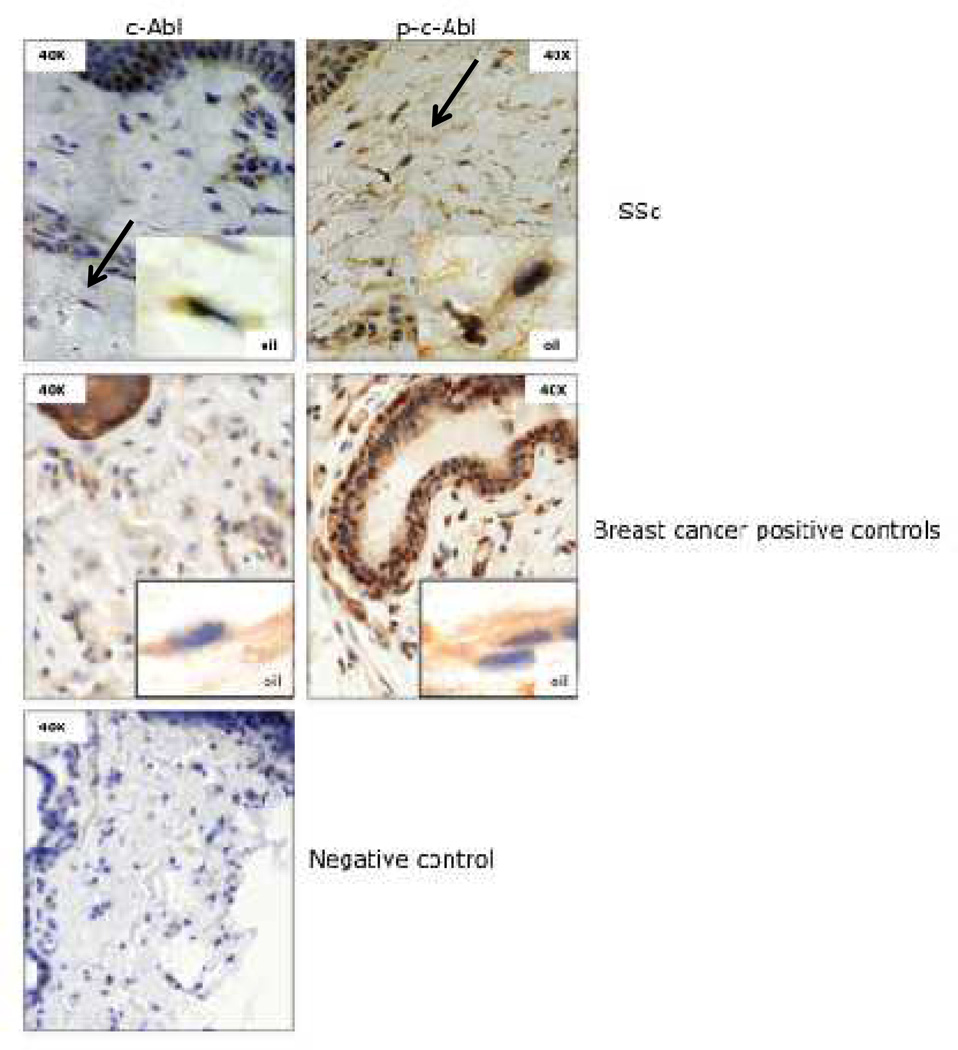
Fibroblastic cells with positive immunostaining for c-Abl and phospho-c-Abl were detected in the dermis of10/13 and 4/6skin biopsies respectively(Fig. 5). In addition to fibroblastic cells in the dermis, epidermal keratinocytes at the basal layer, and perivascular supporting cells also demonstrated strong c-Abl and phospho-c-Abl staining (Fig 5, left and right panel). Three out of 6 skin biopsies from healthy individuals showed positive c-Abl, and 5/6 showed positive phospho-c-Abl immunostaining. There was no significant difference in the numbers of immunopositive fibroblastic cells between SSc and control biopsies. These data suggest that c-Abl is expressed and variably activated in dermal fibroblasts, basal keratinocytes and perivascular supporting cells in the skin.
Figure 5. C-Abl is phosphorylated in SSc skin.
Skin biopsies from involved skin of SSc patients (n=13) and healthy controls (n=6) were immunostained for c-Abl and phospho-c-Abl. Right and left panels (40X magnification), reveal increased dermal thickness in SSc biopsies with increased collagen deposition. Photomicrograph from SSc biopsies with increased c-Abl and p-c-Abl staining. Arrows indicate immunopositive fibroblastic cells. Positive (middle panel) control breast cancer tissue shows strong immunostaining.
Imatinib has dramatic effects on gene expression in SSc fibroblasts but not control fibroblasts
To examine modulation of gene expression by imatinib at the genome-wide level, we performed DNA microarray analysis. For this comparison, we selected a line of fibroblasts explanted from a woman with diffuse SSc that, in preliminary studies, consistently showed elevated baseline collagen gene expression compared to an age-matched healthy control fibroblast line (data not shown).Confluent control and SSc fibroblasts in parallel were serum-deprived overnight and then incubated with 10µM imatinib for 24h, followed by isolation of RNA and hybridization to Illumina chips. Genes whose mean expression showed ≥2-fold change after imatinib were analyzed. Of 20,746 probes (13,091 genes) whose expression levels were above the background, 742 probes (587 genes, 4.5%) showed ≥2-fold change in relative expression in either control or SSc fibroblasts following imatinib (Fig. 6A). Remarkably, of the 587 differentially-expressed genes, 91% showed significant change in response to imatinib in SSc fibroblasts whereas only 12% showed significant change in control fibroblasts (p < 0.001).
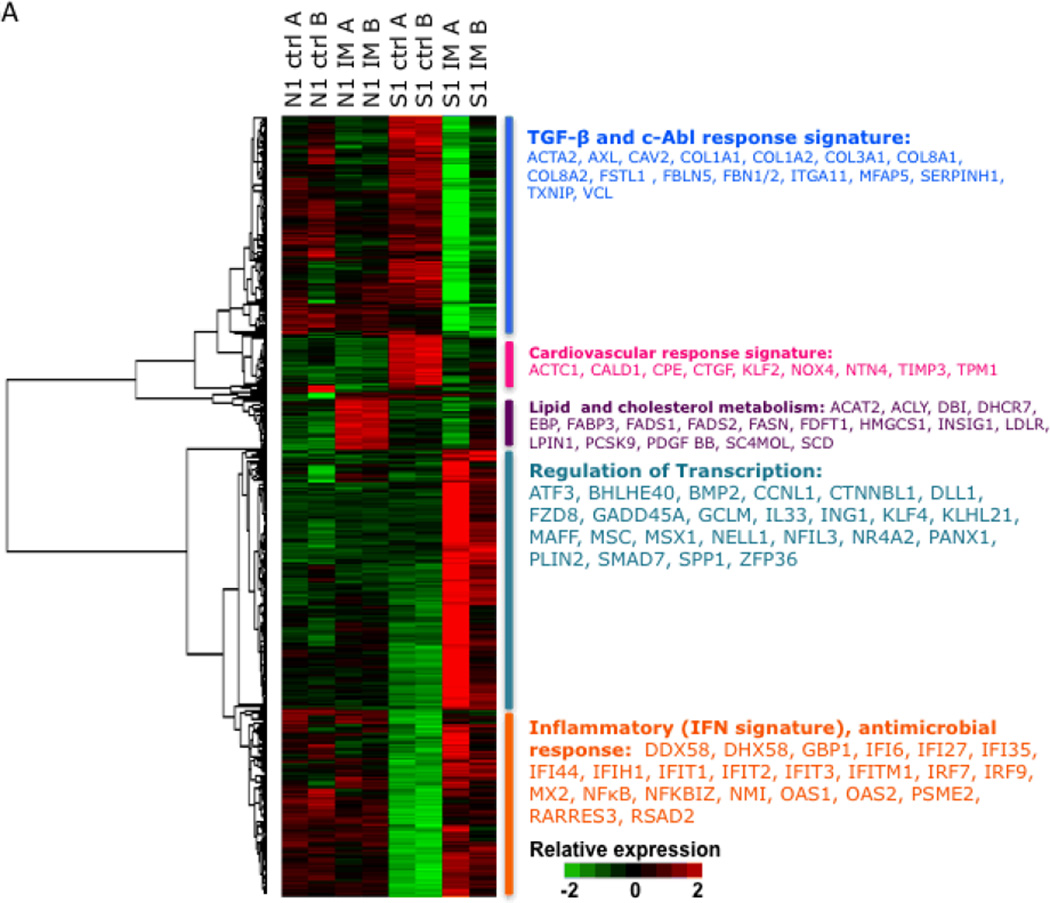
Figure 6.
A: Gene expression pattern of the IM response signature in explanted dermal fibroblasts. Explanted dermal fibroblasts from a healthy individual (N1) and a patient with early diffuse SSc (S1)were left untreated (Ctrl) or incubated with imatinib (IM) for 24h. The heat map demonstrates the pre/post treatment gene expression (N1/S1 ctrl vs. N1/S1 IM) as indicated for the 588 genes whose expression changed upon imatinib treatment ≥2-fold from baseline. Red represents an increase, and green a decrease, in mRNA expression from the mean expression across all samples. Each pair of columns represents the mRNA profile of a sample and its biologic replicate (A and B) while the rows represent the genes present in the imatinib response signature. The dendrogram reveals the five major clusters, and the major functions/pathways of the cluster members are identified to the right.
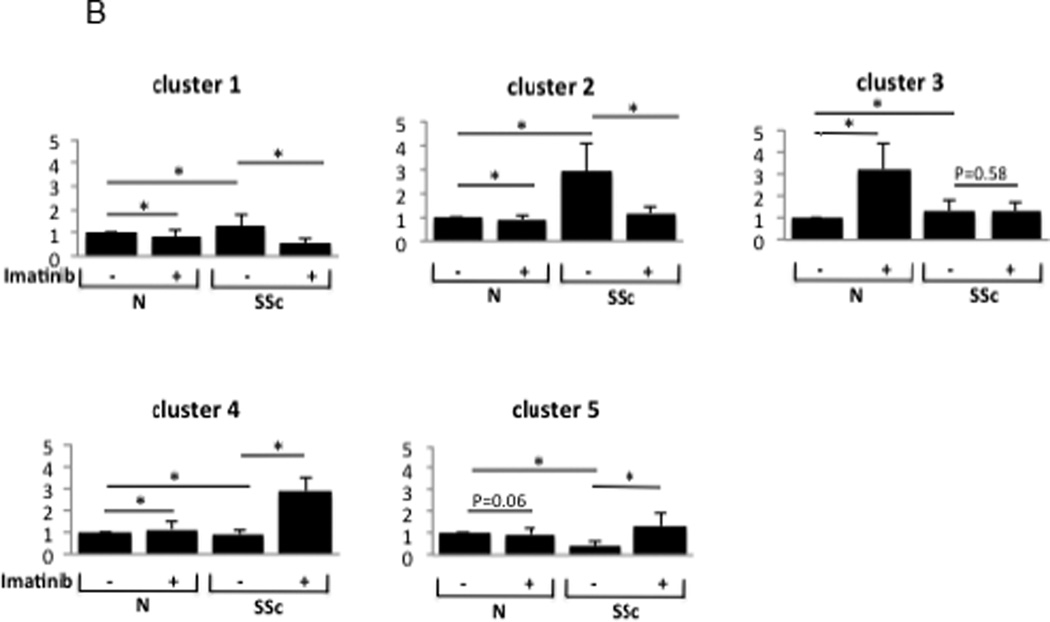
B: Expression changes in genes for five biologically relevant clusters = Ingenuity Pathway Analysis was used to identify the biological theme for the genes with the most significantly altered expression in response to imatinib. To quantify results, fold-change was calculated by comparing expression levels (in control post-, SSc pre-, and SSc post-imatinib) to control pre-imatinib gene expression. Bars represent fold changes for samples compared to healthy control fibroblasts or to untreated SSc fibroblasts.* Indicates P < 0.001.
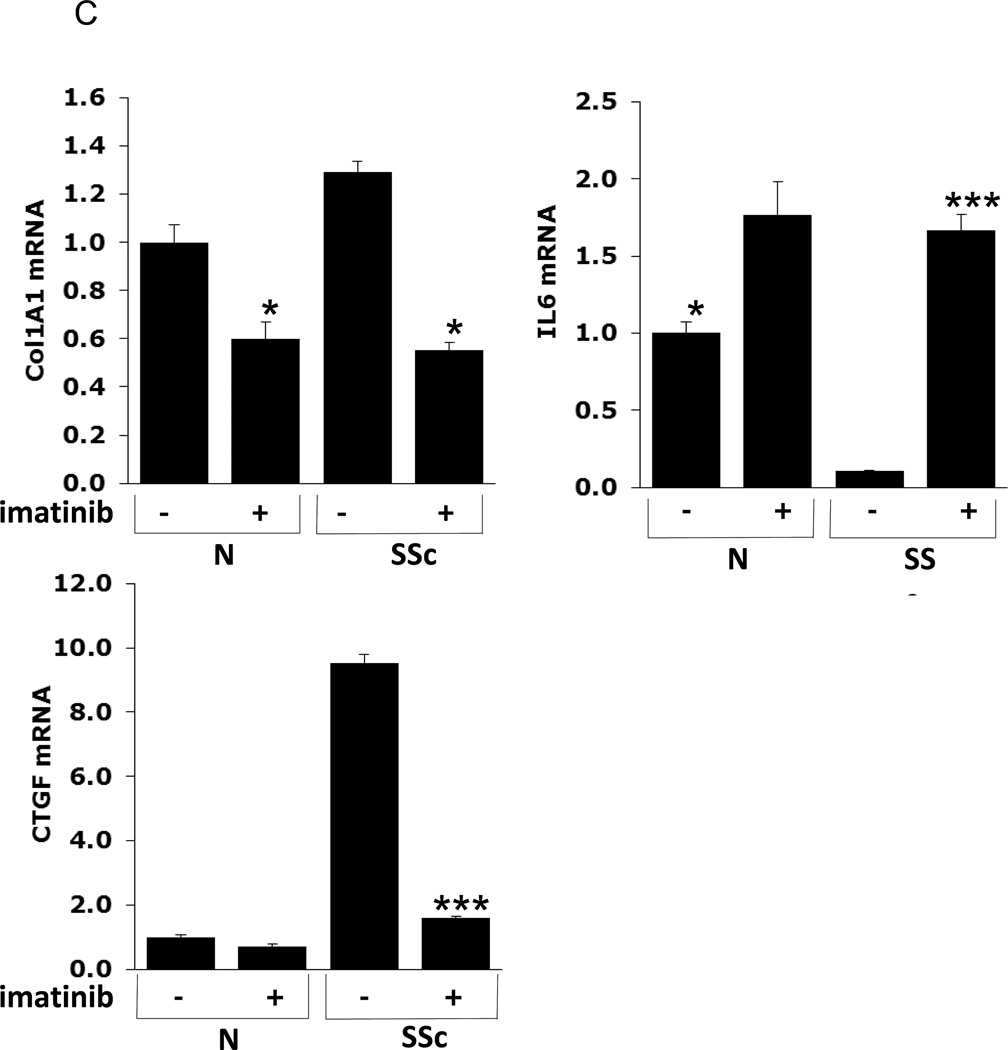
C: Quantitative RT-PCR confirms microarray results. RNA harvested from cultured SSc and healthy dermal fibroblasts (N) pre- (−) and 24h post-imatinib (+) treatment was used to confirm expression levels for Col1A1, interleukin-6 (IL-6), and connective tissue growth factor (CTGF). * Indicates P < 0.05, *** indicates P < 0.001.
The ‘fibroblast imatinib response gene signature’: five clusters
The expression of the imatinib-regulated gene signature in explanted fibroblasts was evaluated. Unsupervised cluster analysis identified five distinct molecular clusters (Fig. 6A). To identify the major biological pathways enriched with genes that showed the greatest changes in expression, we performed two complementary analyses using Ingenuity Pathway Analysis and GeneAnswers (Bioconductor Package). We labeled cluster 1 a TGF-β and c-Abl response gene signature composed of genes that are known to be involved with extracellular matrix homeostasis, including ACTA2: actin, alpha 2, smooth muscle, aorta, CAV2: caveolin 2, COL1A1: collagen, type I, alpha 1, COL1A2: collagen, type I, alpha 2, and FBN1/2: fibrillin 1/2. The baseline expression of these genes was elevated in SSc fibroblasts compared with healthy fibroblasts, consistent with previous studies [22]. Incubation with imatinib reduced the expression of these genes in SSc but not control fibroblasts(Fig. 6A).
Cluster 2 is comprised of genes implicated in cardiovascular system development and function including ACTC1: actin, alpha, cardiac muscle 1, CALD1: caldesmon 1, TPM1: tropomyosin 1, and NOX4: NADPH oxidase 4); as well as genes regulated by TGF-β or PDGF (CTGF: connective tissue growth factor, NOX4and TUBA1A: tubulin, alpha 1a), many of which have been implicated in SSc and fibrosis[34]. The genes in this cluster were overexpressed in the SSc fibroblasts compared to healthy fibroblasts, and their expression was “normalized” by imatinib. In contrast, imatinib had little effect on the expression of these genes in control fibroblasts. Of particular interest was ACTC1, defects in which have been associated with idiopathic dilated cardiomyopathy and familial hypertrophic cardiomyopathy. CALD1 encodes a calmodulin- and actin-binding protein called caldesmon that plays an essential role in the regulation of smooth muscle and non-smooth muscle contraction. Caldesmon phosphorylation promotes actin binding and formation of stress fibers[35].There are no studies to date that have examined the role of caldesmon in Raynaud phenomenon or dermal fibrosis in SSc; however, its biologic function make it an interesting gene for further study. NADPH oxidase 4 catalyzes the formation of reactive oxygen species that may be involved with endothelial angiogenesis, myofibroblast differentiation and pulmonary fibrosis [36–39]. Moreover, studies have reported the potential relevance of NOX4 in SSc dermal and lung fibrosis rendering NOX4 a potential novel therapeutic target [20, 40].
Cluster 3 is comprised of genes involved in lipid and cholesterol metabolism. This cluster includes ACAT2 that encodes acetyl-CoA acetyltransferase 2 that is an enzyme involved in lipid metabolism; ACLY that encodes a enzyme (ATP citrate lyase) known to play a role in lipogenesis and cholesterogenesis; FABP3 that encodes the fatty acid binding protein 3 family of proteins (heart FABP3is released into the circulation after myocardial damage [41–44]); FADS1/2 that encode fatty acid desaturase gene product family that regulate unsaturation of fatty acids; and SCD that encodes stearoyl-CoA desaturase, an enzyme that catalyzes the formation of the unsaturated fatty acid oleic acid that is thought to paly a role in cell membrane fluidity and signal transduction[45, 46]. The expression of these genes was substantially increased by imatinib in control fibroblasts. In striking contrast, in SSc fibroblasts imatinib had no effect on the expression of these genes. These data suggest that there may be alterations in adipogenesis and in cholesterol and lipid metabolism intrinsic to SSc fibroblasts.
The fourth expression cluster included many genes related to transcription(ATF3: activating transcription factor 3, CCNL1: cyclin L1, KLF4: Kruppel-like factor 4, MAFF:v-maf musculoaponeurotic fibrosarcoma oncogene homolog F, MAFG: v-maf musculoaponeurotic fibrosarcoma oncogene family, protein G, MSC: musculin, MSX1: msh homeobox 1, NFIL3: nuclear factor, interleukin 3 regulated, and NR4A2: nuclear receptor subfamily 4, group A, member 2). Interestingly, extracellular signal-regulated kinase1/2 activity has been shown to suppress ATF3 activity in patients with asthma, and ERK2 phosphorylation of NR4A2 is postulated to play a role in Parkinson’s disease [47, 48]. Both of these gene products are activated by phosphorylation, and kinase inhibition by imatinib may upregulate expression. Whereas imatinib treatment resulted in little change in expression of these genes in healthy fibroblasts, in SSc fibroblasts, they demonstrated profound upregulation in response to imatinib.
Genes in cluster 5 are related to inflammation, innate immunity and type I interferon signaling. This cluster includes ADAR: adenosine deaminase, DDX58:DEAD (Asp-Glu-Ala-Asp) box polypeptide 58, DHX58: DEXH (Asp-Glu-X-His) box polypeptide 58, GBP1:guanylate binding protein 1, IFI6/27:interferon, alpha-inducible protein 6/27, IFI35/44: interferon-induced protein 35/44,IFIH1: interferon induced with helicase C domain 1, IFIT1-3: interferon-induced protein with tetratricopeptide repeats 1-3, IFITM1: interferon induced transmembrane protein 1, OAS1&2: 2'-5'-oligoadenylate synthetase 1 and 2.The expression of these genes was reduced in SSc fibroblasts compared to control fibroblasts. Imatinib reduced or did not alter the expression of the genes in this group in the healthy control fibroblasts, but markedly upregulated gene expression in SSc fibroblasts (Fig. 6B). These data may explain the frequent observation of lower extremity edema in SSc patients receiving imatinib for skin and lung fibrosis[19].
We used qRT-PCR to validate key microarray findings. Relative to untreated healthy fibroblasts, levels of Type I Collagen and Connective Tissue Growth Factor (CTGF) mRNA expression were elevated in SSc fibroblasts. Incubation with imatinib abrogated this overproduction (Fig. 6C). We confirmed that interleukin-six (IL-6) mRNA levels were 20-fold higher in imatinib-treated compared to untreated SSc fibroblasts (Fig. 6C). Changes in the levels of expression of each gene examined showed a concordant pattern as determined by microarray.
Comparison of imatinib-induced gene expression changes in vivo and in vitro
A recent study of two patients by Chung et al. identified 987 genes in skin whose expression changed significantly during successful imatinib treatment[23]. We compared the list of these genes to the genes demonstrating ≥2-fold change in expression in imatinib-treated SSc fibroblasts in the present study. The purpose of this analysis was to compare the gene expression response pre- and post-imatinib therapy in biopsies to the in vitro imatinib response in cultured dermal fibroblasts. If changes in gene expression in cultured fibroblasts mirror changes that occur in the biopsies of imatinib treatment responders then the differentially expressed genes from microarray analyses of cultured fibroblasts may be a useful SSc biomarker to predict imatinib response. In the skin biopsies from imatinib-treated SSc patients there were 1,025 probes (987 unique genes)whose expression changed ≥1.5-fold over background. Seventy-eight (10%) of the 587 imatinib-responsive genes in cultured fibroblasts overlapped with the Chung et al. gene list. The hypergeometric test which estimates the probability that a gene will occur on two top-ranking gene lists due to chance alone, predicted only 4.4% overlap (p < 0.0001) [49]. Thus, the gene list from the present study is enriched for imatinib responsive genes and includes ACTA2, CALD1, CAV1, CAV2,COL1A1/2, CTGF, IGFBP1, MYH10 and SERPINB2/PAI. Interestingly, the direction of change in expression was concordant in only 36% of the genes.
Discussion
Because c-Abl inhibition is a promising potential therapy for SSc, we sought to elucidate the role of c-Abl and its modulation by imatinib in the context of fibrogenesis. The results demonstrated that c-Abl is activated in skin fibroblasts in response to TGF-β, and in fibroblasts. Imatinib treatment of SSc dermal fibroblasts abrogated the overproduction of Type I collagen in vitro. Moreover imatinib induced dramatic alterations in gene expression in explanted SSc fibroblasts compared with healthy fibroblasts.
Our results confirm that TGF-β stimulation of neonatal and adult dermal fibroblasts results in phosphorylation and activation of c-Abl. Using genetic targeting, we showed that intact c-Abl is necessary for profibrotic responses in mouse embryonic fibroblasts. Interestingly, low concentrations of TGF-β are sufficient to induce c-Abl kinase activity, while higher concentrations are required for c-Abl phosphorylation, reflecting the greater sensitivity of the kinase assays. Further, we confirmed that that the TGF-β/c-Abl pathway is Smad-independent. Our results demonstrate that imatinib ameliorates the increased expression of collagen in SSc fibroblasts.
We were unable to demonstrate consistent differences in c-Abl and phospho-c-Abl staining in skin from SSc patients and healthy controls. There were positively stained fibroblastic cells in the dermis of SSc biopsies, but the results were inconsistent amongst biopsies. Further, there were some biopsies from healthy individuals that revealed positive staining. This is most likely due to technical limitations of the antibodies used for immunohistochemistry and underscores the need to develop better methodologies for assessing phosphorylation of c-Abl in skin.
Given the heterogeneity in c-Abl activation amongst SSc and healthy fibroblasts, we selected for further analysis an SSc cell line known to overexpress Type I collagen and other extracellular matrix proteins in vitro. Microarray data demonstrate that imatinib induces changes in expression in more genes in SSc fibroblasts than in healthy fibroblasts. Gene ontology analyses revealed that gene expression differed in SSc and healthy fibroblasts in biologically relevant pathways under the control of TFG-β/c-Abl that previous studies have demonstrated may be important in tissue fibrosis in SSc. Additionally, there were marked differences in the expression of genes involved with cardiovascular response, lipid and cholesterol metabolism, regulation of transcription and inflammation that have not been previously reported.
Of note, imatinib induced the expression of genes involved with adipogenesis and lipid and cholesterol metabolism in healthy fibroblasts but not SSc fibroblasts. We have previously demonstrated constitutively low expression of peroxisome proliferator-activated receptor gamma (PPAR-γ)in SSc fibroblasts[50]. Further, SSc skin biopsies showed decreased PPAR-γ activity[50]. The nuclear receptor PPAR-γ plays key roles in regulation of adipogenesis in adipocytes and fibroblasts. Further, we, and others, found an inverse correlation between skin fibrosis and serum adiponectin levels in SSc patients[51]. Together these data suggest that imatinib may have varying effects upon the expression of genes involved with adipogenesis and cholesterol metabolism because there are underlying differences in the activation of these pathways between SSc and healthy fibroblasts.
Imatinib upregulated the expression of multiple genes related to inflammation. These include genes regulated by type I interferons such as Interferon Regulatory Factor (IRF) 7 and 9 and oligoadenylate synthetase 1 and 2 (OAS).Interestingly, a recent study showed that bcr-Abl overexpression in hematopoietic cells blocked interferon Type I responses[52]. We speculate that blockade of c-Abl activation with imatinib may result in disinhibition of some genes that normally dampen the inflammatory response.
There are limitations to the conclusions that may be drawn from microarray experiments that utilize explanted fibroblasts. First, prior studies have demonstrated that SSc fibroblasts lose their SSc signature with increased passaging and that skin biopsies may be a better disease model. We used early passage SSc fibroblasts, but it is possible that the imatinib signature that we identified in SSc and healthy dermal fibroblasts treated with imatinib are not representative. Second, gene expression signatures in fibroblasts may reflect culture techniques and not imatinib treatment, thus it is possible that some of the changes in expression that we are attributing to imatinib may have been due to the addition/removal of serum for instance.
We compared the changes in gene expression induced by imatinib in explanted dermal fibroblasts to genes that showed significant changes in skin biopsies from patients successfully treated with imatinib[23]. The absolute number of genes whose expression changed in response to imatinib in explanted fibroblasts and biopsies was small; however, there were more genes common to both lists than would be expected by chance. Alterations in the expression of genes involved with extracellular matrix synthesis and turnover were most commonly altered indicating that imatinib may have important effects in profibrotic responses and repair.
In summary, these results show that c-Abl is activated by TGF-β in healthy fibroblasts, and is constitutively activated in some SSc fibroblasts. We show that SSc fibroblasts demonstrate a robust response to imatinib with marked changes in the expression of a large number of genes. Furthermore, we identify an imatinib response gene signature in explanted SSc fibroblasts that is enriched with genes involved with regulation of extracellular matrix remodeling, cardiovascular disease, lipid and cholesterol metabolism, and inflammation. These findings suggest that imatinib treatment may affect multiple signaling pathways that may have important clinical implications.
Acknowledgements
We thank Joan Guitart, Junjie Shangguan and Kathleen Kelley for help with immunohistochemistry.
This work was supported in part by grants from the Karen Brown Scleroderma Foundation (MH), NIH K12 HD055884 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development (MH), the Arthritis Foundation Illinois Chapter (MH), NIH P60 AR048098 (CCH), and NIH R01 AR42309 (FF, WI, SB, and JV).
Footnotes
No financial conflicts of interest to declare.
References
- 1.Hinchcliff M, Varga J. Systemic sclerosis/scleroderma: a treatable multisystem disease. Am Fam Physician. 2008;78(8):961–968. [PubMed] [Google Scholar]
- 2.Varga J, Pasche B. Transforming growth factor beta as a therapeutic target in systemic sclerosis. Nat Rev Rheumatol. 2009;5(4):200–206. doi: 10.1038/nrrheum.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trojanowska M. Noncanonical transforming growth factor beta signaling in scleroderma fibrosis. Curr Opin Rheumatol. 2009;21(6):623–629. doi: 10.1097/BOR.0b013e32833038ce. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goldman JM, Melo JV. Targeting the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344(14):1084–1086. doi: 10.1056/NEJM200104053441409. [DOI] [PubMed] [Google Scholar]
- 5.Daniels CE, et al. Imatinib mesylate inhibits the profibrogenic activity of TGF-beta and prevents bleomycin-mediated lung fibrosis. J Clin Invest. 2004;114(9):1308–1316. doi: 10.1172/JCI19603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhattacharyya S, et al. A non-Smad mechanism of fibroblast activation by transforming growth factor-beta via c-Abl and Egr-1: selective modulation by imatinib mesylate. Oncogene. 2009;28(10):1285–1297. doi: 10.1038/onc.2008.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Distler JH, Distler O. Imatinib as a novel therapeutic approach for fibrotic disorders. Rheumatology (Oxford) 2009;48(1):2–4. doi: 10.1093/rheumatology/ken431. [DOI] [PubMed] [Google Scholar]
- 8.Distler JH, et al. Imatinib mesylate reduces production of extracellular matrix and prevents development of experimental dermal fibrosis. Arthritis Rheum. 2007;56(1):311–322. doi: 10.1002/art.22314. [DOI] [PubMed] [Google Scholar]
- 9.Takimoto C, Calvo E. Principles of Oncologic Pharmacotherapy" in (Eds) . 11 ed. 2008. In: Pazdur R, et al., editors. Cancer Management: A Multidisciplinary Approach. 2008. [Google Scholar]
- 10.Druker BJ, et al. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344(14):1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- 11.Joensuu H, et al. Effect of the tyrosine kinase inhibitor STI571 in a patient with a metastatic gastrointestinal stromal tumor. N Engl J Med. 2001;344(14):1052–1056. doi: 10.1056/NEJM200104053441404. [DOI] [PubMed] [Google Scholar]
- 12.Wang S, et al. Imatinib mesylate blocks a non-Smad TGF-beta pathway and reduces renal fibrogenesis in vivo. Faseb J. 2005;19(1):1–11. doi: 10.1096/fj.04-2370com. [DOI] [PubMed] [Google Scholar]
- 13.Wilkes MC, Leof EB. Transforming growth factor beta activation of c-Abl is independent of receptor internalization and regulated by phosphatidylinositol 3-kinase and PAK2 in mesenchymal cultures. J Biol Chem. 2006;281(38):27846–27854. doi: 10.1074/jbc.M603721200. [DOI] [PubMed] [Google Scholar]
- 14.Gordon J. In: Personal communication. Hinchcliff M, editor. 2011. [Google Scholar]
- 15.Daniels CE, et al. Imatinib treatment for idiopathic pulmonary fibrosis: Randomized placebo-controlled trial results. Am J Respir Crit Care Med. 2010;181(6):604–610. doi: 10.1164/rccm.200906-0964OC. [DOI] [PubMed] [Google Scholar]
- 16.Sabnani I, et al. A novel therapeutic approach to the treatment of scleroderma-associated pulmonary complications: safety and efficacy of combination therapy with imatinib and cyclophosphamide. Rheumatology (Oxford) 2009;48(1):49–52. doi: 10.1093/rheumatology/ken369. [DOI] [PubMed] [Google Scholar]
- 17.Spiera RF, et al. Imatinib mesylate (Gleevec) in the treatment of diffuse cutaneous systemic sclerosis: results of a 1-year, phase IIa, single-arm, open-label clinical trial. Ann Rheum Dis. 2011 doi: 10.1136/ard.2010.143974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamaki Z, et al. Efficacy of low-dose imatinib mesylate for cutaneous involvement in systemic sclerosis: a preliminary report of three cases. Mod Rheumatol. 2011 doi: 10.1007/s10165-011-0472-1. [DOI] [PubMed] [Google Scholar]
- 19.Pope J, et al. American College of Rheumatology Annual Scientific Meeting. Philadelphia PA: Arthritis and Rheumatism; 2009. A Proof of Concept Trial of Gleevec (Imatinib) in Active Diffuse Scleroderma (DSSc) p. S227. [Google Scholar]
- 20.Gardner H, et al. Gene profiling of scleroderma skin reveals robust signatures of disease that are imperfectly reflected in the transcript profiles of explanted fibroblasts. Arthritis Rheum. 2006;54(6):1961–1973. doi: 10.1002/art.21894. [DOI] [PubMed] [Google Scholar]
- 21.Whitfield ML, et al. Systemic and cell type-specific gene expression patterns in scleroderma skin. Proc Natl Acad Sci U S A. 2003;100(21):12319–12324. doi: 10.1073/pnas.1635114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sargent JL, et al. A TGFbeta-responsive gene signature is associated with a subset of diffuse scleroderma with increased disease severity. J Invest Dermatol. 2010;130(3):694–705. doi: 10.1038/jid.2009.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung L, et al. Molecular framework for response to imatinib mesylate in systemic sclerosis. Arthritis Rheum. 2009;60(2):584–591. doi: 10.1002/art.24221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mori Y, et al. Selective inhibition of activin receptor-like kinase 5 signaling blocks profibrotic transforming growth factor beta responses in skin fibroblasts. Arthritis Rheum. 2004;50(12):4008–4021. doi: 10.1002/art.20658. [DOI] [PubMed] [Google Scholar]
- 25.Mori Y, Chen SJ, Varga J. Modulation of endogenous Smad expression in normal skin fibroblasts by transforming growth factor-beta. Exp Cell Res. 2000;258(2):374–383. doi: 10.1006/excr.2000.4930. [DOI] [PubMed] [Google Scholar]
- 26.Ksienski D. Imatinib mesylate: past successes and future challenges in the treatment of gastrointestinal stromal tumors. Clin Med Insights Oncol. 2011;5:365–379. doi: 10.4137/CMO.S4259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jing Y, et al. c-Abl tyrosine kinase regulates c-fos gene expression via phosphorylating RNA polymerase II. Arch Biochem Biophys. 2005;437(2):199–204. doi: 10.1016/j.abb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Mori Y, Chen SJ, Varga J. Expression and regulation of intracellular SMAD signaling in scleroderma skin fibroblasts. Arthritis Rheum. 2003;48(7):1964–1978. doi: 10.1002/art.11157. [DOI] [PubMed] [Google Scholar]
- 29.Du P, Kibbe WA, Lin SM. nuID: a universal naming scheme of oligonucleotides for illumina, affymetrix, and other microarrays. Biol Direct. 2007;2:16. doi: 10.1186/1745-6150-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 31.Lin SM, et al. Model-based variance-stabilizing transformation for Illumina microarray data. Nucleic Acids Res. 2008;36(2):e11. doi: 10.1093/nar/gkm1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Einhauer A, Jungbauer A. The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. J Biochem Biophys Methods. 2001;49(1–3):455–465. doi: 10.1016/s0165-022x(01)00213-5. [DOI] [PubMed] [Google Scholar]
- 33.Chen SJ, et al. Stimulation of type I collagen transcription in human skin fibroblasts by TGF-beta: involvement of Smad 3. Journal of Investigative Dermatology. 1999;112(1):49–57. doi: 10.1046/j.1523-1747.1999.00477.x. [DOI] [PubMed] [Google Scholar]
- 34.Varga J, Abraham D. Systemic sclerosis: a prototypic multisystem fibrotic disorder. J Clin Invest. 2007;117(3):557–567. doi: 10.1172/JCI31139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung WK, Ching AK, Wong N. Phosphorylation of Caldesmon by PFTAIRE1 kinase promotes actin binding and formation of stress fibers. Molecular and Cellular Biochemistry. 2011;350(1–2):201–206. doi: 10.1007/s11010-010-0699-8. [DOI] [PubMed] [Google Scholar]
- 36.Pendyala S, et al. Nrf2 regulates hyperoxia-induced Nox4 expression in human lung endothelium: identification of functional antioxidant response elements on the Nox4 promoter. Free Radic Biol Med. 2011;50(12):1749–1759. doi: 10.1016/j.freeradbiomed.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crestani B, Besnard V, Boczkowski J. Signalling pathways from NADPH oxidase-4 to idiopathic pulmonary fibrosis. Int J Biochem Cell Biol. 2011;43(8):1086–1089. doi: 10.1016/j.biocel.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 38.Craige SM, et al. NADPH oxidase 4 promotes endothelial angiogenesis through endothelial nitric oxide synthase activation. Circulation. 2011;124(6):731–740. doi: 10.1161/CIRCULATIONAHA.111.030775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carnesecchi S, et al. A key role for NOX4 in epithelial cell death during development of lung fibrosis. Antioxid Redox Signal. 2011;15(3):607–619. doi: 10.1089/ars.2010.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Andrade JA, Thannickal VJ. Innovative approaches to the therapy of fibrosis. Curr Opin Rheumatol. 2009;21(6):649–655. doi: 10.1097/BOR.0b013e328330da9b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sbarouni E, et al. Increases in serum concentration of human heart-type fatty acid-binding protein following elective coronary intervention. Biomarkers. 2009;14(5):317–320. doi: 10.1080/13547500902887530. [DOI] [PubMed] [Google Scholar]
- 42.Muehlschlegel JD, et al. Heart-type fatty acid binding protein is an independent predictor of death and ventricular dysfunction after coronary artery bypass graft surgery. Anesth Analg. 2010;111(5):1101–1109. doi: 10.1213/ANE.0b013e3181dd9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao J, et al. Human heart-type fatty acid-binding protein for on-site diagnosis of early acute myocardial infarction. Int J Cardiol. 2009;133(3):420–423. doi: 10.1016/j.ijcard.2008.01.049. [DOI] [PubMed] [Google Scholar]
- 44.Giannessi D, et al. Heart-type fatty acid binding protein is an early marker of myocardial damage after radiofrequency catheter ablation. Clin Biochem. 2010;43(15):1241–1245. doi: 10.1016/j.clinbiochem.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Yao-Borengasser A, et al. Stearoyl-coenzyme A desaturase 1 gene expression increases after pioglitazone treatment and is associated with peroxisomal proliferator-activated receptor-gamma responsiveness. J Clin Endocrinol Metab. 2008;93(11):4431–4439. doi: 10.1210/jc.2008-0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ecker J, et al. Lower SCD expression in dendritic cells compared to macrophages leads to membrane lipids with less mono-unsaturated fatty acids. Immunobiology. 2010;215(9–10):748–755. doi: 10.1016/j.imbio.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Roussel L, et al. Steroids and extracellular signal-regulated kinase 1/2 activity suppress activating transcription factor 3 expression in patients with severe asthma. J Allergy Clin Immunol. 2011;127(6):1632–1634. doi: 10.1016/j.jaci.2011.03.034. [DOI] [PubMed] [Google Scholar]
- 48.Zhang T, et al. Nurr1 is phosphorylated by ERK2 in vitro and its phosphorylation upregulates tyrosine hydroxylase expression in SH-SY5Y cells. Neurosci Lett. 2007;423(2):118–122. doi: 10.1016/j.neulet.2007.06.041. [DOI] [PubMed] [Google Scholar]
- 49.Fury W, et al. Overlapping probabilities of top ranking gene lists, hypergeometric distribution, and stringency of gene selection criterion. Conf Proc IEEE Eng Med Biol Soc. 2006;1:5531–5534. doi: 10.1109/IEMBS.2006.260828. [DOI] [PubMed] [Google Scholar]
- 50.Wei J, et al. PPARgamma downregulation by TGFss in fibroblast and impaired expression and function in systemic sclerosis: a novel mechanism for progressive fibrogenesis. PLoS One. 2010;5(11):e13778. doi: 10.1371/journal.pone.0013778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arakawa H, et al. Adiponectin expression is decreased in the involved skin and sera of diffuse cutaneous scleroderma patients. Exp Dermatol. 2011;20(9):764–766. doi: 10.1111/j.1600-0625.2011.01310.x. [DOI] [PubMed] [Google Scholar]
- 52.Katsoulidis E, et al. Suppression of interferon (IFN)-inducible genes and IFN-mediated functional responses in BCR-ABL-expressing cells. J Biol Chem. 2008;283(16):10793–10803. doi: 10.1074/jbc.M706816200. [DOI] [PMC free article] [PubMed] [Google Scholar]