Abstract
Background
Ribonucleotide reductase catalyzes an essential step in the cellular production of deoxyribonucleotide triphosphates and has been associated with clinical outcome in cancer patients receiving nucleoside analog-based chemotherapy.
Materials & methods
In the current study, we sequenced the genes RRM1 and RRM2 in genomic DNA from HapMap cell lines with European (Utah residents with northern and western European ancestry [CEU]; n = 90) or African (Yoruba people in Ibadan, Nigeria [YRI]; n = 90) ancestry.
Results
We identified 44 genetic variants including eight coding SNPs in RRM1 and 15 SNPs including one coding SNP in RRM2. RRM1 and RRM2 mRNA expression levels were significantly correlated with each other in both CEU and YRI lymphoblast cell lines, and in leukemic blasts from acute myeloid leukemia (AML) patients (AML97, n = 89; AML02, n = 187). Additionally, RRM1 expression was higher among patient features indicative of a high relapse hazard. We evaluated SNPs within the RRM1 and RRM2 genes in the HapMap lymphoblast cell lines from CEU and YRI panels for association with expression and cytarabine chemosensitivity. SNPs of potential significance were further evaluated in AML patients. RRM1 SNPs rs1042919 (which occurs in linkage disequilbrium with multiple other SNPs) and promoter SNP rs1561876 were associated with intracellular 1-β-D-arabinofuranosyl-CTP levels, response after remission induction therapy, risk of relapse and overall survival in AML patients receiving cytarabine and cladribine.
Conclusion
These results suggest that SNPs within ribonucleotide reductase might be helpful predictive markers of response to nucleoside analogs and should be further validated in larger cohorts.
Keywords: cytarabine, HapMap, leukemia, nucleoside analogs, pharmacogenomics, ribonucleotide reductase
Ribonucleotide reductase (RR) is a key enzyme involved in the biosysnthesis of deoxynucleotides. Since reduction in the levels of ribonucleotides is a rate-limiting step in DNA synthesis, inhibition of RR has been explored as a potential area for development of anticancer agents. The RR holoenzyme consists of the dimerized large and small subunits, RRM1 and RRM2. RR regulates intracellular pools of deoxy-CTP (dCTP), which in turn, has been implicated in resistance to nucleoside analogs such as cytarabine (ara-C). Cell lines and leukemic blasts with high levels of dCTP are resistant to ara-C [1–3]. Intracellular dCTP levels regulate ara-C metabolism at three levels: by feedback inhibition of DCK resulting in reduced activation of ara-C [4]; by allosteric activation of the inactivating enzyme CDA [3]; and by competing with the active metabolite 1-β-D-arabinofuranosyl-CTP (ara-CTP) for incorporation into DNA [5]. In addition to ara-C, higher intracellular levels of deoxynucleotides due to increased activity of ribonucleotide reductase can increase resistance to other nucleoside analogs such as cladribine using mechanisms mentioned above [6]. In fact biochemical modulation of ara-C by nucleoside analogs such as fludarabine and cladribine (that inhibits RR) has been shown to stimulate ara-CTP accumulation by inhibition of RR in leukemic cells from adult and pediatric patients [7–10].
RRM1 has also been implicated in suppression of cell migration and metastases, and its overexpression in Ras-transformed mouse fibroblasts decreases tumor development and metastases, suggesting a tumor suppressor role [11]. Overexpression of RRM1 has also been shown to be protective against carcinogen-induced lung tumors in mice [11]. RRM2 has been shown to be associated with tumorigenesis and drug resistance in preclinical studies [12]. Due to their key role in DNA synthesis/repair and in cell growth, RRM1 and RRM2 are targeted by several anti-cancer agents including nucleoside analogs (e.g., gemcitabine, cladribine and clofarabine) and hydoxyurea (reviewed by Shao et al. [13]). RRM1 overexpression has been correlated with gemcitabine resistance in human pancreatic cancer [14], non-small-cell lung cancer cell lines and in mice with tumors [15,16].
In the present study, we first sought to identify genetic variation in RRM1 and RRM2 by resequencing the genomic DNA from HapMap European (Utah residents with northern and western European ancestry [CEU]) and African (Yoruba people in Ibadan, Nigeria [YRI]) ancestry panels. Then, we determined the association of SNPs (both from the HapMap database [101] and identified by resequencing) with mRNA expression levels and cytotoxicity to ara-C in HapMap cell lines. Finally, we investigated the clinical significance of RRM1, RRM2 and RRM2B variants in pediatric acute myeloid leukemia (AML) patients who received ara-C along with cladribine treatment in the AML97 study and ara-C without cladribine in the AML02 study.
Materials & methods
Reagents
The Expand Hi Fidelity PCR system was obtained from Roche (IN, USA); the TOPOTA cloning kit and One Shot Mach1-T1 chemically competent E. coli cells were obtained from Invitrogen (CA, USA). Restriction enzymes, pGL3 basic vector and JM109 cells were obtained from Promega (WI, USA). RNeasy Minikit, QIA-prep Plasmid Miniprep kit and Plasmid Plus kit were from Qiagen Inc. All other reagents were of molecular biology grade.
HapMap cell lines
We used Epstein–Barr virus-transformed B-lymphoblastoid HapMap cell lines derived from 30 CEU trios (two parents and a child; n = 90, European descent) and 30 Yoruba trios (n = 90, African descent, referred to as YRI) to identify genetic variants in RRM1 and RRM2. The purpose of using the same cell lines that have been used in the International HapMap project was to allow us to utilize the genotype data generated as part of the HapMap project. Genome-wide gene-expression data using Affymetrix (CA, USA) Exon array was used to extract the expression levels of RRM1 and RRM2 (GSE7761). Cells were grown in an RPMI 1640 medium supplemented with 2 mM L-glutamine (Lonza Walkersville, Inc., MD, USA) and 15% heat-inactivated serum at 37°C under 5% CO2. DNA, RNA and cytoplasmic fractions were extracted from the cell lines using standard protocols. Genomic DNA was used to discover novel genetic variants in the RRM1 and RRM2 genes. Ara-C cellular cytotoxicity was determined in a previous study using alamarBlue® (Biosource International, CA, USA). The area under the survival curve (AUC) was calculated using the trapezoidal rule and AUC values were log2 transformed before statistical analysis as described earlier [17].
Identification of sequence variations in the RRM1 & RRM2 genes
All the coding exons including the flanking intronic sequences as well as 1.5 kb of the 5′-UTR of the genes RRM1 and RRM2 were PCR-amplified with primers designed using Primer Select module of Lasergene v 6.0 software (DNAStar) and synthesized at the University of Minnesota, Biomedical Genomics Center (MN, USA). The primer sequences were verified using UCSC BLAT [102] to exclude amplification of any non-specific DNA sequences. Information on individual primer sequences is provided in the Supplementary Material (see www.futuremedicine.com/doi/suppl/10.2217/pgs.13.131), PCR amplification, amplicon sequencing and analysis of sequences for identification of SNPs was performed as described previously [18].
Studies with primary leukemic cells
Bone marrow samples were obtained at diagnosis from pediatric AML patients enrolled in the St Jude AML97 [19] or St Jude AML02 [20] studies, after obtaining informed consent from their parents/guardians, with assent from the patients, as appropriate. The study was approved by the St Jude institutional review board.
The eligibility for the enrollment, treatment plan and clinical outcome of AML97 have been reported [19]. Because cladribine, an inhibitor of RR, had been shown to augment ara-CTP levels, patients were randomized to receive ara-C along with cladribine either as a daily short infusion or a continuous infusion. Bone marrow aspirates were obtained at diagnosis and after day 1 and 2 of ara-C treatment as described previously [21]. Leukemic cells were separated by Ficoll-Hypaque density-gradient centrifugation and intracellular ara-CTP levels were determined in samples obtained after day 1 and day 2 of ara-C treatment using HPLC as described earlier [21]. The patient population included 58% white patients, 22% black patients and 20% of patients were of other ethnic backgrounds.
Details of eligibility of enrollment, study design and clinical outcome for the AML02 protocol have been reported [20]. Briefly, patients were randomized to receive induction therapy containing either high-dose ara-C (3 g/m2, given every 12 h on days 1, 3 and 5) or low-dose ara-C (100 mg/m2) given every 12 h on days 1–10 plus daunorubicin and etoposide as described earlier [20]. The patient population included 69.6% white patients, 18.7% black patients and 11.7% of patients were of other ethnic backgrounds. In vitro sensitivity of leukemic cells to ara-C was determined in patients enrolled in the AML02 study using the 4-day 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazoliumbromide (MTT) cytotoxicity assay and LC50 values were generated as part of a previously reported study [22,23]. RRM1 and RRM2 mRNA expression levels in diagnostic leukemic blasts were extracted from Affy U133A array data that was available from 207 patients (165 enrolled in AML02 and 42 in AML97).
Genomic DNA was extracted using DNA blood kit (Qiagen, CA, USA) and samples were evaluated for potentially functionally important RRM1 (n = 14), RRM2 (n = 9) and RRM2B (n = 3) SNPs representing major linkage disequilibrium (LD) groups in genomic DNA from AML97 (n = 89) and AML02 patients (n = 187) using Sequenom (CA, USA) based genotyping that uses MALDI-TOF-based chemistry at University of Minnesota, Biomedical Genomics Center.
Statistical analysis
The association of SNP genotype with RRM1 and RRM2 expression and AUC in HapMap cell lines was tested with an analysis of variance model that uses a Toeplitz correlation matrix [24] to account for the correlation of an individual’s measurements with that of parents from HapMap trios. In AML patients, the Kruskal–Wallis test was used to compare expression levels and ara-C LC50 values across groups of AML patients according to SNP genotype, clinical risk group or race. Spearman correlation coefficient was used to characterize the association of the number of minor alleles with minimal residual disease, mRNA expression, and ara-C LC50 values. Event-free survival (EFS) was defined as the time elapsed from protocol enrollment to induction failure, relapse, withdrawal, secondary malignancy or failure with patients free of these events censored at last follow-up. Overall survival (OS) was defined as the time elapsed from protocol enrollment to death, with patients censored at last follow-up. Risk of relapse or resistant disease was defined as time elapsed from protocol enrollment to induction failure, relapse or competing events of withdrawal, secondary malignancy or death. The statistic of Jung, Owzar and George [25] was used to characterize the association of the number of minor alleles with EFS and OS. Cumulative incidence tests [26] or competing risks regression [27] was used to test association of SNPs with risk of relapse. All tests are two-sided and no multiple testing adjustments were performed in these exploratory analyses.
Results
SNPs identified in RRM1 & RRM2
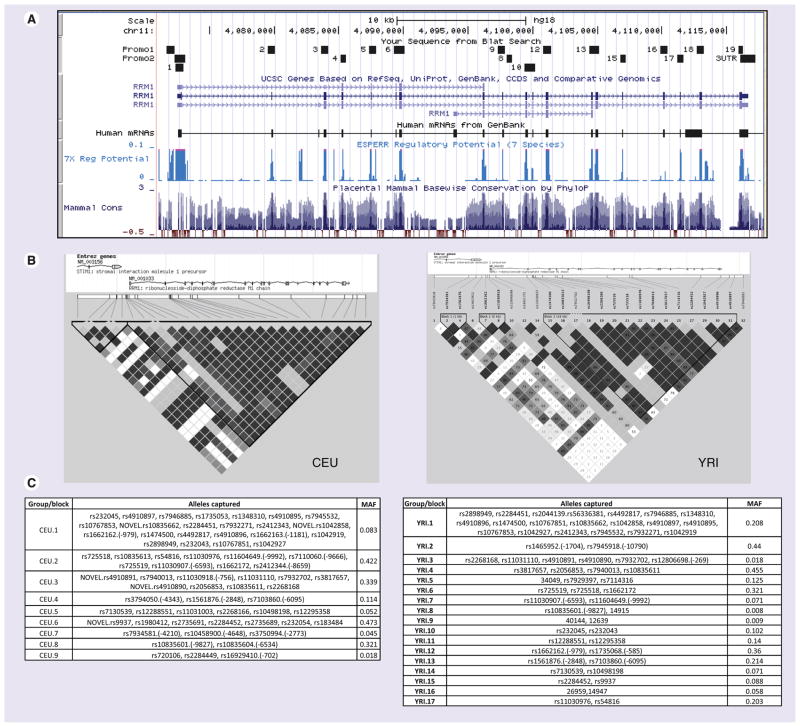
All the coding exons, flanking introns and the proximal promoter in CEU (30 trios with European ancestry, n = 90) and YRI (30 trios with African ancestry, n = 90) samples were sequenced. Figures 1 & 2 show the snapshots from UCSC genome browser [103] of regions sequenced in RRM1 and RRM2 along with key regions of functional importance (CpG site, 7× regulatory potential and conserved transcription factor binding sites). A total of 44 genetic variants in RRM1 were identified by resequencing, while 15 SNPs were identified in RRM2 (Figure 2 & Table 1). Among the 44 RRM1 SNPs, ten were present in the promoter, eight in exons, 25 in intronic regions and one in the 3′-UTR (Table 1). Nineteen SNPs were found exclusively in the African ancestry (YRI panel) and nine exclusively in the European ancestry (CEU panel); 16 SNPs were common to both CEU and YRI groups. Fourteen SNPs overlapped with SNPs present in the HapMap database. Detailed results of the bioinformatic analysis are provided in Table 1. For RRM2, seven SNPs were present in the promoter, one in the exon and seven in intronic regions. Eleven SNPs were found exclusively in the African ancestry (YRI) group and one exclusively in the European ancestry (CEU) group; three SNPs were common to both CEU and YRI groups. Only one SNP (rs1130609) overlapped with SNPs present in the HapMap database (rel#28) [104].
Figure 1. SNPs in RRM1.
(A) Snapshot from the UCSC genome browser [NCBI/hg18 (2006)] for the RRM1 locus. The regions sequenced in HapMap panels are shown by small boxes. (B) Linkage disequilibrium plot of RRM1 gene in European (CEU) and African (YRI) ancestry samples generated in Haploview using genotype data from the present study and from HapMap for both CEU and YRI samples from 10 kb upstream to 5 kb downstream. (C) SNPs that are linked, r2 > 0.8 (and picked by the tagger program), are categorized in the same groups. SNPs without rs numbers are indicated by number corresponding to Table 1.
CEU: Utah residents with northern and western European ancestry; MAF: Minimum allele frequency; YRI: Yoruba people in Ibadan, Nigeria.
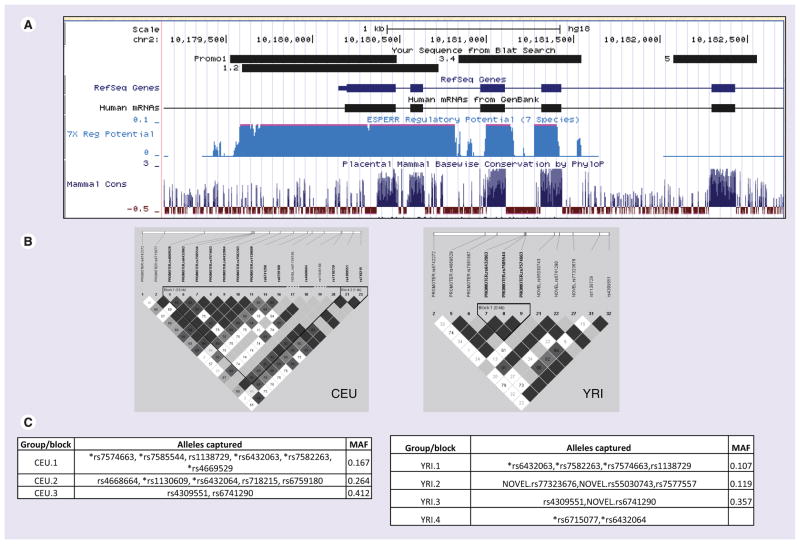
Figure 2. SNPs in RRM2.
(A) Snapshot from the UCSC genome browser [NCBI/hg18 (2006)] for the RRM2 locus. The regions sequenced in HapMap panels are shown by small boxes. (B) Linkage disequilibrium plot of RRM2 gene in European (CEU) and African (YRI) ancestry samples generated in Haploview using genotype data from the present study and from HapMap for both CEU and YRI samples 20 kb upstream to 5 kb downstream. (C) SNPs that are linked, r2 ≥ 0.75 in CEU and r2 ≥ 0.8 in YRI (and picked by the tagger program), are categorized in the same groups. SNPs located upstream of the translation start site (ATG) are denoted by an asterisk. SNPs without rs numbers are indicated by number corresponding to Table 1.
CEU: Utah residents with northern and western European ancestry; MAF: Minimum allele frequency; YRI: Yoruba people in Ibadan, Nigeria.
Table 1.
Genetic variants identified by sequencing RRMI and RRM2 in HapMap lymphoblast cell lines (CEU and YRI).
| rs no. | Change | Chromosomal position UCSC Hg18 (2006) | Position from translation start site (ATG) as +1 | Location in the gene | Amino acid change | Bioinformatics (predictions) | HapMap/Novel | Allele frequency | ||
|---|---|---|---|---|---|---|---|---|---|---|
| CEU | YRI | |||||||||
| RRM1 | ||||||||||
| 1 | rs1662162 | T/C | 4071840 | −979 | Promoter | Gain of V$BCL6, V$NOLF and V$STAT sites; loss of V$PERO site | HAPMAP_CEU, YRI | 0.08 | 0.36 | |
| 2 | rs72555763 | G/T | 4071883 | −936 | Promoter | NOVEL_YRI | 0.01 | |||
| 3 | T/G | 4072057 | −762 | Promoter | Gain of V$HAND, V$HEAT and V$SORY sites; loss of V$KLFS site | NOVEL_CEU | 0.01 | |||
| 4 | rs11030918 | T/C | 4072063 | −756 | Promoter | Loss of V$HEAT site | HAPMAP_CEU, YRI | 0.35 | 0.16 | |
| 5 | rs1735068 | T/G | 4072234 | −585 | Promoter | HAPMAP_CEU, YRI | 0.12 | 0.38 | ||
| 6 | rs72555767 | T/G | 4072245 | −574 | Promoter | Gain of O$VTBP and V$HEAT sites; loss of V$ABDB, V$BRN5, V$BRNF, V$CDXF, V$FKHD, V$RUSH and V$SORY sites | NOVEL_YRI | 0.02 | ||
| 7 | rs72555753 | C/A | 4072411 | −408 | Promoter | Loss of V$GLIF and V$P53F sites | NOVEL_YRI | 0.23 | ||
| 8 | rs12806698 | C/A | 4072550 | −269 | Promoter | Gain of V$DICE and V$GLIF sites; loss of V$CREB and V$PAX5 sites | HAPMAP_CEU | 0.29 | ||
| 9 | rs72555757 | G/T | 4072848 | 30 | 5′-UTR | Gain in V$KLFS, V$RXRF and V$SPZ1 sites; loss of O$XCPE, V$EGRF, V$MAZF, V$MZF1, V$PAX3, V$SP1F and V$ZF02 sites | NOVEL_YRI | 0.01 | ||
| 10 | rs12801088 | A/C | 4072911 | 93 | 5′-UTR | Loss of V$GLIF, V$RREB and V$SMAD sites | NOVEL_CEU, YRI | 0.15 | 0.30 | |
| 11 | C/T | 4079515 | 6697 | Intron 1 | Loss of 5SS_U2 site; loss of SF2/ASF, SF2/ASF (IgM-BRCA1) and SC35 sites | NOVEL_CEU | 0.03 | |||
| 12 | rs11030937 | A/T | 4079695 | 6877 | Intron 1 | Loss of branch site | HAPMAP_YRI | 0.06 | ||
| 13 | rs1348310 | C/G | 4079921 | 7103 | Intron 2 | Gain in SC35 sites | HAPMAP_CEU, YRI | 0.09 | 0.20 | |
| 14 | rs232054 | C/G | 4080003 | 7185 | Intron 2 | Loss of branch site | HAPMAP_CEU, YRI | 0.47 | 0.01 | |
| 15 | T/A | 4083737 | 10919 | Intron 2 | Gain of SF2/ASF and SF2/ASF (IgM-BRCA1) sites | NOVEL_CEU | 0.02 | |||
| 16 | T/C | 4085278 | 12460 | Exon 4 | H108H | NOVEL_YRI | 0.01 | |||
| 17 | rs2228121 | T/C | 4085315 | 12497 | Exon 4 | L121L | NOVEL_YRI | 0.06 | ||
| 18 | rs725518 | G/A | 4085421 | 12603 | Intron 4 | Gain of SF2/ASF and SRp40 sites | HAPMAP_CEU, YRI | 0.42 | 0.32 | |
| 19 | C/T | 4085457 | 12639 | Intron 4 | NOVEL_YRI | 0.01 | ||||
| 20 | C/T | 4087599 | 14781 | Intron 5 | Loss of branch site | NOVEL_YRI | 0.01 | |||
| 21 | T/G | 4087717 | 14899 | Intron 5 | Gain in branch site; loss of SRp40 and SRp55 sites | NOVEL_CEU | 0.01 | |||
| 22 | G/C | 4087733 | 14915 | Intron 5 | NOVEL_YRI | 0.01 | ||||
| 23 | T/G | 4087765 | 14947 | Intron 5 | Gain of donor splice site | NOVEL_YRI | 0.06 | |||
| 24 | G/A | 4089611 | 16793 | Intron 6 | Gain in branch site; loss of SC35 and SRp40 sites | NOVEL_CEU | 0.01 | |||
| 25 | rs2044139 | C/A | 4089893 | 17075 | Intron 7 | Loss of SRp55 site | HAPMAP_CEU, YRI | 0.09 | 0.21 | |
| 26 | T/C | 4097546 | 24728 | Intron 8 | Gain of SRp40 site | NOVEL_YRI | 0.03 | |||
| 27 | rs183484 | C/A | 4097708 | 24890 | Exon 9 | R284R | HAPMAP_CEU | 0.48 | ||
| 28 | A/G | 4097801 | 24983 | Intron 9 | Gain of SRp40 site | NOVEL_YRI | 0.01 | |||
| 29 | T/C | 4098371 | 25553 | Intron 9 | NOVEL_CEU | 0.08 | ||||
| 30 | rs7114316 | T/C | 4099663 | 26845 | Intron 10 | Gain of 5SS_U2 site; gain of SF2/ASF, SF2/ASF (IgM-BRCA1) and SRp40 sites; loss of SC35 site | HAPMAP_YRI | 0.12 | ||
| 31 | rs12288551 | C/G | 4099722 | 26904 | Intron 10 | Loss of SC35 site | HAPMAP_CEU, YRI | 0.05 | 0.14 | |
| 32 | rs12295358 | A/G | 4099733 | 26915 | Intron 10 | Gain of SC35 site | HAPMAP_CEU, YRI | 0.05 | 0.14 | |
| 33 | A/G | 4099777 | 26959 | Intron 10 | Loss of branch site | NOVEL_YRI | 0.06 | |||
| 34 | A/T | 4100018 | 27200 | Exon 11 | L370L | NOVEL_YRI | 0.05 | |||
| 35 | G/A | 4106867 | 34049 | Intron 14 | Gain in branch site | NOVEL_YRI | 0.13 | |||
| 36 | rs2228123 | A/C | 4106955 | 34137 | Exon 15 | K590Q | Benign (0.004) [Polyphen]; tolerated (0.35) [SIFT] | NOVEL_YRI | 0.02 | |
| 37 | T/C | 4109950 | 37132 | Intron 15 | Gain of SF2/ASF and SF2/ASF (IgM-BRCA1) sites | NOVEL_CEU | 0.01 | |||
| 38 | rs4910890 | C/T | 4109983 | 37165 | Intron 15 | NOVEL_CEU, YRI | 0.35 | 0.02 | ||
| 39 | rs4910891 | A/G | 4109984 | 37166 | Intron 15 | Gain of SRp55 site | NOVEL_CEU, YRI | 0.35 | 0.02 | |
| 40 | rs10835662 | A/G | 4110343 | 37525 | Intron 16 | Gain of branch site | NOVEL_CEU, YRI | 0.08 | 0.19 | |
| 41 | C/T | 4112962 | 40144 | Exon 18 | L692L | NOVEL_YRI | 0.01 | |||
| 42 | rs9937 | A/G | 4116033 | 43215 | Exon 19 | T741T | NOVEL_CEU, YRI | 0.47 | 0.09 | |
| 43 | rs1042858 | G/A | 4116042 | 43224 | Exon 19 | A744A | NOVEL_CEU, YRI | 0.08 | 0.24 | |
| 44 | rs1042919 | T/A | 4116340 | 43522 | 3′-UTR | HAPMAP_CEU, YRI | 0.09 | 0.19 | ||
| RRM2 | ||||||||||
| 1 | rs72542776 | C/T | 10179706 | −491 | Promoter | Gain in V$BRAC and V$MOKF sites; loss of V$AP2F and V$NR2F sites | NOVEL_YRI | 0.08 | ||
| 2 | rs72542778 | C/T | 10180005 | −192 | Promoter | Gain in V$RUSH, V$SP1F and V$ZF02 sites; loss of O$XCPE, V$EGRF and V$OAZF sites | NOVEL_YRI | 0.46 | ||
| 3 | rs72542781 | G/A | 10180060 | −137 | Promoter | Gain in V$STAF, site; loss of O$XCPE, V$CTCF, V$ETSF, V$GLIF, V$MAZF and V$PLAG sites | NOVEL_YRI | 0.03 | ||
| 4 | rs72542782 | G/A | 10180118 | −79 | Promoter | Gain in V$HEAT, V$PAX5 and V$PAX6 sites; loss of V$E2FF, V$EGRF, V$NRF1 and V$RXRF sites | NOVEL_YRI | 0.10 | ||
| 5 | rs72542788 | C/G | 10180316 | −61 | 5′-UTR | Loss of V$HESF site | NOVEL_YRI | 0.17 | ||
| 6 | rs68002429 | C/T | 10180343 | −34 | 5′-UTR | Gain in V$SMAD site; loss of V$GREF and V$NRSF sites | NOVEL_YRI | 0.32 | ||
| 7 | rs1130609 | G/T | 10180371 | Exon 1 | S59A | HAPMAP_CEU, YRI | 0.26 | 0.03 | ||
| 8 | rs72542793 | A/C | 10181292 | 916 | Intron 3 | NOVEL_YRI | 0.01 | |||
| 9 | rs5030743 | C/G | 10181325 | 949 | Exon 4 | S110S | NOVEL_YRI | 0.13 | ||
| 10 | rs6432065 | C/T | 10181508 | 1132 | Intron 4 | Loss of SRp55 site | NOVEL_CEU | 0.24 | ||
| 11 | rs6741290 | C/T | 10182160 | 1784 | Intron 4 | NOVEL_CEU, YRI | 0.41 | 0.38 | ||
| 12 | rs72542796 | A/G | 10182185 | 1809 | Intron 4 | Change in SRp55 site | NOVEL_YRI | 0.04 | ||
| 13 | C/T | 10182249 | 1873 | Intron 4 | Loss of SF2/ASF (IgM-BRCA1) and SC35 sites | NOVEL_YRI | 0.03 | |||
| 14 | rs77323676 | C/T | 10184314 | 3938 | Intron 5 | NOVEL_YRI | 0.12 | |||
| 15 | rs61754180 | T/C | 10184662 | 4286 | Intron 6 | Gain of SF2/ASF and SRp40 sites; gain in branchsite | NOVEL_CEU, YRI | 0.03 | 0.01 | |
CEU: Utah residents with northern and western European ancestry; YRI: Yoruba people in Ibadan, Nigeria.
Analysis of LD on SNPs identified in the current study and data from HapMap database [101] was performed using Haploview. We included 10 kb upstream and 5 kb downstream of the gene. For RRM1, the gene immediately upstream is STIM1, which occurs in a head-to-tail configuration with RRM1, with the 3′ end of STIM1 gene 1.6 kb from the 5′ end of the RRM1 gene. Based on the LD plot and results from tagger (Haploview) the strongly linked (r2 > 0.8) RRM1 SNPs in CEU and YRI were grouped into nine (CEU.1–CEU.9) and 17 (YRI.1–YRI.17) distinct groups as shown in Figure 1. For RRM2 there were relatively few SNPs and hence few LD groups, specifically three in CEU (CEU.1: rs6759180, rs4668664 and rs718215; CEU.2 rs6741290 and rs4309551; CEU.3 rs7585544 and rs7582263) and one in YRI (rs6741290 and rs4309551 were in LD). SNPs that did not occur in linkage with other SNPs were termed as a ‘singleton’ (Figure 2).
Association of RRM1 & RRM2 SNPs with mRNA expression & cytotoxicity to nucleoside analogs in HapMap lymphoblast cell lines
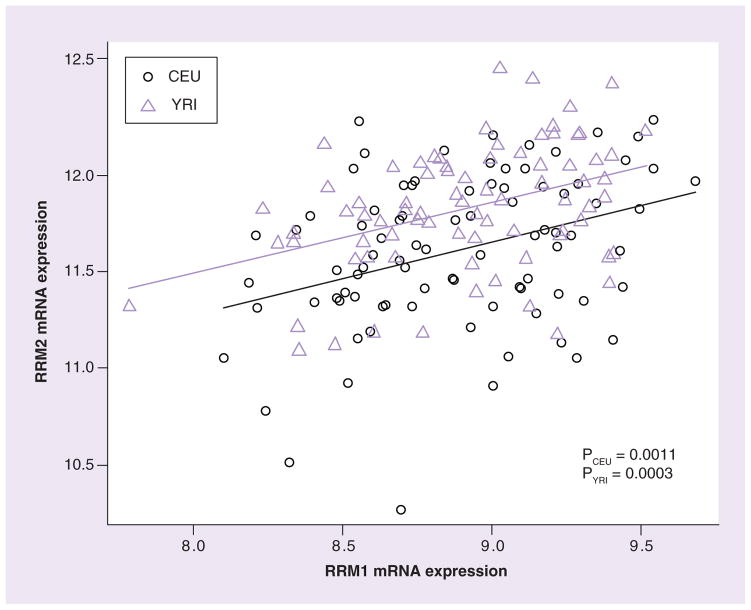
mRNA expression in 87 CEU (representing 29 trios) and 90 YRI (30 trios with African ancestry) samples was obtained from the Gene Expression Omnibus database (GSE7761) [105]. RRM1 and RRM2 mRNA expression levels had wide interindividual variability and were significantly correlated with each other (p ≤ 0.0011) in both CEU and YRI samples (Figure 3). We observed a significantly higher (p < 0.0001) RRM2 mRNA expression in YRI compared with CEU samples. The cytotoxicity of ara-C for both CEU and YRI was previously reported [17]. Although expression levels of RRM1 or RRM2 were not significantly correlated with cellular sensitivity to ara-C in CEU and YRI, we did observe a trend (p = 0.07) for higher expression to be associated with greater AUC (higher AUC is associated with lower drug sensitivity) for cell survival in YRI (data not shown).
Figure 3. Comparison of RRM1 and RRM2 mRNA expression in CEU and YRI cell lines.
RRM1 and RRM2 mRNA expression extracted from GSE7761 in Epstein–Barr virus-transformed lymphoblast cell lines derived from subjects with European (CEU) and African (YRI) ancestries.
CEU: Utah residents with northern and western European ancestry; YRI: Yoruba people in Ibadan, Nigeria.
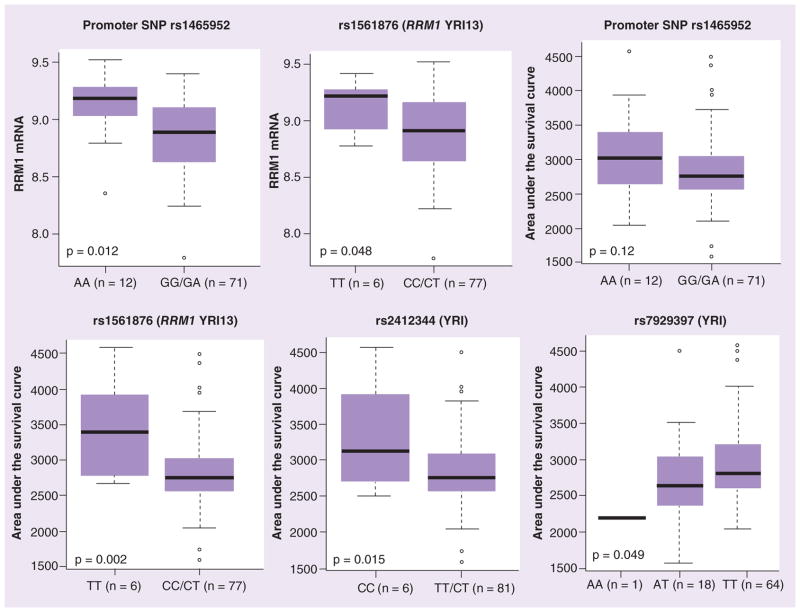
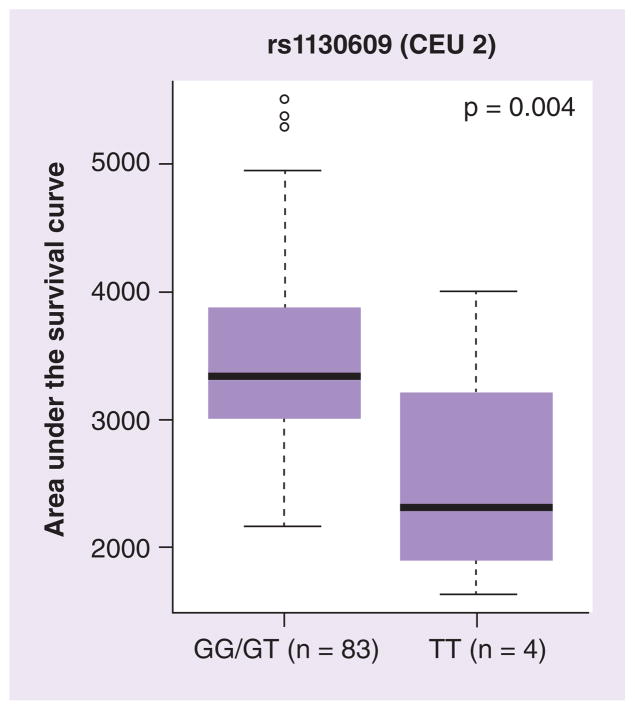
We analyzed the association of SNPs (identified by sequencing as well as from the HapMap database: 10 kb upstream and 5 kb downstream of the gene) with mRNA expression and cellular sensitivity to ara-C in HapMap lymphoblast cell lines (LCLs; Supplementary Table 1). None of the RRM1 SNPs were associated with RRM1 expression in CEU but rs1561876 was associated with ara-C cytotoxicity (GG vs GA vs AA, p = 0.033; Supplementary Table 1). In the YRI panel, promoter SNPs rs1465952 (YRI.2 occurs in LD with rs7945918) and rs1561786 (YRI.14 occurs in LD with rs7103860) were significantly associated with levels of expression of RRM1 (Figure 4). Additionally, rs1465952 showed a trend and rs1561876 was significantly associated with ara-C cytotoxicity in the YRI panel (Supplementary Table 1 & Figure 4). Other SNPs upstream of RRM1 that were significantly associated with ara-C cytotoxicity included rs2412344, rs11030907 and rs7929397 (Figure 4 & Supplementary Table 1). For RRM2, a coding SNP rs1130609 (S59A) was associated with ara-C cytotoxicity in CEU samples (Figure 5) and a promoter SNP rs7574663 was associated with its expression (Supplementary Table 1).
Figure 4. Association of SNPs with mRNA expression and cytarabine cytotoxicity in HapMap lymphoblast cell lines.
Box plots for the association of representative RRM1 SNPs with their expression and cytarabine area under the survival curve in YRI samples. Plots show medians as a line between boxes representing the first and third quartiles; the whiskers represent the range after excluding the outliers. The outliers are defined as data points that fall outside of the first and third quartiles by more than 1.5-times the interquartile range. Circles falling outside the whiskers represent outliers.
YRI: Yoruba people in Ibadan, Nigeria.
Figure 5. Association of RRM2 SNP with cytarabine cytotoxicity in HapMap lymphoblast cell lines.
Box plots for the association of representative RRM2 SNPs with cytarabine area under the survival curve in CEU lymphoblast cell lines. Plots show medians as a line between boxes representing the first and third quartiles; the whiskers represent the range after excluding the outliers. The outliers are defined as data points that fall outside of the first and third quartiles by more than 1.5-times the interquartile range. Circles falling outside the whiskers represent outliers.
CEU: Utah residents with northern and western European ancestry.
Relationship of RRM1 & RRM2 SNPs with treatment response in AML patients
A total of 11 SNPs within RRM1 and seven in RRM2 were genotyped in DNA samples from AML patients (Supplementary Table 2 provides list of SNPs and Supplementary Figure 1 provides LD groups for RRM1 SNPs in AML patients). In addition, three SNPs in the RRM2B gene (rs7830150, rs17414857 and rs1265138) were also genotyped. The frequency of SNPs did not differ significantly between patients enrolled in the AML97 and AML02 studies (p > 0.05). SNPs occurring with the allele frequency of less than 10% were excluded, resulting in ten SNPs in RRM1 (few of these SNPs were in linkage with others), four in RRM2 and two in RRM2B for further analysis. After screening for LD, three additional SNPs in RRM1 were excluded. As was observed for HapMap panels, the LD pattern was very different between white and black AML patients (Supplementary Figure 1).
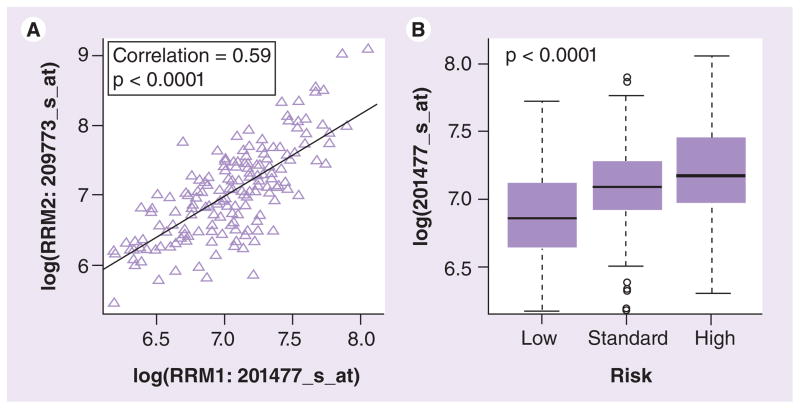
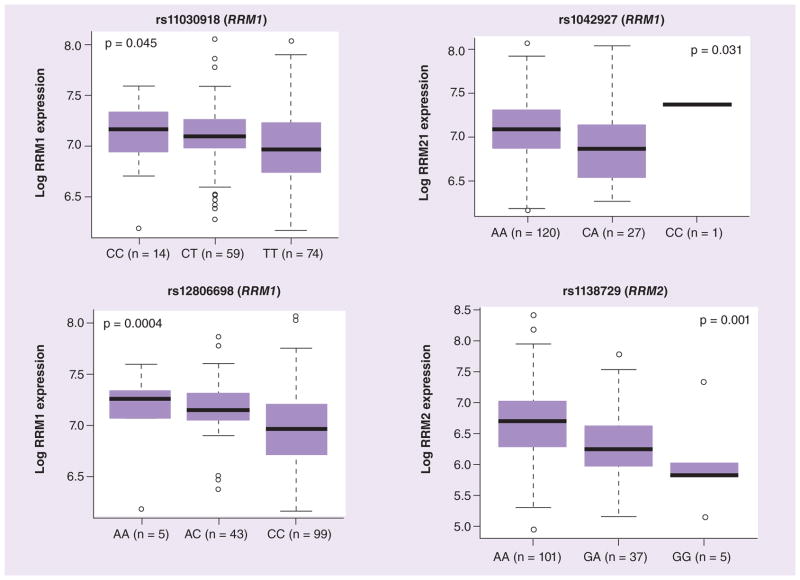
RRM1 and RRM2 mRNA expression data was extracted from Affymetrix U133A microarray data in diagnostic leukemia blasts from the St Jude AML02 study. As observed in HapMap LCLs, we observed significant correlation between RRM1 and RRM2 expression in leukemic blasts obtained at diagnosis from AML patients regardless of race (p < 0.001; Figure 6A & Supplementary Table 3). Higher RRM1 expression was significantly associated with high-risk presenting features, such as -7, FLT3-ITD, t(6,9), megakaryoblastic leukemia, treatment-related AML or AML arising from myelodysplastic syndrome (Figure 6B). However, a similar significant trend was not observed for RRM2 (data not shown). RRM1 mRNA expression was significantly associated with the RRM1 promoter SNPs rs11030918 (p = 0.04), rs12806698 (p = 0.0004) and rs1042927 (p = 0.030; Figure 7). The RRM2 SNP rs1138729 was significantly associated with the mRNA expression of the RRM2 gene (p = 0.001).
Figure 6. Comparison of RRM1 and RRM2 mRNA expression in diagnostic leukemic blasts from acute myeloid leukemia patients.
(A) RRM1 and RRM2 expression was extracted from Affymetrix U133A microarray data and correlation of mRNA expression levels between RRM1 and RRM2 was evaluated. (B) Box plot showing association of RRM1 expression with risk group. Plots show medians as a line between boxes representing the first and third quartiles; the whiskers represent the range after excluding the outliers. The outliers are defined as data points that fall outside of the first and third quartiles by more than 1.5-times the interquartile range. Circles falling outside the whiskers represent outliers.
Figure 7. Association of RRM1 and RRM2 SNPs with mRNA expression in diagnostic AML02 leukemic blasts.
Box plots for the association of representative RRM1 SNPs rs11030918, rs1042927 and rs12806698 with its expression and RRM2 SNP rs1138729 with its mRNA expression levels in diagnostic leukemic blasts from acute myeloid leukemia patients. Plots show medians as a line between boxes representing the first and third quartiles; the whiskers represent the range after excluding the outliers. The outliers are defined as data points that fall outside of the first and third quartiles by more than 1.5-times the interquartile range. Circles falling outside the whiskers represent outliers.
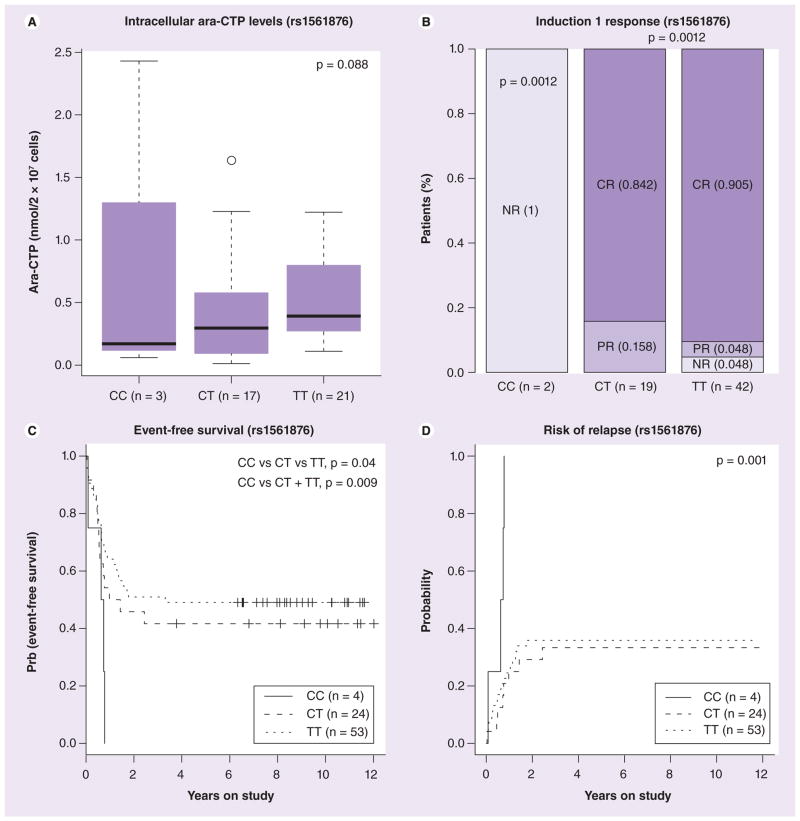
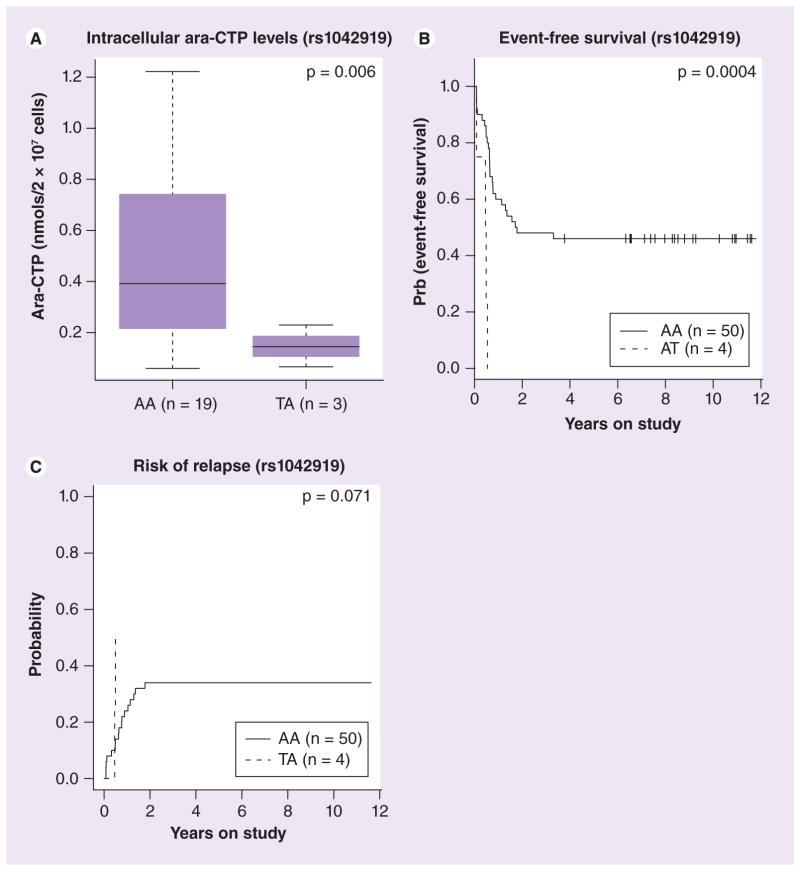
Patients enrolled in AML97 received the RR inhibitor cladribine in combination with ara-C. In a subset of patients within this cohort we had both RRM1 and RRM2 mRNA expression data (extracted from the Affymetrix U133A gene expression array) and intracellular ara-CTP levels in leukemic blasts determined at day 1 and 2 of therapy. We found an inverse correlation between RRM1/2 mRNA expression and intracellular ara-CTP levels in leukemic blasts although it did not reach statistical significance (Supplementary Figure 2). The most interesting RRM1 SNPs included rs1561876 (belonging to CEU.4) that occurs in LD with other SNPs (rs3794050 and rs7103860) in European ancestry (but only with rs7103860 in YRI). The variant allele of this SNP was associated with lower intracellular ara-CTP levels of leukemic blasts determined at day 1 of therapy (p = 0.006 among Caucasian patients and p = 0.088 in all patients); inferior response after the first course of remission induction therapy (p = 0.0012); poor EFS (p = 0.009); and greater risk of relapse (p = 0.001) in AML patients (Figure 8). Another RR M1 SNP rs1042919, belonging to LD group CEU.1 (22 other SNPs occur in LD with this SNP, see Figure 1) was associated with lower intracellular ara-CTP levels in leukemic blasts on day 1 (p = 0.006; Figure 9A) and day 2 of therapy (p < 0.05), and poor outcome as shown by a worse EFS (p = 0.0004; Figure 9). In addition, RRM1 rs2898950 and RRM2B SNP rs1265138 were associated with inferior response after induction therapy (Supplementary Figure 3). For RRM2 SNPs, presence of variant allele for rs1130609 and rs5030743 was associated with poor EFS, although the number of patients with variant allele was small (Supplementary Figure 3).
Figure 8. Association of RRM1 SNP rs1561876 with clinical outcome in AML97 patients.
(A) Box plot representing association of rs1561876 with intracellular levels of ara-CTP (nmol/2 × 107 cells) determined at day 1. Plots show medians as a line between boxes representing the first and third quartiles; the whiskers represent the range after excluding the outliers. The outliers are defined as data points that fall outside of the first and third quartiles by more than 1.5-times the interquartile range. The circle falling outside the whiskers represents an outlier. (B) Association of rs1561876 with response after induction 1 in acute myeloid leukemia (AML) patients. Percentages of patients in each category are indicated in parenthesis. (C) Survival curve for association of rs1561876 with event-free survival in all AML patients. (D) Survival curve for association of rs1561876 with risk of relapse in AML patients.
Ara-CTP: 1-β-D-arabinofuranosyl-CTP; CR: Complete response; NR: No response; PR: Partial response.
Figure 9. Association of RRM1 SNP rs1042919 with clinical outcome in Caucasian patients enrolled in AML97.
(A) Box plot representing association of rs1042919 with intracellular levels of ara-CTP (nmol/2 × 107 cells) determined at day 1 in Caucasian acute myeloid leukemia (AML) patients. Plots show medians as a line between boxes representing the first and third quartiles; the whiskers represent the range after excluding the outliers. (B) Survival curve for association of rs1042919 with event-free survival in white AML patients. (C) Survival curve for association of rs1042919 with risk of relapse in white AML patients.
Ara-CTP: 1-β-D-arabinofuranosyl-CTP.
For the AML02 cohort, we had in vitro ara-C cytotoxicity in a subset of patients (n = 69) determined by treating diagnostic leukemic cells with varying concentration of ara-C. None of the RRM1, RRM2 and RRM2B SNPs were significantly associated with ara-C LC50 in the combined cohort in analyses that ignored risk group. However, in analyses that stratified by risk group, RRM1 SNP rs1042858 (occurs in LD with rs1042927) was associated with higher ara-C LC50 as compared with patients with the AA genotype (p = 0.03; Supplementary Figure 4). RRM1 promoter SNP rs1561876 (belonging to CEU.4 and YRI.13) showed a similar but nonsignificant association with prognosis as observed in AML97. No SNPs in RRM2 and RRM2B were associated significantly with EFS, OS, minimal residual disease or RR in AML02 patients.
Discussion
In the present study we report results of sequencing of RRM1 and RRM2 genes and report on the evaluation of associations between mRNA expression and ara-C cytotoxicity in HapMap LCLs of European (CEU) and African ancestry (YRI). Overall SNPs within RR seem to be of lesser functional significance as few SNPs within the RRM1 gene (predominantly in the YRI panel) were associated with mRNA expression and/or cellular sensitivity to ara-C in Hap-Map LCLs. We further evaluated association of RRM1, RRM2 and RRM2B SNPs in two independent clinical cohorts of patients treated in the St Jude AML97 and St Jude AML02 studies.
The two most clinically interesting SNPs in RRM1 included rs1042919 and rs1561876. The RRM1 SNP rs1042919 represents the group CEU.1, occurs in LD with several other SNPs including rs1042858 (2464G>A; Ala744Ala), rs1042927 (2927A>C) and promoter SNP rs1662163 (−934), and was associated with lower intracellular ara-CTP levels in leukemic cells at day 1 (ara-C alone) and day 2 (ara-C in combination with cladribine). This SNP was also associated with poor survival and greater risk of relapse in patients treated with the AML97 cohort. Previous studies have shown that rs1042927 (also known as 2927) was associated with a significantly decreased survival (p = 0.021) in patients with malignant mesothelioma receiving gemcitabine-based chemotherapy [28]. For coding SNP rs1042858 (commonly known as 2464G>A) presence of the A allele has been previously reported to be associated with greater chemosensitivity to gemcitabine [29], which is in agreement with our observation of the variant G allele being associated with poor clinical response as compared with the A allele.
The second most interesting SNP in RRM1 is promoter SNP rs1561876 (present 2.5 kb upstream of exon 1 of RRM1 and in 3′-UTR of neighboring gene STIM1), which occurs in LD with at least two SNPs within 10 kb upstream of RRM1 in the CEU panel and with only one SNP in the YRI panel. rs1561876 C>T was associated lower ara-CTP levels at day 1 (post ara-C only) and day 2 (ara-C in combination with cladribine) poor response after induction 1 and EFS and greater risk of relapse in all and white patients within the St Jude AML97 cohort. However in Hap-Map LCLs it was associated with lower RRM1 expression and greater ara-C chemosensitivity in the YRI panel (Figure 4). One of the reasons for this observation might be the fact that the LD structure of this gene is very different in European versus African ancestry. When screening for the region 20 kb upstream of RRM1 we identified four additional SNPs in the upstream STIM1 gene that were in LD with rs1561876 in CEU, but none in YRI. STIM1 encodes a type 1 transmembrane protein that mediates Ca2+ influx after depletion of intracellular Ca2+ stores by gating of store-operated Ca2+ influx channels. STIM1 is located 1.5 kb upstream of RRM1 and is oriented in head to tail configuration with RRM1. Another point of consideration is the fact that several genes, including RRM1 and STIM1, are located in the imprinted gene domain of chromosome 11p15.5. This region is an important tumor suppressor gene region with alterations implicated in Beckwith–Wiedemann syndrome, Wilms tumor, rhabdomyosarcoma, adrenocortical carcinoma, and lung, ovarian and breast cancer. We checked correlation of mRNA expression levels of RRM1 and STIM1 and observed significant correlation with both CEU and YRI HapMap LCLs (p < 0.0001 for both CEU and YRI), interestingly within AML patients the RRM1 and STIM1 expression levels were not correlated in diagnostic leukemic blasts (p > 0.4). It is possible that this might be one of the reasons for the observed differences in results between HapMap LCLs and AML patients.
Our results identified the RRM1 SNP rs12806698 (-37A/C) to be associated with expression in AML02 samples, which was found to be consistent with the results from an earlier study [30]. This SNP has also been shown to be associated with response, toxicity sensitivity, survival and response to gemcitabine-based cancer chemotherapy in earlier studies [30–35]. Another SNP rs11030918 (-524C/T) was associated with mRNA expression in AML02 samples and has been related to the response rate in gemcitabine-based therapy [32,34]. It is possible that SNPs mentioned above or other SNPs that occur in LD with these alter mRNA/protein expression levels. The change in expression can influence cellular dCTP levels, which in turn can either, inhibit DCK or compete with ara-C for incorporation into DNA/RNA, both of which will result in inferior response to the chemotherapy. In addition to this it is also possible that altered RR levels/activity can interfere with its biological role in DNA synthesis/repair and cell growth thereby influencing the outcome.
For RRM2, the T allele of the SNP rs1130609 (S59A), was associated with greater ara-C sensitivity in HapMap samples (lower AUC) and diagnostic leukemic blasts (lower LC50) and, accordingly, higher EFS and OS among AML97 patients. We also found one SNP in the gene RRM2B/p53R2 (rs1265138) to be associated with inferior response among AML97 patients (Supplementary Figure 3B). This is an interesting discovery since RRM2B expression has earlier been shown to be associated with better survival in patients with colorectal cancer [36].
For certain SNPs in RRM1/RRM2/RRM2B, we have not seen any association with mRNA expression with respective gene, but have observed significant association with ara-C cytotoxicity or clinical outcome. One of the explanations for this observation is that SNPs (or SNPs in LD) might impact the secondary structure of RNA through post-transcriptional modifications. It is also possible that the presence of synonymous SNPs (e.g., RRM1 rs1042927 occurs in LD with a synonymous SNP rs1042858 [2464G>A; Ala744Ala]) could influence translation kinetics by utilizing less abundant tRNA as has been previously shown for a synonymous SNP of P-gp [37].
Overall we observed more significant association of RR SNPs in the AML97 than in the AML02 cohort. One of the reasons for this observation may be that in the AML97 study patients received cladribine in combination with ara-C whereas in AML02 only ara-C-based therapy was used. Cladribine triphosphate is an inhibitor of RR and is expected to reduce cellular pools of deoxynucleotides thus lessening the feedback inhibition of deoxycytidine kinase. Clofarabine, a second generation nucleoside analog that also inhibits RRs and DNA polymerase is being investigated in combination with ara-C and our future work will further investigate the clinical significance of genetic variation in RRs identified in this study in specimens from patients receiving a combination of clofarabine and ara-C (multicenter AML08 study, clinicalTrials.gov identifier: NCT00703820). Our exploratory study of the association of RRM1/RRM2 genotype with clinical outcomes among pediatric AML patients is one of the largest of its kind to date but nonetheless is limited by the small cohort of subjects with this rare disease. As such, our results should be considered as preliminary until confirmed in a separate cohort with a large sample size. We are currently attempting to obtain additional samples from other clinical trials to perform these confirmatory studies.
Conclusion & future perspective
In conclusion we evaluated SNPs within RRM1 and RRM2 for association with its expression and ara-C sensitivity to ara-C in HapMap LCLs from CEU and YRI panels. SNPs of potential significance were further evaluated in two independent patient cohorts. The most interesting SNPs were identified in the AML97 cohort, where patients received cladribine (RR inhibitor) in combination with ara-C. RRM1 SNPs were associated with multiple clinical end points in AML patients and in most cases presence of a variant allele was associated with lower intracellular levels of ara-CTP in leukemic cells as well as inferior response and greater risk of relapse. The results of our study identified SNPs within RRs that were significantly associated with clinical end points in AML patients (especially in patients receiving RR inhibitor); however, within HapMap LCLs the results were not as impressive. Future work in larger cohorts is needed to further validate our results especially in patients receiving treatment with RR inhibitors. Our results also provide additional SNPs that should be tested in patients undergoing treatment with gemcitabine-containing chemotherapy.
Supplementary Material
Supplementary Figure 1. LD plots for RRM1 SNPs in AML patients demonstrate distinct pattern in whites and blacks.
Supplementary Figure 2. Association of diagnostic blast RRM1 and RRM2 mRNA expression levels with intracellular ara-CTP levels determined 24hr after initiation of therapy. Ara-C given as short daily infusion is indicated in blue and as continuous infusion is in red.
Supplementary Figure 3 Association of RRM SNPs with clinical outcome St. Jude AML 97 patients. Barplot representing association of RRM1 SNP rs2898950 (A) and RRM2B SNP rs1265138 (B) with response after induction 1 in combined cohort. Survival curve for association of RRM2 SNPs rs1130609 (C: Black patients) and rs5030743(D: combined cohort) with overall survival in AML patients.
Supplementary Figure 4. Association of RRM1 (rs1042858 and rs1042927) and RRM2 (rs1130609) SNPs with in vitro ara-C cytotoxicity (LC50) determined in diagnostic leukemic cells from AML patients. Association was determined within three risk groups (high, standard and low) only significant results (p<0.05) are shown.
Supplementary Table 1: Association of SNPs in RRM1 and RRM2 with mRNA expression and ara-C cellular sensitivity.
Supplementary Table 2. Minor allele frequency and genotype frequency of RRM1, RRM2 and RRM2B SNPs in AML patient cohorts
Supplementary Table 3. Correlation between RRM1 and RRM2 in diagnostic leukemic blast from AML97 +AML02 cohorts.
Executive summary.
Aim
RRM1 and RRM2 are critical in regulating cellular deoxy-CTP pools and hence can influence drug sensitivity to nucleoside analogs. Thus, this study was designed to identify genetic variation in ribonucleotide reductase genes (RRM1 and RRM2) and evaluate their relationship with response in acute myeloid leukemia (AML) patients.
Materials & methods
HapMap cell lines from European and African ancestry were used for the discovery phase and specimens from AML patients from the St Jude AML97 and AML02 clinical trials were used to evaluate genetic variation in RRM1 and RRM2.
Results
We identified SNPs within RRM1 and RRM2 that were associated with either its expression and/or cytarabine chemosensitivity in HapMap cell lines.
Evaluation in AML patients identified SNPs associated with intracellular levels of 1-β-D-arabinofuranosyl-CTP and clinical response in AML patients treated with a combination of cladribine and cytarabine.
Conclusion
SNPs within RRM1 and RRM2 might be helpful predictive markers of response to nucleoside analogs and should be further validated in larger cohorts.
Future in vitro functional studies are needed to understand the molecular mechanisms underlying the association of SNPs with the phenotype.
Footnotes
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
This study was supported by NIH grant R01CA132946 (JK Lamba), Minnesota State Partnership funds and by the American Lebanese Syrian Associated Charities (ALSAC), Specialized Center of Research Grant from the Leukemia and Lymphoma Society (ME Dolan), Pharmacogenetics of Anticancer Agents Research Group GM61393 (ME Dolan) and Minnesota State partnership grant. The authors would like to acknowledge the support provided by Biomedical Genomics Center (BMGC) and Minnesota Supercomputing Institute at the University of Minnesota. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Xueyuan Cao, Department of Biostatistics, St Jude Children’s Research Hospital, Memphis, TN, USA.
Amit K Mitra, Department of Experimental & Clinical Pharmacology, PUMA: Institute of Personalized Medicine, College of Pharmacy, University of Minnesota, Minneapolis, Minnesota, MN 55455, USA.
Stanley Pounds, Department of Biostatistics, St Jude Children’s Research Hospital, Memphis, TN, USA.
Kristine R Crews, Department of Pharmaceutical Sciences, St Jude Children’s Research Hospital, Memphis, TN, USA.
Varsha Gandhi, Department of Experimental Therapeutics, MD Anderson Cancer Center, Houston, TX, USA.
William Plunkett, Department of Experimental Therapeutics, MD Anderson Cancer Center, Houston, TX, USA.
M Eileen Dolan, Department of Pathology, Department of Medicine, University of Chicago, IL, USA.
Christine Hartford, Bone Marrow Transplantation & Cellular Therapy, St Jude Children’s Research Hospital, Memphis, TN, USA.
Susana Raimondi, Department of Pathology, St Jude Children’s Research Hospital, Memphis, TN, USA.
Dario Campana, Department of Pathology, St Jude Children’s Research Hospital, Memphis, TN, USA and Department of Oncology, St Jude Children’s Research Hospital, Memphis, TN, USA and National University of Singapore, Singapore.
James Downing, Department of Pathology, St Jude Children’s Research Hospital, Memphis, TN, USA.
Jeffrey E Rubnitz, Department of Oncology, St Jude Children’s Research Hospital, Memphis, TN, USA.
Raul C Ribeiro, Department of Oncology, St Jude Children’s Research Hospital, Memphis, TN, USA.
Jatinder K Lamba, Department of Experimental & Clinical Pharmacology, PUMA: Institute of Personalized Medicine, College of Pharmacy, University of Minnesota, Minneapolis, Minnesota, MN 55455, USA.
References
- 1.Liliemark JO, Plunkett W. Regulation of 1-β-D-arabinofuranosylcytosine 5′-triphosphate accumulation in human leukemia cells by deoxycytidine 5′-triphosphate. Cancer Res. 1986;46(3):1079–1083. [PubMed] [Google Scholar]
- 2.Chabner BA, Hande KR, Drake JC. Ara-C Metabolism: implications for drug resistance and drug interactions. Bull Cancer. 1979;66(1):89–92. [PubMed] [Google Scholar]
- 3.Chiba P, Tihan T, Szekeres T, et al. Concordant changes of pyrimidine metabolism in blasts of two cases of acute myeloid leukemia after repeated treatment with ara-C in vivo. Leukemia. 1990;4(11):761–765. [PubMed] [Google Scholar]
- 4.Tattersall MH, Ganeshaguru K, Hoffbrand AV. Mechanisms of resistance of human acute leukaemia cells to cytosine arabinoside. Br J Haematol. 1974;27(1):39–46. doi: 10.1111/j.1365-2141.1974.tb06772.x. [DOI] [PubMed] [Google Scholar]
- 5.Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp Cell Res. 1989;181(2):305–316. doi: 10.1016/0014-4827(89)90090-6. [DOI] [PubMed] [Google Scholar]
- 6.Lotfi K, Juliusson G, Albertioni F. Pharmacological basis for cladribine resistance. Leuk Lymphoma. 2003;44(10):1705–1712. doi: 10.1080/1042819031000099698. [DOI] [PubMed] [Google Scholar]
- 7.Gandhi V, Estey E, Du M, Nowak B, Keating MJ, Plunkett W. Modulation of the cellular metabolism of cytarabine and fludarabine by granulocyte-colony-stimulating factor during therapy of acute myelogenous leukemia. Clin Cancer Res. 1995;1(2):169–178. [PubMed] [Google Scholar]
- 8.Gandhi V, Estey E, Keating MJ, Plunkett W. Fludarabine potentiates metabolism of cytarabine in patients with acute myelogenous leukemia during therapy. J Clin Oncol. 1993;11(1):116–124. doi: 10.1200/JCO.1993.11.1.116. [DOI] [PubMed] [Google Scholar]
- 9.Gandhi V, Estey E, Keating MJ, Plunkett W. Biochemical modulation of arabinosylcytosine for therapy of leukemias. Leuk Lymphoma. 1993;10(Suppl):S109–S114. doi: 10.3109/10428199309149122. [DOI] [PubMed] [Google Scholar]
- 10.Gandhi V, Estey E, Du M, Keating MJ, Plunkett W. Minimum dose of fludarabine for the maximal modulation of 1-β-D-arabinofuranosylcytosine triphosphate in human leukemia blasts during therapy. Clin Cancer Res. 1997;3(9):1539–1545. [PubMed] [Google Scholar]
- 11.Gautam A, Bepler G. Suppression of lung tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res. 2006;66(13):6497–6502. doi: 10.1158/0008-5472.CAN-05-4462. [DOI] [PubMed] [Google Scholar]
- 12.Cros E, Jordheim L, Dumontet C, Galmarini CM. Problems related to resistance to cytarabine in acute myeloid leukemia. Leuk Lymphoma. 2004;45(6):1123–1132. doi: 10.1080/1042819032000159861. [DOI] [PubMed] [Google Scholar]
- 13.Shao J, Zhou B, Chu B, Yen Y. Ribonucleotide reductase inhibitors and future drug design. Curr Cancer Drug Targets. 2006;6(5):409–431. doi: 10.2174/156800906777723949. [DOI] [PubMed] [Google Scholar]
- 14.Nakahira S, Nakamori S, Tsujie M, et al. Involvement of ribonucleotide reductase M1 subunit overexpression in gemcitabine resistance of human pancreatic cancer. Int J Cancer. 2007;120(6):1355–1363. doi: 10.1002/ijc.22390. [DOI] [PubMed] [Google Scholar]
- 15.Rosell R, Danenberg KD, Alberola V, et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/ cisplatin-treated advanced non-small cell lung cancer patients. Clin Cancer Res. 2004;10(4):1318–1325. doi: 10.1158/1078-0432.ccr-03-0156. [DOI] [PubMed] [Google Scholar]
- 16.Bergman AM, Eijk PP, Ruiz van Haperen VW, et al. In vivo induction of resistance to gemcitabine results in increased expression of ribonucleotide reductase subunit M1 as the major determinant. Cancer Res. 2005;65(20):9510–9516. doi: 10.1158/0008-5472.CAN-05-0989. [DOI] [PubMed] [Google Scholar]
- 17.Hartford CM, Duan S, Delaney SM, et al. Population-specific genetic variants important in susceptibility to cytarabine arabinoside cytotoxicity. Blood. 2009;113(10):2145–2153. doi: 10.1182/blood-2008-05-154302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitra AK, Crews KR, Pounds S, et al. Genetic variants in cytosolic 5′-nucleotidase II are associated with its expression and cytarabine sensitivity in HapMap cell lines and in patients with acute myeloid leukemia. J Pharmacol Exp Ther. 2011;339(1):9–23. doi: 10.1124/jpet.111.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rubnitz JE, Crews KR, Pounds S, et al. Combination of cladribine and cytarabine is effective for childhood acute myeloid leukemia: results of the St Jude AML97 trial. Leukemia. 2009;23(8):1410–1416. doi: 10.1038/leu.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubnitz JE, Inaba H, Dahl G, et al. Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: results of the AML02 multicentre trial. Lancet Oncol. 2010;11(6):543–552. doi: 10.1016/S1470-2045(10)70090-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crews KR, Gandhi V, Srivastava DK, et al. Interim comparison of a continuous infusion versus a short daily infusion of cytarabine given in combination with cladribine for pediatric acute myeloid leukemia. J Clin Oncol. 2002;20(20):4217–4224. doi: 10.1200/JCO.2002.10.006. [DOI] [PubMed] [Google Scholar]
- 22.Holleman A, Cheok MH, Den Boer ML, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351(6):533–542. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 23.Lamba JK, Crews KR, Pounds SB, et al. Identification of predictive markers of cytarabine response in AML by integrative analysis of gene-expression profiles with multiple phenotypes. Pharmacogenomics. 2011;12(3):327–339. doi: 10.2217/pgs.10.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolfinger RD. Heterogeneous variance–covariance structures for repeated measures. J Agric Biol Environ Sci. 1996;1(2):205–230. [Google Scholar]
- 25.Jung SH, Owzar K, George SL. A multiple testing procedure to associate gene expression levels with survival. Stat Med. 2005;24(20):3077–3088. doi: 10.1002/sim.2179. [DOI] [PubMed] [Google Scholar]
- 26.Gray RJ. A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat. 1988;16:14. [Google Scholar]
- 27.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:14. [Google Scholar]
- 28.Erculj N, Kovac V, Hmeljak J, Franko A, Dodic-Fikfak M, Dolzan V. The influence of gemcitabine pathway polymorphisms on treatment outcome in patients with malignant mesothelioma. Pharmacogenet Genomics. 2012;22(1):58–68. doi: 10.1097/FPC.0b013e32834e3572. [DOI] [PubMed] [Google Scholar]
- 29.Kwon WS, Rha SY, Choi YH, et al. Ribonucleotide reductase M1 (RRM1) 2464G>A polymorphism shows an association with gemcitabine chemosensitivity in cancer cell lines. Pharmacogenet Genomics. 2006;16(6):429–438. doi: 10.1097/01.fpc.0000204999.29924.da. [DOI] [PubMed] [Google Scholar]
- 30.Bepler G, Zheng Z, Gautam A, et al. Ribonucleotide reductase M1 gene promoter activity, polymorphisms, population frequencies, and clinical relevance. Lung Cancer. 2005;47(2):183–192. doi: 10.1016/j.lungcan.2004.07.043. [DOI] [PubMed] [Google Scholar]
- 31.Dong S, Guo AL, Chen ZH, et al. RRM1 single nucleotide polymorphism -37C–>A correlates with progression-free survival in NSCLC patients after gemcitabine-based chemotherapy. J Hematol Oncol. 2010;3:10. doi: 10.1186/1756-8722-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feng JF, Wu JZ, Hu SN, et al. Polymorphisms of the ribonucleotide reductase M1 gene and sensitivity to platin-based chemotherapy in non-small cell lung cancer. Lung Cancer. 2009;66(3):344–349. doi: 10.1016/j.lungcan.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 33.Isla D, Sarries C, Rosell R, et al. Single nucleotide polymorphisms and outcome in docetaxel–cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol. 2004;15(8):1194–1203. doi: 10.1093/annonc/mdh319. [DOI] [PubMed] [Google Scholar]
- 34.Kim SO, Jeong JY, Kim MR, et al. Efficacy of gemcitabine in patients with non-small cell lung cancer according to promoter polymorphisms of the ribonucleotide reductase M1 gene. Clin Cancer Res. 2008;14(10):3083–3088. doi: 10.1158/1078-0432.CCR-07-4591. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez J, Boni V, Hernandez A, et al. Association of RRM1 -37A>C polymorphism with clinical outcome in colorectal cancer patients treated with gemcitabine-based chemotherapy. Eur J Cancer. 2011;47(6):839–847. doi: 10.1016/j.ejca.2010.11.032. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Lai L, Wang X, et al. Ribonucleotide reductase small subunit M2B prognoses better survival in colorectal cancer. Cancer Res. 2011;71(9):3202–3213. doi: 10.1158/0008-5472.CAN-11-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sauna ZE, Kimchi-Sarfaty C, Ambudkar SV, Gottesman MM. The sounds of silence: synonymous mutations affect function. Pharmacogenomics. 2007;8(6):527–532. doi: 10.2217/14622416.8.6.527. [DOI] [PubMed] [Google Scholar]
Websites
- 101.International HapMap Project. www.HapMap.org.
- 102.UCSC blat. http://genome.ucsc.edu/cgi-bin/hgBlat?command=start.
- 103.UCSC Genome Bioinformatics. http://genome.ucsc.edu.
- 104.HapMap Data Rel 28. http://hapmap.ncbi.nlm.nih.gov/cgi-perl/gbrowse/hapmap28_B36.
- 105.Gene Expression Omnibus. http://www.ncbi.nlm.nih.gov/geo.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. LD plots for RRM1 SNPs in AML patients demonstrate distinct pattern in whites and blacks.
Supplementary Figure 2. Association of diagnostic blast RRM1 and RRM2 mRNA expression levels with intracellular ara-CTP levels determined 24hr after initiation of therapy. Ara-C given as short daily infusion is indicated in blue and as continuous infusion is in red.
Supplementary Figure 3 Association of RRM SNPs with clinical outcome St. Jude AML 97 patients. Barplot representing association of RRM1 SNP rs2898950 (A) and RRM2B SNP rs1265138 (B) with response after induction 1 in combined cohort. Survival curve for association of RRM2 SNPs rs1130609 (C: Black patients) and rs5030743(D: combined cohort) with overall survival in AML patients.
Supplementary Figure 4. Association of RRM1 (rs1042858 and rs1042927) and RRM2 (rs1130609) SNPs with in vitro ara-C cytotoxicity (LC50) determined in diagnostic leukemic cells from AML patients. Association was determined within three risk groups (high, standard and low) only significant results (p<0.05) are shown.
Supplementary Table 1: Association of SNPs in RRM1 and RRM2 with mRNA expression and ara-C cellular sensitivity.
Supplementary Table 2. Minor allele frequency and genotype frequency of RRM1, RRM2 and RRM2B SNPs in AML patient cohorts
Supplementary Table 3. Correlation between RRM1 and RRM2 in diagnostic leukemic blast from AML97 +AML02 cohorts.