Abstract
Drug abuse and dependence are multifaceted disorders with complex genetic underpinnings. Identifying specific genetic correlates is challenging and may be more readily accomplished by defining endophenotypes specific for addictive disorders. Symptoms and syndromes, including acute drug response, consumption, preference, and withdrawal, are potential endophenotypes characterizing addiction that have been investigated using model organisms. We present a review of major genes involved in serotonergic, dopaminergic, GABAergic, and adrenoreceptor signaling that are considered to be directly involved in nicotine, opioid, cannabinoid, and ethanol use and dependence. The zebrafish genome encodes likely homologs of the vast majority of these loci. We also review the known expression patterns of these genes in zebrafish. The information presented in this review provides support for the use of zebrafish as a viable model for studying genetic factors related to drug addiction. Expansion of investigations into drug response using model organisms holds the potential to advance our understanding of drug response and addiction in humans.
Keywords: Addiction, Zebrafish, Endophenotype, Genetics, Genomics, Behavior
INTRODUCTION
Drug addiction and dependence is a complex human behavior conceptualized as a progression from acute drug exposure to compulsive drug use accompanied by physiological changes such as increased drug tolerance and withdrawal syndromes. Depending upon the individual clinical course, drug dependence commonly includes periods of active use interspersed with abstinence and relapse. Twin and adoption studies have determined that the heritability of addictive disorders is between 40% and 80% depending on the substance (Goldman et al. 2005). Addictive disorders are also dependent on the environment such as allowing or promoting substance access, user stress states and traits, individual coping mechanisms and comorbid medical and psychiatric conditions. Because of this complexity, efforts to identify single genes to explain risk for drug dependence have been challenging.
To overcome this challenge and facilitate the study of complex neurologic diseases, the concept of endophenotypes, or intermediate phenotypes, has been introduced (Gottesman and Gould 2003). Endophenotypes are conceptualized as heritable biological markers associated with illness which co-segregate with illness in affected families and may be found in unaffected relatives of the proband (Gould and Gottesman 2006). Endophenotypes may be state-independent (e.g. require challenge or provocation), reflect mediating factor in behavior, and can be dimensionally or quantitatively measured; these characteristics advance our ability to isolate etiological factors in addictive diseases (Enoch et al. 2003; Gould and Gottesman 2006). Endophenotypes can be neurochemical, neurophysiologic, neuroanatomical, neuroendocrine, neuropsychological or neurocognitive phenomena representing simpler clues to the genetic liability of a disorder (Leboyer et al. 1998). Endophenotypes enable the process of discovering “downstream” clinical phenotypes and “upstream” aberrant genes in polygenic disease vulnerabilities (Gottesman and Gould 2003). Researchers have proposed that drugs of dependence represent a “behavioral vector” comprised of multiple vectors subserving endophenotypes (Farris and Miles 2011). Behavioral vectors are comprised of interacting neuronal networks which are, in turn, controlled by genes expressed within neurons. The concept of “response endophenotypes” has also emerged describing a class of symptomatic and physiologic predictors occurring in response to therapy carrying predictive power for patient outcomes (Leuchter et al. 2009). Characterization of genetic factors influencing endophenotypes of addiction remains difficult but can be facilitated through the use of model systems.
Zebrafish have proved a useful animal model for the study of genetics associated with both complex neurobehavioral phenotypes and drugs of abuse (Clark et al. 2011b; Klee et al. 2011; Mathur and Guo 2010; Stewart et al. 2011). Zebrafish are teleost fish with a haploid genome size of 1.7 gigabases, 25 chromosomes, 12,062 known protein-coding genes, 7,465 predicted protein-coding genes, and 4,431 RNA genes (version 9.0 genome). The lineage between mammals and zebrafish is thought to have split approximately 420 million years ago (Huang et al. 2011). Following that divergence, the zebrafish genome is thought to have undergone an additional duplication and resolution event resulting in the retention of approximately 20% of these additional gene copies (Nusslein-Volhard 2002). Consequently, for a subset of human genes, the zebrafish genome encodes two orthologs (typically annotated as “a” or “b” versions).
Several behavioral zebrafish assays provide a platform to explore acute drug response and the associated reinforcing effect; examples include withdrawal, locomotive activation, and conditioned place preference (Braida et al. 2007; Cachat et al. 2010; Darland and Dowling 2001; Kily et al. 2008; Lau et al. 2006; Ninkovic and Bally-Cuif 2006; Petzold et al. 2009). Novel therapeutic discovery for the treatment of drug dependence can also be facilitated by the use of pre-clinical animal models in the examination of specific neural circuits and functional neural pathways. Drug response in zebrafish may provide clues to the existence of response endophenotypes to drug therapy in humans.
In this article, we characterize the homology in zebrafish of genes associated with nicotine, ethanol, cannabinoid, and opioid drug response. For each of these drugs, a focused set of genes was identified that are associated with mechanistic response. We also describe genes associated with neurotransmitter systems involved in the addiction response cascade such as the dopamine, serotonin, GABA, and noradrenergic systems. Due to the broad scope and often limited certainty of genetic association studies, genes where the only evidence of association with drug response was obtained from genome-wide association studies (GWAS) were excluded from this review.
PRIMARY DRUG TARGET RECEPTORS
Nicotinic Receptors
Societal Impact
Tobacco-related diseases have been declared a global epidemic by the World Health Organization and will cause an estimated 8 million annual deaths annually worldwide by the year 2030(WHO 2008). Within the United States alone, an estimated 222,520 new lung cancer cases were diagnosed and 71,890 lung cancer deaths occurred in 2010 (ACS 2010). Approximately 32% of individuals using tobacco at least once qualify for a diagnosis of drug dependency, compared to about 16% of alcohol and cocaine users (Durrant and Thakker 2003). Cigarette smoking is the predominant risk factor, associated with approximately 90% of all lung cancers.(Carbone 1992). Nicotine dependence, the primary factor driving tobacco abuse, is considered a treatable disease, and tobacco-related deaths are preventable. Significant advances in nicotine dependence research have identified genetic variants associated with addiction (Li 2008) and have led to the development of improved pharmacologic agents that have increased end-of-treatment abstinence rates (Jiménez-Ruiz et al. 2009). Improving our understanding of tobacco dependence etiology, understanding individual risk of dependence, and personalizing pharmacologic treatment requires greater knowledge of the genetic components influencing nicotine response.
Human Receptor Biology
Nicotine is an alkaloid compound found in tobacco responsible for the addictive nature of these products (Jaffe and Kanzler 1979). The compound is a direct ligand for nicotinic acetylcholine receptors (nAChR). The nAChRs are members of the four-transmembrane domain neurotransmitter-gated ion channel superfamily which act as ion channels for sodium and calcium (Karlin 2002). The nAChRs exist as pentameric structures including heteromeric combinations of alpha and beta subunits and alpha-7 homomeric receptors. Eight alpha subunits (α2-α7, α9, α10) and three beta subunits (β2-β4) are expressed in human neural tissue (Jensen et al. 2005). The α4β2 receptor is the most widely expressed nAChR and displays the highest affinity to nicotine binding. However, a diverse set of receptor subunits produce a large number of nAChR subtypes with varying degrees of sensitivity to nicotine binding. Detailed reviews addressing nAChR subtype structures and the associated binding properties of nicotine have been published (Changeux 2010; Gotti et al. 2007; Gotti et al. 2006; Karlin 2002; Rucktooa et al. 2009).
Human Zebrafish Homology
The zebrafish genome contains the complete complement of human neural nAChR subunit genes (Klee et al. 2011). For eight of the twelve human genes exactly one zebrafish ortholog exists. For the remaining four human genes, nicotinic cholinergic receptors alpha 2, 4, 9 and beta 3 (CHRNA2, CHRNA4, CHRNA9, and CHRNB3), two zebrafish homologs were identified. Encoded protein sequence comparisons revealed an average percent identity between human and zebrafish homologs of 73% (range: 64%-84%) with the homologous region spanning an average of 89% (range: 66%-96%) of the human protein (Table 1). As illustrated in the phylogenetic tree (Figure 1), in most cases the greatest similarity between human and zebrafish nAChR genes was found between homologs. However, within the clade containing the human β2 and β4 subunit genes, the association between annotated human and zebrafish homologs is discordant. The most similar gene pairing was the human nicotinic cholinergic receptor beta 2 (CHRNB2) to zebrafish chrnb4, with the zebrafish chrnb2 slightly less similar. The ambiguity in this protein-sequence similarity pairing is reflected in the Bootstrapping analysis of the phylogenetic tree, where the scores for these strata fell below 50% and the Bootstrap consensus tree correctly paired CHRNB2 with chrnb2 and CHRNB4 with chrnb4.
Table 1. Genes linked to addiction through primary drug response.
Genes associated with four drugs of addiction: nicotine, cannabinoids, ethanol, and opioids. Zebrafish homologs were identified for the select set of human genes identified as associated with primary drug response. For each human gene, the corresponding zebrafish homolog is reported, including encoded protein identifiers, the homology codes as identified in the Zebrafish Model Organism Database (ZFIN), and percent identity and coverage extracted from a protein BLAST of the homologous sequence pair.
| Human Gene Symbol |
ZF Gene Name |
Human Protein ID |
ZF Protein ID |
Homology | % Identity | % Coverage | |
|---|---|---|---|---|---|---|---|
| Nicotine | CHRNA2 | chrna2b | NP_000733.2 | XP_697298.3 | AA, CL | 69% | 89% |
| CHRNA2 | chrna2a | NP_000733.2 | NP_001035417.1 | AA, CL | 73% | 92% | |
| CHRNA3 | LOC568467 | NP_000734.2 | XP_001921314.1 | AA, CL | 75% | 93% | |
| CHRNA4 | chrna4 | NP_000735.1 | NP_001041528.1 | AA | 84% | 66% | |
| CHRNA4 | LOC563696 | NP_000735.1 | XP_692148.2 | n/a | 84% | 66% | |
| CHRNA5 | chrna5 | NP_000736.2 | NP_001017885.1 | AA | 75% | 90% | |
| CHRNA6 | chrna6 | NP_004189.1 | NP_001036149.1 | AA, CL | 68% | 96% | |
| CHRNA7 | chrna7 | NP_000737.1 | NP_957513.1 | AA | 78% | 96% | |
| CHRNA9 | LOC568807 | NP_060051.2 | XP_001920894.1 | AA, CL | 68% | 92% | |
| CHRNA9 | LOC798522 | NP_060051.2 | XP_001338964.4 | n/a | 64% | 76% | |
| CHRNA10 | chrna10 | NP_065135.2 | NP_001038269.1 | AA, CL | 68% | 86% | |
| CHRNB2 | LOC100003503 | NP_000739.1 | XP_001343044.3 | AA | 67% | 96% | |
| CHRNB3 | chrnb3a | NP_000740.1 | NP_957514.1 | n/a | 77% | 95% | |
| CHRNB3 | chrnb3b | NP_000740.1 | NP_775394.1 | AA | 77% | 94% | |
| CHRNB4 | LOC568566 | NP_000741.1 | XP_696993.2 | n/a | 64% | 96% | |
| Cannabinoids | CNR1 | cnrl | NP_001153698.1 | NP_997985.1 | AA, CL | 71% | 100% |
| CNR2 | cnr2 | NP_001832.1 | NP_998129.3 | AA, CL | 44% | 88% | |
| CNR2 | LOC100150020 | NP_001832.1 | XP_001920368.1 | n/a | 44% | 84% | |
| CNRIP1 | cnripl | NP_001104571.1 | XP_684894.2 | AA, CL | 60% | 100% | |
| NAPEPLD | napepld | NP_001116310.1 | NP_001074082.2 | AA | 65% | 87% | |
| ABHD4 | abhd4 | NP_071343.2 | NP_001017613.1 | AA, CL | 61% | 99% | |
| DAGLA | dagla | NP_006124.1 | XP_697873.4 | AA, CL | 67% | 93% | |
| DAGLB | daglb | NP_631918.3 | XP_693659.5 | AA | 58% | 52% | |
| GPR55 | LOC793909 | NP_005674.2 | XP_001333765.1 | AA, CL | 43% | 90% | |
| PPARA | pparab | NP_001001928.1 | NP_001096037.1 | AA | 72% | 100% | |
| PPARA | pparaa | NP_001001928.1 | NP_001154805.1 | AA, CL | 66% | 100% | |
| PPARG | pparg | NP_056953.2 | NP_571542.1 | AA | 64% | 94% | |
| TRPA1 | trpala | NP_015628.2 | NP_001007066.1 | AA, CL | 49% | 99% | |
| TRPA1 | trpalb | NP_015628.2 | NP_001007067.1 | AA, CL | 47% | 97% | |
| TRPV1 | trpvl | NP_061197.4 | NP_001119871.1 | AA, CL | 49% | 82% | |
| TRPV4 | trpv4 | NP_067638.3 | NP_001036195.1 | AA, CL | 72% | 93% | |
| GPR18 | gpr18 | NP_001091670.1 | XP_684672.4 | AA, CL | 64% | 97% | |
| GPR119 | LOC100001479 | NP_848566.1 | XP_001337269.1 | n/a | 44% | 89% | |
| FAAH | faah | NP_001432.2 | NP_001103295.1 | AA, CL | 54% | 90% | |
| FAAH2 | faah2a | NP_777572.2 | NP_001002700.1 | AA | 60% | 92% | |
| FAAH2 | faah2b | NP_777572.2 | NP_001070930.1 | AA | 55% | 92% | |
| NAAA | n/a | NP_055250.2 | n/a | n/a | n/a | n/a | |
| MGLL | mgll | NP_009214.1 | NP_956591.1 | AA, CL | 57% | 93% | |
| PTGS2 | ptgs2b | NP_000954.1 | NP_001020675.1 | AA, CL | 75% | 97% | |
| PTGS2 | ptgs2a | NP_000954.1 | NP_705943.1 | AA, CL | 69% | 97% | |
| Ethanol | CYP2E1 | n/a | NP_000764.1 | n/a | n/a | n/a | n/a |
| ADH1B | n/a | NP_000659.2 | n/a | n/a | n/a | n/a | |
| ADH1C | n/a | NP_000660.1 | n/a | n/a | n/a | n/a | |
| ADH4 | n/a | NP_000661.2 | n/a | n/a | n/a | n/a | |
| ALDH2 | aldh2b | NP_000681.2 | NP_998466.2 | AA, CL | 77% | 100% | |
| ALDH2 | aldh2a | NP_000681.2 | NP_956784.1 | AA, CL | 78% | 100% | |
| ALDH2 | LOC100332355 | NP_000681.2 | XP_002662252.1 | n/a | 78% | 97% | |
| GRIN1 | grinlb | NP_000823.4 | NP_001137603.1 | AA | 88% | 99% | |
| GRIN1 | grinla | NP_000823.4 | NP_001070182.2 | AA, CL | 85% | 99% | |
| GRIN2A | LOC563297 | NP_000824.1 | XP_691754.2 | n/a | 68% | 98% | |
| GRIN2B | LOC559976 | NP_000825.2 | NP_001121809.1 | n/a | 41% | 16% | |
| CRH | crh | NP_000747.1 | NP_001007380.1 | AA, CL | 92% | 28% | |
| ARC | n/a | NP_056008.1 | n/a | n/a | n/a | n/a | |
| BDNF | bdnf | NP_733927.1 | NP_571670.2 | AA, IX | 69% | 96% | |
| NPY | npy | NP_000896.1 | NP_571149.1 | AA, CL | 76% | 73% | |
| CREB1 | crebla | NP_604391.1 | NP_959203.1 | AA | 85% | 100% | |
| CREB1 | creblb | NP_604391.1 | NP_001017818.1 | AA, CL | 79% | 53% | |
| Opioids | OPRM1 | oprml | NP_000905.3 | NP_571782.1 | AA, CL | 75% | 90% |
| OPRD1 | oprdla | NP_000902.3 | NP_571333.1 | AA, CL, | 76% | 77% | |
| OPRD1 | oprdlb | NP_000902.3 | NP_997920.1 | AA, CL | 77% | 77% | |
| OPRK1 | oprkl | NP_000903.2 | NP_878306.1 | AA | 68% | 100% | |
| OPRL1 | oprll | NP_000904.1 | NP_991152.1 | AA, CL | 64% | 91% |
Figure 1.
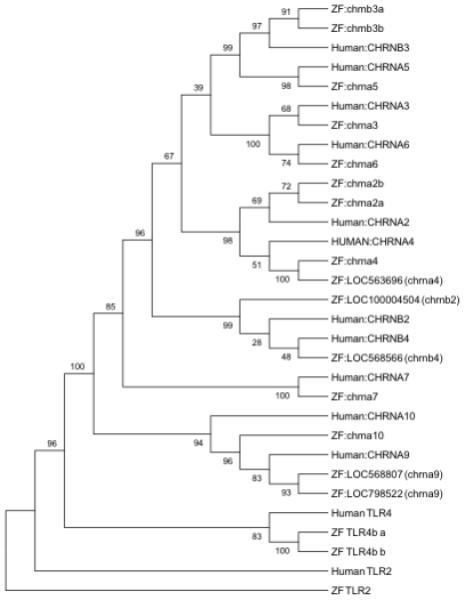
Phylogenetic tree illustrating the protein sequence relationship of human and zebrafish neural nicotinic receptors. All eleven human neural nAChRs are represented in the zebrafish genome by one or more genes. The TLR4 and TLR2 genes are included in the molecular tree to provide an out-group root for relative comparison of the nicotinic receptor genes. Following a Bootstrapping analysis, the only clade falling below a 50% confidence level was the structure of the CHRNB2 and CHRNB4 human genes and corresponding zebrafish homologs. This low confidence for this structure is reflected in the discordant mapping of these homologs.
Zebrafish Gene Localization
Experimental characterization of the neural nicotinic acetylcholine receptor subunit gene expression in the zebrafish has been described for five of the nine alpha subunit genes and two of three beta subunit genes. An understanding of the known expression patterns of these genes can be instrumental in the design of appropriate model animal studies. The zebrafish chrna2b gene expression was characterized using in situ hybridization in 40-48 hour post fertilization (hpf) zebrafish embryos, illustrating a strong spinal cord neuron expression (Thisse and Thisse 2005). The spinal cord expression was independently observed using in situ hybridization in 24-48 hpf zebrafish embryos, and was further observed in the forebrain of 24 hpf embryos (Zirger et al. 2003). The chrna4 gene was characterized in 24, 28, 72, and 96 hpf zebrafish using in situ hybridization (Ackerman et al. 2009). Expression at 24 hpf was observed in neural crest cells, hind brain, spinal neurons, mandibular, hyoid, and brachial pharyngeal arches, and limited expression in the forebrain. Expression at 48 hpf was noted in the medial longitudinal fascicle and reticulospinal neurons of the hindbrain. At 72 and 96 hpf, extensive midbrain expression and limited hindbrain expression was noted. The chrna5 gene expression measured by whole-mount in situ hybridization was observed in adult liver and strongly observed in 3 dpf whole-body larvae(Cheng et al. 2006). The chrna6 gene was extensively characterized using in situ hybridization in 24, 48, 72, and 96 hpf zebrafish larvae (Ackerman et al. 2009). In 24 hpf larvae, expression was observed in the spinal neurons, Rohon-Beard sensory neurons in the trunk, pineal, ventral forebrain, trigeminal ganglion, diencephalon, and hypothalamus. By 48 hpf, expression persisted in the pineal, trigeminal ganglion, and diencephalon, and was also observed heavily in the retina, tectum, and locus coeruleus. At 72 hpf, chrna6 expression remained in the locus coeruleus, retinal ganglion cells, tectum, pineal, trigeminal ganglion, dicencephalon, and was also observed in the pretectal catecholaminergic cluster and amacrine cells of the retina. Finally, at 96 hpf, expression persisted in the retina, pineal, tectum, trigeminal ganglian, diencephalon, locus coeruleus, pretectal catecholaminergic cluster, and was also observed in the cranial sensory neurons. Expression of chrna7 was identified by real-time polymerase chain reaction (RT-PCR) in 8 hpf zebrafish embryos and in the hindbrain of 96 hpf zebrafish larvae using in situ hybridization(Zirger et al. 2003). The expression pattern of the encoded protein of chrnb2b was observed in the spinal cord of 36 hpf larvae using immunohistochemistry (Welsh et al. 2009). Finally, the expression of a non-discriminant form of chrnb3 (chrnb3a/chrnb3b not specified) was observed in the retinal ganglion cell layer in 48 hpf and 72 hpf larvae. While the expression profiles for the nAChR genes are not extensive, they provide context for when and where these genes are observed.
Cannabinoids
Societal Impact
For thousands of years derivatives of Cannabis sativa have been used for recreational, entheogenic, and medicinal purposes. Although delta-9-tetrahydrocannabinol (Δ9-THC) is the most psychoactive phytocannabinoid (El-Alfy et al. 2010), there are over 80 additional cannabinoids present in Cannabis sativa. Each of these cannabinoids potentially has differential physiological activities. Cannabis derivatives are illegal in many jurisdictions, and represent the most widely used illicit drug in the United States, Europe, and Australia(Maldonado et al. 2011). The prevalence of past month Cannabis use in the United States in 2009 was 6.6% (Martínez et al. 2010). In addition, there has been an increase in the popularity of products marketed as “herbal spice” or “herbal incense”. These products often contain synthetic cannabinoids and have rapidly changing formulations to circumvent new laws (Dresen et al. 2010). Chronic Cannabis use has been associated with increased heart rate and risk or heart attack (Mittleman, 2001), impaired physical and mental health (Gruber, 2003), and impaired learning and memory (Pope, 2001). About 9% of Cannabis users will suffer from drug dependency compared to about 16% of alcohol and cocaine users (Wagner and Anthony 2002).
Human Receptor Biology
Cannabinoid receptors are generally found in presynaptic terminals where they impact the release of neurotransmitters, including glutamate, GABA, glycine, acetylcholine, norepinephrine, dopamine, serotonin, and cholecystokinin (CCK). The broad influence of these drugs explains the prominent role of endocannabinoid signaling in anxiety, depression, cognition, addiction, motor function, feeding behavior, and pain (Kano et al. 2009). The first insights into the biology of cannabinoid signaling occurred when the two primary receptors, cannabinoid receptor 1 (CNR1) and cannabinoid receptor 2 (CNR2), were cloned in the early 1990s (Matsuda et al. 1990; Munro et al. 1993). CNR1 and CNR2 are seven transmembrane G-protein coupled receptors (GPCR) conserved in vertebrates. CNR1 is the most abundant GPCR in the brain and is widely distributed throughout the CNS as well as many peripheral tissues (Pertwee et al. 2010). CNR2 was originally identified within peripheral tissues, but more recently has been found within regions of the brain especially glial cells (Cabral et al. 2008; Onaivi 2006). Despite its limited expression in the CNS, CNR2 may play a pivotal role in drug self-administration, most likely through release of dopamine (Xi et al. 2011). Although cannabinoids appear to predominately signal through CNR1 and CNR2, additional receptors are predicted to play a role in cannabinoid signaling. Some of these receptors that have demonstrated for cannabinoid ligands include: G protein-coupled receptors 18, 55, and 119 (GRP18, GPR55, GPR119) (McHugh et al. 2010; Ryberg et al. 2009) (Overton et al. 2006), transient receptor potential cation channel, subfamily V, members 1 and 4 (TRPV1 and TRPV4) (De Petrocellis et al. 2011; Ross 2003), transient receptor potential cation channel, subfamily A, member 1 (TRPA1) (Baraldi et al. 2010), and peroxisome proliferator-activated receptors alpha and gamma (PPARA and PPARG) (Sun et al. 2007) In addition interacting proteins like those from the cannabinoid receptor interacting protein 1 (CNRIP1) gene may be important targets for CNR signaling modulation (Smith et al. 2010).
Endogenous cannabinoids, or endocannabinoids, are a subclass of bioactive lipid mediators that are produced locally on demand (Murakami 2011). Although CNR1 and CNR2 appear to not be conserved outside of vertebrates, cannabinoid responses are observed in invertebrates, suggesting some cannabinoid signaling is conserved in invertebrates (Elphick and Egertová 2001). Because endocannabinoids are produced locally on demand, their synthesis and generally rapid degradation are important aspects of cannabinoid signaling. Some of the enzymes involved in the synthesis and degradation of the two main endocannabinoids, arachidonoyl ethanolamide (anandamide or AEA) and 2-arachidonoyl glycerol (2-AG), have been described (Ueda et al. 2010; Ueda et al. 2011). These include N-acyl phosphatidylethanolamine phospholipase D (NAPEPLD) and abhydrolase domain containing 4 (ABHD4) for synthesis of AEA and diacylglycerol lipase alpha and beta (DAGLA and DAGLB) for synthesis of 2-AG. Genes involved in the degradation of AEA include fatty acid amine hydrolase (FAAH), fatty acid amine hydrolase 2 (FAAH2), and N-acylethanolamine acid amidase (NAAA). 2-AG is degraded primarily by the enzymes encoded by monoglyceride lipase (MGLL), and to a lesser extent prostaglandin-endoperoxide synthase 2 (PTGS2).
Human Zebrafish Homology
A list of cannabinoid receptors and key enzymes known for endocannabinoid synthesis and degradation was assembled and compared to the zebrafish to characterize homologous representation (Table 1). All but one of the genes analyzed has at least one zebrafish homolog. To date no homolog has been identified in the zebrafish genome for N-acylethanolamine acid amidase (NAAA), which is one of multiple enzymes known to breakdown N-arachidonoylethanolamide AEA in mammalian systems. Encoded protein sequence comparisons revealed an average percent identity between human and zebrafish homologs of 59% (range: 43%-75%) with the homologous region spanning an average of 92% (range: 52%-100%) of the human protein. Figure 2 illustrates the evolutionary relationship between human and zebrafish encoded cannabinoid signaling proteins. In all instances, the closest association by sequence identity is found between the human and associated zebrafish homologs. There are four human genes FAAH2, PTGS2, TRPA1, and PPARA, with two homologs in the zebrafish while the remaining fifteen genes having a single homolog.
Figure 2.
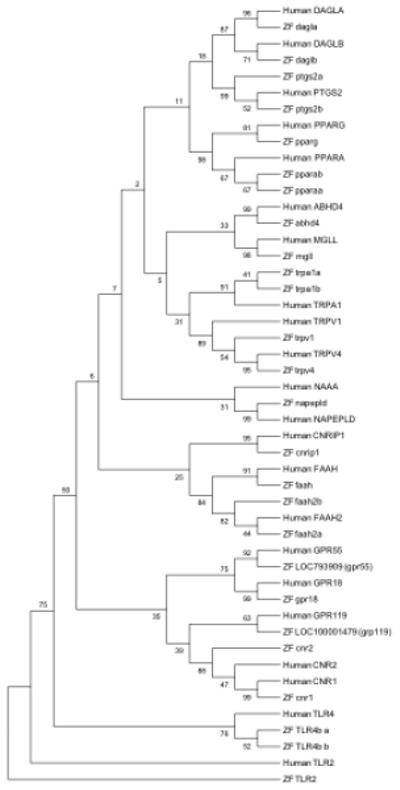
Phylogenetic tree illustrating the sequence relationship of human and zebrafish proteins encoded by genes associated with response to cannabinoid drug use. Nineteen of twenty human genes included in this analysis (excluding out-group) have at least one closely associated zebrafish homolog. The human NAAA is the exception, with no zebrafish homolog. Sub-clade structures, beyond direct ortholog associations, that are significantly (>50%) following Bootstrapping analysis include the association of the DAGLA and DAGLB gene family, the TRPV1 and TRPV4 gene family, the FAAH and FAAH2 gene family, the GRP55 and GRP18 gene family and CNR2 and CNR1 gene family. The out-group consisting of the TLR genes is also significantly distinct from the rest of the cannabinoid associated genes.
Zebrafish Gene Localization
Few investigations have been conducted on the expression of endocannabinoid signaling genes and function of exogenous or endogenous cannabinoids in zebrafish. The first observed expression of cnr1 occurs at 24 hpf in a small subset of the preoptic area of the diencephalon; the distribution of cnr1 transcripts continues through development with significant staining in regions of the zebrafish brain including the telencephalon, hypothalamus, tegmentum, and anterior hindbrain (Lam et al. 2006). cnr2 was initially reported to exist in zebrafish as two paralogs of the human CNR2, but has since been resolved as a single gene (Rodriguez-Martin et al. 2007). RT-PCR analysis of cnr2 in adult zebrafish demonstrated that similarly to other vertebrates, this receptor is primarily peripheral with limited expression in the brain (Rodriguez-Martin et al. 2007). trpa1a and trpa1b expression is primarily restricted to sensory ganglia in developing zebrafish (Caron et al. 2008; Prober et al. 2008). Both the primary synthesis enzyme gene, dagla, and the primary catabolic enzyme gene, mgll, for the endocannabinoid 2-AG have widespread but restricted expression in the brain, similar to cnr1 expression patterns (Watson et al. 2008).
Ethanol
Societal Impact
The earliest evidence of human-controlled alcohol production indicates that a mixed fermented beverage of rice, honey, and fruit (hawthorn fruit and/or grape) was being produced as early as the seventh millennium B.C. (McGovern et al. 2004). Alcohol has had considerable social, religious and medical significance after alcohol producing yeast S. cerevisiae was domesticated on at least two independent occasions from diverse wild populations possessing the ability to produce high concentrations of ethanol (Fay and Benavides 2005; Woolfit and Wolfe 2005). Epidemiological studies have reported J-shaped relations between alcohol consumption and cardiovascular disease and death, indicating health benefits of moderate drinking (1 or less drink per day for women and 1 to 2 drinks per day for men)(Djoussé et al. 2009). However, high alcohol consumption remains strongly associated with negative consequences. World Health Organization (WHO) Global Burden of Disease Project concluded that alcohol accounts for approximately 1.8 million deaths per year or 3.2% of all deaths(Rehm et al. 2004). In the United States, more than half of all adults have a family history of alcoholism or problem drinking while 3 out of 10 adults have met diagnostic criteria for alcoholism and/or engaged in alcohol abuse at some point in their lives. Approximately 16% of all first-time alcohol users will qualify for a diagnosis of drug dependency (Durrant and Thakker 2003). Untreated addiction costs the United States $400 billion annually and recent research indicates that alcoholism and alcohol abuse alone cost the nation’s economy approximately $185 billion each year. This cost includes 15% related to the cost of medical consequences and treatment, more than 70% due to reduced or lost earnings, and the remainder due to lost productivity, accidents, violence, and premature death (Harwood et al. 1998). Research in the United Kingdom has indicated that the most harmful drug to others was alcohol and the most harmful drugs to users were crack cocaine followed by heroin (Nutt et al. 2010).
Human Receptor Biology
Unlike opiates or cannabis, alcohol does not have affinity to a specific receptor. Ethanol has broad pharmacodynamic effects balanced between excitatory and inhibitory neurotransmission. At concentrations associated with behavioral effects in humans, ethanol facilitates gamma-aminobutyric acid (GABA) function and, at the same time, inhibits the N-methyl-D-aspartate (NMDA) receptor which mediates the post-synaptic excitatory effects of glutamate (Krystal et al. 2003; Krystal et al. 2006). Tolerance to ethanol results in up-regulation of the NMDA receptor so that abrupt withdrawal produces a hyperexcitable state that leads to seizures, delirium tremens, and excitotoxic neuronal death(Tsai and Coyle 1998). Long-term exposure to ethanol leads to an imbalance in different excitatory and inhibitory amino acids. When ethanol consumption is reduced or completely stopped, imbalances in amino acid neurotransmitters are behaviorally expressed in the form of ethanol withdrawal. Glutamate, a major excitatory amino acid, and GABA, a major inhibitory amino acid, are responsible, at least partly, for ethanol withdrawal and complications including seizures and delirium tremens (De Witte 2004; Krystal et al. 2003; Krystal et al. 2006; Tsai and Coyle 1998).
Precise mechanisms of these effects of ethanol are not known; however, a number of proteins for which considerable molecular-level evidence for distinct ethanol binding sites have been identified (reviewed by (Harris et al. 2008)). These target proteins include enzymes involved in alcohol pharmacokinetics, alcohol and acetaldehyde dehydrogenases (ADH/ALDH) and Adenylyl cyclase - an enzyme involved in production of the second messenger adenosine 3′,5′-monophosphate (cAMP) from adenosine triphosphate (ATP). Contemporary findings indicate that alcohol is metabolized to acetic acid primarily by ADH/ALDH and studies have implicated sequence variations in those genes in association with alcohol dependence and related phenotypes. Genetics factors influencing the primary ADH/ALDH pathway have been identified in ALDH1b, ALDH1c, and ALDH2 (Chen et al. 2009a; Martínez et al. 2010; Nishimura et al. 2002). Additionally, in the two auxiliary ADH/ALDH pathways, genetic effects have been linked to ADH4 and cytochrome P450, family 2, subfamily E, polypeptide 1 (CYP2E1) (Howard et al. 2003; Luo et al. 2006). Different adenylyl cyclase isozymes show different sensitivities to ethanol enhancement of cAMP production (Yoshimura and Tabakoff 1995) and generation of the adenylyl cyclase chimeras enabled the identification of two “ethanol-responsive domains” that determined the alcohol sensitivity (Yoshimura et al. 2006). Evidence also indicates that ethanol acts on Ion channels, including inhibition of N-methyl-D-aspartate (NMDA)-type glutamate receptors and enhances function of γ-aminobutyric acid type A (GABAA) and glycine receptors (Harris et al. 2008). indings suggested that ethanol may target a cavity existing among the transmembrane regions of γ-aminobutyric acid and F glycine receptors that may accommodate (Wick et al. 1998) and/or a target in the second loop of the extracellular domain of alpha1 glycine receptors (GLRA1) (Crawford et al. 2007).
In addition, alcohol consumption is known to increase beta-endorphin release at the level of midbrain/VTA where it potentially influences drug reinforcement (Jarjour et al. 2009). Moreover, positron emission tomography study demonstrates that a functional opioid receptor mu 1 (OPRM1) A118G polymorphism is a major determinant of striatal dopamine release in responses to alcohol in social drinkers (Ramchandani et al. 2011). Providing that dopamine release in ventral striatum is a common element of drug reward (Volkow et al. 2007), these findings emphasize complex interaction among neurotransmitter systems involved in alcohol dependence-related phenotypes as well as the need for careful selection of the target phenotypes for association studies.
The last but not least, acute and chronic ethanol exposure has been also shown to modulate function of the activity-dependent gene transcription factor, cAMP-responsive element binding (CREB) protein in the brain (reviewed in (Moonat et al. 2010)). The downstream effects on several important CREB-related genes, such as neuropeptide Y (NPY), brain derived neurotrophic factor (BDNF), activity-regulated cytoskeleton-associated protein (ARC) and corticotrophin-releasing hormone (CRH) may play a crucial role in the behavioral effects of ethanol and underlie both alcohol addiction and a genetic predisposition to alcoholism ((Moonat et al. 2010)).
Thus, brief review of evidence supports the notion that ethanol affects various biochemical processes such as neurotransmitter release, enzyme function, and ion channel kinetics and we are only beginning to understand the specific molecular sites to which ethanol molecules bind to produce these myriad effects. However, identified ethanol binding sites and proteins potentially involved in its action allows selection of candidate genetic targets for the purposes of genetic association studies and development of animal models.
Human Zebrafish Homology
Due to the diffuse manner in which ethanol interacts with proteins in humans there are no direct receptors mediating exposure to alcohol, however both the GABA and glycine receptors have been suggested to be directly targeted. The response dynamics are heavily influenced by the genes discussed above, with nine of the fourteen genes (Table 1) having characterized zebrafish homologs (Figure 3). Homology of the GABA genes are independently discussed in the Secondary Neurotransmitter section of this manuscript. Of the protein discussed here ALDH2 has three defined zebrafish homologs, N-methyl D-aspartate 1 glutamate receptor (GRIN1) and CREB1 has two zebrafish homologs, and GLRA1, NPY, BDNF, CRH, GRIN2A and N-methyl D-aspartate 2B glutamate receptor (GRIN2B) have a single zebrafish homolog each. Encoded protein sequence comparisons revealed an average percent identity between human and zebrafish homologs of 77% (range: 41%-92%) with the homologous region spanning an average of 81% (range: 16%-100%) of the human protein (Table 1). Of particular note, is the zebrafish homolog of GRIN2B, which only has 41% identity across 16% of the human encoded protein, bringing the homologous status into doubt. The five genes lacking zebrafish homologs include all three alcohol dehydrogenase genes, cytochrome P450 family 2 gene, and the activity-regulated cytoskeleton-associated protein. BLAST sequence comparisons of these three human alcohol dehydrogenase proteins to the zebrafish proteome reveal the closest homologs were the human proteins to each other, and the closest zebrafish homolog was the alcohol dehydrogenase class-3 protein (NP_571924.2). However, this protein has a well-characterized human ortholog (NP_000662). Consequently, while some alcohol dehydrogenase proteins are conserved, based on our current data these three are lacking zebrafish homologs.
Figure 3.

Phylogenetic tree illustrating the sequence relationship of human and zebrafish proteins encoded by genes associated with alcohol dependence. Of the fourteen human genes, excluding the TLR out-group, nine have at least one zebrafish homolog. The genes ARC, CYP2E1, ADH1B, ADH1C, and ADH4 do not have known zebrafish homologs. The inclusion of the TLR2 and TLR4 out-group genes within the middle of the phylogenetic tree illustrates the disparate association between the genes identified as important in the genetics of alcohol dependence, in stark contrast to the tight association of genes associated with other drugs of abuse (i.e. nicotine). The only significant sub-clade structures in this analysis include the grouping of the GRIN2A and GRIN2B genes, and the grouping of the human ADH1B, AHD1C, and ADH4 genes.
Zebrafish Gene Localization
Extensive analysis was done to define the embryonic and tissue-specific expression patterns of the ALDH2 zebrafish homolog genes using RTPCR (Song et al. 2006). Expression of both zebrafish homologs was detected as early as 2 hpf, with expression also measured at 4, 8, 10, 24, 48, and 72 hpf. The zebrafish gene aldh2b had uniformly high expression at all time points, while aldh2a had highly diminished expression at 8, 10, and 72 hpf. Tissue-specific adult expression was observed for both genes in the brain, eye, gill, heart, intestine, liver, muscle, ovary, pectoral fin, and swim bladder. However, in the caudal fin, only aldh2b was observed to be highly expressed (Song et al. 2006). Independent characterization of the aldh2b gene expression using in situ hybridization revealed defined expression at 16 hpf in the eye and lens placode, at 19 hpf in the alar plate, midbrain epiphysis, lens vesicle, neural crest, optic tectum, and otic vesicle, and at 30-36 hpf in the diencephalon, eye, optic tectum, otic vesicle pigment cell, retina, and YSL. Furthermore, between 42-48 hpf expression was noted in the diencephalon, epidermis, epiphysis, heart, liver, optic tectum, pancreas, pancreatic bud, pharyngeal arch, pigment cell and retina (Thisse et al. 2001). Expression of the protein encoded by aldh2a as measured using Western blot in adult zebrafish was shown to be highly expressed in muscle, heart and brain. The protein was also shown to be expressed, to a lesser extent, in the eye, liver and swim bladder. However, no protein expression was observed when tested in the skin, caudal fin, or gill (Lassen et al. 2005).
BDNF expression measured by in situ hybridization was observed as early as 16 hpf. Whole-body expression continued to be observed up to 24 hpf, where localization of the expression became evident in the forebrain and hindbrain regions (Rauch et al. 2003). Expression at 36 hpf, exhibited more refined localization in the diencephalon, midbrain, rhombomere, tegmentum, and telencephalon (Rauch et al. 2003). At 48 hpf, expression was noted in the cranial ganglion, lateral line system, neuromast, peripheral olfactory organ, retina, retina ganglion cell layer, and telencephalon (Thisse and Thisse 2004). By 5dpf expression was observed in the brain, inner ear, and midbrain. Expression was further characterized in adult zebrafish using RT-PCR, with tissue-specific expression shown the brain, eye, gill, heart, integument, intestine, liver, and musculature (Heinrich and Pagtakhan 2004).
Expression of NPY has not been well described in the zebrafish, with only one report of whole-brain expression measured by RT-PCR (Piccinetti et al. 2010). The expression of CRH, on the other hand, has been extensively studied in the zebrafish. Expression has been measured by RT-PCR from as early as 1 dpf, with transcript abundance steadily increasing to 5 dpf (Chandrasekar et al. 2007) Localization is first noted by in situ hybridization in the hypothalamus at 25 hpf and persists through adulthood (Chandrasekar et al. 2007; Kurrasch et al. 2009; Lohr et al. 2009; Rios et al. 2011). Between 28-32 hpf, expression is also noted in the ventral telencephalon (Chandrasekar et al. 2007; Lohr et al. 2009). By 36 hpf, expression is found in the thalamus (Chandrasekar et al. 2007). At 2 dpf, expression is also observed in the retina, preoptic area, rostal medulla oblongata (Alderman and Bernier 2007). Expression is further noted in the loceus correlus and posterior tuberculum at 3 dpf. Finally, in adult zebrafish, expression is observed in all the developmental regions and the olfactory bulbs (Alderman and Bernier 2007).
CREB1 homolog creb1a expression levels in the zebrafish has been observed as early as 24 hpf with immune-blotting. Detailed immunohistochemical analysis of the adult zebrafish brain has shown that the creb1a encoded protein is expressed in all sixteen known proliferation zones, the optic tract, the corpus cerebelli, the granular zone of the cerebellum, the crista cerebellaris, corpus mammilare, commisura ansulata and oculomotor nerve (Dworkin et al. 2007). There is no reported characterization of the creb1b homolog.
Expression of grin1a was measured in embryonic zebrafish using in situ hybridization and observed to be expressed at 24 hpf in the head and neural tube (Hwang et al. 2009). More detailed analysis revealed that at 24 hpf and 48 hpf, grin1a was expressed in the forebrain, hindbrain, midbrain, spinal cord, and presumptive neural retina (Cox et al. 2005). The second GRIN1 homolog, grin1b, had a more restrictive expression pattern, only being found in the forebrain at 24 hpf, and in the forebrain, hindbrain, and midbrain at 48 hpf (Cox et al. 2005). Similar characterization of grin2a revealed its expression in the retina at 48 hpf but not at 24 hpf (Cox et al. 2005). Analysis of grin2b in whole embryonic tissue by RTPCR revealed its expression is more delayed, only appearing strongly by 96 hpf and then diminishing by 2 months post-fertilization (Cox et al. 2005).
Expression of the glra1 transcript has been characterized in both embryonic and adult zebrafish. Within adult zebrafish brain, the glra1 gene was extensively characterized and shown to be expressed in several different nuclei (Imboden, 2001). Within embryonic zebrafish, glra1 was characterized at 26, 50, 74, 98, and 122 hpf zebrafish, with measurable expression first noted at 50 hpf and increasing at each time poitn there after. Protein expression was also shown to occur in spinal neurons at 50 and 122 hpf zebrafish using IHC (Mongeon, 2008). Expression at 48 hpf but not 24 hpf of glra1 transcripts in the zebrafish spinal neurons was also shown by McDearmid et al (2006). Converesly, whole body RT-PCR showed that glra1 expression can be initially detected at 24 hpf, but at a significantly diminished level from what is observed at 48 hpf (Hirata, 2005). The 24 hpf expression was further characterized by in situ hybridization and shown to occur in the putative hindbrain and spinal cord neurons. glra1 is known to have multiple splice variants which may explain the discrepancy in developmental time of first detect expression, however, characterization of the constitutive and at least one known splice-variant shown expression detected by RT-PCR at 26 hpf, with increasing expression occurring at 52 hpf (Devignot, 2003).
Opioid Receptors
Societal Impact
Heroin addiction is associated with significant morbidity and mortality; past month heroin use prevalence in the United States in 2009 was 0.08% (Martínez et al. 2010). Over the past 20 years, prescription opioid addiction has emerged as an even greater threat (Caudill-Slosberg et al. 2004; Olsen et al. 2006), with a 2.1% past month prevalence of non-medical use of pain-relievers in the United States in 2009 (NSDUH). This has been attributed, in part, to the widespread use of prescription opioids for the treatment of chronic pain (Center for Disease Control and Prevention 2007; Manchikanti 2007). From 1997 to 2006, sales of hydrocodone increased 244%, while sales of methadone and oxycodone increased 1177% and 732%, respectively (Manchikanti and Singh 2008). Commensurate with increased sales of prescription opioids, the milligram per person (mg/person) dose increased 347% from 74 mg/person in 1997 to 329 mg/person in 2006 (Manchikanti and Singh 2008). Epidemiologic studies suggest that increased sales and use of greater quantities of prescription opioids is associated with an increased number of overdose fatalities (Hall et al. 2008; Paulozzi and Ryan 2006; Wysowski 2007), which increased 91% between 1999 and 2002 (Paulozzi et al. 2006). Despite decades of basic science and clinical research, an unmet clinical need exists for targeted clinical strategies aimed at the prevention and treatment of heroin and prescription opioid addiction. The threat posed by these drugs is significant with 23% of first time heroin users qualifying for a diagnosis of drug dependency (Durrant and Thakker 2003). A further understanding of the genetics of the endogenous opioid system in animal models and humans could lead to the development of new pharmacological agents which could improve the clinical outcomes of individuals with opiate addiction.
Human Receptor Biology
Opioids primarily act through three opioid receptors, the mu-opioid receptor (MORP), the kappa-opioid receptor (KORP), and the delta-opioid receptor (DORP), encoded by the human genes OPRM1, OPRK1, and OPRD1, respectively. A fourth receptor, the nociceptin receptor (NOP), is encoded by the human OPRL1 gene, but less is known regarding its physiologic role compared with the three “classical” opioid receptors. These are G-protein-coupled receptors that interact with heterotrimeric (Gi/Go) G-proteins (Dhawan et al. 1996). Endogenous activation of MORP by ß-endorphin, KORP by dynorphin, and DOPR by enkephalin and deltorphin result in increased potassium channel conductance, decreased calcium channel conductance, and inhibition of cyclic adenosine monophosphate production (Dhawan et al. 1996). Efficacious exogenous ligands for these receptors include drugs like morphine and oxycodone for MORP and enadoline, butorphanol, nalbuphine, nalmefene, and pentazocine for KOPR. There are no exogenous agonists for DORP or NOP available for use in humans. The physiologic effects of opioid receptor activation include analgesia, dysphoria, water diuresis, and antipruritic effects. Activation may also modulate responses to other drugs of addiction. For example, KORP activation has been implicated in the mitigation of cocaine craving among individuals with cocaine addiction (Shippenberg et al. 2001). Multiple genetic association studies have identified potentially informative variants modulating the receptor gene expression levels. Functionally, reduced levels of MOPR expression and signaling have been found to be associated with individual variations in heroin addiction (Bart et al. 2004; Drakenberg et al. 2006), responsiveness to systematically administered opioids (Chou et al. 2006; Reyes-Gibby et al. 2007; Sia et al. 2008), and alterations in pain perception (Campa et al. 2008; Fillingim et al. 2005; Lotsch et al. 2009; Sia et al. 2008) in some, but not all, studies (Arias et al. 2006; Walter and Lotsch 2009). Variation in genes encoding for the 3 opioid receptors may also affect the interaction of these receptors with various addictive drugs. For instance, variants in the OPRK1 have been associated with opiate addiction (Gerra et al. 2007; Yuferov et al. 2004) and an increased risk of alcohol dependence (Xuei et al. 2006; Zhang et al. 2008). There is conflicting evidence that variations in the OPRD1 gene are associated with heroin addiction and other drugs of addiction (Franke et al. 1999; Mayer et al. 1997; Xu et al. 2002; Zhang et al. 2008). A SNP in the OPRM1 (A118G) gene has been associated with differences in human smoking behavior, suggesting that opioid receptors can modify behavioral responses to nicotine (Ray et al. 2006).
Zebrafish Receptor Homology
The three classical human opioid receptor genes are fully represented in the zebrafish genome (Table 1). There are single homologs for the human ORPM1, OPRK1, OPRL1 genes, and a pair of homologs for the human OPRD1 gene (Figure 4). There is an average of 72% (range: 64-77%) identical residues between the human and zebrafish-encoded protein sequences, spanning an average of 87% (range: 77-100%) of the human protein sequence. As shown in the phylogenetic tree, all zebrafish homologs pair closest with their respective human gene counterpoints, when jointly analyzing all opioid receptor sequences The zebrafish orpd1a and oprd1b encoded proteins have a high degree of sequence similarity, with 73% identity over 97% of the sequence length. Despite this, studies have shown that the paralogs possess unique response profiles to receptors agonists and antagonists (Gonzalez-Nuñez et al. 2006; Pinal-Seoane et al. 2006) and as discussed below also show divergent expression patterns. This variability indicates that while the paralog sequences likely originated from a common gene, they appear to have evolved to perform unique functions.
Figure 4.
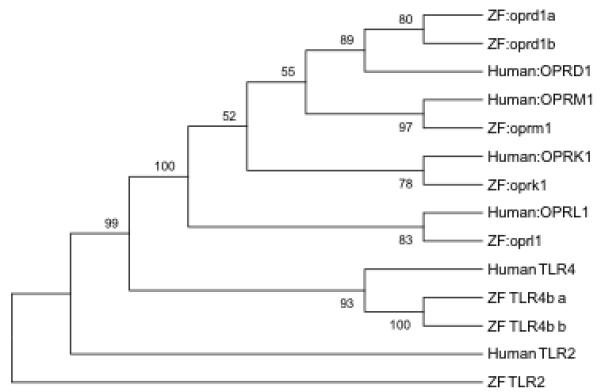
Phylogenetic tree illustrating the sequence relationship of human and zebrafish proteins encoded by genes associated with opioid use. The four human opioid receptor genes all have at least one zebrafish ortholog. All branching points in this tree structure are significant (>50% Bootstrapping). The TLR2 and TLR4 gene out-groups illustrate the tight association of the opioid receptor genes, relative to other known human-zebrafish orthologs.
Zebrafish Receptor Localization
The expression patterns (temporal and spatial) of the opioid receptor genes have been described in a high level of detail. RT-PCR quantification of whole embryo transcript abundance revealed oprm1 is initially expressed at 3 hpf. From that time forward through embryogenesis, the expression level varies, peaking at 22, 48, and 72 hpf (Sanchez-Simon and Rodríguez 2008). Expression at 24 hpf was further verified by RT-PCR and the encoded receptor shown to be function by morpholino induced gene knock-down and subsequent modifications in morphine modulated miRNA regulation (Sanchez-Simon et al. 2010). In situ hybridization localized expression in 24 hpf embryos to multiple CNS structures, including the telencephalon, epiphysis, diencephalon, midbrain, pretectum, isthmus, cerebellum, and hindbrain. By 48 hpf, expression patterns changed and oprm1 expression was noted in the tegmentum, hypophysis, otic vesicle, and pectoral flipper. Expression in 2 week old larvae confirmed continued expression of oprm1 in the CNS but noted an absence of expression in the olfactory epithelium (Bretaud et al. 2007). Expression of oprd1a and oprd1b were highly varied, with oprd1b expression patterns more closely resembling that of oprm1 than oprd1a. RT-PCR whole embryo quantitation revealed the first significant expression of either paralog at 22 hpf. opr1a demonstrated varied expression levels following 22 hpf, with high expression occurring again at 48 and 72 hpf. A similar but dampened overall expression profile was noted for oprd1b (Sanchez-Simon and Rodríguez 2008). In situ hybridization revealed 24 hpf oprd1b expression in the same regions as oprm1 and additional expression noted in the myotomes and spinal cord. At 48 hpf expression was observed in the telencephalon, diencephalon, midbrain, and swim bladder. oprd1a expression at 24hpf was observed in the telencephalon, epiphysis, pretectum, and cerebellum. Expression at 30-36 hpf was detected in the hindbrain, spinal cord, and tegmentum (Thisse 2004), with expression at 48 hpf detected in the tegmentum, hypophysis, and ventral thalamus (Sanchez-Simon and Rodríguez 2008). Adult zebrafish expression of oprd1b as measured by in situ hybridization and revealed strong expression in the hypothalamus, periventricular layer of the optic tectum, and granular layer of the cerebellum(Pinal-Seoane et al. 2006). Expression of oprk1 was first detected by RT-PCR in embryonic zebrafish at 48 hpf, with the highest expression observed in adult fish. However, it is important to note that the overall expression levels were two orders of magnitude lower than that observed for oprm1, oprd1a, and oprd1b. Attempts to localize expression in embryonic zebrafish using in situ hybridization failed, likely owning to the low expression levels (Sanchez-Simon and Rodríguez 2008). In situ hybridization analysis of adult fish revealed high levels of expression in dorsal telencephalic areas, epithalamus, hypothalamic recess nuclei, periventricular layer of the optic tectum, granular layer of the cerebellum, reticular formation, and the facial lobe. Expression at lower levels was also observed in the olfactory bulb, vagal lobe, octavolateral area, and spinal cord(Alvarez et al. 2006).
DOWNSTREAM NEUROTRANSMITTER SIGNALING SYSTEMS
Neurotransmitter systems such as the dopamine, serotonin, adrenoreceptor and GABA system have a facilitating or permissive role in the function of drug addiction. Neurotransmitter receptors of these systems are expressed in neurons that are directly targeted by drugs of addiction or are involved in the control of directly targeted neurons.
Dopamine System
In mammalian and human brains, dopamine (DA) is the main neurotransmitter of the reward circuit, that is formed by neurons projecting from the ventral tegmental area (VTA) to the nucleus accumbens (NAcc) (Schultz 1998; Schultz 2010). The four main functional DA elements of neurons are the dopamine receptors, dopamine transporters, rate-limiting enzymes of the DA metabolic pathway such as tyrosine hydroxylase (TH), and the dopamine catabolic enzyme monoamine oxidase (MAO). Most DA receptors and the DA transporter (DAT) have been associated with addiction. In mammals and humans, activation of the D2 receptors will down-regulate the cAMP concentration in cells(Civelli et al. 1993; Monsma et al. 1990; O'Dowd 1993). There are two genes commonly used to distinguish DA neurons in nervous systems, the tyrosine hydroxylase (TH) and the dopamine transporter (DAT) genes. While tyrosine hydroxylase is a marker for catecholaminergic neurons, the availability of antibodies for the use in the zebrafish produced a detailed expression map of the protein in comparison with neurotransmitter systems for serotonin, histamine and orexin (Guo et al. 1999; Holzschuh et al. 2001; Kaslin et al. 2004). The DAT is both a specific marker for DA neuron labeling and the target of cocaine that affects the reuptake of DA and ultimately leads to an increase in extracellular DA levels.
Human and Zebrafish Homology
All four human dopamine receptor genes (DRD1-4), the dopamine transporters (SLC6A2-3), and associated rate limiting enzymes dopamine beta-hydroxylase (DBH) and TH have zebrafish orthologs (Figure 5). DRD1, DRD3, DBH, SLC6A2, and SLC6A3 have a single zebrafish ortholog. Conversely, TH, DRD2, and DRD4 have a pair of homologs. The average protein sequence identity between homologs is 68% (range: 58-80%) covering an average of 89% (range: 62-99%) of the human protein sequence (Table 2). Comparison of homologs within the context of all eight dopamine related genes resulted in a complete closest association of the zebrafish genes with the respective human homologs (Figure 5).
Figure 5.
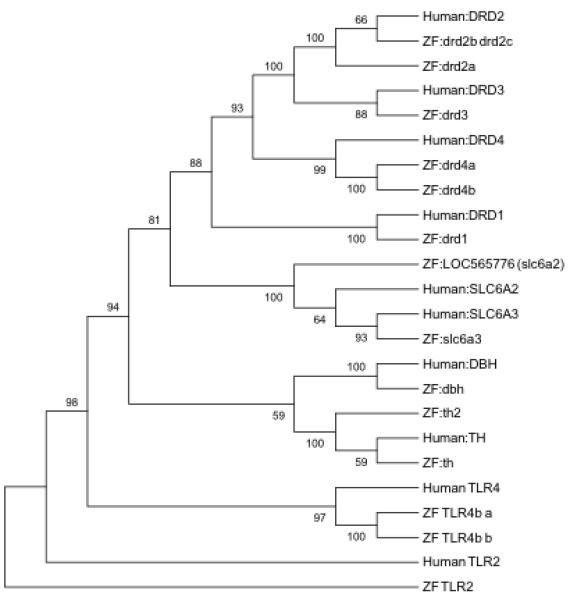
: Phylogenetic tree illustrating the sequence relationship of human and zebrafish proteins encoded by genes associated with downstream dopamine signaling. The eight human non-out-group genes have at least one zebrafish ortholog. All branching points in this tree structure are significant (>50% Bootstrapping). The TLR2 and TLR4 gene out-groups, again illustrate the tight association of the identified genes associated with dopamine signaling.
Table 2. Genes linked to addiction through downstream neurotransmitter signaling systems.
Genes associated with four secondary signaling systems associated with drugs of addiction: dopamine, serotonin, GABA, and adrenoreceptors. Zebrafish homologs were identified for the select set of human genes identified as associated with these secondary signaling systems. For each human gene, the corresponding zebrafish homolog is reported, including encoded protein identifiers, the homology codes as identified in the Zebrafish Model Organism Database (ZFIN), and percent identity and coverage extracted from a protein BLAST of the homologous sequence pair.
| Human Gene Symbol |
ZF Gene Name |
Human Protein ID |
ZF Protein ID | Homology | % Identity | % Coverage | |
|---|---|---|---|---|---|---|---|
| Dopamine | DBH | dbh | NP_000778.3 | NP_001103164.1 | AA, CL | 61% | 94% |
| DRD1 | drdl | NP_000785.1 | NP_001129448.1 | AA | 74% | 96% | |
| DRD2 | drd2a | NP_000786.1 | NP_898891.1 | AA, CL | 69% | 96% | |
| DRD2 | drd2b/drd2c | NP_000786.1 | NP_922918.1 | AA, CL | 69% | 99% | |
| DRD3 | drd3 | NP_000787.2 | NP_898890.1 | AA, CL | 58% | 95% | |
| DRD4 | drd4a | NP_000788.2 | NP_001012634.1 | AA, CL | 66% | 62% | |
| DRD4 | drd4b | NP_000788.2 | NP_001012636.1 | AA, CL | 66% | 63% | |
| SLC6A2 | LOC565776 | NP_001034.1 | XP_694138.3 | n/a | 72% | 98% | |
| SLC6A3 | slc6a3 | NP_001035.1 | NP_571830.1 | AA, CL | 80% | 95% | |
| TH | th | NP_000351.2 | NP_571224.1 | AA, CL | 71% | 89% | |
| TH | th2 | NP_000351.2 | NP_001001829.1 | n/a | 63% | 96% | |
| Serotonin | HTR1A | htrlaa | NP_000515.2 | NP_001116793.1 | AA, CL | 75% | 97% |
| HTR1A | htrlab | NP_000515.2 | NP_001139238.1 | AA, CL | n/a | n/a | |
| HTR1B | LOC561647 | NP_000854.1 | NP_001122181.1 | n/a | n/a | n/a | |
| HTR1D | htrlbd | NP_000855.1 | NP_001139158.1 | n/a | 68% | 94% | |
| HTR1E | LOC100330681 | NP_000856.1 | XP_002665689.1 | n/a | |||
| HTR1F | LOC100005344 | NP_000857.1 | XP_001344430.1 | n/a | 61% | 94% | |
| HTR2A | LOC560808 | NP_000612.1 | XP_689300.5 | AA, CL | 65% | 84% | |
| HTR2B | zgc:194119 | NP_000858.3 | NP_001038208.1 | n/a | 62% | 88% | |
| HTR2C | si:dkey-24g18.2 | NP_000859.1 | NP_001123365.1 | n/a | 55% | 84% | |
| HTR3A | LOC571641 | NP_000860.2 | XP_700338.3 | AA, CL | 62% | 93% | |
| HTR3B | LOC571632 | NP_006019.1 | XP_700329.4 | AA, CL | 48% | 94% | |
| HTR3C | n/a | NP_570126.2 | n/a | n/a | n/a | n/a | |
| HTR3D | n/a | NP_001138615.1 | n/a | n/a | n/a | n/a | |
| HTR3E | n/a | NP_872395.2 | n/a | n/a | n/a | n/a | |
| HTR4 | LOC556843 | NP_000861.1 | XP_684857.4 | n/a | 77% | 83% | |
| HTR5A | ht5a | NP_076917.1 | NP_001119882.2 | AA, CL | 81% | 91% | |
| HTR5A | ht5al | NP_076917.1 | NP_001007122.1 | n/a | 79% | 94% | |
| HTR6 | LOC568269 | NP_000862.1 | XP_696681.4 | AA, CL | 58% | 73% | |
| HTR7 | LOC562111 | NP_000863.1 | XP_690599.5 | n/a | 78% | 75% | |
| MAOA | mao | NP_000231.1 | NP_997992.2 | AA, SS | 69% | 98% | |
| MAOB | mao | NP_000889.3 | NP_997992.2 | n/a | 70% | 99% | |
| SLC6A4 | slc6a4a | NP_001036.1 | NP_001035061.1 | AA | 70% | 98% | |
| SLC6A4 | slc6a4b | NP_001170930.1 | NP_001036.1 | AA | 69% | 86% | |
| TPH1 | tphla | NP_004170.1 | NP_840091.1 | AA, CL | 80% | 89% | |
| TPH1 | tphlb | NP_004170.1 | NP_001001843.2 | AA, CL | 77% | 90% | |
| TPH2 | tph2 | NP_775489.2 | NP_999960.1 | AA, CL | 74% | 93% | |
| GABA | GABRA1 | gabral | NP_001121120.1 | NP_001070794.1 | AA, CL | 85% | 97% |
| GABRA2 | LOC100150704 | NP_001107647.1 | XP_001920091.2 | n/a | 86% | 92% | |
| GABRA2 | LOC556056 | NP_001107647.1 | XP_683856.4 | n/a | 85% | 92% | |
| GABRA4 | zgc:110204 | NP_000800.2 | NP_001017822.1 | n/a | 78% | 73% | |
| GABRA5 | gabra5 | NP_000801.1 | XP_001339511.2 | AA, CL | 78% | 96% | |
| GABRA6 | gabra6b | NP_000802.2 | XP_002667403.2 | AA | 87% | 70% | |
| GABRA6 | gabra6a | NP_000802.2 | NP_957025.1 | AA, CL | 60% | 100% | |
| GABRB1 | LOC100331377 | NP_000803.2 | XP_002664179.1 | n/a | 83% | 94% | |
| GABRB2 | gabrb2 | NP_068711.1 | NP_001019558.2 | AA, CL | 79% | 97% | |
| GABRB3 | LOC566922 | NP_000805.1 | XP_002662540.1 | AA, CL | 83% | 100% | |
| GABBR2 | si:dkey-190|1.2 | NP_005449.5 | NP_001137515.1 | n/a | 80% | 92% | |
| GABBR1 | gabbrla | NP_001461.1 | XP_694497.4 | AA | 76% | 92% | |
| GABBR1 | gabbrlb | NP_001461.1 | XP_003200653.1 | AA, CL | 76% | 92% | |
| GABARAP | gabarapa | NP_009209.1 | NP_001013278.1 | AA, CL | 98% | 99% | |
| GABARAP | gabarapb | NP_009209.1 | n/a | AA, CL | |||
| GABRG1 | LOC556202 | NP_775807.2 | XP_684047.4 | n/a | 80% | 92% | |
| GABRG2 | si:ch211- 145n14.1 |
NP_944494.1 | XP_687331.3 | n/a | 84% | 100% | |
| GABRG3 | LOC567057 | NP_150092.2 | XP_001920261.2 | AA, CL | 83% | 95% | |
| Adrenoreceptors | ADRA1A | LOC798498 | NP_000671.2 | XP_001338938.1 | AA, CL | 60 | 96 |
| ADRA1B | zgc:103685 | NP_000670.1 | NP_001007359.1 | n/a | 68 | 80 | |
| LOC100149100 | NP_000670.1 | XP_001922013.2 | n/a | 59 | 85 | ||
| ADRA1D | LOC568614 | NP_000669.1 | XP_697043.2 | n/a | 59 | 94 | |
| ADRA2A | adra2a | NP_000672.3 | NP_997520 | AA, CL | 59 | 99 | |
| ADRA2B | adra2b | NP_000673.2 | NP_997521.1 | AA, CL | 53 | 91 | |
| ADRA2B | LOC100330650 | NP_000673.2 | XP_002663705.1 | n/a | 58 | 96 | |
| ADRA2C | adra2c | NP_000674.2 | NP_997522.1 | AA, CL | 65 | 88 | |
| n/a | adra2da | n/a | NP_919345.2 | n/a | n/a | n/a | |
| n/a | adra2db | n/a | NP_919346.1 | n/a | n/a | n/a | |
| ADRB1 | adrbl | NP_000675.1 | NP_001122161.1 | AA, CL | 61 | 80 | |
| ADRB2 | adrb2a | NP_000015.1 | NP_001096122.1 | AA, CL | 56 | 95 | |
| ADRB2 | adrb2b | NP_000015.1 | NP_001082940.1 | AA, CL | 58 | 89 | |
| ADRB3 | adrb3a | NP_000016.1 | NP_001121807.1 | AA, CL | 56 | 78 | |
| ADRB3 | adrb3b | NP_000016.1 | NP_001128606.1 | AA, CL | 51 | 75 |
Zebrafish Neurotransmitter Localization
In the zebrafish, dopaminergic neurons are organized in at least 13 different neuron clusters throughout the brain (Panula et al. 2010). The neurons most relevant to drug addiction are the DA neurons of the reward pathway that are located in the VTA (mammalian brain region A10) of the mesencephalon (midbrain) and that produce ascending projections to the NAcc that is located in the striatum in the telencephalon (Corbett and Wise 1980). While the neuroanatomy of the catecholamine/dopamine system in vertebrate animals is somewhat conserved, the identification of homologous DA neuron clusters in zebrafish as compared to mammals is a challenge both from a clinical and evolutionary standpoint. The main difference between the zebrafish and the mammalian system is that the mesencephalic DA neurons, including neurons of the ventral tegmental area (VTA) and the substantia nigra (SN), are not found in the zebrafish brain (Mahler et al. 2010). In the context of addiction, there has been no clear identification of the brain region in the zebrafish that is homologous to the VTA. Rink and Wullimann (Rink and Wullimann 2001) have traced axons from the posterior tuberculum to the subpallium, a region that is homologous to the mammalian and human striatum, thus making the posterior tuberculum/basal diencephalon, a candidate region in which homologous VTA-like DA neurons are located. While the identification of the zebrafish brain region that is homologous to the mammalian VTA is controversial, the posterior tuberculum in the diencephalon just ventral to the mesencephalon is the main candidate region.
To describe the neuroanatomical organization of DA neurons, a genetic labeling approach has been developed in which a catecholamine neuron-specific transcription factor is targeted for driving the expression of Green Fluorescent Protein (Tay et al. 2011). This study demonstrates the power of genetic tools that are available for the zebrafish system - for the first time the entire system of DA neurons has been labeled in a whole vertebrate brain. The expression of the reporter gene provides stunning neuroanatomical details of single neurons and shows neurons of the posterior tuberculum with projections ascending to the subpallium/telencephalon in the larval brain. Similar ascending projection patterns of tubercular neurons were obtained earlier using immunocytochemical and dye tracing methods in adult zebrafish (Rink and Wullimann 2001; Rink and Wullimann 2002a, b). In addition, similar to mammalian midbrain DA neurons, TH1 immunoreactive neurons of the posterior tuberculum express the transcription factor NR4A2. In situ hybridization experiments show that the NR4A2 transcript is expressed in at least subset of TH1 immunoreactive neurons in the posterior tuberculum (Blin et al. 2008; Filippi et al. 2007). Since only a subset of neurons of the posterior tuberculum project to the subpallium (Tay et al. 2011) and since NR4A2b has been detected in a subset of neurons in the posterior tuberculum (Blin et al. 2008) it is possible that a subset of neurons in the posterior tuberculum is homologous to mesencephalic DA neurons in the VTA of mammals.
The expression of the zebrafish DA receptor and associated genes has been well described. The zebrafish drd1, drd2a, drd2b, drd2c, drd3, and drd4b genes show the strongest expression in the tegmentum or diencephalon. The expression of the zebrafish D1 receptor (drd1) gene has been detected in the mid-embryo stage at 30hpf. At this time the gene is expressed in the brain (diencephalon, hindbrain)(Li et al. 2007). In late embryonic stages (48hpf), the gene expression is detectable in the hypothalamus and has spread within the hindbrain. Cells in the retina show expression in larval zebrafish (120hpf)(Li et al. 2007). While the expression of drd1 in the diencephalon, hindbrain, hypothalamus and retina is similar to the expression pattern in mammals, no expression of drd1 has been described in the zebrafish olfactory bulb which shows strong expression in mammals (Li et al. 2007; Mansour et al. 1990; Monsma et al. 1990; Nguyen-Legros et al. 1999). In situ hybridization experiments showed that all three DRD2 receptor gene homologs, drd2a, drd2b and drd2c are expressed in specific brain region such as the diencephalon, the tegmentum, and the hindbrain (Boehmler et al. 2004). The drd2c receptor is more diffusely expressed throughout the brain and spinal cord, whereas the drd2a and drd2b genes are expressed in the epiphysis. Overall, the expression pattern of zebrafish drd2 receptors is similar to the expression pattern of DRD2 receptors in mammals. The drd3 receptor is expressed more diffusely throughout the nervous system in the spinal cord and brain in the embryonic stage(Boehmler et al. 2004). Later in larvae, the expression pattern is more specific in the telencephalon, the tegmentum and the hindbrain as shown by in situ hybridization experiments. In addition, the retina contains cells that are labeled with probes for the drd3 receptor. The expression of the drd4a gene has been detected as early as 24hpf when expression is found in the epiphysis and the spinal cord (Boehmler et al. 2007). Later in development, the expression is detectable by in situ hybridization in the telencephalon, the diencephalon, and the hindbrain. The drd4b gene is described of being expressed in the telencephalon, the diencephalon, the tegmentum, the adenohypophysis, and the otic vesicle from 24hpf. Branchial arches show also expression of the drd4a gene early. More expression of the gene is found in the spinal cord in 5dpf larvae. Compared to the drd4a and drd4c gene, the drd4b genes seems to be more prominently expressed in specific brain regions in the diencephalon and tegmentum than the drd4a and drd4c genes. The drd4c gene is not strongly expressed in the brain of zebrafish embryos and larvae. At 24hpf, the diencephalon and spinal cord are the only regions of the central nervous system that show labeling via in situ hybridization experiments. In mid embryonic stages additional expression has been reported in cranial ganglia and the branchial arch. At 48hpf expression pattern becomes more diffuse and at 5dpf expression seems to be limited mainly to the photoreceptor layer of the retina.
Expression of the zebrafish slc6a3 gene matches the expression of TH-positive cells in 13 identified DA neuron clusters (Holzschuh et al. 2001). The expression of the dat gene is detected as early as 24 hpf in the diencephalon. As the development continues slc6a3 expression is also be detected in the olfactory bulb, the pre-tectum, the locus coeruleus, the hindbrain, the retina and the optic nerve. The TH gene (th1) is widely expressed in the brain and spinal cord of zebrafish embryos and larvae. As demonstrated clearly by Chen et al. (Chen et al. 2009b) in a comparative study, the expression of the second TH gene (th2) is restricted to the hypothalamus. Expression of th1 can be detected in the diencephalon as early as 18 hpf and at 20 hpf also in the locus coeruleus and branchial arch associated neurons (Holzschuh et al. 2001). Later at 72 hpf, expression is also detectable in the olfactory bulbs, the pretectum, and the hindbrain. While the th1 gene expression is abundant, the expression of the th2 gene in the brain is restricted to the pretectum and the hypothalamus, as well as the liver (Chen et al. 2009b).
Serotonin System (5-HT)
Serotonin has been widely recognized to have a strong influence on impulse control, and genetic variation in the biosynthesis, metabolism, and receptor function may have an influence on the development of addiction. For example, serotonin biosynthesis is regulated by the rate-limiting enzyme tryptophan hydroxylase, encoded by two genes which have been associated with heroin addiction(Nielsen et al. 2008). In zebrafish, the system of 5-hydroxytryptamine (5-HT) immunoreactive neurons has been mapped in the developing and the adult zebrafish brain and shows similarities to the mammalian and human 5-HT system (Panula et al. 2010).
The specificity of 5-HT actions is mediated by different 5-HT receptor types in pre- and postsynaptic sites in the central nervous system. The zebrafish 5-HT receptor system is similar to the mammalian and human 5-HT receptor system. The superfamily of vertebrate 5-HT receptors is divided into seven different 5-HT receptor families that together contain at least fourteen different 5-HT receptors (Hannon and Hoyer 2008; Hoyer et al. 2002). With the exception of the 5-HT3 receptor family, all receptors are G-proteins coupled receptors. Activation of 5-HT1 and 5-HT5 receptors ultimately lowers the levels of cAMP in neurons, whereas activation of 5-HT4, 5-HT6, and 5-HT7 receptors increases the cAMP levels in neurons. The 5-HT2 receptors activate the phospholipase C pathway and increase the Ca2+ levels in cells. Similar to nicotinic acetylcholine receptors, the receptors of 5-HT3 family form pentameric cation selective channels that are conducting Na+, K+ and Ca2+ ions (Walstab et al. 2010). Within the zebrafish genome ten 5-HT receptor genes have been predicted and six genes have been characterized experimentally.
The role of individual 5-HT receptors in mammals and humans has been reviewed by Hayes and Greenshaw (Hayes and Greenshaw 2011). We highlight examples of 5-HT receptor function in disease and addiction. There is some indication that 5-HT1A, 5-HT2C, 5-HT3 and 5-HT6 receptors have direct effects on reward-related behaviors, whereas the function of 5-HT1B, 5-HT2A, 5-HT2B, 5-HT4, 5-HT5, 5-HT6, and 5-HT7 is little known or not clearly associated with reward-related behaviors. The 5-HT2A and 5-HT2B receptors have clinical relevance since the 5-HT2A receptor has been associated with mental disorders such as schizophrenia and the 5-HT2B receptor with heart development including heart valves. 5-HT3 receptors have an important clinical relevance, mostly because the 5-HT3B receptor antagonists are used as anti-emetics in chemotherapy patients (Cunningham et al. 1987). Recently, the 5-HT3 receptor has been linked to alcohol consumption (Enoch et al. 2010). The 5-HT5A receptor has been studied as intensively as other 5-HT receptors but the predicted function in regulation of mood, cognitive, and circadian rhythms is based on expression patterns in mammalian brains(Thomas 2006). A genetic link of the human 5-HT5A receptor gene to schizophrenia is controversial (Veenstra-VanderWeele et al. 2000). The 5-HT transporter is a protein of major clinical importance since it is a target of drugs used to treat depression. The 5-HT transporter (5-HTT) is found in presynaptic endings of serotonergic neurons where its function is the re-uptake of released 5-HT. Thus the protein terminates the activation of 5-HT receptors on postsynaptic sites and recycles 5-HT. A critical gene product of the 5-HT system is the enzyme tryptophan hydroxylase (TH) (Dresen et al.) that catalyzes the rate limiting step of the 5-HT synthesis (Chen et al. 2009b). Within the 5-HT system, the breakdown of 5-HT is controlled by the enzyme monoamine oxidase (MAO).
Human and Zebrafish Homology
The genes identified in the zebrafish genome match the organization of 5-HT receptors in the human genome closely. While all seven 5-HT receptor types have been identified in zebrafish, there are differences in the set of receptors within families due to gene duplication. Two genes (ht1aa, ht1ab) have been identified, sequenced and mapped as zebrafish homologs of the 5-HT1A receptor (HTR1A) (Norton et al. 2008). Additionally, a third zebrafish 5-HT1 receptor gene has been designated as 5-HT1bd, because of the similarity to both the human 5-HT1B receptor (HTR1B) and 5-HT1D receptor (HTR1D) (Norton et al. 2008). However, in our analysis, this receptor-encoded protein more closely matches the human protein HTR1D than it does HTR1B. The 5-HT5A receptor (HTR5A) and 5-HT5B receptor (HTR5B) form the 5-HT5 receptor family, but only the 5-HT5A receptor is a functional receptor, whereas the 5-HT5B receptor transcript contains stop codons that lead to a non-functional protein (Thomas 2006). The zebrafish homolog of the non-functional 5-HT5B has not been defined. In addition, homologs of the HTR3C, HTR3D, and HTR3E genes have not been identified in zebrafish. Overall, the encoded zebrafish proteins have an average of 69% (range: 48-81%) identity to the human proteins, covering an average of 90% (range: 73-99%) of the human sequence (Table 2). This is nearly identical to the values observed for the dopaminergic proteins. Comparison of homologs within the context of all twenty-three serotonin related genes resulted in a complete closest association of the zebrafish genes with the respective human homologs (Figure 6).
Figure 6.
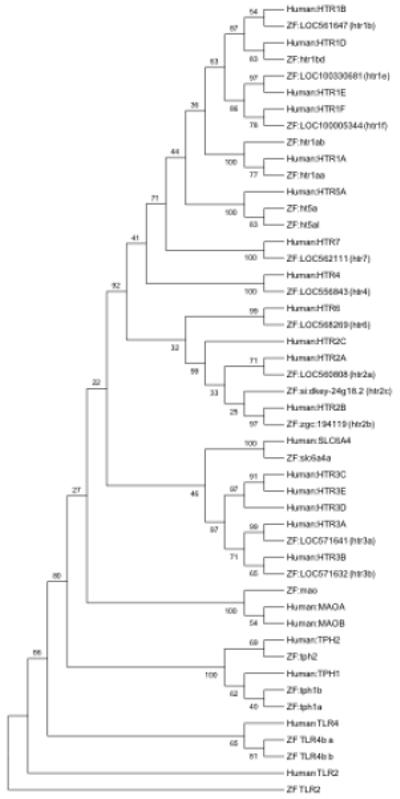
Phylogenetic tree illustrating the sequence relationship of human and zebrafish proteins encoded by genes associated with downstream serotonin signaling. There are three of the twenty-two human genes lacking a defined zebrafish homolog, including HTR3C, HTR3D, and HTR3E. In addition, the human MAOA and MAOB genes share a signal zebrafish homolog (mao). The TLR2 and TLR4 gene out-groups, again illustrate the tight association of the identified genes associated with dopamine signaling. The significance of the sub-clade structures varies, with some significant groups (i.e. HTR1B, HTR1D, HTR1E, and HTR1F genes), but not all branch points are significant. The TLR2 and TLR4 gene out-groups, again illustrate the tight association of the identified genes associated with serotonin signaling.
Zebrafish Neurotransmitter Localization
The anatomical organization of serotonin (5-HT) neurons in the zebrafish brain shows some similarities with the organization of 5-HT neurons in the mammalian brain(Bellipanni et al. 2002; Jacobs and Azmitia 1992; Panula et al. 2010; Steinbusch 1981; Steinbusch and Nieuwenhuys 1981; Törk 1990). In the zebrafish, 5-HT neurons are found in the raphe nuclei and additoinal brain regions. A total of nine clusters have been described in zebrafish using antibodies against 5-HT and antisense probes for the serotonin transporter and tryptophan hydroxylase 2 mRNA(Bellipanni et al. 2002; Kaslin and Panula 2001; Lillesaar et al. 2007; McLean and Fetcho 2004a, b; Rink and Guo 2004). The most prominent immunostaining of 5-HT neurons occurs in the hypothalamus/paraventricular organ; clusters 2-4). In comparison, the dorsal raphe neurons (clusters 5-7) are relatively weakly labeled. The anteriormost cluster (cluster 1) is found in the pretectum/thalamus. 5-HT immunoreactive neurons in rostral clusters (1-7) have ascending projections that spread widely throughout the brain, while the posterior clusters (8, 9) project into the spinal cord.
Unprecedented labeling of projections of serotonergic neurons in the superior and inferior raphe (clusters 5-7) of the zebrafish brain was achieved using a transgenic approach (Lillesaar et al. 2009). The promoter of pet1, a transcription factor gene expressed in serotonergic raphe neurons in rodents (Pfaar et al. 2002; Scott et al. 2005), was utilized to drive the expression of GFP in serotonergic neurons. Other zebrafish 5-HT neurons such as the prominent cluster in the hypothalamus (clusters 2-4) did not express GFP in this transgenic zebrafish line. The genetic approach lead to the identification of a new cluster of previously overlooked zebrafish 5-HT neurons located in the ventro-lateral region of the hindbrain in close proximity to the raphe nuclei. Projections from the ventro-lateral region aim specifically to the general region of the posterior tuberculum, which is potentially homologous/analogous to the dopaminergic VTA of mammals. In the transgenic zebrafish line, neurons of the ventral region of the superior raphe project to the hypothalamus, whereas neurons of the anterior region of the superior raphe project to the telencephalon.
Expression maps generated by in situ hybridization show that ht1aa is expressed broadly in the zebrafish brain, whereas ht1ab expression is limited to the diencephalic and mesencephalic areas of the caudal hypothalamus, the pre-optic area and the 1st rhombomere of the myelencephalon(Norton et al. 2008). The 5-HT1bd gene is diffusely expressed in 3dpf zebrafish larvae and shows a more defined expression pattern in the adult brain and is primarily found in the periventricular area of the telencephalon, the thalamus, the habenula, the preoptic area, the posterior periventricular areas, and the hypothalamus(Norton et al. 2008). In mid-embryonic stages, in situ hybridization shows expression of the 5-HT2C receptor gene primarily in the brain and the spinal cord (H. Schneider, unpublished observations). The 5-HT2C receptor is known for its RNA editing that affects the cell signaling of the receptor. Certain RNA edited forms of the 5-HT2C receptor mRNA have been linked to depression (Burns et al. 1997; Sanders-Bush et al. 2003). In the zebrafish brain, 5-HTT gene (slc6a4a) transcripts are localized in a pretectal diencephalic cell cluster and the raphe nuclei. In situ hybridization shows also expression in the anterior pharynx (Norton et al. 2008). A critical gene of the 5-HT system is the enzyme tryptophan hydroxylase (tph) which catalyzes the rate limiting step of the 5-HT synthesis (Chen et al. 2009c). The tph2 ortholog is the only zebrafish tph gene that is expressed in raphe nuclei where serotonergic neurons are found. The human TPH1 gene has two homologs in the zebrafish genome. The tph1a gene is expressed in 5-HT immunoreactive neurons in the epiphysis, the preoptic area, ventral to the posterior tuberculum and the posterior hypothalamus. The tph1b gene is only transiently expressed in an early embryonic stage (32hpf) in the preoptic area (Chen et al. 2009b) The zebrafish homolog of MAO is expressed in and near areas in the adult zebrafish brain in which serotonergic neurons are found (Sallinen et al. 2009). The expression pattern for the remaining serotonin-related genes has not yet been characterized in the zebrafish.
GABA
γ-aminobutyric acid (GABA) is the primary inhibitory neurotransmitter in the mammalian central nervous system and plays a role in neuronal excitability and muscle tone. GABA neurons are in the mesolimbic dopamine system and also play a role in the regulation of the reinforcement of drugs of abuse(Li et al. 2009). GABA acts through two types of receptors. GABAA receptors are ligand-gated ion channel receptors that consist primarily of alpha, beta, gamma and delta subunits, but epsilon, theta, pi, and rho subunits exist as well (Luscher et al. 2011). GABAB receptors are 7 transmembrane G protein-coupled receptors consisting of two subunits GABAB1 and GABAB2. GABA receptors have binding sites for GABA as well as drugs such as benzodiazepines and barbiturates. The extrasynaptic delta subunit of the GABAA receptor is sensitive to ethanol and is enhanced at concentrations consistent with human consumption (Meera et al. 2010) and there have been two putative binding sites described for the ethanol antagonist Ro 15-4513 at the gamma2 and delta subunits (Hanchar et al. 2006; Linden et al. 2011). Rat studies have suggested that ethanol has a modulation effect on the GABAA receptor α1 and α5 subunits decreasing their expression in the cerebral cortex and cerebellum following ethanol exposure in rats (Charlton et al. 1997).
The GABAA receptors are ionotropic and selectively conduct Cl− through its pore when activated by its ligand GABA. This normally has an inhibitory effect by decreasing the likelihood of an action potential in the neuron. The GABAA receptor specificity is determined by the combination of 5 subunits creating the ion channel. The most common human GABAA receptor subunits include six α, three β and three γ that come together in a pentamer typically consisting of 2 α’s, 2 β’s and 1 γ (Luscher et al. 2011). Additionally, there is a GABAA receptor associated protein (GABARAP) that is important for clustering neurotransmitter receptors through interaction with the cytoskeleton and may play an important role in addiction studies as well (Luo et al. 2006; Mohrlüder et al. 2009). The GABAB receptors are metabotropic receptors that signal to open potassium channels via G proteins when GABA is bound and inhibits neurotransmitter release. The two subunits of the GABAB receptor, GABAB1 and GABAB2, form obligate heterodimers (Pinard et al. 2010; Vlachou and Markou 2010).
The GABA α1 subunit had decreased expression in the ventral tegmental area and hippocampus, but the α5 subunit had increased expression in the hippocampus after treatment with ethanol(Charlton et al. 1997). A human and a rat study both showed a decrease expression of the GABAA receptor subunit gene GABRB2 following chronic ethanol exposure in the frontal cortex and nucleus accumbens, respectively (Lewohl et al. 2000; Rodd et al. 2008). Although with mixed results, association studies in humans have indicated polymorphisms in the GABAA receptor genes may be associated with an increased risk of alcohol dependence including the genes GABRA2, GABRA5, GABRA6, GABRB1,GABRB2, GABRB3, GABRG2 and GABRG3, (Dick et al. 2005; Dick et al. 2004; Edenberg et al. 2004; Edenberg and Foroud 2006; Parsian and Zhang 1999; Radel et al. 2005; Reck et al. 2005; Sander et al. 1999; Sieh et al. 2005; Song et al. 2003; Zhang et al. 2005). Similarly in studies of nicotine addiction, polymorphisms in GABRA2, GABRA4, GABARAP, GABBR1 and GABBR2 genes may be associated with an increased risk of nicotine dependence (Agrawal et al. 2009; Li et al. 2009; Lou et al. 2007). Additionally GABAB receptor agonists, such as baclofen, have had promising results for the treatment of addiction to several classes of drugs of abuse (Tyacke et al. 2010). Notably, a zebrafish study has supporting evidence for GABAB receptor involvement in the nicotine response (Petzold et al. 2009). Associations with GABRG2 and heroin dependence have been detected in a Chinese male population as well (Loh et al. 2007). The GABAA receptor is also implicated in addiction to benzodiazepines as they have a well described binding site on the receptor that confers its anxiolytic effects (Tan et al. 2011).
Human and Zebrafish Homology
The full complement of GABAA receptor genes, GABAB receptor genes, and the GABAA receptor-associated protein encoding gene are represented in the zebrafish. Single homologs exist for GABAA receptors α1, α4, α5, β1, β2, β3, γ1, γ2, and γ3, as well as GABAB receptor 2, and the GABA-A receptor-associated protein genes. A pair of homologs in the zebrafish is present for the GABAA receptors α2 and α6, as well as GABAB receptor 1 genes (Figure 7). The average percent identical residues between encoded proteins for the GABA genes is 81% (range: 60-98%) covering an average of 93% (range: 70-100%) of the human protein sequence (Table 2). Comparison of the complete set of GABA genes reveals separate clustering of the GABAB receptor genes with the GABA-A receptor-associated protein, the GABAA γ-genes, and the GABAA β-genes. The GABAA α-genes split into two clusters, with α1, α2, and α5 clustering together and α4 and α6 clustering together. The are several instances where the strict mapping between homologs is not apparent in the phylogenetic tree. The paralogs gabra6a and gabra6b are discordantly located within the tree. gabra6b is tightly associated with the human homolog GABRA6, however, gabra6a is more distantly associated to these homologs. Within the GABAA-β clade, the associated of the β1, β2, and β3 homologs are non-congruent, with the zebrafish gabrb2 more closely associated to the human GABRB1 and GABRB3, then the corresponding zebrafish homologs (Figure 7).
Figure 7.
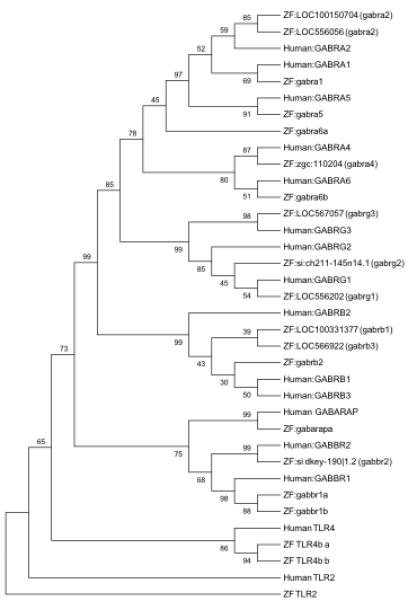
Phylogenetic tree illustrating the sequence relationship of human and zebrafish proteins encoded by genes associated with downstream GABA signaling. The fourteen human GABA associated genes each have at least one zebrafish homolog. Interestingly, there is some evidence that the zebrafish gene gabra6a, the second homolog of the human gene GABRA6, is much more distantly related to this gene, than the gabra6b homolog. It is also interesting to note that the GABRB1, GABRB2, and GABRB3 genes are all closely related and the exact association of these genes with their respective homologs can not be distinguished in this tree structure with statistical certainty (>50% Bootstrapping). The TLR2 and TLR4 gene out-groups, again illustrate the tight association of the identified genes associated with GABA signaling.
Zebrafish Neurotransmitter Localization
The zebrafish GABA-immunoreactive neurons have been described in the developing larvae in the telencephalon, diencephalons, midbrain, hypothalamus, forebrain, hindbrain and the spinal cord (Doldan et al. 1999) as well as the adult in the olfactory bulb, telencephalon, tectum stratum and hypothalamus (Kim et al. 2004) and has also been well described in the retina (Doldan et al. 1999; Sandell et al. 1994). GABAergic neurons have been described throughout the cerebellar corpus, valve, and vestibulolateral lobe and the expression patterns appear to be conserved between fish and mammals (Delgado and Schmachtenberg 2008). Evidence supports the inhibitory role of GABA in the zebrafish as well (Kim et al. 2004).
The zebrafish expression pattern of a subset of the GABA receptor genes has been reported. gabra1 (GABAA receptor subunit α1) in the adult zebrafish has been observed in the cerebellum, cranial ganglion, granular layer corpus cerebelli, granular layer valvula cerebelli, medial caudal lobe, molecular layer corpus cerebelli, molecular layer valvula cerebella. It is expressed throughout the cerebellum of the zebrafish in a pattern similar to the mammalian cerebellar cortex (Delgado and Schmachtenberg 2008). gabra6a (GABAA receptor subunit α6a) is expressed at 42-46 hpf in the ventral and proximal parts of the photoreceptor cell layer, and at 5 dpf it is observed in the photoreceptor cell layer of retina (Thisse 2004). gabra6b (GABAA receptor subunit α6b) is expressed at 7-13 dpf in the cerebellum and granule cell and from 7 dpf through adult in the granular eminence, granular layer corpus cerebelli, granule cell and medial caudal lobe (Volkmann et al. 2008). gabarapa (GABAA receptor associated protein a) is expressed at 24-72 hpf in the forebrain, hindbrain, rhombomere and telencephalon (Komoike et al. 2010). gabbr1a (GABAB receptor subunit 1a) is expressed in adult zebrafish in the cerebellum, cranial ganglion dendrite, cranial ganglion neuronal cell body, granular layer corpus cerebelli, granular layer valvula cerebelli, interneuron cell projection, medial valvula cerebelli, molecular layer corpus cerebelli, molecular layer valvula cerebella and vestibulolateralis lobe. It is expressed in a pattern similar to the mammalian cerebellar cortex (Delgado and Schmachtenberg 2008). The remaining GABA genes (gabra3, gabra5, gabra6a, gabrb2, gabrb3, gabrg3, and gabbr1b) lack reported expression pattern information in the zebrafish.
Adrenoreceptor system
The noradrenergic (NA) system has gained attention for the treatment of addiction as a target for reducing stress-related behavior during withdrawal. The action of the neurotransmitter noradenaline on addiction-related behavior has been the focus of investigations concerning stress-induced drug-seeking behavior and sensitization. For example, clonidine, an alpha2 adrenoreceptor agonist is used for the treatment of opioid withdrawal symptoms (Gonzalez et al. 2002) and alpha1B receptors are involved in the sensitization in the prefrontal cortex in mammals (Tassin 2008). The alpha1 receptors are best known for their regulation of smooth muscle contraction in the vasculature and contraction of smooth muscles in the gastrointestinal tract in mammals. All adrenoreceptors are seven transmembrane G-protein coupled receptors with principal transduction mediated by Gq/11 for alpha1, by Gi/o for alpha2, and by Gs for beta adrenoreceptors (Alexander et al. 2008). In contrast to the alpha2 and beta adrenoreceptors, alpha1 receptor proteins activate the PLC second messenger pathway and regulate intracellular calcium levels in smooth muscles.
Anatomical, physiological and behavioral data link adrenoreceptors to addiction. In mammals, noradrenergic neurons of the locus coeruleus (LC) innervate the VTA and the prefrontal cortex (Tassin 2008). Clonidine, a alpha2 adrenoreceptor agonist is used for the treatment of opioid withdrawal symptoms (Gonzalez et al. 2002) and alpha1B receptors are involved in the sensitization in the prefrontal cortex in mammals (Tassin 2008). The alpha1 receptors are best known for their regulation of smooth muscle contraction in the vasculature and contraction of smooth muscles in the gastrointestinal tract in mammals. In contrast to the alpha2 and beta adrenoreceptors, the alpha1 receptor proteins activate the phospholipase C (PLC) second messenger pathway and regulate intracellular calcium levels in smooth muscles.
Human and Zebrafish Homology
In zebrafish, homologs to all human adrenoreceptors, including the alpha1 receptor genes (ADRA1A, ADRA1B, ADRA1D), the alpha2 receptor genes (ADRA2A, ADRA2B, ADRA2C), and the beta receptor genes (ADRB1, ADRB2, ADRB3), have been identified (Figure 8 and Table 2). Duplication of single adrenoreceptor genes has occurred in the zebrafish so that 15 adrenoreceptor genes are known in zebrafish compared to nine in the human genome (Ruuskanen et al. 2004; Wang et al. 2009). However, the endophenotypes of zebrafish adrenoreceptors and the role of adrenoreceptors in the biology of addictive drugs in zebrafish is unknown. The zebrafish alpha1 receptors are the least characterized of the five different adrenoreceptor types (alpha1, alpha2, beta1, beta2, and beta3). The three alpha1 receptor types (alpha1A, alpha1B, and alpha1D) have been located in the zebrafish genome. For each human receptor gene there is one ortholog in the zebrafish genome, with the exception of the alpha1b for which two zebrafish genes have been predicted. Homologs of the three human alpha2 adrenoreceptor types (alpha2 A, B, and C) are also found in the zebrafish genome (Ruuskanen et al. 2005). In addition, the zebrafish genome contains two additional forms of the alpha2D receptor type that is not found in the human genome (Ruuskanen et al. 2004). The 2B gene is present in two forms the alpha2Ba and the alpha2Bb. Similar to the alpha2 adrenergic receptor system in zebrafish, five beta-adrenergic receptor genes have been identified in the zebrafish: beta1, beta2a, beta2b, beta3a, and beta3b (Wang et al. 2009).
Figure 8.
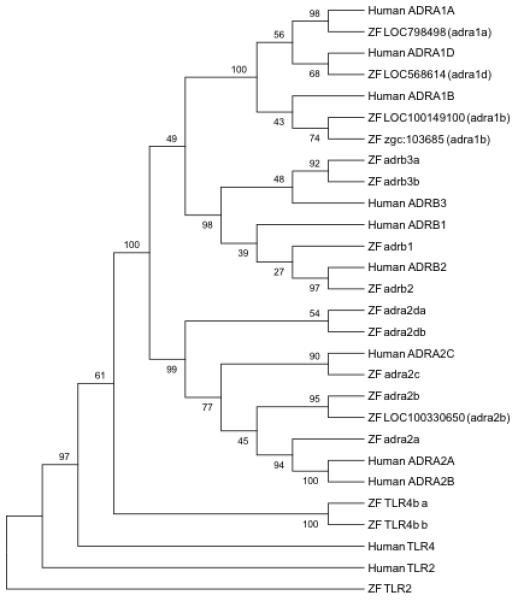
Phylogenetic tree illustrating the sequence relationship of human and zebrafish proteins encoded by genes associated with downstream adrenoreceptor signaling. The nine human genes associated with adrenoreceptor signaling all have at least one zebrafish homolog. In addition, there are a pair of zebrafish paralogs, adra2da and adra2db, that belong to this receptor family, which do not have human homologs. However, these genes do significantly associate with the gene group including ADRA2A, ADRA2B, and ADRA2C. The TLR2 and TLR4 gene out-groups, again illustrate the tight association of the identified genes associated with adrenoreceptor signaling.
Zebrafish Neurotransmitter Localization
The noradrenergic neurons in zebrafish are organized in four neuron clusters in the diencephalon (posterior tuberal nucleus, 12), and the brain stem (locus coeruleus, internal reticular formation and vagal lobe) (review: Panula et al. 2010). Expression of all five different alpha2 receptor genes has been observed in adult zebrafish brain regions (Ruuskanen et al. 2005) and in larvae as early as 50 hpf. Strong expression has been reported for the 2A and 2B forms. In situ hybridization shows strong signal of the alpha2A gene in the adult telencephalon and the optic tectum, but not in the diencephalon and the hindbrain (Ruuskanen et al. 2005). In contrast, the alpha2C receptor gene has been described as the only alpha2 receptor gene with expression in the diencephalon in the adult zebrafish brain (Ruuskanen et al. 2004). Alpha2Ba expression is localized in the hypothalamus in the adult zebrafish brain (Ruuskanen et al. 2005). Like the 2C receptor, the 2Da receptor gene is expressed in the optic tectum of the adult zebrafish brain. The expression of the 2Db receptor is unknown. Two beta adrenoreceptors, the beta1 and beta2a are expressed in the zebrafish brain. The other beta adrenoreceptors are mainly expressed in organs such as muscle, pancreas and liver (beta2b) and blood (beta3) (Wang et al. 2009). Most zebrafish alpha2 receptor expression is found in other organs such as kidney, muscle and ovary. No gene expression studies have been performed in zebrafish for alpha1 adrenoreceptors.
DISCUSSION AND CONCLUSION
Advancement of our understanding of the complex nature of addiction and the development of improved methods for the treatment of addiction may require the focused study of addiction-related endophenotypes using model organisms. In recent years, the use of zebrafish to study the behavioral response to drugs of addiction has increased. Many advantageous characteristics of zebrafish exist enabling efficient and innovative research. The progression of these studies beyond behavioral response and into the identification and characterization of the genetic factors influencing the onset and treatment of addiction holds promise. Here we have provided a detailed description of genes mechanistically impacting four drugs of addiction and four associated neurotransmitter pathways, highlighting the high level of homology between the species. While this review was restricted to eight target groups, the absence of data addressing other forms of drug addiction and other neurotransmitter pathways implicated in addiction response does not suggest a lack of suitability for using the zebrafish to study these areas. Homologues may and likely do exist to many other known and emerging genes associated with addiction. However, the information presented in this review may help investigators capitalize on opportunities to modify zebrafish drug response in the development new drug therapies to counteract drug reinforcement. Zebrafish drug response and the pharmacologic attenuation of drug response may allow us to explore human drug response endophenotypes which could lead to significant advances in the treatment of drug abuse and dependence.
One major benefit of using zebrafish as a model organism is the ease with which genetic features can be manipulated. The ability to obtain a mutation in a specific gene of interest in the zebrafish has been greatly facilitated by new molecular genetic tools. For example, re-sequencing zebrafish after chemical mutagenesis is being actively pursued by the Sanger Center with plans to generate likely null alleles in all zebrafish protein-encoding genes (Zebrafish Mutation Project). This is an open resource with 1627 genes mutated as of July 21, 2011. Alternatively, targeted gene knockouts have been generated through the use of custom restriction enzymes, including zinc finger nucleases (ZFNs) (Doyon et al. 2008; Foley et al. 2009; Meng et al. 2008). In addition, TAL Effector-based nucleases are an extremely promising alternative to ZFNs for germline-based gene knockouts in animals models such as zebrafish (Huang et al. 2011). Finally, insertional mutagenesis approaches are providing an array of additional, molecularly characterized alleles such as those using a retrovirus(Jao et al. 2008) or Tol2 transposons(Clark et al. 2011a; Kawakami et al. 2010).
The zebrafish model system provides a rich platform with which to explore how drugs of addiction impact neurotransmitter systems. Zebrafish drug response could be used to qualitatively and quantitatively explore drug response endophenotypes. Discovery in this area could lead to the development of pharmacotherapeutic approaches to the modulation of drug response which could be used in humans to decrease the death and disability caused by drug addiction and dependence.
METHODS
All sequence data was extracted from the National Center for Biology Information (NCBI). Homologs were identified using the information presented in HomoloGene (http://www.ncbi.nlm.nih.gov/homologene) and ZFIN: the Zebrafish Model Organism Database (http://zfin.org). The values reported in Tables 1 and 2, reflect BLAST searches run using the NCBI BLAST blastp algorithm with default parameters (Altschul et al. 1990). The phylogenic analysis was done using a freely available software suite called MEGA5 (v. 5.05)(Tamura et al. 2011). All alignments were created using the CLUSTALW algorithm (Larkin et al. 2007) with the Gonnet protein weight matrix. A gap opening penalty of 10, gap extension penalty of 0.1 was used for pairwise alignments and a gap opening penalty of 10 and gap extension penalty of 0.2 was used for multiple sequence alignments. The phylogenic trees were created using a maximum parsimony algorithm. The phylogenetic tree branches were tested and scored using a Bootstrapping method with 500 iterations. The final bootstrapped consensus phylogenic trees are reported with branch-point scores. The human TLR2 and TLR4, and zebrafish homologs, were included in all phylogenic analysis as an outgroup to provide a rooted analysis. The genes have well defined characterized homology (Roach et al. 2005; Sullivan et al. 2009) and provide one gene with a single homolog and a second gene with a pair of zebrafish homologs.
ACKNOWLEDGMENTS
Funding supported for VMK: NIAAA1P20AA017830-0; S.C. Johnson Genomics of Addictions Program
REFERENCES
- Ackerman KM, Nakkula R, Zirger JM, Beattie CE, Boyd RT. Cloning and spatiotemporal expression of zebrafish neuronal nicotinic acetylcholine receptor alpha 6 and alpha 4 subunit RNAs. Developmental dynamics : an official publication of the American Association of Anatomists. 2009;238:980–992. doi: 10.1002/dvdy.21912. doi: 10.1002/dvdy.21912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ACS ACS . Cancer Facts & Figures 2010. American Cancer Society; Atlanta: 2010. [Google Scholar]
- Agrawal A, Pergadia ML, Balasubramanian S, Saccone SF, Hinrichs AL, Saccone NL, Breslau N, Johnson EO, Hatsukami D, Martin NG, Montgomery GW, Goate AM, Rice JP, Bierut LJ, Madden PA. Further evidence for an association between the gamma-aminobutyric acid receptor A, subunit 4 genes on chromosome 4 and Fagerstrom Test for Nicotine Dependence. Addiction. 2009;104:471–7. doi: 10.1111/j.1360-0443.2008.02445.x. doi: 10.1111/j.1360-0443.2008.02445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderman SL, Bernier NJ. Localization of corticotropin-releasing factor, urotensin I, and CRF-binding protein gene expression in the brain of the zebrafish, Danio rerio. The Journal of comparative neurology. 2007;502:783–93. doi: 10.1002/cne.21332. doi: 10.1002/cne.21332. [DOI] [PubMed] [Google Scholar]
- Alexander SP, Mathie A, Peters JA. Guide to Receptors and Channels (GRAC) Br J Pharmacol. (3rd edition) 2008;153(Suppl 2):S1–209. doi: 10.1038/sj.bjp.0707746. doi: 0707746 [pii] 10.1038/sj.bjp.0707746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alvarez FA, Rodriguez-Martin I, Gonzalez-Nuñez V, de Velasco EMF, Gonzalez Sarmiento R, Rodríguez RE. New kappa opioid receptor from zebrafish Danio rerio. Neuroscience letters. 2006;405:94–99. doi: 10.1016/j.neulet.2006.06.028. doi: 10.1016/j.neulet.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Arias A, Feinn R, Kranzler HR. Association of an Asn40Asp (A118G) polymorphism in the muopioid receptor gene with substance dependence: a meta-analysis. Drug Alcohol Depend. 2006;83:262–8. doi: 10.1016/j.drugalcdep.2005.11.024. doi: S0376-8716(05)00380-7 [pii] 10.1016/j.drugalcdep.2005.11.024. [DOI] [PubMed] [Google Scholar]
- Baraldi PG, Preti D, Materazzi S, Geppetti P. Transient Receptor Potential Ankyrin 1 (TRPA1) Channel as Emerging Target for Novel Analgesics and Anti-Inflammatory Agents. J. Med. Chem. 2010;53:5085–5107. doi: 10.1021/jm100062h. doi: 10.1021/jm100062h. [DOI] [PubMed] [Google Scholar]
- Bart G, Heilig M, LaForge KS, Pollak L, Leal SM, Ott J, Kreek MJ. Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol Psychiatry. 2004;9:547–9. doi: 10.1038/sj.mp.4001504. doi: 10.1038/sj.mp.4001504 4001504 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellipanni G, Rink E, Bally-Cuif L. Cloning of two tryptophan hydroxylase genes expressed in the diencephalon of the developing zebrafish brain. Mechanisms of development. 2002;119(Suppl 1):S215–20. doi: 10.1016/s0925-4773(03)00119-9. [DOI] [PubMed] [Google Scholar]
- Blin M, Norton W, Bally-Cuif L, Vernier P. NR4A2 controls the differentiation of selective dopaminergic nuclei in the zebrafish brain. Molecular and cellular neurosciences. 2008;39:592–604. doi: 10.1016/j.mcn.2008.08.006. doi: 10.1016/j.mcn.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Boehmler W, Carr T, Thisse C, Thisse B, Canfield VA, Levenson R. D4 Dopamine receptor genes of zebrafish and effects of the antipsychotic clozapine on larval swimming behaviour. Genes, brain, and behavior. 2007;6:155–166. doi: 10.1111/j.1601-183X.2006.00243.x. doi: 10.1111/j.1601-183X.2006.00243.x. [DOI] [PubMed] [Google Scholar]
- Boehmler W, Obrecht-Pflumio S, Canfield V, Thisse C, Thisse B, Levenson R. Evolution and expression of D2 and D3 dopamine receptor genes in zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists. 2004;230:481–493. doi: 10.1002/dvdy.20075. doi: 10.1002/dvdy.20075. [DOI] [PubMed] [Google Scholar]
- Braida D, Limonta V, Pegorini S, Zani A, Guerini-Rocco C, Gori E, Sala M. Hallucinatory and rewarding effect of salvinorin A in zebrafish: kappa-opioid and CB1-cannabinoid receptor involvement. Psychopharmacology. 2007;190:441–448. doi: 10.1007/s00213-006-0639-1. doi: 10.1007/s00213-006-0639-1. [DOI] [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- Bretaud S, Li Q, Lockwood BL, Kobayashi K, Lin E, Guo S. A choice behavior for morphine reveals experience-dependent drug preference and underlying neural substrates in developing larval zebrafish. Neuroscience. 2007;146:1109–1116. doi: 10.1016/j.neuroscience.2006.12.073. doi: 10.1016/j.neuroscience.2006.12.073. [DOI] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Cabral GA, Raborn ES, Griffin L, Dennis J, Marciano-Cabral F. CB2 receptors in the brain: role in central immune function. British journal of pharmacology. 2008;153:240–251. doi: 10.1038/sj.bjp.0707584. doi:10.1038/sj.bjp.0707584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachat J, Canavello P, Elegante M, Bartels B, Hart P, Bergner C, Egan R, Duncan A, Tien D, Chung A, Wong K, Goodspeed J, Tan J, Grimes C, Elkhayat S, Suciu C, Rosenberg M, Chung KM, Kadri F, Roy S, Gaikwad S, Stewart A, Zapolsky I, Gilder T, Mohnot S, Beeson E, Amri H, Zukowska Z, Soignier RD, Kalueff AV. Modeling withdrawal syndrome in zebrafish. Behavioural Brain Research. 2010;208:371–376. doi: 10.1016/j.bbr.2009.12.004. doi: 10.1016/j.bbr.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Campa D, Gioia A, Tomei A, Poli P, Barale R. Association of ABCB1/MDR1 and OPRM1 gene polymorphisms with morphine pain relief. Clin Pharmacol Ther. 2008;83:559–66. doi: 10.1038/sj.clpt.6100385. doi: 6100385 [pii] 10.1038/sj.clpt.6100385. [DOI] [PubMed] [Google Scholar]
- Carbone D. Smoking and cancer. The American journal of medicine. 1992;93:13S–17S. doi: 10.1016/0002-9343(92)90621-h. [DOI] [PubMed] [Google Scholar]
- Caron SJC, Prober D, Choy M, Schier AF. In vivo birthdating by BAPTISM reveals that trigeminal sensory neuron diversity depends on early neurogenesis. Development (Cambridge, England) 2008;135:3259–3269. doi: 10.1242/dev.023200. doi: 10.1242/dev.023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudill-Slosberg MA, Schwartz LM, Woloshin S. Office visits and analgesic prescriptions for musculoskeletal pain in US: 1980 vs. 2000. Pain. 2004;109:514–9. doi: 10.1016/j.pain.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Center for Disease Control and Prevention Unintentional poisoning deaths--United States,1999-2004. MMWR Morb Mortal Wkly Rep. 2007;56:93–6. [PubMed] [Google Scholar]
- Chandrasekar G, Lauter G, Hauptmann G. Distribution of corticotropin-releasing hormone in the developing zebrafish brain. The Journal of comparative neurology. 2007;505:337–51. doi: 10.1002/cne.21496. doi: 10.1002/cne.21496. [DOI] [PubMed] [Google Scholar]
- Changeux J-P. Nicotine addiction and nicotinic receptors: lessons from genetically modified mice. Nature reviews Neuroscience. 2010;11:389–401. doi: 10.1038/nrn2849. doi: 10.1038/nrn2849. [DOI] [PubMed] [Google Scholar]
- Charlton ME, Sweetnam PM, Fitzgerald LW, Terwilliger RZ, Nestler EJ, Duman RS. Chronic ethanol administration regulates the expression of GABAA receptor alpha 1 and alpha 5 subunits in the ventral tegmental area and hippocampus. J Neurochem. 1997;68:121–7. doi: 10.1046/j.1471-4159.1997.68010121.x. [DOI] [PubMed] [Google Scholar]
- Chen Y-C, Peng G-S, Wang M-F, Tsao T-P, Yin S-J. Polymorphism of ethanol-metabolism genes and alcoholism: correlation of allelic variations with the pharmacokinetic and pharmacodynamic consequences. Chemico-biological interactions. 2009a;178:2–7. doi: 10.1016/j.cbi.2008.10.029. doi: 10.1016/j.cbi.2008.10.029. [DOI] [PubMed] [Google Scholar]
- Chen Y-C, Priyadarshini M, Panula P. Complementary developmental expression of the two tyrosine hydroxylase transcripts in zebrafish. Histochemistry and cell biology. 2009b;132:375–381. doi: 10.1007/s00418-009-0619-8. doi: 10.1007/s00418-009-0619-8. [DOI] [PubMed] [Google Scholar]
- Chen YC, Priyadarshini M, Panula P. Complementary developmental expression of the two tyrosine hydroxylase transcripts in zebrafish. Histochem Cell Biol. 2009c;132:375–81. doi: 10.1007/s00418-009-0619-8. doi: 10.1007/s00418-009-0619-8. [DOI] [PubMed] [Google Scholar]
- Cheng W, Guo L, Zhang Z, Soo HM, Wen C, Wu W, Peng J. HNF factors form a network to regulate liver-enriched genes in zebrafish. Developmental biology. 2006;294:482–496. doi: 10.1016/j.ydbio.2006.03.018. doi: 10.1016/j.ydbio.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Chou WY, Yang LC, Lu HF, Ko JY, Wang CH, Lin SH, Lee TH, Concejero A, Hsu CJ. Association of mu-opioid receptor gene polymorphism (A118G) with variations in morphine consumption for analgesia after total knee arthroplasty. Acta Anaesthesiol Scand. 2006;50:787–92. doi: 10.1111/j.1399-6576.2006.01058.x. doi: AAS1058 [pii] 10.1111/j.1399-6576.2006.01058.x. [DOI] [PubMed] [Google Scholar]
- Civelli O, Bunzow JR, Grandy DK. Molecular diversity of the dopamine receptors. Annual review of pharmacology and toxicology. 1993;33:281–307. doi: 10.1146/annurev.pa.33.040193.001433. doi: 10.1146/annurev.pa.33.040193.001433. [DOI] [PubMed] [Google Scholar]
- Clark KJ, Balciunas D, Pogoda H-M, Ding Y, Westcot SE, Bedell VM, Greenwood TM, Urban MD, Skuster KJ, Petzold AM, Ni J, Nielsen AL, Patowary A, Scaria V, Sivasubbu S, Xu X, Hammerschmidt M, Ekker SC. In vivo protein trapping produces a functional expression codex of the vertebrate proteome. Nature methods. 2011a;8:506–515. doi: 10.1038/nmeth.1606. doi: 10.1038/nmeth.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KJ, Boczek NJ, Ekker SC. Stressing zebrafish for behavioral genetics. Reviews in the neurosciences. 2011b;22:49–62. doi: 10.1515/RNS.2011.007. doi: 10.1515/RNS.2011.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett D, Wise RA. Intracranial self-stimulation in relation to the ascending dopaminergic systems of the midbrain: a moveable electrode mapping study. Brain research. 1980;185:1–15. doi: 10.1016/0006-8993(80)90666-6. [DOI] [PubMed] [Google Scholar]
- Cox JA, Kucenas S, Voigt MM. Molecular characterization and embryonic expression of the family of N-methyl-D-aspartate receptor subunit genes in the zebrafish. Dev Dyn. 2005;234:756–66. doi: 10.1002/dvdy.20532. doi: 10.1002/dvdy.20532. [DOI] [PubMed] [Google Scholar]
- Crawford DK, Trudell JR, Bertaccini EJ, Li K, Davies DL, Alkana RL. Evidence that ethanol acts on a target in Loop 2 of the extracellular domain of alpha1 glycine receptors. Journal of neurochemistry. 2007;102:2097–2109. doi: 10.1111/j.1471-4159.2007.04680.x. doi: 10.1111/j.1471-4159.2007.04680.x. [DOI] [PubMed] [Google Scholar]
- Cunningham D, Hawthorn J, Pople A, Gazet JC, Ford HT, Challoner T, Coombes RC. Prevention of emesis in patients receiving cytotoxic drugs by GR38032F, a selective 5-HT3 receptor antagonist. Lancet. 1987;1:1461–1463. doi: 10.1016/s0140-6736(87)92208-2. [DOI] [PubMed] [Google Scholar]
- Darland T, Dowling JE. Behavioral screening for cocaine sensitivity in mutagenized zebrafish. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11691–6. doi: 10.1073/pnas.191380698. doi: 10.1073/pnas.191380698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Petrocellis L, Orlando P, Moriello AS, Aviello G, Stott C, Izzo A, Di Marzo V. Cannabinoid actions at TRPV channels: effects on TRPV3 and TRPV4 and their potential relevance to gastrointestinal inflammation. Acta Physiol (Oxf) 2011 doi: 10.1111/j.1748-1716.2011.02338.x. doi: 10.1111/j.1748-1716.2011.02338.x. [DOI] [PubMed] [Google Scholar]
- De Witte P. Imbalance between neuroexcitatory and neuroinhibitory amino acids causes craving for ethanol. Addictive behaviors. 2004;29:1325–1339. doi: 10.1016/j.addbeh.2004.06.020. doi: 10.1016/j.addbeh.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Delgado L, Schmachtenberg O. Immunohistochemical localization of GABA, GAD65, and the receptor subunits GABAAalpha1 and GABAB1 in the zebrafish cerebellum. Cerebellum. 2008;7:444–50. doi: 10.1007/s12311-008-0047-7. doi: 10.1007/s12311-008-0047-7. [DOI] [PubMed] [Google Scholar]
- Dhawan BN, Cesselin F, Raghubir R, Reisine T, Bradley PB, Portoghese PS, Hamon M. International Union of Pharmacology. XII. Classification of opioid receptors. Pharmacol Rev. 1996;48:567–92. [PubMed] [Google Scholar]
- Dick DM, Edenberg HJ, Xuei X, Goate A, Hesselbrock V, Schuckit M, Crowe R, Foroud T. No association of the GABAA receptor genes on chromosome 5 with alcoholism in the collaborative study on the genetics of alcoholism sample. Am J Med Genet B Neuropsychiatr Genet. 2005;132B:24–8. doi: 10.1002/ajmg.b.30058. [DOI] [PubMed] [Google Scholar]
- Dick DM, Edenberg HJ, Xuei X, Goate A, Kuperman S, Schuckit M, Crowe R, Smith TL, Porjesz B, Begleiter H, Foroud T. Association of GABRG3 with alcohol dependence. Alcohol Clin Exp Res. 2004;28:4–9. doi: 10.1097/01.ALC.0000108645.54345.98. [DOI] [PubMed] [Google Scholar]
- Djoussé L, Lee I-M, Buring JE, Gaziano JM. Alcohol consumption and risk of cardiovascular disease and death in women: potential mediating mechanisms. Circulation. 2009;120:237–244. doi: 10.1161/CIRCULATIONAHA.108.832360. doi: 10.1161/CIRCULATIONAHA.108.832360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doldan MJ, Prego B, Holmqvist BI, de Miguel E. Distribution of GABA-immunolabeling in the early zebrafish (Danio rerio) brain. Eur J Morphol. 1999;37:126–9. doi: 10.1076/ejom.37.2.126.4748. [DOI] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nature biotechnology. 2008;26:702–708. doi: 10.1038/nbt1409. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakenberg K, Nikoshkov A, Horvath MC, Fagergren P, Gharibyan A, Saarelainen K, Rahman S, Nylander I, Bakalkin G, Rajs J, Keller E, Hurd YL. Mu opioid receptor A118G polymorphism in association with striatal opioid neuropeptide gene expression in heroin abusers. Proc Natl Acad Sci U S A. 2006;103:7883–8. doi: 10.1073/pnas.0600871103. doi: 0600871103 [pii] 10.1073/pnas.0600871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dresen S, Ferreirós N, Pütz M, Westphal F, Zimmermann R, Auwärter V. Monitoring of herbal mixtures potentially containing synthetic cannabinoids as psychoactive compounds. J Mass Spectrom. 2010;45:1186–1194. doi: 10.1002/jms.1811. doi: 10.1002/jms.1811. [DOI] [PubMed] [Google Scholar]
- Durrant R, Thakker J. Substance use & abuse : cultural and historical perspectives. Sage Publications; Thousand Oaks, Calif: 2003. [Google Scholar]
- Dworkin S, Heath JK, deJong-Curtain TA, Hogan BM, Lieschke GJ, Malaterre J, Ramsay RG, Mantamadiotis T. CREB activity modulates neural cell proliferation, midbrain-hindbrain organization and patterning in zebrafish. Developmental biology. 2007;307:127–41. doi: 10.1016/j.ydbio.2007.04.026. doi: 10.1016/j.ydbio.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr., O’Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–14. doi: 10.1086/383283. Epub 2004 Mar 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addict Biol. 2006;11:386–96. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- El-Alfy AT, Ivey K, Robinson K, Ahmed S, Radwan M, Slade D, Khan I, ElSohly M, Ross S. Antidepressant-like effect of delta9-tetrahydrocannabinol and other cannabinoids isolated from Cannabis sativa L. Pharmacol Biochem Behav. 2010;95:434–442. doi: 10.1016/j.pbb.2010.03.004. doi: 10.1016/j.pbb.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elphick MR, Egertová M. The neurobiology and evolution of cannabinoid signalling. Philos Trans R Soc Lond, B, Biol Sci. 2001;356:381–408. doi: 10.1098/rstb.2000.0787. doi: 10.1098/rstb.2000.0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch M-A, Gorodetsky E, Hodgkinson C, Roy A, Goldman D. Functional genetic variants that increase synaptic serotonin and 5-HT3 receptor sensitivity predict alcohol and drug dependence. Molecular psychiatry. 2010 doi: 10.1038/mp.2010.94. doi: 10.1038/mp.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch M-A, Schuckit MA, Johnson BA, Goldman D. Genetics of alcoholism using intermediate phenotypes. Alcoholism, clinical and experimental research. 2003;27:169–176. doi: 10.1097/01.ALC.0000052702.77807.8C. [DOI] [PubMed] [Google Scholar]
- Farris SP, Miles MF. Ethanol modulation of gene networks: Implications for alcoholism. Neurobiology of disease. 2011 doi: 10.1016/j.nbd.2011.04.013. doi: 10.1016/j.nbd.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS genetics. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi A, Dürr K, Ryu S, Willaredt M, Holzschuh J, Driever W. Expression and function of nr4a2, lmx1b, and pitx3 in zebrafish dopaminergic and noradrenergic neuronal development. BMC developmental biology. 2007;7:135. doi: 10.1186/1471-213X-7-135. doi: 10.1186/1471-213X-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingim RB, Kaplan L, Staud R, Ness TJ, Glover TL, Campbell CM, Mogil JS, Wallace MR. The A118G single nucleotide polymorphism of the mu-opioid receptor gene (OPRM1) is associated with pressure pain sensitivity in humans. J Pain. 2005;6:159–67. doi: 10.1016/j.jpain.2004.11.008. doi: S1526590004011174 [pii] 10.1016/j.jpain.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Foley JE, Yeh J-RJ, Maeder ML, Reyon D, Sander JD, Peterson RT, Joung JK. Rapid mutation of endogenous zebrafish genes using zinc finger nucleases made by Oligomerized Pool ENgineering (OPEN) PloS one. 2009;4:e4348. doi: 10.1371/journal.pone.0004348. doi: 10.1371/journal.pone.0004348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke P, Nothen MM, Wang T, Neidt H, Knapp M, Lichtermann D, Weiffenbach O, Mayer P, Hollt V, Propping P, Maier W. Human delta-opioid receptor gene and susceptibility to heroin and alcohol dependence. Am J Med Genet. 1999;88:462–4. doi: 10.1002/(sici)1096-8628(19991015)88:5<462::aid-ajmg4>3.0.co;2-s. doi: 10.1002/(SICI)1096-8628(19991015)88:5<462::AID-AJMG4>3.0.CO;2-S [pii] [DOI] [PubMed] [Google Scholar]
- Gerra G, Leonardi C, Cortese E, D’Amore A, Lucchini A, Strepparola G, Serio G, Farina G, Magnelli F, Zaimovic A, Mancini A, Turci M, Manfredini M, Donnini C. Human kappa opioid receptor gene (OPRK1) polymorphism is associated with opiate addiction. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:771–5. doi: 10.1002/ajmg.b.30510. doi: 10.1002/ajmg.b.30510. [DOI] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nature Reviews Genetics. 2005;6:521–532. doi: 10.1038/nrg1635. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Gonzalez G, Oliveto A, Kosten TR. Treatment of heroin (diamorphine) addiction: current approaches and future prospects. Drugs. 2002;62:1331–1343. doi: 10.2165/00003495-200262090-00004. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Nuñez V, Barrallo A, Traynor JR, Rodríguez RE. Characterization of opioid-binding sites in zebrafish brain. The Journal of pharmacology and experimental therapeutics. 2006;316:900–904. doi: 10.1124/jpet.105.093492. doi: 10.1124/jpet.105.093492. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. The American journal of psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Gotti C, Moretti M, Gaimarri A, Zanardi A, Clementi F, Zoli M. Heterogeneity and complexity of native brain nicotinic receptors. Biochemical pharmacology. 2007;74:1102–1111. doi: 10.1016/j.bcp.2007.05.023. doi: 10.1016/j.bcp.2007.05.023. [DOI] [PubMed] [Google Scholar]
- Gotti C, Zoli M, Clementi F. Brain nicotinic acetylcholine receptors: native subtypes and their relevance. Trends in pharmacological sciences. 2006;27:482–491. doi: 10.1016/j.tips.2006.07.004. doi: 10.1016/j.tips.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Gould TD, Gottesman II. Psychiatric endophenotypes and the development of valid animal models. Genes, brain, and behavior. 2006;5:113–119. doi: 10.1111/j.1601-183X.2005.00186.x. doi: 10.1111/j.1601-183X.2005.00186.x. [DOI] [PubMed] [Google Scholar]
- Guo S, Brush J, Teraoka H, Goddard A, Wilson SW, Mullins MC, Rosenthal A. Development of noradrenergic neurons in the zebrafish hindbrain requires BMP, FGF8, and the homeodomain protein soulless/Phox2a. Neuron. 1999;24:555–566. doi: 10.1016/s0896-6273(00)81112-5. [DOI] [PubMed] [Google Scholar]
- Hall AJ, Logan JE, Toblin RL, Kaplan JA, Kraner JC, Bixler D, Crosby AE, Paulozzi LJ. Patterns of abuse among unintentional pharmaceutical overdose fatalities. JAMA. 2008;300:2613–20. doi: 10.1001/jama.2008.802. [DOI] [PubMed] [Google Scholar]
- Hanchar HJ, Chutsrinopkun P, Meera P, Supavilai P, Sieghart W, Wallner M, Olsen RW. Ethanol potently and competitively inhibits binding of the alcohol antagonist Ro15-4513 to alpha4/6beta3delta GABAA receptors. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8546–51. doi: 10.1073/pnas.0509903103. doi: 10.1073/pnas.0509903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon J, Hoyer D. Molecular biology of 5-HT receptors. Behavioural Brain Research. 2008;195:198–213. doi: 10.1016/j.bbr.2008.03.020. doi: 10.1016/j.bbr.2008.03.020. [DOI] [PubMed] [Google Scholar]
- Harris RA, Trudell JR, Mihic SJ. Ethanol’s molecular targets. Science signaling. 2008;1:re7. doi: 10.1126/scisignal.128re7. doi: 10.1126/scisignal.128re7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood HJ, Fountain D, Livermore G. Economic costs of alcohol abuse and alcoholism. Recent developments in alcoholism : an official publication of the American Medical Society on Alcoholism, the Research Society on Alcoholism, and the National Council on Alcoholism. 1998;14:307–330. doi: 10.1007/0-306-47148-5_14. [DOI] [PubMed] [Google Scholar]
- Hayes DJ, Greenshaw AJ. 5-HT receptors and reward-related behaviour: a review. Neuroscience and biobehavioral reviews. 2011;35:1419–1449. doi: 10.1016/j.neubiorev.2011.03.005. doi: 10.1016/j.neubiorev.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Heinrich G, Pagtakhan CJ. Both 5′ and 3′ flanks regulate Zebrafish brain-derived neurotrophic factor gene expression. BMC neuroscience. 2004;5:19. doi: 10.1186/1471-2202-5-19. doi: 10.1186/1471-2202-5-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzschuh J, Ryu S, Aberger F, Driever W. Dopamine transporter expression distinguishes dopaminergic neurons from other catecholaminergic neurons in the developing zebrafish embryo. Mechanisms of development. 2001;101:237–243. doi: 10.1016/s0925-4773(01)00287-8. [DOI] [PubMed] [Google Scholar]
- Howard LA, Ahluwalia JS, Lin S-K, Sellers EM, Tyndale RF. CYP2E1*1D regulatory polymorphism: association with alcohol and nicotine dependence. Pharmacogenetics. 2003;13:321–328. doi: 10.1097/01.fpc.0000054090.48725.a2. doi: 10.1097/01.fpc.0000054090.48725.a2. [DOI] [PubMed] [Google Scholar]
- Hoyer D, Hannon JP, Martin GR. Molecular, pharmacological and functional diversity of 5-HT receptors. Pharmacology, biochemistry, and behavior. 2002;71:533–554. doi: 10.1016/s0091-3057(01)00746-8. [DOI] [PubMed] [Google Scholar]
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nature biotechnology. 2011;29:699–700. doi: 10.1038/nbt.1939. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- Hwang J, Kim H-S, Seok J-W, Kim J-D, Koun S, Park S-Y, Lee J, Kim HS, Kim H-S, Kim KS, Chang K-T, Ryoo ZY, Wang SM, Huh T-l, Lee S. Transcriptome analysis of the zebrafish mind bomb mutant. Molecular genetics and genomics : MGG. 2009;281:77–85. doi: 10.1007/s00438-008-0395-5. doi: 10.1007/s00438-008-0395-5. [DOI] [PubMed] [Google Scholar]
- Jacobs BL, Azmitia EC. Structure and function of the brain serotonin system. Physiological Reviews. 1992;72:165–229. doi: 10.1152/physrev.1992.72.1.165. [DOI] [PubMed] [Google Scholar]
- Jaffe JH, Kanzler M. Smoking as an addictive disorder. NIDA research monograph. 1979:4–23. [PubMed] [Google Scholar]
- Jao L-E, Maddison L, Chen W, Burgess SM. Using retroviruses as a mutagenesis tool to explore the zebrafish genome. Briefings in functional genomics & proteomics. 2008;7:427–443. doi: 10.1093/bfgp/eln038. doi: 10.1093/bfgp/eln038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarjour S, Bai L, Gianoulakis C. Effect of acute ethanol administration on the release of opioid peptides from the midbrain including the ventral tegmental area. Alcoholism, clinical and experimental research. 2009;33:1033–1043. doi: 10.1111/j.1530-0277.2009.00924.x. doi: 10.1111/j.1530-0277.2009.00924.x. [DOI] [PubMed] [Google Scholar]
- Jensen AA, Frølund B, Liljefors T, Krogsgaard-Larsen P. Neuronal nicotinic acetylcholine receptors: structural revelations, target identifications, and therapeutic inspirations. Journal of medicinal chemistry. 2005;48:4705–4745. doi: 10.1021/jm040219e. doi: 10.1021/jm040219e. [DOI] [PubMed] [Google Scholar]
- Jiménez-Ruiz C, Berlin I, Hering T. Varenicline: a novel pharmacotherapy for smoking cessation. Drugs. 2009;69:1319–1338. doi: 10.2165/00003495-200969100-00003. doi: 10.2165/00003495-200969100-00003. [DOI] [PubMed] [Google Scholar]
- Kano M, Ohno-Shosaku T, Hashimotodani Y, Uchigashima M, Watanabe M. Endocannabinoidmediated control of synaptic transmission. Physiol Rev. 2009;89:309–380. doi: 10.1152/physrev.00019.2008. doi: 10.1152/physrev.00019.2008. [DOI] [PubMed] [Google Scholar]
- Karlin A. Emerging structure of the nicotinic acetylcholine receptors. Nature reviews Neuroscience. 2002;3:102–114. doi: 10.1038/nrn731. doi: 10.1038/nrn731. [DOI] [PubMed] [Google Scholar]
- Kaslin J, Nystedt JM, Ostergård M, Peitsaro N, Panula P. The orexin/hypocretin system in zebrafish is connected to the aminergic and cholinergic systems. Journal of Neuroscience. 2004;24:2678–2689. doi: 10.1523/JNEUROSCI.4908-03.2004. doi: 10.1523/JNEUROSCI.4908-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaslin J, Panula P. Comparative anatomy of the histaminergic and other aminergic systems in zebrafish (Danio rerio) The Journal of comparative neurology. 2001;440:342–377. doi: 10.1002/cne.1390. [DOI] [PubMed] [Google Scholar]
- Kawakami K, Abe G, Asada T, Asakawa K, Fukuda R, Ito A, Lal P, Mouri N, Muto A, Suster ML, Takakubo H, Urasaki A, Wada H, Yoshida M. zTrap: zebrafish gene trap and enhancer trap database. BMC developmental biology. 2010;10:105. doi: 10.1186/1471-213X-10-105. doi: 10.1186/1471-213X-10-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kily LJM, Cowe YCM, Hussain O, Patel S, McElwaine S, Cotter FE, Brennan CH. Gene expression changes in a zebrafish model of drug dependency suggest conservation of neuro-adaptation pathways. The Journal of experimental biology. 2008;211:1623–1634. doi: 10.1242/jeb.014399. doi: 10.1242/jeb.014399. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Nam RH, Yoo YM, Lee CJ. Identification and functional evidence of GABAergic neurons in parts of the brain of adult zebrafish (Danio rerio) Neurosci Lett. 2004;355:29–32. doi: 10.1016/j.neulet.2003.10.024. [DOI] [PubMed] [Google Scholar]
- Klee EW, Ebbert JO, Schneider H, Hurt RD, Ekker SC. Zebrafish for the Study of the Biological Effects of Nicotine. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2011 doi: 10.1093/ntr/ntr010. doi: 10.1093/ntr/ntr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komoike Y, Shimojima K, Liang JS, Fujii H, Maegaki Y, Osawa M, Fujii S, Higashinakagawa T, Yamamoto T. A functional analysis of GABARAP on 17p13.1 by knockdown zebrafish. J Hum Genet. 2010;55:155–62. doi: 10.1038/jhg.2010.1. doi: 10.1038/jhg.2010.1. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Petrakis IL, Mason G, Trevisan L, D'Souza DC. N-methyl-D-aspartate glutamate receptors and alcoholism: reward, dependence, treatment, and vulnerability. Pharmacology and Therapeutics. 2003;99:79–94. doi: 10.1016/s0163-7258(03)00054-8. [DOI] [PubMed] [Google Scholar]
- Krystal JH, Staley J, Mason G, Petrakis IL, Kaufman J, Harris RA, Gelernter J, Lappalainen J. Gamma-aminobutyric acid type A receptors and alcoholism: intoxication, dependence, vulnerability, and treatment. Archives of general psychiatry. 2006;63:957–968. doi: 10.1001/archpsyc.63.9.957. doi: 10.1001/archpsyc.63.9.957. [DOI] [PubMed] [Google Scholar]
- Kurrasch DM, Nevin LM, Wong JS, Baier H, Ingraham HA. Neuroendocrine transcriptional programs adapt dynamically to the supply and demand for neuropeptides as revealed in NSF mutant zebrafish. Neural development. 2009;4:22. doi: 10.1186/1749-8104-4-22. doi: 10.1186/1749-8104-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam CS, Rastegar S, Strähle U. Distribution of cannabinoid receptor 1 in the CNS of zebrafish. Neuroscience. 2006;138:83–95. doi: 10.1016/j.neuroscience.2005.10.069. doi: 10.1016/j.neuroscience.2005.10.069. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lassen N, Estey T, Tanguay RL, Pappa A, Reimers MJ, Vasiliou V. Molecular cloning, baculovirus expression, and tissue distribution of the zebrafish aldehyde dehydrogenase 2. Drug metabolism and disposition: the biological fate of chemicals. 2005;33:649–656. doi: 10.1124/dmd.104.002964. doi: 10.1124/dmd.104.002964. [DOI] [PubMed] [Google Scholar]
- Lau B, Bretaud S, Huang Y, Lin E, Guo S. Dissociation of food and opiate preference by a genetic mutation in zebrafish. Genes, brain, and behavior. 2006;5:497–505. doi: 10.1111/j.1601-183X.2005.00185.x. doi: 10.1111/j.1601-183X.2005.00185.x. [DOI] [PubMed] [Google Scholar]
- Leboyer M, Bellivier F, Nosten-Bertrand M, Jouvent R, Pauls D, Mallet J. Psychiatric genetics: search for phenotypes. Trends in neurosciences. 1998;21:102–105. doi: 10.1016/s0166-2236(97)01187-9. [DOI] [PubMed] [Google Scholar]
- Leuchter AF, Cook IA, Hunter AM, Korb AS. A new paradigm for the prediction of antidepressant treatment response. Dialogues in clinical neuroscience. 2009;11:435–446. doi: 10.31887/DCNS.2009.11.4/afleuchter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewohl JM, Wang L, Miles MF, Zhang L, Dodd PR, Harris RA. Gene expression in human alcoholism: microarray analysis of frontal cortex. Alcohol Clin Exp Res. 2000;24:1873–82. [PubMed] [Google Scholar]
- Li MD. Identifying susceptibility loci for nicotine dependence: 2008 update based on recent genome-wide linkage analyses. Human genetics. 2008;123:119–131. doi: 10.1007/s00439-008-0473-0. doi: 10.1007/s00439-008-0473-0. [DOI] [PubMed] [Google Scholar]
- Li MD, Mangold JE, Seneviratne C, Chen GB, Ma JZ, Lou XY, Payne TJ. Association and interaction analyses of GABBR1 and GABBR2 with nicotine dependence in European- and African-American populations. PLoS One. 2009;4:e7055. doi: 10.1371/journal.pone.0007055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Shah S, Huang L, Carr AL, Gao Y, Thisse C, Thisse B, Li L. Cloning and spatial and temporal expression of the zebrafish dopamine D1 receptor. Developmental dynamics : an official publication of the American Association of Anatomists. 2007;236:1339–1346. doi: 10.1002/dvdy.21130. doi: 10.1002/dvdy.21130. [DOI] [PubMed] [Google Scholar]
- Lillesaar C, Stigloher C, Tannhäuser B, Wullimann MF, Bally-Cuif L. Axonal projections originating from raphe serotonergic neurons in the developing and adult zebrafish, Danio rerio, using transgenics to visualize raphe-specific pet1 expression. The Journal of comparative neurology. 2009;512:158–182. doi: 10.1002/cne.21887. doi: 10.1002/cne.21887. [DOI] [PubMed] [Google Scholar]
- Lillesaar C, Tannhäuser B, Stigloher C, Kremmer E, Bally-Cuif L. The serotonergic phenotype is acquired by converging genetic mechanisms within the zebrafish central nervous system. Developmental dynamics : an official publication of the American Association of Anatomists. 2007;236:1072–1084. doi: 10.1002/dvdy.21095. doi: 10.1002/dvdy.21095. [DOI] [PubMed] [Google Scholar]
- Linden AM, Schmitt U, Leppa E, Wulff P, Wisden W, Luddens H, Korpi ER. Ro 15-4513 Antagonizes Alcohol-Induced Sedation in Mice Through alphabetagamma2-type GABA(A) Receptors. Frontiers in neuroscience. 2011;5:3. doi: 10.3389/fnins.2011.00003. doi: 10.3389/fnins.2011.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh EW, Tang NL, Lee DT, Liu SI, Stadlin A. Association analysis of GABA receptor subunit genes on 5q33 with heroin dependence in a Chinese male population. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:439–43. doi: 10.1002/ajmg.b.30429. [DOI] [PubMed] [Google Scholar]
- Lohr H, Ryu S, Driever W. Zebrafish diencephalic A11-related dopaminergic neurons share a conserved transcriptional network with neuroendocrine cell lineages. Development. 2009;136:1007–17. doi: 10.1242/dev.033878. doi: 10.1242/dev.033878. [DOI] [PubMed] [Google Scholar]
- Lotsch J, von Hentig N, Freynhagen R, Griessinger N, Zimmermann M, Doehring A, Rohrbacher M, Sittl R, Geisslinger G. Cross-sectional analysis of the influence of currently known pharmacogenetic modulators on opioid therapy in outpatient pain centers. Pharmacogenet Genomics. 2009;19:429–36. doi: 10.1097/fpc.0b013e32832b89da. [DOI] [PubMed] [Google Scholar]
- Lou XY, Ma JZ, Sun D, Payne TJ, Li MD. Fine mapping of a linkage region on chromosome 17p13 reveals that GABARAP and DLG4 are associated with vulnerability to nicotine dependence in European-Americans. Hum Mol Genet. 2007;16:142–53. doi: 10.1093/hmg/ddl450. doi: 10.1093/hmg/ddl450. [DOI] [PubMed] [Google Scholar]
- Luo X, Kranzler HR, Zuo L, Lappalainen J, Yang B-z, Gelernter J. ADH4 gene variation is associated with alcohol dependence and drug dependence in European Americans: results from HWD tests and case-control association studies. Neuropsychopharmacology. 2006;31:1085–1095. doi: 10.1038/sj.npp.1300925. doi: 10.1038/sj.npp.1300925. [DOI] [PubMed] [Google Scholar]
- Luscher B, Fuchs T, Kilpatrick CL. GABAA receptor trafficking-mediated plasticity of inhibitory synapses. Neuron. 2011;70:385–409. doi: 10.1016/j.neuron.2011.03.024. doi: 10.1016/j.neuron.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler J, Filippi A, Driever W. DeltaA/DeltaD regulate multiple and temporally distinct phases of notch signaling during dopaminergic neurogenesis in zebrafish. Journal of Neuroscience. 2010;30:16621–16635. doi: 10.1523/JNEUROSCI.4769-10.2010. doi: 10.1523/JNEUROSCI.4769-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado R, Berrendero F, Ozaita A, Robledo P. Neurochemical basis of cannabis addiction. Neuroscience. 2011 doi: 10.1016/j.neuroscience.2011.02.035. doi: 10.1016/j.neuroscience.2011.02.035. [DOI] [PubMed] [Google Scholar]
- Manchikanti L. National drug control policy and prescription drug abuse: facts and fallacies. Pain Physician. 2007;10:399–424. [PubMed] [Google Scholar]
- Manchikanti L, Singh A. Therapeutic opioids: a ten-year perspective on the complexities and complications of the escalating use, abuse, and nonmedical use of opioids. Pain Physician. 2008;11:S63–88. [PubMed] [Google Scholar]
- Mansour A, Meador-Woodruff JH, Bunzow JR, Civelli O, Akil H, Watson SJ. Localization of dopamine D2 receptor mRNA and D1 and D2 receptor binding in the rat brain and pituitary: an in situ hybridization-receptor autoradiographic analysis. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1990;10:2587–600. doi: 10.1523/JNEUROSCI.10-08-02587.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez C, Galván S, Garcia-Martin E, Ramos MI, Gutiérrez-Martín Y, Agúndez JAG. Variability in ethanol biodisposition in whites is modulated by polymorphisms in the ADH1B and ADH1C genes. Hepatology (Baltimore, Md.) 2010;51:491–500. doi: 10.1002/hep.23341. doi: 10.1002/hep.23341. [DOI] [PubMed] [Google Scholar]
- Mathur P, Guo S. Use of zebrafish as a model to understand mechanisms of addiction and complex neurobehavioral phenotypes. Neurobiology of disease. 2010;40:66–72. doi: 10.1016/j.nbd.2010.05.016. doi: 10.1016/j.nbd.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda LA, Lolait SJ, Brownstein MJ, Young AC, Bonner TI. Structure of a cannabinoid receptor and functional expression of the cloned cDNA. Nature. 1990;346:561–564. doi: 10.1038/346561a0. doi: 10.1038/346561a0. [DOI] [PubMed] [Google Scholar]
- Mayer P, Rochlitz H, Rauch E, Rommelspacher H, Hasse HE, Schmidt S, Hollt V. Association between a delta opioid receptor gene polymorphism and heroin dependence in man. Neuroreport. 1997;8:2547–50. doi: 10.1097/00001756-199707280-00025. [DOI] [PubMed] [Google Scholar]
- McGovern PE, Zhang J, Tang J, Zhang Z, Hall GR, Moreau RA, Nuñez A, Butrym ED, Richards MP, Wang C-S, Cheng G, Zhao Z, Wang C. Fermented beverages of pre- and proto-historic China. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:17593–17598. doi: 10.1073/pnas.0407921102. doi: 10.1073/pnas.0407921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh D, Hu SS, Rimmerman N, Juknat A, Vogel Z, Walker JM, Bradshaw HB. N-arachidonoyl glycine, an abundant endogenous lipid, potently drives directed cellular migration through GPR18, the putative abnormal cannabidiol receptor. BMC Neurosci. 2010;11:44. doi: 10.1186/1471-2202-11-44. doi: 10.1186/1471-2202-11-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean DL, Fetcho JR. Ontogeny and innervation patterns of dopaminergic, noradrenergic, and serotonergic neurons in larval zebrafish. The Journal of comparative neurology. 2004a;480:38–56. doi: 10.1002/cne.20280. doi: 10.1002/cne.20280. [DOI] [PubMed] [Google Scholar]
- McLean DL, Fetcho JR. Relationship of tyrosine hydroxylase and serotonin immunoreactivity to sensorimotor circuitry in larval zebrafish. The Journal of comparative neurology. 2004b;480:57–71. doi: 10.1002/cne.20281. doi: 10.1002/cne.20281. [DOI] [PubMed] [Google Scholar]
- Meera P, Olsen RW, Otis TS, Wallner M. Alcohol- and alcohol antagonist-sensitive human GABAA receptors: tracking delta subunit incorporation into functional receptors. Molecular pharmacology. 2010;78:918–24. doi: 10.1124/mol.109.062687. doi: 10.1124/mol.109.062687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nature biotechnology. 2008;26:695–701. doi: 10.1038/nbt1398. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrlüder J, Schwarten M, Willbold D. Structure and potential function of gammaaminobutyrate type A receptor-associated protein. The FEBS journal. 2009;276:4989–5005. doi: 10.1111/j.1742-4658.2009.07207.x. doi: 10.1111/j.1742-4658.2009.07207.x. [DOI] [PubMed] [Google Scholar]
- Monsma FJ, Mahan LC, McVittie LD, Gerfen CR, Sibley DR. Molecular cloning and expression of a D1 dopamine receptor linked to adenylyl cyclase activation. Proceedings of the National Academy of Sciences of the United States of America. 1990;87:6723–6727. doi: 10.1073/pnas.87.17.6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moonat S, Starkman BG, Sakharkar A, Pandey SC. Neuroscience of alcoholism: molecular and cellular mechanisms. Cellular and molecular life sciences : CMLS. 2010;67:73–88. doi: 10.1007/s00018-009-0135-y. doi: 10.1007/s00018-009-0135-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro S, Thomas KL, Abu-Shaar M. Molecular characterization of a peripheral receptor for cannabinoids. Nature. 1993;365:61–65. doi: 10.1038/365061a0. doi: 10.1038/365061a0. [DOI] [PubMed] [Google Scholar]
- Murakami M. Lipid Mediators in Life Science. Exp Anim Tokyo. 2011;60:7–20. doi: 10.1538/expanim.60.7. [DOI] [PubMed] [Google Scholar]
- Nguyen-Legros J, Versaux-Botteri C, Vernier P. Dopamine receptor localization in the mammalian retina. Molecular neurobiology. 1999;19:181–204. doi: 10.1007/BF02821713. doi: 10.1007/BF02821713. [DOI] [PubMed] [Google Scholar]
- Nielsen DA, Barral S, Proudnikov D, Kellogg S, Ho A, Ott J, Kreek MJ. TPH2 and TPH1: association of variants and interactions with heroin addiction. Behavior genetics. 2008;38:133–150. doi: 10.1007/s10519-007-9187-7. doi: 10.1007/s10519-007-9187-7. [DOI] [PubMed] [Google Scholar]
- Ninkovic J, Bally-Cuif L. The zebrafish as a model system for assessing the reinforcing properties of drugs of abuse. Methods. 2006;39:262–74. doi: 10.1016/j.ymeth.2005.12.007. doi: 10.1016/j.ymeth.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Nishimura FT, Fukunaga T, Kajiura H, Umeno K, Takakura H, Ono T, Nishijo H. Effects of aldehyde dehydrogenase-2 genotype on cardiovascular and endocrine responses to alcohol in young Japanese subjects. Autonomic neuroscience : basic & clinical. 2002;102:60–70. doi: 10.1016/s1566-0702(02)00206-0. [DOI] [PubMed] [Google Scholar]
- Norton WHJ, Folchert A, Bally-Cuif L. Comparative analysis of serotonin receptor (HTR1A/HTR1B families) and transporter (slc6a4a/b) gene expression in the zebrafish brain. The Journal of comparative neurology. 2008;511:521–542. doi: 10.1002/cne.21831. doi: 10.1002/cne.21831. [DOI] [PubMed] [Google Scholar]
- Nury H, Bocquet N, Le Poupon C, Raynal B, Haouz A, Corringer P-J, Delarue M. Crystal structure of the extracellular domain of a bacterial ligand-gated ion channel. Journal of molecular biology. 2010;395:1114–1127. doi: 10.1016/j.jmb.2009.11.024. doi: 10.1016/j.jmb.2009.11.024. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C. 1st edn Oxford University Press; Oxford, England: 2002. Zebrafish - A Practical Approach. [Google Scholar]
- Nutt DJ, King LA, Phillips LD, Drugs ISCo. Drug harms in the UK: a multicriteria decision analysis. Lancet. 2010;376:1558–1565. doi: 10.1016/S0140-6736(10)61462-6. doi: 10.1016/S0140-6736(10)61462-6. [DOI] [PubMed] [Google Scholar]
- O’Dowd BF. Structures of dopamine receptors. Journal of neurochemistry. 1993;60:804–816. doi: 10.1111/j.1471-4159.1993.tb03224.x. [DOI] [PubMed] [Google Scholar]
- Olsen Y, Daumit GL, Ford DE. Opioid prescriptions by U.S. primary care physicians from 1992 to 2001. J Pain. 2006;7:225–35. doi: 10.1016/j.jpain.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Onaivi ES. Neuropsychobiological Evidence for the Functional Presence and Expression of Cannabinoid CB2 Receptors in the Brain. Neuropsychobiology. 2006;54:231–246. doi: 10.1159/000100778. doi: 10.1159/000100778. [DOI] [PubMed] [Google Scholar]
- Overton HA, Babbs AJ, Doel SM, Fyfe MCT, Gardner LS, Griffin G, Jackson HC, Procter MJ, Rasamison CM, Tang-Christensen M, Widdowson PS, Williams GM, Reynet C. Deorphanization of a G protein-coupled receptor for oleoylethanolamide and its use in the discovery of small-molecule hypophagic agents. Cell Metab. 2006;3:167–175. doi: 10.1016/j.cmet.2006.02.004. doi: 10.1016/j.cmet.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Panula P, Chen Y-C, Priyadarshini M, Kudo H, Semenova S, Sundvik M, Sallinen V. The comparative neuroanatomy and neurochemistry of zebrafish CNS systems of relevance to human neuropsychiatric diseases. Neurobiology of disease. 2010;40:46–57. doi: 10.1016/j.nbd.2010.05.010. doi: 10.1016/j.nbd.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Parsian A, Zhang ZH. Human chromosomes 11p15 and 4p12 and alcohol dependence: possible association with the GABRB1 gene. Am J Med Genet. 1999;88:533–8. doi: 10.1002/(sici)1096-8628(19991015)88:5<533::aid-ajmg18>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Paulozzi LJ, Budnitz DS, Xi Y. Increasing deaths from opioid analgesics in the United States. Pharmacoepidemiol Drug Saf. 2006;15:618–27. doi: 10.1002/pds.1276. [DOI] [PubMed] [Google Scholar]
- Paulozzi LJ, Ryan GW. Opioid analgesics and rates of fatal drug poisoning in the United States. Am J Prev Med. 2006;31:506–11. doi: 10.1016/j.amepre.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Pertwee RG, Howlett AC, Abood ME, Alexander SPH, Di Marzo V, Elphick MR, Greasley PJ, Hansen HS, Kunos G, Mackie K, Mechoulam R, Ross RA. International Union of Basic and Clinical Pharmacology. LXXIX. Cannabinoid receptors and their ligands: beyond CB1 and CB2. Pharmacol. Rev. 2010;62:588–631. doi: 10.1124/pr.110.003004. doi: 10.1124/pr.110.003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold AM, Balciunas D, Sivasubbu S, Clark KJ, Bedell VM, Westcot SE, Myers SR, Moulder GL, Thomas MJ, Ekker SC. Nicotine response genetics in the zebrafish. Proceedings of the National Academy of Sciences. 2009;106:18662–18667. doi: 10.1073/pnas.0908247106. doi: 10.1073/pnas.0908247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaar H, von Holst A, Vogt Weisenhorn DM, Brodski C, Guimera J, Wurst W. mPet-1, a mouse ETS-domain transcription factor, is expressed in central serotonergic neurons. Development genes and evolution. 2002;212:43–46. doi: 10.1007/s00427-001-0208-x. doi: 10.1007/s00427-001-0208-x. [DOI] [PubMed] [Google Scholar]
- Piccinetti CC, Migliarini B, Olivotto I, Coletti G, Amici A, Carnevali O. Appetite regulation: the central role of melatonin in Danio rerio. Hormones and behavior. 2010;58:780–5. doi: 10.1016/j.yhbeh.2010.07.013. doi: 10.1016/j.yhbeh.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Pinal-Seoane N, Martin IR, Gonzalez-Nuñez V, de Velasco EMF, Alvarez FA, Sarmiento RG, Rodríguez RE. Characterization of a new duplicate delta-opioid receptor from zebrafish. Journal of molecular endocrinology. 2006;37:391–403. doi: 10.1677/jme.1.02136. doi: 10.1677/jme.1.02136. [DOI] [PubMed] [Google Scholar]
- Pinard A, Seddik R, Bettler B. GABAB receptors: physiological functions and mechanisms of diversity. Advances in pharmacology (San Diego, Calif.) 2010;58:231–255. doi: 10.1016/S1054-3589(10)58010-4. doi: 10.1016/S1054-3589(10)58010-4. [DOI] [PubMed] [Google Scholar]
- Prober DA, Zimmerman S, Myers BR, McDermott BM, Kim S-H, Caron S, Rihel J, Solnica-Krezel L, Julius D, Hudspeth AJ, Schier AF. Zebrafish TRPA1 channels are required for chemosensation but not for thermosensation or mechanosensory hair cell function. Journal of Neuroscience. 2008;28:10102–10110. doi: 10.1523/JNEUROSCI.2740-08.2008. doi: 10.1523/JNEUROSCI.2740-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radel M, Vallejo RL, Iwata N, Aragon R, Long JC, Virkkunen M, Goldman D. Haplotype-based localization of an alcohol dependence gene to the 5q34 {gamma}-aminobutyric acid type A gene cluster. Arch Gen Psychiatry. 2005;62:47–55. doi: 10.1001/archpsyc.62.1.47. [DOI] [PubMed] [Google Scholar]
- Ramchandani VA, Umhau J, Pavon FJ, Ruiz-Velasco V, Margas W, Sun H, Damadzic R, Eskay R, Schoor M, Thorsell A, Schwandt ML, Sommer WH, George DT, Parsons LH, Herscovitch P, Hommer D, Heilig M. A genetic determinant of the striatal dopamine response to alcohol in men. Molecular psychiatry. 2011;16:809–817. doi: 10.1038/mp.2010.56. doi: 10.1038/mp.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch G, Lyons D, Middendorf I, Friedlander B, Arana N, Reyes T, Talbot W. Submission and Curation of Gene Expression Data. 2003 http://zfin.org. [Google Scholar]
- Ray R, Jepson C, Patterson F, Strasser A, Rukstalis M, Perkins K, Lynch KG, O’Malley S, Berrettini WH, Lerman C. Association of OPRM1 A118G variant with the relative reinforcing value of nicotine. Psychopharmacology (Berl) 2006;188:355–63. doi: 10.1007/s00213-006-0504-2. doi: 10.1007/s00213-006-0504-2. [DOI] [PubMed] [Google Scholar]
- Reck BH, Mukhopadhyay N, Tsai HJ, Weeks DE. Analysis of alcohol dependence phenotype in the COGA families using covariates to detect linkage. BMC Genet. 2005;6:S143. doi: 10.1186/1471-2156-6-S1-S143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm Room, Monteiro Gmel, Graham Rehn. Comparative quantification of health risks: Global and regional burden of disease due to selected major risk factors. WHO, Geneva. 2004:959–1108. [Google Scholar]
- Reyes-Gibby CC, Shete S, Rakvag T, Bhat SV, Skorpen F, Bruera E, Kaasa S, Klepstad P. Exploring joint effects of genes and the clinical efficacy of morphine for cancer pain: OPRM1 and COMT gene. Pain. 2007;130:25–30. doi: 10.1016/j.pain.2006.10.023. doi: S0304-3959(06)00590-2 [pii] 10.1016/j.pain.2006.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink E, Guo S. The too few mutant selectively affects subgroups of monoaminergic neurons in the zebrafish forebrain. Neuroscience. 2004;127:147–154. doi: 10.1016/j.neuroscience.2004.05.004. doi: 10.1016/j.neuroscience.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Rink E, Wullimann MF. The teleostean (zebrafish) dopaminergic system ascending to the subpallium (striatum) is located in the basal diencephalon (posterior tuberculum) Brain research. 2001;889:316–330. doi: 10.1016/s0006-8993(00)03174-7. [DOI] [PubMed] [Google Scholar]
- Rink E, Wullimann MF. Connections of the ventral telencephalon and tyrosine hydroxylase distribution in the zebrafish brain (Danio rerio) lead to identification of an ascending dopaminergic system in a teleost. Brain research bulletin. 2002a;57:385–387. doi: 10.1016/s0361-9230(01)00696-7. [DOI] [PubMed] [Google Scholar]
- Rink E, Wullimann MF. Development of the catecholaminergic system in the early zebrafish brain: an immunohistochemical study. Brain research. Developmental brain research. 2002b;137:89–100. doi: 10.1016/s0165-3806(02)00354-1. [DOI] [PubMed] [Google Scholar]
- Rios Y, Melmed S, Lin S, Liu NA. Zebrafish usp39 mutation leads to rb1 mRNA splicing defect and pituitary lineage expansion. PLoS genetics. 2011;7:e1001271. doi: 10.1371/journal.pgen.1001271. doi: 10.1371/journal.pgen.1001271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Rowen L, Kaur A, Purcell MK, Smith KD, Hood LE, Aderem A. The evolution of vertebrate Toll-like receptors. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9577–82. doi: 10.1073/pnas.0502272102. doi: 10.1073/pnas.0502272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89:481–98. doi: 10.1016/j.pbb.2008.01.023. Epub 2008 Feb 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Martin I, Herrero-Turrion MJ, Marron Fdez de Velasco E, Gonzalez-Sarmiento R, Rodríguez RE. Characterization of two duplicate zebrafish Cb2-like cannabinoid receptors. Gene. 2007;389:36–44. doi: 10.1016/j.gene.2006.09.016. doi: 10.1016/j.gene.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Ross RA. Anandamide and vanilloid TRPV1 receptors. British journal of pharmacology. 2003;140:790–801. doi: 10.1038/sj.bjp.0705467. doi: 10.1038/sj.bjp.0705467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rucktooa P, Smit AB, Sixma TK. Insight in nAChR subtype selectivity from AChBP crystal structures. Biochemical pharmacology. 2009;78:777–787. doi: 10.1016/j.bcp.2009.06.098. doi: 10.1016/j.bcp.2009.06.098. [DOI] [PubMed] [Google Scholar]
- Ruuskanen JO, Peitsaro N, Kaslin JVM, Panula P, Scheinin M. Expression and function of alpha-adrenoceptors in zebrafish: drug effects, mRNA and receptor distributions. Journal of neurochemistry. 2005;94:1559–1569. doi: 10.1111/j.1471-4159.2005.03305.x. doi: 10.1111/j.1471-4159.2005.03305.x. [DOI] [PubMed] [Google Scholar]
- Ruuskanen JO, Xhaard H, Marjamäki A, Salaneck E, Salminen T, Yan Y-L, Postlethwait JH, Johnson MS, Larhammar D, Scheinin M. Identification of duplicated fourth alpha2-adrenergic receptor subtype by cloning and mapping of five receptor genes in zebrafish. Molecular biology and evolution. 2004;21:14–28. doi: 10.1093/molbev/msg224. doi: 10.1093/molbev/msg224. [DOI] [PubMed] [Google Scholar]
- Ryberg E, Larsson N, Sjögren S, Hjorth S, Hermansson N-O, Leonova J, Elebring T, Nilsson K, Drmota T, Greasley PJ. The orphan receptor GPR55 is a novel cannabinoid receptor. British journal of pharmacology. 2009;152:1092–1101. doi: 10.1038/sj.bjp.0707460. doi: 10.1038/sj.bjp.0707460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallinen V, Sundvik M, Reenilä I, Peitsaro N, Khrustalyov D, Anichtchik O, Toleikyte G, Kaslin J, Panula P. Hyperserotonergic phenotype after monoamine oxidase inhibition in larval zebrafish. Journal of neurochemistry. 2009;109:403–415. doi: 10.1111/j.1471-4159.2009.05986.x. doi: 10.1111/j.1471-4159.2009.05986.x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Simon FM, Rodríguez RE. Developmental expression and distribution of opioid receptors in zebrafish. Neuroscience. 2008;151:129–137. doi: 10.1016/j.neuroscience.2007.09.086. doi: 10.1016/j.neuroscience.2007.09.086. [DOI] [PubMed] [Google Scholar]
- Sanchez-Simon FM, Zhang XX, Loh HH, Law PY, Rodriguez RE. Morphine regulates dopaminergic neuron differentiation via miR-133b. Molecular pharmacology. 2010;78:935–42. doi: 10.1124/mol.110.066837. doi: 10.1124/mol.110.066837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell JH, Martin SC, Heinrich G. The development of GABA immunoreactivity in the retina of the zebrafish (Brachydanio rerio) J Comp Neurol. 1994;345:596–601. doi: 10.1002/cne.903450409. doi: 10.1002/cne.903450409. [DOI] [PubMed] [Google Scholar]
- Sander T, Ball D, Murray R, Patel J, Samochowiec J, Winterer G, Rommelspacher H, Schmidt LG, Loh EW. Association analysis of sequence variants of GABA(A) alpha6, beta2, and gamma2 gene cluster and alcohol dependence. Alcohol Clin Exp Res. 1999;23:427–31. [PubMed] [Google Scholar]
- Sanders-Bush E, Fentress H, Hazelwood L. Serotonin 5-ht2 receptors: molecular and genomic diversity. Molecular interventions. 2003;3:319–330. doi: 10.1124/mi.3.6.319. doi: 10.1124/mi.3.6.319. [DOI] [PubMed] [Google Scholar]
- Schultz W. Predictive reward signal of dopamine neurons. Journal of neurophysiology. 1998;80:1–27. doi: 10.1152/jn.1998.80.1.1. [DOI] [PubMed] [Google Scholar]
- Schultz W. Dopamine signals for reward value and risk: basic and recent data. Behavioral and brain functions : BBF. 2010;6:24. doi: 10.1186/1744-9081-6-24. doi: 10.1186/1744-9081-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MM, Krueger KC, Deneris ES. A differentially autoregulated Pet-1 enhancer region is a critical target of the transcriptional cascade that governs serotonin neuron development. Journal of Neuroscience. 2005;25:2628–2636. doi: 10.1523/JNEUROSCI.4979-04.2005. doi: 10.1523/JNEUROSCI.4979-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shippenberg TS, Chefer VI, Zapata A, Heidbreder CA. Modulation of the behavioral and neurochemical effects of psychostimulants by kappa-opioid receptor systems. Ann N Y Acad Sci. 2001;937:50–73. doi: 10.1111/j.1749-6632.2001.tb03558.x. [DOI] [PubMed] [Google Scholar]
- Sia AT, Lim Y, Lim EC, Goh RW, Law HY, Landau R, Teo YY, Tan EC. A118G single nucleotide polymorphism of human mu-opioid receptor gene influences pain perception and patient-controlled intravenous morphine consumption after intrathecal morphine for postcesarean analgesia. Anesthesiology. 2008;109:520–6. doi: 10.1097/ALN.0b013e318182af21. doi: 10.1097/ALN.0b013e318182af2100000542-200809000-00023 [pii] [DOI] [PubMed] [Google Scholar]
- Sieh W, Basu S, Fu AQ, Rothstein JH, Scheet PA, Stewart WC, Sung YJ, Thompson EA, Wijsman EM. Comparison of marker types and map assumptions using Markov chain Monte Carlo-based linkage analysis of COGA data. BMC Genet. 2005;6:S11. doi: 10.1186/1471-2156-6-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TH, Sim-Selley LJ, Selley DE. Cannabinoid CB1 receptor-interacting proteins: novel targets for central nervous system drug discovery? British journal of pharmacology. 2010;160:454–466. doi: 10.1111/j.1476-5381.2010.00777.x. doi: 10.1111/j.1476-5381.2010.00777.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Koller DL, Foroud T, Carr K, Zhao J, Rice J, Nurnberger JI, Jr., Begleiter H, Porjesz B, Smith TL, Schuckit MA, Edenberg HJ. Association of GABA(A) receptors and alcohol dependence and the effects of genetic imprinting. Am J Med Genet B Neuropsychiatr Genet. 2003;117B:39–45. doi: 10.1002/ajmg.b.10022. [DOI] [PubMed] [Google Scholar]
- Song W, Zou Z, Xu F, Gu X, Xu X, Zhao Q. Molecular cloning and expression of a second zebrafish aldehyde dehydrogenase 2 gene (aldh2b) DNA sequence : the journal of DNA sequencing and mapping. 2006;17:262–269. doi: 10.1080/10425170600885609. [DOI] [PubMed] [Google Scholar]
- Steinbusch HW. Distribution of serotonin-immunoreactivity in the central nervous system of the rat-cell bodies and terminals. Neuroscience. 1981;6:557–618. doi: 10.1016/0306-4522(81)90146-9. [DOI] [PubMed] [Google Scholar]
- Steinbusch HW, Nieuwenhuys R. Localization of serotonin-like immunoreactivity in the central nervous system and pituitary of the rat, with special references to the innervation of the hypothalamus. Advances in experimental medicine and biology. 1981;133:7–35. doi: 10.1007/978-1-4684-3860-4_1. [DOI] [PubMed] [Google Scholar]
- Stewart A, Wong K, Cachat J, Gaikwad S, Kyzar E, Wu N, Hart P, Piet V, Utterback E, Elegante M, Tien D, Kalueff AV. Zebrafish models to study drug abuse-related phenotypes. Reviews in the neurosciences. 2011;22:95–105. doi: 10.1515/RNS.2011.011. doi: 10.1515/RNS.2011.011. [DOI] [PubMed] [Google Scholar]
- Sullivan C, Charette J, Catchen J, Lage CR, Giasson G, Postlethwait JH, Millard PJ, Kim CH. The gene history of zebrafish tlr4a and tlr4b is predictive of their divergent functions. Journal of immunology. 2009;183:5896–908. doi: 10.4049/jimmunol.0803285. doi: 10.4049/jimmunol.0803285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Alexander SPH, Garle MJ, Gibson CL, Hewitt K, Murphy SP, Kendall DA, Bennett AJ. Cannabinoid activation of PPAR alpha; a novel neuroprotective mechanism. British journal of pharmacology. 2007;152:734–743. doi: 10.1038/sj.bjp.0707478. doi: 10.1038/sj.bjp.0707478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular biology and evolution. 2011;28:2731–9. doi: 10.1093/molbev/msr121. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan KR, Rudolph U, Luscher C. Hooked on benzodiazepines: GABAA receptor subtypes and addiction. Trends Neurosci. 2011;34:188–97. doi: 10.1016/j.tins.2011.01.004. Epub 2011 Feb 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin J-P. Uncoupling between noradrenergic and serotonergic neurons as a molecular basis of stable changes in behavior induced by repeated drugs of abuse. Biochemical pharmacology. 2008;75:85–97. doi: 10.1016/j.bcp.2007.06.038. doi: 10.1016/j.bcp.2007.06.038. [DOI] [PubMed] [Google Scholar]
- Tay TL, Ronneberger O, Ryu S, Nitschke R, Driever W. Comprehensive catecholaminergic projectome analysis reveals single-neuron integration of zebrafish ascending and descending dopaminergic systems. Nature communications. 2011;2:171. doi: 10.1038/ncomms1171. doi: 10.1038/ncomms1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse B, Pflumino S, Furthauer M, Loppin B, Heyer V, Degrave A, Woehl R, Lux A, Steffan T, Charbonnier XQ, Thisse C. Expression of the zebrafish genome during embryogenesis. 2001 http://zfin.org. [Google Scholar]
- Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. 2004 http://zfin.org. [Google Scholar]
- Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. 2004 [Google Scholar]
- Thisse C, Thisse B. High Throughput Expression Analysis of ZF-Models Consortium Clones. 2005 http://zfin.org. [Google Scholar]
- Thomas DR. 5-ht5A receptors as a therapeutic target. Pharmacology and Therapeutics. 2006;111:707–714. doi: 10.1016/j.pharmthera.2005.12.006. doi: 10.1016/j.pharmthera.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Törk I. Anatomy of the serotonergic system. Annals of the New York Academy of Sciences. 1990;600:9–34. doi: 10.1111/j.1749-6632.1990.tb16870.x. discussion 34-5. [DOI] [PubMed] [Google Scholar]
- Tsai G, Coyle JT. The role of glutamatergic neurotransmission in the pathophysiology of alcoholism. Annual review of medicine. 1998;49:173–184. doi: 10.1146/annurev.med.49.1.173. doi: 10.1146/annurev.med.49.1.173. [DOI] [PubMed] [Google Scholar]
- Tyacke RJ, Lingford-Hughes A, Reed LJ, Nutt DJ. GABAB receptors in addiction and its treatment. Adv Pharmacol. 2010;58:373–96. doi: 10.1016/S1054-3589(10)58014-1. [DOI] [PubMed] [Google Scholar]
- Ueda N, Tsuboi K, Uyama T. Enzymological studies on the biosynthesis of Nacylethanolamines. Biochim Biophys Acta. 2010;1801:1274–1285. doi: 10.1016/j.bbalip.2010.08.010. doi: 10.1016/j.bbalip.2010.08.010. [DOI] [PubMed] [Google Scholar]
- Ueda N, Tsuboi K, Uyama T, Ohnishi T. Biosynthesis and degradation of the endocannabinoid 2-arachidonoylglycerol. Biofactors. 2011;37:1–7. doi: 10.1002/biof.131. doi: 10.1002/biof.131. [DOI] [PubMed] [Google Scholar]
- Veenstra-VanderWeele J, Anderson GM, Cook EH. Pharmacogenetics and the serotonin system: initial studies and future directions. European journal of pharmacology. 2000;410:165–181. doi: 10.1016/s0014-2999(00)00814-1. [DOI] [PubMed] [Google Scholar]
- Vlachou S, Markou A. GABAB receptors in reward processes. Advances in pharmacology (San Diego, Calif.) 2010;58:315–371. doi: 10.1016/S1054-3589(10)58013-X. doi: 10.1016/S1054-3589(10)58013-X. [DOI] [PubMed] [Google Scholar]
- Volkmann K, Rieger S, Babaryka A, Koster RW. The zebrafish cerebellar rhombic lip is spatially patterned in producing granule cell populations of different functional compartments. Dev Biol. 2008;313:167–80. doi: 10.1016/j.ydbio.2007.10.024. doi: 10.1016/j.ydbio.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang G-J, Swanson JM, Telang F. Dopamine in drug abuse and addiction: results of imaging studies and treatment implications. Archives of neurology. 2007;64:1575–1579. doi: 10.1001/archneur.64.11.1575. doi: 10.1001/archneur.64.11.1575. [DOI] [PubMed] [Google Scholar]
- Wagner FA, Anthony JC. From first drug use to drug dependence; developmental periods of risk for dependence upon marijuana, cocaine, and alcohol. Neuropsychopharmacology. 2002;26:479–488. doi: 10.1016/S0893-133X(01)00367-0. doi: 10.1016/S0893-133X(01)00367-0. [DOI] [PubMed] [Google Scholar]
- Walstab J, Rappold G, Niesler B. 5-HT(3) receptors: role in disease and target of drugs. Pharmacology and Therapeutics. 2010;128:146–169. doi: 10.1016/j.pharmthera.2010.07.001. doi: 10.1016/j.pharmthera.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Walter C, Lotsch J. Meta-analysis of the relevance of the OPRM1 118A>G genetic variant for pain treatment. Pain. 2009;146:270–5. doi: 10.1016/j.pain.2009.07.013. doi: S0304-3959(09)00396-0 [pii] 10.1016/j.pain.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Wang Z, Nishimura Y, Shimada Y, Umemoto N, Hirano M, Zang L, Oka T, Sakamoto C, Kuroyanagi J, Tanaka T. Zebrafish beta-adrenergic receptor mRNA expression and control of pigmentation. Gene. 2009;446:18–27. doi: 10.1016/j.gene.2009.06.005. doi: 10.1016/j.gene.2009.06.005. [DOI] [PubMed] [Google Scholar]
- Watson S, Chambers D, Hobbs C, Doherty P, Graham A. The endocannabinoid receptor, CB1, is required for normal axonal growth and fasciculation. Molecular and cellular neurosciences. 2008;38:89–97. doi: 10.1016/j.mcn.2008.02.001. doi: 10.1016/j.mcn.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Welsh L, Tanguay RL, Svoboda KR. Uncoupling nicotine mediated motoneuron axonal pathfinding errors and muscle degeneration in zebrafish. Toxicology and applied pharmacology. 2009;237:29–40. doi: 10.1016/j.taap.2008.06.025. doi: 10.1016/j.taap.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO WHO . WHO Report on the Global Tobacco Epideminc. WHO Press; Geneva Switzerland: 2008. [Google Scholar]
- Wick MJ, Mihic SJ, Ueno S, Mascia MP, Trudell JR, Brozowski SJ, Ye Q, Harrison NL, Harris RA. Mutations of gamma-aminobutyric acid and glycine receptors change alcohol cutoff: evidence for an alcohol receptor? Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6504–9. doi: 10.1073/pnas.95.11.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfit M, Wolfe K. The gene duplication that greased society's wheels. Nature genetics. 2005;37:566–567. doi: 10.1038/ng0605-566. doi: 10.1038/ng0605-566. [DOI] [PubMed] [Google Scholar]
- Wysowski DK. Surveillance of prescription drug-related mortality using death certificate data. Drug Saf. 2007;30:533–40. doi: 10.2165/00002018-200730060-00007. [DOI] [PubMed] [Google Scholar]
- Xi ZX, Peng XQ, Li X, Song R, Zhang HY, Liu QR, Yang HJ, Bi GH, Li J, Gardner EL. Brain cannabinoid CB receptors modulate cocaine’s actions in mice. Nature neuroscience. 2011;14:1160–6. doi: 10.1038/nn.2874. doi: 10.1038/nn.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Liu XH, Nagarajan S, Gu XY, Goldman D. Relationship of the delta-opioid receptor gene to heroin abuse in a large Chinese case/control sample. Am J Med Genet. 2002;110:45–50. doi: 10.1002/ajmg.10374. doi: 10.1002/ajmg.10374. [DOI] [PubMed] [Google Scholar]
- Xuei X, Dick D, Flury-Wetherill L, Tian HJ, Agrawal A, Bierut L, Goate A, Bucholz K, Schuckit M, Nurnberger J, Jr., Tischfield J, Kuperman S, Porjesz B, Begleiter H, Foroud T, Edenberg HJ. Association of the kappa-opioid system with alcohol dependence. Mol Psychiatry. 2006;11:1016–24. doi: 10.1038/sj.mp.4001882. doi: 4001882 [pii] 10.1038/sj.mp.4001882. [DOI] [PubMed] [Google Scholar]
- Yoshimura M, Pearson S, Kadota Y, Gonzalez CE. Identification of ethanol responsive domains of adenylyl cyclase. Alcoholism, clinical and experimental research. 2006;30:1824–32. doi: 10.1111/j.1530-0277.2006.00219.x. doi: 10.1111/j.1530-0277.2006.00219.x. [DOI] [PubMed] [Google Scholar]
- Yoshimura M, Tabakoff B. Selective effects of ethanol on the generation of cAMP by particular members of the adenylyl cyclase family. Alcoholism, clinical and experimental research. 1995;19:1435–40. doi: 10.1111/j.1530-0277.1995.tb01004.x. [DOI] [PubMed] [Google Scholar]
- Yuferov V, Fussell D, LaForge KS, Nielsen DA, Gordon D, Ho A, Leal SM, Ott J, Kreek MJ. Redefinition of the human kappa opioid receptor gene (OPRK1) structure and association of haplotypes with opiate addiction. Pharmacogenetics. 2004;14:793–804. doi: 10.1097/00008571-200412000-00002. doi: 00008571-200412000-00002 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Mol Psychiatry. 2008;13:531–43. doi: 10.1038/sj.mp.4002035. doi: 4002035 [pii] 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhong X, Ye Y. Multivariate linkage analysis using the electrophysiological phenotypes in the COGA alcoholism data. BMC Genet. 2005;6:S118. doi: 10.1186/1471-2156-6-S1-S118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirger JM, Beattie CE, McKay DB, Boyd RT. Cloning and expression of zebrafish neuronal nicotinic acetylcholine receptors. Gene expression patterns : GEP. 2003;3:747–754. doi: 10.1016/s1567-133x(03)00126-1. [DOI] [PubMed] [Google Scholar]


