Abstract
Retroviruses, including HIV, can activate innate immune responses, but the host sensors for retroviruses are largely unknown. Here we show that HIV infection activates cyclic-GMP-AMP (cGAMP) synthase (cGAS) to produce cGAMP, which binds to and activates the adaptor protein STING to induce type-I interferons and other cytokines. Inhibitors of HIV reverse transcriptase, but not integrase, abrogated interferon-β induction by the virus, suggesting that the reverse transcribed HIV DNA triggers the innate immune response. Knockout or knockdown of cGAS in mouse or human cell lines blocked cytokine induction by HIV, murine leukemia virus (MLV) and Simian immunodeficiency virus (SIV). These results indicate that cGAS is an innate immune sensor of HIV and other retroviruses.
Although tremendous advances have been made in our understanding of innate immune recognition of many microbial pathogens (1–3), relatively little is known about innate immune responses against retroviral infections (4). Retroviruses were thought to trigger weak or no innate immune responses, which were typically measured through the production of inflammatory cytokines and type-I interferons. However, recent research has shown that retroviruses such as HIV can trigger innate immune responses, which are normally masked by viral or host factors (5–8). For example, TREX1 is a cytosolic exonuclease that degrades DNA derived from HIV or endogenous retroelements, thereby preventing the accumulation of cytosolic DNA which would otherwise trigger innate immunity (9, 10). Loss of function mutations of TREX1 in humans have been closely linked to Aicardi Goutieres Syndrome (AGS), a lupus-like disease characterized by elevated expression of inflammatory cytokines and interferon-stimulated genes (11).
We have recently identified the enzyme cyclic GMP-AMP (cGAMP) synthase (cGAS) as a cytosolic DNA sensor that triggers the production of type-I interferons and other cytokines (12, 13). DNA binds and activates cGAS, which catalyzes the synthesis of a unique cGAMP isomer from ATP and GTP. This cGAMP isomer, termed 2′3′-cGAMP, which contains both 2′-5′ and 3′-5′ phosphodiester linkages, functions as a second messenger that binds and activates the endoplasmic reticulum protein STING (14–17). STING then activates the protein kinases IKK and TBK1, which in turn activate the transcription factors NF-κB and IRF3 to induce interferons and other cytokines (18). Knockdown of cGAS inhibits IFNβ induction by DNA viruses such as herpes simplex virus-1 (HSV-1) and vaccinia virus (13). Because retroviruses generate complementary DNA from the viral RNA by reverse transcription, we hypothesized that cGAS might detect retroviral DNA and trigger innate immune responses.
We used a single-round HIV-1 virus in which its envelope protein was replaced with the glycoprotein of vesicular stomatitis virus (VSV-G), which allows it to infect a large variety of human and mouse cell types (9). This virus also expresses GFP, which can be used to monitor viral infection. Infection of the human monocytic cell line THP1 with HIV-GFP led to dimerization of IRF3 (fig S1A), a hallmark of its activation. Phosphorylation of STAT1 at Tyr-701 was also detected after HIV infection (fig. S1A), indicating that the interferon signaling pathway was activated in the virus infected cells (19). HIV infection led to the induction of IFNβ and the chemokine CXCL10 (fig. S1B), concomitant with the generation of the HIV Gag episomal DNA (fig. S1C). The levels of IFNβ production were proportional to the multiplicity of infection by HIV (fig. S1D). Treatment of HIV-GFP virus with DNase I did not impair its ability to induce IFNβ (fig. S1E), whereas treatment of herring testis DNA (HT-DNA) with DNase I inhibited IFNβ induction (fig. S1F), indicating that IFNβ induction by HIV-GFP was not due to any contaminating DNA. Differentiation of THP1 from monocytes to macrophages by treating the cells with phorbol-12-myristate-13-acetate (PMA) inhibited HIV-GFP infection or replication (fig. S1G) and strongly inhibited IFNβ induction (fig. S1H). Thus, unless otherwise indicated, THP1 cells used in our study were not treated with PMA prior to HIV infection.
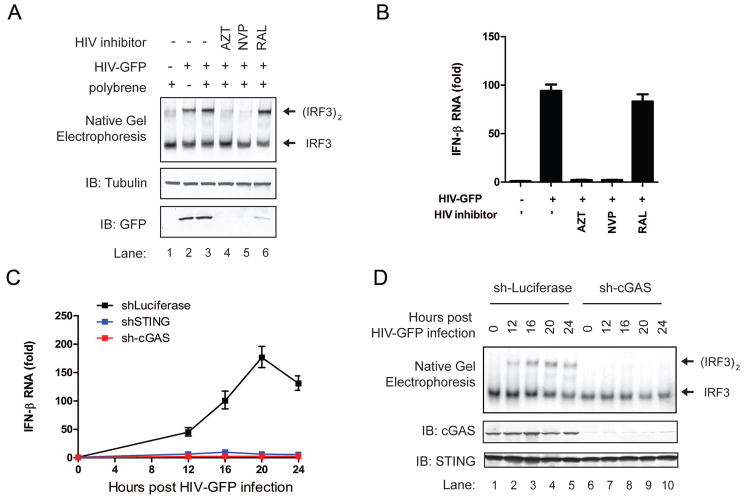
To test if reverse transcription is required for HIV to activate the innate immune response, we treated THP1 cells with the HIV reverse transcriptase inhibitors, azidothymidine (AZT) and nevirapine (NVP). Both inhibitors blocked IRF3 activation and IFNβ induction by HIV (Figure 1A and 1B). In contrast, the HIV integrase inhibitor raltegravir (RAL) did not affect the activation of this pathway. AZT and NVP, even at high concentrations, did not inhibit IFNβ induction by HT-DNA (fig. S2A-S2C), indicating that the inhibitory effects of AZT and NVP were due to their specific inhibition of HIV reverse transcription. These results suggest that the reverse transcribed HIV DNA is the trigger of IRF3 activation and IFNβ production.
Figure 1. The cGAS-STING pathway mediates innate immune responses against HIV.
(A and B) THP1 cells were treated with the HIV reverse transcriptase inhibitors (AZT and NVP, each at 5μM) or integrase inhibitor (RAL at 10μM) for 30 min before infection with HIV-GFP. 24 h after infection, cell extracts were analyzed by native gel electrophoresis or SDS-PAGE followed by immunoblotting with indicated antibodies (A) and total RNA was isolated for q-RT-PCR (B). (C and D) THP1 cells stably expressing an shRNA against human cGAS, STING or luciferase (control) were infected with HIV-GFP for the indicated time followed by measurement of IFNβ RNA by q-RT-PCR (C) and immunoblotting with the indicated antibodies (D). Error bars indicate standard deviations of triplicate measurements. Unless otherwise indicated, data shown in this and all other figures are representative of at least two independent experiments.
Strikingly, shRNA-mediated knockdown of cGAS or STING in THP1 cells strongly inhibited the induction of IFNβ and CXCL10 and the activation of IRF3 by HIV-GFP (Figure 1C and 1D; fig. S2D and S2E). Control experiments showed that shRNA against luciferase did not inhibit the activation of the pathway, and that the shRNA vectors knocked down the intended targets specifically. In particular, the cGAS shRNA knocked down cGAS but not STING (Figure 1D), and the induction of IFNβ in these cells was rescued by delivering cGAMP into the cells (fig. S2F), indicating that the cGAS shRNA did not have off-target effects in the STING pathway.
Previous studies have shown that VSV-G pseudotyped HIV-1 strongly induces IFNβ in TREX1-deficient mouse embryonic fibroblasts (MEF) but not in the wild-type (WT) MEF (9). We generated Trex1−/− MEF cell lines stably expressing shRNA against cGAS, STING or luciferase (as a control; fig. S3A and S3B). HIV infection induced IFNβ and CXCL10 RNA in the control cells (sh-luciferase) but not in cGAS or STING depleted cells (fig. S3C and S3D). In contrast, knockdown of cGAS or STING did not affect the induction of IFNβ or CXCL10 by the double-stranded RNA analogue poly[I:C].
To obtain definitive evidence for the role of cGAS in the innate sensing of cytosolic DNA and retroviruses, we employed the TALEN technology to disrupt the gene that encodes cGAS (Mb21d1), specifically the region that encodes the catalytic domain, in L929 cells (see Methods and fig. S4A) (20). Although L929 cells contain three copies of chromosome 9 that harbors the cGAS gene, DNA sequencing of the TALEN expressing cells identified multiple clones that had deletions in all three chromosomes; three of these clones were chosen for further studies (fig. S4B). All three clones contained deletions in the cGAS locus that generated frame-shift mutations (21).
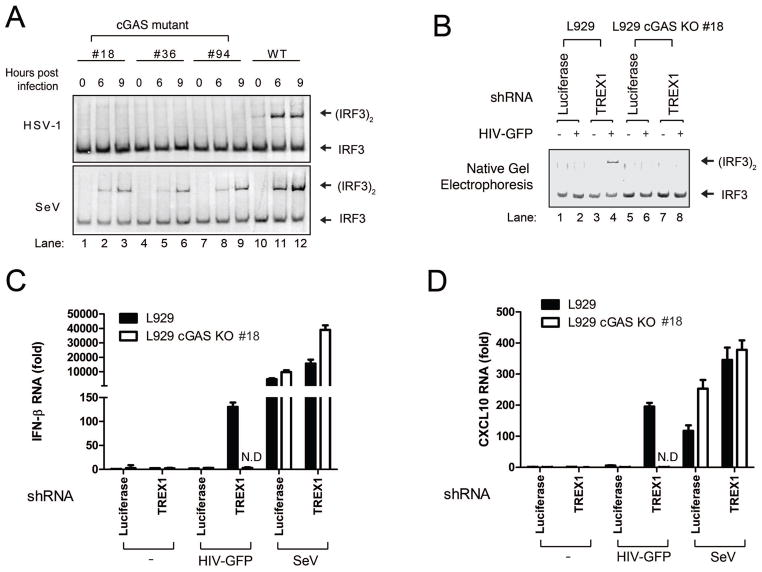
All three cGAS mutant cell lines failed to activate IRF3 in response to HT-DNA transfection or herpes simplex virus (HSV-1; a double-stranded DNA virus) infection (Figures 2A and fig. S4C). As controls, these cells activated IRF3 normally in response to transfection with poly[I:C] or infection with Sendai virus, an RNA virus. The cGAS mutant cells were also defective in inducing CXCL10 in response to HT-DNA, but this defect was rescued by transfecting the cells with the mouse cGAS expression plasmid (fig. S4D).
Figure 2. cGAS is essential for innate immune responses triggered by HIV.
(A) L929 cell lines harboring various deletions in exon 2 of the cGAS locus were generated by TALEN (see fig. S4). These cells and the parental L929 cells were infected with herpes simplex virus (HSV-1) or Sendai virus (SeV), followed by measurement of IRF3 dimerization. (B) L929 cGAS KO clone#18 and parental L929 cells were stably transfected with shRNA vector targeting TREX1 or luciferase (as a control). These cells were infected with HIV-GFP for 20 h, followed by IRF3 dimerization assay. (C and D) Similar to (B) except that RNA levels of IFNβ (C) and CXCL10 (D) were measured by q-RT-PCR. As a control, some cells were also infected with Sendai virus for 12 h. The error bars indicate standard deviations of triplicate measurements. N.D: non-detectable.
We chose cGAS mutant clone #18 and the parental L929 cells to investigate the role of cGAS in innate immune recognition of HIV infection. In L929 cells stably expressing an shRNA against TREX1, but not the control luciferase, HIV-GFP infection induced IRF3 dimerization and the production of IFNβ and CXCL10 (Figures 2B–2D; fig. S4E and S4F). In contrast, the L929 cGAS mutant cells failed to mount any detectable immune response to HIV infection even when TREX1 was depleted, demonstrating the essential role of cGAS in immune responses against HIV. The depletion of cGAS did not affect IFNβ or CXCL10 induction by Sendai virus (Figure 2C and 2D).
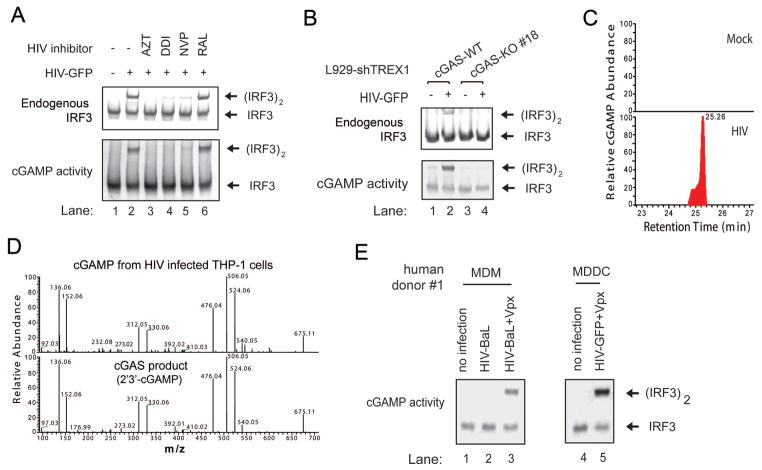
We have previously shown that HEK293T cells do not express detectable levels of cGAS and STING and thus fail to activate IRF3 in response to DNA transfection or DNA virus infection (13). Consistent with an important role of cGAS and STING in retrovirus detection, HIV-GFP infection activated IRF3 and STAT1 in THP1 but not HEK293T cells (fig. S5A). In contrast, Sendai virus activated IRF3 and STAT1 in both cell lines. To determine if HIV infection leads to the production of endogenous cGAMP in human cells, we prepared lysates from HIV-infected THP1 and HEK293T cells, heated the lysates at 95°C to denature most proteins, which were removed by centrifugation (12). The supernatant that potentially contained cGAMP was delivered to THP1 cells that had been permeabilized with the bacterial toxin perfringolysin-O (PFO), and then IRF3 dimerization was assayed by native gel electrophoresis (fig. S5B). The heat-resistant supernatant from HIV-infected THP1, but not HEK293T cells, contained the cGAMP activity that stimulated IRF3 activation in the recipient cells. Furthermore, inhibition of HIV reverse transcription by AZT, DDI (didanosine) or NVP blocked the generation of the cGAMP activity, whereas the HIV integrase inhibitor RAL had no effect (Figure 3A). HIV-GFP infection in L929-shTrex1 cells also led to generation of the cGAMP activity, which was dependent on cGAS (Figure 3B). Taken together, these results indicate that HIV infection induces the production of endogenous cGAMP in a manner that depends on cGAS and reverse transcription of HIV RNA to cDNA.
Figure 3. HIV infection induces the production of cyclic GMP-AMP in human cells.
(A) THP1 cells were treated with inhibitors of HIV reverse transcriptase (AZT, DDI and NVP) or integrase (RAL) before infection with HIV-GFP for 24h. Cell extracts were prepared for native gel electrophoresis to detect endogenous IRF3 dimer (top panel). Aliquots of the cell extracts were treated at 95°C for 5 min to denature proteins which were subsequently removed by centrifugation. The cGAMP activity in the supernatant was measured after its delivery into PFO-permeabilized THP1 cells followed by IRF3 dimerization assay (bottom panel). (B) L929 cGAS KO clone #18 and the parental cells, both stably expressing an shRNA against TREX1, were infected with HIV-GFP for 22 h. Endogenous IRF3 activation and cGAMP production in the cells were measured as described in (A). (C) The heat-resistant supernatants from THP-1 cells without (mock) or with HIV-GFP infection were fractionated by HPLC using a C18 column, and the abundance of cGAMP was quantitated by mass spectrometry using SRM. (D) Comparison of the MS/MS spectra of cGAMP isolated from HIV-infected THP-1 cells and that synthesized in vitro by recombinant human cGAS protein. Higher-energy collision dissociation (HCD) was used to fragment the precursor ion ([M+H]+=675.107) and normalized collision energy was set at 25. (E) Monocyte-derived macrophages (MDM) or dendritic cells (MDDC) from a human donor were either untreated or treated with Vpx-VLP for 24 h before infection with HIV. MDMs were infected with HIV-BaL for 3 days, whereas MDDCs were infected with HIV-GFP for 1 day. The cGAMP activity in these cells were measured as described in (A). FACS analysis of HIV infection and measurement of cGAMP abundance by mass spectrometry are shown in fig. S6. The data are representative of three independent experiments involving three human donors.
To test if HIV infection produces retroviral cDNA in the cytoplasm to activate cGAS, we infected HEK293T cells with HIV-GFP and prepared cytosolic extracts that were then incubated with purified cGAS protein in the presence of ATP and GTP (fig. S5C). Cytosolic extracts from HIV-infected cells, but not from uninfected cells, were able to stimulate cGAS to produce the cGAMP activity that activated IRF3 in permeabilized THP1 cells. Treatment of HEK293T cells with AZT inhibited the generation of the cGAS stimulatory activity. Further analyses showed that the cytoplasm of HIV-infected cells contained the HIV Gag DNA (fig. S5D) and GFP protein (fig. S5E), both of which were inhibited by AZT.
Quantitative measurement of cGAMP abundance by mass spectrometry using selective reaction monitoring (SRM) provided the direct evidence that cGAMP was produced in HIV-infected, but not mock-treated, THP1 cells (Figure 3C). Tandem mass spectrometry of the endogenous cGAMP from HIV-infected THP1 cells revealed that it was identical to the cGAS product, 2′3′-cGAMP (Figure 3D) (15).
To test whether HIV infection in primary human immune cells leads to cGAMP production, we infected monocyte-derived macrophages (MDM) and monocyte-derived dendritic cells (MDDC) with the clinical HIV-1 isolate HIV-BaL and HIV-GFP, respectively. Previous research has shown that human macrophages and dendritic cells express SAMHD1, a nuclease that hydrolyzes dNTP, thereby inhibiting HIV reverse transcription. HIV-2 and simian immunodeficiency virus (SIV) contain the protein Vpx, which targets SAMHD1 for ubiquitin-mediated proteasomal degradation, thus removing this host restriction factor. To facilitate HIV infections in human MDMs and MDDCs, we delivered the SIV Vpx into these cells using a virus-like particle (VLP) before HIV infection (fig. S6A, S6B and S6D). In the presence of Vpx, infection of MDMs and MDDCs with HIV-BaL and HIV-GFP, respectively, led to the generation of cGAMP activity (Figure 3E). Quantitative mass spectrometry analysis further confirmed the production of 2′3′-cGAMP in HIV-infected MDDCs that expressed Vpx (fig. S6C). The cGAMP activity was consistently observed in MDDCs and MDMs of additional human donors, and this activity was higher in the cells infected with HIV than those treated with Vpx alone (fig. S6D, S6E and S6F). These results demonstrate that HIV infection in human macrophages and dendritic cells lead to the generation of cGAMP under conditions that are permissive to viral replication.
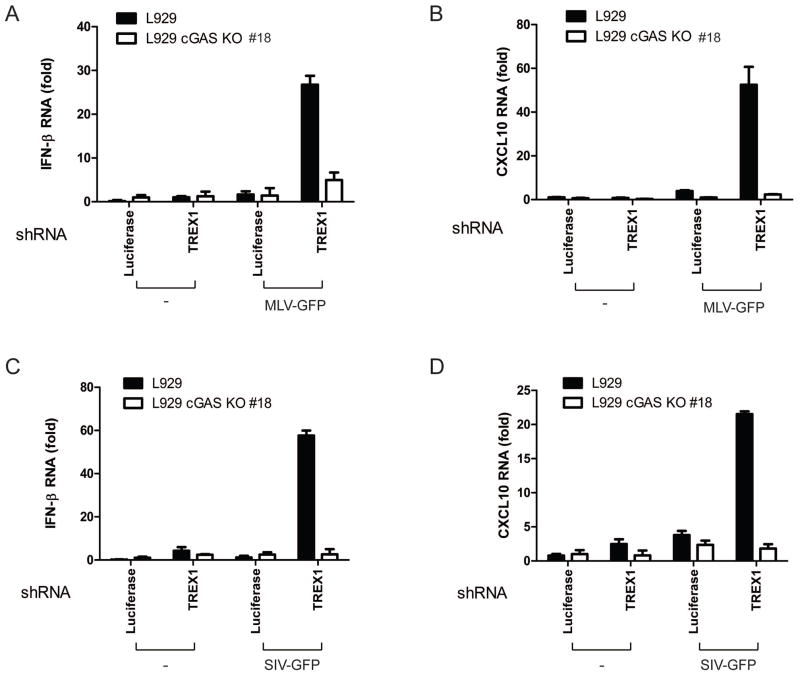
Finally, we tested whether cGAS is required for innate immune responses against other retroviruses by infecting L929 and L929-cGAS KO cell lines with murine leukemia virus (MLV) and SIV. Similar to HIV, MLV and SIV induced IFNβ and CXCL10 RNA in L929 cells depleted of endogenous TREX1, but such induction was completely abolished in the cGAS KO cells (Figure 4A–4D). In further support of an essential role of the cGAS-STING pathway in innate immune sensing of retroviruses, knockdown of cGAS or STING in Trex1−/− MEF cells strongly inhibited IFNβ induction by MLV and SIV (fig. S7A–S7D).
Figure 4. Murine leukemia virus and simian immunodeficiency virus activate innate immune responses through cGAS.
(A and B) L929 cGAS KO clone#18 and the parental L929 cells stably expressing shRNA against TREX1 or luciferase (control) were infected with MLV-GFP (MOI=2) for 20 h, followed by measurement of IFNβ (A) and CXCL10 (B) RNA by q-RT-PCR. (C and D) Similar to (A and B), except that cells were infected with SIV-GFP (MOI=1.5) for 20 h.
Here we demonstrate that cGAS is essential for innate immune responses against HIV, SIV and MLV, suggesting that cGAS is a general innate immune sensor of retroviral DNA. Although HIV primarily infects human CD4 T cells, it can also enter macrophages and dendritic cells, normally without triggering an overt innate immune response by concealing the viral nucleic acids within the capsid and by limiting the accumulation of viral DNA through co-opting host factors such as TREX1 and SAMHD1 (8). The absence of a rigorous innate immune response to HIV in dendritic cells is thought to be a major factor that hampers productive T cell responses and vaccine development (7). Our finding that HIV and other retroviruses can induce the production of cGAMP through cGAS under permissive conditions suggests that cGAMP might be used to bypass the block of innate immune responses against HIV. As such, cGAMP could be a candidate vaccine adjuvant for HIV and other pathogens that are adept at subverting the host innate immune system.
Supplementary Material
Acknowledgments
We thank Dr. X. Chen for assistance with mass spectrometry. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. Materials including the cGAS knockout cell lines may be requested upon signing of a material transfer agreement. This work was supported by NIH grants to Z.J.C. (R01-AI093967) and N.Y. (RO1-AI098569). Z. J. C is an investigator of Howard Hughes Medical Institute.
References and Notes
- 1.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010 Jan 15;327:291. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010 Mar 19;140:805. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Ronald PC, Beutler B. Plant and animal sensors of conserved microbial signatures. Science. 2010 Nov 19;330:1061. doi: 10.1126/science.1189468. [DOI] [PubMed] [Google Scholar]
- 4.Medzhitov R, Littman D. HIV immunology needs a new direction. Nature. 2008 Oct 2;455:591. doi: 10.1038/455591a. [DOI] [PubMed] [Google Scholar]
- 5.Manel N, Littman DR. Hiding in plain sight: how HIV evades innate immune responses. Cell. 2011 Oct 14;147:271. doi: 10.1016/j.cell.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manel N, et al. A cryptic sensor for HIV-1 activates antiviral innate immunity in dendritic cells. Nature. 2010 Sep 9;467:214. doi: 10.1038/nature09337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luban J. Innate immune sensing of HIV-1 by dendritic cells. Cell host & microbe. 2012 Oct 18;12:408. doi: 10.1016/j.chom.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan N, Regalado-Magdos AD, Stiggelbout B, Lee-Kirsch MA, Lieberman J. The cytosolic exonuclease TREX1 inhibits the innate immune response to human immunodeficiency virus type 1. Nature immunology. 2010 Nov;11:1005. doi: 10.1038/ni.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008 Aug 22;134:587. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crow YJ, et al. Mutations in the gene encoding the 3′-5′ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nature genetics. 2006 Aug;38:917. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 12.Wu J, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013 Feb 15;339:826. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013 Feb 15;339:786. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao P, et al. Cyclic [G(2′,5′)pA(3′,5′)p] Is the Metazoan Second Messenger Produced by DNA-Activated Cyclic GMP-AMP Synthase. Cell. 2013 May 23;153:1094. doi: 10.1016/j.cell.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, et al. Cyclic GMP-AMP Containing Mixed Phosphodiester Linkages Is An Endogenous High-Affinity Ligand for STING. Molecular cell. 2013 Jun 3; doi: 10.1016/j.molcel.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diner EJ, et al. The Innate Immune DNA Sensor cGAS Produces a Noncanonical Cyclic Dinucleotide that Activates Human STING. Cell Rep. 2013 May 30;3:1355. doi: 10.1016/j.celrep.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ablasser A, et al. cGAS produces a 2′-5′-linked cyclic dinucleotide second messenger that activates STING. Nature. 2013 Jun 20;498:380. doi: 10.1038/nature12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barber GN. Cytoplasmic DNA innate immune pathways. Immunological reviews. 2011 Sep;243:99. doi: 10.1111/j.1600-065X.2011.01051.x. [DOI] [PubMed] [Google Scholar]
- 19.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nature reviews Molecular cell biology. 2002 Sep;3:651. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 20.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011 Jul;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clone #18 has frame-shift mutations in all three chromosomes. In addition to frame-shifts, clone #36 harbored a 9-bp deletion in one chromosome that removed 3 amino acids (215MFK217) in the catalytic domain, whereas clone #94 had 12-bp deletion in one chromosome and 18-bp deletion in another that removed 4 (214VMFK217) and 6 (212FDVMFK217) amino acids in the catalytic domain, respectively. For details, see Materials and Methods in Supplementary Online Materials.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.