Abstract
In this paper, we describe a nonlinear threshold chemistry based on enzymatic inhibition and demonstrate how it can be coupled with microfluidics to convert a chemical concentration (analog input) into patterns of ON or OFF reaction outcomes (chemical digital readout). Quantification of small changes in concentration is needed in a number of assays, such as that for cystatin C, where a 1.5-fold increase in concentration may indicate the presence of acute kidney injury or the progression of chronic kidney disease. We developed an analog-to-digital chemical signal conversion that gives visual readout and applied it to an assay for cystatin C as a model target. The threshold chemistry is based on enzymatic inhibition and gives sharper responses with tighter inhibition. The chemistry described here uses acetylcholinesterase (AChE) and produces an unambiguous color change when the input is above a pre-determined threshold concentration. An input gives a pattern of ON/OFF responses when subjected to a monotonic sequence of threshold concentrations, revealing the input concentration at the point of transition from OFF to ON outcomes. We demonstrated that this threshold chemistry can detect a 1.30–fold increase in concentration at 22 °C, and that it is robust to experimental fluctuations: it provided the same output despite changes in temperature (22–34 °C) and readout time (10-fold range). We applied this threshold chemistry to diagnostics by coupling it with a traditional sandwich immunoassay for serum cystatin C. Because one quantitative measurement comprises several assays, each with its own threshold concentration, we used a microfluidic SlipChip device to process 12 assays in parallel, detecting a 1.5-fold increase (0.64 mg/L (49 nM) to 0.96 mg/L (74 nM)) of cystatin C in serum. We also demonstrated applicability to analysis of patient serum samples and the ability to image results using a cell phone camera. This work indicates that combining developments in nonlinear chemistries with microfluidics may lead to the development of user-friendly diagnostic assays with simple readouts.
INTRODUCTION
This paper describes the conversion of a chemical analog input (concentration) via chemical reactions into a digital output, which is represented by a pattern of ON/OFF binary digits (bits) (Figure 1). To achieve such responses, we implemented a threshold chemistry that produces a color change above a predetermined threshold concentration of input. Visual inspection of the pattern of bits with the naked eye or with a simple imaging device such as a cell phone indicates whether the input concentration is above or below the threshold concentration. In this paper, we report theory, experiments, and evaluation of this chemistry in the context of a realistic problem: quantifying small changes in the concentration of serum cystatin C (which inversely correlates with kidney performance).1 We are not presenting a clinical device that is ready for market, but we have chosen to assess the threshold chemistry within a realistic context, with a clinically relevant analyte and a clinically relevant change in concentration.
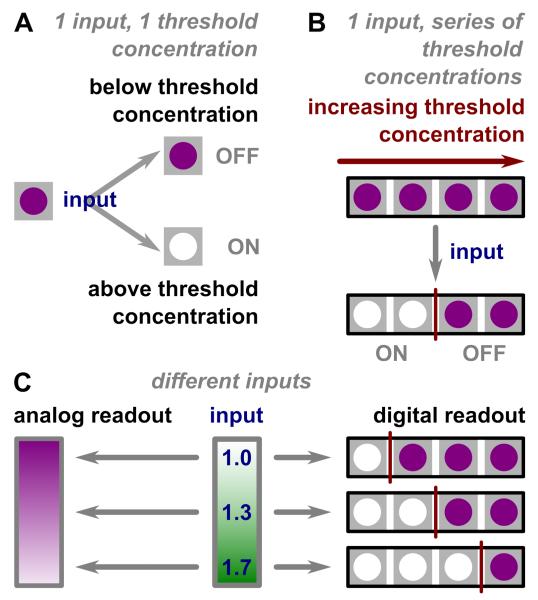
Figure 1.
Illustration of analog-to-digital conversion. A) A depiction of ON/OFF bits. If the input concentration is higher than the threshold concentration, the threshold chemistry gives an ON readout, shown as the color change from purple to white. If the input concentration is lower than the threshold concentration, the color remains purple, or OFF. B) An illustration of a series of ON/OFF bits. An input, when subjected to a series of increasing threshold concentrations, gives a series of ON/OFF responses, with the point of transition (red) between these ON and OFF states depending on the input concentration. C) An illustration of analog readout versus digital readout. An analog readout gives a gradient change in color intensity depending on input concentration (blue lettering). A digital readout comprises a series of bits; each bit is either clear (ON) or intensely colored (OFF), and the series of bits changes as the input concentration changes.
Simple readerless qualitative assays, such as pregnancy test strips, work well with various analytes and are widely utilized. Qualitative assays have two advantages: their simple readout eliminates the need for a machine (reader) to interpret the results, and they are usually robust to changes in temperature and assay time. Quantitative assays, however, typically rely on kinetic measurements for quantification, and therefore require careful control of temperature and assay time to accurately detect small changes in concentration. Simple quantitative assays2,3 can be readerless and can detect changes in concentration over a large range, but the change in readout is apparent only if those changes are one order of magnitude or more. With traditional analog readout, such as in an enzyme-linked immunoassay, the amount of output chemical is approximately proportional to input analyte concentration, and smaller changes in output lead to subtle changes in color that are hard to detect accurately with the naked eye. To quantify smaller changes, readers must be used to detect absorbance, fluorescence, and voltage.4-6
Quantification of small increases in concentration is needed for a number of assays. For example, a 1.5-fold increase in serum cystatin C concentration soon after a traumatic incident may indicate acute kidney injury (AKI),7 which carries increased morbidity and mortality. In chronic kidney disease (CKD), increases in cystatin C as small as 1.5-fold (from < 0.7 mg/L to > 0.9 mg/L) may indicate the progression from normal kidney function to stage 3 CKD, which entails complications such as anemia, bone-mineral disease, and life-threatening electrolyte abnormalities (Table S1 in Supporting Information1,8). We therefore chose an assay for cystatin C as a model system.
Here, we asked whether exploiting chemical nonlinearities can produce chemistry that gives robust, readerless quantification of small changes of concentrations despite changes in temperature or time. We tested the idea of “digitizing” the analog chemical signal such that the concentration is presented as a series of ON/OFF bits. At the single-molecule level, such analog-to-digital chemical conversion can be performed with digital PCR9,10 and digital immunoassays11 by using microfluidic devices to split a sample into small compartments containing individual molecules. This digital approach can provide quantitative information despite changes in temperature,12 but typically requires a reader. Instead of physically compartmentalizing the individual molecules of the sample, we wished to explore the potential to chemically digitize the analog signal (i.e., concentration) itself. We implemented the analog-to-digital conversion of the input concentration by using threshold chemistry, in which a reaction gives an OFF response if the input concentration of the signal is lower than the threshold concentration and gives an ON response—a prominent color change—otherwise (Figure 1A). In this framework, each reaction comprises one “bit.” Multiple bits with different threshold concentrations can be processed in parallel. Arranged in increasing order of threshold concentration, these bits combine to give a distinct point of transition from OFF bits to ON bits, enabling simple visual readout (Figure 1B). Different input concentrations cause the transition to occur at different locations along the series of bits (Figure 1C).
In principle, any threshold chemistry could be used to implement this approach. Thresholds are common in chemical reaction networks in Nature,13-16 and several synthetic threshold chemistries have been developed.17-19 In practice, we set four requirements for the threshold chemistry to be used for analog-to-digital conversion: i) it must be able to detect increases in concentration that are small enough to match the sensitivity requirements of the assay; ii) it must be tunable to different threshold concentrations; iii) it must provide visual readout detectable by the naked eye; and iv) it must be adaptable for different assays. Many established threshold chemistries were not sharp enough for this purpose: they are capable of detecting 2- to 10-fold increases in concentration,17-19 while a useful assay for cystatin C, for example, requires detection of 1.5-fold or smaller increases. A threshold chemistry based on quenching the product of a 1 : 1 conversion (1 product molecule from 1 input molecule)20 can work only at concentrations high enough to enable the product to be seen without amplification. Natural threshold chemistries13,14 and those inspired by nature15,16 can be much sharper but require complex systems of enzymes that are not easily adapted to assays. An elegant system has been developed to use the metabolic “branch point effect” to generate digital outputs that are results of inputs of mM glucose concentration and are detected by an electrical reader.21 However, we wished to develop a general system that is compatible with current immunoassays, is sensitive to concentrations below μM, and gives visual readout. In principle, the metabolic “branch point effect” system could be engineered further to perform analog-to-digital chemical conversion, but we found a solution of a different type that can resolve small steps (1.22-fold in theory and 1.30-fold in experiments) at nM concentrations and has the potential to be adapted to other assays that give the biotin group as the reporter.
RESULTS AND DISCUSSION
The threshold chemistry evaluated here is based on enzymatic inhibition: the enzyme is incubated with the competitive inhibitor first, and then, it is allowed to react with the substrate. The concentration of available active enzyme (output) increases dramatically when the total concentration of enzyme (input) changes from just below the concentration of the inhibitor to just above it. This increase in output is larger when the inhibitor binds more tightly to the enzyme.23 We first analytically specify the requirements for the threshold chemistry to be functional by considering the binding between the enzyme and the inhibitor. We consider only this step because it is where the threshold is set up, and we do not explore upstream steps (analyte and enzyme) or downstream steps (enzymatic reaction and other reactions for readout). Suppose we have two inputs (i.e., two enzyme concentrations) that need to be distinguished from one another. The inhibitor concentration, which is the threshold concentration (thresh_conc), is set to be the average value of the inputs. We define the dimensionless parameters α (where 0 < α < 1) and γ (where 0 < γ < 1) to describe the resolution and sharpness of the threshold chemistry, such that the two inputs, (1 − α)* thresh_conc and (1 + α) * thresh_conc, give relative outputs of (1 − γ) and 1, respectively (Figure 2B). The resolution (ratio of the two inputs that gives distinguishable outputs) is (1 + α)/(1 + α) and the corresponding ratio of outputs is 1/(1 − γ)
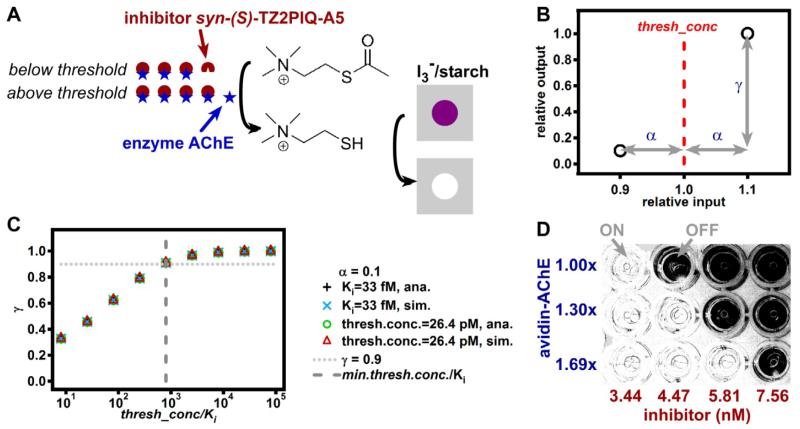
Figure 2.
Threshold chemistry achieved by competitive inhibition. A) A schematic drawing of the threshold chemistry and detection reactions. When the concentration of the enzyme acetylcholinesterase (AChE) exceeds that of the inhibitor (syn-(S)-TZ2PIQ-A522), the enzyme hydrolyzes acetylthiocholine to give thiocholine, which reduces the purple suspension of I3−/starch complex and results in a clear mixture. B) Graphical definition of the parameters α and γ (Equation 1). C) A plot comparing γ values calculated from different thresh_conc/Ki via analytical calculations (ana.) and numerical simulations (sim.) with γ = 0.9 at min_thresh_conc/Ki = 801 as predicted by Equation 2. D) A photograph showing detection of 1.30-fold increases in enzyme concentration with multiple threshold concentrations and visual readout in a 96-well plate. Details of image handling can be found in the Supporting Information.
The threshold chemistry is considered to be sharp when the ratio of outputs is large even with a small ratio of inputs. The parameters α and γ (Equation 1 and Figure 2B) are dictated by the requirements of the assay. If a small increase in concentration needs to be detected, then α must be small. If a large difference in output needs to be detected, for instance, to enable easy visual readout, then the ratio of outputs and γ need to be large.
| (1) |
With a certain required resolution (1 + α)/(1 + α), the minimum threshold concentration (min_thresh_conc) that can give a ratio of outputs ≥ 1/(1 − γ) is given by Equation 2 (see the Supporting Information for derivation), where Ki is the dissociation constant of the enzyme/inhibitor complex. The model used to obtain this result consists of enzyme/inhibitor equilibrium followed by Michaelis-Menten kinetics.
| (2) |
According to Equation 2, one would like to choose as small a ki as possible, so that when the parameters α and γ are predetermined, the threshold chemistry has as large a working range of concentrations as possible (lowest min_thresh_conc), or so that when the min_thresh_conc is predetermined, one has as sharp a threshold as possible (large γ with small α).
To evaluate this idea, we chose acetylcholinesterase (AChE) as the enzyme (Figure 2A). We used a potent inhibitor, syn-(S)-TZ2PIQ-A5, with Ki= 33 fM.22 when the concentration of the enzyme is higher than that of the inhibitor, it hydrolyzes acetylthiocholine to give thiocholine, which reduces the purple suspension of I3−/starch complex to give a clear mixture, enabling threshold responses (Figures 1, 2). To achieve a resolution of 1.22 = (1 + 0.1)/(1 − 0.1) (with α = 0.1) and a ratio of outputs of 10 = 1/(1 − 0.9) (with γ = 0.9), we have min_thresh_conc = 26.4 pM , and min_thresh_conc/Ki = 801 (Equation 2). To verify those predictions, we fixed the resolution (by maintaining α = 0.1), varied thresh_conc/Ki (by changing thresh_conc or Ki) and calculated γ in each case (by using both rate laws and simulation with ordinary differential equations) (Figure 2C). Only when thresh_conc/Ki ≥ min_thresh_conc/Ki = 801 did we find γ ≥ 0.9, satisfying the requirement for producing a large distinction in outputs, suitable for visual readout. Note that these calculations assume idealized kinetics, ignoring a number of potential confounding factors present in experiments (e.g., changes of enzymatic activity in the presence of proteins present in realistic samples, or competition between the inhibitor and the substrate used for readout of enzyme activity).
We experimentally detected 1.30-fold increases at nM AChE concentrations with visual readout (Figure 2D). The concentration of input, avidin-conjugated AChE (avidin-AChE), increased by 1.30-fold going across each row of wells on a 96-well plate, while the threshold concentration (set by concentration of the inhibitor) increased 1.30-fold going down each column. In each well, avidin-AChE and the inhibitor were incubated before the addition of the substrate mixture (acetylthiocholine/I3-/starch). The reactions produced ON (clear) or OFF (dark) responses, visible by the naked eye, depending on whether the enzyme concentration was larger or smaller than the inhibitor concentration.
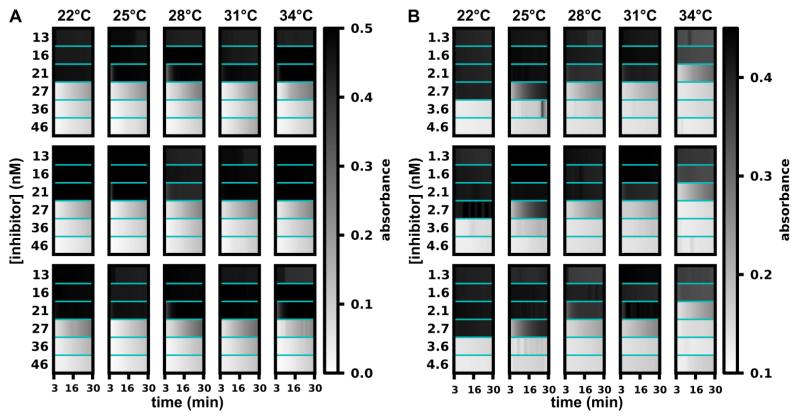
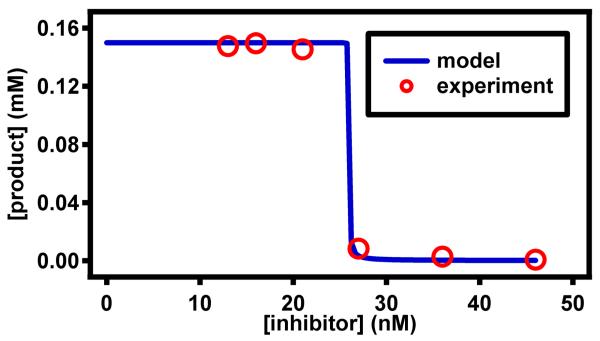
Furthermore, we found that this threshold chemistry is robust to changes in temperature and readout time (Figure 3 and 4). We used a plate reader to control the temperature and measure absorbance from well-plate reactions of acetylcholinesterase with the chromogenic substrate 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB) (absorbance ∝ [product]).24 This experiment was designed to follow the principle of the analog-to-digital conversion (Figure 1B): subjecting one concentration of input (enzyme concentration) to multiple thresholds (set by inhibitor concentrations). To test thresholds at high (1.3-fold) resolution, at each temperature, six reactions were set up with the inhibitor concentration following a 1.3× dilution series from 13 to 46 nM. Reactions with inhibitor concentrations that were lower than the enzyme concentration gave ON results (13, 16, 21 nM), while the rest gave OFF results (27, 36, 46 nM). This threshold was robust: the contrast between ON and OFF reactions was consistent over a range of readout times (3–30 min), a range of temperatures (22–34 °C), and three repeats (Figure 3A). This result, predicted by equation 2, was somewhat surprising because this equation does not capture a number of complications, including the temperature dependence of enzyme-substrate and enzyme-inhibitor binding, and also the competition between inhibitor present at a low concentration and readout substrate present at a high concentration. We also found that when the enzyme and the inhibitor were diluted 10 times, the threshold chemistry yielded narrower working ranges in temperature and readout (25 °C/3–10 min; 28-34 °C/3-30 min) (Figure 3B). We observed agreement between results from robustness experiments and the model (Figure 4). We used 28 °C because this falls in the middle of the range of temperatures that we tested experimentally. The plot of the model in Figure 4 was generated using analytical calculations of the enzyme/inhibitor equilibrium (Equation S5) before the enzymatic reaction, followed by use of the Michaelis-Menten model to simulate the enzymatic reaction (with inhibition) via numerically solving ordinary differential equations. The concentrations used in the model match those from the robustness experiments (Figure 3A), and the rate constants were obtained from previously published work.22,25
Figure 3.
Robustness of threshold chemistry at different temperatures and readout times. A) False-color maps depict 90 time traces of absorbance from 90 threshold reactions of acetylcholinesterase with a chromogenic substrate (see text for details) as the inhibitor concentrations were changed in a series of 1.3× dilutions (13, 16, 21, 27, 36, and 46 nM) across a 12-degree range of temperatures (22 °C, 25 °C, 28 °C, 31 °C, and 34 °C). B) False-color maps depict 90 time traces of absorbance from 90 threshold reactions of acetylcholinesterase with a chromogenic substrate as the inhibitor concentrations were arranged in a series of 1.3× dilutions (1.3, 1.6, 2.1, 2.7, 3.6, and 4.6 nM; 10 times less [enzyme] and [inhibitor] compared to A) across a 12-degree range of temperatures. Experiment repeats are arranged in rows. Note that in this figure, higher absorbance (dark) is equivalent to higher enzymatic activity, which is opposite from the visual readout system used in other experiments.
Figure 4.
Comaparison of experimental results from robustness experiments and the model. The experimental results are the average of 3 repeats of the results shown in Figure 3A, at 3 minutes and 28 °C. The plot of the model was generated using analytical calculations (Equation S5) before the enzymatic reaction and the Michaelis-Menten model to simulate the enzymatic reaction (with inhibition). Concentrations used in the model match those of the robustness experiments (Figure 3A).
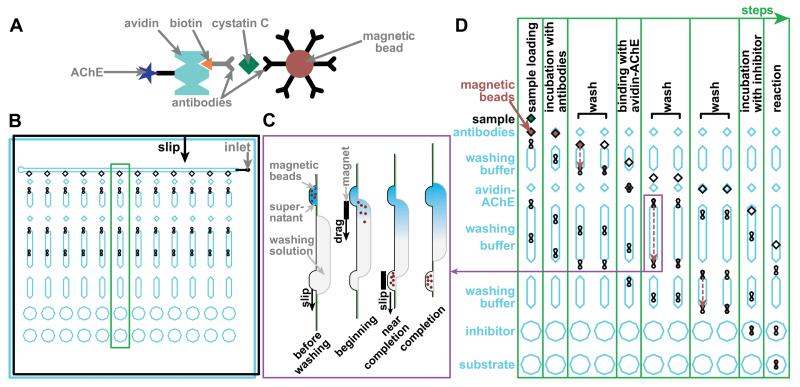
To test this chemistry in the assay for cystatin C, we used a bead-based sandwich assay, in which the analyte cystatin C molecule was bound by two different antibodies (Figure 5A). The capture antibody was conjugated to magnetic beads, while the detection antibody was conjugated to biotin, which can bind to avidin-AChE. The amount of AChE immobilized on the beads should therefore correlate to the amount of cystatin C. The magnetic beads allowed for washing and physical transfer of the whole complex from well to well throughout the multi-step process.
Figure 5.
Illustrations of the immunoassay and SlipChip design. A) A drawing of the complex used in the magnetic bead-based immunoassay for cystatin C. B) A drawing of the overall SlipChip design. The chip has a top plate (black lines) and bottom plate (blue lines) that face each other. Each of the 12 columns is designed to perform an assay at a different threshold concentration, for a total of up to 12 threshold concentrations (or fewer, if duplicate assays are desired). Reagents are pre-loaded into the chip in rows into the layer shown in blue before the sample is introduced into the inlet. To perform each step of the assay, the top plate is slipped according to the “slip” arrow (solid black arrow), performing the assay on all 12 columns in parallel. The green box indicates the column shown in D. C) Schematic of the washing mechanism, based on dragging magnetic beads through a channel containing washing solution (side view). When the top plate is slipped relative to the bottom plate, beads are moved to the beginning of the channel containing washing solution. A magnet is used to move the beads to the other end of the channel, and then, to another well on the top plate. The plate is then slipped once more to separate the beads and the channel. D) A time series illustrating the assay. Only one of the 12 columns is shown (11 times), illustrating the 8 steps of the assay. The movement of the plates of the SlipChip (7 slips total) is shown. The blue (bottom) plate is stationary, and the top (black) plate moves along the direction of the arrow shown in B. The movement of magnetic beads (3 transfers) is shown with red dashed arrows.
This analog-to-digital conversion replaces a single analog experiment with multiple ON/OFF digital experiments, and uses an approach that enables one to perform multiple experiments in parallel easily while minimizing consumption of samples and reagents. Such multiplexing and miniaturization are enabled by microfluidics, and we used a SlipChip26 microfluidic device to evaluate the chemistry and the feasibility of the overall approach. The SlipChip we used was composed of two plates that face one another, each having recessed features (wells, ducts, etc.) to contain aqueous reagents. Complex manipulations of fluids can be programmed into a SlipChip as a pattern of these features. The user can then execute the program by moving these features relative to one another by slipping the plates. This enables multistep processing for a number of applications.12,26,27
We designed the SlipChip (Figure 5B, C) based on previously published designs for performing immunoassays,27 with an extra step of inhibition, a modified washing mechanism (Figure 5C), different surface coating (fluorinated ethylene propylene), and different loading and assembly protocols. Each of the 12 columns of wells on the chip performs one assay with a certain predetermined threshold concentration. As the top plate moves relative to the bottom plate, the steps of the assay are performed sequentially in each column (Figure 5D), and all of the columns are processed at the same time so that the assays run in parallel.
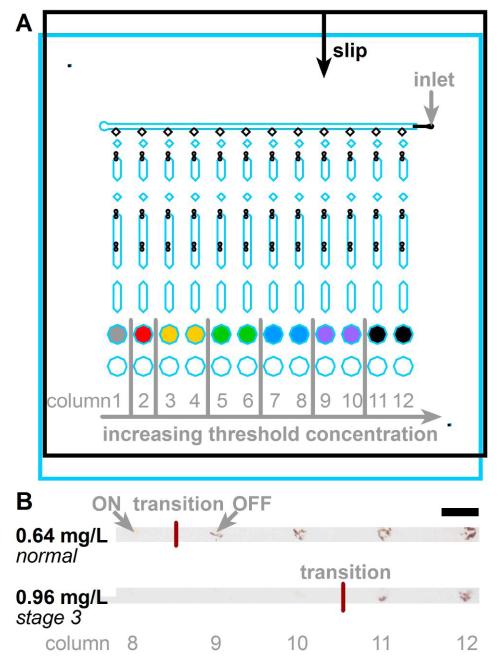
To test whether this combination of threshold chemistry (Figure 2) and microfluidics (Figure 5) could enable detection of a 1.5-fold increase in the concentration of cystatin C in serum, we set up a device with 1.5-fold steps of increasing inhibitor concentration going from left to right (Figure 6A). In these experiments, we used purified cystatin C dissolved in cystatin C-free serum to avoid confounding the experiments by potential variability of cystatin C concentration in serum of patients. In each column, the output was OFF (dark) if the input (concentration of cystatin C) was below the threshold concentration, and the output turned ON (clear) otherwise. The transition position was defined as the point in the bottom row at which a clear-to-dark step was observed. A cystatin C concentration of 0.64 mg/L, which is in the range of normal kidney function, transitioned between columns 8 and 9, producing 4 wells with OFF outputs. The concentration of 0.96 mg/L, which corresponds to stage 3 chronic kidney disease, transitioned between columns 10 and 11, producing 2 wells with OFF outputs (Figure 6B). Experimental details are described in Supporting Information.
Figure 6.
Assays designed to detect a 1.5-fold increase in cystatin C (0.64–0.96 mg/L), when a patient may progress from normal kidney function to acute kidney injury (AKI) or stage 3 chronic kidney disease (CKD). A) Schematic showing the arrangement of threshold concentrations on the device. The inhibitor droplets were loaded such that concentrations increased from left to right (0, 5.3, 7.9, 12, 18, 27, and 40 nM; identical colors indicate identical concentrations). B) Photographs showing ON-to-OFF transition for solutions of 0.64 mg/L (top) and 0.96 mg/L (bottom) cystatin C spiked into cystatin C-free serum. Scale bar: 2 mm. Images were taken with a consumer digital camera; details of image handling are included in the Supporting Information.
We performed two additional sets (4 chips) of experiments, which also showed a difference in the position of the transition between these two concentrations. All three sets of experiments gave aggregated results of (3, 4, 5) wells with OFF outputs for 0.64 mg/L and (2, 2, 2) wells with OFF outputs for 0.96 mg/L. Thus, an increase of 1.5-fold (0.64 to 0.96 mg/L) resulted in a statistically significant difference in outputs (1-tailed p-value of 0.0129) for this combination of threshold chemistry and microfluidics.
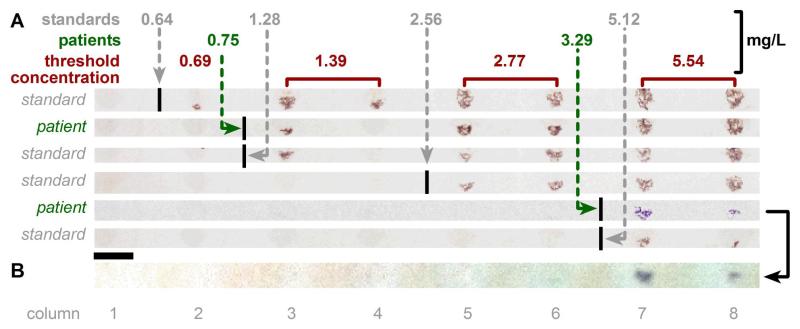
Next, we tested whether we could systematically adjust the threshold concentrations over a large dynamic range, and whether this approach would still perform adequately when used with patient samples. The patient samples were collected and characterized for purposes other than this project (for a prospective observational study investigating novel biomarkers of renal tubular injury following adult cardiac surgery),28,29 archived, and de-identified. Once collected, the samples were centrifuged and the supernatant was kept and immediately frozen at −80 °C. The samples that we used were known to have high (3.29 mg/L) and low (0.75 mg/L) cystatin C values. We chose to test samples from patients with variable renal function in order to gain values across the spectrum of disease and across the range of the assay. For controls, we measured concentrations independently using a commercially available ELISA kit with the platform of microtiter plates. With the same chip design, the inhibitor concentrations within rows were modified to detect bigger steps (2-fold) in concentration, with a larger overall range of detection (0.64-5.12 mg/L) and correspondingly coarser resolution. The 12 columns on the chip enabled us to set up some of the thresholds in duplicate. This was most useful at low concentrations (and correspondingly, closer transitions), where we observed some noise (out-of-order transitions) for the 1.39 mg/L concentration. Overall, we observed good agreement between ON/OFF transitions for the two samples from patients and those for four standard solutions of known concentration (solution of purified cystatin C dissolved in cystatin C-free serum) (Figure 7A). The contrast of the visual readout was strong enough to be recorded with the camera of a cell phone (iPhone 4) (Figure 7B). We emphasize that there is a trade-off between the resolution, dynamic range, and the number of experiments (and therefore the complexity of the microfluidic device) required to implement this analog-to-digital conversion.
Figure 7.
Results from standard solutions and clinical serum samples over a wide dynamic range. A) Photographs of results, with each row corresponding to one assay (one SlipChip device). B) Photograph of the result of the 3.29 mg/L patient sample recorded with an iPhone 4 camera. Only the first 8 columns of the device described in Figure 5 are shown in Figure 7. The sequence of inhibitor concentrations was set up as in Figure 6A, but with larger steps in concentration (0, 9.4, 19, 38, 75, 150, and 300 nM). Columns 3–4, 5–6, 7–8, 9–10, and 11–12 were set up as duplicates such that each pair contained the same inhibitor concentration. Scale bar: 2 mm. Details of image handling are included in the Supporting Information.
We now discuss the features of this approach, its potential shortcomings, and the remaining challenges to its implementation, as well as its implications for diagnostic testing using kidney injury as an example. In terms of chemistry, the use of tight AChE-inhibitor binding combined with iodine-starch based readout is attractive because it can be implemented for any sandwich immunoassay, as long as the detection antibody can be conjugated to biotin. The limits of performance of such threshold chemistry would be defined by Equation 2. When the input concentration is sufficiently larger or smaller than the threshold concentration, variation in input concentration does not affect the reaction time, because the reaction time is either very fast or very slow, respectively. However, the sensitive range around the threshold is one disadvantage of this chemistry (and any other threshold chemistry). We use the parameter α (Equation 1 and Figure 2B) to specify how far away from the threshold concentration the input concentration needs to be for robust performance. Input concentrations sufficiently larger than the threshold concentration (Io (1 + α) < input, where Io is the threshold concentration) would make the reactions finish much faster than those from input concentrations slightly smaller than the threshold concentration (input < Io (1 − α) (Figure 2). The reactions from the former cases would all finish before the reactions from the latter case have hardly started, and the readout time would not depend on concentration in those cases. However, when the input concentration is minimally below or above the threshold concentration (o (1 − α) < input < Io (1 + α)), the chemistry may result in intermediate outputs, which may not be easily distinguished from those sufficiently smaller or larger than the threshold. Quantitatively, when the input increases from Io (1 − α) to Io (1 + α), the output increases 1 / (1 − γ) times (10 times when γ = 0.9). In general, any threshold chemistry, by definition, has a sensitive range around the threshold and gives robust output outside the sensitive range. In particular, this threshold chemistry would also give slower reactions (regardless of the threshold) when the absolute concentrations are lower. Some improvement can be achieved by increasing the enzymatic turnover rate or change in absorbance per turnover. An even better approach would be to develop additional nonlinear amplification chemistries that would give even higher resolution and faster readout time. For example, due to the tight and specific binding in DNA and RNA hybridization, we see significant potential in nucleic acid amplification strategies based on autocatalytic enzymatic (e.g., polymerase chain reaction and various flavors of isothermal chemistries12,30,31) amplification or enzyme-free amplification strategies based on DNA-circuits and DNA computation.32-35 Such approaches, combined with microfluidics, could extend analog-to-digital conversion to quantification of nucleic acids. Furthermore, aptamers and related nucleic acid-based reagents36 already used for detection of proteins and small molecules may enable combining threshold approaches and amplification of nucleic acids. This could open the door for the detection of a wide range of molecules beyond nucleic acids. Finally, we see opportunities for exquisite molecular control provided by an exciting array of chemical amplification approaches using autocatalysts, such as diarylketone,37 piperidine,38 fluoride,39 hydrogen peroxide, and glucose.40 These amplification chemistries, when coupled with controls to provide threshold behavior, chemistries to correlate with target analytes, and appropriate devices, will provide quantification techniques for a wide range of analytes and uses.
Here, we have used microfabricated devices produced in a research laboratory and operated by a trained user. For diagnostic use, a mass-produced device would be required, with on-board reagent storage and stabilization. For measuring an analyte with only a few discrete clinically relevant cut-offs, such as cystatin C, a device that performs approximately a dozen parallel assays is sufficient. However, larger numbers of assays would be needed for measurements that require higher dynamic range or finer resolution than those presented here, to generate redundant assays, and to implement on-chip controls and calibration.
We have not optimized the speed of the assay (each chip assay took about 4.5 hours, with the major time-consuming steps being analyte binding and the readout reaction). Now that we have demonstrated that the analog-to-digital conversion chemistry can be used in the context of a realistic assay, such optimization of timing would be appropriate for subsequent work.
While visual readout and immediate interpretation are attractive for designs with only a few assays, the ability to read the assay with a cell phone (Figure 7B) is appealing for several reasons: i) interpreting results of controls and calibrations and inferring the validity of the assay; ii) performing statistical interpretation of the data to provide confidence intervals for assay results; and iii) transmitting the data to physicians and archiving data in electronic medical record (EMR) databases. For example, acute kidney injury is detectable by measuring cystatin C levels in a range compatible with this approach. Diagnosing AKI under limited-resource settings may be needed in events of trauma (injuries of the kidney or injuries that lead to sudden, serious blood loss, blockage of the urinary tract, or poisoning) if laboratory-based medical testing is impractical or unavailable. Such situations may arise in the event of accidents in remote areas or injuries after natural disasters such as earthquakes that damage infrastructure (where crush injuries and the resultant rhabdomyolysis result in kidney injury). Furthermore, chronic kidney disease offers another reason to measure cystatin C under limited-resource settings, such as in developing countries. In the United States, as well, awareness of chronic kidney disease remains low among the estimated 20 million people afflicted (7.8 % awareness for individuals with stage 3 disease41), even though cystatin C immunoassays are available in hospitals. In both acute kidney injury and chronic kidney disease, identifying small increases in the concentration of cystatin C (1.5-fold or smaller) is crucial for accurate diagnosis. The availability of an assay that can quantify these small increases would enable timely detection of acute kidney injury and encourage screening to increase awareness of chronic kidney disease, facilitating care. Robustness of digitized assays to temperature and reaction time (Figure 3) may aid implementation of such assays in limited-resource settings, because it could enable simple devices for quantitative measurements without requiring precise control of temperature or timing of the measurement.
CONCLUSIONS
We draw five conclusions from this work: i) Analog-to-digital signal conversion can be implemented using threshold chemistry (as discussed above, the analog-to-digital conversion described in this paper is distinct from “digital” single-molecule measurements9-11); ii) This threshold chemistry shows robustness to variations in temperature and readout time; iii) The threshold chemistry can be implemented within the realistic context of a cystatin C assay: combined with a microfluidic device, it detected a 1.5-fold increase in the concentration of cystatin C in serum at nM concentrations, and the assay performed properly with clinical samples; iv) An advantage of this method is that it provides quantitative information interpretable by the naked eye or a cell-phone camera, without requiring quantitative instrumentation; v) A disadvantage of analog-to-digital signal conversion is that one assay is replaced with multiple assays (the number of assays depends on the resolution and the dynamic range desired). Advances in microfluidics could enable parallel manipulations, reduce the amount of sample required, and make digital-to-analog conversion feasible with no extra reagent cost or user intervention. Clinically validated and field-usable readerless quantitative assays suitable for providing medical care under limited-resource settings, while not a goal of this paper, would be enabled by additional developments in automated user-friendly microfluidics and in threshold chemistries.
Supplementary Material
ACKNOWLEDGMENT
This work was supported in part by the NIH Director’s Pioneer Award DP10D003584, NIH Career Development Award K23DK081616 (Jay Koyner), DARPA Cooperative Agreement No. HR0011-11-2-0006, and ONR Grant No. N00014-08-1-0936. The authors thank Jordany Maignan and Roman Manetsch for providing pure syn-(S)-TZ2PIQ-A5 (AChE inhibitor), Elena K. Davydova and Jason E. Kreutz for aiding in the selection of cystatin C, Liang Ma for the idea of depositing plugs, Weishan Liu for advice about manipulation of magnetic beads, Andrew Chiang for references about thresholds, Yu-Hsiang Hsu for suggestions for the imaging of chips, Stefano Begolo for suggestions about image processing, and Bridget Samuels and Whitney Robles for contributions to the writing and editing of this manuscript.
Footnotes
Supporting Information. Derivation of Equation 2, correlation between stages of CKD and cystatin C concentration, buffers and solutions, conjugation, working ranges of temperature and detection time, threshold chemistry in bulk with visual readout, device fabrication, treatment of chip surface, setup and execution of assay, handling of images, and details of estimation of threshold concentration from assay results of standards and patient samples. This material is available free of charge via the Internet at http://pubs.acs.org.
Disclosure: Rustem F. Ismagilov and Liang Li have a financial interest in SlipChip Corp.
REFERENCES
- (1).Stevens LA, Coresh J, Schmid CH, Feldman HI, Froissart M, Kusek J, Rossert J, Van Lente F, Bruce RD, 3rd, Zhang YL, Greene T, Levey AS. Am. J. Kidney Dis. 2008;51:395–406. doi: 10.1053/j.ajkd.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Cheng C-M, Martinez AW, Gong J, Mace CR, Phillips ST, Carrilho E, Mirica KA, Whitesides GM. Angew. Chem., Int. Ed. 2010;49:4771–4774. doi: 10.1002/anie.201001005. [DOI] [PubMed] [Google Scholar]
- (3).Martinez AW, Phillips ST, Whitesides GM, Carrilho E. Anal. Chem. 2010;82:3–10. doi: 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- (4).Nie Z, Deiss F, Liu X, Akbulut O, Whitesides GM. Lab Chip. 2010;10:3163–3169. doi: 10.1039/c0lc00237b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ellerbee AK, Phillips ST, Siegel AC, Mirica KA, Martinez AW, Striehl P, Jain N, Prentiss M, Whitesides GM. Anal. Chem. 2009;81:8447–8452. doi: 10.1021/ac901307q. [DOI] [PubMed] [Google Scholar]
- (6).Faulstich K, Gruler R, Eberhard M, Lentzsch D, Haberstroh K. Handheld and Portable Reader Devices for Lateral Flow Immunoassays. In: Wong R, Tse H, editors. Lateral Flow Immunoassay. Springer; New York: 2009. [Google Scholar]
- (7).Spahillari A, Parikh CR, Sint K, Koyner JL, Patel UD, Edelstein CL, Passik CS, Thiessen-Philbrook H, Swaminathan M, Shlipak MG, Consortium T-A. Am. J. Kidney Dis. 2012;60:922–929. doi: 10.1053/j.ajkd.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Levey AS, Coresh J, Balk E, Kausz AT, Levin A, Steffes MW, Hogg RJ, Perrone RD, Lau J, Eknoyan G. Ann. Intern. Med. 2003;139:137–147. doi: 10.7326/0003-4819-139-2-200307150-00013. [DOI] [PubMed] [Google Scholar]
- (9).Sykes PJ, Neoh SH, Brisco MJ, Hughes E, Condon J, Morley AA. BioTechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- (10).Vogelstein B, Kinzler KW. Proc. Natl. Acad. Sci. U. S. A. 1999;96:9236–9241. doi: 10.1073/pnas.96.16.9236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Rissin DM, Kan CW, Campbell TG, Howes SC, Fournier DR, Song L, Piech T, Patel PP, Chang L, Rivnak AJ, Ferrell EP, Randall JD, Provuncher GK, Walt DR, Duffy DC. Nat. Biotech. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Shen F, Davydova EK, Du W, Kreutz JE, Piepenburg O, Ismagilov RF. Anal. Chem. 2011;83:3533–3540. doi: 10.1021/ac200247e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Ferrell JE. Curr. Opin. Cell Biol. 2002;14:140–148. doi: 10.1016/s0955-0674(02)00314-9. [DOI] [PubMed] [Google Scholar]
- (14).Kastrup CJ, Runyon MK, Lucchetta EM, Price JM, Ismagilov RF. Acc. Chem. Res. 2008;41:549–558. doi: 10.1021/ar700174g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Goldbeter A, Koshland DE. Proc. Natl. Acad. Sci. U. S. A. 1981;78:6840–6844. doi: 10.1073/pnas.78.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Goldbeter A, Koshland DE. J. Biol. Chem. 1984;259:14441–14447. [PubMed] [Google Scholar]
- (17).Pia̧tek AM, Bomble YJ, Wiskur SL, Anslyn EV. Org. Biomol. Chem. 2004;126:6072–6077. doi: 10.1021/ja040006e. [DOI] [PubMed] [Google Scholar]
- (18).Tsalta CD, Meyerhoff ME. Anal. Chem. 1987;59:837–841. doi: 10.1021/ac00133a011. [DOI] [PubMed] [Google Scholar]
- (19).Vallée-Bélisle A, Ricci F, Plaxco KW. J. Am. Chem. Soc. 2011;134:2876–2879. doi: 10.1021/ja209850j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Palmer JL, Timmerman MW. Threshold color control system. U.S. Patent 5,036,000. 1991
- (21).Rafael SP, Vallée-Bélisle A, Fabregas E, Plaxco K, Palleschi G, Ricci F. Anal. Chem. 2011;84:1076–1082. doi: 10.1021/ac202701c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Krasiński A, Radić Z, Manetsch R, Raushel J, Taylor P, Sharpless KB, Kolb HC. J. Am. Chem. Soc. 2005;127:6686–6692. doi: 10.1021/ja043031t. [DOI] [PubMed] [Google Scholar]
- (23).Watanabe M. Genes Environ. 2008;30:17–24. [Google Scholar]
- (24).Ellman GL, Courtney KD, Andres V, jr, Featherstone RM. Biochem. Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- (25).Salih E. Biochem J. 1992;285:451–460. doi: 10.1042/bj2850451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Du W, Li L, Nichols KP, Ismagilov RF. Lab Chip. 2009;9:2286–2292. doi: 10.1039/b908978k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Liu W, Chen D, Du W, Nichols KP, Ismagilov RF. Anal. Chem. 2010;82:3276–3282. doi: 10.1021/ac100044c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Koyner JL, Bennett MR, Worcester EM, Ma Q, Raman J, Jeevanandam V, Kasza KE, Connor MFO, Konczal DJ, Trevino S, Devarajan P, Murray PT. Kidney Int. 2008;74:1059–1069. doi: 10.1038/ki.2008.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Koyner JL, Vaidya VS, Bennett MR, Ma Q, Worcester E, Akhter SA, Raman J, Jeevanandam V, O’Connor MF, Devarajan P, Bonventre JV, Murray PT. Clin. J. Am. Soc. Nephrol. 2010;5:2154–2165. doi: 10.2215/CJN.00740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Shen F, Du W, Kreutz JE, Fok A, Ismagilov RF. Lab Chip. 2010;10:2666–2667. doi: 10.1039/c004521g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Shen F, Sun B, Kreutz JE, Davydova EK, Du W, Reddy PL, Joseph LJ, Ismagilov RF. J. Am. Chem. Soc. 2011;133:17705–17712. doi: 10.1021/ja2060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Stojanovic MN. In: Prog. Nucleic Acid Res. Mol. Biol. Conn PM, editor. Vol. 82. Academic Press; 2008. pp. 199–217. [DOI] [PubMed] [Google Scholar]
- (33).Dirks RM, Pierce NA. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15275–15278. doi: 10.1073/pnas.0407024101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Qian L, Winfree E. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- (35).Allen PB, Arshad SA, Li B, Chen X, Ellington AD. Lab Chip. 2012;12:2951–2958. doi: 10.1039/c2lc40373k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, Flather D, Forbes A, Foreman T, Fowler C, Gawande B, Goss M, Gunn M, Gupta S, Halladay D, Heil J, Heilig J, Hicke B, Husar G, Janjic N, Jarvis T, Jennings S, Katilius E, Keeney TR, Kim N, Koch TH, Kraemer S, Kroiss L, Le N, Levine D, Lindsey W, Lollo B, Mayfield W, Mehan M, Mehler R, Nelson SK, Nelson M, Nieuwlandt D, Nikrad M, Ochsner U, Ostroff RM, Otis M, Parker T, Pietrasiewicz S, Resnicow DI, Rohloff J, Sanders G, Sattin S, Schneider D, Singer B, Stanton M, Sterkel A, Stewart A, Stratford S, Vaught JD, Vrkljan M, Walker JJ, Watrobka M, Waugh S, Weiss A, Wilcox SK, Wolfson A, Wolk SK, Zhang C, Zichi D. PLoS ONE. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Kottani R, Majjigapu JRR, Kurchan A, Majjigapu K, Gustafson TP, Kutateladze AG. J. Am. Chem. Soc. 2006;128:14794–14795. doi: 10.1021/ja066692u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Baker MS, Phillips ST. J. Am. Chem. Soc. 2011;133:5170–5173. doi: 10.1021/ja108347d. [DOI] [PubMed] [Google Scholar]
- (39).Baker MS, Phillips ST. Org. Biomol. Chem. 2012;10:3595–3599. doi: 10.1039/c2ob25363a. [DOI] [PubMed] [Google Scholar]
- (40).Karton-Lifshin N, Shabat D. New J. Chem. 2012;36:386–393. [Google Scholar]
- (41).Plantinga LC, Boulware LE, Coresh J, Stevens LA, Miller ER, Saran R, Messer KL, Levey AS, Powe NR. Arch. Intern. Med. 2008;168:2268–2275. doi: 10.1001/archinte.168.20.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.