Abstract
Many biological processes such as cell proliferation, differentiation, and cell death depend precisely on the timely synthesis and degradation of key regulatory proteins. While protein synthesis can be regulated at multiple levels, protein degradation is mainly controlled by the ubiquitin—proteasome system (UPS), which consists of two distinct steps: (1) ubiquitylation of targeted protein by E1 ubiquitin-activating enzyme, E2 ubiquitin-conjugating enzyme and E3 ubiquitin ligase, and (2) subsequent degradation by the 26S proteasome. Among all E3 ubiquitin ligases, the SCF (SKP1-CUL1-F-box protein) E3 ligases are the largest family and are responsible for the turnover of many key regulatory proteins. Aberrant regulation of SCF E3 ligases is associated with various human diseases, such as cancers, including skin cancer. In this review, we provide a comprehensive overview of all currently published data to define a promoting role of SCF E3 ligases in the development of skin cancer. The future directions in this area of research are also discussed with an ultimate goal to develop small molecule inhibitors of SCF E3 ligases as a novel approach for the treatment of human skin cancer. Furthermore, altered components or substrates of SCF E3 ligases may also be developed as the biomarkers for early diagnosis or predicting prognosis.
Keywords: Carcinogenesis, F-box proteins, RING proteins, SCF E3 ligases, Skin, Ubiquitin ligases
INTRODUCTION
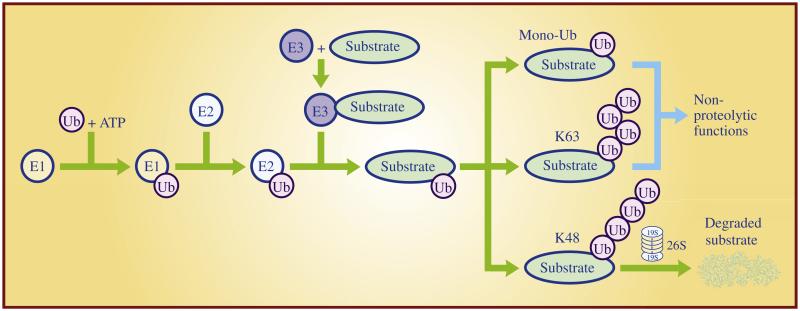
The ubiquitin—proteasome system provides one of the key mechanisms regulating cellular protein homeostasis. Ubiquitylation occurs through three sequential steps in cells. Ubiquitin (Ub) is first activated by the E1 ubiquitin-activating enzyme in an ATP-dependent manner. The activated Ub is subsequently transferred to an E2 ubiquitin-conjugating enzyme and then attached to a target protein, catalyzed by an E3 ubiquitin ligase. The targeted protein can be mono-ubiquitylated with one ubiquitin attachment or polyubiquitylated with multiple ubiquitin attachments in different linkages. Mono-ubiquitylation controls numerous cellular processes such as receptor transport, virus budding, signal transduction, and DNA damage repair (Ikeda and Dikic, 2008), whereas polyubiquitylation could either alter protein function if the ubiquitin linkage is through K63 (Chen and Sun, 2009), or lead to degradation by the 26S proteasome if the linkage is through K48 (Hershko and Ciechanover, 1998) (Fig. 1). The polyubiquitin chains can be disassembled by deubiquitylation enzymes (DUBs, also known as isopeptidases) for recycling (D’Andrea and Pellman, 1998). Therefore, ubiquitylation is a dynamic, reversible, and covalent modification of targeted proteins.
Fig. 1. The ubiquitin—proteasome system.
Ubiquitin (Ub) is activated by the E1 ubiquitin-activating enzyme in an ATP-dependent reaction. Activated Ub is then transferred to an E2 ubiquitin-conjugating enzyme. The E3 ubiquitin ligase recognizes a substrate and catalyzes the ubiquitin transfer from the E2 to the substrate. A single run of this ubiquitylation reaction is referred as monoubiquitination (mono-Ub), which changes substrate function, while multiple runs of this reaction cause polyubiquitylation of the targeted substrate. Dependent on the isopeptide linkage of the polyubiquitin chain, polyubiquitylated substrate can be either degraded by 26S proteasome if in a K48 linkage or activated for nonproteolytic function if in a K63 linkage.
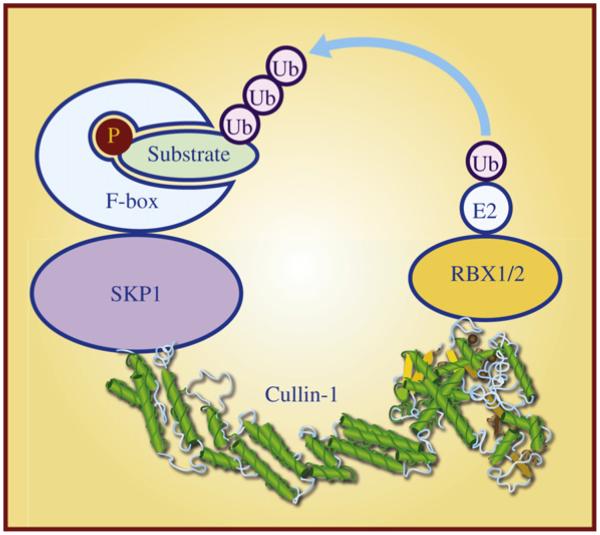
The human genome encodes two E1 ubiquitin-activating enzymes, at least 38 E2 ubiquitin-conjugating enzymes, and more than 600 distinct E3 ubiquitin ligases (Ye and Rape, 2009). The largest family of E3 ubiquitin ligases is the SCF E3 ligases, which is conserved among eukaryotic cells from yeast to humans, and consists of four structural and functional components: (1) an adaptor protein SKP1, (2) a scaffold protein Cullin-1 (CUL1), (3) a substrate-recognizing F-box protein, and (4) a RING protein RBX1 or RBX2 (Fig. 2). All SCF E3 ligases share a similar structure in which CUL1 binds to SKP1 and an F-box protein at the N-terminus and a RING protein RBX1 or RBX2 at the C-terminus (Zheng et al., 2002). Substrate specificity of SCF E3 ligases is determined largely by the F-box protein, which usually recognizes phosphorylated target protein (Jin et al., 2004), whereas CUL1-RBX1 or CUL1-RBX2 constitutes the core ligase activity, catalyzing the transfer of the ubiquitin from an E2 to the substrate (Wu et al., 2000) (Fig. 2). By promoting targeted degradation of many key regulatory proteins, SCF E3 ligases play the critical roles in many biological processes, including cell cycle progression, DNA replication, signal transduction, and development (Nakayama and Nakayama, 2006; Deshaies and Joazeiro, 2009). SCF E3 ligases are altered in many human cancers, and thereby have been emerging as attractive anticancer targets (Sun, 2006; Jia and Sun, 2011).
Fig. 2. SCF E3 ubiquitin ligase and its four components.
SCF E3 ubiquitin ligase is composed of four subunits: a scaffold protein Cullin-1, an adaptor protein SKP1, a substrate receptor F-box protein, and a RING finger protein RBX1 or RBX2. Cullin-1 binds to SKP1 and F-box protein at its N-terminus and to the RING protein RBX1/2 at its C-terminus. F-box protein usually recognizes phosphorylated substrate and presents it for ubiquitylation by SCF E3 ubiquitin ligases.
Notably, aberrant regulation of SCF E3 ligases is clinically related to a broad range of human cancers including skin cancer (Jia and Sun, 2011). Importantly, skin cancer is a common human cancer with over 1 million new cases yearly in the USA (Siegel et al., 2012). Melanoma and non-melanoma are two major types of skin cancer with non-melanoma cancer including carcinoma of basal cells, squamous cells, and merkel cells (Leiter and Garbe, 2008). Ultraviolet (UV) radiation from the sun exposure is the primary cause of skin cancer (Rosso et al., 1998; Einspahr et al., 2003; Saladi and Persaud, 2005). Additional factors, including smoking, ionizing radiation, environmental carcinogens, and human papillomavirus (HPV) infections also contribute significantly to the development of skin cancer (Saladi and Persaud, 2005). Here, we will review and summarize the up-to-date experimental data to support the notion that SCF E3 ligases play a critical role in promoting skin carcinogenesis, and thus are attractive therapeutic targets for the treatment of skin cancer.
ROLE OF THE RING COMPONENT IN SKIN CANCER
In mammals, the RING component of SCF has two family members, namely RBX1 (RING box protein 1), also known as ROC1 (regulator of Cullins-1), and the closely related RBX2 or ROC2 (Wei and Sun, 2010), also known as SAG (sensitive to apoptosis gene) (Duan et al., 1999; Sun et al., 2001). The function of RBX is to bring the ubiquitin-loaded E2 into close proximity to the targeted substrate. Both RBX1 and RBX2/SAG are highly conserved during evolution from yeast to human (Sun et al., 2001; Wei and Sun, 2010). Earlier studies from multiple research groups including us have reported that targeted deletion of Hrt1, the only yeast homolog of RBX1/RBX2, causes yeast death, which can be completely rescued by either human RBX1 orSAG (Ohta et al., 1999; Seol et al., 1999; Swaroop et al., 2000). Functionally, both family members are actively involved in the ubiquitylation and degradation of both cytoplasmic and nuclear proteins (Jia et al., 2009; Wei and Sun, 2010).
RBX1/ROC1, was initially cloned in 1999 by four different laboratories as an essential component required for full activity of SCF E3 ligases (Kamura et al., 1999; Ohta et al., 1999; Seol et al., 1999; Tan et al., 1999). Although RBX1 is overexpressed in various cancers, including carcinomas derived from liver, kidney, lung, and breast (Clifford et al., 2001; Jia et al., 2009; Yang et al., 2013), very little is currently known about the potential pathological involvement of RBX1 in skin cancer. To this end, a recent association study correlated the expression of ROC1 with one of its known substrates, cyclin D1 in 62 cases of primary melanomas and 58 cases of compound melanocytic nevi (Nai and Marques, 2011). The authors found a higher ROC1 expression in nevi than in melanomas, which was just opposite for cyclin D1 expression (Nai and Marques, 2011). The study established an inverse relationship between the expressions of ROC1 and cyclin D1, and implied that ROC1 may play an antiproliferative role in part by targeting cyclin D1 for degradation in nevi, which is attenuated during melanomagenesis.
SAG/RBX2/ROC2 was originally cloned in our laboratory as an antioxidant protein (Duan et al., 1999; Swaroop et al., 1999) and later characterized as the second member of the RING component of SCF E3 ubiquitin ligases (Swaroop et al., 2000). SAG regulates cell proliferation, apoptosis, vasculogenesis, and tumorigenesis by targeting the degradation of many critical cellular regulators including c-Jun (Gu et al., 2007a; Gu et al., 2007b), IκBα (Gu et al., 2007a; Tan et al., 2010), HIF-1α (Tan et al., 2008), NF1 (Tan et al., 2011b), procaspase (Tan et al., 2006), p27 (He et al., 2008), NOXA (Jia et al., 2010), and DEPTOR (Zhao et al., 2011).
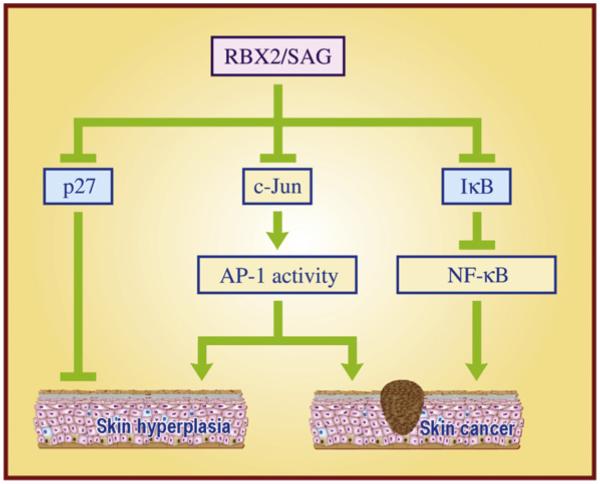
Our early study using a mouse JB6 epidermal cell culture model, representing a preneoplastic-to-neoplastic progression (Colburn et al., 1979; Sun et al., 1994), clearly showed that ectopic expression of SAG inhibits TPA-induced neoplastic progression, whereas conversely, siRNA silencing of SAG promotes it (Gu et al., 2007b). Further mechanistic studies revealed that SAG promotes the targeted degradation of c-Jun, leading to AP-1 inactivation, thus abrogating TPA-induced neoplastic transformation, a process that requires AP-1 activation (Gu et al., 2007b). We further extended this in vitro cell culture observation into an in vivo mouse transgenic model through which we found that SAG regulated skin tumorigenesis in a stage-dependent manner. Specifically, we generated two independent lines of FVB/N mice with SAG transgenic expression specifically in skin epidermis, driven by a well-characterized K14 promoter (Gu et al., 2007a). In a classic DMBA/TPA two-stage skin carcinogenesis model, SAG transgenic expression suppressed hyperplasic proliferation at the early stage, leading to a longer latent period for tumor formation, but promoted tumor growth at the late stage, giving rise to larger sized tumors. Consequently, SAG transgenic expression led to the formation of fewer number of, but bigger size of tumors. Mechanistic studies revealed that the early stage suppression was mediated largely by SAG-induced c-Jun degradation and subsequent AP1 inactivation, whereas the later stage promotion of tumorigenesis was mediated in part by SAG-induced IκB degradation and subsequent NF-κB activation, leading to the blockage of cellular apoptosis (Gu et al., 2007a) (Fig. 3).
Fig. 3. The role of RBX2/SAG in skin cancer.
Transgenic SAG expression inhibits DMBA/TPA-induced carcinogenesis by inactivating AP1 via targeting the c-Jun oncoprotein for degradation at the early stage, but promotes skin carcinogenesis by activating NF-κB via targeting the IκB tumor suppressor for degradation at the later stage. In addition, SAG regulates UVB-mediated skin hyperplasia via promoting the degradation of both p27 and c-Jun.
We also used the JB6 epidermal cell culture model to further determine the responsiveness to UV-irradiation and found that ectopic expression of SAG significantly increases the resistance of epidermal JB6-C1.41 cells to UVB-induced apoptosis, as indicated by the loss of apoptotic cells, and by the absence of both caspase-3 activity and sub-G1 cell populations (He et al., 2008). Using the same transgenic model, we found that SAG transgenic expression promotes UVB-induced skin hyperplasia, but not tumor formation. Subsequent mechanistic studies revealed that SAG, on one hand, promotes c-Jun degradation to inactivate AP1 DNA binding and transactivation (anti-proliferating effect), whereas, on the other hand, it promotes p27 degradation (pro-proliferating effect) (He et al., 2008). The net outcome of hyperplasia as a result of these two opposite effects suggests that p27 plays a bigger role than c-Jun in the response of SAG transgenic skin to UVB exposure (Fig. 3). Given the fact that SAG is over-expressed in a number of human cancers which correlates with poor patient prognosis (Wei and Sun, 2010; Jia and Sun, 2011) and that SAG promotes the development of skin cancer (Gu et al., 2007a), targeting SAG E3 ligase might be a valid therapeutic option for the effective treatment of human skin cancers (Sun and Li, 2012).
ROLE OF CULLIN-1 IN SKIN CANCER
CUL1, the first member of the Cullin family (Sarikas et al., 2011), was first identified in 1996 as being required for cell cycle exit in Caenorhabditis elegans (Kipreos et al., 1996) and for the G1-to-S-phase transition in budding yeast (Mathias et al., 1996). CUL1 acts as the molecular scaffold to constitute the intact, functional SCF E3 ligases (also known as Cullin-RING ligase-1, CRL1) (Sarikas et al., 2011; Zhao and Sun, 2012a). Aberrant expression of CUL1 was found in a number of human cancers which is closely associated with poor patient prognosis (Salon et al., 2007; Bai et al., 2011). Thus, CUL1 and associated SCF E3 ligases appear to be attractive anti-cancer targets (Nalepa et al., 2006; Jia and Sun, 2011; Zhao and Sun, 2012a).
Few studies from the same laboratory were reported to define a pathological role of CUL1 in melanoma. Through the immunohistochemical staining, the authors showed that both cytoplasmic and nuclear CUL1 levels are increased at the early stages of melanoma development (between dysplastic nevi and primary melanoma), but no significant difference was observed between primary and metastatic melanoma (Chen et al., 2010). CUL1 expression was also characterized as one of the significant biomarkers which may be used to discriminate melanoma from dysplastic nevi (Zhang and Li, 2012). On the other hand, CUL1 levels correlate with neither age, gender, tumor thickness, ulceration, tumor subtype and sites, nor the patient survival (Chen et al., 2010). Nevertheless, this correlation/association study should be followed with skin-specific transgenic and knockout mouse models to elucidate potential causal relationship between altered CUL1 and melanomagenesis.
The in vitro cell culture work showed that knockdown of CUL1 inhibits melanoma cell growth by arresting cells at the G1 phase, likely through p27 accumulation, resulting at least in part from blockage of the functional SKP2-SCF E3 ligase (Chen and Li, 2010). However, no rescue experiment via simultaneous knockdown of CUL1 and p27 was performed to confirm the causal role of p27 in this experimental setting. Finally, although it is out of the scope of this review, it is worth noting that skin-specific deletion of Cul-4a, another member of the Cullin family, rendered mice much more resistant to UVB-induced skin carcinogenesis, implying that Cul-4A associated CRL4 E3 ligase complex is also an attractive therapeutic target for skin cancer (Liu et al., 2009).
ROLE OF THE F-BOX PROTEINS IN SKIN CANCER
The F-box proteins are the substrate recognizing subunits of SCF E3 ligases, thus determining the substrate specificity of SCF E3 ligases (Deshaies and Joazeiro, 2009). Although the human genome encodes 69 F-box proteins (Jin et al., 2004), only three F-box proteins are well-defined, namely β-transducin repeats-containing proteins (b-TrCP), S-phase kinase-associated protein 2 (SKP2) and FBXW7. We will focus on these three F-box proteins to review their potential involvement in skin cancer.
β-TrCPs
Human β-TrCP, originally identified as a cellular ubiquitin ligase that is bound by the HIV-1 Vpu viral protein for targeted degradation of cellular CD4 (Margottin et al., 1998), has two family members, β-TrCP1 and β-TrCP2 (Koike et al., 2000). The β-TrCPs are highly conserved from Xenopus to human and contain at the N-terminus an F-box domain which facilitates their binding to SKP1 and CUL1 and at the C-terminus a WD40 repeat domain for substrate recognition (Fuchs et al., 2004). SCFβ-TrCP E3 ligase promotes the degradation of many key regulatory proteins, including IκB, β-catenin, cyclin D1, p53, MCL1, Procaspase-3, WEE1, and CDC25, among many others (Skaar et al., 2009) as well as DEPTOR (Duan et al., 2011; Gao et al., 2011; Zhao et al., 2011), a newly identified naturally occurring inhibitor of mTORC1 and mTORC2 (Peterson et al., 2009). In most cases, β-TrCPs functions as oncogenes, whereas in a few others, they have displayed tumor suppressive functions. These opposite functions are determined by the cell context as well as spatially and temporally dependent degradation of tumor suppressors or onco-proteins (Frescas and Pagano, 2008; Skaar et al., 2009).
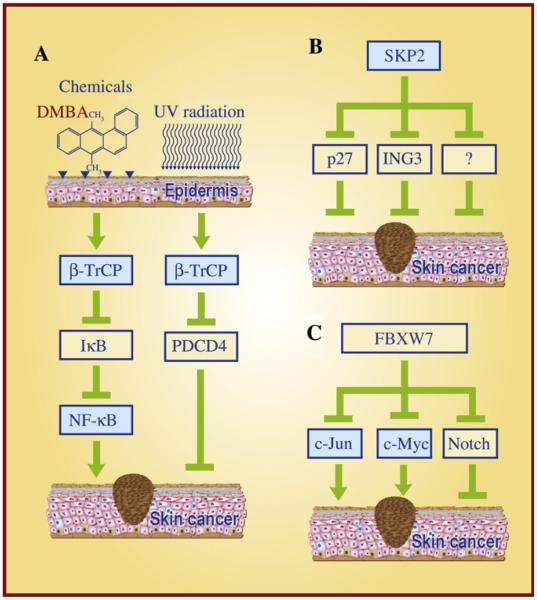
Although neither mutations nor complete loss of expression of β-TrCP were detected in malignant melanomas (Reifenberger et al., 2002) or in cutaneous basal cell carcinomas (Wolter et al., 2003), overexpression of β-TrCP1 (Gu et al., 2007a) and β-TrCP2 (also known as HOS) (Bhatia et al., 2002) was found in DMBA/TPA-induced mouse skin papillomas. Interestingly, overexpression of β-TrCPs coincides with the accelerated degradation of IκB in vivo, suggesting that NF-κB activation mediated by β-TrCPs contributes to skin papillomagenesis (Bhatia et al., 2002; Gu et al., 2007a). Furthermore, TPA exposure decreases the protein levels of tumor suppressor Pdcd4 in mouse skin papillomas and keratinocytes, which appears to be mediated by β-TrCP (Schmid et al., 2008) as well. It has been previously shown that upon phosphorylation by S6K1 or ERK, PDCD4 is recognized by β-TrCP, followed by targeted degradation (Dorrello et al., 2006; Schmid et al., 2008). Thus, targeted degradation of IκBα and PDCD4 by β-TrCP may contribute to the development of skin squamous carcinoma (Fig. 4A).
Fig. 4. The important role of F-box proteins in skin cancer.
A: β-TrCP acts as an oncogenic protein that mediates skin carcinogenesis induced by DMBA/TPA and UVB irradiation, in part via targeting the degradation of IκB to activate NF-κB and promoting PDCD4 destruction to enhance protein synthesis. B: SKP2 acts as an oncogenic protein during skin carcinogenesis largely by targeting degradation of p27, ING3 and other yet-to-be identified tumor suppressors. C: FBXW7 acts as a tumor suppressor by targeting the degradation of the c-Jun and c-Myc oncoproteins, but may act as an oncogenic protein by targeting Notch degradation during skin carcinogenesis.
In a transgenic mouse model, targeted expression of a dominant negative β-TrCP-ΔF in mouse epidermis by K5 promoter attenuates UVB-induced edema, hyperplasia and inflammatory response, and enhances UVB-induced apoptosis in mouse skin, suggesting that β-TrCP confers skin proliferation and apoptosis resistance in response to UVB irradiation (Bhatia et al., 2011a). Consistently, a cell culture study showed that inhibition of β-TrCP by a dominant-negative β-TrCP2-ΔF in immortalized human keratinocytes increases UVB-induced apoptosis in a manner independent of NF-κB and p53 (Bhatia et al., 2008).
In addition to its response to chemical and physical carcinogens, β-TrCP also responds to active oncogenes. Specifically, ectopic expression of oncogenic BRAF (V660E) in melanocytes enhances β-TrCP expression with concomitant increase in IκBα degradation (Liu et al., 2007). Conversely, blockage of BRAF signaling by a small molecular inhibitor or by BRAF siRNA knockdown in melanoma cells reduces β-TrCP expression with subsequent IκBα stabilization and NF-κB inactivation, leading to apoptosis sensitization (Liu et al., 2007). On the other hand, a recent study showed that silymarin, a plant flavonoid derived from Silybum marianum, actively suppresses migration and invasion of human melanoma cells in part through β-TrCP-mediated degradation of β-catenin, suggesting that β-TrCP may act as a negative regulator of migration and invasion of melanoma cells (Vaid et al., 2011). Thus, the role of β-TrCP will depend largely on its targeted substrates. Nevertheless, the studies using both cell culture and transgenic mouse models support the notion that β-TrCP plays a pivotal role in promoting skin carcinogenesis induced by chemical carcinogens and UV irradiation and thereby may serve as a potential anticancer target for skin cancer therapy.
SKP2
SKP2 was first identified in 1995 as the second essential element of the cyclin A-CDK2 S phase kinase (Zhang et al., 1995) and subsequently characterized as an SKP1-binding protein to regulate cell cycle progression through ubiquitinmediated proteolysis (Bai et al., 1996). SKP2 is a typical oncogene, which promotes the S phase entry by targeting p27 degradation (Carrano et al., 1999; Sutterluty et al., 1999; Tsvetkov et al., 1999). SKP2 is overexpressed in many types of human cancers with corresponding reduction of p27 and poor prognosis (Frescas and Pagano, 2008). During human melanomagenesis starting from melanocytic proliferation to melanocytic nevi, then to melanoma in situ and primary melanoma, and finally to metastatic melanoma, a progressive increase of SKP2 and loss of p27 were observed (Li et al., 2004). Consistently, another study showed that reduced expression of p27 is associated with melanoma development and progression (Alonso et al., 2004). However, there is some degree of controversy in the literatures with regard to the significance of subcellular expression of SKP2 in melanomas. Li et al. (2004) reported a progressive increase and decrease of the nuclear SKP2 and p27, respectively, during melanomagenesis from melanocytic nevi to metastatic melanoma. But neither SKP2 nor p27 nuclear expression has significant impact on patient prognosis. However, cytoplasmic expression of SKP2 correlates with worse 10-year overall survival in patients with primary melanoma (Li et al., 2004). In another independent study, Chen et al. (2011) reported that cytoplasmic, but not nuclear, expression of SKP2 was gradually increased during melanomagenesis and correlated with a poorer five-year survival of patients with primary melanoma. In contrast, Woenckhaus et al. (2005) reported that nuclear SKP2 expression is inversely correlated with p27 levels and clinically associated with increasing malignancy and poorer patient survival. Expression of cytoplasmic SKP2, however, was detectable, but decreased during the progression from nevi to superficial melanomas and to nodular melanoma, but increased again in melanoma metastases (Woenckhaus et al., 2005). It is not clear whether the discrepancy is derived from the use of different batches of SKP2 antibodies or different methods in tissue sample preparation. Nevertheless, none of these three studies performed a systematic correlation analysis on the cytoplasmic expression between SKP2 and p27. It is known that unlike nuclear p27, which functions as a tumor suppressor, the SKP2-resistant cytoplasmic p27 acts as an oncogene (Serres et al., 2011). Thus, the role of cytoplasmic SKP2 and p27 in melanomagenesis remains to be determined.
In addition to melanoma, an inverse relationship between SKP2 and p27 can also be found in Merkel cell carcinoma (MCCs), an aggressive form of skin malignancy often associated with virus infections (Bhatia et al., 2011b). Specifically, most MCCs with increased SKP2 have a decreased p27, and vice versa (Erickson et al., 2003), suggesting a SKP2 involvement in Merkel cell tumorigenesis. Likewise, in Kaposi’s sarcoma with cutaneous lesions, SKP2 nuclear overexpression was found in all stages of this deadly disease with significantly higher levels in skin tumors (Penin et al., 2002). Consistently, the progression of Kaposi’s sarcoma is associated with decreased p27, although increased SKP2 is not always directly correlated with p27 reduction (Penin et al., 2002). Nevertheless, this finding suggests SKP2’s involvement in Kaposi’s sarcoma progression. Finally, in cutaneous T cell lymphomas, an increased SKP2 and decreased p27 was reported with the mechanism involving upregulation of SKP2 by mutant pro-IL-16 (Curiel-Lewandrowski et al., 2011).
Mechanistic studies using the in vitro cell culture models were performed to validate the critical role of the SKP2-p27 axis in the growth and invasion of melanoma cells. Simultaneous siRNA-based knockdown of SKP2 and mutant BRAF increases the p27 levels and effectively inhibits the growth and invasion of melanoma cells (Sumimoto et al., 2006). Similarly, siRNA knockdown of SKP2 causes p27 accumulation and suppresses the growth of melanoma cells in monolayer culture and in nude mice (Katagiri et al., 2006). In addition to p27, SKP2 was found to target the degradation of ING3 (inhibitor of growth family member 3), a putative tumor suppressor whose expression is found to be reduced remarkably in melanomas, which correlated with poorer patient survival (Wang et al., 2007). Mechanistically, SKP2, but not β-TrCP, binds to ING3 and promotes its ubiquitylation and degradation. SKP2 knockdown stabilizes ING3 by blocking its degradation in melanoma cells, leading to a G1 arrest and sensitization to UVB-induced apoptosis. Finally, SKP2 was found to be necessary for Myc-induced keratinocyte proliferation in an in vivo study using the K5-Myc-transgenic mice in the SKP2-null genetic background (Sistrunk et al., 2011). Taken together, these findings indicate that the SKP2-mediated reduction, as a result of enhanced degradation of p27, ING3, and other yet-to-be identified tumor suppressors, likely contributes to the development of melanoma (Fig. 4B).
FBXW7
FBXW7 (also known as Sel-10, hCdc4, hAgo, or Fbw7), another well-studied F-box protein, was first identified in budding yeast in 1973, designated as Cdc4 (Hartwell et al., 1973). FBXW7 is classified as a tumor suppressor based upon the following observations: (1) almost all FBXW7 substrates are oncogenic proteins, including c-Myc, c-Jun, cyclin E, and Notch (Welcker and Clurman, 2008) with one exception being the tumor suppressor NF1 (neurofibromatosis type 1) (Tan et al., 2011b); (2) mutations and deletions of FBXW7 were found in many human cancers (Akhoondi et al., 2007) (Welcker and Clurman, 2008; Wang et al., 2012); and (3) Fbxw7 is a p53-dependent haploinsufficient tumor suppressor in mice (Mao et al., 2004).
FBXW7 mutations or deletions have not been reported in human skin cancer. However, a recent study showed that an allele-specific deletion of Fbxw7 can be detected in mouse skin tumors induced by DMAB/TPA (Perez-Losada et al., 2012). Furthermore, in this DMBA/TPA skin carcinogenesis study, mice with the p53+/−;Fbxw7+/− background developed many more papillomas in number than those with p53+/−;Fbxw7+/+ genetic background, clearly demonstrating that heterozygous deletion of Fbxw7 increases the susceptibility to papilloma development (Perez-Losada et al., 2012).
Oncoprotein c-Jun is required for the development of skin cancer and can be stabilized by UV radiation (Anzi et al., 2008). It has been recently shown that UV exposure reduces Fbxw7α transcripts, which is closely correlated with c-Jun induction. Consistently, blockage of Fbxw7 UV-responsiveness abrogates c-Jun induction by UV, whereas knockdown of Fbxw7 increases c-Jun basal levels (Anzi et al., 2008). Thus, UV-induced skin cancer could be mediated in part by inhibition of Fbxw7 expression, followed by c-Jun stabilization. A recent study showed that Fbxw7 is involved in c-Myc regulation by Vitamin D. Specifically, ablation of Fbxw7 blocks c-Myc degradation mediated by the Vitamin D receptor and promotes hyper-proliferation of skin epithelia (Salehi-Tabar et al., 2012).
Additionally, it is worthy of mention that Notch, a well-characterized Fbxw7 substrate, is an oncogenic protein in various tissues but may serve as a tumor suppressor in skin (Nicolas et al., 2003; Koch and Radtke, 2007). Deregulated expression of Notch receptors and ligands is observed in skin cancer, including squamous cell carcinomas of the head and neck and metastatic melanoma (Miele, 2006). Integrin-linked kinase (ILK) was found to phosphorylate Notch1-IC (Notch1 intracellular domain) and facilitate its proteasomal degradation by Fbxw7 (Mo et al., 2007). The same study also showed that Notch1-IC is down-regulated, whereas ILK is upregulated in melanoma and basal cell carcinoma (Mo et al., 2007). Finally, a very recent study showed that Fbxw7 regulates the proliferation and differentiation of keratinocytes by promoting the degradation of oncogenic c-Myc and tumor suppressive Notch. Although loss of Fbxw7 fails to predispose keratinocytes to the formation of squamous cell carcinoma upon Ras expression, inactivation of the Notch signaling does confer a more aggressive tumorigenic phenotype (Ishikawa et al., 2012). Taken together, it appears that Fbxw7 could either suppress or promote skin carcinogenesis by targeted degradation of c-Jun and c-Myc or Notch, respectively (Fig. 4C). The fact that partial loss of Fbxw7 under the p53 heterozygous background did accelerate skin tumor formation in the DMBA-TPA skin tumor model supports the notion that Fbxw7 is a p53-dependent tumor suppressor, at least in the mouse skin experimental setting (Perez-Losada et al., 2012).
CONCLUSION AND FURTHER PERSPECTIVES
In summary, the limited studies so far have been conducted to elucidate the critical role of SCF E3 ubiquitin ligases in skin carcinogenesis. Most available data come from the studies on SAG/RBX2 and three most known F-box proteins, β-TrCP, SKP2 and Fbxw7. Limited mouse transgenic (SAG and β-TrCP2ΔF) (Gu et al., 2007a; Bhatia et al., 2011a) and knockout (Skp2−/− and Fbxw7+/−) (Sistrunk et al., 2011; Perez-Losada et al., 2012) models were used in skin carcinogenesis induced by either DMBA/TPA or UVB irradiation. With regard to the human skin tumor tissues, most studies are limited to immunohistochemistry staining to define a potential correlation in expression of SCF E3 components and their limited substrates (mainly SKP2 vs. p27) in different stages of skin cancer, particularly during melanomagenesis. Some cell line studies were conducted to reveal potential molecular mechanisms. Overall, some components of SCF E3 ligases are overexpressed to activate SCF E3 ligases during skin carcinogenesis. Thus, SCF E3 ligases may serve as attractive targets for the treatment of skin cancer. As a matter of fact, MLN4924, a small molecule inhibitor of NEDD8 activating enzyme that inhibits SCF E3 ligases by preventing Cullin-1 neddylation (Soucy et al., 2009; Brownell et al., 2010), is currently in Phase I clinical trials for the treatment of several human malignancies, including melanoma (Soucy et al., 2010; Nawrocki et al., 2012). Multiple preclinical studies showed that MLN4924 effectively suppresses the growth of human cancer cells via (1) inducing apoptosis mainly seen in leukemia and lymphoma as well as in several solid tumor lines through accumulation of IκBα to inactivate NFκB (Soucy et al., 2009; Milhollen et al., 2010; Swords et al., 2010; Milhollen et al., 2011; Tan et al., 2011a; Luo et al., 2012); (2) inducing senescence seen in various solid tumor lines through p21 accumulation (Lin et al., 2010a; Lin et al., 2010b; Jia et al., 2011); and (3) inducing autophagy in various solid tumor lines through accumulation of DEPTOR and HIF1α to block the mTOR pathway (Luo et al., 2012; Zhao and Sun, 2012b; Zhao et al., 2012).
Future studies in this area of research should be directed to the following aspects:
To further characterize the causal role of SCF components, particularly F-box proteins, in skin carcinogenesis: new mouse models should be established to target relevant components for skin specific transgenic expression (gain-of-function) and knockout (loss-of-function), ideally in an inducible and tissue-specific manner. These models can be used in the studies of skin carcinogenesis, triggered by chemical carcinogens (e.g., DMBA/TPA) or UVB irradiation to understand the causal relationship under the physiological or pathological settings. The models will also be useful for potential chemoprevention and therapeutic intervention studies.
To generate compound mice in which manipulation of a SCF E3 component is combined with the activation of an oncogene (such as Kras) or inactivation of a tumor suppressor (such as p53 or Pten loss): the majority of individual components of SCF E3s may not be a bona fide dominant tumor suppressor or oncogene by itself, but they may cooperate with dominant oncogenes or tumor suppressors to regulate skin tumorigenesis. For example, our recent work showed that Sag regulates embryonic vasculogenesis (Tan et al., 2011b) and skin carcinogenesis (Gu et al., 2007a). It would be of great interest to determine if Sag regulates tumor angiogenesis by generating a Sag skin-specific knockout mouse model in combination with a K14-HPV16 E6/E7 transgenic model, in which skin-specific expression of HPV16 E6 and E7 oncogenes inactivate both the p53 and Rb tumor suppressors (Munger et al., 1992) in epidermal cells to induce the formation of angiogenic invasive squamous cell carcinoma (Arbeit et al., 1994; Coussens et al., 1996; Bergers et al., 1998; Ribatti et al., 2007).
To further define the component(s) of SCF E3 ligases or their substrates related to skin cancer as potential biomarkers for early detection and prognosis prediction: current work is much limited to the SKP2-p27 axis in various human skin cancers, particularly in melanoma. Given the fact that the cancer occurs in the skin surface, it is advantageous to take skin biopsies for individualized diagnosis and therapy, if such biomarkers can be identified and developed in the near future.
Finally, to test clinical effectiveness of MLN4924, an indirect inhibitor of SCF E3 ligases (Brownell et al., 2010) currently in Phase I clinical trials (Soucy et al., 2010; Nawrocki et al., 2012), as a promising chemopreventive or therapeutic agent against skin cancer using the mouse models established above: furthermore, although it is a daunting task to discover the small molecules that inactivate SCF E3 ligases via disrupting the protein-protein interaction (e.g., binding of F-box proteins and their substrates; binding of E2 and E3, or binding among individual components) (Nalepa et al., 2006; Sun, 2006; Jia and Sun, 2011; Zhao and Sun, 2012a), the progress has been made with identification of several such inhibitors (Chen et al., 2008; Aghajan et al., 2010; Orlicky et al., 2010). Future efforts should be devoted to the development of these inhibitors or the discovery of more potent novel inhibitors with an ultimate goal to treat human cancer, including skin cancer, via targeting SCF E3 ligases.
ACKNOWLEDGEMENTS
This work was supported by the National Cancer Institute grants (Nos. CA118762, CA156744, CA170995 and CA171277) to Y.S. and the National Institute of General Medical Sciences grant (No. GM094777) to W.W.
REFERENCES
- Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, Damoiseaux R, Hao R, Del Moral PM, Verma R, Li Y, Li C, Houk KN, Jung ME, Zheng N, Huang L, Deshaies RJ, Kaiser P, Huang J. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat. Biotechnol. 2010;28:738–742. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, Mueller-Holzner E, Corcoran M, Dagnell M, Nejad SZ, Nayer BN, Zali MR, Hansson J, Egyhazi S, Petersson F, Sangfelt P, Nordgren H, Grander D, Reed SI, Widschwendter M, Sangfelt O, Spruck C. Fbxw7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007;67:9006–9012. doi: 10.1158/0008-5472.CAN-07-1320. [DOI] [PubMed] [Google Scholar]
- Alonso SR, Ortiz P, Pollan M, Perez-Gomez B, Sanchez L, Acuna MJ, Pajares R, Martinez-Tello FJ, Hortelano CM, Piris MA, Rodriguez-Peralto JL. Progression in cutaneous malignant melanoma is associated with distinct expression profiles: a tissue microarray-based study. Am. J. Pathol. 2004;164:193–203. doi: 10.1016/s0002-9440(10)63110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzi S, Finkin S, Shaulian E. Transcriptional repression of c-Jun’s E3 ubiquitin ligases contributes to c-Jun induction by UV. Cell Signal. 2008;20:862–871. doi: 10.1016/j.cellsig.2007.12.020. [DOI] [PubMed] [Google Scholar]
- Arbeit JM, Munger K, Howley PM, Hanahan D. Progressive squamous epithelial neoplasia in K14-human papillomavirus type 16 transgenic mice. J. Virol. 1994;68:4358–4368. doi: 10.1128/jvi.68.7.4358-4368.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Bai J, Zhou Y, Chen G, Zeng J, Ding J, Tan Y, Zhou J, Li G. Overexpression of Cullin1 is associated with poor prognosis of patients with gastric cancer. Hum. Pathol. 2011;42:375–383. doi: 10.1016/j.humpath.2010.09.003. [DOI] [PubMed] [Google Scholar]
- Bergers G, Hanahan D, Coussens LM. Angiogenesis and apoptosis are cellular parameters of neoplastic progression in transgenic mouse models of tumorigenesis. Int. J. Dev. Biol. 1998;42:995–1002. [PubMed] [Google Scholar]
- Bhatia N, Demmer TA, Sharma AK, Elcheva I, Spiegelman VS. Role of beta-TrCP ubiquitin ligase receptor in UVB mediated responses in skin. Arch. Biochem. Biophys. 2011a;508:178–184. doi: 10.1016/j.abb.2010.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia N, Demmer TA, Spiegelman VS. Inhibition of beta-TrCP function potentiates UVB-induced apoptosis in hTERT-immortalized normal human keratinocytes. Photochem. Photobiol. 2008;84:376–381. doi: 10.1111/j.1751-1097.2007.00272.x. [DOI] [PubMed] [Google Scholar]
- Bhatia N, Herter JR, Slaga TJ, Fuchs SY, Spiegelman VS. Mouse homologue of HOS (mHOS) is overexpressed in skin tumors and implicated in constitutive activation of NF-kappaB. Oncogene. 2002;21:1501–1509. doi: 10.1038/sj.onc.1205311. [DOI] [PubMed] [Google Scholar]
- Bhatia S, Afanasiev O, Nghiem P. Immunobiology of Merkel cell carcinoma: implications for immunotherapy of a polyomavirus-associated cancer. Curr. Oncol. Rep. 2011b;13:488–497. doi: 10.1007/s11912-011-0197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol. Cell. 2010;37:102–111. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]
- Carrano AC, Eytan E, Hershko A, Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Chen G, Cheng Y, Martinka M, Li G. Cul1 expression is increased in early stages of human melanoma. Pigment Cell Melanoma Res. 2010;23:572–574. doi: 10.1111/j.1755-148X.2010.00725.x. [DOI] [PubMed] [Google Scholar]
- Chen G, Cheng Y, Zhang Z, Martinka M, Li G. Cytoplasmic Skp2 expression is increased in human melanoma and correlated with patient survival. PLoS ONE. 2011;6:e17578. doi: 10.1371/journal.pone.0017578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Li G. Increased Cul1 expression promotes melanoma cell proliferation through regulating p27 expression. Int. J. Oncol. 2010;37:1339–1344. doi: 10.3892/ijo_00000786. [DOI] [PubMed] [Google Scholar]
- Chen Q, Xie W, Kuhn DJ, Voorhees PM, Lopez-Girona A, Mendy D, Corral LG, Krenitsky VP, Xu W, Moutouh-de Parseval L, Webb DR, Mercurio F, Nakayama KI, Nakayama K, Orlowski RZ. Targeting the p27 E3 ligase SCF(Skp2) results in p27- and Skp2-mediated cell-cycle arrest and activation of autophagy. Blood. 2008;111:4690–4699. doi: 10.1182/blood-2007-09-112904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Clifford SC, Astuti D, Hooper L, Maxwell PH, Ratcliffe PJ, Maher ER. The pVHL-associated SCF ubiquitin ligase complex: molecular genetic analysis of elongin B and C, Rbx1 and HIF-1αlpha in renal cell carcinoma. Oncogene. 2001;20:5067–5074. doi: 10.1038/sj.onc.1204602. [DOI] [PubMed] [Google Scholar]
- Colburn NH, Former BF, Nelson KA, Yuspa SH. Tumour promoter induces anchorage independence irreversibly. Nature. 1979;281:589–591. doi: 10.1038/281589a0. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Hanahan D, Arbeit JM. Genetic predisposition and parameters of malignant progression in K14-HPV16 transgenic mice. Am. J. Pathol. 1996;149:1899–1917. [PMC free article] [PubMed] [Google Scholar]
- Curiel-Lewandrowski C, Yamasaki H, Si CP, Jin X, Zhang Y, Richmond J, Tuzova M, Wilson K, Sullivan B, Jones D, Ryzhenko N, Little F, Kupper TS, Center DM, Cruikshank WW. Loss of nuclear pro-IL-16 facilitates cell cycle progression in human cutaneous T cell lymphoma. J. Clin. Invest. 2011;121:4838–4849. doi: 10.1172/JCI41769. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- D’Andrea A, Pellman D. Deubiquitinating enzymes: a new class of biological regulators. Crit. Rev. Biochem. Mol. Biol. 1998;33:337–352. doi: 10.1080/10409239891204251. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dorrello NV, Peschiaroli A, Guardavaccaro D, Colburn NH, Sherman NE, Pagano M. S6K1- and betaTRCP-mediated degradation of PDCD4 promotes protein translation and cell growth. Science. 2006;314:467–471. doi: 10.1126/science.1130276. [DOI] [PubMed] [Google Scholar]
- Duan H, Wang Y, Aviram M, Swaroop M, Loo JA, Bian J, Tian Y, Mueller T, Bisgaier CL, Sun Y. SAG, a novel zinc RING finger protein that protects cells from apoptosis induced by redox agents. Mol. Cell Biol. 1999;19:3145–3155. doi: 10.1128/mcb.19.4.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Skaar JR, Kuchay S, Toschi A, Kanarek N, Ben-Neriah Y, Pagano M. mTOR generates an auto-amplification loop by triggering the betaTrCP- and CK1alpha-dependent degradation of DEPTOR. Mol. Cell. 2011;44:317–324. doi: 10.1016/j.molcel.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einspahr JG, Bowden GT, Alberts DS. Skin cancer chemoprevention: strategies to save our skin. Recent Results Cancer Res. 2003;163:151–164. doi: 10.1007/978-3-642-55647-0_14. discussion 264–266. [DOI] [PubMed] [Google Scholar]
- Erickson LA, Papotti M, Volante M, Jin L, Lewis JE, Lloyd RV. Merkel cell carcinomas: expression of S-phase kinase-associated protein 2 (Skp2), p27, and proliferation markers. Endocr. Pathol. 2003;14:221–229. doi: 10.1007/s12022-003-0014-2. [DOI] [PubMed] [Google Scholar]
- Frescas D, Pagano M. Deregulated proteolysis by the F-box proteins SKP2 and beta-TrCP: tipping the scales of cancer. Nat. Rev. Cancer. 2008;8:438–449. doi: 10.1038/nrc2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SY, Spiegelman VS, Kumar KG. The many faces of beta-TrCP E3 ubiquitin ligases: reflections in the magic mirror of cancer. Oncogene. 2004;23:2028–2036. doi: 10.1038/sj.onc.1207389. [DOI] [PubMed] [Google Scholar]
- Gao D, Inuzuka H, Tan MK, Fukushima H, Locasale JW, Liu P, Wan L, Zhai B, Chin YR, Shaik S, Lyssiotis CA, Gygi SP, Toker A, Cantley LC, Asara JM, Harper JW, Wei W. mTOR drives its own activation via SCF(betaTrCP)-dependent degradation of the mTOR inhibitor DEPTOR. Mol. Cell. 2011;44:290–303. doi: 10.1016/j.molcel.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Bowden GT, Normolle D, Sun Y. SAG/ROC2 E3 ligase regulates skin carcinogenesis by stage-dependent targeting of c-Jun/AP1 and IkappaB-alpha/NF-kappaB. J. Cell Biol. 2007a;178:1009–1023. doi: 10.1083/jcb.200612067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Q, Tan M, Sun Y. SAG/ROC2/Rbx2 is a novel activator protein-1 target that promotes c-Jun degradation and inhibits 12-O-tetradecanoylphorbol-13-acetate-induced neoplastic transformation. Cancer Res. 2007b;67:3616–3625. doi: 10.1158/0008-5472.CAN-06-4020. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Mortimer RK, Culotti J, Culotti M. Genetic control of the cell division cycle in yeast: V. Genetic analysis of cdc mutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Gu Q, Zheng M, Normolle D, Sun Y. SAG/ROC2/RBX2 E3 ligase promotes UVB-induced skin hyperplasia, but not skin tumors, by simultaneously targeting c-Jun/AP-1 and p27. Carcinogenesis. 2008;29:858–865. doi: 10.1093/carcin/bgn021. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Bio-chem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa Y, Hosogane M, Okuyama R, Aoyama S, Onoyama I, Nakayama KI, Nakayama K. Opposing functions of Fbxw7 in keratinocyte growth, differentiation and skin tumorigenesis mediated through negative regulation of c-Myc and Notch. Oncogene. 2012 doi: 10.1038/onc.2012.213. http://dx.doi.org/10.1038/onc.2012.213. [DOI] [PubMed] [Google Scholar]
- Jia L, Li H, Sun Y. Induction of p21-dependent senescence by an NAE inhibitor, MLN4924, as a mechanism of growth suppression. Neoplasia. 2011;13:561–569. doi: 10.1593/neo.11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Soengas MS, Sun Y. ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res. 2009;69:4974–4982. doi: 10.1158/0008-5472.CAN-08-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Sun Y. SCF E3 ubiquitin ligases as anticancer targets. Curr. Cancer Drug Targets. 2011;11:347–356. doi: 10.2174/156800911794519734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Yang J, Hao X, Zheng M, He H, Xiong X, Xu L, Sun Y. Validation of SAG/RBX2/ROC2 E3 ubiquitin ligase as an anticancer and radiosensitizing target. Clin. Cancer Res. 2010;16:814–824. doi: 10.1158/1078-0432.CCR-09-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13:2928–2933. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri Y, Hozumi Y, Kondo S. Knockdown of Skp2 by siRNA inhibits melanoma cell growth in vitro and in vivo. J. Dermatol. Sci. 2006;42:215–224. doi: 10.1016/j.jdermsci.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. Cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Koch U, Radtke F. Notch and cancer: a double-edged sword. Cell. Mol. Life Sci. 2007;64:2746–2762. doi: 10.1007/s00018-007-7164-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike J, Sagara N, Kirikoshi H, Takagi A, Miwa T, Hirai M, Katoh M. Molecular cloning and genomic structure of the betaTRCP2 gene on chromosome 5q35.1. Biochem. Biophys. Res. Commun. 2000;269:103–109. doi: 10.1006/bbrc.2000.2241. [DOI] [PubMed] [Google Scholar]
- Leiter U, Garbe C. Epidemiology of melanoma and nonmelanoma skin cancerethe role of sunlight. Adv. Exp. Med. Biol. 2008;624:89–103. doi: 10.1007/978-0-387-77574-6_8. [DOI] [PubMed] [Google Scholar]
- Li Q, Murphy M, Ross J, Sheehan C, Carlson JA. Skp2 and p27kip1 expression in melanocytic nevi and melanoma: an inverse relationship. J. Cutan. Pathol. 2004;31:633–642. doi: 10.1111/j.0303-6987.2004.00243.x. [DOI] [PubMed] [Google Scholar]
- Lin HK, Chen Z, Wang G, Nardella C, Lee SW, Chan CH, Yang WL, Wang J, Egia A, Nakayama KI, Cordon-Cardo C, Teruya-Feldstein J, Pandolfi PP. Skp2 targeting suppresses tumorigenesis by Arf-p53-independent cellular senescence. Nature. 2010a;464:374–379. doi: 10.1038/nature08815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res. 2010b;70:10310–10320. doi: 10.1158/0008-5472.CAN-10-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Suresh Kumar KG, Yu D, Molton SA, McMahon M, Herlyn M, Thomas-Tikhonenko A, Fuchs SY. Oncogenic BRAF regulates beta-Trcp expression and NF-kappaB activity in human melanoma cells. Oncogene. 2007;26:1954–1958. doi: 10.1038/sj.onc.1209994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Lee S, Zhang J, Peters SB, Hannah J, Zhang Y, Yin Y, Koff A, Ma L, Zhou P. CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol. Cell. 2009;34:451–460. doi: 10.1016/j.molcel.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Z, Yu G, Lee HW, Li L, Wang L, Yang D, Pan Y, Ding C, Qian J, Wu L, Chu Y, Yi J, Wang X, Sun Y, Jeong LS, Liu J, Jia L. The Nedd8-activating enzyme inhibitor MLN4924 induces autophagy and apoptosis to suppress liver cancer cell growth. Cancer Res. 2012;72:3360–3371. doi: 10.1158/0008-5472.CAN-12-0388. [DOI] [PubMed] [Google Scholar]
- Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, Brown K, Bryson S, Balmain A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell. 1998;1:565–574. doi: 10.1016/s1097-2765(00)80056-8. [DOI] [PubMed] [Google Scholar]
- Mathias N, Johnson SL, Winey M, Adams AE, Goetsch L, Pringle JR, Byers B, Goebl MG. Cdc53p acts in concert with Cdc4p and Cdc34p to control the G1-to-S-phase transition and identifies a conserved family of proteins. Mol. Cell. Biol. 1996;16:6634–6643. doi: 10.1128/mcb.16.12.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele L. Notch signaling. Clin. Cancer Res. 2006;12:1074–1079. doi: 10.1158/1078-0432.CCR-05-2570. [DOI] [PubMed] [Google Scholar]
- Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 2011;71:3042–3051. doi: 10.1158/0008-5472.CAN-10-2122. [DOI] [PubMed] [Google Scholar]
- Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, Manfredi M, Narayanan U, Rolfe M, Staudt LM, Soucy TA, Yu J, Zhang J, Bolen JB, Smith PG. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-kappaB-dependent lymphoma. Blood. 2010;116:1515–1523. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]
- Mo JS, Kim MY, Han SO, Kim IS, Ann EJ, Lee KS, Seo MS, Kim JY, Lee SC, Park JW, Choi EJ, Seong JY, Joe CO, Faessler R, Park HS. Integrin-linked kinase controls Notch1 signaling by down-regulation of protein stability through Fbw7 ubiquitin ligase. Mol. Cell. Biol. 2007;27:5565–5574. doi: 10.1128/MCB.02372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger K, Scheffner M, Huibregtse JM, Howley PM. Interactions of HPV E6 and E7 oncoproteins with tumour suppressor gene products. Cancer Surv. 1992;12:197–217. [PubMed] [Google Scholar]
- Nai G, Marques M. Role of ROC1 protein in the control of cyclin D1 protein expression in skin melanomas. Pathol. Res. Pract. 2011;207:174–181. doi: 10.1016/j.prp.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 2006;6:369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- Nalepa G, Rolfe M, Harper JW. Drug discovery in the ubiquitin-proteasome system. Nat. Rev. Drug Discov. 2006;5:596–613. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- Nawrocki ST, Griffin P, Kelly KR, Carew JS. MLN4924: a novel first-in-class inhibitor of NEDD8-activating enzyme for cancer therapy. Expert Opin. Investig. Drugs. 2012;21:1563–1573. doi: 10.1517/13543784.2012.707192. [DOI] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer JA, Mill P, van Noort M, Hui CC, Clevers H, Dotto GP, Radtke F. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat. Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penin RM, Fernandez-Figueras MT, Puig L, Rex J, Ferrandiz C, Ariza A. Over-expression of p45(SKP2) in Kaposi’s sarcoma correlates with higher tumor stage and extracutaneous involvement but is not directly related to p27(KIP1) down-regulation. Mod. Pathol. 2002;15:1227–1235. doi: 10.1097/01.MP.0000036589.99516.D6. [DOI] [PubMed] [Google Scholar]
- Perez-Losada J, Wu D, DelRosario R, Balmain A, Mao JH. Allele-specific deletions in mouse tumors identify Fbxw7 as germline modifier of tumor susceptibility. PLoS ONE. 2012;7:e31301. doi: 10.1371/journal.pone.0031301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, Gray NS, Sabatini DM. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifenberger J, Knobbe CB, Wolter M, Blaschke B, Schulte KW, Pietsch T, Ruzicka T, Reifenberger G. Molecular genetic analysis of malignant melanomas for aberrations of the WNT signaling pathway genes CTNNB1, APC, ICAT and BTRC. Int. J. Cancer. 2002;100:549–556. doi: 10.1002/ijc.10512. [DOI] [PubMed] [Google Scholar]
- Ribatti D, Nico B, Crivellato E, Roccaro AM, Vacca A. The history of the angiogenic switch concept. Leukemia. 2007;21:44–52. doi: 10.1038/sj.leu.2404402. [DOI] [PubMed] [Google Scholar]
- Rosso S, Zanetti R, Pippione M, Sancho-Garnier H. Parallel risk assessment of melanoma and basal cell carcinoma: skin characteristics and sun exposure. Melanoma Res. 1998;8:573–583. doi: 10.1097/00008390-199812000-00013. [DOI] [PubMed] [Google Scholar]
- Saladi RN, Persaud AN. The causes of skin cancer: a comprehensive review. Drugs Today (Barc) 2005;41:37–53. doi: 10.1358/dot.2005.41.1.875777. [DOI] [PubMed] [Google Scholar]
- Salehi-Tabar R, Nguyen-Yamamoto L, Tavera-Mendoza LE, Quail T, Dimitrov V, An BS, Glass L, Goltzman D, White JH. Vitamin D receptor as a master regulator of the c-MYC/MXD1 network. Proc. Natl. Acad. Sci. USA. 2012;109:18827–18832. doi: 10.1073/pnas.1210037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salon C, Brambilla E, Brambilla C, Lantuejoul S, Gazzeri S, Eymin B. Altered pattern of Cul-1 protein expression and neddylation in human lung tumours: relationships with CAND1 and cyclin E protein levels. J. Pathol. 2007;213:303–310. doi: 10.1002/path.2223. [DOI] [PubMed] [Google Scholar]
- Sarikas A, Hartmann T, Pan ZQ. The cullin protein family. Genome Biol. 2011;12:22. doi: 10.1186/gb-2011-12-4-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid T, Jansen AP, Baker AR, Hegamyer G, Hagan JP, Colburn NH. Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res. 2008;68:1254–1260. doi: 10.1158/0008-5472.CAN-07-1719. [DOI] [PubMed] [Google Scholar]
- Seol JH, Feldman RMR, Zachariae WZ, Shevchenko A, Correll CC, Lyapina S, Chi Y, Galova M, Claypool J, Sandmeyer S, Nasmyth K, Shevchenko A, Deshaies RJ. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serres MP, Zlotek-Zlotkiewicz E, Concha C, Gurian-West M, Daburon V, Roberts JM, Besson A. Cytoplasmic p27 is oncogenic and cooperates with Ras both in vivo and in vitro. Oncogene. 2011;30:2846–2058. doi: 10.1038/onc.2011.9. [DOI] [PubMed] [Google Scholar]
- Siegel R, DeSantis C, Virgo K, Stein K, Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, Lin C, Leach C, Cannady RS, Cho H, Scoppa S, Hachey M, Kirch R, Jemal A, Ward E. Cancer treatment and survivorship statistics, 2012. CA Cancer J. Clin. 2012;62:220–241. doi: 10.3322/caac.21149. [DOI] [PubMed] [Google Scholar]
- Sistrunk C, Macias E, Nakayama K, Kim Y, Rodriguez-Puebla ML. Skp2 is necessary for Myc-induced keratinocyte proliferation but dispensable for Myc oncogenic activity in the oral epithelium. Am. J. Pathol. 2011;178:2470–2477. doi: 10.1016/j.ajpath.2011.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar JR, D’Angiolella V, Pagan JK, Pagano M. SnapShot: F box proteins II. Cell. 2009;137:1358 e1. doi: 10.1016/j.cell.2009.05.040. [DOI] [PubMed] [Google Scholar]
- Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. The NEDD8 Conjugation Pathway and Its Relevance in Cancer Biology and Therapy. Genes Cancer. 2010;1:708–716. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Sumimoto H, Hirata K, Yamagata S, Miyoshi H, Miyagishi M, Taira K, Kawakami Y. Effective inhibition of cell growth and invasion of melanoma by combined suppression of BRAF (V599E) and Skp2 with lentiviral RNAi. Int. J. Cancer. 2006;118:472–476. doi: 10.1002/ijc.21286. [DOI] [PubMed] [Google Scholar]
- Sun Y. E3 ubiquitin ligases as cancer targets and biomarkers. Neoplasia. 2006;8:645–654. doi: 10.1593/neo.06376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Hegamyer G, Colburn NH. Molecular cloning of five messenger RNAs differentially expressed in preneoplastic or neoplastic JB6 mouse epidermal cells: one is homologous to human tissue inhibitor of metalloproteinases-3. Cancer Res. 1994;54:1139–1144. [PubMed] [Google Scholar]
- Sun Y, Li H. Functional characterization of SAG/RBX2/ROC2/RNF7, an antioxidant protein and an E3 ubiquitin ligase. Protein Cell. 2012 doi: 10.1007/s13238-012-2105-7. http://dx.doi.org/10.1007/s13238-012-2105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Tan M, Duan H, Swaroop M. SAG/ROC/Rbx/Hrt, a zinc RING finger gene family: molecular cloning, biochemical properties, and biological functions. Antioxid. Redox Signal. 2001;3:635–650. doi: 10.1089/15230860152542989. [DOI] [PubMed] [Google Scholar]
- Sutterluty H, Chatelain E, Marti A, Wirbelauer C, Senften M, Muller U, Krek W. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- Swaroop M, Bian J, Aviram M, Duan H, Bisgaier CL, Loo JA, Sun Y. Expression, purification, and biochemical characterization of SAG, a RING finger redox sensitive protein. Free Radic. Biol. Med. 1999;27:193–202. doi: 10.1016/s0891-5849(99)00078-7. [DOI] [PubMed] [Google Scholar]
- Swaroop M, Wang Y, Miller P, Duan H, Jatkoe T, Madore S, Sun Y. Yeast homolog of human SAG/ROC2/Rbx2/Hrt2 is essential for cell growth, but not for germination: Chip profiling implicates its role in cell cycle regulation. Oncogene. 2000;19:2855–2866. doi: 10.1038/sj.onc.1203635. [DOI] [PubMed] [Google Scholar]
- Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O’Dwyer M, Nawrocki ST, Giles FJ, Carew JS. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood. 2010;115:3796–3800. doi: 10.1182/blood-2009-11-254862. [DOI] [PubMed] [Google Scholar]
- Tan M, Gallegos JR, Gu Q, Huang Y, Li J, Jin Y, Lu H, Sun Y. SAG/ROC-SCFbeta-TrCP E3 ubiquitin ligase promotes pro-caspase-3 degradation as a mechanism of apoptosis protection. Neoplasia. 2006;8:1042–1054. doi: 10.1593/neo.06568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Gu Q, He H, Pamarthy D, Semenza GL, Sun Y. SAG/ROC2/RBX2 is a HIF-1 target gene that promotes HIF-1αlpha ubiquitination and degradation. Oncogene. 2008;27:1404–1411. doi: 10.1038/sj.onc.1210780. [DOI] [PubMed] [Google Scholar]
- Tan M, Li Y, Yang R, Xi N, Sun Y. Inactivation of SAG E3 ubiquitin ligase blocks embryonic stem cell differentiation and sensitizes leukemia cells to retinoid acid. PLoS ONE. 2011a;6:e27726. doi: 10.1371/journal.pone.0027726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Zhao Y, Kim SJ, Liu M, Jia L, Saunders TL, Zhu Y, Sun Y. SAG/RBX2/ROC2 E3 ubiquitin ligase is essential for vascular and neural development by targeting NF1 for degradation. Dev. Cell. 2011b;21:1062–1076. doi: 10.1016/j.devcel.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Zhu Y, Kovacev J, Zhao Y, Pan ZQ, Spitz DR, Sun Y. Disruption of Sag/Rbx2/Roc2 induces radiosensitization by increasing ROS levels and blocking NF-κB activation in mouse embryonic stem cells. Free Radic. Biol. Med. 2010;49:976–983. doi: 10.1016/j.freeradbiomed.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, Pan Z-Q. Recruitment of a ROC1-CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of IκBα. Mol. Cell. 1999;3:527–533. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- Tsvetkov LM, Yeh K-H, Lee S-J, Sun H, Zhang H. p27kip1ubiquitination and degradation is regulated by the SCFskp2 complex through phosphorylated Thr187 in p27. Curr. Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- Vaid M, Prasad R, Sun Q, Katiyar SK. Silymarin targets beta-catenin signaling in blocking migration/invasion of human melanoma cells. PLoS ONE. 2011;6:e23000. doi: 10.1371/journal.pone.0023000. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Y, Dai DL, Martinka M, Li G. Prognostic significance of nuclear ING3 expression in human cutaneous melanoma. Clin. Cancer Res. 2007;13:4111–4116. doi: 10.1158/1078-0432.CCR-07-0408. [DOI] [PubMed] [Google Scholar]
- Wang Z, Inuzuka H, Zhong J, Wan L, Fukushima H, Sarkar FH, Wei W. Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett. 2012;586:1409–1418. doi: 10.1016/j.febslet.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Sun Y. Small RING finger proteins RBX1 and RBX2 of SCF E3 ubiquitin ligases: the role in cancer and as cancer targets. Genes Cancer. 2010;1:700–707. doi: 10.1177/1947601910382776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Woenckhaus C, Maile S, Uffmann S, Bansemir M, Dittberner T, Poetsch M, Giebel J. Expression of Skp2 and p27KIP1 in naevi and malignant melanoma of the skin and its relation to clinical outcome. Histol. Histopathol. 2005;20:501–508. doi: 10.14670/HH-20.501. [DOI] [PubMed] [Google Scholar]
- Wolter M, Scharwachter C, Reifenberger J, Koch A, Pietsch T, Reifenberger G. Absence of detectable alterations in the putative tumor suppressor gene BTRC in cerebellar medulloblastomas and cutaneous basal cell carcinomas. Acta Neuropathol. 2003;106:287–290. doi: 10.1007/s00401-003-0745-7. [DOI] [PubMed] [Google Scholar]
- Wu K, Fuchs SY, Chen A, Tan P, Gomez C, Ronai Z, Pan ZQ. The SCF(HOS/beta-TRCP)-ROC1 E3 ubiquitin ligase utilizes two distinct domains within CUL1 for substrate targeting and ubiquitin ligation. Mol. Cell. Biol. 2000;20:1382–1393. doi: 10.1128/mcb.20.4.1382-1393.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Li L, Liu H, Wu L, Luo Z, Li H, Zheng S, Gao H, Chu Y, Sun Y, Liu J, Jia L. Induction of autophagy and senescence by knockdown of ROC1 E3 ubiquitin ligase to suppress the growth of liver cancer cells. Cell Death Differ. 2013;20:235–247. doi: 10.1038/cdd.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Li G. Novel multiple markers to distinguish melanoma from dysplastic nevi. PLoS ONE. 2012;7:e45037. doi: 10.1371/journal.pone.0045037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Sun Y. Cullin-RING ligases (CRLs) as attractive anti-cancer targets. Curr. Pharm. Des. 2012a doi: 10.2174/13816128113199990300. PMID:23151137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Sun Y. Targeting the mTOR-DEPTOR pathway by CRL E3 ubiquitin ligases: therapeutic application. Neoplasia. 2012b;14:360–367. doi: 10.1593/neo.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xiong X, Jia L, Sun Y. Targeting Cullin-RING ligases by MLN4924 induces autophagy via modulating the HIF1-REDD1-TSC1-mTORC1-DEPTOR axis. Cell Death Dis. 2012;3:e386. doi: 10.1038/cddis.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Xiong X, Sun Y. DEPTOR, an mTOR Inhibitor, is a physiological substrate of SCFβTrCP E3 ubiquitin ligase and regulates survival and autophagy. Mol. Cell. 2011;44:304–316. doi: 10.1016/j.molcel.2011.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, Conaway RC, Conaway JW, Harper JW, Pavletich NP. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]