Abstract
Aims/objective
Influence of genetic variations on the response of clopidogrel, an antiplatelet drug is implicated. In the present study, the prevalence of single nucleotide polymorphisms of MDR1 (C3435T), CYP2C19 [CYP2C19*2 CYP2C19*3, CYP2C19*17] and P2Y12 (i-T744C) in Indian population and their effects on clopidogrel response was analyzed.
Methods and results
To analyze the prevalence of polymorphisms, 102 healthy individuals were recruited. Clopidogrel response was assessed by ADP induced platelet aggregation in clopidogrel naïve acute myocardial infarction (AMI) patients (n = 26) screened from 100 AMI cases, before loading dose of 300 mg, at 24 h before next dose and 6 days after on 75 mg per day and platelet aggregation inhibition (PAI) was calculated between these time intervals. Genotyping was carried out by PCR-based restriction enzyme digestion method for C3435T of MDR1 and i-T744C of P2Y12, by multiplex PCR for CYP2C19*2 (G681A) and CYP2C19*3 (G636A) and by nested PCR for CYP2C19*17 (C806T). The effect of the above mentioned genetic variations on PAI was analyzed. Variant allele of CYP2C19*3 was not observed while the prevalence of 3435T of MDR1 (0.524), CYP2C19*2 (681A, 0.352); i-744C of P2Y12 (0.088), as well as wild type allele CYP2C19*17 (C806, 0.897) associated with decrease clopidogrel response were observed. Trend toward poor response to clopidogrel was observed at 24 h with the variant genotypes of CYP2C19*2 and i-T744C of P2Y12 as compared to wild type.
Conclusion
The present study did show a trend toward impaired response of clopidogrel to inhibit platelet aggregation with variant genotypes of CYP2C19*2 and iT744C of P2Y12 compared to respective wild type genotype at 24 h.
Keywords: Clopidogrel, CYP2C19, Gene polymorphisms, MDR1, P2Y12
1. Background
It has become extensively clear over the past decade that millions of loci within the human genome can vary from person to person which affect gene and subsequent protein expression and influence our metabolism. Among our many happening metabolic processes, the action of drug is dependent on various metabolic processes that includes its activation and metabolism; thus determining its efficacy. For a drug like clopidogrel, a highly prescribed antiplatelet agent, a genetic testing which is able to predict variability in drug response has found its place in the field of pharmacogenomics.
Clopidogrel, alone or combined with aspirin, is routinely used to treat patients with a variety of vascular disorders. It is a thienopyridine derivative; a prodrug activated in the liver by cytochrome P450 (CYP) enzymes mainly by CYP2C19. This active metabolite of the drug then binds irreversibly to the platelet adenosine diphosphate (ADP) receptor P2Y12 which inhibits platelet degranulation, glycoprotein IIb/IIIa (GPIIbIIIa) receptor activation and thus platelet aggregation.1–3 The pharmacodynamic response to clopidogrel varies widely from subject to subject, and about 25% of patients treated with standard clopidogrel doses display low ex vivo inhibition of ADP-induced platelet aggregation. Generally patients with <30% inhibition of platelet aggregation in response to clopidogrel are considered weak responders to the antiplatelet treatment while those with <10% are considered as resistant to its effect.4,5
This poor response to clopidogrel is associated with an increased risk of recurrent ischemic events. The mechanisms underlying clopidogrel resistance are unclear. In addition to non-adherence to treatment, poor bioavailability, accelerated platelet turnover; certain genetic factors have been demonstrated to be associated with this phenomenon. The single nucleotide polymorphisms (SNPs) of Multidrug Resistance Protein 1 (MDR1), CYP2C19, and P2Y12 genes are proposed which influence the response to clopidogrel.6,7 MDR1 encodes a drug-efflux transporter called P-glycoprotein which functions as a physiologic intestinal barrier against drug absorption.8 The best studied polymorphism in this gene is a transversion of C to T at position 34359,10 associated with decreased expression. CYP2C19 of cytochrome P450 isoenzymes system is the predominant enzyme involved in clopidogrel metabolic activation that carries out 45% of the first step in the biotransformation of clopidogrel.11 Genetic variations of this enzyme are CYP2C19*1 wild type, CYP2C19*2, [G681A] with splicing defect, and CYP2C19*3 [G636A] with stop codon. CYP2C19*2 and CYP2C19*3 comprises about 95% of the poor metabolizer type.12–16 In contrast, an allelic variant reported as CYP2C19*17 (C806T; 5′-flanking region of the gene), has been associated with increased enzyme function.17,18 P2Y12 belongs to the Gi class of a group of G protein-coupled (GPCR) purinergic receptors and is a chemoreceptor for ADP.19 Two functional haplotypes of P2Y12 designated as H1 and H2, tagged by 4 SNPs in absolute linkage disequilibrium (i-C139T, i-T744C, i-ins801A, G52T) have been identified. H1 haplotype is the one showing a ‘C’ in position 139 of intron 1, a ‘T’ in position 744 of intron 1, absence of ‘i-ins801A’ in intron 1, as well as a ‘G’ in position 52 of exon 2. H2 haplotype comprises of variations 139T, 744C and presence of i-ins801A in intron 1 along with 52T of exon 2. Of these two haplotypes, haplotype (H2) has been reported to be associated with enhanced platelet reactivity.20
In addition, it has been observed that the genotypic frequencies of MDR1, CYP2C19 and P2Y12 polymorphisms present some degree of variations among different ethnic groups. As a result, the prevalence of the predicted metabolic phenotypes associated with these polymorphisms may vary significantly among different world-wide populations. Indians have been documented to have high 3435T allele frequency (0.61–0.62) as compared to the C3435 allele of MDR1.21,22 For Indian population Adithan et al23 have specifically documented the frequency of CYP2C19*1, *2 and *3 alleles as 0.598 (59.8%), 0.379 (37.9%) and 0.022 (2.2%), respectively for Tamilians (n = 112) and 0.703 (70.3%), 0.297 (29.7%) and 0 for North Indians (n = 121), respectively. Another study by Panchabhai et al24 have evaluated the poor and extensive metabolizer type of CYP2C19 enzyme activity by analyzing the drug metabolite in blood. Their study population was Gujarati and Marwadi living in Mumbai. It was seen that 10.36% of this population were poor metabolizers (PM) whereas 89.63% were extensive metabolizers (EM). The prevalence as per CYP2C19*2 or *3 was not carried out in this study. In fact, it was suggested that a genotyping evaluation would better help in individualize drug therapy. Moreover, pharmacogenetic testing is more in demand since the inclusion of an FDA “black box” warning recommending genetic testing prior to prescription of clopidogrel.25
Therefore, the aim of the present study was to analyze the prevalence of the variant genotypes of the above said polymorphisms i.e. C3435T of MDR1, CYP2C19*2 (G681A), CYP2C19*3 – (G636A) and CYP2C19*17 (C806T) as well as i-T744C of P2Y12, and analyze their effect on the antiplatelet activity of standard-dose of clopidogrel assessed by ex vivo ADP induced platelet aggregation.
2. Material and methods
2.1. Study population
The study population included 102 healthy individuals and 26 acute myocardial infarction (AMI) patients. These subjects were Maharashtrian, Gujarati and Marwadi in decreasing number representing the Western region of India based out of Mumbai.
The healthy individuals included were volunteers in the study, recruited for genotyping of these polymorphisms. They were with systolic blood pressure/diastolic blood pressure (SBP/DBP) = 135/85 mmHg or less, with no clinical symptoms of any other organic disease. Their blood sample was collected after an overnight, 12 h fast. The subjects having fasting glucose levels >110 mg/dL, serum transaminases, Blood Urea Nitrogen (BUN), Creatinine levels and lipid profile beyond normal range, abnormal ECGs and abnormal Carotid Doppler were excluded from the control group. These criteria are followed for including healthy individuals under controls. They form a part of our study population to analyze the overall prevalence of the polymorphisms. Of these subjects, blood sample was also collected of randomly selected 10 healthy individuals for platelet aggregation study.
To analyze the effect of these polymorphism on platelet aggregation only those AMI cases (n = 26) were selected and recruited who were not on clopidogrel before and were prescribed clopidogrel later. They were clopidogrel naïve patients screened from 100 AMI patients in the span of two years (2009–2011). The platelet aggregation of these patients was compared before and after the intake of the drug and thus platelet aggregation inhibition (PAI) was calculated and the effect of genotypes on PAI was analyzed. AMI patients were those who suffered an AMI with prolonged chest pain more than 30 min and ST segment elevation more than 0.1 mv on at least two adjacent ECG leads with elevated cardiac enzymes from ICCU ward. These AMI patients were with first episode of chest pain and had neither past history of such clinical symptoms nor were on any known medications for the same. AMI patients on prior clopidogrel treatment at the time of admission were excluded as their basal platelet aggregation could not be obtained which was essential to compare the extent of PAI after the intake of clopidogrel. Other exclusion criteria were valvular heart disease, known cardiomyopathy, malignancy, renal or liver diseases as well as subjects with systemic inflammatory disease to exclude any other complications and keep the patient population homogenous. These AMI patients at presentation were given 300 mg loading dose of clopidogrel and aspirin (150 mg) along with the standard treatment of nitrates antiarrhythmics, antifailures, statins and proton pump inhibitors (PPI) and subsequently were on maintenance dose of 75 mg clopidogrel per day. PPIs, frequently used in patients receiving clopidogrel and aspirin, are also metabolized by CYP2C19 and CYP3A4.7 In our study, pantoprazole 40 was used as PPI for all the patients. In contrast to the reported negative omeprazole-clopidogrel drug interaction, the intake of pantoprazole or esomeprazole has been demonstrated, not to interfere with the activation of clopidogrel and was therefore not associated with impaired response.26,27 The subjects were recruited in study only after obtaining informed consent. Information regarding their demographic status, clinical history, family history and medications were noted down in detail. The ethical committee of Sir H. N. Hospital and Research Centre and Rajawadi Municipal Hospital approved the study protocol. The analyzes of the samples were carried out at Sir H. N. Medical Research Society at Sir H. N. Hospital and Research Centre, Mumbai.
2.2. Sample collection
Blood samples of 102 healthy individuals were collected in the plain and EDTA anti-coagulated vacutainer after overnight, 12 h fast for analyzing biochemical parameters and complete blood count respectively. Of these subjects, 10 healthy individuals were randomly selected and their blood sample was also collected in Na-Citrate vacutainer (3.2%) for platelet aggregation test. Peripheral blood samples of AMI patients were collected at presentation for routine analysis such as electrolytes, cardiac enzymes, complete blood count and prothrombin time. Remaining serum was stored at −80 °C and EDTA blood sample at 4 °C for further analysis. Peripheral blood of each AMI patient was collected in Na-Citrate vacutainer (3.2%) for platelet aggregation test at three different point of time – (1) baseline or 0 h – before clopidogrel loading dose (300 mg) administration, (2) 24 h after loading dose (300 mg) administration and before the next dose of 75 mg and (3) at 6 days when the patient was on 75 mg maintenance dose of clopidogrel.
2.3. Platelet aggregation by turbidometric method
Platelet aggregation test was carried out at the earlier mentioned three different point of time within 4 h of blood collection. Platelet rich plasma (PRP) and platelet poor plasma (PPP) were separated from each Na-citrated blood and then the test was carried out in duplicates using ADP (10 μM) as agonist in a two chambered Chronolog Platelet Aggregometer (model 490-2D). With the help of AggroLink software package, platelet aggregation was expressed as the maximal percent change in light transmittance from baseline in PRP with PPP used as a reference. All the patients were administered aspirin along with clopidogrel. The predominant antiplatelet effect of aspirin is mediated through its irreversible inactivation of cyclooxygenase (COX) 1, which is responsible for the formation of thromboxane A2, a potent vasoconstrictor and platelet aggregator, from arachidonic acid.27 In the present study, final concentration of 10 μM ADP was chosen for platelet aggregation because at this concentration ADP induces complete platelet aggregation in a manner independent of thromboxane A2 production.14,28,29
2.4. Genotyping of polymorphisms
DNA was extracted from peripheral leukocytes by modified Miller et al's salting out procedure.30 Genotyping was carried out by polymerase chain reaction (PCR) – based restriction enzyme digestion method for C3435T of MDR131 and i-T744C of P2Y12.32 A multiplex PCR was standardized for CYP2C19*2 and CYP2C19*3 using primer sequence of Pang et al33 while CYP2C19*17 was genotyped using nested PCR.34 The primers used for PCR of each polymorphism are given in Table 1. The content of the master mix and PCR conditions are depicted in Table 2. Taq polymerase and dNTPs were from Fermentas–MBI. PCR was carried out in thermal cycler “T Gradient” from “Biometra” (Genetix Biotech Asia Pvt. Ltd.). Each sample was run in duplicates with a positive and negative control for each for a batch of samples. Amplified PCR product was then run on Agarose gel electrophoresis. Finally, the PCR products were subjected to restriction digestion with the enzymes Sau 3A1, Sma I, Bam HI, Nsi I and Rsa I for identifying genotypes of C3435T of MDR1, CYP2C19*2, CYP2C19*3, CYP2C19*17 and i-T744C of P2Y12 respectively. Fig. 1 shows the restriction digestion pattern of C3435T of MDR1, Fig. 2 shows CYP2C19*2 and CYP2C19*3 genotypes of multiplex PCR, Figs. 3 and 4 show that of CYP2C19*17, and i-T744C of P2Y12, respectively on 10% polyacrylamide gel electrophoresis.
Table 1.
Primers sequence.
| Gene | Primer sequence |
|---|---|
|
MDR131 (C3435T) |
5′ TGT TTT CAG CTG CTT GAT GG 3′ 5′ AAG GCA TGT ATG TTG GCC TC 3′ |
| CYP2C19*233 (G681A) | 5′ CAG AGC TTG GCA TAT TGT ATC 3′ 5′ GTA AAC ACA CAA CTA GTC AAT G 3′ |
| CYP2C19*333 (G636A) |
5′ AAA TTG TTT CCA ATC ATT TAG CT 3′ 5′ ACT TCA GGG CTT GGT CAA TA 3′ |
| CYP2C19*1734 (C806T) |
SET 1 5′ GCC CTT AGC ACC AAA TTC TC 3′ 5′ ATT TAA CCC CCT AAA AAA ACA CG 3′ |
| SET 2 5′ AAA TTT GTG TCT TCT GTT CTC AAT G 3′ 5′ AGA CCC TGG GAG AAC AGG AC 3′ | |
|
P2Y1232 (i-T744C) |
5′ TCA CTT ATC TCT GGT GAA ATA AAA AGA TTA CGT A 3′ 5′ GTC AGA AAT GGC CTG TGT ATA TAT GGT CAT GAG 3′ |
Table 2.
Details of the PCR protocol for genotyping polymorphisms.
| Genotype | Content of the PCR reaction | PCR conditions | PCR amplified product |
|---|---|---|---|
| MDR1 (C3435T) | 1 U Taq DNA polymerase, 2.5 μl of 10X Buffer with 25 mM MgCl2, 0.5 μl of 10 mM dNTPs, 10 pmoles of primers and 100 ng of genomic DNA | Initial denaturation at 95 °C for 5 min followed by 30 cycles of denaturation at 95 °C for 45 s, annealing at 62 °C for 30 s and extension at 72 °C for 30 s followed by final extension at 72 °C for 5 min. | 197 base pair (bp) |
| CYP2C19*2 and CYP2C19*3 polymorphisms by multiplex PCR | The 50 μl reaction mixture contained 2 U Taq DNA polymerase, 5 μl 10X Buffer with 25 mM MgCl2, 1 μl 10 mM dNTPs and 15 pmoles of each specific primers and 100 ng of genomic DNA | The PCR amplification conditions consisted 3 steps – an initial denaturation step at 95 °C for 5 min followed by 30 cycles of denaturation at 95 °C for 45 s, annealing at 58 °C for 50 s and extension at 72 °C for 1 min followed by final extension at 72 °C for 3 min | 321 bp of CYP2C19*2 (G681A) and 271 bp of CYP2C19*3 (G636A) |
| CYP2C19*17 polymorphism by nested PCR |
First PCR The first 25 μl reaction mixture contained 1 U Taq DNA polymerase, 2.5 μl of 10X buffer with 25 mM MgCl2, 0.5 μl of 10 mM dNTPs and 10 pmoles of each specific primers and 100 ng of genomic DNA Second PCR The next 50 μl reaction mixture contained 1U Taq DNA polymerase, 5 μl of 10X Buffer with 25 mM MgCl2, 0.5 μl of 10 mM dNTPs and 10 pmoles of each specific primers. For each sample to be genotyped 1 μl of earlier amplified PCR product was added later. |
An initial denaturation step at 95 °C for 5 min followed by 30 cycles of denaturation 95 °C for 45 s, annealing at 62 °C for 30 s and denaturation at 72 °C for 30 s followed by final extension at 72 °C for 5 min. Second PCR Initial denaturation step at 95 °C for 5 min followed by 30 cycles of denaturation at 95 °C for 45 s, annealing at 62 °C for 30 s and extension at 72 °C for 30 s followed by final extension at 72 °C for 3 min |
473bp 143bp |
|
P2Y12 (i-T744C) |
2 U Taq DNA polymerase (Fermentas - MBI), 2.5 μl 10X Buffer with 25 mM MgCl2, 0.5 μl of 10 mM dNTPs and 10 pmoles of each specific primers and 100 ng of genomic DNA. | Initial denaturation step at 95 °C for 5 min followed by 30 cycles of denaturation at 95 °C for 45 s, annealing at 60 °C for 2 min and extension at 72 °C for 2 min followed by final extension at 72 °C for 5 min | 220bp |
Fig. 1.
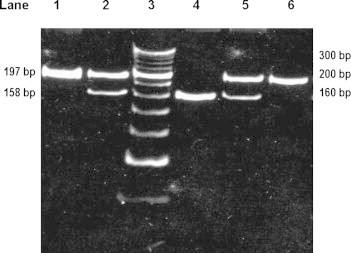
The restriction digestion pattern of MDR1 [C3435T] on 10% polyacrylamide gel electrophoresis. Lane 1: PCR product of 197 bp. Lane 2 and 4: heterozygous genotype (C3435T) showing two bands of 197 bp and 158 bp. Lane 3: O′ Range Ruler, 20 bp, ready to use DNA Ladder from MBI Fermentas. Lane 4: homozygous mutant genotype (T3435T) showing one band of 158 bp. Lane 6: wild type genotype (C3435C) showing one band of 197 bp.
Fig. 2.
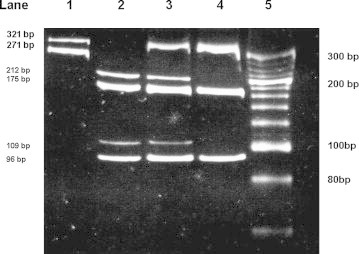
The restriction digestion pattern of CYP2C19*2 and CYP2C19*3 on 10% polyacrylamide gel electrophoresis. Lane 1: PCR products of 321 bp and 271 bp for CYP2C19*2 and CYP2C19*3 respectively. Lane 2–Lane 4: wild type genotype (*1 × *1) of CYP2C19*2 showing two bands 212 bp and 109 bp and wild type genotype (*1 × *1) of CYP2C19*3 showing two bands 175 bp and 196 bp. Lane 3: Heterozygous genotype (*1 × *2) of CYP2C19*2 showing three bands 321 bp, 212 bp and 109 bp and wild type genotype (*1 × *1) of CYP2C19*3 showing two bands 175 bp and 196 bp. Lane 4: homozygous mutant genotype (*2 × *2) showing one band of 321 bp and wild type genotype (*1 × *1) of CYP2C19*3 showing two bands 175 bp and 196 bp. Lane 5: O′ Range Ruler, 20 bp, ready to use DNA Ladder from MBI Fermentas.
Fig. 3.
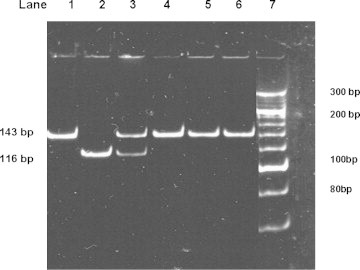
The restriction digestion pattern of CYP2C19*17 on 10% polyacrylamide gel electrophoresis. Lane 1: PCR products of 143 bp. Lane 2: wild type genotype (*1 × *1) showing one band of 116 bp. Lane 3: Heterozygous genotype (*1 × *17) showing two bands 143 bp and 116 bp. Lane 4 to 6: homozygous mutant genotype (*17 × *17) showing one band of 143 bp. Lane 7: O′ Range Ruler, 20 bp, ready to use DNA Ladder from MBI Fermentas.
Fig. 4.
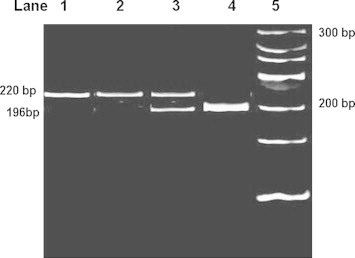
The restriction digestion pattern of P2Y12 [i-T744C] on 10% polyacrylamide gel electrophoresis. Lane 1: PCR products of 220 bp. Lane 2: wild type genotype (i-T744T) showing one band of 220 bp. Lane 3: heterozygous genotype (i-T744C) showing two bands 220 bp and 196 bp. Lane 4: Homozygous mutant genotype (i-C744C) showing one band of 196 bp. Lane 5: O′ Range Ruler, 20 bp, ready to use DNA Ladder from MBI Fermentas.
2.5. Statistical analysis
Genotype frequencies were estimated by the gene-counting method. Allelic frequencies were calculated from genotype frequencies. Genotypes were tested for deviations from Hardy–Weinberg equilibrium. The change in platelet aggregation within a group from baseline to 24 h or 6th day was assessed using Wilcoxon Signed Rank test. The comparison of change in platelet aggregation between groups was assessed by Mann–Whitney U test.
3. Results
3.1. Characteristics of the study population
Demographic data and routine pathology data of 102 healthy individuals and 26 AMI patients is given in Table 3 and Table 4 respectively. There was no significant difference in the age, body mass index and blood pressure amongst AMI patients. Six out of 26 patients were hypertensive while 4 were diabetic.
Table 3.
Demographic data.
| Parameters | Healthy individuals (n = 102) | AMI patients (n = 26) |
|---|---|---|
| M:F | 47: 55 | 19:7 |
| Age [years] | 43.59 ± 10.74 | 51 ± 10 |
| BMI [kg/m2] | 24.4 ± 4.53 | 23.98 ± 2.12 |
| SBP [mmHg] | 118 ± 9.23 | 135 ± 29 |
| DBP [mmHg] | 78.96 ± 6.73 | 82 ± 17 |
| Smoking | 14 | 14 |
| Alcohol consumption | 15 | 8 |
| Hypertension | – | 6 |
| Diabetes mellitus | – | 4 |
Table 4.
Routine pathology data.
| Parameters | Healthy individuals (n = 102) | AMI patients (n = 26) |
|---|---|---|
| Total cholesterol (TC) (mg/dl) | 188.5 ± 35.38 | 172.5 ± 44.4 |
| HDL-cholesterol (HDL-C) (mg/dl) | 44.63 ± 9.29 | 38.4 ± 7.65 |
| TC/HDL-C | 4.31 ± 0.86 | 4.47 ± 0.74 |
| LDL cholesterol (LDL-C) (mg/dl) | 124.8 ± 25.5 | 105.9 ± 40.1 |
| LDL-C/HDL-C | 2.8 ± 0.71 | 2.8 ± 0.6 |
| Triglycerides (mg/dl) | 102.6 ± 41.5 | 127.4 ± 61.02 |
| VLDL cholesterol (mg/dl) | 22.24 ± 13.11 | 25.5 ± 12.2 |
| Glucose (mg/dl) | 95.1 ± 10.1 | 154.2 ± 68.3 |
| SGOT U/L | 16 ± 3.25 | 51.4 ± 30.12 |
| SGPT U/L | 20.1 ± 2.3 | 38.2 ± 15.8 |
| BUN (mg/dl) | 8.1 ± 2.37 | 10.1 ± 7.32 |
| Creatinine (mg/dl) | 0.7 ± 0.12 | 0.98 ± 0.24 |
| Uric acid (mg/dl) | 4.0 ± 1.3 | 4.95 ± 1.84 |
| Total WBC count/μL | 6975 ± 1602 | 11 210 ± 2926 |
| Platelet count × 103/μL | 259 ± 109.5 | 328 ± 137 |
3.2. Allelic frequencies
The genotype and allele frequencies for all the polymorphisms studied are given in Table 5 and Table 6 for healthy individuals and AMI patients, respectively. The study population did not show the presence of the variant allele of CYP2C19*3. The observed genotype distributions did not deviate from Hardy–Weinberg equilibrium.
Table 5.
Genotype and allele frequencies of healthy individuals (n = 102) for all the polymorphisms studied.
| Genotype frequency |
Allele frequency |
||||
|---|---|---|---|---|---|
| Wild type | Heterozygous | Homozygous | Wild type allele | Mutant allele | |
|
MDR1 (C3435T) |
26.4% (27) | 42.1% (43) | 31.4% (32) | 3435C = 0.475 | 3435T = 0.524 |
| CYP2C19*2 (G681A) |
46% (47) | 37% (38) | 16.7% (17) | 681G = 0.647 | 681A = 0.352 |
| CYP2C19*3 (G636A) |
100% (102) | 0 | 0 | 636G = 1.0 | – |
| CYP2C19*17 (C806T) |
81.4% (83) | 16.7% (17) | 1.96% (2) | 806C = 0.897 | 806T = 0.102 |
|
P2Y12 (i-T744C) |
85.2% (87) | 11.8% (12) | 2.94% (3) | i744T = 0.911 | i744C = 0.088 |
Table 6.
Genotype and allele frequencies of AMI patients (n = 26) for all the polymorphisms studied.
| Genotype frequency |
Allele frequency |
||||
|---|---|---|---|---|---|
| Wild type | Heterozygous | Homozygous | Wild type allele | Mutant allele | |
|
MDR1 (C3435T) |
15.4% (04) | 53.9% (14) | 30.8% (08) | 3435C = 0.423 | 3435T = 0.576 |
| CYP2C19*2 (G681A) |
27% (07) | 61.5% (16) | 11.5% (03) | 681G = 0.576 | 681A = 0.423 |
| CYP2C19*3 (G636A) |
100% (26) | 0 | 0 | 636G = 1.0 | – |
| CYP2C19*17 (C806T) |
73.1% (19) | 26.9% (07) | 0 | 806C = 0.865 | 806T = 0.134 |
|
P2Y12 (i-T744C) |
80.7% (21) | 3.8% (01) | 15.4% (04) | i744T = 0.826 | i744C = 0.173 |
As the sample size of healthy individuals (n = 102) and that of AMI patients (n = 26) differed, the significance of difference of prevalence of the genotypes and alleles (using Chi Square statistics with Yates Corrections) was not calculated. However, it was observed that the prevalence of wild type allele frequency of CYP2C19*17 (C806, 0.897 > 0.865) and the variant allele frequency of 3435T of MDR1 (0.576 > 0.524), CYP2C19*2 (681A, 0.423 > 0.352) as well as i744C of P2Y12 (0.173 > 0.088) of AMI patients although marginal, were more as compared that of healthy individuals (Tables 5 and 6).
3.3. Platelet aggregation
The platelet aggregation of healthy individuals (n = 10) was in the range of 60–80% (70.4 ± 8.87) almost similar to average baseline platelet aggregation (0 h) of 26 AMI patients (69.16%). The platelet aggregation of all AMI patients (n = 26) dropped to 45.6% at 24 h subsequent to the administration of 300 mg loading dose with PAI of only 23.56%. Further, platelet aggregation dropped to 43.4% at 6 days on further 75 mg maintenance dose per day of clopidogrel demonstrating PAI of only 25.76%.
Among 26 AMI patients, none demonstrated homozygous mutant (T806T) genotype of CYP2C19*17. All the patients were associated with either wild type or heterozygous genotypes associated with decreased enzyme function. Also there were only 4 patients each with wild type (C3435C) genotype of MDR1 and rests were with variant genotypes associated decrease intestinal absorption. Therefore, further analysis of effect of polymorphisms on response to clopidogrel was carried out with respect to CYP2C19*2 genotypes and patients were grouped according to common genotypes as depicted in Table 7. Group I, II and III were with wild type (G681G), heterozygous (G681A) and homozygous mutant (A681A) genotype of CYP2C19*2 respectively and wild type genotype (i-T744T) of P2Y12. Subsequently, subjects were grouped as per variant genotypes of i-T744C of P2Y12. Group IV was one case having homozygous mutant genotype (A681A) of CYP2C19*2 and heterozygous genotype (i-T744C) of P2Y12 while three cases with heterozygous genotype (G681A) of CYP2C19*2 and homozygous mutant genotype (i-C744C) of P2Y12 were included in Group V. There was only one case with wild type genotype (G681G) of CYP2C19*2 and homozygous mutant genotype (i-C744C) of P2Y12 (Group VI). The PAI at 24 h was in the decreasing order; 37%, 22%, 18.5%, 18.0%, 14.3% and 11.0% for Group I, II, III, IV, V and VI respectively.
Table 7.
Platelet aggregation, platelet aggregation inhibition (PAI) of AMI patients (n = 26) grouped as per genotypes.
| Groups | Platelet aggregation (%) |
Platelet aggregation inhibition (PAI) (%) |
Genotypes |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| CYP2C19 |
P2Y12 |
CYP2C19 |
MDR 1 |
|||||||
| 0 h | 24 h | 6 days | At 0 h | At 6 days | *2 | *3 | -i T744C | *17 | C3435T | |
| Group I (n = 6) | 67.8 | 30.7 | 40.3 | 37 | 27.3 | W | W | W | W | HMM |
| W | W | W | W | HMM | ||||||
| W | W | W | W | HMM | ||||||
| W | W | W | W | HMM | ||||||
| W | W | W | W | HZ | ||||||
| W | W | W | HZ | HZ | ||||||
| Group II (n = 13) | 67.15 | 45.15 | 42.15 | 22 | 24 | HZ | W | W | W | HZ |
| HZ | W | W | W | HZ | ||||||
| HZ | W | W | W | HZ | ||||||
| HZ | W | W | W | HMM | ||||||
| HZ | W | W | W | HZ | ||||||
| HZ | W | W | HZ | W | ||||||
| HZ | W | W | W | HMM | ||||||
| HZ | W | W | W | W | ||||||
| HZ | W | W | W | HMM | ||||||
| HZ | W | W | HZ | HZ | ||||||
| HZ | W | W | W | W | ||||||
| HZ | W | W | W | HZ | ||||||
| HZ | W | W | W | HZ | ||||||
| Group III (n = 2) | 71.5 | 53 | 51 | 18.5 | 20.5 | HMM | W | W | HZ | HZ |
| HMM | W | W | HZ | HZ | ||||||
| Group IV (n = 1) | 79 | 61 | 47 | 18 | 32 | HMM | W | HZ | W | HZ |
| Group V (n = 3) | 76.7 | 62.3 | 50 | 14.3 | 26.7 | HZ | W | HMM | HZ | HMM |
| HZ | W | HMM | HZ | HZ | ||||||
| HZ | W | HMM | W | W | ||||||
| Group VI (n = 1) | 64 | 53 | 33 | 11 | 31 | W | W | HMM | W | HZ |
W–wild type genotype, HZ–heterozygous genotype, HMM–homozygous mutant genotype.
Statistical analysis of Group I and Group II with 6 and 13 cases, respectively demonstrated that decrease in platelet aggregation i.e. PAI of Group I from 0 h to 24 h (37%, p = 0.002) and 0 to 6th day (27%, p = 0.027) was significant. The same was also observed for Group II for PAI from baseline to 24 h (22.0%, p = 0.001) as well as from baseline to 6th day (24.3%, p = 0.001). PAI at 24 h of Group II (22.0%) was less by 15% as compared to Group I (37.0%) but this decrease in PAI did not reach statistical significance of p< 0.05 (p = 0.072). PAI at 6 days from baseline of Group I and II were almost similar 27% and 24.3%, respectively and did not reach statistical significance of p < 0.05 (p = 0.639). There were 3 hypertensives and one diabetic patient in Group I while in Group II there were 2 hypertensives and 2 diabetic patients. After excluding these patients, the difference in the PAI between Group I and II was 16.0% almost similar to that obtained on including these patients (15.0%). There was no significant difference in the BMI or age between the two patient groups (data not shown).
Table 7 also depicts the CYP2C19*17 and C3435T genotypes of MDR1. In Group I, out of 6 patients, 5 had wild type genotype (C806C) for CYP2C19*17 while just one showed heterozygous genotype (C806T) for the same. Whereas in Group II, 11 out of 13 had wild type genotype for CYP2C19*17 and just 2 showed heterozygous genotype for the same. On the other hand, in Group I out of 6 patients 2 were heterozygous (C3435T) and 4 were homozygous mutant (T3435T) thus all were with variant genotypes of C3435T polymorphism of MDR1. In Group II, out of 13 patients, 7 were heterozygous and 3 each showed homozygous mutant and wild type genotype (C3435C) for C3435T polymorphism of MDR1.
Remaining 7 patients could not be included in Group I or II and therefore in Table 7 they are grouped separately. Their PAI at 24 h was less as compared to Group I. Of these notably, 3 patients of Group V with not only heterozygous genotype for CYP2C19*2 but also homozygous mutant genotype (i-C744C) of P2Y12 demonstrated further (22.0%–14.3%) 7.7% less PAI as compared to Group II at 24 h. One patient with wild type genotype of CYP2C19*2 and homozygous mutant genotype (i-C744C) of P2Y12 demonstrated only 11% PAI at 24 h. At 6th day, the PAI for all seven patients was either similar or near to Group I patients.
4. Discussion
The present study demonstrated the prevalence of polymorphisms of drug-efflux transporter protein responsible for drug absorption (MDR1), one of the metabolizing enzymes of clopidogrel (CYP2C19) and its target (P2Y12), in our study population. Its effect was demonstrated on antiplatelet activity of clopidogrel in AMI patients assessed ex vivo by ADP-induced platelet aggregation.
In our study population variant allele frequency of 3435T of MDR1 affecting intestinal absorption was found to be 52.4% (0.524) nearing to that reported for Indian population (0.61–0.62).21,22 In African,35 Asian and Caucasian36 population, allele frequency of 3435T of MDR1 has been reported to be 21.0%, 42.0% and 55.0%, respectively. The variant allele frequency of CYP2C19*2 (681A) associated with poor metabolizer type, was observed to be 35.2% (0.352) in the present study which is similar to that reported by Adithan et al23 for Tamilian population (37.9%) and Mizutani et al36 who have reported 30–35% in Asians and is greater than reported for African (17–20%)35 and Caucasians (13–18%).36 In the present study, the presence of CYP2C19*3 variant allele (636A), also associated with poor metabolizer type, was not observed which has also been reported by Pang et al33 in Malaysian subjects. However, Adithan et al22 have reported CYP2C9*3 variant allele frequency as 2.2% in Tamilian population. In African,35 Asians and Caucasians,36 the variant allele frequency of CYP2C19*3 has been reported to be <1%, 5–10% and <1%, respectively. Our study population demonstrated low variant allelic frequency of 10.2% (0.102) for CYP2C19*17 which in fact is associated with increased enzyme function. The variant allelic frequency of CYP2C19*17 is reported to be 18%, 2–4% and 18–20% in African,35 Asian and Caucasian36 population respectively. With respect to P2Y12 (i-T744C) polymorphism, although the prevalence of wild type allele was maximum, variant allele associated with enhanced platelet reactivity was also observed in our study population. The frequency of i-744C variant allele of P2Y12 was 8.8% (0.088). Thus, overall our study population was associated with low response to clopidogrel with respect to the prevalence of wild type allele (C806) frequency of 0.897 of CYP2C19*17 and variant allele frequency of MDR1 (3435T) of 0.524, CYP2C19*2 (681A) of 0.352 as well as P2Y12 (-i744C) of 0.088.
None of the AMI patient out of 26 demonstrated a combination of wild type genotype of MDR1 (C3435C), CYP2C19*2 (G681G), CYP2C19*3 (G636G), with heterozygous (C806T) or homozygous (T806T) variant genotype of CYP2C19*17 as well as wild type genotype (iT744T) of P2Y12 favoring good absorption, metabolization and effective action on target receptor of clopidogrel, respectively. This would explain overall low PAI i.e. just 23.56% in the study population at 24 h after clopidogrel administration which can be grouped under weak responders to clopidogrel (<30%).
Although our entire study population did not demonstrate CYP2C19*3 variant genotypes associated with poor metabolizer type, with respect to CYP2C19*17 only 7 AMI patients were heterozygous (C806T) associated with increased enzyme function and rest were all with wild type allele associated with normal enzyme function. Also there were only four AMI patients associated with wild type (C3435C) genotype of MDR1 with optimum absorption.
In the present study, a trend toward poor response to clopidogrel with variant genotypes of CYP2C19*2 as opposed to their wild type counterparts, as assessed ex vivo was observed. These results of our study are similar to those of Hulot et al,14 Brandt et al15 and Gusti et al37 who have underlined the role of CYP2C19 polymorphisms in the clopidogrel metabolic activation process. However, in contrast to the study by Hulot et al14 our study didn't find any significant effect of CYP2C19*2 on PAI at 6th day when the AMI patients were on maintenance dose of 75 mg clopidogrel per day, indicating that a part of low pharmacodynamic response of clopidogrel due to the CYP2C19*2 polymorphism may be reversed to some extent by repeated dosing.
Fontana et al20 have identified a P2Y12 receptor haplotype H2 to be strongly associated with an increase in ADP-induced platelet aggregation. In 2008, Malek et al32 reported that the co-existence of the variant i-744C allele of P2Y12 and the variant 681A allele of CYP2C19*2 is associated with persisting platelet activity in patients with acute coronary syndrome (ACS) on clopidogrel treatment. Similar to this finding, in the present study, there were three patients with homozygous mutant genotype (i-C744C) for P2Y12 receptor in combination with heterozygous genotype for CYP2C19*2 (Group V) who showed additional 7.7% decreased PAI as compared to patients of Group II with wild type genotype (i-T744T) of P2Y12 receptor and heterozygous genotype (G681A) of CYP2C19*2. Further Malek et al32 have reported that the carriers of i-744C allele of P2Y12 without variant allele of CYP2C19*2 did not have significantly decreased platelet response to ADP suggesting that i-T744C polymorphism of P2Y12 could also independently elevate platelet reactivity. In the present study we had only one case with homozygous mutant (i-C744C) genotype of P2Y12 and wild type of CYP2C19*2 which was also associated with increased response to ADP with PAI of only 11% at 24 h.
Small study population for analyzing the platelet aggregation effect is the major limitation of our study which was further affected by variations in genotypes. To see the extent of clopidogrel to inhibit the platelet aggregation, it was very important that these patients were without any prior antiplatelet intake. For this reason, patients with prior antiplatelet treatment were excluded and these 26 AMI clopidogrel naïve patients were screened from 100 AMI patients in the span of two years from January 2009 to March 2011.
5. Conclusion
In our study the occurrence of variant alleles of MDR1 (3435T), CYP2C19*2 (681A), P2Y12 (i-744C) as well as wild type allele of CYP2C19*17 (C806) associated with decreased clopidogrel response were observed. The present study did show a trend toward impaired response of clopidogrel to inhibit platelet aggregation with variant genotypes of CYP2C19*2 and iT744C of P2Y12 compared to respective wild type genotype at 24 h. Similar studies of larger sample size and of longer follow-up may strengthen our view for individualized antiplatelet treatment based on genotypic analysis for patients with ACS and those undergoing PCI who receive loading dose of 300 mg of clopidogrel.
Funding source
The work is being supported by the grants from Sir H. N. Medical Research Society.
Conflicts of interest
All authors have none to declare.
Acknowledgment
Authors would like to acknowledge Sir H. N. Hospital and Research Centre, and Rajawadi Municipal Hospital for recruitment of the patients and Sir H. N. Medical Research Society for the financial support.
References
- 1.Yusuf S., Zhao F., Mehta S.R. Effects of Clopidogrel in addition to aspirin in patients with acute coronary syndromes without ST-segment elevation. N Engl J Med. 2001;345:494–502. doi: 10.1056/NEJMoa010746. [DOI] [PubMed] [Google Scholar]
- 2.Sabatine M.S., Cannon C.P., Gibson C.M. Addition of clopidogrel to aspirin and fibrinolytic therapy for myocardial infarction with ST-segment elevation. N Engl J Med. 2005;352:1179–1189. doi: 10.1056/NEJMoa050522. [DOI] [PubMed] [Google Scholar]
- 3.King S.B., 3rd, Smith S.C., Jr., Hirshfeld J.W., Jr. 2007 focused update of the ACC/AHA/SCAI 2005 Guideline Update for Percutaneous Coronary Intervention: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines: 2007 Writing Group to Review New Evidence and Update the ACC/AHA/SCAI 2005 Guideline Update for Percutaneous Coronary Intervention, Writing on behalf of the 2005 Writing Committee. Circulation. 2008;117:261–295. doi: 10.1161/CIRCULATIONAHA.107.188208. [DOI] [PubMed] [Google Scholar]
- 4.Gurbel P.A., Bliden K.P., Hiatt B.L., O'Connor C.M. Clopidogrel for coronary stenting: response variability, drug resistance, and the effect of pretreatment platelet reactivity. Circulation. 2003;107:2908–2913. doi: 10.1161/01.CIR.0000072771.11429.83. [DOI] [PubMed] [Google Scholar]
- 5.Muller I., Besta F., Schulz C., Massberg S., Schomig A., Gawaz M. Prevalence of Clopidogrel non-responders among patients with stable angina pectoris scheduled for elective coronary stent placement. Thromb Haemost. 2003;89:783–787. [PubMed] [Google Scholar]
- 6.Nguyen T.A., Diodati J.G., Pharand C. Resistance to clopidogrel: a review of the evidence. J Am Coll Cardiol. 2005;45:1157–1164. doi: 10.1016/j.jacc.2005.01.034. [DOI] [PubMed] [Google Scholar]
- 7.Wiviott S.D., Antman E.M. Clopidogrel resistance: a new chapter in a fast-moving story. Circulation. 2004;109:3064–3067. doi: 10.1161/01.CIR.0000134701.40946.30. [DOI] [PubMed] [Google Scholar]
- 8.Taubert D., von Beckerath N., Grimberg G. Impact of P-glycoprotein on clopidogrel absorption. Clin Pharmacol Ther. 2006;80:486–501. doi: 10.1016/j.clpt.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Fung K.L., Gottesman M.M. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. (BBA)—Proteins and Proteomics. 2009;1794:860–871. doi: 10.1016/j.bbapap.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang K., Wong L.P., Lee E.J.D., Chong S.S., Lee C.G.L. Genomic evidence for recent positive selection at the human MDR1 gene locus. Hum Mol Genet. 2004;13:783–797. doi: 10.1093/hmg/ddh099. [DOI] [PubMed] [Google Scholar]
- 11.Kazui M., Nishiya Y., Ishizuka T. Identification of the human cytochrome P450 enzymes involved in the two oxidative steps in the bioactivation of clopidogrel to its pharmacologically active metabolite. Drug Metab Dispos. 2010;38:92–99. doi: 10.1124/dmd.109.029132. [DOI] [PubMed] [Google Scholar]
- 12.De Morais S.M., Wilkinson G.R., Blaisdell J., Meyer U.A., Nakamura K., Goldstein J.A. Identification of a new genetic defect responsible for the polymorphism of (S)-mephenytoin metabolism in Japanese. Mol Pharmacol. 1994;46:594–598. [PubMed] [Google Scholar]
- 13.De Morais S.M., Wilkinson G.R., Blaisdell J., Nakamura K., Meyer U.A., Goldstein J.A. The major genetic defect responsible for the polymorphism of S-mephenytoin metabolism in humans. J Biol Chem. 1994;269:15419–15422. [PubMed] [Google Scholar]
- 14.Hulot J.S., Bura A., Villard E. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108:2244–2247. doi: 10.1182/blood-2006-04-013052. [DOI] [PubMed] [Google Scholar]
- 15.Brandt J.T., Close S.L., Iturria S.J. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5:2429–2436. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 16.Trenk D., Hochholzer W., Fromm M.F. Cytochrome P450 2C19 681G>A polymorphism and high on-Clopidogrel platelet reactivity associated with adverse 1-year clinical outcome of elective percutaneous coronary intervention with drug eluting or bare-metal stents. J Am Coll Cardiol. 2008;51:1925–1934. doi: 10.1016/j.jacc.2007.12.056. [DOI] [PubMed] [Google Scholar]
- 17.Sim S.C., Risinger C., Dahl M.L. A common novel CYP2C19 gene variant causes ultrarapid drug metabolism relevant for the drug response to proton pump inhibitors and antidepressants. Clin Pharmacol Ther. 2006;79:103–113. doi: 10.1016/j.clpt.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 18.Sibbing D., Koch W., Gebhard D. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–518. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 19.Hollopeter G., Jantzen H.M., Vincent D. Identification of the platelet ADP receptor targeted by antithrombotic drugs. Nature. 2001;409:202–207. doi: 10.1038/35051599. [DOI] [PubMed] [Google Scholar]
- 20.Fontana P., Dupont A., Gandrille S. Adenosine diphosphate – induced platelet aggregation is associated with P2Y12 gene sequence variations in healthy subjects. Circulation. 2003;108:989–995. doi: 10.1161/01.CIR.0000085073.69189.88. [DOI] [PubMed] [Google Scholar]
- 21.Balram C., Sharma A., Sivathasan C., Lee E.J. Frequency of C3435T single nucleotide MDR1 genetic polymorphism in an Asian population: phenotypic–genotypic correlates. Br J Clin Pharmacol. 2003;56:78–83. doi: 10.1046/j.1365-2125.2003.01820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashavaid T., Raje H., Shalia K., Shah B. Effect of gene polymorphisms on the levels of calcineurin inhibitors in Indian renal transplant recipients. Indian J Nephrol. 2010;20:146–151. doi: 10.4103/0971-4065.70846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adithan C., Gerard N., Vasu S., Rosemary J., Shashindran C.H., Krishnamoorthy R. Allele and genotype frequency of CYP2C19 in a Tamilian population. Br J Clin Pharmacol. 2003;56:331–333. doi: 10.1046/j.1365-2125.2003.01883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panchabhai T.S., Noronha S.F., Davis S., Shinde V.M., Kshirsagar N.A., Gogtay J. Evaluation of the activity of CYP2C19 in Gujrati and Marwadi subjects living in Mumbai (Bombay) BMC Clin Pharmacol. 2006;24:6–8. doi: 10.1186/1472-6904-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellis K.J., Souffer G.A., McLeod H.L., Lee C.R. Clopidogrel pharmacogenomics and risk of inadequate platelet inhibition: US FDA recommendations. Pharmacogenomics. 2009;10:1799–1817. doi: 10.2217/pgs.09.143. [DOI] [PubMed] [Google Scholar]
- 26.Siller-Matula J.M., Spiel A.O., Lang I.M., Kreiner G., Christ G., Jilma B. Effects of pantoprazole and esomeprazole on platelet inhibition by clopidogrel. Am Heart J. 2009;157:148e1–148e5. doi: 10.1016/j.ahj.2008.09.017. [DOI] [PubMed] [Google Scholar]
- 27.Cattaneo M. Aspirin and clopidogrel: efficacy, safety and the issue of drug resistance. Arterioscler Thromb Vasc Biol. 2004;24:1980–1987. doi: 10.1161/01.ATV.0000145980.39477.a9. [DOI] [PubMed] [Google Scholar]
- 28.Dörr G., Schmidt G., Gräfe M., Regitz-Zagrosek V., Fleck E. Effects of combined therapy with clopidogrel and acetylsalicylic acid on platelet glycoprotein expression and aggregation. J Cardiovasc Pharmacol. 2002;39:523–532. doi: 10.1097/00005344-200204000-00008. [DOI] [PubMed] [Google Scholar]
- 29.Wilkinson G.R. Drug metabolism and variability among patients in drug response. N Engl J Med. 2005;352:2211–2221. doi: 10.1056/NEJMra032424. [DOI] [PubMed] [Google Scholar]
- 30.Miller S.A., Dykes D.D., Polesky H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cizmarikova M., Wagnerova M., Schonova L., Habalova V., Kohut A., Linkova A. MDR1 (C3435T) polymorphism: relation to the risk of breast cancer and therapeutic outcome. Pharmacogenomic J. 2010;10:62–69. doi: 10.1038/tpj.2009.41. [DOI] [PubMed] [Google Scholar]
- 32.Malek L.A., Kisiel B., Spiewak M. Coexisting polymorphisms of P2Y12 and CYP2C19 genes as a risk factor for persistent platelet activation with clopidogrel. Circ J. 2008;72:1165–1169. doi: 10.1253/circj.72.1165. [DOI] [PubMed] [Google Scholar]
- 33.Pang Y.S., Wong L.P., Lee T.C., Mustafa A.M., Mohamed Z., Lang C.C. Genetic polymorphism of Cytochrome P450 2C19 in healthy Malaysian subjects. Br J Clin Pharmacol. 2004;58:332–335. doi: 10.1111/j.1365-2125.2004.02144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baldwin R.M., Ohlsson S., Pedersen R.S. Increased omeprazole metabolism in carriers of the CYP2C19*17 allele; a pharmacokinetic study in healthy volunteers. Br J Clin Pharmacol. 2008;65:767–774. doi: 10.1111/j.1365-2125.2008.03104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie H.-G., Kim R.B., Wood A.J.J., Stein C.M. Molecular basis of ethnic differences in drug disposition and response. Annu Rev Pharmacol Toxicol. 2001;41:815–850. doi: 10.1146/annurev.pharmtox.41.1.815. [DOI] [PubMed] [Google Scholar]
- 36.Mizutani T. PM frequencies of major CYPs in Asians and Caucasians. Drug Metabol Rev. 2003;35(2–3):99–106. doi: 10.1081/dmr-120023681. [DOI] [PubMed] [Google Scholar]
- 37.Giusti B., Gori A.M., Marcucci R. Cytochrome P450 2C19 loss-of-function polymorphism, but not CYP3A4 IVS10 + 12G/A and P2Y12 T744C polymorphisms, is associated with response variability to dual antiplatelet treatment in high-risk vascular patients. Pharmacogenet Genomics. 2007;17:1057–1064. doi: 10.1097/FPC.0b013e3282f1b2be. [DOI] [PubMed] [Google Scholar]


