Abstract
Melanoma is the most aggressive form of skin cancer with estimated 48,000 deaths per year worldwide. The polyphenol curcumin derived from the plant Curcuma longa is well known for its anti-inflammatory and anti-cancerogenic properties. Accordingly, dietary intake of this compound may be suitable for melanoma prevention. However, how this compound affects basic cellular mechanisms in developing melanoma still remains elusive. Therefore, the aim of this study was to investigate for the first time the impact of oral curcumin administration on the miRNA signature of engrafting melanoma. For this purpose, the effects of a 4% curcumin diet were tested on melanoma, which were established by injection of murine B78H1 cells in the flank of C57BL/6 mice. Curcumin diet or standard chow (control) was administered two weeks prior to injection of tumor cells until termination of the experiment. High throughput chip-based array analysis was deployed to detect alterations in the miRNA signature of the tumors. Curcumin treatment significantly reduced the growth of the flank tumors. Furthermore the miRNA expression signature in tumors was substantially altered by curcumin intake with mmu-miR-205-5p over 100 times higher expressed when compared to controls. The expression levels of identified key miRNAs in the tumor samples were confirmed by quantitative real-time polymerase chain reaction (qRT-PCR). A comparable expression pattern of these miRNAs was also detected in other curcumin-treated melanoma cell lines under in vitro conditions. Putative targets of curcumin-induced up-regulated miRNAs were enriched in ‘o-glycan biosynthesis’, ‘endoplasmatic reticulum protein processing’ and different cancer-related pathways. Western Blot analyses revealed that of these targets anti-apoptotic B-cell CLL/lymphoma 2 (Bcl-2) and proliferating cell nuclear antigen (PCNA) were significantly down-regulated in curcumin-treated tumors. These findings demonstrate a profound alteration of the miRNA expression signature in engrafting curcumin-treated melanoma with mmu-miR-205-5p being up-regulated most significantly.
Introduction
Small non-coding RNAs, so-called microRNAs (miRNAs, miRs), post-transcriptionally attenuate many cellular processes [1], [2]. These evolutionary conserved miRNAs are about 22 nucleotides long and decrease protein expression in proliferating cells by binding to corresponding mRNAs leading either to transcriptional silencing or to mRNA degradation. Deregulation of the miRNA profile is found in many diseases including cancer [3]. For instance, miRNAs with anti-proliferative and anti-angiogenic properties such as hsa-miR-15, hsa-miR-16 or hsa-miR-221/-222 targeting vascular endothelial factor (VEGF), anti-B-cell CLL/lymphoma 2 (Bcl-2) and stem cell receptor c-kit, respectively, play an important role in either preventing or initiating carcinogenesis [4], [5], [6]. Because most mammalian mRNAs have been shown to be targeted by miRNAs, disease-associated expression changes are mirrored by changes of the miRNA profiles [7], [8]. As miRNAs can be purified from blood samples, it was suggested to utilize them as biomarkers for common illnesses, including cardiovascular diseases and cancer [9], [10]. Moreover, miRNA expression profiles may also be suitable as markers to evaluate the safety and efficacy of anti-cancer agents or to predict therapy response [11], [12], [13].
Melanoma is a highly metastatic skin cancer derived from malignant melanocytes. Since curing malignant melanoma remains difficult, evading risk factors is of upmost importance. Herein the avoidance of prolonged sun exposure and sunburns is most important and can be supported by additional dietary chemoprevention with green tea flavonoids, proanthocyanides, and vitamin E [14]. Also, the polyphenol curcumin (diferuloylmethan) derived from the rhizome of Curcuma longa has been thoroughly described for its chemopreventive effects by down-regulation of cellular pathways involved in protein-biosynthesis, mitochondrial activity and free radical scavenging [15], [16]. Oral administration of curcumin was shown to reduce skin inflammation, to support skin healing and even to suppress the development of chemically induced skin cancer in different animal models [17], [18], [19]. Besides, phase I and phase II clinical trials have demonstrated promising effects of oral curcumin administration in patients with colorectal neoplasia, advanced pancreatic and breast cancer either with or without additional chemotherapy [20], [21], [22], [23].
Recently, curcumin has been shown to influence miRNAs of different tumor entities, including pancreatic, breast and lung cancer cells [24], [25], [26]. For instance, down-regulation of oncomir hsa-miR-21 was found in colorectal cancer cells after stimulation with curcumin [27]. In addition, miRNAs from the hsa-let-7 and hsa-mir-200 families were up-regulated and hsa-miR-21 down-regulated by a synthetic curcumin derivate [28], [29].
In the present study we investigated the effects of oral curcumin intake on melanoma growth with the goal to identify potential changes of the tumor miRNA signature. We demonstrate a growth-inhibitory effect of dietary curcumin and profound changes in miRNA expression with mmu-miR-205-5p being up-regulated most significantly.
Materials and Methods
Curcumin
C3 complex, referred to in this article as curcumin, consisting of 77% curcumin, 17% demethoxycurcumin, and 3% bis-demethoxycurcumin was purchased in powdered form from Sabinsa Corporation.
Ethics Statement
All animal care and experimental procedures were approved by the local governmental animal care committee (Landesamt für Verbraucherschutz, Abteilung C Lebensmittel- und Veterinärwesen, Saarbrücken, Germany; Permit Number: 07/2010) and were conducted in accordance with the European legislation on protection of animals (Guide line 2010/63/EU) and the NIH Guidelines for the Care and Use of Laboratory Animals (http://oacu.od.nih.gov/regs/index.htm. 8th Edition; 2011). All experiments were performed under isoflurane anesthesia, and all efforts were made to minimize animal suffering.
Animals and curcumin diet
Male C57BL/6 mice with a body weight (b.w.) of 20-22g were used for the study and housed in groups of 3–4 animals. The animals were kept in a temperature- and humidity-controlled 12 h dark/light environment of the animal care facility of the Institute for Clinical and Experimental Surgery at the University of Saarland. They were allowed free access to tap water and animal chow (ssniff Spezialdiäten GmbH). The diet consisted of either standard mouse chow (control group) or chow enriched with 4% of curcumin (treatment group) prepared from the same batch. This equals a curcumin intake of 8 g/kg b.w. or 160 mg per day. The dietary intervention started 14 days before tumor induction and continued until the end of the in vivo experiments.
Flank model
For continuous measurements of tumor size over 28 days, 1×105 B78H1 cells were injected subcutaneously in each flank of male C57BL/6 mice (curcumin diet: n = 7, control: n = 6). B78H1 cells are murine amelanotic melanoma cells, which are Tap-2 and MHC-I negative clones derived from B16 cells in the laboratory of S. Silagi [30]. The B78H1 cells were a kind gift from the Laboratory of Immunology and Biology of Metastasis of the Department of Experimental Pathology at the University of Bologna [31]. Once a week tumor volumes were assessed by means of the high-resolution ultrasound imaging system Vevo 770 (VisualSonics, Inc.). For this purpose, the mice were anesthetized with 4% isoflurane (Baxter) and fixed in supine position on a heated stage. Anesthesia was maintained at 2% isoflurane and ultrasound coupling gel (Aquasonic 100; Parker Laboratories Inc.) was generously applied to the flanks before images were obtained. The stage was driven by a motorized mechanism so that the real-time microvisualization (RMV) 707B-scanhead (Visual Sonics; 30 MHz) was conducted linearly across the skin of the animals. To acquire parallel two-dimensional (2D) images, recording was done at regular spatial intervals, uniformly spaced at 100 µm over the visual tumor and its close vicinity, with a field of view of 17×17 mm2. A predefined parallel geometry of the 2D images allowed 3D image reconstructions and subsequent determination of tumor volumes. After killing the animals with an overdose of pentobarbital, tumors were extracted and divided up in samples for miRNA isolation and analyses, quantitative real-time polymerase chain reaction (qRT-PCR), Western Blotting and immunohistochemistry.
miRNA isolation and analysis
Total RNA was isolated after phenol/guanidine-based lysis of 50 µg samples from flank tumors by silicamembrane-based purification with the miRNeasy Mini Kit (Qiagen) following the manufacturer's instructions. Isolated RNA was analyzed on mouse Sure Print G3 miRNA V17.0 microarray chips (8x60K; # G4859B) from Agilent Technologies based on miRBase, release 17.0, representing 1079 mouse miRNAs with 40 replicates [32]. Briefly, hybridization of Cyanine3-labeled miRNA samples to the high-definition arrays was carried out at 55°C for 20 h under constant rotation (20 rpm). Array-chips were scanned in a high resolution microarray scanner from Agilent Technologies using Scan Control 8.5.1 software, 3 microns resolution in double path mode. Signal intensity values were extracted from the raw data file using feature extraction software (Agilent Technologies). We used the computed gTotalProbeSignal, which is the average of all the background corrected signals for each replicated probe. Then we summed up the gTotalProbeSignals to calculate the total expression value for each miRNA per sample. Quantile normalization was applied to normalize expression values across the arrays using the preprocessCore package of the programming language R. We performed a log2 transformation of the data, which were then deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE47211 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc= GSE47211) [33]. For cluster analysis, we applied complete linkage hierarchical clustering using the Euclidian distance to compute the dissimilarity of miRNA (rows) and samples (columns) independently of each other. To compare expression values of miRNAs between the control and treatment groups, we applied the independent two-tailed t-test to find significantly deregulated miRNAs. The computed P-values were adjusted for multiple testing using the FDR (false discovery rate) approach by Benjamini and Hochberg [34]. MiRNAs with a FDR adjusted P-value smaller than 0.05 were considered statistically significant.
Pathway analysis of miRNA
For in silico data mining predicted mouse miRNA targets were downloaded from miRDB (http://mirdb.org/miRDB/). Pathways putatively regulated by miRNAs which are altered by curcumin consumption were found by over-representation analysis (ORA). This was carried out by means of the online analysis-tool GeneTrail [35]. As test sets we used the targets of the up-regulated and the down-regulated miRNAs found differentially expressed comparing treatment and control group. As reference set we used the targets of all mouse miRNAs on the microarray. GeneTrail exhibited a search in disease relevant cellular pathways in the Kyoto Encyclopedia of Genes and Genomes (KEGG) for enrichment of putative miRNA targets [36]. The P-values for the KEGG pathways were FDR adjusted and considered significant if smaller than 0.05.
qRT-PCR of tumor samples
For the validation of the miRNA microarray results, qRT-PCR was performed to analyze the expression of the four selected miRNAs mmu-miR-205-5p, mmu-miR-205-3p, mmu-miR-142-5p and mmu-miR-130b-3p using miScript PCR System (Qiagen). For this purpose, RNA of tumor samples was reversely transcribed into cDNA with the miScript II RT Kit (Qiagen). The samples were analyzed by StepOnePlus™ RT-PCR System (Applied Biosystems) with miScript primer assays (Qiagen), according to the instructions of the manufacturer. RNU6B was used as endogenous control.
Western Blot analysis
To investigate the expression of putative targets of mmu-miR-205-5p, which was identified to be the most up-regulated miRNA under curcumin treatment, the whole protein fractions were purified from the organic phase of the phenol/guanidine-based RNA-isolation of four randomly chosen curcumin-treated and four control tumors. Then, 7.5 µg of protein were loaded per lane, separated discontinuously on 12% sodium dodecylsulfate polyacrylamide gels and transferred to nitrocellulose membranes (BioRad). After blocking of non-specific binding sites, membranes were incubated overnight at 4°C with a mouse monoclonal anti-Bcl-2 antibody (1∶1.000; Cell Signaling), a mouse monoclonal anti-proliferating cell nuclear antigen (PCNA) antibody (1∶1.000; DAKO) and a rabbit polyclonal anti-E2F1 antibody (1∶200; Santa Cruz) followed by the corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies (1∶5.000; Dianova). Protein expression was visualized using luminol-enhanced chemiluminescence (ECL Plus; GE Healthcare) and autoradiography film (Hyperfilm; GE Healthcare) as specified by the manufacturer. Signals were densitometrically assessed with the gel analysis tool of the software ImageJ.
Immunohistochemistry
Formalin-fixed specimens of curcumin-treated and control tumors were embedded in paraffin. To analyze the microvessel density of the tumors by immunohistochemical detection of the endothelial cell marker CD31, 2 µm-thick sections were cut and stained with a monoclonal rat anti-mouse CD31 antibody (1∶30; Dianova) as primary antibody followed by cyanin-3-coupled goat anti-rat IgG (1∶50; Dianova) as secondary antibody. Counterstaining of cell nuclei was performed with Hoechst (1∶500; Sigma-Aldrich). Subsequently, sections were examined using a BZ-8000 microscope (Keyence) for the quantitative analysis of the microvessel density within the tumors, given in mm−2.
qRT-PCR of melanoma cell lines
To investigate whether the curcumin-induced expression pattern of the key miRNAs identified in the in vivo experiments is also transferable to other melanoma cell lines, murine B78H1 cells as well as human SK-MEL-28 (ATCC® HTB-72™) and MeWo cells (ATCC® HTB-65™) were treated with 20 µM curcumin or vehicle (0.1% DMSO; control) at 37°C and 5% CO2 for 48 h. Subsequently, the cells were harvested and the expression of mmu-miR-205-5p, mmu-miR-205-3p (or hsa-miR-205-3p for human cells), mmu-miR-142-5p and mmu-miR-130b-3p was analyzed by qRT-PCR, as described above.
Flow cytometric analysis of B78H1 cells
To investigate the effect of curcumin treatment on proliferation and apoptosis of unsynchronized B78H1 cells, cell cycle analyses were performed by means of flow cytometry. For this purpose, B78H1 cells were treated with 20 µM and 40 µM curcumin or vehicle (0.1% DMSO; control) at 37°C and 5% CO2 for 48 h. Subsequently, the cells were fixed in methanol and stored at −20°C overnight. After thawing and washing, the cells were then treated with RNase A (1∶400, 10 mg/mL stock solution; Macherey-Nagel), stained with propidium iodide (1∶100, 1 mg/mL stock solution; Sigma-Aldrich) and analyzed by means of a FACScan™ (BD Biosystems). Four independent experiments were performed and each sample was analyzed in duplicate.
Statistical analysis
Values are shown as mean ± standard error of the mean (SEM), as fold expression compared to control (qRT-PCR) or as relative expression (Western Blot analysis). Data were first analyzed for normal distribution and equal variance. Differences between two experimental groups were calculated by the unpaired Student's t-test. To test for time effects during the duration of the experiment within each experimental group, analysis of variance (ANOVA) for repeated measures was applied. The post hoc analyses included the correction of the α-error according to Bonferroni probabilities to compensate for multiple comparisons (SigmaStat; Jandel Corporation). A value of P<0.05 was considered statistically significant.
Results
Analysis of tumor growth and vascularization
We analyzed the growth of amelanotic B78H1 tumors in the flanks of curcumin-treated and untreated C57BL/6 mice by means of high-resolution ultrasound imaging. For this purpose, we subcutaneously injected 1×105 B78H1 cells in both flanks of male C57BL/6 mice for the measurement of tumor growth over a period of 28 days and to obtain tumor samples for miRNA profiling. Repetitive ultrasound imaging of the developing flank tumors revealed a markedly decreased tumor volume in curcumin-treated animals at day 28 when compared to untreated controls (Figure 1). However, additional immunohistochemical analyses showed that the density of CD31-positive microvessels in curcumin-treated tumors (64±9 mm−2) did not significantly differ from that of controls (82±8 mm−2; P = 0.142).
Figure 1. Significant growth inhibition of B78H1 melanoma by curcumin diet.
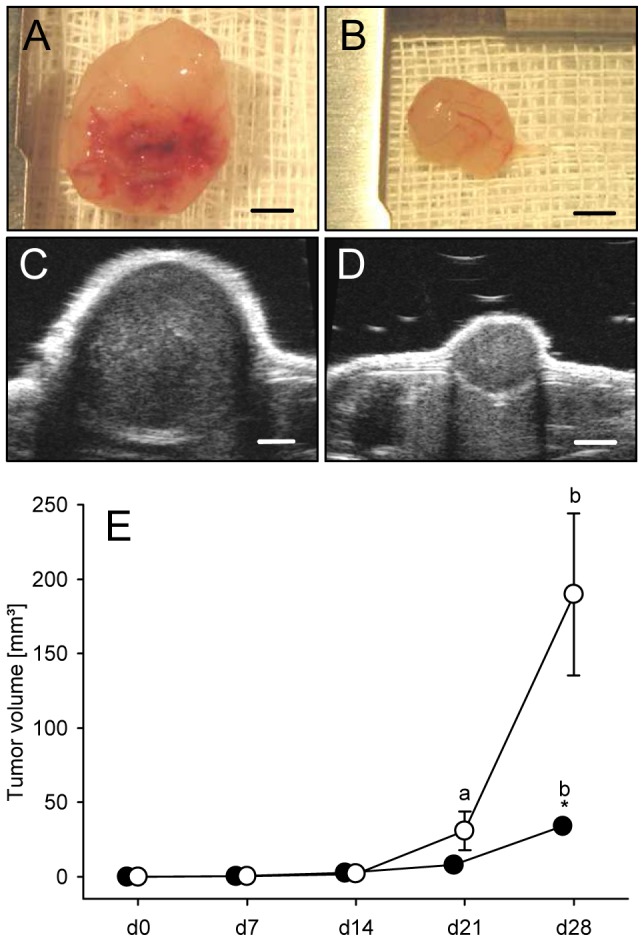
A, B: Representative images of tumors from a control (A) and a curcumin-treated animal (B) at day 28. Scale bars: 2.7 mm. C, D: High resolution ultra-sound images of B78H1 tumors of either a control (C) or a curcumin-treated mouse (D) at day 28. Scale bars: 2.0 mm. E: The volume (mm3) of control tumors (white circles) and curcumin-treated tumors (black circles), as assessed by repetitive high-resolution ultrasound imaging. Means ± SEM. a P<0.05 vs. d0, d7 and d14 within the individual group; b P<0.05 vs. d0, d7, d14 and d21 within the individual group; *P = 0.008 vs. control tumors.
Analysis of miRNA expression
At day 28, total RNA including miRNA was extracted from flank tumors of curcumin-treated and untreated control animals. Following total RNA isolation from the flank tumors, we analyzed the expression of 1079 mouse miRNAs on a mouse Sure Print G3 miRNA V17.0 microarray from Agilent Technologies. We applied an independent two-tailed t-test to search for miRNAs that were significantly altered by curcumin intake. We identified 147 miRNAs to be significantly differentially regulated by curcumin administration with an adjusted P-value lower than 0.05. Out of the 86 up-regulated miRNAs, we found 49 miRNAs more than two-fold up-regulated, and out of the 61 down-regulated miRNAs, we found 34 miRNAs lower than 0.5-fold down-regulated (Table S1). The ten most up-regulated miRNAs included mmu-miR-205-5p, mmu-miR-222-3p, mmu-miR-205-3p, mmu-miR-146b-5p, mmu-miR-21-5p, mmu-miR-21-3p, mmu-miR-221-3p, mmu-miR-140-3p, mmu-miR-142-5p, and mmu-miR-140-5p and the ten most down-regulated miRNAs comprised mmu-miR-211-5p, mmu-miR-3096-5p, mmu-miR-711, mmu-miR-466h-5p, mmu-miR-130b-3p, mmu-miR-3082-5p, mmu-miR-1199-5p, mmu-miR-669b-5p, mmu-miR-1187, and mmu-miR-1224-5p (Table 1). The expression levels of mmu-miR-205-5p, mmu-miR-205-3p, mmu-miR-142-5p and mmu-miR-130b-3p, which have previously been described in the literature as miRNAs with potential anti-cancer properties [37], [38], were confirmed by qRT-PCR (Figure 2). Of interest, we also found these key miRNAs regulated by curcumin in cultured murine B78H1 cells and human SK-MEL-28 cells (Figure 3). In contrast, MeWo cells, which are derived from melanoma lymph node metastases, did not exhibit a strong regulation of these miRNAs (Figure 3).
Table 1. Top-ten miRNAs regulated by curcumin diet.
| miRNA | Δ expression (fold change) | p vs. co (t-test) | sequence mmu | sequence hsa |
| ↑ | ||||
| 205-5p | 135.506 | 0.025 | UCCUUCAUUCCACCGGAGUCUG | UCCUUCAUUCCACCGGAGUCUG |
| 222-3p | 9.392 | 0.003 | AGCUACAUCUGGCUACUGGGU | AGCUACAUCUGGCUACUGGGU |
| 205-3p | 9.137 | 0.010 | GAUUUCAGUGGAGUGAAGCUCA | GAUUUCAGUGGAGUGAAGUUC |
| 146b-5p | 7.379 | 0.005 | UGAGAACUGAAUUCCAUAGGCU | UGAGAACUGAAUUCCAUAGGCU |
| 21-5p | 7.148 | 0.028 | UAGCUUAUCAGACUGAUGUUGA | UAGCUUAUCAGACUGAUGUUGA |
| 21-3p | 6.877 | 0.003 | CAACAGCAGUCGAUGGGCUGUC | CAACACCAGUCGAUGGGCUGU |
| 221-3p | 6.658 | 0.004 | AGCUACAUUGUCUGCUGGGUUUC | AGCUACAUUGUCUGCUGGGUUUC |
| 140-3p | 6.647 | 0.003 | UACCACAGGGUAGAACCACGG | UACCACAGGGUAGAACCACGG |
| 142-5p | 6.217 | 0.005 | CAUAAAGUAGAAAGCACUACU | CAUAAAGUAGAAAGCACUACU |
| 140-5p | 5.524 | 0.004 | CAGUGGUUUUACCCUAUGGUAG | CAGUGGUUUUACCCUAUGGUAG |
| ↓ | ||||
| 211-5p | 0.028 | 0.039 | UUCCCUUUGUCAUCCUUUGCCU | UUCCCUUUGUCAUCCUUCGCCU |
| 3096-5p | 0.178 | 0.013 | UGGCCAAGGAUGAGAACU | nd |
| 711 | 0.222 | 0.016 | GGGACCCGGGGAGAGAUGUAAG | nd |
| 466h-5p | 0.229 | 0.049 | UGUGUGCAUGUGCUUGUGUGUA | nd |
| 130b-3p | 0.242 | 0.013 | CAGUGCAAUGAUGAAAGGGCAU | CAGUGCAAUGAUGAAAGGGCAU |
| 3082-5p | 0.242 | 0.036 | GACAGAGUGUGUGUGUCUGUGU | nd |
| 1199-5p | 0.261 | 0.023 | UCUGAGUCCCGGUCGCGCGG | nd |
| 669b-5p | 0.294 | 0.049 | AGUUUUGUGUGCAUGUGCAUGU | nd |
| 1187 | 0.298 | 0.049 | UAUGUGUGUGUGUAUGUGUGUAA | nd |
| 1224-5p | 0.299 | 0.010 | GUGAGGACUGGGGAGGUGGAG | GUGAGGACUCGGGAGGUGG |
The top-ten regulated miRNAs of B78H1 melanoma by curcumin diet are listed including the delta (Δ) fold change expression, P-values and sequences of both mouse (mmu) and human (hsa) species if available. Differences in the miRNA sequence between both species are indicated by bold letters. Note the high level of inter-species conservation of miRNAs.
Figure 2. qRT-PCR validation of key miRNA expression in B78H1 melanoma regulated by curcumin diet.
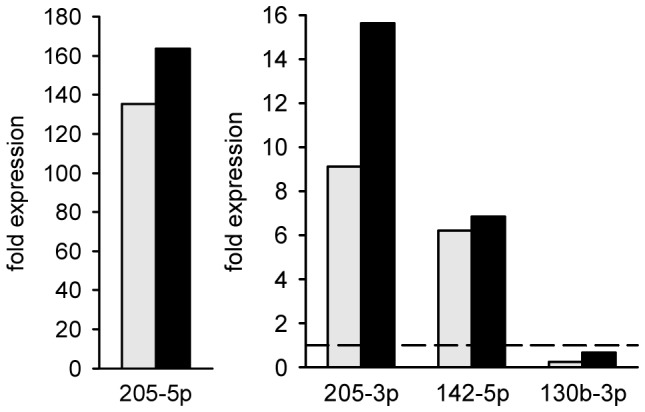
The diagrams display bar charts on the fold expression (compared to control) of mmu-miR-205-5p, mmu-miR-205-3p, mmu-miR-142-5p and mmu-miR-130b-3p in curcumin-treated B78H1 melanoma, as assessed by miRNA array (grey bars) and qRT-PCR (black bars). Broken line indicates expression level of control.
Figure 3. Expression of key miRNAs in different curcumin-treated melanoma cell lines.
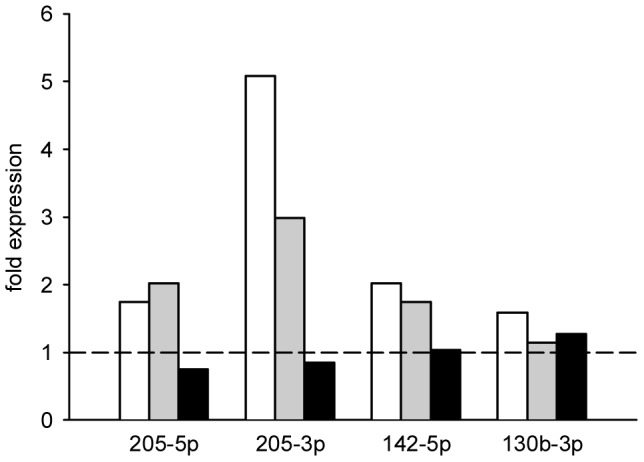
The diagram displays bar charts on the fold expression (compared to vehicle-treated control cells) of mmu-miR-205-5p, mmu-miR-205-3p, mmu-miR-142-5p and mmu-miR-130b-3p of curcumin-treated murine B78H1 (white bars), human SK-MEL-28 (grey bars) and human MeWo melanoma cells (black bars), as assessed by qRT-PCR. The cultured cells were treated with 20 µM curcumin or vehicle (0.1% DMSO) for 48 h. Broken line indicates expression level of control.
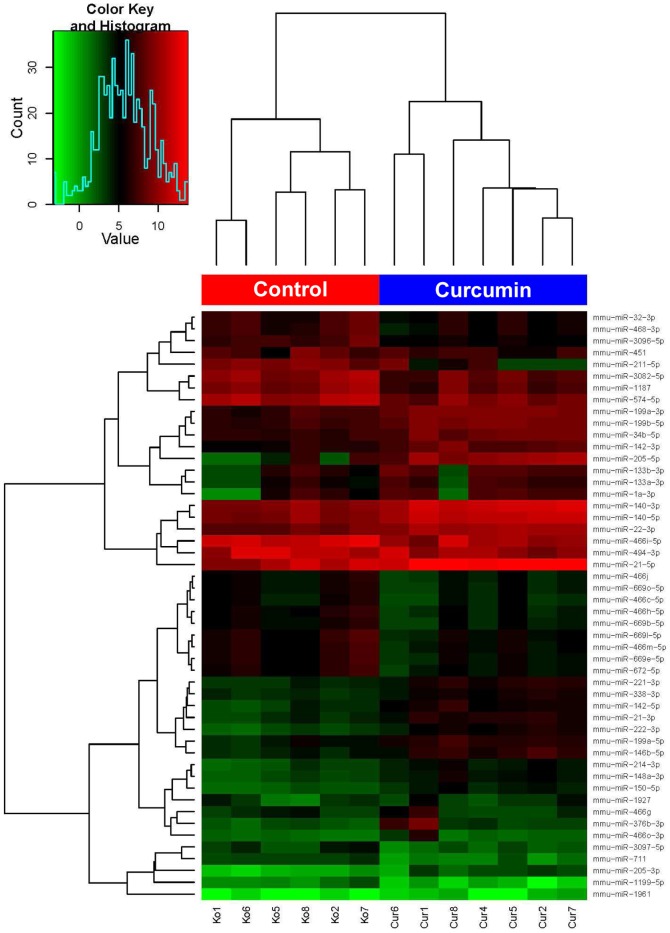
The median log expression of the ten most deregulated miRNAs is given in Figure 4. The most up-regulated miRNA, mmu-mir-205-5p, was 135 times higher and the most down-regulated miRNA, mmu-mir-211-5p, was 35.7 times lower expressed in the curcumin-treated samples (Table 1). These data show that curcumin intake has a profound impact on the miRNA expression signature of the tumors. To further evaluate the effect of curcumin, we performed complete linkage hierarchical clustering using 50 miRNAs that showed the highest expression variance across all samples. As shown in Figure 5, we found two major clusters of miRNA expression profiles for 7 curcumin and 6 control samples. The obvious clustering between the two groups provides further evidence for the impact of the curcumin diet on the miRNA expression profile of the tumors.
Figure 4. Expression profile of top-ten miRNA of B78H1 melanoma regulated by curcumin diet.
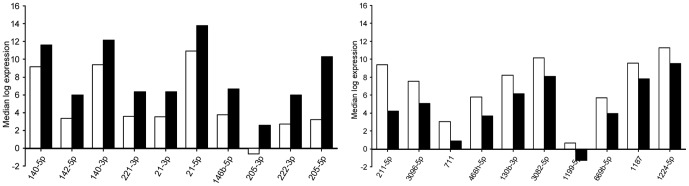
The diagram displays bar charts on the median log expression of the top-ten up-regulated (A) and top-ten down-regulated (B) miRNA of the B78H1 melanoma after curcumin intake (black bars) compared to the control group (white bars).
Figure 5. Curcumin diet causes clustering of biological replicates by changing B78H1 melanoma miRNA signature.
Clustering of male C57BL/6 mice fed for 6 weeks either curcumin diet or control diet based on miRNA expression levels in the B78H1 melanoma. This diagram is based on 50 miRNAs with the highest variance across all samples. MiRNA expression variation is represented by different colors (see color key). Complete linkage hierarchical clustering was performed using the Euclidian distance to calculate the dissimilarity of miRNA and samples independently of one another. Clustering of samples is illustrated by upper dendrogram, clustering of miRNAs by the left dendrogram. Note that, based on the evaluated C57BL/6 mice, control and curcumin animals clearly divide into two main clusters.
Using our gene set analysis tool GeneTrail, we performed in silico pathway analyses separately for the predicted targets of the total of the 86 up-regulated and the total of 61 down-regulated miRNAs. Putative targets of curcumin-induced up-regulated miRNAs were enriched in the KEGG pathways “o-glycan biosynthesis”, “endoplasmatic reticulum protein processing” as well as in several cancer-related pathways (Table 2). Predicted targets of up-regulated miRNAs were also found to be enriched in the insulin-pathway and furthermore linked to heart-associated and neurological contexts (Table 2). Additionally, we detected over-representation in the Gene Ontology (GO) terms “cellular proliferation”, “cell death” and “regulation of apoptosis”. Predicted targets of down-regulated miRNAs were found to be enriched in a larger number of KEGG pathways including mTOR- and ErbB-signalling pathway, and also several cancer related pathways (Table 2).
Table 2. Pathways putatively regulated by curcumin-altered miRNAs as found by over-representation analysis.
| KEGG pathways with accumulated predicted targets | P-value | Expected number of targets | Observed number of targets | x-fold of target accumulation | |
| Up-regulated miRNAs ↑ | |||||
| mucine type O-glycan biosynthesis | 0.007 | 15 | 24 | 1.60 | |
| renal cell carcinoma | 0.024 | 37 | 50 | 1.35 | |
| pathways in cancer | 0.026 | 171 | 198 | 1.16 | |
| protein processing in endoplasmatic reticulum | 0.029 | 85 | 104 | 1.22 | |
| acute myeloid leukemia | 0.029 | 31 | 42 | 1.35 | |
| Down-regulated miRNAs ↓ | |||||
| Cholinergic synapsis | 0.021 | 42 | 58 | 1.38 | |
| ErbB signaling pathway | 0.021 | 34 | 50 | 1.47 | |
| Focal adhesion | 0.021 | 80 | 102 | 1.28 | |
| Neuroactive ligand-receptor interaction | 0.021 | 94 | 117 | 1.24 | |
| Pancreatic cancer | 0.021 | 29 | 43 | 1.48 | |
| Renal cell carcinoma | 0.021 | 28 | 41 | 1.46 | |
| Dilated cardiomyopathie | 0.021 | 31 | 45 | 1.45 | |
| Arrhythmogenic right ventricular cardiomyopathy | 0.022 | 28 | 41 | 1.46 | |
| Long-term potentiation | 0.022 | 27 | 39 | 1.44 | |
| Retrograde endocannabinoid sginaling | 0.022 | 38 | 52 | 1.37 | |
| Type II diabetes mellitus | 0.022 | 18 | 28 | 1.56 | |
| mTOR signaling pathway | 0.022 | 26 | 38 | 1.47 | |
| Hypertrophic cardiomyopathy (HCM) | 0.025 | 30 | 42 | 1.42 | |
| Prostate cancer | 0.025 | 35 | 48 | 1.38 | |
| Insulin signaling pathway | 0.029 | 56 | 72 | 1.29 | |
| Neurotrophin signaling pathway | 0.030 | 48 | 63 | 1.31 | |
| Morphine addiction | 0.030 | 34 | 47 | 1.38 | |
| Colorectal cancer | 0.030 | 25 | 36 | 1.42 | |
| Dopaminergic synapse | 0.043 | 51 | 65 | 1.28 |
Listed are pathways being putatively regulated by miRNAs of B78H1 melanoma which are altered by curcumin diet, as found by over-representation analysis with the online analysis-tool GeneTrail. As test sets we used the predicted targets of differentially regulated miRNAs and as reference set the putative targets of all murine miRNAs printed on the microarray. GeneTrail exhibited a search in disease-relevant cellular pathways as found in the Kyoto Encyclopedia of Genes and Genomes (KEGG) for enrichment of putative targets. MiRNAs up-regulated by curcumin diet (upper part of table) were separately tested from down-regulated miRNAs (lower part of table). The expected number of target genes, modulated in a pathway, is shown as well as the observed ones, including the according P-value and x-fold enrichment of targets.
Target validation
Based on our in silico analyses, we selected anti-apoptotic Bcl-2 and the transcription factor E2F1 for PCNA to validate some of the predicted targets of mmu-miR-205-5p, which was the most highly regulated miRNA by curcumin treatment. We found that Bcl-2 expression was significantly down-regulated in curcumin-treated tumors when compared to controls (Figure 6). In contrast, E2F1 expression was not markedly altered by curcumin treatment (Figure 6). However, the expression of the downstream proliferation marker PCNA was significantly reduced in the tumors under curcumin diet (Figure 6).
Figure 6. Western Blot analysis of the expression of target proteins of mmu-miR-205-5 in B78H1 melanoma.
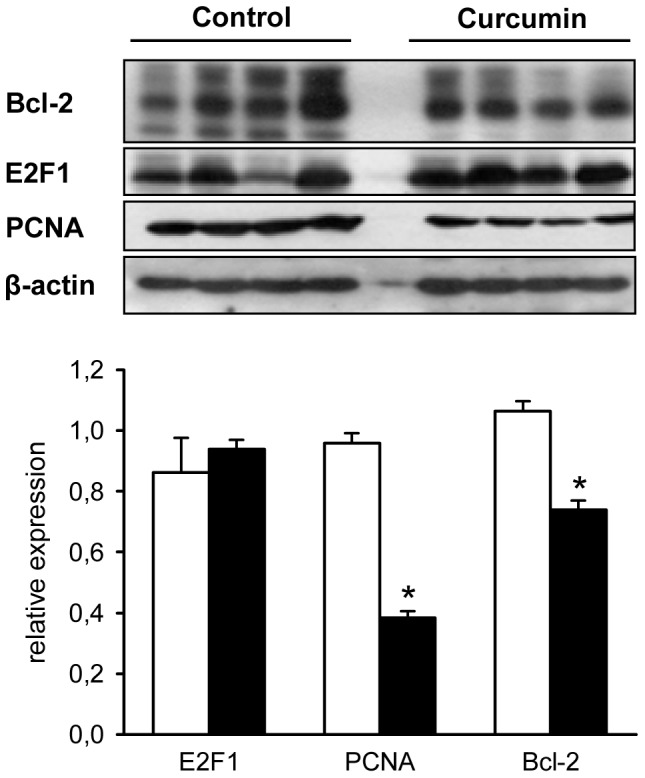
Relative expression of Bcl-2, E2F1 and PCNA in B78H1 melanoma of control (white bars, n = 4) and curcumin-treated animals (black bars, n = 4), as assessed by Western Blot analysis. Means ± SEM. *P<0.05 vs. control.
Cell cycle analysis
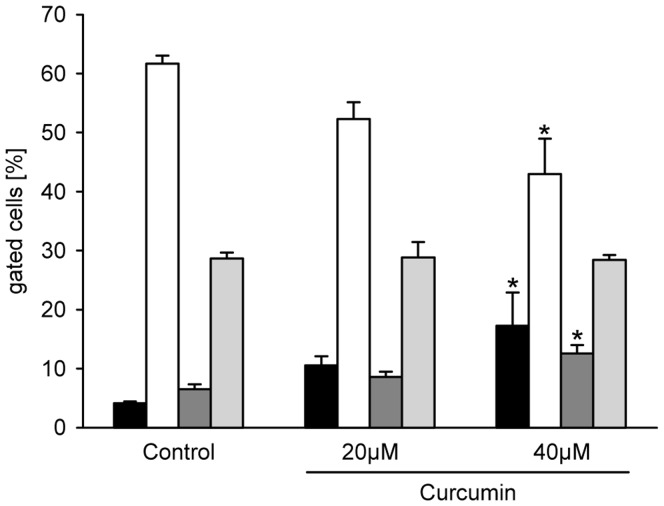
Flow cytometric cell cycle analyses showed dose-dependently a higher percentage of apoptotic cells in curcumin-treated B78H1 cells when compared to vehicle-treated controls (Figure 7). Moreover, curcumin treatment dose-dependently resulted in an S phase delay, indicating an additional inhibitory action on cell proliferation (Figure 7).
Figure 7. Cell cycle analysis of curcumin-treated B78H1 cells.
Gated cells [%] in G1 phase (white bars), S phase (dark grey bars), G2 phase (light grey bars) and apoptosis (black bars), as measured by flow cytometry. The cultured cells were treated with 20 µM and 40 µM curcumin or vehicle (0.1% DMSO) for 48 h. Means ± SEM. *P<0.05 vs. control.
Discussion
In the present study we investigated the effect of dietary curcumin consumption on the miRNA expression signature of engrafting mouse melanomas, which were established by injection of murine B78H1 cells in the flank of C57BL/6 mice. Because this approach does not include the transformation of normal cells into tumor cells (tumor initiation), the herein observed curcumin effects may be primarily of relevance for therapy or secondary prevention, but not for primary prevention of cancer.
We first analyzed potential anti-cancer properties of oral curcumin treatment that have previously been reported in the literature. These include in particular the inhibition of cell proliferation and tumor growth [18], [19], [39]. In line with these findings, we observed in our flank model a reduction of tumor size in curcumin-treated animals.
Moreover, curcumin has been described as an anti-angiogenic agent [16]. Therefore, we additionally measured the microvessel density of curcumin-treated and control tumors by immunohistochemical detection of the endothelial cell marker CD31. We found that the density of CD31-positive microvessels in curcumin-treated tumors did not significantly differ from that of controls. However, this does not necessarily indicate that curcumin did not exert an anti-angiogenic action on the treated tumors. In fact, according to Hlatky et al. [40] and Nico et al. [41], tumors under anti-angiogenic treatment may follow a ‘shrink to fit’ adaptation. This means that the tumor cell population may decrease in direct proportion to the loss of its supporting vasculature, resulting in a constant ratio between tumor cells and microvessels and, thus, a constant microvessel density. Accordingly, we herein found that tumor cell proliferation was markedly reduced in curcumin-treated animals when compared to controls.
In the next step we analyzed the miRNA signature of the isolated flank tumors, because there is increasing proof for miRNAs playing a pivotal role in the etiology of cancer. MiRNAs can act as tumor suppressors but also as oncogenes (onco-mir). Here, we provide strong evidence that the overall miRNA expression pattern in melanomas of curcumin-treated mice is markedly altered. The top ten miRNAs being significantly up-regulated by curcumin treatment comprised mmu-miR-205-5p (135-fold), mmu-miR-205-3p (9-fold) and mmu-miR-142-5p (6-fold). Their expression levels were confirmed by qRT-PCR. Up-regulation of mmu-miR-205-5p and -3p has been shown to reverse epithelial-to-mesenchymal transition in various tumors with E-cadherin as a central target [42], [43]. Hsa-miR-205 was found to be 100 times down-regulated in human metastatic melanoma compared to non-metastatic ones [38]. Down-regulation of miR-142-5p is associated with recurrence and poor overall survival in gastric and pancreatic cancer patients [12], [44]. Furthermore, we also identified several miRNAs, which were significantly down-regulated in curcumin-treated melanomas, such as mmu-miR-130b-3p. In former studies, this miRNA has been found to be up-regulated in cutaneous malignant melanoma [37]. Li et al. [45] could demonstrate that hsa-miR-130b induces epithelial-to-mesenchymal transition in endometrial cancer. Taken together, these results indicate that the curcumin-regulated miRNAs mmu-miR-205-5p, mmu-miR-205-3p, mmu-miR-142-5p and mmu-miR-130b-3p represent promising targets as well as markers to determine the aggressiveness and metastatic activity of malignant tumors.
In an additional experimental setting, we analyzed whether these key miRNAs underlie a similar regulation in different melanoma cell lines. For this purpose, we determined their expression by means of qRT-PCR in curcumin-treated and vehicle-treated murine B78H1 cells as well as human SK-MEL-28 and MeWo cells. Curcumin treatment resulted in higher expression levels in cultured B78H1 and SK-MEL-28 cells when compared to controls. In contrast, MeWo cells did not show a strong regulation of the key miRNAs. This may be due to the fact that MeWo cells are derived from melanoma lymph node metastases. Thus, they may exhibit a completely different miRNA expression profile when compared to cells originating from primary tumors.
Of interest, some of the most highly altered miRNAs were not regulated in a manner as expected based on the results of other studies. For example, down-regulation of hsa-miR-211-5p has been shown to be associated with melanoma progression [38], [46] and not as observed in the present study with inhibition of tumor growth in curcumin-treated melanoma. These contradictory findings may be explained by the fact that an altered miRNA expression does not necessarily imply a direct influence of curcumin on the miRNA expression of the tumor but can also be based on indirect feedback regulation. Furthermore, the effects of curcumin treatment may vary between tumors of different cell etiology or besides based on different experimental set-ups. For example, several investigators showed a down-regulation of up-regulated hsa-mir-21 after curcumin treatment in human pancreatic, oesophageal and prostate cancer cells [28], [29], [47], [48]. In contrast, Sun et al. [24] found an up-regulation of hsa-miR-21 expression induced by curcumin treatment in pancreatic cells similar to our findings.
Our over-representation analysis (ORA) identified several putative cellular pathways, which are modulated by up- and down-regulated miRNAs. Predicted targets of miRNAs were found to be enriched in metabolic pathways like insulin-pathway but also in heart-associated and neurological contexts, indicating a potential role for curcumin in managing metabolic syndrome. In addition, many of the pathways discovered by ORA were connected to cell proliferation and apoptotic cell death in cancer, emphasizing the impact of curcumin in the treatment of malignancies.
Based on the current literature and our in silico analyses, predicted targets of mmu-miR-205-5p include E2F1, E2F3, E2F5, Bcl-2, CD31, hypoxia-inducible transcription factor (HIF)-1α, and VEGF [49], [50]. From these targets we selected anti-apoptotic Bcl-2 and E2F1, which is a transcription factor for PCNA, for further validation by Western Blotting. Of interest, we found that Bcl-2 expression was significantly down-regulated in curcumin-treated tumors when compared to controls, whereas E2F1 expression was not markedly altered. The expression of PCNA, however, was significantly reduced under curcumin diet. This indicates that the expression of this proliferation marker may be regulated by other miRNAs than mmu-miR-205-5p or by other transcription factors than E2F1. In this context, mmu-miR-222 represents a potential candidate, which was also found to be up-regulated in the present study and directly regulates the expression of PCNA. Our results on the down-regulation of Bcl-2 and PCNA expression were further supported by flow cytometric cell cycle analyses of curcumin-treated B78H1 cells, indicating that curcumin affects both apoptosis and proliferation of this melanoma cell line.
In conclusion, we demonstrate for the first time a clear impact of dietary curcumin on the miRNA profile in murine melanoma with mmu-miR-205-5p being up-regulated most extensively. Linkage hierarchical clustering of the 50 miRNAs with the highest expression variance revealed a grouping into two main clusters, one comprising the treated animals and the other the controls. Putative targets of curcumin-induced up-regulated miRNAs are enriched in several cancer-related pathways. These data suggest miRNAs playing a significant role in melanoma growth inhibition by oral curcumin administration.
Supporting Information
Significant differential expression of miRNA after curcumin diet (DOC).
(DOCX)
Acknowledgments
We are highly appreciating the technical assistance of Janine Becker. We thank Prof. Klaus Römer for kindly providing the E2F1 antibody.
Funding Statement
This work was supported by funds from the German José Carreras Leukemia Foundation (DJCLS R 09/20) to UM. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Fabian MR, Sonenberg N, Filipowicz W (2010) Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 79: 351–379. [DOI] [PubMed] [Google Scholar]
- 2. Chitwood DH, Timmermans MC (2010) Small RNAs are on the move. Nature 467: 415–419. [DOI] [PubMed] [Google Scholar]
- 3. Taby R, Issa JP (2010) Cancer epigenetics. CA Cancer J Clin 60: 376–392. [DOI] [PubMed] [Google Scholar]
- 4. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, et al. (2005) miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A 102: 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Litz J, Krystal GW (2006) Imatinib inhibits c-Kit-induced hypoxia-inducible factor-1alpha activity and vascular endothelial growth factor expression in small cell lung cancer cells. Mol Cancer Ther 5: 1415–1422. [DOI] [PubMed] [Google Scholar]
- 6. Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC (2007) Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res 100: 1164–1173. [DOI] [PubMed] [Google Scholar]
- 7. Friedman JM, Jones PA (2009) MicroRNAs: critical mediators of differentiation, development and disease. Swiss Med Wkly 139: 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poliseno L, Haimovic A, Segura MF, Hanniford D, Christos PJ, et al. (2012) Histology-specific microRNA alterations in melanoma. J Invest Dermatol 132: 1860–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fichtlscherer S, Zeiher AM, Dimmeler S (2011) Circulating microRNAs: biomarkers or mediators of cardiovascular diseases? Arterioscler Thromb Vasc Biol 31: 2383–2390. [DOI] [PubMed] [Google Scholar]
- 10. Keller A, Leidinger P, Gislefoss R, Haugen A, Langseth H, et al. (2011) Stable serum miRNA profiles as potential tool for non-invasive lung cancer diagnosis. RNA Biol 8: 506–516. [DOI] [PubMed] [Google Scholar]
- 11. Izzotti A, Cartiglia C, Steele VE, De Flora S (2012) MicroRNAs as targets for dietary and pharmacological inhibitors of mutagenesis and carcinogenesis. Mutat Res 751: 287–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ohuchida K, Mizumoto K, Kayashima T, Fujita H, Moriyama T, et al. (2011) MicroRNA expression as a predictive marker for gemcitabine response after surgical resection of pancreatic cancer. Ann Surg Oncol 18: 2381–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Izzotti A, Larghero P, Cartiglia C, Longobardi M, Pfeffer U, et al. (2010) Modulation of microRNA expression by budesonide, phenethyl isothiocyanate and cigarette smoke in mouse liver and lung. Carcinogenesis 31: 894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jensen JD, Wing GJ, Dellavalle RP (2010) Nutrition and melanoma prevention. Clin Dermatol 28: 644–649. [DOI] [PubMed] [Google Scholar]
- 15. Singh M, Singh N (2009) Molecular mechanism of curcumin induced cytotoxicity in human cervical carcinoma cells. Mol Cell Biochem 325: 107–119. [DOI] [PubMed] [Google Scholar]
- 16. Kunnumakkara AB, Anand P, Aggarwal BB (2008) Curcumin inhibits proliferation, invasion, angiogenesis and metastasis of different cancers through interaction with multiple cell signaling proteins. Cancer Lett 269: 199–225. [DOI] [PubMed] [Google Scholar]
- 17. Okunieff P, Xu J, Hu D, Liu W, Zhang L, et al. (2006) Curcumin protects against radiation-induced acute and chronic cutaneous toxicity in mice and decreases mRNA expression of inflammatory and fibrogenic cytokines. Int J Radiat Oncol Biol Phys 65: 890–898. [DOI] [PubMed] [Google Scholar]
- 18. Phillips JM, Clark C, Herman-Ferdinandez L, Moore-Medlin T, Rong X, et al. (2011) Curcumin inhibits skin squamous cell carcinoma tumor growth in vivo. Otolaryngol Head Neck Surg 145: 58–63. [DOI] [PubMed] [Google Scholar]
- 19. Limtrakul P, Lipigorngoson S, Namwong O, Apisariyakul A, Dunn FW (1997) Inhibitory effect of dietary curcumin on skin carcinogenesis in mice. Cancer Lett 116: 197–203. [DOI] [PubMed] [Google Scholar]
- 20. Carroll RE, Benya RV, Turgeon DK, Vareed S, Neuman M, et al. (2011) Phase IIa clinical trial of curcumin for the prevention of colorectal neoplasia. Cancer Prev Res (Phila) 4: 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kanai M, Yoshimura K, Asada M, Imaizumi A, Suzuki C, et al. (2011) A phase I/II study of gemcitabine-based chemotherapy plus curcumin for patients with gemcitabine-resistant pancreatic cancer. Cancer Chemother Pharmacol 68: 157–164. [DOI] [PubMed] [Google Scholar]
- 22. Dhillon N, Aggarwal BB, Newman RA, Wolff RA, Kunnumakkara AB, et al. (2008) Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin Cancer Res 14: 4491–4499. [DOI] [PubMed] [Google Scholar]
- 23. Bayet-Robert M, Kwiatkowski F, Leheurteur M, Gachon F, Planchat E, et al. (2010) Phase I dose escalation trial of docetaxel plus curcumin in patients with advanced and metastatic breast cancer. Cancer Biol Ther 9: 8–14. [DOI] [PubMed] [Google Scholar]
- 24. Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, et al. (2008) Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther 7: 464–473. [DOI] [PubMed] [Google Scholar]
- 25. Yang J, Cao Y, Sun J, Zhang Y (2010) Curcumin reduces the expression of Bcl-2 by upregulating miR-15a and miR-16 in MCF-7 cells. Med Oncol 27: 1114–1118. [DOI] [PubMed] [Google Scholar]
- 26. Zhang J, Du Y, Wu C, Ren X, Ti X, et al. (2010) Curcumin promotes apoptosis in human lung adenocarcinoma cells through miR-186* signaling pathway. Oncol Rep 24: 1217–1223. [DOI] [PubMed] [Google Scholar]
- 27. Mudduluru G, George-William JN, Muppala S, Asangani IA, Kumarswamy R, et al. (2011) Curcumin regulates miR-21 expression and inhibits invasion and metastasis in colorectal cancer. Biosci Rep 31: 185–197. [DOI] [PubMed] [Google Scholar]
- 28. Bao B, Ali S, Banerjee S, Wang Z, Logna F, et al. (2012) Curcumin analogue CDF inhibits pancreatic tumor growth by switching on suppressor microRNAs and attenuating EZH2 expression. Cancer Res 72: 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bao B, Ali S, Kong D, Sarkar SH, Wang Z, et al. (2011) Anti-tumor activity of a novel compound-CDF is mediated by regulating miR-21, miR-200, and PTEN in pancreatic cancer. PLoS One 6: e17850. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30. Newcomb EW, Silverstein SC, Silagi S (1978) Malignant mouse melanoma cells do not form tumors when mixed with cells of a non-malignant subclone: relationships between plasminogen activator expression by the tumor cells and the host's immune response. J Cell Physiol 95: 169–177. [DOI] [PubMed] [Google Scholar]
- 31. Lollini PL, De Giovanni C, Del Re B, Nicoletti G, Prodi G, et al. (1987) Interferon-mediated enhancement of metastasis. Are MHC antigens involved? Clin Exp Metastasis 5: 277–287. [DOI] [PubMed] [Google Scholar]
- 32. Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36: D154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Benjamini Y, Hochberg Y (1995) Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Statist Soc B 57: 289–300. [Google Scholar]
- 35. Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, et al. (2007) GeneTrail—advanced gene set enrichment analysis. Nucleic Acids Res 35: W186–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanabe M, Kanehisa M (2012) Using the KEGG database resource. Curr Protoc Bioinformatics Chapter 1: Unit1 12. [DOI] [PubMed]
- 37. Sand M, Skrygan M, Sand D, Georgas D, Gambichler T, et al. (2013) Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell Tissue Res 351: 85–98. [DOI] [PubMed] [Google Scholar]
- 38. Xu Y, Brenn T, Brown ER, Doherty V, Melton DW (2012) Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br J Cancer 106: 553–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Leyon PV, Kuttan G (2003) Studies on the role of some synthetic curcuminoid derivatives in the inhibition of tumour specific angiogenesis. J Exp Clin Cancer Res 22: 77–83. [PubMed] [Google Scholar]
- 40. Hlatky L, Hahnfeldt P, Folkman J (2002) Clinical application of antiangiogenic therapy: microvessel density, what it does and doesn't tell us. J Natl Cancer Inst 94: 883–893. [DOI] [PubMed] [Google Scholar]
- 41. Nico B, Benagiano V, Mangieri D, Maruotti N, Vacca A, et al. (2008) Evaluation of microvascular density in tumors: pro and contra. Histol Histopathol 23: 601–607. [DOI] [PubMed] [Google Scholar]
- 42. Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, et al. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601. [DOI] [PubMed] [Google Scholar]
- 43. Greene SB, Gunaratne PH, Hammond SM, Rosen JM (2010) A putative role for microRNA-205 in mammary epithelial cell progenitors. J Cell Sci 123: 606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang X, Yan Z, Zhang J, Gong L, Li W, et al. (2011) Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for recurrence risk in gastric cancer patients following surgical resection. Ann Oncol 22: 2257–2266. [DOI] [PubMed] [Google Scholar]
- 45. Li BL, Lu C, Lu W, Yang TT, Qu J, et al. (2013) miR-130b is an EMT-related microRNA that targets DICER1 for aggression in endometrial cancer. Med Oncol 30: 484. [DOI] [PubMed] [Google Scholar]
- 46. Mazar J, DeYoung K, Khaitan D, Meister E, Almodovar A, et al. (2010) The regulation of miRNA-211 expression and its role in melanoma cell invasiveness. PLoS One 5: e13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Subramaniam D, Ponnurangam S, Ramamoorthy P, Standing D, Battafarano RJ, et al. (2012) Curcumin induces cell death in esophageal cancer cells through modulating Notch signaling. PLoS One 7: e30590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, et al. (2010) Gemcitabine sensitivity can be induced in pancreatic cancer cells through modulation of miR-200 and miR-21 expression by curcumin or its analogue CDF. Cancer Res 70: 3606–3617. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49. Dar AA, Majid S, de Semir D, Nosrati M, Bezrookove V, et al. (2011) miRNA-205 suppresses melanoma cell proliferation and induces senescence via regulation of E2F1 protein. J Biol Chem 286: 16606–16614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alla V, Kowtharapu BS, Engelmann D, Emmrich S, Schmitz U, et al. (2012) E2F1 confers anticancer drug resistance by targeting ABC transporter family members and Bcl-2 via the p73/DNp73-miR-205 circuitry. Cell Cycle 11: 3067–3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Significant differential expression of miRNA after curcumin diet (DOC).
(DOCX)