Abstract
Objectives
The introduction of two new non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the past 5 years and the identification of novel NNRTI-associated mutations have made it necessary to reassess the extent of phenotypic NNRTI cross-resistance.
Methods
We analysed a dataset containing 1975, 1967, 519 and 187 genotype–phenotype correlations for nevirapine, efavirenz, etravirine and rilpivirine, respectively. We used linear regression to estimate the effects of RT mutations on susceptibility to each of these NNRTIs.
Results
Sixteen mutations at 10 positions were significantly associated with the greatest contribution to reduced phenotypic susceptibility (≥10-fold) to one or more NNRTIs, including: 14 mutations at six positions for nevirapine (K101P, K103N/S, V106A/M, Y181C/I/V, Y188C/L and G190A/E/Q/S); 10 mutations at six positions for efavirenz (L100I, K101P, K103N, V106M, Y188C/L and G190A/E/Q/S); 5 mutations at four positions for etravirine (K101P, Y181I/V, G190E and F227C); and 6 mutations at five positions for rilpivirine (L100I, K101P, Y181I/V, G190E and F227C). G190E, a mutation that causes high-level nevirapine and efavirenz resistance, also markedly reduced susceptibility to etravirine and rilpivirine. K101H, E138G, V179F and M230L mutations, associated with reduced susceptibility to etravirine and rilpivirine, were also associated with reduced susceptibility to nevirapine and/or efavirenz.
Conclusions
The identification of novel cross-resistance patterns among approved NNRTIs illustrates the need for a systematic approach for testing novel NNRTIs against clinical virus isolates with major NNRTI-resistance mutations and for testing older NNRTIs against virus isolates with mutations identified during the evaluation of a novel NNRTI.
Keywords: HIV-1, drug resistance, linear regression, etravirine, rilpivirine
Introduction
Before the approvals of etravirine and rilpivirine, the prevailing dogma was that most non-nucleoside reverse transcriptase inhibitor (NNRTI)-resistance mutations conferred clinically significant high-level NNRTI class resistance. Most phenotypic studies were performed using site-directed mutants1–5 and few efforts were made to quantify the phenotypic effects of NNRTI-resistance mutations in clinical isolates. Since the approvals of etravirine and rilpivirine, views about NNRTI resistance and cross-resistance have evolved. Etravirine has been shown to have a higher genetic barrier to resistance than nevirapine and efavirenz.6,7 In addition, novel etravirine- and rilpivirine-associated mutations have been identified.6,8–10
In a previous study, we compared several statistical learning approaches to estimate the effects of RT mutations on susceptibility to nevirapine, efavirenz and delavirdine using the PhenoSense assay.11 In that study, we analysed 740 genotype–phenotype correlations for nevirapine and efavirenz. Here we analysed a dataset that has more than 1900 genotype–phenotype correlations for nevirapine and efavirenz, 519 for etravirine and 187 for rilpivirine. As part of this analysis, we specifically examined the effects of the novel etravirine- and rilpivirine-associated mutations on nevirapine and efavirenz.
Methods
HIV-1 isolates and phenotypes
We analysed HIV-1 isolates in the Stanford HIV Drug Resistance Database (HIVDB)12 on which we performed in vitro NNRTI susceptibility testing using the PhenoSense assay (Monogram, South San Francisco, CA, USA).13 Susceptibility results were expressed as the fold change in susceptibility, defined as the 50% effective concentration (EC50) of the tested isolate divided by the EC50 of a standard wild-type control isolate. Fold susceptibility results above the limit of quantification (200-fold decreased susceptibility) were right-censored. Forty percent of the susceptibility test results were from published studies; 60% were from collaborating clinics. One-half of the nevirapine and efavirenz genotype–phenotype correlations, 80% of the etravirine correlations and 70% of the rilpivirine correlations have not been previously described in the literature. With the exception of 17 patients who received rilpivirine in an ongoing clinical trial,14 few isolates in this dataset were from patients receiving etravirine or rilpivirine. The Stanford University Human Subjects Committee approved this study. The data described here are available at http://hivdb.stanford.edu/cgi-bin/GenoPhenoDS.cgi.
To minimize bias that would result from over-representing highly similar viruses, we excluded viruses obtained from the same individual that contained the same pattern of major NNRTI-resistance mutations: L100I, K101P, K103N, V106A/M, Y181C/I/V, Y188C/H/L, G190A/E/Q/S and M230L. Because mixtures of wild-type and mutant variants at major NNRTI-resistance positions may confound genotype–phenotype correlations, we excluded viruses with sequences containing electrophoretic mixtures at these same NNRTI-resistance mutations.
Definitions of mutations
Mutations were defined as differences from the consensus subtype B sequence (http://hivdb.stanford.edu/pages/documentPage/consensus_amino_acid_sequences.html). Non-polymorphic mutations were defined here as mutations that occur in ≤1.0% of viruses not subjected to antiretroviral drug (ARV) selection pressure regardless of subtype. We used the Rega subtyping method15 to assign an HIV-1 subtype to isolates with available nucleotide sequences. We obtained subtypes for the remaining isolates from the studies from which the phenotype data had been obtained or from the phenotype report.
Study-defined NNRTI-resistance mutations included: (i) common established NNRTI-resistance mutations (A98G, L100I, K101E/P, K103N/S, V106A/M, V108I, V179D, Y181C/I/V, Y188C/H/L, G190A/S, P225H and F227L); (ii) less well characterized uncommon NNRTI-resistance mutations including G190E/Q, M230L and K238T; and (iii) recently described etravirine- and rilpivirine-associated mutations, including V90I, K101H, V106I, E138A/G/K/Q, V179F/T, H221Y and F227C. With the exceptions of V90I, V106I, E138A and V179D, each of these mutations is non-polymorphic in each of the HIV-1 subtypes.12
Contribution of RT mutations to decreased NNRTI susceptibility
We performed one analysis to study the effects of established NNRTI-resistance mutations on nevirapine, efavirenz, etravirine and rilpivirine susceptibility. We performed a second, exploratory, analysis to identify novel NNRTI-resistance mutations. For the first analysis, we quantified the effects of the established study-defined mutations on susceptibility using least squares regression. For the second analysis, we used least angle regression (LARS), a model selection algorithm for the efficient prediction of a response variable from a large collection of explanatory variables,16 to identify novel mutations from the large pool of mutations that occurred commonly in the genotype–phenotype dataset. The first analysis constituted our primary analysis because it was suitable for all four NNRTIs. The second analysis was not suitable for etravirine or rilpivirine because the number of genotype–phenotype correlations for these NNRTIs was too low.
For both regression analyses, the presence or absence of each mutation was an explanatory variable and the log10-fold change in susceptibility was the response variable. In each regression model, the mutation coefficients were proportional to the contribution of the mutation to susceptibility. Five-fold cross-validation was performed on randomly chosen subdivisions of the complete dataset, which were stratified according to the proportions of susceptible, partially resistant and resistant isolates in the entire dataset. Mutations for which the mean regression coefficients from the 50 runs (10 runs of 5-fold cross-validation) exceeded 3 SD above or below zero were considered to be associated with a statistically significant change in susceptibility.
For the primary analysis, least squares regression was applied to the 35 study-defined mutations. For the secondary analysis, 469 RT mutations that occurred three or more times in the dataset were included in LARS as potential explanatory variables. During the learning stage, LARS used four-fifths of the training set (64% of the entire set) to select mutations for inclusion in the model and one-fifth of the training set to validate the mutations using the LASSO (least absolute shrinkage and selection operator) option. The test set (i.e. the remaining one-fifth of the entire set) was used to estimate the prediction performance.
Prediction performance
During each 5-fold cross-validation, the 20% of samples that constituted the test set was used to assess the prediction performance of the model. Prediction performance was then estimated using two approaches. First, the predicted log10-fold change in susceptibility in each cross-validation run was compared with the actual log10-fold change in susceptibility. The mean of the squared difference between these values constituted the mean-squared error.
The second approach involved choosing three intervals for each NNRTI: susceptible, partial resistance and resistance. The PhenoSense assay has clinical cut-offs for etravirine derived from the DUET trials,17,18 in which patients with NNRTI-resistant viruses were treated with an etravirine-containing regimen. Based on these cut-offs, <3-fold reduced susceptibility was considered susceptible, a 3- to 10-fold decrease was considered partially resistant and a >10-fold decrease was considered fully resistant.19 The PhenoSense assay does not have clinical cut-offs for nevirapine, efavirenz and rilpivirine. However, to simplify the description of our data, we also used the 3- and 10-fold cut-offs for nevirapine, efavirenz and rilpivirine, as well as etravirine. Although the clinical significance of ARV susceptibility lies on a continuum, a categorical approach provides a consistent frame of reference for assessing prediction that complements the continuous mean-squared error.
Site-directed mutants
Two site-directed mutant recombinant infectious molecular clones were created: (i) NL43G190E, a clone with G190E placed into a wild-type genetic backbone; and (ii) 21176G190E→G, a clone derived from a cryopreserved plasma virus sample (from PID 21176) in which glutamine (E) at position 190 was back-mutated to glycine (G). HIV-1 cDNA was generated from RNA extracted from the cryopreserved plasma sample 21176. cDNA was amplified using the thermostable Pfu DNA Polymerase (Agilent Technologies, Santa Clara, CA) and the PCR product was subcloned into the vector pNLPFB20 using Msc1 and PflM1 restriction sites spanning RT positions 24–311. Site-directed mutagenesis was performed using the QuickChange XL Site Directed Mutagenesis Kit (Agilent Technologies). Recombinant molecular clones were transfected into CEM8166 cells. NL43G190E induced syncytia after a delayed period of 7–10 days but could not be successfully expanded. 21176G190E→G induced syncytia within 4 days and was successfully expanded and submitted for PhenoSense susceptibility testing. Sanger sequencing yielded the expected sequence for each of the post-transfection clones.
Results
Summary of NNRTI susceptibility results
In vitro drug susceptibility results were available for 2047 isolates, including 1927 clinical isolates from 1686 individuals and 120 laboratory isolates or site-directed mutants. The numbers of results for nevirapine, efavirenz, etravirine and rilpivirine were 1975, 1967, 519 and 187, respectively. Of the 2047 isolates, 1752 were eligible for analysis. One hundred and nine isolates were excluded because they shared the same pattern of major NNRTI-resistance mutations with an isolate from the same individual; 186 isolates were excluded because their sequence had one or more major NNRTI-resistance mutations as part of an electrophoretic mixture. Ninety-six percent of isolates were classified as subtype B. Table 1 summarizes the number of viral isolates analysed for each NNRTI according to their levels of phenotypic resistance.
Table 1.
Number of HIV-1 isolates with genotypic and phenotypic data grouped by NNRTI and resistance levela
| NNRTI | Susceptibleb [n (%)] | Partially resistantb [n (%)] | Resistantb [n (%)] | Number of isolates |
|---|---|---|---|---|
| Nevirapine | 858 (51) | 62 (4) | 761 (45) | 1681 |
| Efavirenz | 973 (58) | 147 (9) | 554 (33) | 1674 |
| Etravirine | 331 (70) | 78 (17) | 62 (13) | 471 |
| Rilpivirine | 109 (64) | 26 (15) | 35 (21) | 170 |
aNumber of isolates determined following the exclusion of similar isolates from the same patient and isolates containing electrophoretic mixtures with major NNRTI-resistance mutations.
bSusceptible, <3-fold decreased susceptibility. Partially resistant, 3- to 10-fold decreased susceptibility. Resistant, ≥10-fold decreased susceptibility.
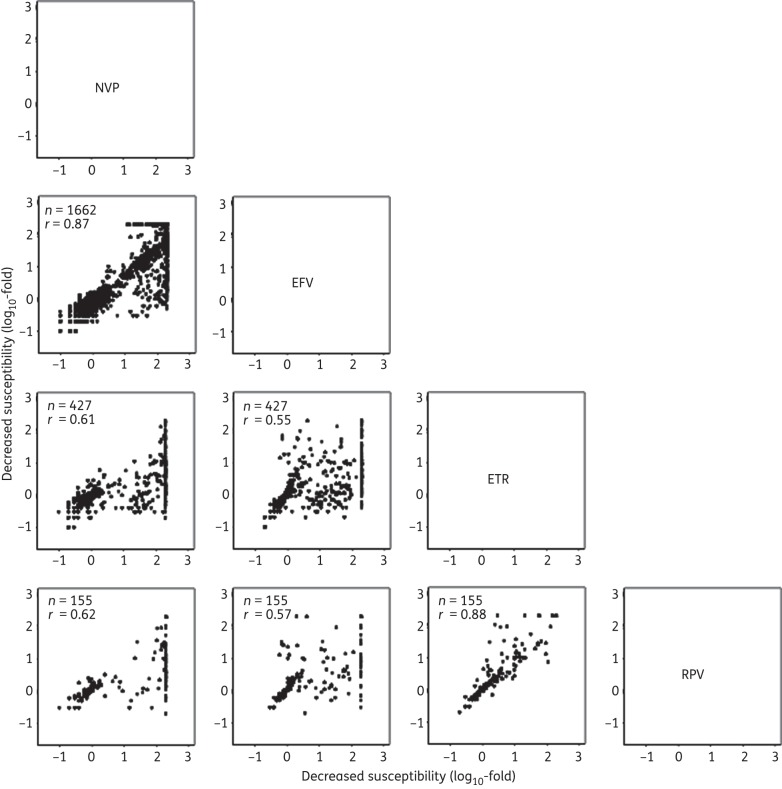
The phenotypic results of nevirapine and efavirenz (r = 0.87) and of etravirine and rilpivirine (r = 0.88) were highly correlated (Figure 1). The etravirine susceptibility results were less well correlated with the susceptibility results for efavirenz (r = 0.55) and nevirapine (r = 0.61). The rilpivirine susceptibility results were also less well correlated with the susceptibility results for efavirenz (r = 0.57) and nevirapine (r = 0.62).
Figure 1.
Correlations between the log10-fold reductions in susceptibility of each isolate for each pair of NNRTIs. The x-axis indicates the log10-fold susceptibility reduction of the NNRTI shown above the plots, and the y-axis indicates the log10-fold susceptibility reduction of the NNRTI to the right of the plots. The number of isolates (n) for which phenotype results were available and the correlation coefficient (r) are contained within each plot. NVP, nevirapine; EFV, efavirenz; ETR, etravirine; RPV, rilpivirine.
The proportions of the 35 study-defined NNRTI-resistance mutations in the genotype–phenotype dataset were strongly correlated with their proportions in sequences from 11 200 individuals in the HIVDB who had viruses with at least one study-defined NNRTI-resistance mutation (r = 0.98; Table 2).
Table 2.
Correlation of the frequency of the study-defined mutations in the genotype–phenotype dataset and in clinical isolates in the HIVDB
| Study-defined mutation | Genotype–phenotype isolates, % (n = 1752) | Clinical isolates in the HIVDB, % (n = 11 200)a |
|---|---|---|
| K103N | 44 | 52 |
| Y181C | 26 | 30 |
| G190A | 14 | 23 |
| V90I | 11 | 13 |
| A98G | 9.7 | 12 |
| K101E | 9.6 | 11 |
| V108I | 9.6 | 9.2 |
| H221Y | 9.4 | 8.7 |
| L100I | 7.4 | 6.6 |
| Y188L | 7.4 | 5.9 |
| V106I | 5.5 | 5 |
| E138A | 4.2 | 4.8 |
| G190S | 3.8 | 3.8 |
| V179D | 3.2 | 3.6 |
| K101P | 3.1 | 3.5 |
| P225H | 3.1 | 3.4 |
| K238T | 3.1 | 3.4 |
| F227L | 3.1 | 3 |
| E138G | 2.9 | 2.8 |
| M230L | 2.5 | 2 |
| K101H | 2.5 | 1.8 |
| E138K | 2.3 | 1.8 |
| V106A | 2.1 | 1.5 |
| K103S | 2 | 1.4 |
| Y181I | 1.6 | 1.3 |
| V179F | 1.5 | 1.1 |
| E138Q | 1.3 | 1 |
| G190E | 1 | 1 |
| Y181V | 1 | 0.7 |
| V106M | 0.8 | 0.7 |
| Y188H | 0.7 | 0.5 |
| G190Q | 0.6 | 0.4 |
| Y188C | 0.6 | 0.4 |
| V179T | 0.5 | 0.4 |
| F227C | 0.3 | 0 |
aThe frequency of the mutations in isolates (n = 11 200) from NNRTI-experienced individuals in the HIVDB who had a sequence with at least one study-defined NNRTI-resistance mutation.
Quantitative effects of study-defined mutations on reduced NNRTI susceptibility
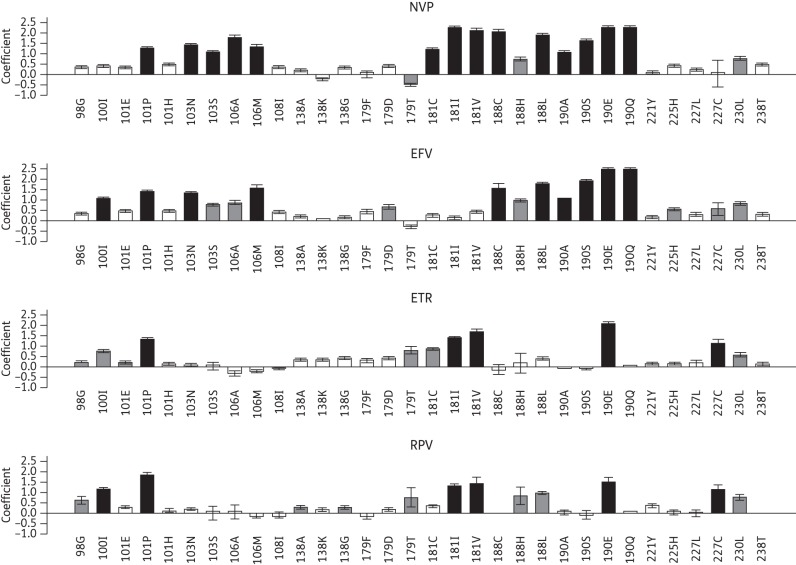
Of the 35 study-defined NNRTI-resistance mutations, 32 mutations at 16 positions had a regression coefficient of ≥0.3 log10 (a contribution to decreased susceptibility of ∼2-fold or greater) for one or more NNRTIs. Figure 2 plots the magnitude of the regression coefficients of these 32 mutations for each NNRTI.
Figure 2.
Graphical representation of the regression coefficients of the least squares regression model for predicting fold changes in NNRTI susceptibility using the study-defined NNRTI-resistance mutations. The y-axis indicates the regression coefficients on a log10 scale. Positive coefficients indicate mutations that contribute to reduced susceptibility. Negative coefficients indicate mutations that contribute to increased drug susceptibility. Black bars indicate mutations for which the mean regression coefficient was ≥1.0 log10-fold. Grey bars indicate mutations for which the mean regression coefficient was between 0.5 log10-fold and 1.0 log10-fold. The error bars indicate the standard deviation of the mean of the coefficients determined in 10 replicates of 5-fold cross-validation. NVP, nevirapine; EFV, efavirenz; ETR, etravirine; RPV, rilpivirine.
Sixteen mutations at eight positions had a mean regression coefficient ≥1.0 log10 (a contribution to decreased susceptibility of ∼10-fold or greater) for nevirapine, efavirenz, etravirine and/or rilpivirine: L100I, K101P, K103N/S, V106A/M, Y181C/I/V, Y188C/L, G190A/E/Q/S and F227C (the black bars in Figure 2). With the exception of L100I and F227C, each mutation had a mean regression coefficient ≥1.0 log10 for nevirapine. With the exception of K103S, V106A, Y181C/I/V and F227C, each had a mean regression coefficient ≥1.0 log10 for efavirenz. Five mutations, K101P, Y181I/V, G190E and F227C, had a mean regression coefficient ≥1.0 log10 for etravirine. Six mutations, L100I, K101P, Y181I/V, G190E and F227C, had a mean regression coefficient ≥ 1.0 log10 for rilpivirine.
Six additional mutations had a mean regression coefficient between 0.5 and 1.0 log10 (a contribution to decreased susceptibility of 3- to 10-fold; the grey bars in Figure 2) to one or more NNRTIs: A98G, V179D/T, Y188H, P225H and M230L. The remaining 10 mutations in Figure 2 had a mean regression coefficient between 0.3 and 0.5 log10 (a contribution to decreased susceptibility of 2- to 3-fold) for one or more NNRTIs: K101E/H, V108I, E138A/G/K, V179F, H221Y, F227L and K238T. The three mutations not shown in Figure 2 (V90I, V106I and E138Q) had mean regression coefficients <0.3 log10.
G190E was present in nine clinical viruses, none of which had other major NNRTI-resistance mutations (Table 3). Four of the nine had the NRTI-resistance mutation L74V, which is known to compensate for the decreased fitness associated with G190E21,22 and three had the accessory NNRTI-resistance mutation E138A. The site-directed G190E→G reversion mutant 21176G190E→G was susceptible to each of the NNRTIs.
Table 3.
Phenotypes and genotypes of isolates with G190E and a site-directed mutant with G190G
| Isolatea | Reference | Fold resistance |
Mutationsb | |||
|---|---|---|---|---|---|---|
| NVP | EFV | ETR | RPV | |||
| 21 176 | Stanford University | 200 | 200 | 200 | 200 | V60I, D67G, S68G, K70R, L74V, I135V, E138A, K173E, I178M, G190E, R211A, K219N, V245E, E248D, D250E, A272P, T286A |
| 21176G190E→G | 0.7 | 1.2 | 2.3 | 2.0 | V60I, D67G, S68G, K70R, L74V, I135V, E138A, K173E, I178M, R211A, K219N, V245E, E248D, D250E, A272P, T286A | |
| 24 746 | Stanford University | 200 | 200 | 94 | 28 | K20R, D67N, K70R, K122E, I135V, E138A, S162C, M184MV, G190E, T200A, F214M, T215F, K219Q, L228H, V245E |
| 59 209 | Stanford University | 200 | 200 | 110 | 7.3 | V35L, D67N, K70R, K101R, K103R, D177E, I178M, G190E, G196E, T200A, E204N, R211K, T215F, K219E, L228H, V245E, A272P, T286A, E297K |
| 16 894 | S.-Y. Rhee | 200 | 200 | NA | NA | K20R, V35I, E36EK, M41L, K43N, D67N, I135V, G190E, T200A, L210W, T215Y, K219N, A272P, K277R |
| 63 800 | S.-Y. Rhee | 200 | 200 | NA | NA | S48T, D67N, T69DN, K70GR, L74V, K101KR, K104KR, D121Y, K122E, D123E, E169D, D177E, M184V, G190E, T200A, Q207K, F214L, K219EQ, K275KR, V276VI, P294T, E297KR |
| 55 467 | S.-Y. Rhee | 200 | 200 | NA | NA | D67G, S68G, L74V, Y115F, K122Q, D123E, I135T, I142V, D177E, M184V, V189I, G190E, T200A, T215F, K219E, V245E, E297R |
| 127 280 | G. L. Melikian | 200 | 200 | NA | NA | K46KIM, K65R, V75L, V90VI, K102KR, K103KR, Y115F, D123E, E169D, D177E, V179VI, G190E, T200A, R211K |
| 33 143 | W. Huang | 200 | 200 | NA | NA | K20R, M41L, K43Q, E44D, V60I, D67N, L74V, K102R, V118I, K122E, D123N, I135 V, S162Y, I178F, G190E, G196R, Q197K, T200A, Q207N, L210W, R211K, T215Y, K219N, E224N, A272P, T286A, E297K |
| 91 738 | J. D. Baxter | 200 | 181 | NA | NA | M41L, D67N, R72RG, D86DN, K122E, F160FL, K166R, D177E, V179G, G190E, T200A, Q207E, H208HD, L210W, T215Y, D218DE, K219N, K220KR, L228H |
NVP, nevirapine; EFV, efavirenz; ETR, etravirine; RPV, rilpivirine; NA, not available.
aThe creation of the cloned site-directed mutant 21176G190E→G is described in the Methods section. The NRTI susceptibility results indicated slightly higher levels of decreased susceptibility than the 21 176 clone containing G190E, suggesting that the dramatic reductions in NNRTI resistance were not a result of decreased replication.
Prediction accuracy of the least squares regression model
Table 4 shows the mean-squared errors and classification accuracy of the least squares regression model for each NNRTI calculated from the 10 replicates of 5-fold cross-validation. The classification accuracy for nevirapine, efavirenz, etravirine and rilpivirine was 94%, 90%, 83% and 83%, respectively. The mean-squared error for nevirapine, efavirenz, etravirine and rilpivirine was 0.26 log10-fold (∼1.8-fold), 0.15 log10-fold (∼1.4-fold), 0.14 log10-fold (∼1.4-fold) and 0.17 log10-fold (∼1.5-fold), respectively. The large proportion of viruses with high-level nevirapine resistance resulted in the nevirapine model having the highest classification accuracy and the highest mean-squared error.
Table 4.
Prediction accuracy of least squares regression: mean-squared error and classification accuracy
| NNRTI | Mean-squared error (SD)a | Classification accuracy (SD)b |
|---|---|---|
| Nevirapine | 0.26 (0.02) | 0.94 (0.01) |
| Efavirenz | 0.15 (0.02) | 0.90 (0.02) |
| Etravirine | 0.14 (0.02) | 0.83 (0.03) |
| Rilpivirine | 0.17 (0.07) | 0.83 (0.06) |
SD, standard deviation.
aMean-squared error of 10 replicates of 5-fold cross-validation results of least squares regression using the 35 study-defined NNRTI-resistance mutations.
bClassification accuracy is defined as the proportion of phenotypes for which the classification categories of predicted and actual fold change were concordant (susceptible versus partial resistant versus resistant).
Figure 3 shows three confusion matrices in which each cell contains the proportion of concordances (diagonal), complete discordances (upper right and lower left) and partial discordances between the classification categories of actual and predicted fold decreased susceptibility. For each NNRTI, the classification accuracy shown in Table 3 was calculated as the sum of the values in the shaded diagonal. The proportion of viruses with complete discordances was 0.9%, 0.9%, 1.4% and 2.0% for nevirapine, efavirenz, etravirine and rilpivirine, respectively.
Figure 3.
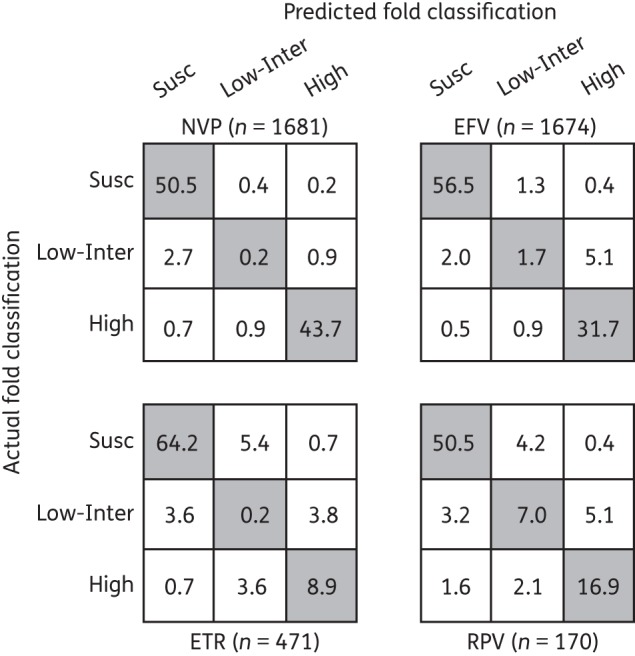
Confusion matrices of nevirapine (NVP), efavirenz (EFV), etravirine (ETR) and rilpivirine (RPV) containing the mean proportions of isolates for which the classification of predicted and actual fold reduction in susceptibility were the same (shaded diagonal cells), were completely discordant (lower left and upper right cells) or were partially discordant (remaining four cells) determined during 10 trials of 5-fold cross-validation.
Approximately one-half of complete discordances were ‘false-negative’ predictions of susceptibility of NNRTI-resistant isolates containing rare mutations or combinations of mutations that were not in our models but are known to decrease NNRTI susceptibility. For example, three isolates in our dataset had the mutations K103R + V179D, a synergistic combination associated with >10-fold decreased nevirapine and efavirenz susceptibility that occurs despite the fact that K103R alone does not reduce NNRTI susceptibility.5 Other rare mutations not included in our models were: G190C, which was present in two viruses in our dataset;4 Y181S, which was present in one virus;10,23 and L234I (in combination with M230L), which was present in one virus.7 The addition of M184I to least squares regression models did not improve the prediction accuracy for any of the NNRTIs.
Feature selection using LARS
Of the 469 mutations present three or more times in the dataset, LARS identified 28 as significant predictors of reduced nevirapine or efavirenz susceptibility. These 28 mutations included 25 of the 35 study-defined mutations (i.e. those used by the least-squares regression models). Ten of the 35 study-defined mutations—V90I, V106I, E138A/G/K/Q, V179F/T and F227C/L—were not found to be significant predictors of reduced nevirapine or efavirenz susceptibility in the LARS model. Conversely, three non-study-defined mutations—I31L, K101Q and V179E—were associated with slightly reduced nevirapine or efavirenz susceptibility. LARS also identified four mutations associated with increased susceptibility to nevirapine or efavirenz: M41L, V118I, M184V and T215Y. Two of these mutations, V118I and T215Y, have been reported to increase NNRTI susceptibility.24,25 The prediction accuracy of the LARS models was similar to least-squares regression (Table S1, available as Supplementary data at JAC Online).
Discussion
This study is the most comprehensive analysis of the genetic predictors of phenotypic NNRTI resistance in a large set of predominantly clinical, publicly available virus sequences. Sixteen mutations at eight positions—L100I, K101P, K103N/S, V106A/M, Y181C/I/V, Y188C/L, G190A/E/Q/S and F227C—had a mean regression coefficient ≥1.0 log10 (≥10-fold contribution to reduced susceptibility) for one or more NNRTIs. Fourteen, 10, 5 and 6 of these 16 mutations were associated with nevirapine, efavirenz, etravirine and rilpivirine, respectively. An additional 16 mutations at 11 positions—A98G, K101E/H, V108I, E138A/G/K, V179D/F/T, Y188H, H221Y, P225H, F227L, M230L and K238T—had a mean regression coefficient between 0.3 log10-fold (≥2-fold contribution to decreased susceptibility) and 1.0 log10-fold. The validity of our findings is supported by the low mean-squared errors and high classification accuracies for all four NNRTIs.
G190E, a mutation known to cause high-level nevirapine and efavirenz resistance4 was found also to markedly reduce etravirine and rilpivirine susceptibility. G190E occurs in 1% of viruses from patients receiving efavirenz.12,26 It is also selected in vitro by etravirine and rilpivirine7,9 and has been reported in one patient receiving rilpivirine.27 Our regression analysis, combined with our site-directed mutagenesis results, suggests that viruses with this mutation should be considered highly resistant to etravirine and rilpivirine, as well as nevirapine and efavirenz.
This study provides additional data on the extent of cross-resistance conferred by mutations at position 138, which have become important because of their frequent occurrence in patients with rilpivirine-associated virological failure.8,28 E138A and two non-polymorphic mutations at this position, E138K/G, were significantly but modestly associated with reduced etravirine susceptibility. E138K did not confer cross-resistance to nevirapine or efavirenz. E138G conferred minimal cross-resistance to nevirapine but not efavirenz. The finding that E138K/G modestly decreased etravirine susceptibility but had little if any impact on nevirapine or efavirenz susceptibility is consistent with two site-directed mutagenesis studies28,29 and one other regression analysis.10
K101H, associated with etravirine resistance in the analysis of the DUET trials was significantly associated with reduced nevirapine and efavirenz susceptibility but had little or no effect on etravirine or rilpivirine susceptibility. K101H is a non-polymorphic mutation that is selected in 2% of patients receiving nevirapine or efavirenz, supporting its role as a nevirapine- and efavirenz-resistance mutation.30 A clinical isolate belonging to the Monogram PhenoSense panel that contained K101H alone was also reported as being associated with 4-fold decreased efavirenz susceptibility and 12-fold decreased nevirapine susceptibility.31 This finding underscores the importance of assessing the effects of novel NNRTI-resistance mutations against older drugs.
Because LARS is particularly useful for selecting a subset of predictors when the set of possible predictors is large, we were able to analyse the effect on nevirapine and efavirenz of all 469 mutations that occurred three or more times in the dataset. Despite its use of a parsimonious variable selection algorithm, LARS nonetheless identified 25 of 35 study-defined mutations as contributors to decreased nevirapine and/or efavirenz susceptibility. The 10 mutations that were not significantly associated with nevirapine and/or efavirenz reduced susceptibility in the LARS model included the predominantly etravirine-associated mutations V90I, V106I, E138A/K/G/Q, V179F/T and F227C/L. Reassuringly, it identified just three non-study-defined mutations (I31L, K101Q and V179E). The three mutations have previously been reported to be selected by NNRTIs12 and to be more commonly found in nevirapine- and efavirenz-resistant viruses.5
Among the virus isolates in our dataset, 116 were site-directed mutants, including 57 (3.4%) of the 1680 of the isolates for which nevirapine and efavirenz susceptibility results were available, 49 (10.4%) of the 471 isolates for which etravirine susceptibility results were available and 20 (11.8%) of the 170 isolates for which rilpivirine susceptibility results were available. The phenotypic testing of site-directed NNRTI-resistant mutants may yield reductions in susceptibility that are lower or greater than those observed in clinical isolates. Lower reductions in susceptibility would occur if the effect of the site-directed mutation depended heavily on the presence of unrecognized backbone RT mutations that co-evolve with that mutation. Greater reductions in susceptibility would occur in clinical isolates with multiple NRTI-resistance mutations, because these mutations often modestly increase susceptibility to NNRTIs.32 The inclusion of site-directed NNRTI-resistant viruses therefore represents an important, albeit modest, addition to our analysis.
Our analysis has several limitations. First, the numbers of genotype–phenotype correlations for etravirine and rilpivirine were much lower than the numbers of correlations for nevirapine and efavirenz. For example, G190Q was not present in any of the isolates submitted for etravirine or rilpivirine susceptibility testing and F227C was present in just two isolates. Nonetheless, G190Q appears to reduce etravirine and rilpivirine susceptibility33 as well as nevirapine and efavirenz susceptibility. F227C appears to reduce nevirapine and efavirenz susceptibility34 as well as etravirine and rilpivirine susceptibility. In addition, the effect of uncommon mutations or those with minimal effects on etravirine or rilpivirine susceptibility may not have been detected. The limited number of etravirine and rilpivirine genotype–phenotype correlations also made it impossible to use LARS to identify potentially novel etravirine or rilpivirine resistance mutations.
Second, we do not know the NNRTI treatment history of most of the individuals from whom the virus isolates were obtained. Although more than 500 isolates had been obtained since 2008 and nearly 200 since 2011, the years in which etravirine and rilpivirine were approved for clinical use, few of these isolates were from patients who had received etravirine or rilpivirine. Finally, because the PhenoSense assay tests recombinant viruses created from patient amplicons with a C-terminus at codon 311,13 we were unable to assess the effects of the known NNRTI-resistance mutations L318F and N348I, as well as potential novel NNRTI-resistance mutations in the RT connection domain.35
NNRTI cross-resistance patterns have been shown to be clinically significant primarily for NNRTI-experienced individuals receiving etravirine in combination with other active ARVs.17,18 This has led to renewed interest in developing novel non-cross-resistant NNRTIs with higher genetic barriers to resistance.36,37 Our analysis provides a quantitative assessment of the effects of NNRTI-resistance mutations on the in vitro activity of nevirapine, efavirenz, etravirine and rilpivirine. Our analysis also underscores the need for systematically testing new NNRTIs against clinical virus isolates containing a broad spectrum of major non-polymorphic NNRTI-resistance mutations and of testing older NNRTIs against virus isolates containing mutations associated with resistance to novel NNRTIs.
Funding
This work was supported by NIAID at NIH (AI068581: ‘Public HIV Drug Resistance Database’ to S.-Y. R., T. F. L. and R. W. S.).
Transparency declarations
D. P. and K. W. are employees and stockholders of Gilead Sciences, Inc.; for all other authors, none to declare.
Supplementary data
Table S1 is available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
References
- 1.Bacheler L, Jeffrey S, Hanna G, et al. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J Virol. 2001;75:4999–5008. doi: 10.1128/JVI.75.11.4999-5008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrnes VW, Emini EA, Schleif WA, et al. Susceptibilities of human immunodeficiency virus type 1 enzyme and viral variants expressing multiple resistance-engendering amino acid substitutions to reverse transcriptase inhibitors. Antimicrob Agents Chemother. 1994;38:1404–7. doi: 10.1128/aac.38.6.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrigan PR, Mo T, Wynhoven B, et al. Rare mutations at codon 103 of HIV-1 reverse transcriptase can confer resistance to non-nucleoside reverse transcriptase inhibitors. AIDS. 2005;19:549–54. doi: 10.1097/01.aids.0000163930.68907.37. [DOI] [PubMed] [Google Scholar]
- 4.Huang W, Gamarnik A, Limoli K, et al. Amino acid substitutions at position 190 of human immunodeficiency virus type 1 reverse transcriptase increase susceptibility to delavirdine and impair virus replication. J Virol. 2003;77:1512–23. doi: 10.1128/JVI.77.2.1512-1523.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parkin NT, Gupta S, Chappey C, et al. The K101P and K103R/V179D mutations in human immunodeficiency virus type 1 reverse transcriptase confer resistance to nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother. 2006;50:351–4. doi: 10.1128/AAC.50.1.351-354.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vingerhoets J, Tambuyzer L, Azijn H, et al. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled Phase III clinical studies. AIDS. 2010;24:503–14. doi: 10.1097/QAD.0b013e32833677ac. [DOI] [PubMed] [Google Scholar]
- 7.Vingerhoets J, Azijn H, Fransen E, et al. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J Virol. 2005;79:12773–82. doi: 10.1128/JVI.79.20.12773-12782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rimsky L, Vingerhoets J, Van Eygen V, et al. Genotypic and phenotypic characterization of HIV-1 isolates obtained from patients on rilpivirine therapy experiencing virologic failure in the phase 3 ECHO and THRIVE studies: 48-week analysis. J Acquir Immune Defic Syndr. 2012;59:39–46. doi: 10.1097/QAI.0b013e31823df4da. [DOI] [PubMed] [Google Scholar]
- 9.Azijn H, Tirry I, Vingerhoets J, et al. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob Agents Chemother. 2010;54:718–27. doi: 10.1128/AAC.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Westen GJ, Hendriks A, Wegner JK, et al. Significantly improved HIV inhibitor efficacy prediction employing proteochemometric models generated from antivirogram data. PLoS Comput Biol. 2013;9:e1002899. doi: 10.1371/journal.pcbi.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhee SY, Taylor J, Wadhera G, et al. Genotypic predictors of human immunodeficiency virus type 1 drug resistance. Proc Natl Acad Sci. 2006;103:17355–60. doi: 10.1073/pnas.0607274103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhee SY, Gonzales MJ, Kantor R, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petropoulos CJ, Parkin NT, Limoli KL, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–8. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porter DP, Kulkarni R, Fralich T, et al. Primary and secondary analyses of emergent drug resistance through week 48 from STaR study: rilpivirine/emtricitabine/tenofovir vs efavirenz/emtricitabine/tenofovir DF single - tablet regimens. Abstracts of the 2013 International Workshop on HIV & Hepatitis Virus Drug Resistance and Curative Strategies; Toronto, Canada. 2013. Antiviral Ther 2013; 18 Suppl 1: A38. [Google Scholar]
- 15.de Oliveira T, Deforche K, Cassol S, et al. An automated genotyping system for analysis of HIV-1 and other microbial sequences. Bioinformatics. 2005;21:3797–800. doi: 10.1093/bioinformatics/bti607. [DOI] [PubMed] [Google Scholar]
- 16.Efron B, Hastie T, Johnstone I, et al. Least angle regression. Ann Stat. 2004;32:407–99. [Google Scholar]
- 17.Lazzarin A, Campbell T, Clotet B, et al. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-2: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet. 2007;370:39–48. doi: 10.1016/S0140-6736(07)61048-4. [DOI] [PubMed] [Google Scholar]
- 18.Madruga JV, Cahn P, Grinsztejn B, et al. Efficacy and safety of TMC125 (etravirine) in treatment-experienced HIV-1-infected patients in DUET-1: 24-week results from a randomised, double-blind, placebo-controlled trial. Lancet. 2007;370:29–38. doi: 10.1016/S0140-6736(07)61047-2. [DOI] [PubMed] [Google Scholar]
- 19.Coakley E, Chappey C, Benhamida J, et al. Biological and clinical cut-off analyses for etravirine in the PhenoSense HIV assay; Abstracts of the Seventeenth International HIV Drug Resistance Workshop, Sitges, Spain, 2008. Abstract 122. [Google Scholar]
- 20.Balamane M, Varghese V, Melikian GL, et al. Panel of prototypical recombinant infectious molecular clones resistant to nevirapine, efavirenz, etravirine, and rilpivirine. Antimicrob Agents Chemother. 2012;56:4522–4. doi: 10.1128/AAC.00648-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleim JP, Rosner M, Winkler I, et al. Selective pressure of a quinoxaline nonnucleoside inhibitor of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase (RT) on HIV-1 replication results in the emergence of nucleoside RT-inhibitor-specific (RT Leu-74→Val or Ile and Val-75→Leu or Ile) HIV-1 mutants. Proc Natl Acad Sci USA. 1996;93:34–8. doi: 10.1073/pnas.93.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boyer PL, Gao HQ, Hughes SH. A mutation at position 190 of human immunodeficiency virus type 1 reverse transcriptase interacts with mutations at positions 74 and 75 via the template primer. Antimicrob Agents Chemother. 1998;42:447–52. doi: 10.1128/aac.42.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van der Borght K, Van Craenenbroeck E, Lecocq P, et al. Cross-validated stepwise regression for identification of novel non-nucleoside reverse transcriptase inhibitor resistance associated mutations. BMC Bioinformatics. 2011;12:386. doi: 10.1186/1471-2105-12-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shulman NS, Bosch RJ, Mellors JW, et al. Genetic correlates of efavirenz hypersusceptibility. AIDS. 2004;18:1781–5. doi: 10.1097/00002030-200409030-00006. [DOI] [PubMed] [Google Scholar]
- 25.Clark SA, Shulman NS, Bosch RJ, et al. Reverse transcriptase mutations 118I, 208Y, and 215Y cause HIV-1 hypersusceptibility to non-nucleoside reverse transcriptase inhibitors. AIDS. 2006;20:981–4. doi: 10.1097/01.aids.0000222069.14878.44. [DOI] [PubMed] [Google Scholar]
- 26.Alcaro S, Alteri C, Artese A, et al. Docking analysis and resistance evaluation of clinically relevant mutations associated with the HIV-1 non-nucleoside reverse transcriptase inhibitors nevirapine, efavirenz and etravirine. ChemMedChem. 2011;6:2203–13. doi: 10.1002/cmdc.201100362. [DOI] [PubMed] [Google Scholar]
- 27.Rimsky L, Van Eygen V, Hoogstoel A, et al. 96-week resistance analyses of rilpivirine in treatment-naive, HIV-1-infected adults from the ECHO and THRIVE Phase III trials. Antivir Ther. 2013 doi: 10.3851/IMP2636. doi:10.3851/IMP2636. [DOI] [PubMed] [Google Scholar]
- 28.Tambuyzer L, Nijs S, Daems B, et al. Effect of mutations at position E138 in HIV-1 reverse transcriptase on phenotypic susceptibility and virologic response to etravirine. J Acquir Immune Defic Syndr. 2011;58:18–22. doi: 10.1097/QAI.0b013e3182237f74. [DOI] [PubMed] [Google Scholar]
- 29.Kulkarni R, Babaoglu K, Lansdon EB, et al. The HIV-1 reverse transcriptase M184I mutation enhances the E138K-associated resistance to rilpivirine and decreases viral fitness. J Acquir Immune Defic Syndr. 2012;59:47–54. doi: 10.1097/QAI.0b013e31823aca74. [DOI] [PubMed] [Google Scholar]
- 30.Shahriar R, Rhee SY, Liu TF, et al. Nonpolymorphic human immunodeficiency virus type 1 protease and reverse transcriptase treatment-selected mutations. Antimicrob Agents Chemother. 2009;53:4869–78. doi: 10.1128/AAC.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Z, Xu W, Koh YH, et al. A novel nonnucleoside analogue that inhibits human immunodeficiency virus type 1 isolates resistant to current nonnucleoside reverse transcriptase inhibitors. Antimicrob Agents Chemother. 2007;51:429–37. doi: 10.1128/AAC.01032-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitcomb JM, Huang W, Limoli K, et al. Hypersusceptibility to non-nucleoside reverse transcriptase inhibitors in HIV-1: clinical, phenotypic and genotypic correlates. AIDS. 2002;16:F41–7. doi: 10.1097/00002030-200210180-00002. [DOI] [PubMed] [Google Scholar]
- 33.Haddad M, Stawiski E, Benhamida J, et al. Improved genotypic algorithm for predicting etravirine susceptibility: comprehensive list of mutations identified through correlation with matched phenotype; Alexandria, VA, USA: Foundation for Retrovirology and Human Health; Abstracts of the Seventeenth Conference on Retroviruses and Opportunistic Infections, San Francisco, CA, USA, 2010. Abstract 474. [Google Scholar]
- 34.Tambuyzer L, Azijn H, Rimsky LT, et al. Compilation and prevalence of mutations associated with resistance to non-nucleoside reverse transcriptase inhibitors. Antivir Ther. 2009;14:103–9. [PubMed] [Google Scholar]
- 35.Paredes R, Puertas MC, Bannister W, et al. A376S in the connection subdomain of HIV-1 reverse transcriptase confers increased risk of virological failure to nevirapine therapy. J Infect Dis. 2011;204:741–52. doi: 10.1093/infdis/jir385. [DOI] [PubMed] [Google Scholar]
- 36.Vernazza P, Wang C, Pozniak A, et al. Efficacy and safety of lersivirine (UK-453,061) versus efavirenz in antiretroviral treatment-naive HIV-1-infected patients: week 48 primary analysis results from an ongoing, multicenter, randomized, double-blind, phase IIb trial. J Acquir Immune Defic Syndr. 2013;62:171–9. doi: 10.1097/QAI.0b013e31827a2ba2.. [DOI] [PubMed] [Google Scholar]
- 37.Lu M, Felock PJ, Munshi V, et al. Antiviral activity and in vitro mutation development pathways of MK-6186, a novel nonnucleoside reverse transcriptase inhibitor. Antimicrob Agents Chemother. 2012;56:3324–35. doi: 10.1128/AAC.00102-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.