Abstract
The genetic diversity of 123 wild strains of Pleurotus eryngii var. tuoliensis, which were collected from nine geographical locations in Yumin, Tuoli, and Qinghe counties in the Xinjiang Autonomous Region of China, was analysed using two molecular marker systems (inter-simple sequence repeat and start codon targeted). At the variety level, the percentage of polymorphic loci and Nei’s gene diversity index for P. eryngii var. tuoliensis was 96.32% and 0.238, respectively. At the population level, Nei’s gene diversity index ranged from 0.149 to 0.218 with an average of 0.186, and Shannon's information index ranged from 0.213 to 0.339 with an average of 0.284. These results revealed the abundant genetic variability in the wild resources of P. eryngii var. tuoliensis. Nei’s gene diversity analysis indicated that the genetic variance was mainly found within individual geographical populations, and the analysis of molecular variance revealed low but significant genetic differentiation among local and regional populations. The limited gene flow (Nm = 1.794) was inferred as a major reason for the extent of genetic differentiation of P. eryngii var. tuoliensis. The results of Mantel tests showed that the genetic distance among geographical populations of P. eryngii var. tuoliensis was positively correlated with the geographical distance and the longitudinal distances (rGo = 0.789 and rLn = 0.873, respectively), which indicates that geographical isolation is an important factor for the observed genetic differentiation. Nine geographical populations of P. eryngii var. tuoliensis were divided into three groups according to their geographical origins, which revealed that the genetic diversity was closely related to the geographical distribution of this wild fungus.
Introduction
The Pleurotus mushroom, which is commercially called Bai Ling Gu, is a precious edible fungus of high nutrient and medicinal value [1,2]. In the wild, this mushroom parasitically or saprophytically grows together with Ferula plants in the Umbelliferae family [3]. It has a restricted habitat in the northwest part of China, i.e., Xinjiang Autonomous Region [4]. According to the traditional criteria of fungal classification, taxonomic status of this wild fungus is not clear yet [4-6]. The results of rDNA sequence analysis, mating, and cultivation tests indicated that the Pleurotus mushroom found specifically in China is a branch of Pleurotus eryngii, which evolved independently in China [7]. The scientific name was then defined as Pleurotus eryngii var. tuoliensis through morphological identification and internal transcribed spacer (ITS) analysis [8].
P. eryngii species complex has the most abundant population diversity in the genus Pleurotus. It consists of at least six varieties [9]. Because it is important both economically and ecologically, this species complex has attracted more and more attention worldwide [10-13]. Analyses via molecular markers revealed a high level of heterogeneity within P. eryngii var. eryngii, var. ferulae, and var. nebrodensis [14]. The genetic diversity of P. eryngii var. eryngii and var. ferulae was more abundant than that of var. nebrodensis, var. Elaeoselinum, and var. Thapsia [15]. The analysis of the population structure of P. eryngii var. eryngii and var. ferulae revealed a greater variation within geographical populations than that between geographical populations [16].
Inter-simple sequence repeats (ISSR) markers have been widely applied to analyses of genetic variance and population structure in many types of organisms [17-21]. Because a molecular marker system usually targets to detect a particular genome region, the level of genetic diversity might not be objectively revealed using a single marker system. Previous studies have shown that ISSR analyses only showed a relatively low genetic diversity among peanut cultivars despite abundant morphological, physiological, and agronomic variance [22]. Nevertheless, a molecular marker system termed start codon targeted (SCoT) polymorphism, which is a simple and novel DNA marker system, could detect more polymorphisms compared with several other molecular marker systems [23,24]. The SCoT marker system was employed not only in the study of genetic relationships but also in revealing the geographical origins of cultivars [24-28]. Xiong et al. [29] suggested that the SCoT marker system could be used as an effective supplement to ISSR and RAPD. Additional genetic variances can be reflected through the comprehensive use of various molecular marker systems [30,31].
In this study, the genetic variation of 123 samples of P. eryngii var. tuoliensis was studied using ISSR and SCoT markers. The goals of this study were to investigate the genetic structure characteristic and the level of genetic variation of P. eryngii var. tuoliensis, and to thereby infer the factors that influence the genetic differentiation in the populations.
Materials and Methods
Ethics Statement
Although the wild mushroom of P. eryngii var. tuoliensis is not considered an endangered species, commercial collecting nowadays is forbidden for protecting the local ecological environment, but collecting for research purposes is allowed. Anyone who collects the wild mushroom of P. eryngii var. tuoliensis for commercial purpose will be fined by the local Agricultural Bureaus (Agricultural Bureaus of Yumin County, Agricultural Bureaus of Tuoli County, and Agricultural Bureaus Qinghe County of Xinjiang Autonomous Region of China).
Sampling
Based on the natural occurrence of fruiting bodies, we confirmed the number of sampling sites in Yumin, Tuoli, and Qinghe. A total of 123 wild samples were collected from nine sites in three regions of Xinjiang from March to May of 2009 (Figure 1). These sampling regions spanned approximately 650 km from the east to the west and approximately 150 km from the south to the north. The sample size and the geographical coordinates for each geographical population are presented in Table 1.
Figure 1. Sampling sites of Pleurotus eryngii var. tuoliensis in China.
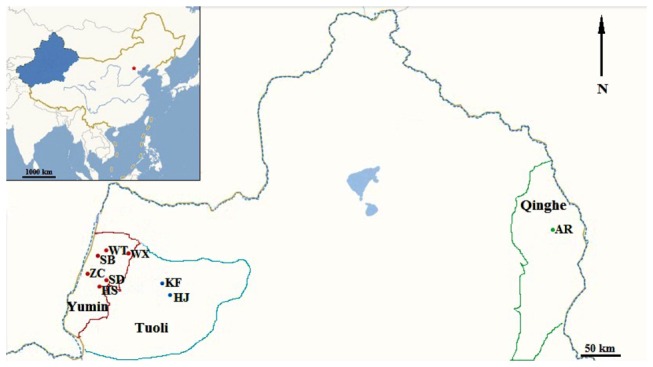
The Xinjiang Autonomous Region is highlighted in blue in the map of China shown in the upper left. WX, ZC, SB, SD, HS, and WT in the Yumin region are marked as red rotundities; KF and HJ in the Tuoli region are marked as blue rotundities; and AR in the Qinghe region is marked as a green rotundity.
Table 1. Geographical and climatic data of the P. eryngii var. tuoliensis populations.
| Geographic Region | Annual Temperature (°C ) * | Annual Rainfall (mm) * | Geographic Population | Sample Size | Latitude (North) | Longitude (East) |
|---|---|---|---|---|---|---|
| Yumin | 7.7 | 291 | WX | 16 | 46.35 | 83.31 |
| ZC | 24 | 46.10 | 82.84 | |||
| SB | 9 | 46.28 | 82.90 | |||
| SD | 8 | 45.98 | 82.99 | |||
| HS | 17 | 45.85 | 82.94 | |||
| WT | 32 | 46.45 | 82.96 | |||
| Tuoli | 6.0 | 245 | KF | 6 | 45.95 | 83.72 |
| HJ | 3 | 45.85 | 83.86 | |||
| Qinghe | 1.3 | 189 | AR | 8 | 46.50 | 90.34 |
| Total samples | 123 |
* , The values were the average of 30 years (1981-2010). Data from: http://www.cma.gov.cn/2011qxfw/2011qsjgx/
Sample isolation
Pure culture of each strain was obtained by isolating tissue culture from the fruiting bodies. A small piece of tissue was removed aseptically and transferred into a culture tube containing potato dextrose agar (PDA), and incubated in the dark at 25°C for 7-9 days. These samples were next stored at 4°C until needed.
DNA extract
Mycelia for DNA extraction were cultured on PDA Petri dishes with cellophane at 25°C for 7 days. The total DNA was extracted using a DP305-Plant Genome Extraction Kit (Tiangen, China). The purity and quality of the genomic DNA were determined through spectrophotometry and electrophoresis on 1.0% agarose gel. The DNA solution was stored at -20°C.
ISSR analysis
Seven ISSR primers were selected in a pre-experiment (Table 2). The amplification reactions were performed in a final volume of 20 μL containing 20 ng of the template DNA, 10X Ex Taq buffer, 0.2 mM dNTPs, 0.5 mM of each primer, and 1 U of Ex Taq DNA polymerase (TaKaRa). The PCR reaction conditions were the following: initial denaturation step at 94°C for 4 min, 35 cycles of 50 s at 94°C, annealing at 55°C for 50 s, and extension at 72°C for 2 min, and final extension at 72°C for 7 min. The amplified products were resolved on a 1.0% agarose gel and stained with ethidium bromide. The results were observed through an ultraviolet gel imaging system. The molecular weights of DNA bands on the agarose gel were estimated using a DNA ladder, i.e., 2-log (TaKaRa).
Table 2. ISSR and SCoT primer sequences for P. eryngii var. tuoliensis.
| Molecular Marker | Primer | Sequence (5'→3') |
|---|---|---|
| ISSR | P1 | TGCACACACACACAC |
| P2 | GTGACACACACACAC | |
| P4 | GGATGCAACACACACACAC | |
| P10 | GAGAGAGAGAGAGAGAC | |
| P19 | ACACACACACACACACCT | |
| P24 | CACGAGAGAGAGAGAGA | |
| P864 | ATGATGATGATGATGATG | |
| SCoT | S13 | ACGACATGGCGACCATCG |
| S14 | ACGACATGGCGACCACGC | |
| S19 | ACCATGGCTACCACCGGC | |
| S27 | ACCATGGCTACCACCGTG | |
| S28 | CCATGGCTACCACCGCCA | |
| S29 | CCATGGCTACCACCGGCC | |
| S30 | CCATGGCTACCACCGGCG | |
| S31 | CCATGGCTACCACCGCCT |
SCoT analysis
Eight out of 36 SCoT primers were selected, which produced clear and reproducible profiles [23]. Sequences of the SCoT primer are listed in Table 2. The PCR mix consisted of a total volume of 20 μL containing 2 μL of 10X Ex Taq buffer, 0.25 mM dNTPs, 0.5 mM of each primer, 1 U of Ex Taq DNA polymerase (TaKaRa), and 20 ng of the template DNA. The PCR reaction was performed using the following thermal cycling protocol: 94°C for 5 min, 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 2 min. The reaction was completed through incubation at 72°C for 10 min. The SCoT products were resolved through electrophoresis on 1.5% agarose gels.
Data analysis
The gels were scored for the presence or absence of reproducible bands. Each band was regarded as a locus with two alternative alleles. The data from the ISSR and the SCoT analyses were converted into a binary data matrix as discrete variables (1 = presence and 0 = absence). To expand the range of DNA polymorphisms assayed in a genome and to increase the reliability of the results, the diversity analyses were performed in this study using combined data of ISSR and SCoT.
The genetic variability at the variety or population level was analysed under the assumption of Hardy-Weinberg equilibrium. The percentage of polymorphic bands (PPB), the numbers of effective alleles (Ne), Shannon’s information index (I), and Nei’s gene diversity index (H) were calculated using the GenAIEx 6.41 software [32].
According to the method developed by Nei, the gene diversity statistics, including the total allelic diversity (Ht), the mean allelic diversity within populations (Hs), the proportion of the total allelic diversity found among populations (Gst), and the gene flow among populations (Nm), were obtained using the POPGENE (v 1.31) [33]. The total genetic variation among the samples was calculated using the phi-statistic through the analysis of molecular variance (AMOVA). This analysis was performed using the computer program GENALEX [32]. The total genetic variation is partitioned at three levels—within populations (Phi-PT), among populations within regions (Phi-PR), and among regional populations (Phi-RT) [34].
The pairwise Nei’s genetic distances between populations were calculated using the POPGENE software. In order to examine whether local populations that are geographically close are genetically similar, correlation of genetic distances with geographical, latitudinal and longitudinal distances respectively was analyzed through three Mantel tests using the GENALEX program [32].
The cluster analysis for the populations was performed with the NTSYSpc-2.1e program [35] based on the Nei’s genetic distances using the UPGMA (Unweighted Pair Group Method with Arithmetic Mean).
Results
Genetic variation at the variety and population level
The analysis of the genetic diversity among 123 samples of P. eryngii var. tuoliensis showed that the percentage of polymorphic bands, the number of effective alleles, Nei’s gene diversity index, and Shannon’s information index were 96.32%, 1.381, 0.238, and 0.377, respectively. The results from these analyses suggested that there are high levels of genetic variations within this variety.
The percentage of polymorphic bands among the nine geographical populations ranged from a low of 33.44% for the HJ population in Tuoli to a high of 77.59% for the WT population in Yumin with an average of 57.75%. In addition, Nei’s gene diversity index ranged from a minimum of 0.149 to a maximum of 0.218 with an average of 0.186, whereas Shannon’s Information index ranged from 0.213 to 0.339 with an average of 0.284 (Table 3). The two parameters were highest for the ZC population in Yumin and lowest for the HJ population in Tuoli.
Table 3. Analysis of the genetic variation of P. eryngii var. tuoliensis.
| Population | Percentage of Polymorphic Loci | No. of Effective Alleles | Shannon’s Information Index | Gene Diversity |
|---|---|---|---|---|
| WX | 60.54% | 1.303 (0.020) | 0.284 (0.015) | 0.184 (0.11) |
| ZC | 75.59% | 1.355 (0.020) | 0339 (0.014) | 0.218 (0.010) |
| SB | 51.17% | 1.289 (0.020) | 0.263 (0.016) | 0.174 (0.011) |
| SD | 53.18% | 1.312 (0.021) | 0.280 (0.016) | 0.186 (0.011) |
| HS | 69.90% | 1.326 (0.019) | 0.316 (0.015) | 0.203 (0.010) |
| WT | 77.59% | 1.332 (0.019) | 0.324 (0.014) | 0.206 (0.010) |
| KF | 45.48% | 1.301 (0.021) | 0.259 (0.017) | 0.175 (0.012) |
| HJ | 33.44% | 1.268 (0.022) | 0.213 (0.017) | 0.149 (0.012) |
| AR | 52.84% | 1.303 (0.020) | 0.276 (0.016) | 0.182 (0.011) |
| Population level | 57.75% | 1.310 (0.007) | 0.284 (0.005) | 0.186 (0.004) |
| Species level | 96.32% | 1.381 (0.019) | 0.377 (0.012) | 0.238 (0.009) |
The values in parentheses brace denote the standard errors.
Genetic differentiation between populations and levels of gene flow
The results from Nei’s genetic diversity analysis showed that the average total genetic diversity (Ht) over all loci was 0.206 for P. eryngii var. tuoliensis and that the genetic diversity within populations (Hs) was 0.161. The relative degree of gene differentiation among the nine populations (Gst) was 0.218. In other words, 21.8% and 78.2% of the genetic diversity was between populations and within populations, respectively, which reveals that a large amount of the genetic diversity was found within populations.
The results of AMOVA suggested significant genetic differences within populations. The results obtained from the Nei’s genetic diversity analysis were consistent. Specifically, 13% of the total genetic variance (PhiRT) was found between populations from different regions. The next level, i.e., among populations within regions, contributed 8% of the total genetic variance (PhiPR), and the remaining 79% genetic variance was obtained from within individual populations (PhiPT). All of the three levels contributed significantly to the overall genetic variation, as determined through the permutation analyses (Table 4).
Table 4. Summary of the AMOVA results for 123 specimens of P. eryngii var. tuoliensis.
| Source | d.f. | SS | MS | Estimated Variance | Percentage % | Phi Statistic | Value | P |
|---|---|---|---|---|---|---|---|---|
| Among Regions | 2 | 272.85 | 136.43 | 5.24 | 13% | PhiRT | 0.130 | 0.01 |
| Among Populations within Regions | 6 | 481.74 | 80.29 | 3.29 | 8% | PhiPR | 0.094 | 0.01 |
| Within Populations | 114 | 3622.32 | 31.78 | 31.78 | 79% | PhiPT | 0.212 | 0.01 |
| Total | 122 | 4376.91 | 40.31 | 100% |
d.f., degree of freedom; SS, sum of squared observations; MS, mean of squared observations; PhiRT, proportion of the total genetic variance that is due to the variance between regions; PhiPR, proportion of the total genetic variance that is due to the variance among populations within a region; PhiPT, proportion of the total genetic variance that is due to the variance among individuals within a variant.
The gene flow (Nm) estimate obtained by Gst was 1.794 for P. eryngii var. tuoliensis. Compared with those of other basidiomycetes [36,37], the level of gene flow that was present between nine populations was relatively low.
Relationship between genetic distance and geographical distribution
The genetic distances between populations of P. eryngii var. tuoliensis varied from 0.021 to 0.134 (Table 5). The smallest genetic distance was observed between the ZC population and the HS population, both of which are located in the same region (Yumin). The largest genetic distance was found between the SD population of Yumin and the HJ population of Tuoli. The results showed that the genetic distance between the populations located in the same region was smaller than that obtained between populations from different regions. The results from the Mantel tests showed a significant positive correlation between genetic distance and geographical distance among all of the tested populations except for the AR population, which is located approximately 500 km from the other 8 populations (Figure 2a; rGo = 0.789, P = 0.02). Moreover, we found that the longitudinal differences, rather than the latitudinal differences, were associated with the genetic distance (Figure 2b; rLt = 0.157, P = 0.24; Figure 2c; rLn = 0.873, P = 0.01).
Table 5. Pairwise Nei’s genetic distances between geographical populations of P. eryngii var. tuoliensis.
| WX | ZC | SB | SD | HS | WT | KF | HJ | |
|---|---|---|---|---|---|---|---|---|
| ZC | 0.046 | |||||||
| SB | 0.054 | 0.043 | ||||||
| SD | 0.055 | 0.033 | 0.064 | |||||
| HS | 0.054 | 0.021 | 0.058 | 0.025 | ||||
| WT | 0.038 | 0.024 | 0.033 | 0.050 | 0.038 | |||
| KF | 0.093 | 0.085 | 0.099 | 0.118 | 0.090 | 0.084 | ||
| HJ | 0.101 | 0.107 | 0.129 | 0.134 | 0.111 | 0.102 | 0.043 | |
| AR | 0.083 | 0.074 | 0.093 | 0.092 | 0.087 | 0.076 | 0.112 | 0.105 |
Figure 2. Mantel tests between the genetic distance and the geographical parameters.
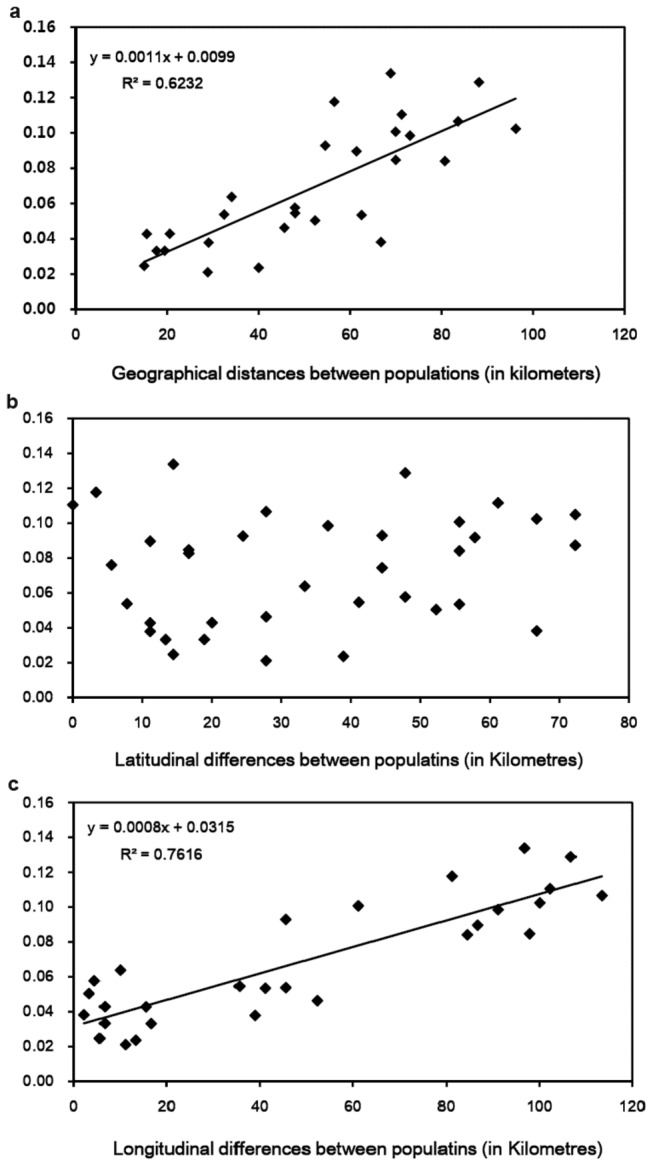
(a) Mantel test between Nei’s genetic distance and the geographical distance among populations. (b) Mantel test between Nei’s genetic distance and the latitudinal distance. (c) Mantel test between Nei’s genetic distance and the longitudinal distance. In 2a, 2b, and 2c, the X-axis represents the geographical parameter, and the Y-axis represents Nei’s genetic distances between the populations.
Cluster analysis
Nine geographical populations of P. eryngii var. tuoliensis were divided into three groups according to their geographical origins, which indicate that the genetic diversity is closely related to the geographical distribution. The populations from six locations in Yumin were group together in the first cluster, and the second cluster consisted of the two populations from Tuoli. The AR population located in Qinghe was first clustered with the Yumin group, whereas the Tuoli group was far from the above two groups and clustered last (Figure 3).
Figure 3. UPGMA dendrogram of the nine populations of P. eryngii var. tuoliensis based on Nei’s genetic distance.
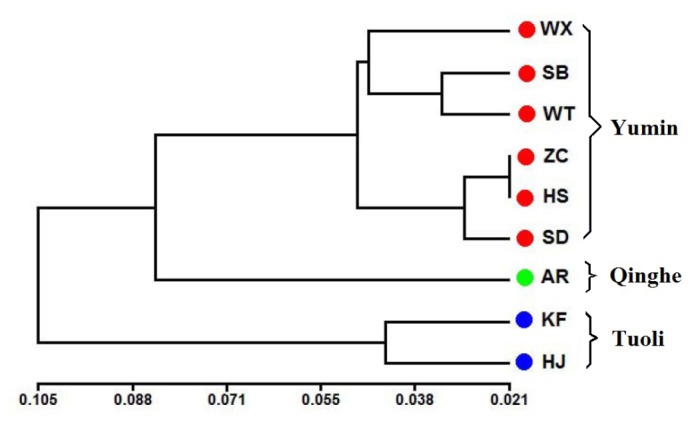
The scale bar means the genetic distance.
Discussion
Two dominant molecular markers (ISSR and SCoT) were used to evaluate the genetic diversity of the wild mushroom of P. eryngii var. tuoliensis in the study. Compared to results obtained with co-dominant markers, this type of markers usually underestimate the within-population variation while produce similar or somewhat higher population differentiation parameters [38-40]. These differences were expected since dominant markers can only produce two alleles in each locus, while they cannot distinguish between homozygotes and heterozygotes [41]. Despite these disadvantages, dominant markers are characterized by estimating unbiased genetic variation and no sequence requirement, which makes them appropriate to analyze those species of little or no genetic information [39,42].
In fungi, high levels of genetic variability are usually observed in wild populations that reproduce sexually, have broad ecological niches, and/or have a wide geographical distribution [43]. The percentage of polymorphic bands (PPB) and Nei’s gene diversity index (H) are two important parameters that are used to measure the genetic variation at the species level [44]. Our results showed a particularly high level of genetic polymorphism. Similar polymorphism levels were previously observed in other edible mushrooms, such as Lentinula edodes (99.6%) [45] and Auricularia polytricha (99.8%) [46]. The mushroom of P. eryngii var. tuoliensis, which independently evolved in China, is a major branch of the P. eryngii species complex and is only distributed in the north area of Xinjiang. The geographical distribution of this variety was not as wide as that observed for P. eryngii var. eryngii or var. ferulae [47]. In general, widespread species were more diverse compared to stenochoric species [48]. However, our analyses indicated that the level of genetic variation of P. eryngii var. tuoliensis is higher than those of the other varieties (PPB = 84.6% in var. ferulae; PPB = 38.5% in var. nebrodensis, var. Elaeoselinum, and var. Thapsia) in the species complex, with the exception of P. eryngii var. eryngii [15]. This result may be mainly attributed to the special climate of Xinjiang, which exhibits dry and windy climate conditions and obvious temperature variations. The intraspecies genetic diversity was closely related to the horizontal transmission system of propagules (airborne spores) and to the adaptation to stressful and temporally heterogeneous environments [13]. The dry and windy climate in Xinjiang contributed to the spore dispersal. Moreover, the Spearman rank correlation revealed that the gene diversity exhibited a strong positive association with the aridity index and the temperature [11]. Stressful (e.g., dry and high temperature) and temporally heterogeneous environments tend to increase the genetic diversity in basidiomycetes of higher classification status [49,50].
The genetic variance was partitioned within populations rather than among populations of P. eryngii var. tuoliensis (Gst = 0.218), and this result is similar to the findings obtained previously in several basidiomycete species. For example, the intercontinental populations of the model basidiomycete Schizophyllum commune were differentiated significantly with a Gst value of 0.214 [43]. For the wood-decay fungus Phlebia centrifuga, eight populations across northern Europe showed a Gst value of 0.072 [51]. The Gst value for geographical populations of Tricholoma matsutake respectively from southwestern, and northeastern China was found to be approximately 0.10 [34,52]. The natural populations of heterothallic basidiomycetes are composed of individual secondary mycelia, which are somatically incompatible and genetically and physiologically distinct. The population structure was regulated through the somatic compatibility systems and mating systems. Some studies have led to the generalization that outcrossing species maintain the majority of the genetic variability within populations, whereas inbreeding species hold most variability among populations [43].
Efficient patterns of dispersal, establishment and spread of the fungi appear to be other factors that influence the local genetic structure of populations. Fungal populations are established by two different dispersal mechanisms. The structure of local populations that are mainly maintained through basidiospore dispersal is characterised by a large number of genetically different individuals within a bit substrate, as has been observed with Pleurotus ostreatus [53] and Mycena rosea [54]. The populations are maintained not by direct basidiospore dispersal but through rapid vegetative spread in the leaf litter or soil. These populations often contain a few but widespread genotypes or just one genotype, as was found for Armillaria bulbosa [55] and Lycoperdon pyriforme [56], respectively. The partitioning pattern of the genetic variation of P. eryngii var. tuoliensis, which corresponds to those of the Italian populations of P. eryngii and P. ferulae, as shown by the fact that most of the variations are contained within local populations [16], would indicate that basidiospore dispersal is the main dissemination mechanism for populations of P. eryngii var. tuoliensis.
Our results found significant genetic differentiation between populations of P. eryngii var. tuoliensis (Table 4). This finding is inconsistent with the results obtained from a previous study about P. eryngii and P. ferulae [16]. It was inferred that the degree of differentiation between populations could be affected by the level of gene flow. In some sense, the population structure was coordinated through two forces: gene flow and genetic drift. A higher level of gene flow would lead to a more homogeneous population structure and a lower extent of genetic differentiation. The gene flow within populations of P. eryngii var. tuoliensis was obviously lower than those of P. eryngii and P. ferulae, which exhibit Nm values of 3.50 and 5.43, respectively [16]. The level of gene flow might not be strong enough to counter the local genetic differentiation between certain populations, although in theory, the differentiation caused by genetic drift could be prevented when the number of migrants per generation (Nm) exceeds one [57].
The results from the Mantel tests and the cluster analysis supported the hypothesis that the genetic relationship among populations was closely associated with their geographic distributions (Tables 2 and 3). Compared with the latitudinal differences, the longitudinal differences played a more important role in the local genetic differentiation between populations. This observation is in agreement with the results obtained by Lewinsohn et al. [58] for the P. eryngii species complex in Israel. Although geographically Yumin and Tuoli distribute 73 km apart and share similar climates, the degree of population divergence between these two regions was higher than that obtained between Qinghe and the other two regions, which are approximately 500 km away from Qinghe. This unexpected finding might be attributed to the peculiar geographical positions of Yumin and Tuoli. Yumin and Tuoli lie in the northwestern Xinjiang region, which is prone to strong winds. During the fruiting season of P. eryngii var. tuoliensis (March to May), the northwest winds prevail in these regions. The dry windy climate might contribute to the long-distance transmission of spores. The gene flow mediated by spore dispersal might weaken the effect of the geographical distance isolation to some extent. The microclimate in Yumin is similar to that in Tuoli, but the populations within these two regions are effectively separated by hills. Studies of biogeography have indicated that certain particular ecological factors can cause population divergence at the microscale. Natural geographical barriers, including mountains and/or rivers, are one of the most important factors [59,60].
Acknowledgments
We highly appreciate Mr. Peng Wei from Xinjiang Academy of Agricultural Sciences and the local mushroom guiders for their help in the sample collection.
Funding Statement
This work was supported by China Agriculture Research System (CARS-24), Ministry of Agriculture of the People’s Republic of China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Guo LQ, Lin JY, Lin JF (2007) Non-volatile components of several novel species of edible fungi in China. Food Chem 100: 643-649. doi: 10.1016/j.foodchem.2005.09.087. [DOI] [Google Scholar]
- 2. Lv H, Kong Y, Yao Q, Zhang B, Leng FW et al. (2009) Nebrodeolysin, a novel hemolytic protein from mushroom Pleurotus nebrodensis with apoptosis-inducing and anti-HIV-1 effects. Phytomedicine 16: 198-205. doi: 10.1016/j.phymed.2008.07.004. PubMed: 18722099. [DOI] [PubMed] [Google Scholar]
- 3. Chen ZC (1991) Study on Pleurotus ferulae . Arid Zone Res 2: 94-95. [Google Scholar]
- 4. Mou CJ, Cao YQ, Ma JL (1987) A new variety of Pleurotus eryngii and its cultural characters. Acta Mycol Sin 6: 153-156. [Google Scholar]
- 5. Huang NL (1996) Pleurotus eryngii var. nebrodensis . In: Huang NL. Cultivation of 18 species of rare and delicious mushroom. Beijing: Chinese Agriculture Press; pp 17-21. [Google Scholar]
- 6. Mao XL (2001) The macrofungi in China. Zhengzhou: Henan Science and Technology Press; pp 64-66. [Google Scholar]
- 7. Kawai G, Babasaki K, Neda H (2008) Taxonomic position of a Chinese Pleurotus “Bai-Ling-Gu”: it belongs to Pleurotus eryngii (DC.:Fr.) Quél. and evolved independently in China. Mycoscience 49: 75-87
- 8. Huang CY, Chen Y, Deng WQ, Li TH, Gao W et al. (2011) Correction of scientific name for cultivated Bai-Ling-Gu in China. J Plant Genet Resour 5: 825-827. [Google Scholar]
- 9. Estrada AE, Mdel Jimenez-Gasco M, Royse DJ (2010) Pleurotus eryngii species complex: sequence analysis and phylogeny based on partial EF1a and RPB2 genes. Fungal Biol 114: 421-428. doi: 10.1016/j.funbio.2010.03.003. PubMed: 20943152. [DOI] [PubMed] [Google Scholar]
- 10. Zervakis G, Sourdis J, Balis C (1994) Genetic variability and systematic of eleven Pleurotus species based on isozyme analysis. Mycol Res 98: 329-341. doi: 10.1016/S0953-7562(09)80461-9. [DOI] [Google Scholar]
- 11. Lewinsohn D, Wasser SP, Resbetnikov SV, Hadar Y, Beharav A et al. (2005) Morphological, ecological, and genetic characterization of the Pleurotus eryngii species complex in Israel. Int J Med Mushrooms 7: 429-430. doi: 10.1615/IntJMedMushr.v7.i3.700. [DOI] [Google Scholar]
- 12. Zhang JX, Huang CY, Ng TB, Wang HX (2006) Genetic polymorphism of ferula mushroom growing on Ferula sinkiangensis . Appl Microbiol Biotechnol 71: 304-309. doi: 10.1007/s00253-005-0139-y. PubMed: 16200340. [DOI] [PubMed] [Google Scholar]
- 13. Ravash R, Shiran B, Alavi AA, Bayat F, Rajaee S et al. (2010) Genetic variability and molecular phylogeny of Pleurotus eryngii species-complex isolates from Iran, and notes on the systematics of Asiatic populations. Mycol Prog 9: 181-194. doi: 10.1007/s11557-009-0624-2. [DOI] [Google Scholar]
- 14. De Gioia T, Sisto D, Rana GL, Figliuolo G (2005) Genetic structure of the Pleurotus eryngii species-complex. Mycol Res 109: 71-80. doi: 10.1017/S0953756204001637. PubMed: 15736864. [DOI] [PubMed] [Google Scholar]
- 15. Zervakis GI, Venturella G, Papadopoulou K (2001) Genetic polymorphism and taxonomic infrastructure of the Pleurotus eryngii species-complex as determined by RAPD analysis, isozyme profiles and ecomorphological characters. Microbiology 147: 3183-3194. PubMed: 11700370. [DOI] [PubMed] [Google Scholar]
- 16. Urbanelli S, Della Rosa V, Fanelli C, Fabbri AA, Reverberi M (2003) Genetic diversity and population structure of the Italian fungi belonging to the taxa Pleurotus eryngii and Pleurotus ferulae (DC.:FrFr.). Heredity (Edinb) 90: 253-259. doi: 10.1038/sj.hdy.6800239. PubMed: 12634809. [DOI] [PubMed] [Google Scholar]
- 17. Kauserud H, Schumacher T (2003) Genetic structure of Fennoscandian populations of the threatened wood-decay fungus Fomitopsis rosea (Basidiomycota). Mycol Res 2: 155-163. PubMed: 12747326. [DOI] [PubMed] [Google Scholar]
- 18. George S, Sharma J, Yadon VL (2009) Genetic diversity of the endangered and narrow endemic Piperia yadonii (Orchidaceae) assessed with ISSR polymorphisms. Am J Bot 11: 2022-2030. PubMed: 21622322. [DOI] [PubMed] [Google Scholar]
- 19. Wang HZ, Wu ZX, Lu JJ, Shi NN, Zhao Y et al. (2009) Molecular diversity and relationships among Cymbidium goeringii cultivars based on inter-simple sequence repeat (ISSR) markers. Genetica 136: 391-399. doi: 10.1007/s10709-008-9340-0. PubMed: 19085060. [DOI] [PubMed] [Google Scholar]
- 20. Singh YT, Mazumdar-Leighton S, Saikia M, Pant P, Kashung S et al. (2012) Genetic variation within native populations of endemic silkmoth Antheraea assamensis (Helfer) from Northeast India indicates need for in situ conservation. PLOS ONE 7: e49972. doi: 10.1371/journal.pone.0049972. PubMed: 23185503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang XM, Yang R, Feng SF, Hou XQ, Zhang YQ et al. (2012) Genetic variation in Rheum palmatum and Rheum tanguticum (Polygonaceae), two medicinally and endemic species in china using ISSR markers. PLOS ONE 7: e51667. doi: 10.1371/journal.pone.0051667. PubMed: 23289054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Raina SN, Rani V, Kojima T, Ogihara Y, Singh KP et al. (2001) RAPD and ISSR fingerprints as useful genetic markers for analysis of genetic diversity, varietal identification, and phylogenetic relationships in peanut (Arachis hypogaea) cultivars and wild species. Genome 44: 763-772. doi: 10.1139/g01-064. PubMed: 11681599. [DOI] [PubMed] [Google Scholar]
- 23. Collard BCY, Mackill DJ (2009) Start codon targeted (SCoT) polymorphism: a simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol Biol Rep 27: 86-93. doi: 10.1007/s11105-008-0060-5. [DOI] [Google Scholar]
- 24. Xiong FQ, Zhong RC, Han ZQ, Jiang J, He LQ, et al. (2011) Start codon targeted polymorphism for evaluation of functional genetic variation and relationships in cultivated peanut (Arachis hypogaea L.) genotypes. Mol Biol Rep 38: 3487-3494 [DOI] [PubMed]
- 25. Gorji AM, Poczai P, Polgar Z, Taller J (2011) Efficiency of arbitrarily amplified dominant markers (SCoT, ISSR and RAPD) for diagnostic fingerprinting in tetraploid potato. Am J Potato Res 88: 226-237. doi: 10.1007/s12230-011-9187-2. [DOI] [Google Scholar]
- 26. Guo DL, Zhang JY, Liu CH (2012) Genetic diversity in some grape varieties revealed by SCoT analyses. Mol Biol Rep 39: 5307-5313. doi: 10.1007/s11033-011-1329-6. PubMed: 22170600. [DOI] [PubMed] [Google Scholar]
- 27. Luo C, He XH, Chen H, Ou SJ, Gao MP et al. (2010) Analysis of diversity and relationships among mango cultivars using Start Codon Targeted (SCoT) markers. Biochem Syst Ecol 38: 1176-1184. doi: 10.1016/j.bse.2010.11.004. [DOI] [Google Scholar]
- 28. Luo C, He XH, Chen H, Hu Y, Ou SJ (2012) Genetic relationship and diversity of Mangifera indica L.: revealed through SCoT analysis. Genet Resour Crop Evol 7: 1505-1515. [Google Scholar]
- 29. Xiong FQ, Tang HJ, Chen ZL, Pan LH, Zhuang WJ (2009) SCoT: A novel gene targeted marker technique based on the translation start codon. Mol Plant Breed 3: 635-638. [Google Scholar]
- 30. Biswas MK, Xu Q, Deng XX (2010) Utility of RAPD, ISSR, IRAP and REMAP markers for the genetic analysis of Citrus spp. Sci Hort 124: 254-261. doi: 10.1016/j.scienta.2009.12.013. [DOI] [Google Scholar]
- 31. Luo C, He XH, Chen H, Ou SJ, Gao MP et al. (2011) Genetic diversity of mango cultivars estimated using SCoT and ISSR markers. Biochem Syst Ecol 39: 676-684. doi: 10.1016/j.bse.2011.05.023. [DOI] [Google Scholar]
- 32. Peakall R, Smouse PE (2006) Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6: 288-295. doi: 10.1111/j.1471-8286.2005.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yeh FC, Boyle TJB (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129: 157. [Google Scholar]
- 34. Xu JP, Sha T, Li YC, Zhao ZW, Yang ZL (2008) Recombination and genetic differentiation among natural populations of the ectomycorrhizal mushroom Tricholoma matsutake from southwestern China. Mol Ecol 17: 1238-1247. doi: 10.1111/j.1365-294X.2007.03665.x. PubMed: 18302686. [DOI] [PubMed] [Google Scholar]
- 35. Rohlf FJ (2000) NTSYS-pc: numerical taxonomy and multivariate analysis system. Version 2.1. New York: Exeter Publications. [Google Scholar]
- 36. Heinzelmann R, Rigling D, Prospero S (2012) Population genetics of the wood-rotting basidiomycete Armillaria cepistipes in a fragmented forest landscape. Fungal Biol 9: 985-994. [DOI] [PubMed] [Google Scholar]
- 37. Cao Y, Zhang Y, Yu Z, Mi F, Liu C et al. (2013) Structure, gene flow, and recombination among geographic populations of a Russula virescens Ally from southwestern China. PLOS ONE 8: e73174. doi: 10.1371/journal.pone.0073174. PubMed: 24069176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lynch M, Milligan BG (1994) Analysis of population genetic structure with RAPD markers. Mol Ecol 3: 91-99. doi: 10.1111/j.1365-294X.1994.tb00109.x. PubMed: 8019690. [DOI] [PubMed] [Google Scholar]
- 39. Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13: 1143-1155. doi: 10.1111/j.1365-294X.2004.02141.x. PubMed: 15078452. [DOI] [PubMed] [Google Scholar]
- 40. Soldati MC, Fornes L, van Zonneveld M, Thomas E, Zelener N (2013) An assessment of the genetic diversity of Cedrela balansae C. dC. (Meliaceae) in Northwestern Argentina by means of combined use of SSR and AFLP molecular markers. Biochem Syst Ecol 47: 45-55. doi: 10.1016/j.bse.2012.10.011. [DOI] [Google Scholar]
- 41. Xu JP (2006) Fundamentals of fungal molecular population genetic analyses. Curr Issues Mol Biol 8: 75-90. PubMed: 16875415. [PubMed] [Google Scholar]
- 42. Stewart CN, Excoffier L (1996) Assessing population genetic structure and variability with RAPD data: Application to Vaccinium macrocarpon (American Cranberry). J Evolution Biol 9: 153-171. doi: 10.1046/j.1420-9101.1996.9020153.x. [DOI] [Google Scholar]
- 43. James TY, Porter D, Hamrick JL, Vilgalys R (1999) Evidence for limited intercontinental gene flow in the cosmopolitan mushroom, Schizophyllum commune . Evolution 53: 1665-1677. doi: 10.2307/2640430. [DOI] [PubMed] [Google Scholar]
- 44. Hamrick JL, Godt MJ (1989) Allozyme diversity in plant species. In: Brown AHD, Clegg MT, Kahler AL, Weir BS. Plant population genetics, breeding and genetic resources. Sinauer: Sunderland, MA: pp 43-63. [Google Scholar]
- 45. Xiao Y, Liu W, Dai YH, Fu C, Bian YB (2010) Using SSR markers to evaluate the genetic diversity of Lentinula edodes’ natural germplasm in China. World J Microbiol Biotechnol 26: 527-536. doi: 10.1007/s11274-009-0202-4. [DOI] [Google Scholar]
- 46. Du P, Cui BK, Dai YC (2011) High genetic diversity in wild culinary-medicinal wood ear mushroom, Auricularia polytricha (Mont.) Sacc., in tropical China revealed by ISSR analysis. Int J Med Mushrooms 13: 289-297 [DOI] [PubMed]
- 47. Urbanelli S, Fanelli C, Fabbri AA, Della Rosa V, Maddau L, et al. (2002) Molecular genetic analysis of two taxa of the Pleurotus eryngii complex: Pleurotus eryngii (DC.:Fr.) Quel. var. eryngii and P. eryngii (DC.:Fr.) Quel. var. ferulae. Biol J Linn Soc Lond 75: 125-136. [Google Scholar]
- 48. Hamrick JL, Godt MJW, Murawski DA, Loveless MD (1991) Correlations between species traits and allozyme diversity: implications for conservation biology. In: Falk DA, Holsinger KE. Genetics and conservation of rare plants. New York: Oxford University Press; pp 75-86. [Google Scholar]
- 49. Nevo E, Beiles A (1988) Genetic parallelism of protein polymorphism in nature: Ecological test of the neutral theory of molecular evolution. Biol J Linn Soc Lond 35: 229-245. doi: 10.1111/j.1095-8312.1988.tb00468.x. [DOI] [Google Scholar]
- 50. Nevo E (1998) Molecular evolution and ecological stress at global, regional and local scales: the Israeli perspective. J Exp Zool 282: 95-119. doi: 10.1002/(SICI)1097-010X(199809/10)282:1/2. [DOI] [Google Scholar]
- 51. Franzén I, Vasaitis R, Penttilä R, Stenlid J (2007) Population genetics of the wood-decay fungus Phlebia Centrifuga P Karst. in fragmented and continuous habitats. Mol Ecol 16: 3326-3333. doi: 10.1111/j.1365-294X.2007.03394.x. PubMed: 17688536. [DOI] [PubMed] [Google Scholar]
- 52. Ma DL, Yang GT, Mu LQ, Song YT (2010) Application of SRAP in the genetic diversity of Tricholoma matsutake in northeastern China. Afr J Biotechnol 9: 6244-6250. [Google Scholar]
- 53. Kay E, Vilgalys R (1992) Spatial distribution and genetic relationships among individuals in a natural population of the oyster mushroom Pleurotus ostreatus . Mycologia 84: 173-182. doi: 10.2307/3760248. [DOI] [Google Scholar]
- 54. Boisselier-Dubayle MC, Perreau-Bertrand J, Lambourdiere J (1996) Genetic variation in wild populations of Mycena rosea . Mycol Res 100: 753-758. doi: 10.1016/S0953-7562(96)80210-3. [DOI] [Google Scholar]
- 55. Smith ML, Bruhn JN, Anderson JB (1992) The fungus Armillaria bulbosa is among the largest and oldest living organisms. Nature 356: 428-431. doi: 10.1038/356428a0. [DOI] [Google Scholar]
- 56. Huss MJ (1993) Spatial distribution among mycelial individuals of Lycoperdon pyriforme occurring on decaying logs. Mycol Res 97: 1119-1125. doi: 10.1016/S0953-7562(09)80513-3. [DOI] [Google Scholar]
- 57. Slatkin M (1987) Gene flow and the geographic structure of natural populations. Science 236: 787-792. doi: 10.1126/science.3576198. PubMed: 3576198. [DOI] [PubMed] [Google Scholar]
- 58. Lewinsohn D, Nevo E, Wasser SP, Hadar Y, Beharav A (2001) Genetic diversity in populations of the Pleurotus eryngii complex in Israel. Mycol Res 105: 941-951. doi: 10.1016/S0953-7562(08)61950-4. [DOI] [Google Scholar]
- 59. Cozzolino S, Cafasso D, Pellegrino G, Musacchio A, Widmer A (2003) Fine-scale phylogeographical analysis of Mediterranean Anacamptis palustris (Orchidaceae) populations based on chloroplast minisatellite and microsatellite variation. Mol Ecol 12: 2783-2792. doi: 10.1046/j.1365-294X.2003.01958.x. PubMed: 12969480. [DOI] [PubMed] [Google Scholar]
- 60. Gascon C, Malcolm JR, Patton JL, da Silva MN, Bogart JP et al. (2000) Riverine barriers and the geographic distribution of Amazonian species. Proc Natl Acad Sci U S A 97: 13672-13677. doi: 10.1073/pnas.230136397. PubMed: 11095705. [DOI] [PMC free article] [PubMed] [Google Scholar]


