Abstract
Natural killer cell responses play a crucial role in virus clearance by the innate immune system. Although the killer immunoglobulin-like receptor (KIR) in combination with its cognate human leukocyte antigen (HLA) ligand, especially KIR2DL3-HLA-C1, is associated with both treatment-induced and spontaneous clearance of hepatitis C virus (HCV) infection in Caucasians, these innate immunity genes have not been fully clarified in Japanese patients. We therefore investigated 16 KIR genotypes along with HLA-B and -C ligands and a genetic variant of interleukin (IL) 28B (rs8099917) in 115 chronic hepatitis C genotype 1 patients who underwent pegylated-interferon-α2b (PEG-IFN) and ribavirin therapy. HLA-Bw4 was significantly associated with a sustained virological response (SVR) to treatment (P = 0.017; odds ratio [OR] = 2.50, ), as was the centromeric A/A haplotype of KIR (P = 0.015; OR 3.37). In contrast, SVR rates were significantly decreased in patients with KIR2DL2 or KIR2DS2 (P = 0.015; OR = 0.30, and P = 0.025; OR = 0.32, respectively). Multivariate logistic regression analysis subsequently identified the IL28B TT genotype (P = 0.00009; OR = 6.87, 95% confidence interval [CI] = 2.62 - 18.01), KIR2DL2/HLA-C1 (P = 0.014; OR = 0.24, 95% CI = 0.08 - 0.75), KIR3DL1/HLA-Bw4 (P = 0.008, OR = 3.32, 95% CI = 1.37 - 8.05), and white blood cell count at baseline (P = 0.009; OR = 3.32, 95% CI = 1.35 - 8.16) as independent predictive factors of an SVR. We observed a significant association between the combination of IL28B TT genotype and KIR3DL1-HLA-Bw4 in responders (P = 0.0019), whereas IL28B TT along with KIR2DL2-HLA-C1 was related to a non-response (P = 0.0067). In conclusion, combinations of KIR3DL1/HLA-Bw4, KIR2DL2/HLA-C1, and a genetic variant of the IL28B gene are predictive of the response to PEG-IFN and ribavirin therapy in Japanese patients infected with genotype 1b HCV.
Introduction
Hepatitis C virus (HCV) infection is a major cause of chronic liver disease worldwide. Chronic HCV infection often develops into chronic hepatitis, which may progress to liver cirrhosis and/or hepatocellular carcinoma (HCC)[1]. HCC is a leading cause of death from malignant neoplasms in Japan[2]. Since approximately 70% of Japanese HCC patients are infected with HCV, the successful eradication of this virus, defined as a sustained virological response (SVR), is considered important to decrease the incidence of HCC.
Natural killer (NK) cells are key components of the innate antiviral immune response that are controlled by a balance of activation and inhibitory receptors. NK cell activation receptors include C-type lectin-like receptors (NKG2C, NKG2D, and NKG2E), natural cytotoxicity receptors (NKp30, NKp44, and NKp46), and CD16, while known inhibitory receptors include killer cell immunoglobulin-like receptors (KIRs) and the CD94/NKG2 family, which also contains a C-type lectin-like receptor (NKG2A) [3,4]. Sixteen KIR genes and pseudogenes have been identified that are encoded by a family of genes located on human chromosome 19q13.4. One particular feature of KIRs is their substantial genetic diversity. Some inhibitory KIRs recognize human leukocyte antigen (HLA) class I molecules as their ligands; KIR2DL1 recognizes HLA-C group 2 (HLA-C2) allotypes having lysine at amino acid position 80, whereas KIR2DL2 and KIR2DL3 recognize HLA-C group 1 (HLA-C1) allotypes having asparagine at amino acid position 80 [5]. KIR2DL2 and KIR2DL3 also recognize HLA-B*4601 acquiring the-C1 epitope by gene conversion [6]. Furthermore, KIR3DL1 recognizes subsets of HLA-A and HLA-B allotypes having the -Bw4 epitope determined by amino acid positions 77-83 [7].
It has been well documented that certain KIR-HLA receptor-ligand combinations are associated with susceptibility to infectious diseases, such as HCV, as well as with disease progression and treatment response [8-15]. Recent reports have also identified a relationship between interleukin (IL) 28B gene polymorphisms and treatment and spontaneous resolution of HCV infection[16-19]. Dring et al. observed that the presence of IL28B gene polymorphisms and KIR genotypes synergized to increase the risk of chronic HCV infection[20], although this finding is under debate[21]. Suppiah et al. [22] recently reported that genotyping for IL28B, HLA-C, and KIR genes was useful for predicting HCV treatment response in patients of European descent. As these gene associations have not yet been studied in the Japanese population, we evaluated whether HLA-KIR interactions, in addition to an IL28B polymorphism, would influence the outcome of pegylated-interferon-α (PEG-IFN) and ribavirin therapy in Japanese patients with chronic hepatitis C.
Materials and Methods
Ethics statement
This study was approved by the ethical committee of Shinshu University School of Medicine, Matsumoto, Japan, and written informed consent was obtained from all participants. The study was conducted in accordance with the principles of the Declaration of Helsinki.
Subjects
One hundred and fifteen consecutive IFN-treatment-naïve patients with chronic hepatitis C were enrolled in this study. All subjects were seen at Shinshu University Hospital or one of its affiliated hospitals. The clinical and demographic characteristics of our cohort are shown in Table 1. Diagnosis of chronic hepatitis C was based on previously reported criteria [23]: 1) presence of serum HCV antibodies and detectable viral RNA; 2) absence of detectable hepatitis B surface antigen and antibody to the human immunodeficiency virus; and 3) exclusion of other causes of chronic liver disease or a history of decompensated cirrhosis or HCC. Serum levels of HCV RNA were determined using Cobas Amplicor assays (sensitivity: 50 IU/mL; Roche Diagnostic Systems, Tokyo, Japan). HCV genotypes were determined using INNO-LiPA HCV II kits (Innogenetics, Gent, Belgium). Alanine aminotransferase (ALT), aspartate aminotransferase (AST), and other relevant biochemical tests were performed using standard methods[24]. Liver fibrosis was assessed using the AST to platelet ratio index (APRI) in this study. APRI has been recognized as a noninvasive test to estimate the degree of liver fibrosis in chronic liver disease with HCV infection[25]. APRI was calculated for all study subjects as follows: AST/upper limit of normal (45 IU/L) × 100/platelet count (109/L). Patients received PEG-IFN-α2b (Pegintron; MSD KK, Tokyo, Japan; 1.5 μg/kg of body weight by subcutaneous injection once per week) and ribavirin (Rebetol; MSD KK; 600-1000 grams daily, according to body weight) for 48 weeks, as described previously[26]. Patients achieving a sustained HCV response were defined as those whose serum HCV RNA was undetectable 24 weeks after completing therapy. Patients who did not meet this criterion, who included non-responders and relapsers, were regarded as treatment failures.
Table 1. Clinical features of sustained and non-sustained virological response patients with chronic hepatitis C.
| Characteristic | All | SVR | Non-SVR | P |
|---|---|---|---|---|
| (n = 115) | (n = 56) | (n = 59) | ||
| Age (yr) | 60 (24 - 80) | 59 (25 - 80) | 60 (24 - 75) | 0.43 |
| Male | 66 (57) | 34 (61) | 32 (54) | 0.48 |
| Alanine aminotransferase (IU/L) | 46 (17 - 389) | 48 (17 - 389) | 45 (17 - 309) | 0.81 |
| Aspartate aminotransferase (IU/L) | 43 (17 - 246) | 42 (17 - 231) | 43 (17 - 246) | 0.49 |
| White blood cells (/μL) | 4410 (2280 - 8240) | 4740 (2700 - 8170) | 4070 (2280 - 8240) | 0.011 |
| Hemoglobin (g/dL) | 14.4 (9.2 - 18.2) | 15.1 (11.0 - 18.2) | 13.9 (9.2 - 17.4) | 0.002 |
| Platelet count (104/μL) | 15.9 (6.7 - 33.6) | 16.6 (8.3 - 26.2) | 15.6 (6.7 - 33.6) | 0.30 |
| APRI | 0.89 (0.21 - 5.40) | 0.59 (0.22 - 5.40) | 0.66 (0.21 - 5.06) | 0.41 |
| HCV RNA (log10 IU/mL) | 6.4 (5.0 - 7.3) | 6.1 (5.0 - 6.8) | 6.5 (5.0 - 7.3) | < 0.001 |
Data are expressed as median (range) or n (%) as appropriate. SVR, sustained virological response; HCV, hepatitis C virus
HLA, KIR, and IL28B (rs8099917) Genotyping
Genomic DNA was isolated from whole blood samples using QuickGene-800 assays (Fujifilm, Tokyo, Japan). We genotyped HLA-B, HLA-C, and KIR using a Luminex multi-analyzer profiling system with a LAB type® HD and KIR SSO genotyping kit (One Lambda, Inc., Canoga Park, CA), which is based on PCR sequence-specific oligonucleotide probes[27]. Subjects were identified as having the B/x or A/A genotype as defined previously[28]. Genotypes for the centromeric (Cen) and telomeric (Tel) parts of the KIR locus were determined according to the presence or absence of one or more B haplotype-defining KIR genes. Thus, Cen-A1 and Tel-A1 were the centromeric and telomeric motifs, respectively, of the canonical A KIR haplotype in the present study, Cen-B1 and Cen-B2 were alternative centromeric motifs of common B KIR haplotypes, and Tel-B1 was the common telomeric motif of B haplotypes[29]. For much of this analysis, Cen-B1 and -B2 were grouped together as Cen-B, whereas Cen-A1 was shortened to Cen-A and Tel-A1 to Tel-A, as reported previously[30,31]. Genotyping of an IL28B SNP (rs8099917) was performed using a TaqMan 5’ exonuclease assay with primers supplied by Applied Biosystems[32]. Probe fluorescence signals were detected using a TaqMan assay for Real-Time PCR (7500 Real Time PCR System, Applied Biosystems) according to the manufacturer’s instructions.
Statistical Analysis
The Mann-Whitney U test was employed to analyze continuous variables. Pearson’s chi-squared test was used for the analysis of categorical data. We adopted Fisher’s exact test when the number of subjects was less than 5. The Bonferroni correction for multiple testing was applied to our data of KIR-HLA combinations using the number of comparisons performed by our primary factors of interest in Table 2 (i.e., 8 tests = 4 combinations × 2 comparisons between two groups). A P value of < 0.05 was considered to be statistically significant. Association strength was estimated by calculating the odds ratio (OR) and 95% confidence interval (CI). Our model was checked by regression diagnostic plots to verify normality, linearity of data, and constant variance. Stepwise logistic regression analysis with a forward approach was performed to identify independent factors associated with an SVR after continuous variables were separated into 2 categorical variables by each median value. Statistical analyses were performed using SPSS software version 21.0J (IBM, Tokyo, Japan). Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) were calculated to determine the reliability of the predictors of therapy response.
Table 2. Frequency of IL28B genotype, KIR3DL1/HLA-Bw4, and KIR2DL2/HLA-C1 combinations in 56 patients with a sustained virological response (SVR) and 59 patients with a non-SVR to pegylated interferon and ribavirin therapy of chronic hepatitis C.
| KIR3DL1/HLA-Bw4 | KIR2DL2/HLA-C1 | SVR | Non-SVR | P (Pc) | OR (95% CI) |
|---|---|---|---|---|---|
| (n = 56) | (n = 59) | ||||
| +/+ | +/+ | 5 (9%) | 7 (12%) | 0.61 | |
| +/+ | Other | 31 (55%) | 19 (32%) | 0.012 (0.1) | 2.61 (1.22 - 5.58) |
| Other | +/+ | 1 (2%) | 10 (17%) | 0.014 (0.12) | 0.09 (0.01 - 0.72) |
| Other | Other | 19 (34%) | 23 (39%) | 0.57 | |
| IL28B | KIR3DL1/HLA-Bw4 | SVR | Non-SVR | P (Pc) | OR (95% CI) |
| (n = 56) | (n = 59) | ||||
| TT | +/+ | 27 (48%) | 13 (22%) | 0.003 (0.024) | 3.29 (1.47 - 7.39) |
| TT | Other | 17 (30%) | 14 (24%) | 0.42 | |
| TG/GG | +/+ | 9 (16%) | 13 (22%) | 0.42 | |
| TG/GG | Other | 3 (5%) | 19 (32%) | 0.00062 (0.0005) | 0.12 (0.03 - 0.43) |
| IL28B | KIR2DL2/HLA-C1 | SVR | Non-SVR | P (Pc) | OR (95% CI) |
| (n = 56) | (n = 59) | ||||
| TT | Other | 38 (68%) | 18 (31%) | 0.000062 (0.0005) | 4.81 (2.19 - 10.58) |
| TT | +/+ | 6 (11%) | 9 (15%) | 0.47 | |
| TG/GG | Other | 12 (21%) | 24 (41%) | 0.026 (0.21) | 0.40 (0.17 - 0.91) |
| TG/GG | +/+ | 0 (0%) | 8 (14%) | 0.013 (0.1) | - |
Data are expressed as n (%).
Results
Patient Characteristics and Treatment Outcome
All patients in our test cohort were infected with HCV genotype 1b. Of the 115 patients receiving PEG-IFN-α2b and ribavirin therapy, 56 (49%) achieved an SVR. The remaining 59 patients were non-responders, 28 of whom experienced a relapse and 31 who were null responders. The median white blood cell count (P = 0.011) and hemoglobin value (P = 0.002) in the SVR group were significantly higher than those in the non-SVR group prior to treatment. HCV viral load at baseline was significantly associated with treatment outcome (P < 0.001).
Association of HLA and KIR with a Sustained Virological Response
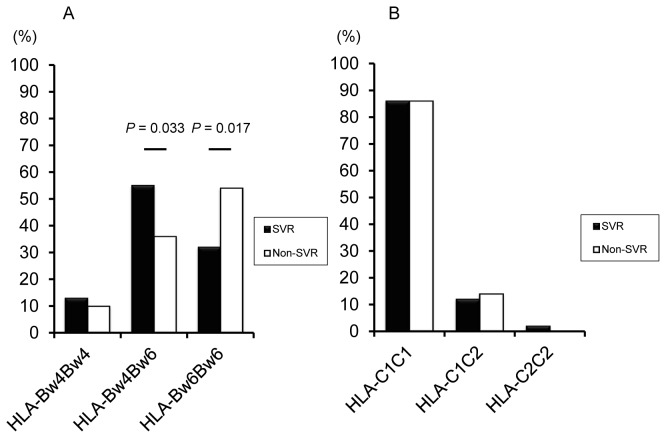
We first determined the frequency of HLA-Bw and HLA-C alleles in SVR and non-SVR patients (Figure 1). The frequency of HLA-Bw4Bw6 in responders was significantly higher than that in non-responders (55% [31/56] vs. 36% [21/59]; P = 0.033; OR = 2.24, 95% CI = 1.06 - 4.75). Conversely, patients with the HLA-Bw6 homozygote had a higher non-SVR rate (32% [18/56] vs. 54% [32/59]; P = 0.017; OR = 0.40, 95% CI = 0.19 - 0.85). Overall, HLA-Bw4 was associated with an SVR among patients (68% [38/56] vs. 46% [27/59]; P = 0.017; OR = 2.50, 95% CI = 1.17 - 5.35). The frequencies of HLA-C were not statistically significant. We further checked whether particular HLA-Bw or HLA-C alleles were beneficial to treatment outcome. The HLA-B*35:01 allele was more frequently found in patients with an SVR than in those without (13% [15/102] vs. 4% [5/118]; P = 0.014 [Pc = 0.36]; OR = 3.49, 95% CI = 1.23 - 9.97).
Figure 1. Frequency of HLA-Bw and -C alleles in 56 patients with a sustained virological response (SVR) and 59 patients with a non-SVR to pegylated interferon and ribavirin therapy of chronic hepatitis C.
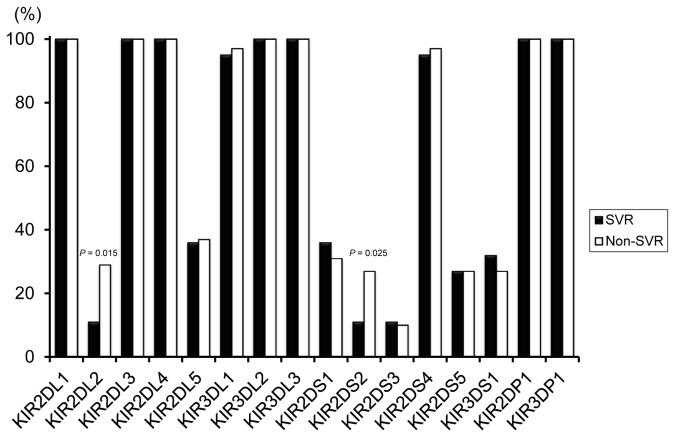
The distribution of KIR genes and their association with treatment outcome are shown in Figure 2. No statistically significant differences were found for any allele combination apart from KIR2DL2 and KIR2DS2; patients with these genes had significantly decreased SVR frequencies compared with those without (P = 0.015 [Pc = 0.48]; OR = 0.30, 95% CI = 0.11 - 0.82 and P = 0.025 [Pc = 0.8]; OR = 0.32, 95% CI = 0.12 - 0.90, respectively).
Figure 2. Frequency of each KIR gene in 56 patients with a sustained virological response (SVR) and 59 patients with a non-SVR to pegylated interferon and ribavirin therapy of chronic hepatitis C.
KIR genotype profiles were determined by the presence or absence of each KIR locus in patients (Figure 3). Since strong linkage disequilibrium is a prominent feature in the KIR region, KIR gene profiles were classified based on Cen and Tel motifs. When we evaluated SVR according to genotype and Cen and Tel frequencies, we observed that virologic clearance with Cen-A/A was significantly higher than that without (54% [50/92] vs. 26% [6/23], P = 0.015; OR = 3.37, 95% CI = 1.22 - 9.33). There were no significant differences regarding AA genotype and Tel.
Figure 3. KIR gene profile frequencies in 56 patients with a sustained virological response (SVR) and 59 patients with a non-SVR to pegylated interferon and ribavirin therapy of chronic hepatitis C.
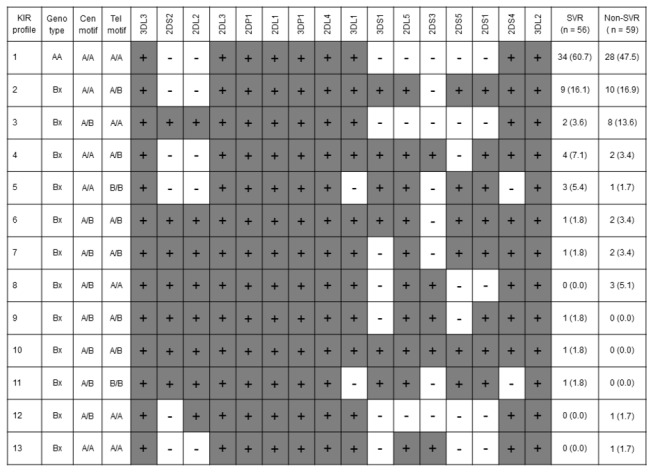
Numerical data represent the number of individuals (%). The presence of KIR genes is indicated by gray shading. Cen, centromeric; Tel, telomeric.
We next analyzed combinations of activation/inhibitory KIRs and their HLA ligands for possible associations with an SVR. Among the combinations of KIR3DL1-HLA-Bw4, KIR2DL2-HLA-C1, and KIR2DL1-HLA-C2, patients who carried the inhibitory KIR3DL1 receptor and its ligand HLA-Bw4 had a significantly higher response rate than those without KIR3DL1 or HLA-Bw4 (58% [36/62] vs. 38% [20/53]; P = 0.030 [Pc = 0.12]; OR = 2.29, 95% CI = 1.08 - 4.84). In contrast, the KIR2DL2-HLA-C1 combination resulted in a significantly lower SVR rate (26% [6/23] vs. 54% [50/92]; P = 0.015 [Pc = 0.06]; OR = 0.30, 95% CI = 0.11 - 0.82). Although several studies have found that KIR2DL3-HLA-C1 carriers are associated with treatment-induced and spontaneous clearance of HCV in Caucasians, no such association was found in our cohort (data not shown).
Patients with KIR3DL1-HLA-Bw4 but without KIR2DL2-HLA-C1 had a higher SVR rate (55% [31/56] vs. 32% [19/59]; P = 0.012 [Pc = 0.1]; OR = 2.61, 95% CI = 1.22 - 5.58) (Table 2). Conversely, the frequency of the KIR2DL2-HLA-C1 positive, but KIR3DL1-HLA-Bw4 negative condition was significantly higher in non-responders (17% [10/59] vs. 2% [1/56]; P = 0.014 [Pc = 0.12]; OR = 0.09, 95% CI = 0.01 - 0.72).
Prediction of a Sustained Virological Response by KIR-HLA and IL28B
Examination of the IL28B rs8099917 SNP in our cohort revealed significant differences in SVR frequencies. The SVR rate in patients with the IL28B TT genotype was significantly higher in those with TG or GG genotypes (62% [44/71] vs. 27% [12/44), P = 0.0003; OR = 4.35, 95% CI = 1.92 - 9.85). In subjects with IL28B TT and KIR3DL1-HLABw4, virologic clearance was significantly increased over other combinations (68% [27/40] vs. 39% [29/75]; P = 0.003 [Pc = 0.024]; OR 3.29, 95% CI = 1.47 - 7.39).
We next evaluated several factors found in association with an SVR to PEG-IFN and ribavirin therapy for independence by logistic regression analysis. Fifty-six responders were compared with 59 non-responders by means of a forward stepwise likelihood ratio logistic regression method; estimated OR coefficients, 95% CI, and P values are summarized in Table 3 for the variables that remained in equation at the last step. IL28B TT genotype (P = 0.00009; OR = 6.87, 95% CI = 2.62 - 18.01), KIR2DL2-HLA-C1 (P = 0.014; OR = 0.24, 95% CI = 0.08 - 0.75), white blood cell count ≥ 4410/μL (P = 0.009; OR = 3.32, 95% CI= 1.35 - 8.16), and KIR3DL1-HLA-Bw4 (P = 0.008; OR = 3.32, 95% CI = 1.37 - 8.05) were all identified as independent parameters that significantly influenced an SVR.
Table 3. Logistic regression analysis of variables contributing to a sustained virological response to pegylated interferon and ribavirin.
| Factor | Odds ratio | 95% confidence interval | P |
|---|---|---|---|
| IL28B TT genotype | 6.87 | 2.62 - 18.01 | 0.00009 |
| KIR2DL2/HLA-C1 | 0.24 | 0.08 - 0.75 | 0.014 |
| White blood cells ≥ 4410/μL | 3.32 | 1.35 - 8.16 | 0.009 |
| KIR3DL1/HLA-Bw4 | 3.32 | 1.37 - 8.05 | 0.008 |
Only variables achieving statistical significance (P < 0.05) in multivariate logistic regression analysis are shown.
The frequency of the IL28B TT genotype with KIR3DL1-HLA-Bw4 in responders was significantly higher than in non-responders (48% [27/56] vs. 22% [13/59]; P = 0.003 [Pc = 0.024]; OR = 3.29, 95% CI = 1.47 - 7.39) (Table 2). Patients with the IL28B TT genotype without KIR2DL2-HLA-C1 had a significantly higher SVR rate (68% [38/56] vs. 31% [18/59]; P = 0.000062 [Pc = 0.0005]; OR = 4.81, 95% CI = 2.19 - 10.58). The frequency of a non-SVR was significantly higher in patients with the IL28B non-TT genotype both with and without KIR2DL2-HLA-C1 (14% [8/59] vs. 0% [0/8]; P = 0.013 [Pc = 0.1] and 41% [24/59] vs. 21% [12/56]; P = 0.026 [Pc = 0.21]; OR = 0.40, 95% CI = 0.17 - 0.91, respectively). The ability to predict an SVR by IL28B genotype and KIR3DL1-HLA-Bw4 and KIR2DL2-HLA-C1 was next evaluated. Corresponding values for sensitivity, specificity, PPV, and NPV are listed in Table S1 in File S1. A combination of the IL28B TT genotype and KIR3DL1-HLA-Bw4 demonstrated high predictive specificity (78%), as did the combination of IL28B TT genotype and KIR2DL2-HLA-C1 (86%).
Lastly, we analyzed combinations of the three factors of IL28B genotype, KIR3DL1-HLA-Bw4, and KIR2DL2-HLA-C1 for prediction of treatment outcome (Table S2 in File S1). The frequencies of IL28B TT, KIR2DL2-HLA-C1-negative, with and without KIR3DL1-HLA-Bw4 were significantly higher among responders (38% [21/56] vs. 19% [11/59]; P = 0.024 [Pc = 0.29]; OR = 2.62, 95% CI = 1.12 - 6.12 and 30% [17/56] vs. 12% [7/59]; P = 0.015 [Pc = 0.18]; OR = 3.24, 95% CI = 1.22 - 8.57, respectively).
Discussion
The present study examined HLA, KIR, and IL28B gene variant associations with an SVR following PEG-IFN and ribavirin therapy in Japanese patients with chronic hepatitis C. We found a significant association of HLA-Bw alleles with treatment outcome, although the frequency of HLA-C alleles did not differ significantly between responders and non-responders. Functional analyses have demonstrated that NK cells in HLA-C1C1 subjects exhibit a more rapid and stronger antiviral response that those in HLA-C2C2 subjects due to differing responses of HLA-C-inhibited NK subsets[33]. HLA-C2C2 homozygousity is strongly associated with treatment failure in HCV patients of European ancestry [11,22], but we could not assess its role in our study because this genotype was found in only 1 of 115 patients.
We uncovered a significant association between the presence of KIR2DL2 or KIR2DS2 and lower SVR rates. Several reports have shown that KIR2DL3-HLAC1 in Caucasians [11,22] and KIR2DL5 in Brazilians [34] are associated with treatment outcome of antiviral therapy. Since our results showed no such statistical significances, these conflicting interpretations may reflect differences in patient selection, genetic background, sample size, and/or treatment regimen. Further studies are required to clarify this discrepancy in the Japanese population.
A study by Dring et al. examined KIR haplotypes in patients with HCV infection and showed that a centromeric KIR haplotype was increased in chronic HCV infection as compared with resolved cases [20]. We therefore determined KIR haplotypes and Cen-A/B and Tel-A/B in our patients as well, and found an interesting association between Cen-A/A and an SVR to antiviral therapy (P = 0.015; OR 3.37). Since Cen-A/B is determined by KIR2DL3 and KIR2DS2 and/or KIR2DL2, this finding is consistent with our results demonstrating a relationship between KIR2DS2 and KIR2DL2 genotypes and treatment failure.
The most significant finding in this study was the association between KIR-HLA receptor-ligand pairings and treatment outcome in chronic hepatitis C. Among the inhibitory KIR-HLA receptor-ligand pairs, patients with KIR3DL1-HLA-Bw4 exhibited a significantly higher SVR rate when compared to those without this pair (P = 0.03; OR 2.29). Conversely, virologic clearance in patients with KIR2DL2-HLA-C1 was significantly lower than in those without (P = 0.015; OR = 0.30). Stratification analysis of the 4 groups of KIR3DL1-HLA-Bw4 (presence or absence) and KIR2DL2-HLA-C1 (presence or absence) revealed a higher frequency of responders with KIR3DL1-HLA-Bw4 presence, KIR2DL2-HLA-C1 absence compared with those possessing KIR2DL2-HLA-C1 presence, KIR3DL1-HLA-Bw4 absence (62% vs. 9%; P = 0.0044; OR =16.32). When these KIR-HLA pairs were both either positive or negative, SVR rates were similar at 42% and 45%, respectively. Together with the results of logistic regression analysis, we clearly showed that KIR3DL1-HLA-Bw4 was positively associated with an SVR (OR = 3.32) and that KIR2DL2-HLA-C1 had a negative association (OR = 0.24) with treatment outcome. As almost one half of the Japanese population have the functional KIR3DL1-HLA-Bw4 combination, this inhibitory receptor-ligand interaction is potentially important in understanding NK cell diversification. The NK-cell surface expression of KIR3DL1 is higher in individuals having Bw4 than in those lacking it [35]. Therefore, these cells might be more weakly controlled by inhibitory signals than other NK cells, more easily activated by viral infection, and more readily promoted for cytolysis and IFN-gamma production.
This study confirmed that the IL28B TT genotype is a strong predictor of an SVR in Japanese patients[18,32]. Furthermore, SVR frequencies were positively correlated with a combination of the IL28B TT genotype and KIR3DL1-HLA-Bw4 (P = 0.0019) and negatively associated with the IL28B TT genotype and KIR2DL2-HLA-C1 (P = 0.0067). These combinations were also highly specific for virologic response prediction. In light of these findings, patients with poor expected treatment outcome may be advised to wait for the use of combinations of direct acting antiviral agents[36]. Akuta et al. reported that a combination of amino acid substitutions in the core region of HCV and IL28B genotype was a useful predictor of PEG-IFN, ribavirin, and telaprevir therapy results in Japan[37]. Since we could not collect sera before treatment for all patients, we were not able to assess the effect of amino acid substitutions in the HCV core region. Furthermore, interferon-free combinations of direct-acting antiviral agents have become an area of considerable clinical interest. Chu et al. have reported that IL28B genotype appears to affect early viral kinetics in patients with chronic hepatitis C receiving interferon-free treatment [38]. Recently, two groups have discovered IFN lambda 4 (IFNL4), a new gene that may account for associations of spontaneous and IFN-based treatment clearance of HCV [39,40]. The IFN-λ 4 protein is generated by individuals who carry the ∆G allele of the ss469415590 variant, and the presence of this protein is strongly associated with impaired clearance of HCV. Linkage disequilibrium is strong between the IFNL4-∆G allele and the unfavorable rs12979860-T allele (IL28B) in subjects of European or Asian ancestry, whereas this linkage disequilibrium is moderate in individuals of African ancestry [39]. We have confirmed that the linkage disequilibrium between the IFNL4-∆G allele and IL28B SNP (rs8099917) is high and that the IFNL4-∆G allele is strongly associated with treatment failure of PEG-IFN and ribavirin therapy in patients with Japanese chronic hepatitis C [41]. Hence, the clinical impacts of HLA-KIR genetic variants, IL28B genotype, and the IFNL4 allele should be explored.
In conclusion, the present study showed significant associations of KIR3DL1-HLA-Bw4, KIR2DL2-HLA-C1, and IL28B combinations with an SVR to PEG-IFN and ribavirin therapy in Japanese patients with genotype 1 HCV. The clinical significance of IL28B genotyping combined with HLA/KIR pairs to predict treatment outcome warrants further validation for triple therapy.
Supporting Information
Table S1, Sensitivity, specificity, and predictive values of IL28B TT genotype and KIR3DL1/HLA-Bw4 or KIR2DL2/HLA-C1 for a sustained virological response in 115 patients with chronic hepatitis C. Data are expressed as % (n). PPV, positive predictive value; NPV, negative predictive value. Table S2, Frequency of IL28B genotype and KIR3DL1/HLA-Bw4 and KIR2DL2/HLA-C1 combinations in 56 patients with a sustained virological response (SVR) and 59 patients with a non-SVR to pegylated interferon and ribavirin therapy of chronic hepatitis C. Data are expressed as n (%).
(DOC)
Acknowledgments
The authors thank Yuki Akahane, Asami Yamazaki, and Toyo Amaki for their technical assistance, and Trevor Ralph for his editorial assistance.
Funding Statement
This work was supported by a grant from the Ministry of Health, Labor, and Welfare of Japan. Takeji Umemura and Eiji Tanaka report receiving grant support from MSD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kiyosawa K, Sodeyama T, Tanaka E, Gibo Y, Yoshizawa K et al. (1990) Interrelationship of blood transfusion, non-A, non-B hepatitis and hepatocellular carcinoma: analysis by detection of antibody to hepatitis C virus. Hepatology 12: 671-675. doi: 10.1002/hep.1840120409. PubMed: 2170265. [DOI] [PubMed] [Google Scholar]
- 2. Umemura T, Ichijo T, Yoshizawa K, Tanaka E, Kiyosawa K (2009) Epidemiology of hepatocellular carcinoma in Japan. J Gastroenterol 44 Suppl 19: 102-107. doi: 10.1007/s00535-008-2251-0. PubMed: 19148802. [DOI] [PubMed] [Google Scholar]
- 3. Moretta A, Bottino C, Vitale M, Pende D, Cantoni C et al. (2001) Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol 19: 197-223. doi: 10.1146/annurev.immunol.19.1.197. PubMed: 11244035. [DOI] [PubMed] [Google Scholar]
- 4. Lanier LL (2005) NK cell recognition. Annu Rev Immunol 23: 225-274. doi: 10.1146/annurev.immunol.23.021704.115526. PubMed: 15771571. [DOI] [PubMed] [Google Scholar]
- 5. Mandelboim O, Reyburn HT, Valés-Gómez M, Pazmany L, Colonna M et al. (1996) Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med 184: 913-922. doi: 10.1084/jem.184.3.913. PubMed: 9064351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barber LD, Percival L, Valiante NM, Chen L, Lee C et al. (1996) The inter-locus recombinant HLA-B*4601 has high selectivity in peptide binding and functions characteristic of HLA-C. J Exp Med 184: 735-740. doi: 10.1084/jem.184.2.735. PubMed: 8760827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cella M, Longo A, Ferrara GB, Strominger JL, Colonna M (1994) NK3-specific natural killer cells are selectively inhibited by Bw4-positive HLA alleles with isoleucine 80. J Exp Med 180: 1235-1242. doi: 10.1084/jem.180.4.1235. PubMed: 7931060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X et al. (2004) HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 305: 872-874. doi: 10.1126/science.1097670. PubMed: 15297676. [DOI] [PubMed] [Google Scholar]
- 9. Paladino N, Flores AC, Marcos CY, Fainboim H, Theiler G et al. (2007) Increased frequencies of activating natural killer receptors are associated with liver injury in individuals who do not eliminate hepatitis C virus. Tissue Antigens 69 Suppl 1: 109-111. doi: 10.1111/j.1399-0039.2006.762_7.x. PubMed: 17445180. [DOI] [PubMed] [Google Scholar]
- 10. Romero V, Azocar J, Zúñiga J, Clavijo OP, Terreros D et al. (2008) Interaction of NK inhibitory receptor genes with HLA-C and MHC class II alleles in Hepatitis C virus infection outcome. Mol Immunol 45: 2429-2436. doi: 10.1016/j.molimm.2008.01.002. PubMed: 18289678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Knapp S, Warshow U, Hegazy D, Brackenbury L, Guha IN et al. (2010) Consistent beneficial effects of killer cell immunoglobulin-like receptor 2DL3 and group 1 human leukocyte antigen-C following exposure to hepatitis C virus. Hepatology 51: 1168-1175. doi: 10.1002/hep.23477. PubMed: 20077564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seich Al Basatena NK, Macnamara A, Vine AM, Thio CL, Astemborski J et al. (2011) KIR2DL2 enhances protective and detrimental HLA class I-mediated immunity in chronic viral infection. PLoS Pathog 7: e1002270 PubMed: 22022261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. López-Vázquez A, Rodrigo L, Martínez-Borra J, Pérez R, Rodríguez M et al. (2005) Protective effect of the HLA-Bw4I80 epitope and the killer cell immunoglobulin-like receptor 3DS1 gene against the development of hepatocellular carcinoma in patients with hepatitis C virus infection. J Infect Dis 192: 162-165. doi: 10.1086/430351. PubMed: 15942906. [DOI] [PubMed] [Google Scholar]
- 14. Marangon AV, Silva GF, de Moraes CF, Grotto RM, Pardini MI et al. (2011) KIR genes and their human leukocyte antigen ligands in the progression to cirrhosis in patients with chronic hepatitis C. Hum Immunol 72: 1074-1078. doi: 10.1016/j.humimm.2011.08.017. PubMed: 21920398. [DOI] [PubMed] [Google Scholar]
- 15. Vidal-Castiñeira JR, López-Vázquez A, Díaz-Peña R, Alonso-Arias R, Martínez-Borra J et al. (2010) Effect of killer immunoglobulin-like receptors in the response to combined treatment in patients with chronic hepatitis C virus infection. J Virol 84: 475-481. doi: 10.1128/JVI.01285-09. PubMed: 19846535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV et al. (2009) Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461: 399-401. doi: 10.1038/nature08309. PubMed: 19684573. [DOI] [PubMed] [Google Scholar]
- 17. Suppiah V, Moldovan M, Ahlenstiel G, Berg T, Weltman M et al. (2009) IL28B is associated with response to chronic hepatitis C interferon-alpha and ribavirin therapy. Nat Genet 41: 1100-1104. doi: 10.1038/ng.447. PubMed: 19749758. [DOI] [PubMed] [Google Scholar]
- 18. Tanaka Y, Nishida N, Sugiyama M, Kurosaki M, Matsuura K et al. (2009) Genome-wide association of IL28B with response to pegylated interferon-alpha and ribavirin therapy for chronic hepatitis C. Nat Genet 41: 1105-1109. doi: 10.1038/ng.449. PubMed: 19749757. [DOI] [PubMed] [Google Scholar]
- 19. Thomas DL, Thio CL, Martin MP, Qi Y, Ge D et al. (2009) Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461: 798-801. doi: 10.1038/nature08463. PubMed: 19759533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dring MM, Morrison MH, McSharry BP, Guinan KJ, Hagan R et al. (2011) Innate immune genes synergize to predict increased risk of chronic disease in hepatitis C virus infection. Proc Natl Acad Sci U S A 108: 5736-5741. doi: 10.1073/pnas.1016358108. PubMed: 21402922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knapp S, Warshow U, Ho KM, Hegazy D, Little AM, et al. (2011) A polymorphism in IL28B distinguishes exposed, uninfected individuals from spontaneous resolvers of HCV infection. Gastroenterology 141: 320-325, e321-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suppiah V, Gaudieri S, Armstrong NJ, O'Connor KS, Berg T et al. (2011) IL28B, HLA-C, and KIR variants additively predict response to therapy in chronic hepatitis C virus infection in a European Cohort: a cross-sectional study. PLOS Med 8: e1001092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Umemura T, Wang RY, Schechterly C, Shih JW, Kiyosawa K et al. (2006) Quantitative analysis of anti-hepatitis C virus antibody-secreting B cells in patients with chronic hepatitis C. Hepatology 43: 91-99. doi: 10.1002/hep.20917. PubMed: 16323211. [DOI] [PubMed] [Google Scholar]
- 24. Umemura T, Zen Y, Hamano H, Kawa S, Nakanuma Y et al. (2007) Immunoglobin G4-hepatopathy: association of immunoglobin G4-bearing plasma cells in liver with autoimmune pancreatitis. Hepatology 46: 463-471. doi: 10.1002/hep.21700. PubMed: 17634963. [DOI] [PubMed] [Google Scholar]
- 25. Wai CT, Greenson JK, Fontana RJ, Kalbfleisch JD, Marrero JA et al. (2003) A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology 38: 518-526. doi: 10.1016/S0270-9139(03)80785-1. PubMed: 12883497. [DOI] [PubMed] [Google Scholar]
- 26. Yoneda S, Umemura T, Katsuyama Y, Kamijo A, Joshita S et al. (2011) Association of serum cytokine levels with treatment response to pegylated interferon and ribavirin therapy in genotype 1 chronic hepatitis C patients. J Infect Dis 203: 1087-1095. doi: 10.1093/infdis/jiq165. PubMed: 21398397. [DOI] [PubMed] [Google Scholar]
- 27. Umemura T, Joshita S, Ichijo T, Yoshizawa K, Katsuyama Y et al. (2012) Human leukocyte antigen class II molecules confer both susceptibility and progression in Japanese patients with primary biliary cirrhosis. Hepatology 55: 506-511. doi: 10.1002/hep.24705. PubMed: 21953406. [DOI] [PubMed] [Google Scholar]
- 28. Cooley S, Trachtenberg E, Bergemann TL, Saeteurn K, Klein J et al. (2009) Donors with group B KIR haplotypes improve relapse-free survival after unrelated hematopoietic cell transplantation for acute myelogenous leukemia. Blood 113: 726-732. doi: 10.1182/blood-2008-07-171926. PubMed: 18945962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yawata M, Yawata N, Abi-Rached L, Parham P (2002) Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit Rev Immunol 22: 463-482. PubMed: 12803322. [PubMed] [Google Scholar]
- 30. Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L et al. (2010) Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLOS ONE 5: e15115. doi: 10.1371/journal.pone.0015115. PubMed: 21206914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cooley S, Weisdorf DJ, Guethlein LA, Klein JP, Wang T et al. (2010) Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 116: 2411-2419. doi: 10.1182/blood-2010-05-283051. PubMed: 20581313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Umemura T, Joshita S, Yoneda S, Katsuyama Y, Ichijo T et al. (2011) Serum interleukin (IL)-10 and IL-12 levels and IL28B gene polymorphisms: pretreatment prediction of treatment failure in chronic hepatitis C. Antivir Ther 16: 1073-1080. doi: 10.3851/IMP1869. PubMed: 22024523. [DOI] [PubMed] [Google Scholar]
- 33. Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B (2008) Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest 118: 1017-1026. PubMed: 18246204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carneiro VL, Lemaire DC, Bendicho MT, Souza SL, Cavalcante LN et al. (2010) Natural killer cell receptor and HLA-C gene polymorphisms among patients with hepatitis C: a comparison between sustained virological responders and non-responders. Liver Int 30: 567-573. doi: 10.1111/j.1478-3231.2010.02212.x. PubMed: 20456039. [DOI] [PubMed] [Google Scholar]
- 35. Yawata M, Yawata N, Draghi M, Little AM, Partheniou F et al. (2006) Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med 203: 633-645. doi: 10.1084/jem.20051884. PubMed: 16533882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chayama K, Takahashi S, Toyota J, Karino Y, Ikeda K et al. (2012) Dual therapy with the nonstructural protein 5A inhibitor, daclatasvir, and the nonstructural protein 3 protease inhibitor, asunaprevir, in hepatitis C virus genotype 1b-infected null responders. Hepatology 55: 742-748. doi: 10.1002/hep.24724. PubMed: 21987462. [DOI] [PubMed] [Google Scholar]
- 37. Akuta N, Suzuki F, Hirakawa M, Kawamura Y, Yatsuji H et al. (2010) Amino acid substitution in hepatitis C virus core region and genetic variation near the interleukin 28B gene predict viral response to telaprevir with peginterferon and ribavirin. Hepatology 52: 421-429. doi: 10.1016/S0168-8278(10)61091-4. PubMed: 20648473. [DOI] [PubMed] [Google Scholar]
- 38. Chu TW, Kulkarni R, Gane EJ, Roberts SK, Stedman C et al. (2012) Effect of IL28B genotype on early viral kinetics during interferon-free treatment of patients with chronic hepatitis C. Gastroenterology 142: 790-795. doi: 10.1053/j.gastro.2011.12.057. PubMed: 22248659. [DOI] [PubMed] [Google Scholar]
- 39. Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H et al. (2013) A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat Genet 45: 164-171. doi: 10.1038/ng.2521. PubMed: 23291588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bibert S, Roger T, Calandra T, Bochud M, Cerny A et al. (2013) IL28B expression depends on a novel TT/-G polymorphism which improves HCV clearance prediction. J Exp Med 210: 1109-1116. doi: 10.1084/jem.20130012. PubMed: 23712427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nozawa Y, Umemura T, Katsuyama Y, Shibata S, Kimura T et al. (2013) Genetic polymorphism in IFNL4 and response to Peg-Interferon-α and ribavirin in Japanese chronic hepatitis C patients. Tissue Antigens (in press). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Sensitivity, specificity, and predictive values of IL28B TT genotype and KIR3DL1/HLA-Bw4 or KIR2DL2/HLA-C1 for a sustained virological response in 115 patients with chronic hepatitis C. Data are expressed as % (n). PPV, positive predictive value; NPV, negative predictive value. Table S2, Frequency of IL28B genotype and KIR3DL1/HLA-Bw4 and KIR2DL2/HLA-C1 combinations in 56 patients with a sustained virological response (SVR) and 59 patients with a non-SVR to pegylated interferon and ribavirin therapy of chronic hepatitis C. Data are expressed as n (%).
(DOC)