Background: Phytochromes are red/far-red photoreceptors using a bilin chromophore.
Results: Compared with Cph1, the Cph2 bilin-binding site differs around the propionates, but utilizes an otherwise conserved tongue for sealing the chromophore from solvent.
Conclusion: The tongue signals via a tryptophan switch within the tongue-GAF domain interface.
Significance: The first structure of a Cph2-type phytochrome indicates a common mechanism for photoswitching in all canonical phytochromes.
Keywords: Cyanobacteria, Protein Conformation, Signal Transduction, Signaling, Structural Biology, Biliprotein, c-di-GMP Signaling, Photochromicity, Phytochrome, Red Light Photoreceptor
Abstract
Phytochromes are highly versatile photoreceptors, which occur ubiquitously in plants as well as in many light-responsive microorganisms. Here, photosynthetic cyanobacteria utilize up to three different phytochrome architectures, where only the plant-like and the single-domain cyanobacteriochromes are structurally characterized so far. Cph2 represents a third group in Synechocystis species and affects their capability of phototaxis by controlling c-di-GMP synthesis and degradation. The 2.6-Å crystal structure of its red/far-red responsive photosensory module in the Pr state reveals a tandem-GAF bidomain that lacks the figure-of-eight knot of the plant/cph1 subfamily. Its covalently attached phycocyanobilin chromophore adopts a highly tilted ZZZssa conformation with a novel set of interactions between its propionates and the GAF1 domain. The tongue-like protrusion from the GAF2 domain interacts with the GAF1-bound chromophore via its conserved PRXSF, WXE, and W(G/A)G motifs. Mutagenesis showed that the integrity of the tongue is indispensable for Pr → Pfr photoconversion and involves a swap of the motifs' tryptophans within the tongue-GAF1 interface. This “Trp switch” is supposed to be a crucial element for the photochromicity of all multidomain phytochromes.
Introduction
Phytochromes are red/far-red light absorbing photoreceptors first discovered in plants (1), and later in many photosynthetic and non-photosynthetic bacteria (2) as well as in several fungi (3). A hallmark of these photochromic light sensors is their bilin chromophore covalently bound to a cGMP phosphodiesterase/adenylyl cyclase/FhlA (GAF)2 domain. Upon red light illumination of its Pr state, this chromophore undergoes a Z → E isomerization around the C14–C15 double bond of the C-D ring methine bridge. In the ZZEssa conformation of the newly formed far-red-sensitive Pfr state the D-ring is flipped and the protein environment is altered to accommodate this rotation. These alterations ultimately control signal transfer either to effector proteins (inter-molecular (4)) or effector domains like histidine kinase domains (intra-molecular (5)). How these allosteric effects are triggered by D-ring rotation at the molecular level is still unknown thus prompting alternative scenarios for downstream signaling by phytochromes including an additional A-ring rotation upon Pr → Pfr photoconversion (6).
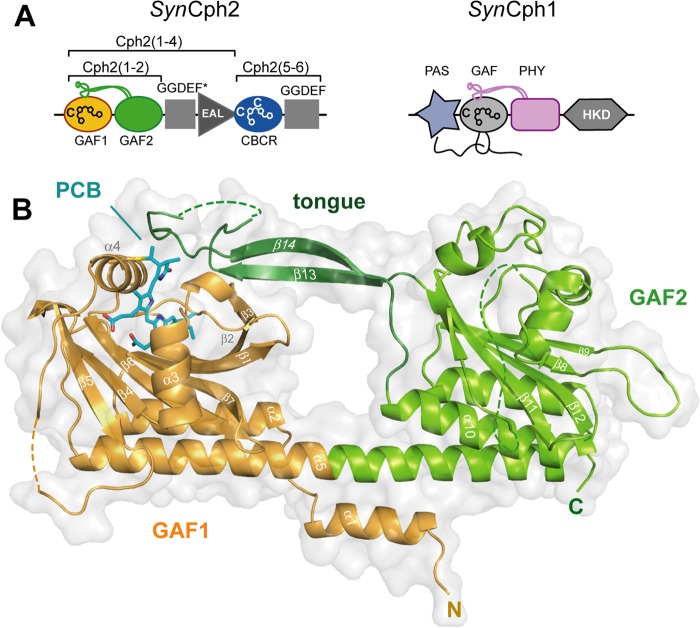
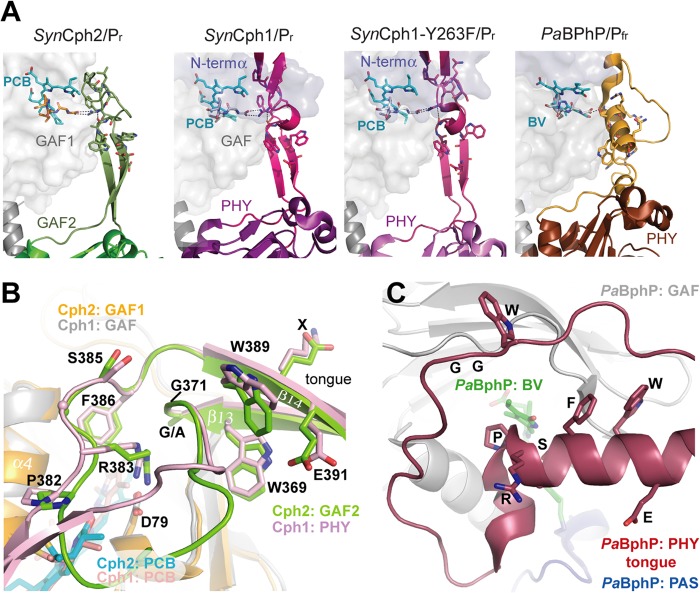
Phytochromes can be classified into subfamilies according to the domain architecture of their photosensory module. Group I includes canonical plant and eubacterial phytochromes such as Cph1 from the cyanobacterium Synechocystis sp. PCC 6803 (SynCph1 (7)) as well as fungal and eubacterial bacteriophytochromes. These phytochromes use a three-domain PAS-GAF-PHY (PAS, period/ARNT/single-minded; PHY, phytochrome) photosensory module, where an unusual figure-of-eight knot is formed by the N terminus of the PAS domain and a loop protruding from the chromophore-binding GAF domain (Fig. 1A). The succeeding PHY domain belongs also to the GAF superfamily and stabilizes the chromophore, by sealing it from solvent access, through its tongue-like extension (8, 9). Together, the GAF domain, N terminus, and tongue region form a complex, tripartite pocket enclosing the bilin chromophore. The Cph2 subfamily (Group II) lacks the PAS domain and utilizes instead only a tandem-GAF module as exemplified by the N-terminal photosensory module in Cph2 from Synechocystis sp. (SynCph2) (10) and several other cyanobacterial phytochromes (11). Finally, the cyanobacteriochromes (CBCRs, Group III) utilize a single, bilin-binding GAF domain (11, 12). CBCRs cover a broad range of the visible/near UV spectrum but are restricted to cyanobacteria. Whereas, the photosensory modules of several prokaryotic phytochromes from Groups I (8, 9) and III (13, 14) have been characterized structurally, little is known about Cph2 homologues in Group II.
FIGURE 1.
Structure of the SynCph2(1–2) photosensor. A, domain organization of SynCph1 and SynCph2. The tongue-like regions protruding from the GAF2 and PHY domains are shown in green and purple, respectively; the knot in SynCph1 as black lines. SynCph1 and SynCph2 differ in their effector domains: a histidine kinase domain (HKD) and c-di-GMP turnover (GGDEF and EAL) and regulatory (GGDEF*) domains. B, crystal structure of the SynCph2(1–2) module. GAF2 (green) contains a tongue-like region (dark green) for sealing the PCB (cyan)-binding pocket. The GAF1 domain is in orange.
Interestingly, the founding member of the Cph2 subfamily, SynCph2, is a bimodular photoreceptor (15, 16) (molecular mass 144.7 kDa) that functions as a light-dependent master regulator for cyanobacterial motility by controlling cytosolic levels of the eubacterial second messenger c-di-GMP (17). c-di-GMP is known to be a universal regulator for pili-based motility in eubacteria (18) with domains of the GGDEF and EAL type being responsible for its synthesis and degradation (19). SynCph2 inhibits phototaxis, when cyanobacterial cells are exposed to blue but not to white or red light (20, 21). Accordingly, the complex architecture of SynCph2 (Fig. 1A) harbors not only at its N terminus the red/far-red sensitive tandem-GAF module (SynCph2(1–2) (10)) that precedes a GGDEF*-EAL effector module, but also CBCR and catalytically active GGDEF domains. This C-terminal CBCR-GGDEF module is capable of switching between blue and green light sensitive states and controls the catalytic activity of the GGDEF domain, thereby inhibiting motility by increasing c-di-GMP levels (17). The GGDEF*-EAL module likely acts as c-di-GMP degrading phosphodiesterase, as its EAL domain is catalytically active, whereas the apparently inactive GGDEF* domain is predicted to act as an allosteric, c-di-GMP-dependent regulator.
Here, we report the crystal structure of the photosensory module SynCph2(1–2) in its Pr conformation. An antiparallel dimer is formed in which both the N-terminal PAS domain and the associated knot structures are missing. Other significant differences relative to known phytochrome structures are apparent in the phycocyanobilin (PCB) binding pocket. On the other hand, the interactions between the tongue region of GAF2 and the bilin-binding site of GAF1 are conserved, implying a universal route for signal transduction in Group I and II phytochromes that involves a toggling of conserved Trp motifs within the tongue/GAF1 interface.
EXPERIMENTAL PROCEDURES
Crystallization and Data Collection
Recombinant SynCph2(1–2) was produced as a histidine-tagged fusion with phycocyanobilin co-assembled in vivo. The expression and purification of SynCph2(1–2) was performed as previously described (10). Crystals for the native datasets were grown using the hanging-drop vapor-diffusion method. The drops contained 1 μl of 20 mg/ml of SynCph2(1–2)/Pr-state in crystallization buffer (10 mm HEPES, 100 mm NaCl, pH 8.0) and 1 μl of reservoir solution (0.1 m HEPES, pH 7.0, 1.0 m NH4COOH, 0.1% (w/v) lysine, 0.16% (w/v) arginine, 0.05% (w/v) glutamate). Crystals were streak-seeded with a solution of crushed SynCph2(1–2) crystals. After pipetting the pre-irradiated (far-red light, λmax = 735 nm) protein solution under blue-safe light (λmax = 482 nm) conditions the plate was irradiated with far-red light to ensure 100% occupancy of the Pr state. The crystals grew at 18 °C in the dark, were visualized under blue light, and frozen in reservoir buffer supplemented with 20% (v/v) glycerol. Selenomethionine-labeled SynCph2(1–2) crystals appeared with streak seeding using a reservoir solution of 0.1 m HEPES, pH 6.5, 0.5 m NH4COOH and an initial protein concentration of 10 mg/ml.
Native datasets were recorded at beamline ID14–1 (European Synchrotron Radiation Facility (ESRF), Grenoble, France) and at beamline 14.1 (Berliner Elektronenspeicherring-Gesellschaft für Synchrotronstrahlung (BESSY-II), Berlin, Germany). The 3.9-Å SAD dataset from the selenomethionine-labeled crystal was recorded at ID 14-4 (ESRF, Grenoble, France).
Initial SAD phasing was performed with PHENIX (22) using the native 3.20-Å dataset. Subsequent structural refinement with a 2.60-Å dataset was executed with PHENIX and COOT (23). Stereochemical restraints were configured for the PCB chromophore and its covalent attachment. Figures were created by using PYMOL 0.99 (24).
Single-crystal Resonance Raman Spectroscopy
Resonance Raman (RR) spectra were recorded offline at the Cryobench (European Synchrotron Radiation Facility (ESRF), Grenoble (France)) with a 785 nm near-IR laser using an inVia (Renishaw) Raman spectrometer (4 cm−1 spectral resolution) (25). Resonance Raman spectra of the crystal (1 m MgOAc2 as cryo-protectant) were collected under cryo-conditions (100 K) in the 200–2000 cm−1 range using the SynchroScan method with a laser power of 300 milliwatts (50 milliwatts at the sample position) and 20-s exposure time. 30–50 spectra were accumulated and baseline corrected to remove contributions from fluorescence and normalized to the 1642 cm−1 peak. Resonance Raman spectra were recorded before x-ray exposure, and then the crystal was irradiated like during data collection (ID 14–1, 100% transmission, 4 s exposure time per image, 350 images, 0.5° oscillation range; flux: ∼4.8 × 1010 photons/s; the crystal size was ∼30% of beam size so that a dose of ∼1.4 × 1010 photons/s reached the crystal). The crystal was characterized spectroscopically and then annealed by short thawing for 4 s in the dark, whereupon another Raman spectrum was taken. The crystal remained under cryo-conditions during the whole data collection/spectra recording process.
Single-crystal UV-visible Absorption Spectroscopy
UV-visible spectra of the crystal were measured online at 100 K with a HR2000 spectrometer (OceanOptics) and a DH-2000-BAL light source (Mikropack) at beamline ID 14-1 (ESRF, Grenoble (26)). Spectra were taken before x-ray irradiation, immediately after irradiation (60 s exposure; 100% transmission; flux: ∼4.2 × 1010 photons/s; crystal size ∼30% of x-ray beam diameter), after a 3-s cryo-annealing and subsequent freezing, and after a 60-s incubation under cryo-conditions following x-ray irradiation. Initial two exposure pictures were taken following a 3-s exposure per image with a 1.0° oscillation.
Quantum Mechanical/Molecular Mechanical (QM/MM) Calculations
The initial structural models were set up by adding missing hydrogen atoms and solvent water to the experimental x-ray structure. Two models were considered in this work: (a) the crystal structure of SynCph2(1–2) itself and (b) a modified structure of SynCph2(1–2) with a planar PCB chromophore as found in the structure of the SynCph1 photosensory module (8).
The insertion of hydrogen atoms was done with the HBUILD routine of CHARMM (31). All titratable amino acids were protonated according to pH 7. In the case of the histidines, the protonation states were chosen upon visual inspection of their environment. In particular, His130 and His160, lying in the vicinity of the chromophore, were modeled with a hydrogen atom at the ϵ-nitrogen and δ-nitrogen, respectively. In addition, based on spectroscopic evidence (27, 28) the PCB chromophore is nitrogen-protonated, whereas the propionic side chains are deprotonated yielding a total charge of −1e.
The geometry optimization of the PCB chromophore and binding site of Cph2 and Cph1 phytochromes, comprising all atoms within a 22-Å radius from the chromophore, were performed at the QM/MM level (29). Although the QM fragment, consisting of the PCB chromophore, the Cys129 side chain, and the pyrrole water that is bound to rings A, B, and C nitrogens, were described with the B3LYP density functional (30), the remaining protein atoms as well as solvation waters with the CHARMM22 force field (31). The coupling between the QM and MM regions was described with the electrostatic embedding model combined with a charge shifted scheme (32), as implemented in the ChemShell software. The covalent bond cut at the QM/MM border on the Cys129 was saturated with a hydrogen link atom. The optimization was performed using a limited memory quasi-Newton L-BFGS algorithm working with hybrid delocalized internal coordinates (33).
Production and Spectral Characterization of SynCph2(1–2) Variants
SynCph2(1–2) mutants were produced by QuikChange site-directed mutagenesis (Stratagene) of the pCDFDuet-1/SynCph2(1–2). They were co-transformed into BL21 Gold (DE3) (Novagen) together with the p171 vector that includes genes for PCB production (34). The recombinant overproduction and purification of the proteins were performed as previously described (10).
Orthologous swapping variants of SynCph2(1–2) in the tongue and propionate region were produced by adding additional restriction sites into pCDFDuet-1/SynCph2(1–2). Thus the stretches Trp369-Glu391 in the tongue variants and Val97-Val126 in the propionate variants, respectively, were replaced by regions of Cph2-like phytochromes from Nostoc punctiforme PCC 73102 (YP_001868577.1), Oscillatoria sp. PCC 6506 (WP_007353933.1), and Cyanothece sp. PCC 7822 (YP_003886347.1) using synthetic gene fragments cloned into pCDFDuet-1/SynCph2(1–2).
UV-visible absorbance spectra were measured with a Jasco V-660 spectrometer at 1-nm resolution at room temperature. In a cell of 1-cm path length the sample was irradiated for 30 s with red or far-red LED sources (B5–436-30D, λmax 664 nm and SMC735, λmax 735 nm; Roithner, Vienna). Difference spectra were calculated via A(Pr) − A(PPhotoequilibrium). Determined pure Pfr spectra of the wild type are discussed in Ref. 10.
Near UV-visible circular dichroism (CD) spectra were measured following irradiation of the sample with a J-810 spectropolarimeter (Jasco) at room temperature in a 1-mm path length cell. Pr spectra were averaged over three scans. Short-lived intermediate or Pfr states were averaged over six scans where the sample was irradiated in between.
RESULTS AND DISCUSSION
Overall Structure of the SynCph2 Photosensor
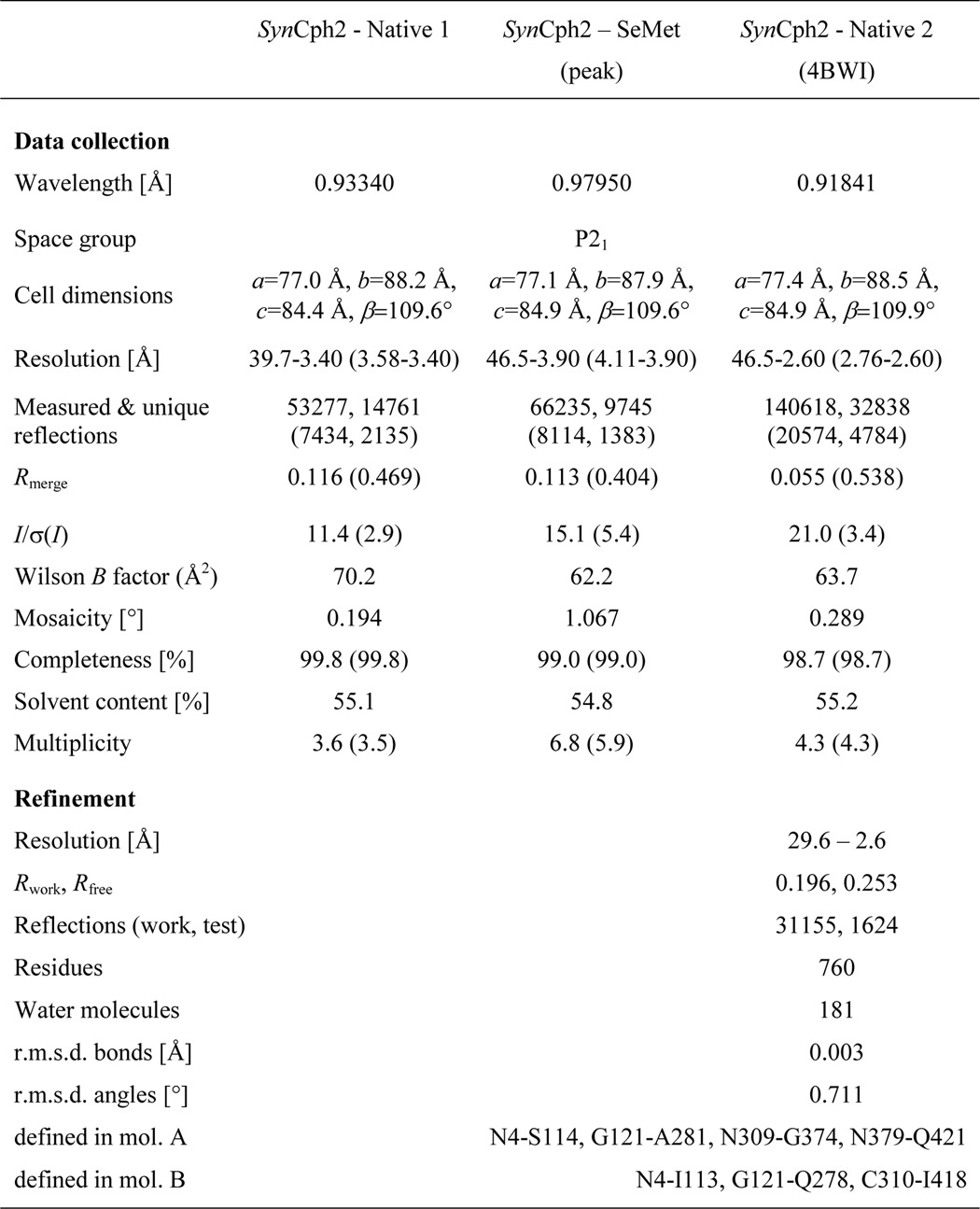
The N-terminal photosensory module of SynCph2 (M1-T424, (10)) in its Pr state crystallizes as an antiparallel dimer whose structure was solved at 2.6-Å resolution by a combination of molecular replacement and single-wavelength anomalous diffraction (SAD)-phasing (final Rfactor/Rfree, 19.6/25.3%, see Table 1).
TABLE I.
Data collection and refinement statistics
Values in parentheses denote the highest resolution shell.
The SynCph2(1–2) photosensor shows the characteristic bilobal shape of a tandem-GAF module with the N-terminal, PCB-binding GAF domain (Thr23-Thr187; GAF1) being linked to the second GAF domain (Lys197-Gln420; GAF2) via a 71-Å long α-helix (Thr168-Tyr215). The GAF domains of SynCph2(1–2) hence mimic the overall organization of canonical photosensory GAF-PHY-modules of phytochromes, including the tongue-like protrusion (Glu362-Asn400) that juts of the GAF2 domain and contacts the chromophore-binding pocket of the GAF1 domain (Fig. 1B).
The GAF1 domain with its buried PCB chromophore resembles the bilin-binding domains of canonical phytochromes such as SynCph1 and RpBphP3 (root mean square deviation (r.m.s.) deviations of 0.98 and 1.02 Å for 175 and 164 Cα atoms, respectively). In contrast, the core of the GAF2 domain diverges significantly from known PHY domains yet remains more closely related to PHY domains from biliverdin-dependent bathy phytochromes such as RpBphP3 or PaBPhP (2.26 and 2.82 Å for 117 and 135 Cα atoms, respectively) than to PCB-dependent phytochromes like Cph1 (3.40 Å for 150 Cα atoms). Structural differences are mostly around the five-pleaded central β-sheet, including the elongated loops connecting β-strands 8–9 and 11–12 as well as the α-helices covering the β-sheet opposite the α-helix linking GAF1 and GAF2 (Fig. 1B). Furthermore, in both dimer subunits an additional stretch of 27 amino acids (Ala282-Thr308) is structurally disordered as indicated by missing electron density. Interestingly, this stretch is unique for Cph2 as it is absent from PHY domains of Group I phytochromes.
Crystals of SynCph2 are cyan-colored due to the incorporated PCB cofactor and give the same UV-visible and resonance Raman spectra as in solution. Accordingly, this proves that SynCph2(1–2) adopts a native-like conformation for its bilin chromophore in the crystalline state (Fig. 2) (35).
FIGURE 2.
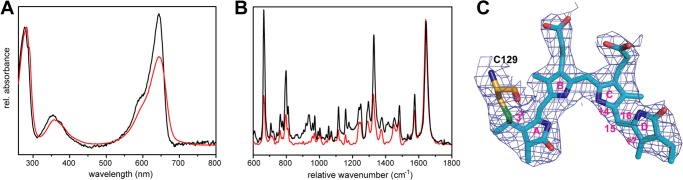
Spectral characteristics of SynCph2(1–2) in soluble and crystallized forms. A, UV-visible absorbance spectra of SynCph2(1–2) in solution at room temperature (red curve) and a crystal (black) at 100 K in the Pr conformation. B, Raman spectra of the protein in frozen solution (red curve) at 100 K and in crystalline form (black) at 100 K. C, omit electron density of the PCB chromophore and its covalent attachment to Cys129 (contouring level 1.0 σ).
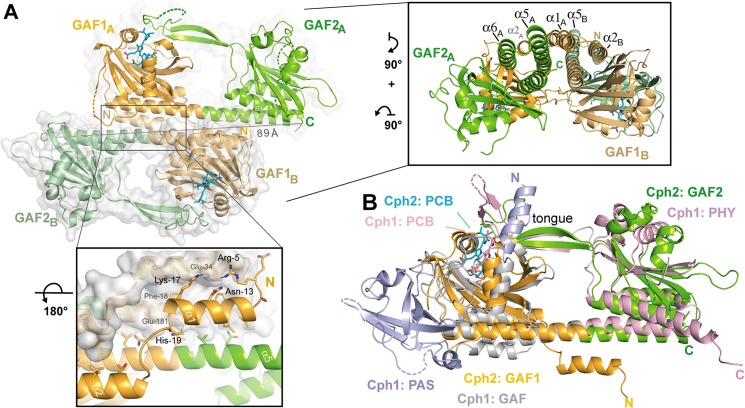
Despite its monomeric state in solution (10) SynCph2(1–2) forms an antiparallel dimer in crystals similar to the photosensory module of SynCph1 (Fig. 3). Its interface area is predicted by the PISA server to be ∼1904 Å2 and thus considerably smaller than that of SynCph1 (2545 Å2) (8) due to the missing PAS domain. Its staggered and antiparallel orientation differs from other known phytochrome structures such as PaBPhP, in which a parallel association is observed over the whole length of the GAF-PHY linker helices. The latter mode of association is thought to be crucial for signal transduction to histidine kinase domains (9, 36) and illustrated by the cryoEM structure of full-length bacteriophytochrome DrBphP, where the photosensory module provides the largest dimerization interface. Like in parallel dimers of the photosensory phytochrome modules, the antiparallel arrangement of SynCph2(1–2) is mainly stabilized by association of its linker α5-helices between GAF1 and GAF2 (Figs. 1B and 3A). Interestingly, the α5-helices resemble those of the Y263F SynCph1 mutant (37), whereas in the wild type SynCph1 (Fig. 3B) and PaBphP structures these helices are significantly kinked. Some structural plasticity between the GAF and PHY domains as observed for PaBphP and SynCph1 (8, 9, 37) may be involved in long-distance signal transmission. Accordingly, the GAF2 domains of the two copies of SynCph2(1–2) in the crystal structure are off-rotated by 6.4° relative to each other, whereas the tongue regions are almost invariantly associated with the GAF1 domains.
FIGURE 3.
Quaternary structure of the knotless SynCph2(1–2) photosensor and comparison to SynCph1. A, the antiparallel dimer of SynCph2(1–2). The distance between C-terminal residues His421 (molecule A) and Ile418 (molecule B) is 89 Å; lower inlet, the N-terminal helix forms part of the dimer interface; inlet on the right side, perpendicular view on the SynCph2(1–2) dimer. The interface between the monomers is built of a helix bundle composed of the linker α-helix and shorter helices, especially the N-terminal helix. B, structural superposition of SynCph2 and SynCph1 (r.m.s. deviation 2.64 Å for 249 Cα atoms); the PAS, GAF, and PHY domains of SynCph1 are depicted in blue, gray, and pale red, respectively.
Another striking feature of SynCph2(1–2) is its missing figure-of-eight knot, a hallmark of Group I phytochromes. In SynCph1 the N-terminal helix threads through the loop region connecting β9 with α8 and is part of the PCB binding pocket. The Group II SynCph2(1–2) phytochrome lacks this knot along with the associated PAS domain, so that the binding pocket is partly solvent-exposed at the A- and B-rings of the PCB chromophore (Fig. 3B). Instead, the N-terminal α1-helix of SynCph2(1–2) is part of a six-helix bundle comprising α1, α5, α6, and α10 of one monomer and α2 and α5 of the other monomer (Fig. 3A). In this interface, the amphiphilic α1-helix makes first several polar interactions via its residues Arg5, Lys17, and His19 with Glu34 and Glu181 from α2 and α5, respectively. Second, for hydrophobic interactions the α1-helix packs its residues Leu7, Phe10, Leu11, Val14, and Phe18 against a surface patch generated by Val30, Ile31, Phe38, Leu177, and Leu178 of α2 and α5. Accordingly, one may hence postulate that the dimerization of PAS-less phytochromes like SynCph2(1–2) depends on their N-terminal helix.
The different output domains, especially the GGDEF*–EAL module downstream of the SynCph2(1–2) sensor, imply that the antiparallel association of the photosensory domains could be functionally relevant in SynCph2 and not just a crystallization artifact as found for SynCph1 (8). For example, FimX from Pseudomonas aeruginosa, a twitching motility regulator, also carries GGDEF-EAL domains. Here, the overall quaternary organization corresponds to an elongated antiparallel dimer, where the GGDEF-EAL modules are not involved in dimerization (38). Such an arrangement of the photosensory SynCph2(1–2) module may be of physiological relevance for signaling toward the GGDEF*–EAL module. This contrasts to eubacterial phytochromes such as SynCph1, which are part of two-component signaling systems. Only these phytochromes have to form parallel homodimers (36) similar to the structurally characterized sensor histidine kinases YF1 and VicK (39, 40) to allow proper dimerization and thereby catalytic activity of their histidine kinase domain-like regions.
The PCB-binding Site of SynCph2(1–2)
Overall, the bilin chromophore is well defined in the GAF1 domain including its thioether linkage to Cys129 (Fig. 2C). Despite structural differences in the protein environment relative to other Group I phytochromes, mainly around pyrrole rings A and B, the conformation of the chromophore resembles SynCph1. First, the new chiral center at C31, as formed by covalent attachment to Cys129, has an R configuration. This may not be a general feature for bilin chromophores covalently tethered to GAF domains; for example, in the structure of the Y263F mutant of SynCph1 both stereochemical configurations have been found for the C31–Cys thioether linkage (37). Second, and as expected, for the SynCph2(1–2) Pr state, the chromophore adopts the ZZZssa conformation (Fig. 2C, supplemental Table S1), but in a strongly non-planar conformation, when compared with other phytochrome structures harboring bilin chromophores.
Despite the latter, the local environment of the D-ring is well conserved between SynCph2 and SynCph1 as exemplified by the residues Tyr47, Val57, Asp79, Tyr133, Leu134, Met137, and His160. Differences here are mainly found for Lys45 or Leu70, but appear to only play a minor role in the Pr → Pfr conversion, as the SynCph2 K45M mutant shows only a ∼3 nm hypsochromic shift in its Pfr state. Other residues of the C- and D-ring binding site like Tyr133 are functionally similar to their structural counterparts in SynCph1 (Tyr263) as indicated by the similar photochemistry of their mutants, for example, Y133F or H160A (supplemental Fig. S1 and Table S2) (37).
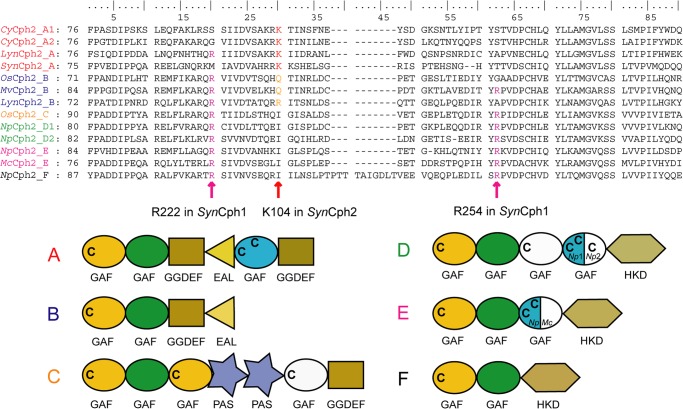
The loop connecting β5 and α4 in SynCph2, which is otherwise part of the knot region of Group I phytochromes, is partly disordered due to a lack of further interactions. Furthermore, the SynCph2 loop appears to interact with the B-ring propionate differently than in SynCph1. SynCph2 forms an additional β-strand (β5, Arg102-Glu108), of which Lys104 makes a surface-exposed salt bridge with the B-ring propionate (Fig. 4A). In SynCph1 as well as other Group I phytochromes this propionate interacts with Arg254 (loop region) in Pr, but probably swaps partners upon photoconversion to interact with Arg222, part of β-strand 9 (41) (Fig. 4B). Both of these arginines are missing in SynCph2. Here, Lys104 apparently substitutes for Arg254 in SynCph1. However, an equivalent interaction partner for the B-ring propionate in the Pfr state seems to be missing. Thus the propionate signaling route envisaged for Group I phytochromes (8) is unlikely for SynCph2. Multiple sequence alignments of Group II phytochromes (Fig. 5) show that Lys104 is conserved in other cyanobacterial phytochromes with the same complex domain architecture of SynCph2. In Group II phytochromes with different architectures, for example, GAF-GAF-GGDEF*-EAL (17) or GAF-GAF-histidine kinase domain, this residue is replaced by polar or isoleucine residues. Interestingly, Cph2 orthologues with downstream histidine kinase effector domains harbor both SynCph1 arginines (Arg222 and Arg254). It seems, therefore, that the propionate interactions may vary according to the route required for signal transmission to the effector domains.
FIGURE 4.
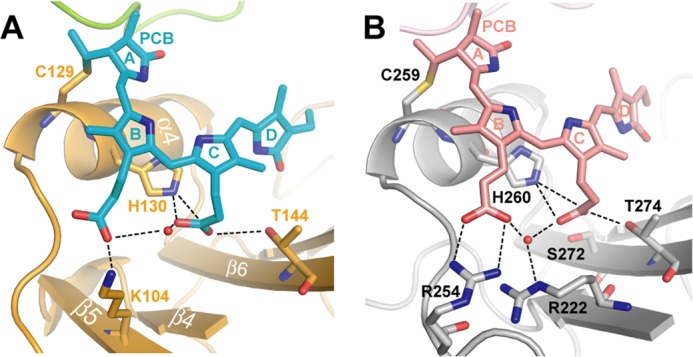
Close-up view of the chromophore binding sites of the SynCph2(1–2) (A) and SynCph1 (B) photosensor. The PCB chromophore within SynCph2(1–2) is structurally distorted compared with the almost planar A-, B-, and C-rings of SynCph1. The hydrogen bond network of the B- and C-ring propionates is indicated with dashed lines.
FIGURE 5.
Multiple sequence alignment of Cph2-like Group II phytochromes. GAF1 sequences forming the knot/propionate binding site region. Abbreviations: SynCph2_A (organism: Synechocystis sp. PCC 6803; GenBank entry NP_442466.1; sequence identity: 100%), CyCph2_A1 (Cyanothece sp. PCC 7424; YP_002378924.1; 38%), CyCph2_A2 (Cyanothece sp. PCC 7822; YP_003886347.1; 39%), LynCph2_A (Lyngbya sp. PCC 8106; WP_009783799.1; 33%), OsCph2_B (Oscillatoria sp. PCC 6506; WP_007353933.1; 36%), MvCph2_B (Microcoleus vaginatus FGP-2; WP_006623727.1; 34%), LynCph2_B (Lyngbya sp. PCC 8106; WP_009783371.1; 33%), OsCph2_C (Oscillatoria sp. PCC 6506; WP_007357482.1; 30%), NpCph2_D1 (N. punctiforme PCC 73102; YP_001870049.1; 31%), NpCph2_D2 (N. punctiforme PCC 73102; YP_001868068.1; 33%), NpCph2_E (N. punctiforme PCC 73102; YP_001868577.1; 34%), McCph2_E (Microcoleus chthonoplastes PCC 7420; WP_006100748.1; 31%), NpCph2_F (N. punctiforme PCC 73102; YP_001865364.1; 31%). Pairwise sequence homologies to SynCph2(1–2) were performed with the BLOSUM62 exchange matrix. Letters at the end of the abbreviation describe the domain architecture of the proteins described below: C indicates conserved cysteines; green, GAF domain of the “PHY”-type; blue, GAF domain of the “CBCR”-type; white, GAF domain with sequence similarities with “CBCR”-type but with one or no conserved cysteine. Mixed colors, gene-dependent domain organization as indicated by the organism abbreviation.
The unusual interactions of the B- and C-ring propionates are not crucial for Pr → Pfr photoconversion, however. Alanine mutants of Lys104 and the neighboring amino acids Arg103 and Lys105 show wild type Pr → Pfr photoconversion. Only a ∼3 nm hypsochromic shift is observed for the Pr and Pfr states of K104A. Even swapping variants of SynCph2(1–2), where the whole region around the B-ring propionate (Val97-Val126) is exchanged with regions of other phytochromes of the Cph2 subfamily, show almost normal Pfr (supplemental Fig. 1 and Table S2).
A study of the B- and C-ring propionate amide adducts in SynCph1 reveals that a free B-ring propionate is essential for wild type Pr and that full photoconversion to Pfr requires a conformational switch at the C-ring propionate (42). Apart from the missing arginines in SynCph2 B-ring propionate interactions, the C-ring propionate interactions are similar in SynCph1 and SynCph2 and involve residues Thr144 and His130, as well as a water molecule.
SynCph2(1–2) Induces the Tilted PCB Conformation
An unusual feature of the PCB chromophore compared with other phytochrome and even phycobilisome-PCB structures is the strong tilt between its constituent rings (see supplemental Table S1). The chromophore shows tilts of 19.2°, 32.5°, and 59.8° between rings A-B, B-C, and C-D, respectively (in comparison SynCph1: 9.8°, 1.5°, and 26.3°). The tilt between rings B-C and C-D is hence the largest among known phytochromes and PCB-binding phycobilisomes. Only non-conjugated chromophores in phycobiliproteins showed so far similar or higher tilts (see supplemental Table S1). Given earlier suggestions that the phytochrome chromophore conformation might generally be damaged by x-ray radiation (6), we validated the observed PCB conformation by a combination of single crystal UV-visible and resonance Raman spectroscopy with QM/MM geometry calculations based on structural models of SynCph2(1–2).
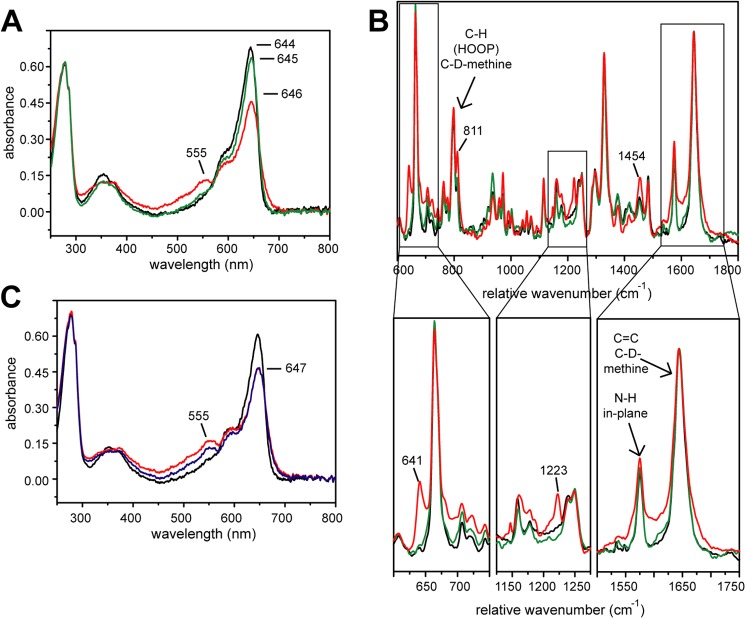
Upon x-ray irradiation of crystals the spectral signatures of the Pr state of SynCph2(1–2) are changed. First, in the UV-visible spectrum an additional shoulder at 555 nm appears, whereas the signal at 644 nm decreases and the maximum shifts to 646 nm (Fig. 6A) accompanied by a broadening of the 646 nm peak. Second, in single-crystal Raman spectra (Fig. 6B) x-ray exposure of SynCph2(1–2) crystals cause additional Raman signals at 641 cm−1 and 1223 cm−1, a signal increase at 811 cm−1 and 1454 cm−1, and a slight peak broadening between 1550 and 1700 cm−1. Only cryo-annealing restores the pre-x-ray UV-visible and resonance Raman spectra. Under cryogenic conditions, after x-ray exposure, a slow relaxation process takes place within minutes (Fig. 6C), where the shoulder at 555 nm in the UV-visible spectrum decreases but the signal at ∼644 nm remains unchanged. Again cryo-annealing allows complete reversal to the Pr state before x-ray irradiation.
FIGURE 6.
Temporary radiation effect on SynCph2(1–2). Black, no x-ray; red, after x-ray; green, after annealing; blue, 60 s after exposure. A, UV-visible absorbance spectra of a crystal before x-ray, after 60 s exposure (ID 14-1 (ESRF, Grenoble), 100% transmission; flux: ∼4.2 × 1010 photons/s (approximately ∼1.4 × 1010 photons/s reached the crystal)), and after 3 s of annealing in the dark (annealing process was not complete). B, Raman spectra of a crystal before x-ray, after 30 min x-ray irradiation (ID 14–1 (ESRF, Grenoble), 100% transmission, 4-s exposure time per image, 350 images, 0.5° oscillation range; flux: ∼4.8 × 1010 photons/s) and after 4 s of annealing in the dark; below: partial scale-up of the Raman spectra. C, UV-visible absorbance spectra of a crystal before x-ray, after exposition for 60 s, and 60 s after exposure.
These data are in line with previous observations on the photosensory module of SynCph1, where long x-ray radiation induced additional absorbance peaks in the UV-visible spectra (43). In SynCph2(1–2) the x-ray-induced species relaxes in at least two steps. At 100 K it decays to a further species without recovery of the Pr maximum (Fig. 6C). Apparently, the activation energy needed for conversion into native-like Pr cannot be overcome and the chromophore of SynCph2(1–2) remains hence arrested in an intermediate state. Further relaxation to Pr, which means recovery of the spectral characteristics of SynCph2(1–2) crystals, proceeds only at higher temperature.
The resonance Raman spectra indicate that alterations occur near the ring B and C nitrogens and could be caused by the pyrrole water that is hydrogen bonded to the pyrrole nitrogens. However, because the absolute positions of the Raman bands, in particular those in the fingerprint region between 1500 and 1700 cm−1 (Fig. 6B), are virtually identical in irradiated- and non-irradiated samples, one may infer that the main conformation of the tetrapyrrole chromophore is undistorted upon irradiation. Thus, x-ray damage is not per se responsible for the very large tilts observed between rings B and C.
To assess the significance of the tilt between rings B and C, for example, excluding that the tilts are caused by the nature of the parameters used during crystallographic refinement, we optimized the structure of the PCB binding site of SynCph2(1–2) using a QM/MM approach as described under “Experimental Procedures.” The geometry of PCB converges to a conformation, which is only slightly different than in the crystal structure, as indicated by a r.m.s. deviation of only 0.4 Å. In particular, the large twist observed at the B-C methine bridge (Table 2) is maintained (23°) and exceeds those observed for any other phytochrome. In the optimized structure, the protein environment around the chromophore remains largely conserved. Analogous results are obtained for a SynCph2 planar model, where the chromophore of SynCph2 is substituted by the nearly planar PCB chromophore from the SynCph1 structure. QM/MM calculations based on this model again provide a non-planar PCB chromophore that is strongly twisted at the B-C methine bridge (18°). These calculations indicate that the large tilt between the B- and C-rings is indeed enforced by the SynCph2 protein matrix rather than by radiation damage.
TABLE 2.
Ring tilts of PCB from x-ray crystallography and QM/MM calculations
Cph2 refers to the crystal structure of SynCph2(1–2). The planar geometries relate to a modified SynCph2(1–2) structure, where the chromophore was substituted by the PCB chromophore from the SynCph1 structure (PDB 2VEA).
| Tilt A-B | Tilt B-C | Tilt C-D | |
|---|---|---|---|
| X-ray SynCph2 (PDB 4BWI) | 19.2° | 32.5° | 59.8° |
| X-ray SynCph1 (PDB 2VEA) | 9.8° | 1.5° | 26.3° |
| X-ray SynCph2 QM/MM optimized | 17.8° | 23.1° | 40.9° |
| SynCph2-planar QM/MM optimized | 19.5° | 17.5° | 41.6° |
Interactions of the Tongue Region with the PCB-binding Pocket
The hairpin-like tip of the GAF2 tongue region, which contacts the GAF1 domain, is highly conserved among Group I and II phytochromes. However, the stem regions of the tongue connect differently to the GAF and/or PHY domains in different homologs (Fig. 7A). These differences are exemplified by the structure of PaBphP in the Pfr state (Fig. 7C), where a mostly α-helical conformation is adopted.
FIGURE 7.
The tongue-like region of SynCph2(1–2). A, structure and interactions between the tongues and the chromophore-bearing GAF domains of phytochromes. B, comparison of the tongue regions of SynCph2 and SynCph1, superimposition of the GAF domains (r.m.s. deviation = 1.095 Å for 125 Cα); GAF1 and GAF2 of SynCph2 are displayed in orange and green; PAS, GAF, and PHY of SynCph1 in blue, gray, and pale red, respectively. The chromophores are shown in cyan (Cph2) and red (Cph1). The positions of the PRXSF, W(G/A)G, and WXE motifs in the tongue regions are conserved between SynCph1 and SynCph2, the residues are numbered along the SynCph2 count. C, tongue region of PaBphP (PDB code 3NHQ), the PAS (blue), GAF (gray) and PHY (red) domains and biliverdin in green. In contrast to SynCph2 and SynCph1 the tongue region consists of an extended loop region and α-helical elements. The spatial positions of the conserved motifs differ in PaBphP. The orientation of the tongue region correlates to that of SynCph2.
Although in SynCph2 the β-strands (β13, β14) forming the stem of the tongue are longer than in SynCph1 (the GAF2 domain is tilted further away from GAF1 than the PHY domain in SynCph1), the overall positions of the conserved W(G/A)G, PRXSF, and WXE motifs relative to the GAF1 domain are well conserved between the two (Fig. 7B). Only in the Pfr state of PaBphP do these motifs differ significantly (Fig. 7C), allowing only the α-helical part of the tongue to cover the PCB binding pocket and displacing the W(G/A)G and WXE motifs by 11–15 Å relative to their counterparts in SynCph2 and SynCph1.
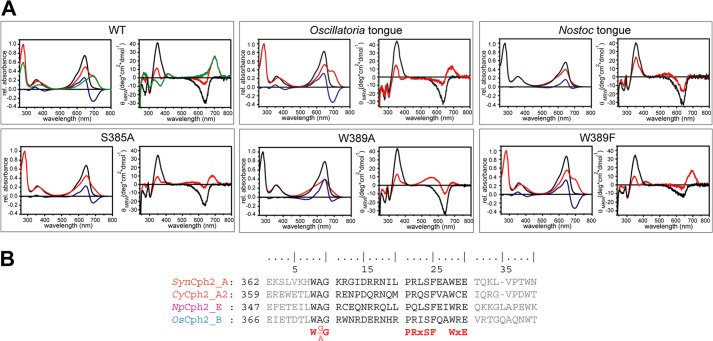
In SynCph2 the W(G/A)G motif (Trp369-Gly371) extends from β13 into the hairpin tip region of the tongue. This region is distal from the GAF1 domain by being located on an elongated loop region in PaBphP/Pfr (Fig. 7C). In the SynCph2/Pr structure, Trp369 of the W(G/A)G motif forms an H-bond via the indole nitrogen with Glu391 from the WXE motif on β14. The role of the former anchor-like WGG motif is underlined by the finding that the W369A mutant remains arrested in an intermediate state upon red light illumination. The same is observed for the W389A mutation within the second anchor of the tongue, the WXE motif, but not for the E391A mutant, a fact suggesting that the interaction of the latter with Trp369 is not crucial for photoconversion. Given the large distance of 9–14 Å to the PCB binding pocket in the Pr state of SynCph1 and SynCph2, the involvement of the Trp motifs in Pfr formation conversion is elusive. As mutant Group I phytochromes in which the PHY domain has been deleted, are known to be unable to form bona fide Pfr, one may conclude that the bulky side chains of these tryptophans are required to stabilize not only the Pr state, but also an alternative conformation of the tongue when switched to the Pfr state. Indeed, mutations of Trp369 or Trp389 to phenylalanine in SynCph2 allow Pfr formation (Fig. 9), albeit with compromised efficiency for the W369F mutant (supplemental Fig. S2). This underlines the importance of the general bulky aromatic character of these tryptophans for the structural reorganization of the tongue during Pr → Pfr photoconversion. When comparing the SynCph2 and SynCph1 Pr structures with the PaBphP/Pfr structure, it can be observed that in both states a tryptophan is sandwiched at almost the same position in the tongue-GAF1 interface. However, in Pr the indole moiety comes from the W(G/A)G, but in Pfr from the WXE motif. In this way, the swap of the bulky tryptophan side chains in the tongue-GAF1 interface may support the bistability and hence photochromicity of Group I and II phytochromes.
FIGURE 9.
Mutants and hybrids of the SynCph2 tongue region. A, UV-visible absorbance (left) and CD spectra (right) of SynCph2(1–2) wild type and mutants after far-red (Pr-state, black line) and red light illumination (red). Difference spectra (APr − APhotoequilibrium) are shown in blue. Green curves in the wild type spectra are calculated for pure Pfr (10). The partial lack of a wild type-like Pfr CD spectrum of Oscillatoria tongue is due to its instability. B, multiple sequence alignment of Group II phytochromes for GAF2 sequence stretches defining the tongue region. Tongue regions swapped between hybrid variants are depicted in black, conserved motifs are highlighted in red. Abbreviations: SynCph2_A (organism: Synechocystis sp. PCC 6803; GenBank entry: NP_442466.1; sequence identity: 100%), CyCph2_A2 (Cyanothece sp. PCC 7822; YP_003886347.1; 39%), OsCph2_B (Oscillatoria sp. PCC 6506; WP_007353933.1; 36%), NpCph2_E (N. punctiforme PCC 73102; YP_001868577.1; 34%).
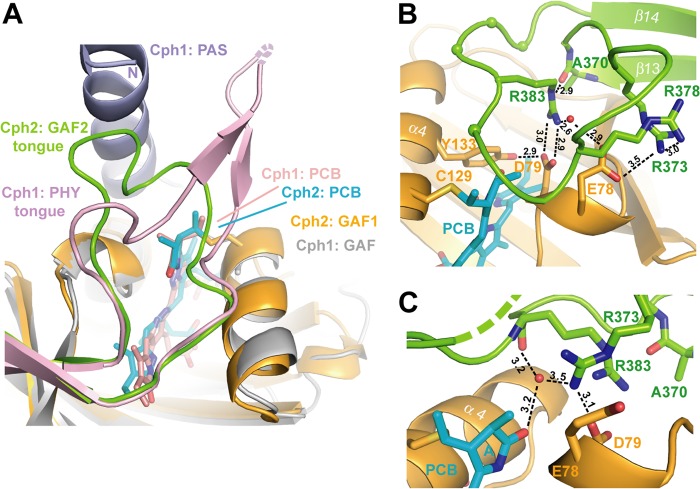
The third conserved motif within the tip of the tongue, the PRXSF motif, shields the entrance to the PCB binding pocket by adopting an elongated conformation above it and allowing for Arg383 to project into the GAF1 domain where it forms a salt bridge with Asp79 of the DIP motif. The conformation of the PRXSF motif, including the outward orientation of the serine residue and the backbone conformation until the WXE motif, is absolutely conserved between SynCph2 and SynCph1 (Figs. 7B and 8A). Mutants of the salt bridge involving the PRXSF motif like R383D or D79R remain hence either arrested in an intermediate state (10), or are photochemically impotent (supplemental Fig. S1).
FIGURE 8.
The tip of the tongue region of SynCph2. A, comparison of the photosensory modules of SynCph2 and SynCph1 at the tongue-GAF1 interface, color-coding corresponds to Fig. 7. B, the tongue region seals the PCB-binding pocket. In molecule B the whole tongue is defined by electron density. Only Arg383 of the PRXSF motif (spheres) is depicted as stick representation. Hydrogen bond distances are given in Å. C, chain A shows that the tip of the tongue is rather motile as several residues are missing and Arg373 adopts a different conformation.
Pro382 from the PRXSF motif is directly placed above the thioether bridge between Cys129 and ring A of the PCB chromophore. Together with Pro81 of the DIP motif, Pro382 forms a clamp hindering large conformational changes of the A-ring. Interestingly, earlier NMR studies on the Pfr-like state of an isolated GAF domain of another Group II phytochrome postulated A-ring rotation as part of the photocycle (6). Given that this ubiquitous structural restraint in Group I and II phytochromes is missing in that case, it is feasible that the tongue-GAF interaction serves to exclude alternative isomerization pathways for bound bilin chromophores. Clearly, a disturbance of the A-ring clamp as given by the SynCph2 P382G mutant causes a lack of photoconversion to Pfr (supplemental Fig. S2) and again suggests that a rigid element is needed here to allow conformational Pr → Pfr switching. Interestingly, a threonine mutant of Pro382 photoconverts to Pfr like the wild type but exhibits a faster dark reversion (supplemental Fig. S2). Accordingly, this backbone version requires only partial restriction of its conformational space to maintain the function of the tongue motif.
Finally, the S385A and F386A mutants, like most of the other mutants of the tongue motifs, fail to form Pfr upon red light illumination. Phe386 is buried in the tongue-GAF1 interface close to the hydrophobic environment of ring D and thus stabilizes at least the Pr state. In contrast, the surface-exposed Ser386 makes no interactions to any other residue in the SynCph2 structure. When comparing the Pr state structures of SynCph2 and SynCph1 with the Pfr state of PaBphP, it is apparent that the PRXSF motif adopts an inverted orientation in the latter; the serine instead of the arginine points into the binding pocket to build a hydrogen bond to the aspartate of the DIP motif. The role of this serine is further underlined by the fact that a mutation to alanine destabilizes the Pfr state of PaBphP (9). Together with the role of the tryptophan motifs one can suggest that during conversion into Pfr the salt bridge is released and the tip of the tongue undergoes huge conformational changes (Fig. 10). According to this model these residues are crucial for conformational stabilization of the Pfr state and thus for complete photoconversion. Interestingly, in the case of the SynCph2(1–2) module, such a large conformational change is indicated by an increased α-helical content and hydrodynamic diameter upon Pfr formation (10).
FIGURE 10.
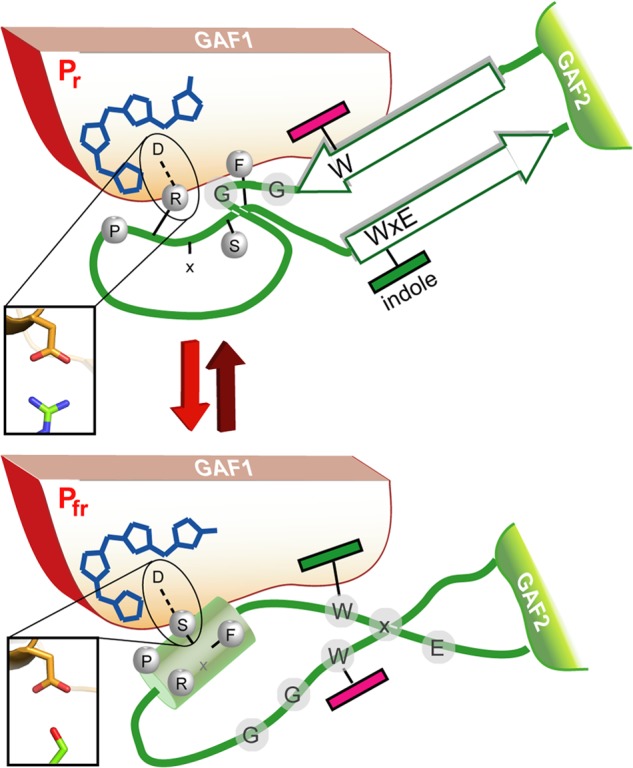
A model for the Trp motif switch within phytochromes. Upon red-light triggered formation of the Pfr state and breakage of the tongue/GAF1 Asp-Arg salt bridge (inlet) the tip of the tongue refolds with concomitant disordering of the stalk-like β-hairpin and formation of an Asp-Ser salt bridge (inlet).
Apart from the conserved motifs mentioned above for the tongue tip there is some length variation of different phytochromes, which is illustrated by the 5-amino acid larger tongue tip for SynCph1 than for SynCph2. The tip of the loop in SynCph2 adopts alternative conformational states within the crystal. In chain A of SynCph2 the Asp79-Arg383 salt bridge is part of a hydrogen-bonding network consisting of Tyr133, together with a second arginine from the tip of the tongue (Arg373) as well as a water molecule. The latter stabilizes the tongue-GAF1 interaction by coordinating the amide nitrogen of Arg383 and the ring A carbonyl group (Fig. 8C). In chain B Arg373 flips out and interacts with Arg378 and Glu78. This interaction stabilizes the loop region, which can be clearly defined by electron density (Fig. 8B). However, in both molecules, Arg383 H-bonds to the backbone carbonyl of Ala370 of the W(G/A)G motif. The tongue-GAF1 interaction is further stabilized by an interaction absent in SynCph1. Lys367, located in β13 of the tongue contacts the edge of the GAF1 domain via an extended hydrogen bond network between His74 and Ser54.
The Tongue Region as a Functional Module within Phytochromes
Given the strict dependence of the Pfr conformation on conserved motifs within the tongue region, we wondered whether the tongue extension presents on its own a functional module that is interchangeable between different phytochromes of the Cph2 family. We generated swapping variants of SynCph2(1–2) in the tongue region by replacing the stretch between the W(G/A)G motif and the WXE motif (Trp369-Glu391) with regions of orthologous Group II phytochromes from N. punctiforme, Oscillatoria sp. and Cyanothece sp. (see Fig. 9 and supplemental Fig. S2). The Oscillatoria sp. and Cyanothece sp. tongue-containing variants exhibit normal photoconversion, whereas the Cyanothece sp. tongue-containing variant showed only partial Pfr formation after red light illumination. Interestingly, the variant with the N. punctiforme tongue displayed incomplete photoconversion. This tongue harbors a degenerated PRXSF motif due to an arginine to glutamine substitution. Nevertheless, a R383Q mutant in SynCph2(1–2) showed normal Pr/Pfr photoconversion indicating that in the Pr state glutamine can form analogous H-bonds with Asp79 as the arginine and that in the Pfr state other residues are at least equally crucial for the proper association of the tongue with the GAF1-bilin binding site.
The Tongue, a Subdomain for Pfr Stabilization and/or Signaling?
So far, the light-triggered Z → E isomerization around the C15-C16 double bond is insufficient to rationalize the strong bathochromic shift of the Pfr state of Group I and II phytochromes by about 100–130 nm when compared with simple bilin-chromopeptides. Clearly, the complex bilin-binding site of phytochromes stabilizes these Pfr photo states by an unresolved mechanism, e.g. by electrostatic interaction, steric control of bilin conformation, or even formation of enol-like tautomers (44). Our structural and mutagenesis data on SynCph2 indicate that the conserved PRXSF, W(G/A)G, and WXE motifs of the tongue region are crucial for Pfr formation. Other structural elements, which contribute to the bilin-binding site of a canonical phytochrome such as SynCph1 (8, 9), are either missing (N terminus) or replaced like the subsite for ring B and C propionates. The W(G/A)G and WXE motifs are shared by Group I and II phytochromes and affect Pr → Pfr photoconversion due to their distance and link to the bilin binding pocket via a conserved H-bonding network. Together with structural data on the Pfr state of bathy phytochromes (9) we suggest conformational switching of the tongue region via swap of the tryptophans in the tongue-GAF1 interface during Pr → Pfr conversion (Fig. 10). This folding event on the local protein level is predicted to be part of the slow, light-independent lumi-R → Pfr conversion that occurs in the millisecond range. Accordingly, bathy phytochromes may prefer Pfr as the paternal state to Pr due to different energetics of the interaction between their chromophore/GAF1-adduct and the tongue region. Slow protein conformational changes such as the suggested Trp switch are instrumental for further signal transfer either to the C-terminal effector domains as in bacterial phytochromes or for modulating protein-protein interactions as in plant phytochromes. Interestingly, the tongue region of known phytochrome structures passes into a helix that directly connects the PHY/GAF2 domains with downstream effector modules and thus provides a short route for intra-molecular signaling. This may suggest that different effector domains such as histidine kinases, diguanylate cyclases, and c-di-GMP-specific phosphodiesterases share a common mechanism for intramolecular signal transduction.
Acknowledgments
We thank the beamline staff of the ESRF, Grenoble, and BESSY-II, Berlin, for support, Holger Steuber for data collection, Yann Geisselbrecht, Holger Steuber, and Maik Veelders for support in structure solution and refinement, Petra Gnau and Ralf Pöschke for technical support, and Georgios Psakis for discussions.
This work was supported by Deutsche Forschungsgemeinschaft Grants ES152/9-1 and MR81/3-1 and the LOEWE Center for Synthetic Microbiology (to L.-O. E.).

This article contains supplemental Figs. S1 and S2 and Tables S1 and S2.
The atomic coordinates and structure factors (code 4BWI) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- GAF
- cGMP phosphodiesterase/adenylyl cyclase/FhlA
- PCB
- phycocyanobilin
- PAS
- period/ARNT/single-minded
- PHY
- phytochrome
- CBCR
- cyanobacteriochrome
- r.m.s. deviation
- root mean square deviation
- QM-MM
- quantum mechanical/molecular mechanical
- PDB
- Protein Data Bank.
REFERENCES
- 1. Butler W. L., Norris K. H., Siegelman H. W., Hendricks S. B. (1959) Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proc. Natl. Acad. Sci. U.S.A. 45, 1703–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hughes J., Lamparter T., Mittmann F., Hartmann E., Gärtner W., Wilde A., Börner T. (1997) A prokaryotic phytochrome. Nature 386, 663. [DOI] [PubMed] [Google Scholar]
- 3. Rodriguez-Romero J., Hedtke M., Kastner C., Müller S., Fischer R. (2010) Fungi, hidden in soil or up in the air. Light makes a difference. Annu. Rev. Microbiol. 64, 585–610 [DOI] [PubMed] [Google Scholar]
- 4. Nagatani A. (2010) Phytochrome. Structural basis for its functions. Curr. Opin. Plant Biol. 13, 565–570 [DOI] [PubMed] [Google Scholar]
- 5. Vierstra R. D., Zhang J. (2011) Phytochrome signaling. Solving the Gordian knot with microbial relatives. Trends Plant Sci. 16, 417–426 [DOI] [PubMed] [Google Scholar]
- 6. Ulijasz A. T., Cornilescu G., Cornilescu C. C., Zhang J., Rivera M., Markley J. L., Vierstra R. D. (2010) Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature 463, 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rockwell N. C., Su Y. S., Lagarias J. C. (2006) Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57, 837–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Essen L. O., Mailliet J., Hughes J. (2008) The structure of a complete phytochrome sensory module in the Pr ground state. Proc. Natl. Acad. Sci. U.S.A. 105, 14709–14714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang X., Kuk J., Moffat K. (2008) Crystal structure of Pseudomonas aeruginosa bacteriophytochrome. Photoconversion and signal transduction. Proc. Natl. Acad. Sci. U.S.A. 105, 14715–14720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Anders K., von Stetten D., Mailliet J., Kiontke S., Sineshchekov V. A., Hildebrandt P., Hughes J., Essen L. O. (2011) Spectroscopic and photochemical characterization of the red-light sensitive photosensory module of Cph2 from Synechocystis PCC 6803. Photochem. Photobiol. 87, 160–173 [DOI] [PubMed] [Google Scholar]
- 11. Montgomery B. L., Lagarias J. C. (2002) Phytochrome ancestry. Sensors of bilins and light. Trends Plant Sci. 7, 357–366 [DOI] [PubMed] [Google Scholar]
- 12. Rockwell N. C., Lagarias J. C. (2010) A brief history of phytochromes. ChemPhysChem 11, 1172–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Narikawa R., Ishizuka T., Muraki N., Shiba T., Kurisu G., Ikeuchi M. (2013) Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc. Natl. Acad. Sci. U.S.A. 110, 918–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burgie E. S., Walker J. M., Phillips G. N., Jr., Vierstra R. D. (2013) A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCF-cyanobacteriochromes. Structure 21, 88–97 [DOI] [PubMed] [Google Scholar]
- 15. Park C.-M., Shim J.-Y., Yang S.-S., Kang J.-G., Kim J.-I., Luka Z., Song P.-S. (2000) Chromophore-apoprotein interactions in Synechocystis sp. PCC6803 phytochrome Cph1. Biochemistry 39, 6349–6356 [DOI] [PubMed] [Google Scholar]
- 16. Wu S. H., Lagarias J. C. (2000) Defining the bilin lyase domain. Lessons from the extended phytochrome superfamily. Biochemistry 39, 13487–13495 [DOI] [PubMed] [Google Scholar]
- 17. Savakis P., De Causmaecker S., Angerer V., Ruppert U., Anders K., Essen L. O., Wilde A. (2012) Light-induced alteration of c-di-GMP level controls motility of Synechocystis sp. PCC 6803. Mol. Microbiol. 85, 239–251 [DOI] [PubMed] [Google Scholar]
- 18. Jain R., Behrens A.-J., Kaever V., Kazmierczak B. I. (2012) Type IV pilus assembly in Pseudomonas aeruginosa over a broad range of cyclic di-GMP concentrations. J. Bacteriol. 194, 4285–4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Simm R., Morr M., Kader A., Nimtz M., Römling U. (2004) GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53, 1123–1134 [DOI] [PubMed] [Google Scholar]
- 20. Fiedler B., Börner T., Wilde A. (2005) Phototaxis in the cyanobacterium Synechocystis sp. PCC 6803. Role of different photoreceptors. Photochem. Photobiol. 81, 1481–1488 [DOI] [PubMed] [Google Scholar]
- 21. Wilde A., Fiedler B., Börner T. (2002) The cyanobacterial phytochrome Cph2 inhibits phototaxis towards blue light. Mol. Microbiol. 44, 981–988 [DOI] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 25. Carpentier P., Royant A., Ohana J., Bourgeois D. (2007) Advances in spectroscopic methods for biological crystals. 2. Raman spectroscopy. J. Appl. Crystallogr. 40, 1113–1122 [Google Scholar]
- 26. McGeehan J., Ravelli R. B., Murray J. W., Owen R. L., Cipriani F., McSweeney S., Weik M., Garman E. F. (2009) Colouring cryo-cooled crystals. Online microspectrophotometry. J. Synchrotron Radiat. 16, 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kneip C., Hildebrandt P., Schlamann W., Braslavsky S. E., Mark F., Schaffner K. (1999) Protonation state and structural changes of the tetrapyrrole chromophore during the Pr → Pfr phototransformation of phytochrome. A resonance raman spectroscopic study. Biochemistry 38, 15185–15192 [DOI] [PubMed] [Google Scholar]
- 28. Strauss H. M., Hughes J., Schmieder P. (2005) Heteronuclear solution-state NMR studies of the chromophore in cyanobacterial phytochrome Cph1. Biochemistry 44, 8244–8250 [DOI] [PubMed] [Google Scholar]
- 29. Senn H. M., Thiel W. (2007) QM/MM studies of enzymes. Curr. Opin. Chem. Biol. 11, 182–187 [DOI] [PubMed] [Google Scholar]
- 30. Becke A. D. (1993) Density-functional thermochemistry. III. The role of exact exchange. J. Chem. Phys. 98, 5648–5652 [Google Scholar]
- 31. MacKerell A. D., Bashford D., Bellott, Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiórkiewicz-Kuczera J., Yin D., Karplus M. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 32. Bakowies D., Thiel W. (1996) Hybrid models for combined quantum mechanical and molecular mechanical approaches. J. Phys. Chem. 100, 10580–10594 [Google Scholar]
- 33. Billeter S. R., Turner A. J., Thiel W. (2000) Linear scaling geometry optimisation and transition state search in hybrid delocalised internal coordinates. Phys. Chem. Chem. Phys. 2, 2177–2186 [Google Scholar]
- 34. Landgraf F. T., Forreiter C., Hurtado Picó A., Lamparter T., Hughes J. (2001) Recombinant holophytochrome in Escherichia coli. FEBS Lett. 508, 459–462 [DOI] [PubMed] [Google Scholar]
- 35. von Stetten D., Günther M., Scheerer P., Murgida D. H., Mroginski M. A., Krauss N., Lamparter T., Zhang J., Anstrom D. M., Vierstra R. D., Forest K. T., Hildebrandt P. (2008) Chromophore heterogeneity and photoconversion in phytochrome crystals and solution studied by resonance Raman spectroscopy. Angew. Chem. Int. Ed. Engl. 47, 4753–4755 [DOI] [PubMed] [Google Scholar]
- 36. Li H., Zhang J., Vierstra R. D., Li H. (2010) Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc. Natl. Acad. Sci. U.S.A. 107, 10872–10877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mailliet J., Psakis G., Feilke K., Sineshchekov V., Essen L. O., Hughes J. (2011) Spectroscopy and a high-resolution crystal structure of Tyr-263 mutants of cyanobacterial phytochrome Cph1. J. Mol. Biol. 413, 115–127 [DOI] [PubMed] [Google Scholar]
- 38. Navarro M. V., De N., Bae N., Wang Q., Sondermann H. (2009) Structural analysis of the GGDEF-EAL domain-containing c-di-GMP receptor FimX. Structure 17, 1104–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Diensthuber R. P., Bommer M., Gleichmann T., Möglich A. (2013) Full-length structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure 21, 1127–1136 [DOI] [PubMed] [Google Scholar]
- 40. Wang C., Sang J., Wang J., Su M., Downey J. S., Wu Q., Wang S., Cai Y., Xu X., Wu J., Senadheera D. B., Cvitkovitch D. G., Chen L., Goodman S. D., Han A. (2013) Mechanistic insights revealed by the crystal structure of a histidine kinase with signal transducer and sensor domains. Plos Biol. 11, e1001493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Song C., Psakis G., Lang C., Mailliet J., Gärtner W., Hughes J., Matysik J. (2011) Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proc. Natl. Acad. Sci. U.S.A. 108, 3842–3847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shang L., Rockwell N. C., Martin S. S., Lagarias J. C. (2010) Biliverdin amides reveal roles for propionate side chains in bilin reductase recognition and in holophytochrome assembly and photoconversion. Biochemistry 49, 6070–6082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mailliet J., Psakis G., Schroeder C., Kaltofen S., Dürrwang U., Hughes J., Essen L. O. (2009) Dwelling in the dark: procedures for the crystallography of phytochromes and other photochromic proteins. Acta Crystallogr. D 65, 1232–1235 [DOI] [PubMed] [Google Scholar]
- 44. Lagarias J. C., Rapoport H. (1980) Chromopeptides from phytochromes. The structure and linkage of the Pr form of the phytochrome chromophore. J. Am. Chem. Soc. 102, 4821–4828 [Google Scholar]