Background: Mutations in transmembrane domains can affect activity of viral fusion proteins, but little is known about potential interactions between these domains.
Results: Isolated paramyxovirus fusion protein transmembrane domains interact as trimers.
Conclusion: Viral fusion protein transmembrane domains self-associate.
Significance: Transmembrane domain associations may regulate stability of the prefusion conformation.
Keywords: Fusion Protein, Membrane Fusion, Negative-strand RNA Viruses, Virus, Virus Entry, Transmembrane Domain
Abstract
Paramyxovirus fusion (F) proteins promote membrane fusion between the viral envelope and host cell membranes, a critical early step in viral infection. Although mutational analyses have indicated that transmembrane (TM) domain residues can affect folding or function of viral fusion proteins, direct analysis of TM-TM interactions has proved challenging. To directly assess TM interactions, the oligomeric state of purified chimeric proteins containing the Staphylococcal nuclease (SN) protein linked to the TM segments from three paramyxovirus F proteins was analyzed by sedimentation equilibrium analysis in detergent and buffer conditions that allowed density matching. A monomer-trimer equilibrium best fit was found for all three SN-TM constructs tested, and similar fits were obtained with peptides corresponding to just the TM region of two different paramyxovirus F proteins. These findings demonstrate for the first time that class I viral fusion protein TM domains can self-associate as trimeric complexes in the absence of the rest of the protein. Glycine residues have been implicated in TM helix interactions, so the effect of mutations at Hendra F Gly-508 was assessed in the context of the whole F protein. Mutations G508I or G508L resulted in decreased cell surface expression of the fusogenic form, consistent with decreased stability of the prefusion form of the protein. Sedimentation equilibrium analysis of TM domains containing these mutations gave higher relative association constants, suggesting altered TM-TM interactions. Overall, these results suggest that trimeric TM interactions are important driving forces for protein folding, stability and membrane fusion promotion.
Introduction
Entry of enveloped viruses requires fusion between the viral envelope and a target cell membrane. This critical early event in infection is promoted by specific viral glycoproteins, termed fusion (F)2 proteins, which undergo conformational rearrangements that ultimately drive membrane fusion (1). Paramyxoviruses are nonsegmented negative-strand RNA viruses and include important human pathogens such as measles, mumps, respiratory syncytial virus, human metapneumovirus (HMPV), and the zoonotic Hendra and Nipah viruses (2). Attachment and entry of paramyxoviruses are generally promoted by two membrane glycoproteins: the attachment protein, which facilitates binding to cellular receptors, and the F protein, which promotes fusion between the viral envelope and a target cell membrane (3).
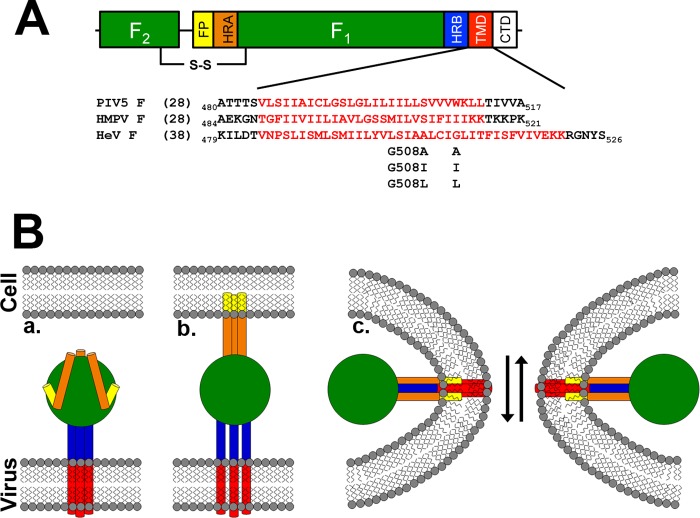
Like other class I viral fusion proteins, paramyxovirus F proteins are synthesized as inactive precursors (F0), which are proteolytically processed to the mature and fusogenic (F1 + F2) form (see Fig. 1A). This cleavage event can be promoted by furin in the trans-Golgi network, as in the case of parainfluenza virus 5 F (PIV5 F (4)); by exogenous proteases, as seen with HMPV F (5); or by cathepsin L after endocytosis of the F protein, a unique pathway utilized by the Hendra and Nipah F proteins (6). The processed F1 + F2 form (see Fig. 1B, part a) exists in a metastable state that can be triggered to undergo the massive conformational changes associated with fusion. Paramyxovirus F proteins, along with other class I viral fusion proteins, share conserved elements that play fundamental roles in the fusion process. Crystal structures of the prefusion form of the PIV5 F protein (7, 8) and of the postfusion forms of the Newcastle disease virus and HPIV3 F proteins (9–11), combined with research to date on a number of paramyxovirus F proteins, suggest a model for fusion that elucidates the role of these conserved regions (2, 3). The fusion peptide, a hydrophobic region at the N terminus of the F1 subunit, inserts into the target membrane, and the adjacent heptad repeat A (HRA) region forms a trimeric coiled-coil (see Fig. 1B, part b). A second heptad repeat region (HRB) proximal to the transmembrane (TM) domain separates and refolds with the HRA coiled-coil, forming an extremely stable six-helix bundle structure that is critical for fusion promotion (see Fig. 1B, part c).
FIGURE 1.
Schematic of a paramyxovirus F protein and model of membrane fusion. A, diagram of a mature paramyxovirus F protein, with TM sequences indicated. FP = fusion peptide; HR = heptad repeat; TMD = transmembrane domain; CTD = cytoplasmic tail. B, model for F protein-promoted membrane fusion. Part a, the HRB domains adjacent to the TM form a coiled-coil in the prefusion state, whereas the HRA domains are present as a series of small helices. Part b, triggering of fusion leads to insertion of the fusion peptide into the target membrane and formation of an HRA coiled-coil, and separation of the HRB helices and TM domains. Part c, fusion pore opening occurs concomitant with formation of a six-helix bundle with the HRA segments on the interior of the bundle, placing the HRB and TM segments on the exterior.
A number of studies have observed that modifications to the TM domain can result in modulation of class I viral fusion protein activity (12–18), including impairment of fusion pore opening or enlargement (13–15, 19–21) or alterations in the efficiency of fusion promotion (18, 22, 23). Recent studies have also demonstrated an important role for the TM domains of class II fusion proteins (24) and reovirus fusion-associated small transmembrane (FAST) proteins (25) in the fusion process. Specific sequences within the TM domain are not obligately required as replacement with alternative TM domains can in some cases produce a functional fusion protein (26–29). However, fusion alterations observed following the introduction of point mutations within some TM domains indicate that specific sequence elements or residues are important. Replacement of the paramyxovirus PIV5 F protein TM domain with polyleucine resulted in a misfolded protein (22), confirming that the TM domain serves as more than a simple membrane anchor. However, the mechanisms by which TM domains contribute to the folding and fusion process remain poorly understood, partially due to the difficulties in obtaining direct structural information about these regions.
TM-TM interactions are critical for a number of important biological processes (30, 31), and TM domains from the HIV env protein have been shown recently to interact (32), although the oligomeric form could not be assessed. Several lines of evidence suggest a potential role for TM-TM interactions in paramyxovirus F protein folding and function. Results of a cysteine-scanning study of the PIV5 F TM domain were consistent with interacting TM helices (22), and a prefusion form of both the PIV5 (7) and the respiratory syncytial virus F protein (33) was only obtained when the TM domain was replaced with a GCN4t or fibritin trimerization domain, respectively. We therefore utilized a well developed method of studying TM-TM interactions utilizing sedimentation equilibrium ultracentrifugation to analyze potential TM interactions in paramyxovirus F proteins and confirmed our results using isolated TM domain peptides. Our results show that the isolated TM domains of Hendra F, PIV5 F, and HMPV F associate in monomer-trimer equilibria, suggesting that TM-TM interactions may play an important role in F protein folding, stability, and function. As glycine-containing motifs and polar residues have been proposed to promote TM-TM interactions (30, 31), the role of these residues was examined in the Hendra F TM domain. Mutational analyses indicate that a glycine residue in the Hendra F TM is important for the stable surface expression of the cleaved prefusion form and for TM-TM association. In contrast, mutational studies did not indicate a significant role for noncharged polar residues. Our results support a model whereby TM-TM interactions are important for stability of the prefusion form of the molecule, thus potentially regulating the triggering of the conformational changes in F required for membrane fusion.
EXPERIMENTAL PROCEDURES
Cell Lines and Culture
Vero and BSR (provided by Karl-Klaus Conzelmann, Max Pettenkofer Institut) cells were grown in DMEM (Invitrogen) containing 10% FBS and 1% penicillin and streptomycin. Every third passage, BSR cells were selected for expression of T7 polymerase by the addition of 0.5 mg/ml G-418 sulfate (Invitrogen) to the growth medium.
Plasmids, Peptides, and Mutagenesis
Plasmids containing Hendra F and G were generously provided by Dr. Lin-Fa Wang (Australian Animal Health Laboratory), and all Hendra F mutants were generated as described previously (34–36). Staphylococcal nuclease (SN) fused to the glycophorin A (GpA) TM domain in the pET-11a expression vector (37) was generously provided by Dr. Karen Fleming (Johns Hopkins University). The TM domains of wild-type or mutant Hendra F, HMPV F and PIV5 F were identified using bioinformatics (using the TMHMM Server v. 2.0), with the C-terminal charged residues left in place to aid in solubility. The TMs were PCR-amplified from pCAGGS-Hendra F, -HMPV F, or -PIV5 F, respectively, and ligated into pET-11a using the XmaI (5′) and BamHI (3′) restriction sites. All constructs were sequenced in their entirety prior to use. A peptide corresponding to residues 485–512 of the PIV F protein with the sequence (VLSIIAICLGSLGLILIILLSVVVWKLL) was synthesized by Peptide Protein Research Ltd. The peptide was dissolved in the appropriate buffer for analytical ultracentrifugation, dialyzed, and analyzed at a concentration of 35 μm.
Recombinant Protein Expression and Purification
Constructs expressing chimeric proteins containing the TM of either wild-type or mutant Hendra F, HMPV F or PIV5 F fused to SN in pET-11a were transformed into Rosetta-gami cells (EMD Chemicals, Gibbstown, NJ) and plated on LB plates containing 100 μg/ml ampicillin. Colonies were grown in 25 ml of 2×YT under the selection of 15 μg/ml kanamycin, 12.5 μg/ml tetracycline, 50 μg/ml streptomycin, 34 μg/ml chloramphenicol, and 100 μg/ml ampicillin overnight at 37 °C. Cultures were transferred to 500 ml of 2×YT containing100 μg/ml ampicillin and grown to an A600 of 0.6–1.0 at 37 °C (approximately 2 h), induced with 1 mm isopropyl β-d-1-thiogalactopyranoside (Sigma), grown for 4 h, and harvested by centrifugation. Cells were resuspended in a 1:20 culture volume of lysis buffer (50 mm HEPES, 2 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, pH 8.0), and recombinant protein was purified using the detergent Thesit-290 (Sigma) and a single salt (1 m NH4OAc) extraction as described previously (38), except using 50 mm HEPES instead of 20 mm Tris-HCl. Following extraction, the supernatant containing the recombinant protein was dialyzed (6000–8000 molecular weight cut-off) overnight at 4 °C against lysis buffer containing 0.1 m NH4OAc and 0.2% v/v Thesit and clarified by centrifugation at 15,000 × g for 15 min. Recombinant protein was then FPLC-purified by cation-exchange chromatography using a 1-ml HiTrap SP FF column (GE Healthcare (39)) and eluted in lysis buffer containing 0.1 m NH4OAc, 1 m NaCl, and 0.2% v/v Thesit. A second round of FPLC was performed to exchange the protein into a solution containing 200 mm NaCl, 20 mm Na2HPO4/NaH2PO4 (pH = 7), 29% D2O, and the Zwittergent detergent 3-(N,N-dimethylmyristyl-ammonio)propanesulfonate (C14SB) (Sigma/Fluka (40)). Recombinant protein was eluted using the C14SB-containing solution with 1 m NaCl and dialyzed using Slide-A-Lyzer MINI dialysis units (10,000 molecular weight cut-off; Pierce). Protein concentrations were determined by spectrophotometry, using ϵ280 = 17,420 m−1 cm−1.
Hendra F TM Synthesis and Purification
A thrombin cleavage site was introduced between the SN coding region and the Hendra TM coding region. Recombinant protein was expressed as described above. Following the first FPLC purification step, protein was eluted in buffer containing C14SB, and dialysis was performed to reduce salt concentration to 200 mm. Thrombin (ZymoGenetics, Seattle, WA) was added at a 1:100 concentration, and the sample was incubated at room temperature for 4 h. The sample was repurified through a 1-ml HiTrap SP FF column as described above.
Analytical Ultracentrifugation
Sedimentation equilibrium measurements were obtained at three different rotor speeds using a Beckman XL-A analytical ultracentrifuge equipped with an An-60 Ti rotor operated at 25 °C. Three protein concentrations corresponding to 280 nm absorbances between 0.25 and 0.8 were utilized for determination of the best fit models for three TM domains (see Fig. 2). For analysis of relative association constants, duplicate samples with an absorbance of 0.25 at 280 nm were utilized. Attainment of sedimentation equilibrium was monitored by comparing radial scans. Equilibrium was considered to be obtained when scans taken 6 h apart were indistinguishable. Further verification of attainment of equilibrium included approach to equilibrium from higher and lower rotor speeds and approach to equilibrium by temperature shift. On attainment of apparent sedimentation equilibrium by these different paths, concentration distributions and equilibrium constants were statistically indistinguishable. In addition, samples cycled between sedimentation equilibrium states at two different rotor speeds gave closely reproducible concentration profiles, indicating the absence of hysteresis. Together, these tests suggest that chemical and transport equilibria were attained. The buffer density was matched to that of C14SB detergent (ρ = 1.04 g/ml) using D2O, as described previously (40). Partial specific volumes of each protein were estimated using SEDNTERP (69), and data analysis was performed using KaleidaGraph (Synergy Software, Reading, PA) and HeteroAnalysis (41). For analysis, molecular weight values were fixed, and baselines were experimentally determined from absorbance near the meniscus after overspeeding.
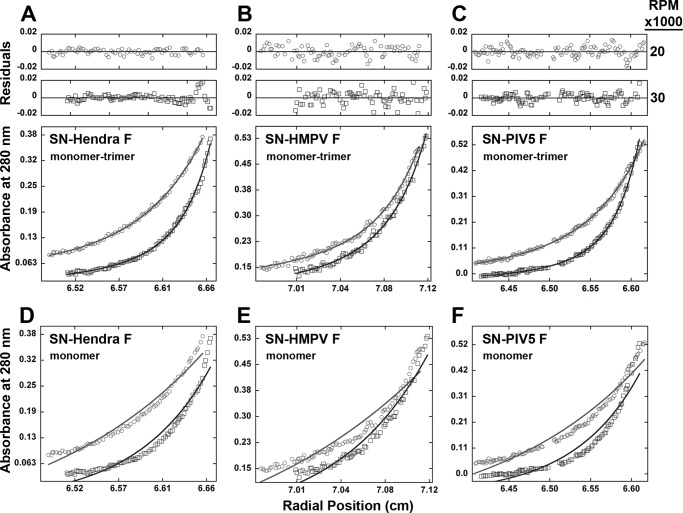
FIGURE 2.
Sedimentation equilibrium analysis of the chimeric SN-F-TM proteins demonstrates a monomer-trimer equilibrium. Protein samples were prepared in C14SB detergent. A280 data were collected at 20,000, 25,000, and 30,000 rpm on a Beckman XL-A analytical ultracentrifuge, with only the 20,000 and 30,000 displayed for clarity. A–C, monomer-trimer fits, with data (points) shown along with the predicted theoretical fit for a monomer-trimer. D–F, monomer-only fits, with data (points) shown along with the predicted theoretical fit for a monomer-trimer. A and D, SN-Hendra F TM. B and E, SN-HMPV F TM. C and F, SN-PIV5 F TM.
Biotinylation of Cell Surface Proteins
Subconfluent Vero cells in 60-mm dishes were transiently transfected with 3 μg of either wild-type or mutant Hendra F in pCAGGS using Lipofectamine and Lipofectamine Plus (Invitrogen) according to the manufacturer's instructions. Eighteen to twenty-four hours after transfection, cells were starved and metabolically labeled, and cell surface proteins were biotinylated as described previously (42). Anti-peptide polyclonal antibodies (43) were used to immunoprecipitate Hendra F, and protein was analyzed via 15% SDS-PAGE. Surface-expressed Hendra F was visualized using a Typhoon Imaging System (GE Healthcare), and band densitometry was performed using ImageQuant 5.2 software (GE Healthcare).
Luciferase Reporter Gene Assay
Vero cells in 6-well plates were transiently transfected with 0.3 μg of either wild-type or mutant Hendra F, 0.9 μg of wild-type Hendra G, and 0.8 μg of a plasmid encoding luciferase under control of the T7 promoter using Lipofectamine and Lipofectamine Plus (Invitrogen). Eighteen hours after transfection, Vero cells were overlaid for 3 h with BSR cells, which stably express the T7 polymerase (44), and luciferase activity was measured using an Lmax luminometer (Molecular Devices, Sunnyvale, CA). Background (Hendra G alone) values were subtracted, and luciferase activity was expressed as the percentage of wild-type Hendra F.
RESULTS
Construction of Chimeric Proteins and Analysis of TM-TM Interactions
To analyze TM interactions, we utilized chimeric proteins containing the SN protein linked to the TM segment of interest, a system that has been used extensively for analysis of glycophorin A (37, 45, 46), synaptobrevin and syntaxin (39), and the epidermal growth factor (ErB) receptor (47). SN is a monomeric protein under the conditions utilized for analytical ultracentrifugation (37). The use of SN-TM chimeras gives high yield expression in Escherichia coli, allows the use of lower protein concentrations during analysis due to the larger molar extinction coefficient versus an isolated TM peptide, and permits centrifugation at lower speeds due to the higher molecular weight of the chimeric protein (37). We confirmed that the SN-glycophorin A TM construct, kindly provided by Dr. Karen Fleming (Johns Hopkins University), is in monomer-dimer equilibrium, as had been previously shown using this assay (37). Chimeric proteins were expressed, purified, and exchanged into C14SB detergent (40). Samples at three concentrations were brought to sedimentation equilibrium in a Beckman XL-A analytical ultracentrifuge, radial absorbance data were obtained at 20,000, 30,000, and 40,000 rpm, and the data to determine best fit were analyzed using both KaleidaGraph and HeteroAnalysis (41).
Analysis of a chimeric protein containing the wild-type Hendra F TM domain region (residues 484–521; Fig. 1A) at three concentrations and speeds indicated that a monomer-trimer equilibrium was the best fit model (Fig. 2A). The small, symmetrically distributed residuals (Fig. 2A, upper panels) indicated that this model was consistent with the mass distributions present. The molecular weights of the species and the third-power concentration dependence of the higher molecular weight species on the concentration of monomer support this conclusion. Other single-species fits, such as monomer-only (Fig. 2, D–F) or two species models, fit the data significantly less well or gave nonrandom residual distributions. Inclusion of additional species (such as monomer-dimer-trimer) gave amplitudes for the additional terms that were within error equal to zero (results not shown). These results demonstrate for the first time that an F protein TM domain, separate from the rest of the protein, is in a monomer-trimer equilibrium. Analysis using pentaethylene glycol monooctyl ether (C8E5) (40) instead of C14SB yielded similar results, indicating that the trimeric interactions between the F protein TM domains are not dependent on the identity of the detergent. The Hendra F TM domain contains a cysteine residue (Cys-506), but analysis performed in the presence of 2 mm β-mercaptoethanol did not lead to alterations in the best fit monomer-trimer model, indicating that disulfide bond formation was not a significant contributing factor to the TM-TM interactions in this system (data not shown). Consistent with this, a previous study found that no difference in oligomeric forms on a nonreducing gel was seen between the WT PIV5 F protein and a mutant lacking the one TM cysteine residue (22). Analysis of the SN-TM protein by SDS-PAGE, both before (Fig. 3) and after centrifugation, showed both a monomer and a fainter oligomer band, and no proteolytic degradation was observed after centrifugation.
FIGURE 3.
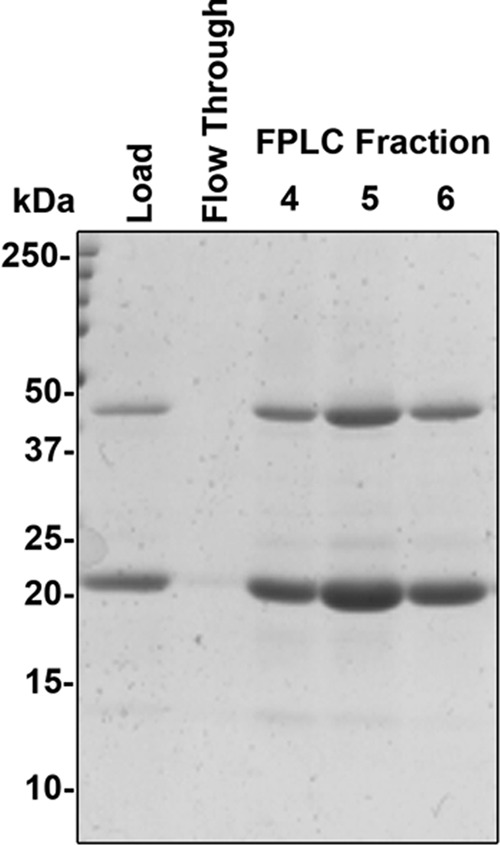
Purified SN-Hendra F protein. Samples from load, flow-through, and the three FPLC fractions containing the majority of the Hendra SN-TM protein were separated on a 15% SDS-PAGE gel and visualized by Coomassie Blue Staining. Bands corresponding to the monomeric and an oligomeric size are apparent.
To determine whether these results were specific for the TM domain from Hendra F, TM regions from PIV5 F (residues 485–512) and HMPV F (residues 489–516) were analyzed by the same method. Data for both were again best fit by a monomer-trimer model in the majority of speeds and concentrations (Fig. 2, B and C), with a slightly better fit for monomer-trimer-hexamer at the lowest speeds for HMPV. As Hendra, PIV5, and HMPV represent diverse branches of the paramyxovirus family, these results suggest that trimeric TM-TM interactions are a common paradigm for paramyxovirus F proteins.
A recent study of the PIV5 F protein, which also employed sedimentation equilibrium analysis, found evidence for a fusion peptide-TM domain interaction, but did not observe TM-TM association in dodecylphosphocholine micelles (48). To verify that TM association was not related to the addition of the SN chimera, a PIV5 TM peptide was synthesized and examined. A higher rotor speed (40,000 rpm) was needed to generate a useful concentration gradient than was used for the SN-TM construct, consistent with the much smaller size of the peptide. A single-species monomer model gave a poor fit to the data, whereas a monomer-trimer model provided data with a significantly better fit (Fig. 4). To further explore this, a SN-Hendra TM construct was synthesized with a thrombin cleavage site added between SN and the Hendra TM domain. After initial purification, the sample was digested by thrombin and the TM peptide was further purified. Bands corresponding to the expected sizes of both the monomeric peptide (molecular weight 4297) and a trimeric form were apparent by SDS-PAGE analysis (Fig. 5). Sedimentation equilibrium analysis indicated a poor fit for monomer-only (Fig. 6), a much improved fit for monomer-trimer, and a best fit for monomer-trimer-hexamer, suggesting some interaction between the trimers. These results indicate that trimer formation in the SN chimeric proteins was due to interactions of the TM domains and confirm that isolated paramyxovirus F protein TM domains can interact to form trimers.
FIGURE 4.
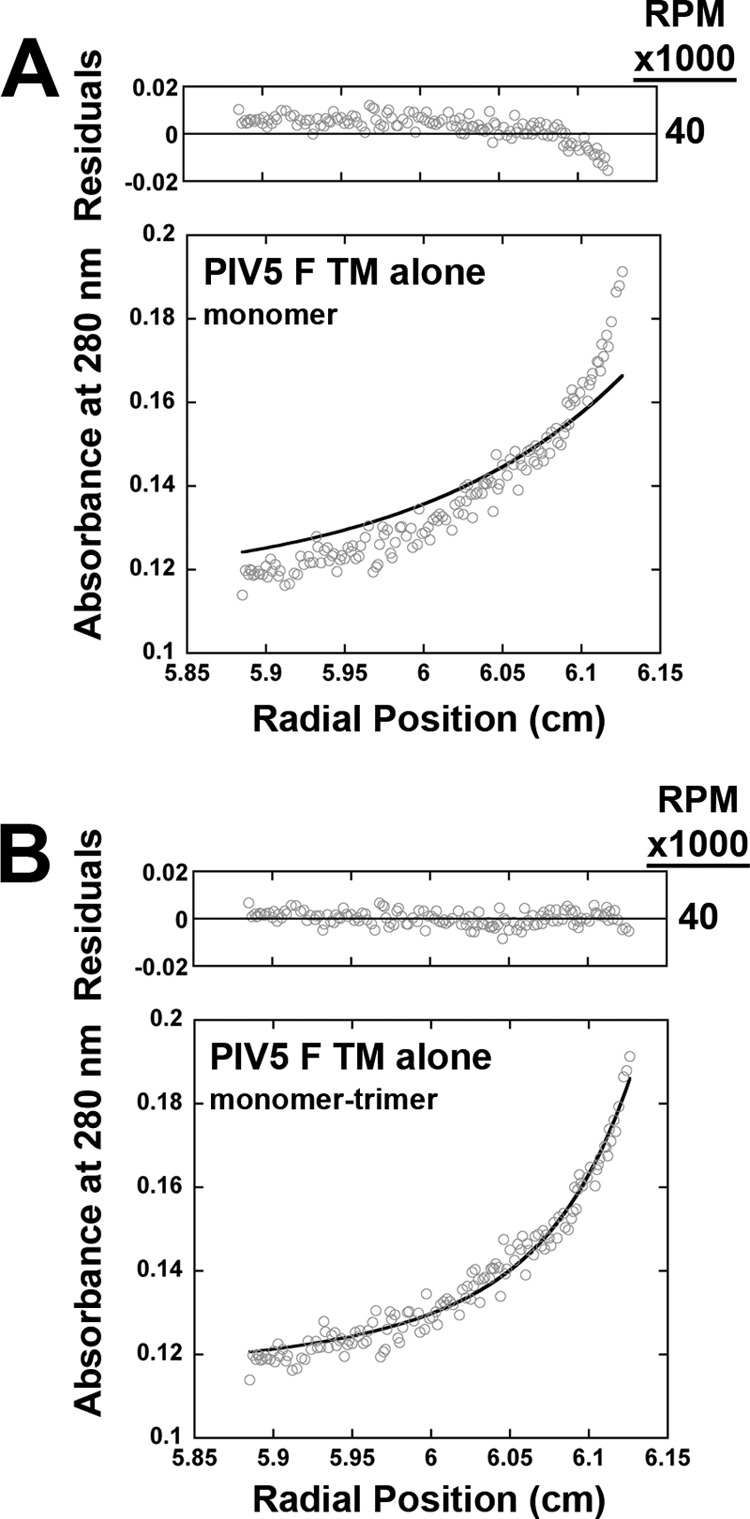
Sedimentation equilibrium analysis of a PIV5 F TM domain peptide confirms a monomer-trimer equilibrium. The PIV5 TM peptide was prepared in C14SB detergent (protein:lipid molar ratio 1:280), and A280 data were collected at 40,000 rpm on a Beckman XL-1 analytical ultracentrifuge. A and B, the predicted curves (dark lines) for monomer (A) and monomer-trimer (B) fits are shown and superimposed with actual data points.
FIGURE 5.

Purification of the Hendra F TM domain peptide. Samples from before (pre-digest) and after (post-digest) thrombin cleavage and the three FPLC fractions containing the Hendra TM peptide were separated on a 4–12% NuPAGE Bis-Tris gel and visualized by Simply Blue SafeStain (Invitrogen). Bands correspond to the predicted molecular weight of a monomer (molecular weight 4297) and trimer.
FIGURE 6.
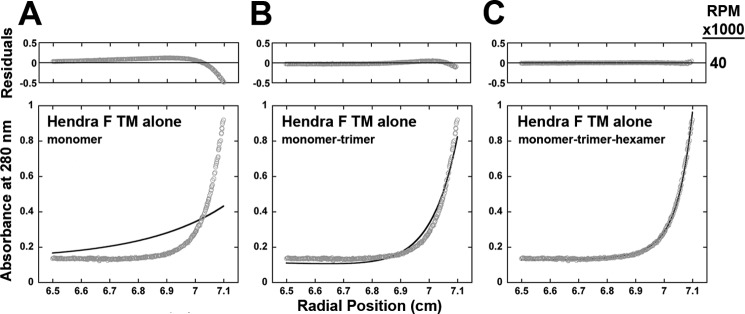
Sedimentation equilibrium analysis of a Hendra F TM domain peptide indicates a best fit monomer-trimer-hexamer equilibrium. The Hendra TM peptide was prepared in C14SB detergent (protein:lipid molar ratio 1:50), and A280 data were collected at 40,000 rpm on a Beckman XL-1 analytical ultracentrifuge using a two-channel centerpiece. A–C, the predicted curves (dark lines) for monomer (A), monomer-trimer (B), and monomer-trimer-hexamer (C) fits are shown and superimposed with actual data points.
Role of the Hendra F TM Residue Gly-508
Our results strongly support a trimeric association between isolated TM domains of paramyxovirus F proteins, but the potential impact of TM association on F protein folding and function and the identity of the residues that promote this association were unclear. We therefore performed mutational analyses of residues or motifs previously implicated in promotion of TM-TM interactions (30, 31). Motifs containing one or more glycine residues (GXXXG or GXXXY, where Y is a small residue) have been demonstrated to promote TM interactions, and these motifs have been suggested to promote interactions between the TM domains of HIV env (16, 32, 49). One glycine residue is present in the TM domain of Hendra F, forming a portion of an AXXXG motif. To test the role of this motif in the whole protein, Gly-508 was mutated to alanine, isoleucine, or leucine, and protein expression and localization were analyzed by biotinylation of cell surface proteins as described previously (35). Mutation of Gly-508 to alanine resulted in a small decrease in F1 cell surface expression (72.3 ± 20.4% of wild-type F levels), whereas mutation to isoleucine or leucine caused significant reductions (19.9 ± 4.4 and 11.8 ± 2.2% of wild-type F levels, respectively) in F1 expression as compared with wild type (Fig. 7A). Interestingly, cell surface expression levels of the uncleaved F0 form were similar to wild type. The fraction of the mutant proteins present on the cell surface was also reduced as compared with WT F (surface (F0 + F1)/total (F0 + F1) = 1.0 ± 0.2 for WT F, 0.8 ± 0.1 for G508A, 0.6 ± 0.2 for G508I, and 0.7 ± 0.3 for G508L). These results suggest that the G508I and G508L mutations affect the stable expression of the cleaved prefusion form on the cell surface, potentially through alterations in stability or endosomal trafficking. Several regions within Hendra F have been previously implicated in maintenance of prefusion metastability (3); however, such a role for the TM domain has yet to be described. As mutations within TM domains can affect membrane fusion promotion, cell-cell fusion levels were examined using a luciferase reporter gene assay (6). Each mutant promoted cell-cell fusion at levels consistent with F1 cell surface expression (Fig. 7B), indicating that substitutions at Gly-508 do not disrupt F-mediated membrane fusion beyond the effect on cell surface expression.
FIGURE 7.
Mutations at Hendra F Gly-508 can modulate expression of the cleaved, fusogenic form. A, cell surface expression of transiently transfected wild type and Hendra F mutants in Vero cells following a 3-h metabolic label. Surface proteins were biotinylated prior to immunoprecipitation, and the total and surface populations were separated by streptavidin pulldown. Proteins were analyzed via 15% SDS-PAGE and visualized using autoradiography. B, cell surface expression (CSE) and fusion for the wild type and Gly-508 mutants. F1 band quantitation via densitometry was normalized to wild-type levels plus or minus one standard deviation. Cell surface expression represents the average of three independent experiments. Fusion was analyzed using a reporter gene assay, n = 9; ± S.E.
To assess the effect of these mutations on TM-TM interactions, the G508A, G508I, and G508L mutations were introduced into the SN-TM chimera system, the recombinant proteins were expressed and purified, and sedimentation equilibrium analysis was performed. To maintain a constant protein:lipid ratio, all samples were examined at the same protein concentration. An apparent molar association constant of 8.38 ± 0.84 × 108 m−2 was calculated for the SN-Hendra F TM at the lowest speed (Table 1), and this value decreased with increasing rotor speed, displaying an ∼2-fold reduction at the highest speed, suggesting that the equilibrium is pressure-dependent (50, 51). Pressure effects are expected when the oligomeric form displays alterations in specific volume from what would be calculated as the sum of the specific volumes of each monomer, and thus this finding is consistent with recent work indicating a potential bulge in the PIV5 TM helix region upon oligomerization (22). Similar results were obtained with multiple preparations and with samples analyzed either immediately following purification or after prolonged storage. When TM-TM interactions were analyzed, the G508A mutant displayed a moderate increase in TM-TM association (Table 1; relative association constant as compared with the WT TM at 20,000 rpm of 3.29 ± 0.51), whereas the G508I or G508L mutations resulted in an even stronger TM-TM association (relative association constants at 20,000 rpm of 14.91 ± 3.56 and 6.46 ± 1.13, respectively). These results indicate that interactions involving the GXXXA motif are not the driving force behind the association of Hendra F TM domains as TM association increased when the glycine residue was removed. In addition, these results demonstrate that substitutions at Gly-508 can alter the stability or trafficking of the cleaved F1 form, likely through modulating TM-TM interactions, with modest effects observed with an alanine substitution and more dramatic effects observed upon introduction of larger, more hydrophobic amino acids (Ile and Leu).
TABLE 1.
Best fit model and relative association constants
SN-Hendra F TM constructs were purified and analyzed by sedimentation equilibrium analysis.
| Sample | 20,000 rpm |
25,000 rpm |
30,000 rpm |
|||
|---|---|---|---|---|---|---|
| Best fit model | Relative association constanta | Best fit model | Relative association constanta | Best fit model | Relative association constanta | |
| WT F TM | 1:3 | 1 | 1:3 | 0.54 ± 0.04 | 1:3 | 0.45 ± 0.02 |
| G508A | 1:3 | 3.29 ± 0.51 | 1:3 | 0.82 ± 0.07 | 1:3 | 0.71 ± 0.02 |
| G508I | 1:3 | 14.91 ± 3.56 | 1:3 | 2.56 ± 0.19 | 1:3 | 1.49 ± 0.11 |
| G508L | 1:3 | 6.46 ± 1.13 | 1:3 | 2.01 ± 0.14 | 1:3 | 1.71 ± 0.16 |
a Relative association constant as compared with WT F at 20,000 rpm, 8.38 ± 0.84 × 108 m−2.
Role of Noncharged Polar Residues in the Hendra F TM Domain
Noncharged polar residues can provide a driving force for association of TM domains (52, 53), with serine and threonine residues playing critical roles in some systems (54). The Hendra F TM domain contains four serine or threonine residues at positions Ser-493, Ser-501, Thr-511, and Ser-514, with two additional serine residues at positions 487 and 490 predicted to be at or near the TM-ectodomain border. To analyze the role of the TM Ser and Thr residues in folding and function of the F protein, individual alanine substitutions were created. Interestingly, although alanine replacements at Ser-501, Thr-511, or Ser-514 did not significantly impact cell surface expression or fusion, an S493A mutation, predicted to be at the TM-ectodomain border, gave surface expression at 70% of wild-type levels, but presented a dramatic loss in fusion promotion (Fig. 8, A and B), indicating that this mutation directly affects membrane fusion. A recent study of the PIV5 F protein has also implicated residues in the N-terminal end of the TM domain in fusion promotion (22). TM-TM interactions were not significantly altered when the S493A mutation was introduced into the SN-Hendra F TM construct, as shown by the preservation of a monomer-trimer fit with association constants similar to that of the wild-type protein. Thus, these data suggest that the reductions in fusion observed for the S493A mutation are not directly due to changes in TM-TM association. Although single polar residues may impact TM-TM association, cooperative interaction of several polar residues has been implicated in helix packing (55–58). To test this, double (T511A/S514A), triple (S501A/T511A/S514A), and quadruple (S493A/S501A/T511A/S514A) mutations were created and assessed for their effects on cell surface expression and fusion. Cell surface expression and fusion were moderately reduced for the T511A/S514A and S501A/T511A/S514A mutants, whereas the S493A/S501A/T511A/S514A mutant displayed greatly reduced cell surface expression, suggesting that at least one noncharged polar residue is needed for protein stability (Fig. 8A). Fusion promoted by S493A/S501A/T511A/S514A was similar to the low levels observed with the S493A single mutant, consistent with defects in fusion driven primarily by substitutions at Ser-493 (Fig. 8B). These data suggest that substitution of polar residues within the Hendra F TM domain can impact overall protein folding and stability, potentially by altering TM-TM interactions. Additionally, these data indicate that a single polar residue near the N terminus of the TM (S493A) is the only one that significantly modulates membrane fusion.
FIGURE 8.
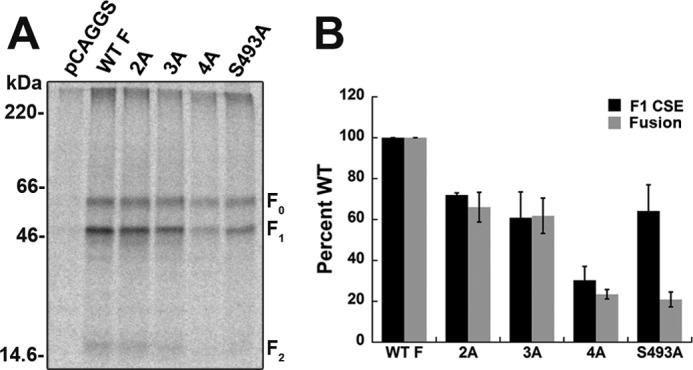
Mutations at Hendra F Ser-493 inhibit fusion. A, cell surface expression of transiently transfected wild type and Hendra F mutants in Vero cells following a 3-h metabolic label. 2A = T511A/S514A; 3A = S501A/T511A/S514A; and 4A = S493A/S501A/T511A/S514A. Proteins were analyzed via 15% SDS-PAGE and visualized using autoradiography. B, cell surface expression (CSE) and fusion for the wild type and TM mutants. F1 band quantitation via densitometry was normalized to wild-type levels plus or minus one standard deviation. Cell surface expression represents the average of three independent experiments. Fusion was analyzed using a reporter gene assay, n = 5–9; ± S.E.
DISCUSSION
Historically, the role of TM domains in viral fusion proteins has been difficult to address. The work presented here demonstrates that isolated TM domains from class I fusion proteins are present in a monomer-trimer equilibrium. Furthermore, our data indicate that TM-TM interactions contribute to the folding, stability, and fusion function of these important viral proteins. As similar interactions were observed using the TM domains from Hendra F, PIV5 F and HMPV F (Fig. 2), it is likely that trimeric TM-TM interactions are characteristic of F proteins throughout the paramyxovirus family. Because all class I viral fusion proteins, including influenza HA and Ebola GP, are trimeric molecules, TM-TM interactions may represent a critical and conserved structural feature of viral fusion proteins. Consistent with our findings, Bissonnette et al. (22) found evidence for TM-TM interactions using cysteine scanning of the PIV5 F protein, several groups have reported multiple oligomeric species of the influenza HA TM domain (59, 60), and a recent study confirmed TM oligomerization of the HIV env protein (32). Interestingly, a recent study of the PIV5 F protein, which also employed sedimentation equilibrium analysis, found evidence for a fusion peptide-TM domain interaction, but did not observe TM-TM association in dodecylphosphocholine micelles (48). However, our results with both synthesized PIV5 TM peptides (Fig. 4) and isolated Hendra TM peptides (Fig. 6) confirm trimeric association of the TM regions in the absence of the SN construct. The PIV5 peptide utilized by Donald et al. (48) included mutation of an internal cysteine residue in the PIV5 TM sequence to alanine and lacked the two C-terminal leucine residues present in our peptide, and these changes may account for the difference in findings.
The factors that drive TM-TM interactions remain poorly understood (30, 31), but GXXXG motifs or similar motifs with small (Gly, Ala, or Ser) residues spaced four apart are hypothesized to provide a framework to facilitate interactions of some TM helices. TM glycine residues are important for the fusion activity of the vesicular stomatitis virus G protein (13) and for the trafficking and function of the HIV env protein (16, 23, 49), potentially due to alterations in TM-TM interactions (32). In our studies, mutation of the Hendra F TM Gly-508 residue resulted in both an increase in the relative association of TM domains and a decrease in cell surface expression of the fusogenic F1 protein. An alanine substitution, yielding an AXXXA motif, resulted in modest effects, potentially due either to a requirement for backbone flexibility for optimal helix interactions or to steric effects. Although replacement of the G/A in these motifs with bulkier side chains often results in decreased association in dimeric systems, a significant increase in trimer association was noted when such substitutions were made in the Hendra F TM (Table 1). In addition, the G508I or G508L mutations in the context of the whole protein led to greatly decreased cell surface expression of the fusogenic F1 form (Fig. 7). An alteration in TM association driven by introduction of the bulky Ile or Leu side chain may result in changes to the transmembrane domain region and subsequent alterations to the ectodomain structure, thereby altering protein stability. Alternatively, these modifications to TM structure may affect trafficking through the endocytic pathway. Integrin TM domains, like viral fusion protein TM domains, must undergo changes in association for proper function, and their association has been shown to be facilitated by complementary small and large side chains on neighboring helices (62), consistent with the idea that bulky residues can drive TM helix interactions.
Polar residues can also promote TM-TM interactions (52–54) and are present at higher than average percentages in paramyxovirus F TM domains. Interestingly, only Hendra F Ser-493 is critical for the fusion process (Fig. 8), but no alterations were observed in TM-TM association when this residue was replaced with alanine. As Ser-493 is located near the N-terminal end of the TM segment, the fusion effects may reflect a critical role for side chain interactions with the environment at the lipid interface. No other significant folding or fusion defects were observed with replacement of one, two, or three TM serine or threonine residues (Fig. 8), although significant folding and fusion defects were observed when all four TM S/T residues (including S493A) were changed to alanine. Thus, cooperative interaction of several polar residues, previously implicated in helix packing (55–58), may regulate F protein TM interactions, but a series of residues spread throughout the TM domain would likely be involved. Recent work has demonstrated that dimerization of TM domains is driven by factors extending beyond recognizable sequence motifs (66). Our results suggest that paramyxovirus TM trimerization is also likely driven by multiple sequence and structural determinants, consistent with the apparent lack of conservation observed between different F protein TMs.
TM-TM interactions could potentially drive both F protein folding and fusion. One possibility is that these interactions stabilize the HRB coiled-coil in the prefusion form (Fig. 1B) (7). Support for this comes from peptide studies indicating that stable HRB interactions do not occur for the isolated domain (63) and from the requirement for trimerization domains in place of the PIV5 F and respiratory syncytial virus F TM domains to obtain prefusion structures (7, 33). Consistent with the hypothesis that TM-TM interactions stabilize the prefusion form, and particularly the HRB coiled-coil, we have recently reported that the addition of the HRB segment to the Hendra F TM domain results in destabilization of the TM trimer (64). A recent study also suggested a requirement for the herpes simplex virus gB fusion protein TM domain in stabilization of a prefusion structure (65). Combined, these data strongly support a critical role for the TM domain in maintaining prefusion stability and suggest that this function may extend beyond the paramyxoviruses.
TM-TM interactions could also influence triggering of membrane fusion. Interactions with the viral attachment protein are hypothesized to trigger fusion for most paramyxovirus F proteins (2, 3) except for HMPV F (67), and altered association of the measles attachment protein with the cleaved fusogenic F1 + F2 protein was observed when specific mutations were introduced into the F TM domain (17). Alterations in TM-TM association due to attachment protein interactions or alterations in the lipid environment could regulate the dissociation of the heptad repeat B coiled-coil needed to reach the final fusogenic structure (Fig. 1B). Information on factors that stabilize or destabilize F protein TM interactions may therefore provide critical new insight into the triggering process. TM domain β-branched residues such as valine or isoleucine have been implicated in backbone dynamics and fusogenicity of peptides (68) and the fusion function of the reovirus fusion-associated small transmembrane proteins (61). β-Branched residues are present throughout the paramyxovirus F protein TM domains, and we have recently demonstrated that mutation of these residues can alter membrane fusion without affecting surface expression (64); however, the effect of these mutations on TM interactions remains to be determined. The work presented here presents a new system for the study of TM mutations and interactions in viral fusion proteins, providing a foundation to begin dissecting the role of trimeric TM interactions in the folding, stability, and fusion activity of these important viral proteins.
Acknowledgments
We are grateful to Katie Routt for technical assistance and Dr. Karen Fleming for the kind gift of the pet11A-SN-GpA construct. We also thank the members of the Dutch laboratory and Dr. Trevor Creamer for critical reviews of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI051517 and U54 AI057157 (through the NIAID) from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense (to R. E. D.), R01GM070662 (to M. G. F.), and 2P20 RR020171 from the National Center for Research Resources (to R. E. D and M. G. F.).
- F
- fusion protein
- HMPV
- human metapneumovirus
- PIV 5
- parainfluenza virus 5
- TM
- transmembrane
- SN
- Staphylococcal nuclease
- HRA
- heptad repeat A
- HRB
- heptad repeat region
- C14SB
- 3-(N,N-dimethylmyristyl-ammonio)propanesulfonate
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Harrison S. C. (2008) Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lamb R. A., Parks G. D. (2007) Paramyxoviridae: the viruses and their replication. in Fields Virology (Knipe D. M., Howley P. M., eds), pp. 1449–1496, Lippincott, Williams and Wilkins [Google Scholar]
- 3. Smith E. C., Popa A., Chang A., Masante C., Dutch R. E. (2009) Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J. 276, 7217–7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Garten W., Hallenberger S., Ortmann D., Schäfer W., Vey M., Angliker H., Shaw E., Klenk H. D. (1994) Processing of viral glycoproteins by the subtilisin-like endoprotease furin and its inhibition by specific peptidylchloroalkylketones. Biochimie 76, 217–225 [DOI] [PubMed] [Google Scholar]
- 5. van den Hoogen B. G., de Jong J. C., Groen J., Kuiken T., de Groot R., Fouchier R. A., Osterhaus A. D. (2001) A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 7, 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pager C. T., Dutch R. E. (2005) Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79, 12714–12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yin H. S., Wen X., Paterson R. G., Lamb R. A., Jardetzky T. S. (2006) Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439, 38–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Welch B. D., Liu Y., Kors C. A., Leser G. P., Jardetzky T. S., Lamb R. A. (2012) Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein. Proc. Natl. Acad. Sci. U.S.A. 109, 16672–16677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen L., Gorman J. J., McKimm-Breschkin J., Lawrence L. J., Tulloch P. A., Smith B. J., Colman P. M., Lawrence M. C. (2001) The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure 9, 255–266 [DOI] [PubMed] [Google Scholar]
- 10. Colman P. M., Lawrence M. C. (2003) The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4, 309–319 [DOI] [PubMed] [Google Scholar]
- 11. Yin H. S., Paterson R. G., Wen X., Lamb R. A., Jardetzky T. S. (2005) Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc. Natl. Acad. Sci. U.S.A. 102, 9288–9293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Armstrong R. T., Kushnir A. S., White J. M. (2000) The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J. Cell Biol. 151, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cleverley D. Z., Lenard J. (1998) The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. U.S.A. 95, 3425–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin X., Derdeyn C. A., Blumenthal R., West J., Hunter E. (2003) Progressive truncations C terminal to the membrane-spanning domain of simian immunodeficiency virus Env reduce fusogenicity and increase concentration dependence of Env for fusion. J. Virol. 77, 7067–7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Melikyan G. B., Markosyan R. M., Roth M. G., Cohen F. S. (2000) A point mutation in the transmembrane domain of the hemagglutinin of influenza virus stabilizes a hemifusion intermediate that can transit to fusion. Mol. Biol. Cell 11, 3765–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miyauchi K., Curran R., Matthews E., Komano J., Hoshino T., Engelman D. M., Matsuda Z. (2006) Mutations of conserved glycine residues within the membrane-spanning domain of human immunodeficiency virus type 1 gp41 can inhibit membrane fusion and incorporation of Env onto virions. Jpn. J. Infect. Dis. 59, 77–84 [PubMed] [Google Scholar]
- 17. Mühlebach M. D., Leonard V. H., Cattaneo R. (2008) The measles virus fusion protein transmembrane region modulates availability of an active glycoprotein complex and fusion efficiency. J. Virol. 82, 11437–11445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taylor G. M., Sanders D. A. (1999) The role of the membrane-spanning domain sequence in glycoprotein-mediated membrane fusion. Mol. Biol. Cell 10, 2803–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kemble G. W., Danieli T., White J. M. (1994) Lipid-anchored influenza hemagglutinin promotes hemifusion, not complete fusion. Cell 76, 383–391 [DOI] [PubMed] [Google Scholar]
- 20. Li Z., Blissard G. W. (2008) Functional analysis of the transmembrane (TM) domain of the Autographa californica multicapsid nucleopolyhedrovirus GP64 protein: substitution of heterologous TM domains. J. Virol. 82, 3329–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Melikyan G. B., Brener S. A., Ok D. C., Cohen F. S. (1997) Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J. Cell Biol. 136, 995–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bissonnette M. L., Donald J. E., DeGrado W. F., Jardetzky T. S., Lamb R. A. (2009) Functional analysis of the transmembrane domain in paramyxovirus F protein-mediated membrane fusion. J. Mol. Biol. 386, 14–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shang L., Yue L., Hunter E. (2008) Role of the membrane-spanning domain of human immunodeficiency virus type 1 envelope glycoprotein in cell-cell fusion and virus infection. J. Virol. 82, 5417–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fritz R., Blazevic J., Taucher C., Pangerl K., Heinz F. X., Stiasny K. (2011) The unique transmembrane hairpin of flavivirus fusion protein e is essential for membrane fusion. J. Virol. 85, 4377–4385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Clancy E. K., Duncan R. (2009) Reovirus FAST protein transmembrane domains function in a modular, primary sequence-independent manner to mediate cell-cell membrane fusion. J. Virol. 83, 2941–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Melikyan G. B., Lin S., Roth M. G., Cohen F. S. (1999) Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion. Mol. Biol. Cell 10, 1821–1836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Odell D., Wanas E., Yan J., Ghosh H. P. (1997) Influence of membrane anchoring and cytoplasmic domains on the fusogenic activity of vesicular stomatitis virus glycoprotein G. J. Virol. 71, 7996–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schroth-Diez B., Ponimaskin E., Reverey H., Schmidt M. F., Herrmann A. (1998) Fusion activity of transmembrane and cytoplasmic domain chimeras of the influenza virus glycoprotein hemagglutinin. J. Virol. 72, 133–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilk T., Pfeiffer T., Bukovsky A., Moldenhauer G., Bosch V. (1996) Glycoprotein incorporation and HIV-1 infectivity despite exchange of the gp160 membrane-spanning domain. Virology 218, 269–274 [DOI] [PubMed] [Google Scholar]
- 30. Langosch D., Arkin I. T. (2009) Interaction and conformational dynamics of membrane-spanning protein helices. Protein Sci. 18, 1343–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Moore D. T., Berger B. W., DeGrado W. F. (2008) Protein-protein interactions in the membrane: sequence, structural, and biological motifs. Structure 16, 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reuven E. M., Dadon Y., Viard M., Manukovsky N., Blumenthal R., Shai Y. (2012) HIV-1 gp41 transmembrane domain interacts with the fusion peptide: implication in lipid mixing and inhibition of virus-cell fusion. Biochemistry 51, 2867–2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McLellan J. S., Chen M., Leung S., Graepel K. W., Du X., Yang Y., Zhou T., Baxa U., Yasuda E., Beaumont T., Kumar A., Modjarrad K., Zheng Z., Zhao M., Xia N., Kwong P. D., Graham B. S. (2013) Structure of RSV fusion glycoprotein trimer bound to a prefusion-specific neutralizing antibody. Science 340, 1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carter J. R., Pager C. T., Fowler S. D., Dutch R. E. (2005) Role of N-linked glycosylation of the Hendra virus fusion protein. J. Virol. 79, 7922–7925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gardner A. E., Dutch R. E. (2007) A conserved region in the F2 subunit of paramyxovirus fusion proteins is involved in fusion regulation. J. Virol. 81, 8303–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gardner A. E., Martin K. L., Dutch R. E. (2007) A conserved region between the heptad repeats of paramyxovirus fusion proteins is critical for proper F protein folding. Biochemistry 46, 5094–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fleming K. G., Ackerman A. L., Engelman D. M. (1997) The effect of point mutations on the free energy of transmembrane α-helix dimerization. J. Mol. Biol. 272, 266–275 [DOI] [PubMed] [Google Scholar]
- 38. Sulistijo E. S., Jaszewski T. M., MacKenzie K. R. (2003) Sequence-specific dimerization of the transmembrane domain of the “BH3-only” protein BNIP3 in membranes and detergent. J. Biol. Chem. 278, 51950–51956 [DOI] [PubMed] [Google Scholar]
- 39. Kroch A. E., Fleming K. G. (2006) Alternate interfaces may mediate homomeric and heteromeric assembly in the transmembrane domains of SNARE proteins. J. Mol. Biol. 357, 184–194 [DOI] [PubMed] [Google Scholar]
- 40. Burgess N. K., Stanley A. M., Fleming K. G. (2008) Determination of membrane protein molecular weights and association equilibrium constants using sedimentation equilibrium and sedimentation velocity. Methods Cell Biol. 84, 181–211 [DOI] [PubMed] [Google Scholar]
- 41. Cole J. L. (2004) Analysis of heterogeneous interactions. Methods Enzymol. 384, 212–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schowalter R. M., Chang A., Robach J. G., Buchholz U. J., Dutch R. E. (2009) Low-pH triggering of human metapneumovirus fusion: essential residues and importance in entry. J. Virol. 83, 1511–1522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pager C. T., Wurth M. A., Dutch R. E. (2004) Subcellular localization and calcium and pH requirements for proteolytic processing of the Hendra virus fusion protein. J. Virol. 78, 9154–9163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Buchholz U. J., Finke S., Conzelmann K. K. (1999) Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73, 251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Doura A. K., Fleming K. G. (2004) Complex interactions at the helix-helix interface stabilize the glycophorin A transmembrane dimer. J. Mol. Biol. 343, 1487–1497 [DOI] [PubMed] [Google Scholar]
- 46. Lemmon M. A., Flanagan J. M., Hunt J. F., Adair B. D., Bormann B. J., Dempsey C. E., Engelman D. M. (1992) Glycophorin A dimerization is driven by specific interactions between transmembrane α-helices. J. Biol. Chem. 267, 7683–7689 [PubMed] [Google Scholar]
- 47. Stanley A. M., Fleming K. G. (2005) The transmembrane domains of ErbB receptors do not dimerize strongly in micelles. J. Mol. Biol. 347, 759–772 [DOI] [PubMed] [Google Scholar]
- 48. Donald J. E., Zhang Y., Fiorin G., Carnevale V., Slochower D. R., Gai F., Klein M. L., DeGrado W. F. (2011) Transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proc. Natl. Acad. Sci. U.S.A. 108, 3958–3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Miyauchi K., Curran A. R., Long Y., Kondo N., Iwamoto A., Engelman D. M., Matsuda Z. (2010) The membrane-spanning domain of gp41 plays a critical role in intracellular trafficking of the HIV envelope protein. Retrovirology 7, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harrington W. F., Kegeles G. (1973) Pressure effects in ultracentrifugation of interacting systems. Methods Enzymol. 27, 306–345 [DOI] [PubMed] [Google Scholar]
- 51. Molina-Garcia A. D. (1999) Hydrostatic pressure in ultracentrifugation. in Analytical Ultracentrifugation V (Cölfen H., ed), pp. 57–61, Springer, Berlin/Heidelberg, Germany [Google Scholar]
- 52. Gratkowski H., Lear J. D., DeGrado W. F. (2001) Polar side chains drive the association of model transmembrane peptides. Proc. Natl. Acad. Sci. U.S.A. 98, 880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou F. X., Merianos H. J., Brunger A. T., Engelman D. M. (2001) Polar residues drive association of polyleucine transmembrane helices. Proc. Natl. Acad. Sci. U.S.A. 98, 2250–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dawson J. P., Weinger J. S., Engelman D. M. (2002) Motifs of serine and threonine can drive association of transmembrane helices. J. Mol. Biol. 316, 799–805 [DOI] [PubMed] [Google Scholar]
- 55. Adamian L., Jackups R., Jr., Binkowski T. A., Liang J. (2003) Higher-order interhelical spatial interactions in membrane proteins. J. Mol. Biol. 327, 251–272 [DOI] [PubMed] [Google Scholar]
- 56. Adamian L., Liang J. (2002) Interhelical hydrogen bonds and spatial motifs in membrane proteins: polar clamps and serine zippers. Proteins 47, 209–218 [DOI] [PubMed] [Google Scholar]
- 57. Curran A. R., Engelman D. M. (2003) Sequence motifs, polar interactions and conformational changes in helical membrane proteins. Curr. Opin. Struct. Biol. 13, 412–417 [DOI] [PubMed] [Google Scholar]
- 58. Senes A., Engel D. E., DeGrado W. F. (2004) Folding of helical membrane proteins: the role of polar, GxxxG-like and proline motifs. Curr. Opin. Struct. Biol. 14, 465–479 [DOI] [PubMed] [Google Scholar]
- 59. Chang D. K., Cheng S. F., Kantchev E. A., Lin C. H., Liu Y. T. (2008) Membrane interaction and structure of the transmembrane domain of influenza hemagglutinin and its fusion peptide complex. BMC Biol. 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tatulian S. A., Tamm L. K. (2000) Secondary structure, orientation, oligomerization, and lipid interactions of the transmembrane domain of influenza hemagglutinin. Biochemistry 39, 496–507 [DOI] [PubMed] [Google Scholar]
- 61. Clancy E. K., Duncan R. (2011) Helix-destabilizing, β-branched, and polar residues in the baboon reovirus p15 transmembrane domain influence the modularity of FAST proteins. J. Virol. 85, 4707–4719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Berger B. W., Kulp D. W., Span L. M., DeGrado J. L., Billings P. C., Senes A., Bennett J. S., DeGrado W. F. (2010) Consensus motif for integrin transmembrane helix association. Proc. Natl. Acad. Sci. U.S.A. 107, 703–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Joshi S. B., Dutch R. E., Lamb R. A. (1998) A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 248, 20–34 [DOI] [PubMed] [Google Scholar]
- 64. Smith E. C., Culler M. R., Hellman L. M., Fried M. G., Creamer T. P., Dutch R. E. (2012) Beyond anchoring: the expanding role of the Hendra virus fusion protein transmembrane domain in protein folding, stability, and function. J. Virol. 86, 3003–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Vitu E., Sharma S., Stampfer S. D., Heldwein E. E. (2013) Extensive mutagenesis of the HSV-1 gB ectodomain reveals remarkable stability of its postfusion form. J. Mol. Biol. 425, 2056–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li E., Wimley W. C., Hristova K. (2012) Transmembrane helix dimerization: beyond the search for sequence motifs. Biochim. Biophys. Acta 1818, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schowalter R. M., Smith S. E., Dutch R. E. (2006) Characterization of human metapneumovirus F protein-promoted membrane fusion: critical roles for proteolytic processing and low pH. J. Virol. 80, 10931–10941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Langosch D., Crane J. M., Brosig B., Hellwig A., Tamm L. K., Reed J. (2001) Peptide mimics of SNARE transmembrane segments drive membrane fusion depending on their conformational plasticity. J. Mol. Biol. 311, 709–721 [DOI] [PubMed] [Google Scholar]
- 69. Hayes D., Laue T., Philo J. (1995) Program Sednterp: sedimentation interpretation program, Alliance Protein Laboratories, Thousand Oaks, CA [Google Scholar]