Background: Lysosomes are required for autophagic degradation, which can be suppressed by lysosome inhibitors.
Results: Inhibition of lysosome function resulted in autophagy activation via down-regulation of MTORC1.
Conclusion: Lysosomes can affect autophagy initiation in addition to its role in autophagy degradation.
Significance: The finding expands lysosome function to include regulation of autophagy activation and indicates a dual effect of lysosome inhibitors in autophagy.
Keywords: Autophagy, Cell Biology, Lysosomes, mTOR, Signal Transduction
Abstract
Autophagy can be activated via MTORC1 down-regulation by amino acid deprivation and by certain chemicals such as rapamycin, torin, and niclosamide. Lysosome is the degrading machine for autophagy but has also been linked to MTORC1 activation through the Rag/RRAG GTPase pathway. This association raises the question of whether lysosome can be involved in the initiation of autophagy. Toward this end, we found that niclosamide, an MTORC1 inhibitor, was able to inhibit lysosome degradation and increase lysosomal permeability. Niclosamide was ineffective in inhibiting MTORC1 in cells expressing constitutively activated Rag proteins, suggesting that its inhibitory effects were targeted to the Rag-MTORC1 signaling system. This places niclosamide in the same category of bafilomycin A1 and concanamycin A, inhibitors of the vacuolar H+-ATPase, for its dependence on Rag GTPase in suppression of MTORC1. Surprisingly, classical lysosome inhibitors such as chloroquine, E64D, and pepstatin A were also able to inhibit MTORC1 in a Rag-dependent manner. These lysosome inhibitors were able to activate early autophagy events represented by ATG16L1 and ATG12 puncta formation. Our work established a link between the functional status of the lysosome in general to the Rag-MTORC1 signaling axis and autophagy activation. Thus, the lysosome is not only required for autophagic degradation but also affects autophagy activation. Lysosome inhibitors can have a dual effect in suppressing autophagy degradation and in initiating autophagy.
Introduction
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved mechanism by which cytoplasmic materials can be transported to and degraded in the lysosome. Autophagy serves to provide nutrients to starving cells or to remove superfluous subcellular organelles, aggregated proteins, or intracellular pathogens (1). As an important pathophysiological regulation mechanism during development, aging, and pathogenesis, autophagy can be induced or suppressed by a variety of signaling events. However, many of the signaling pathways have not been well defined.
In general, autophagy can be induced by MTOR complex 1 (MTORC1)2-dependent pathway or -independent pathway (2, 3). MTORC1 negatively regulates autophagic initiation, and many agents can thus induce autophagy by suppressing MTORC1 (2). A commonly used physiological autophagy stimulus is the deprivation of nutrients such as amino acids, which inactivates MTORC1. Rapamycin and Torin 1 are well defined small molecule chemicals that inhibit MTORC1 (4). Niclosamide is another chemical recently found to be able to induce autophagy and inhibit MTORC1 (5). The latter was attributed to the ability of niclosamide to cause cytoplasmic acidification by releasing protons from lysosomes (6).
The lysosome has recently been found to play a uniquely important role in MTORC1 activation (7). Activation of MTORC1 by amino acids depends on the Rag/RRAG family of GTPase, which is found on the lysosomal membrane (8, 9). The Rag proteins are composed of RagA, RagB, RagC, and RagD. The functionally equivalent RagA and RagB can form heterodimers with RagC or RagD, which are also functionally redundant. The heterodimers with the GTP-loaded RagA or RagB are the active form that can recruit MTORC1 to the lysosome where it can be activated by RHEB. Expression of a constitutively activated RagA mutant (RagAQ66L or RagAGTP/GTP) can activate MTORC1 independently of amino acids (8, 10).
Heterodimeric Rag proteins interact with another pentemeric complex called Regulator/LAMTOR, which is tethered to the lysosome membranes via one of the subunits, p18/LAMTOR1 (11, 12). Both complexes can interact with the vacuolar H+-ATPase (v-ATPase) on the lysosome (10). Under a nutrient-replete condition, amino acids accumulated in the lumen of the lysosome will weaken the interaction of v-ATPase with LAMTOR, thus activating its guanine nucleotide exchange factor activity toward RagA and RagB (12). Upon GTP loading to RagA/B, the interaction of Rag with LAMTOR complex is reduced, allowing the interaction of Rag heterodimers with MTORC1 and thus recruiting the latter to the lysosomal surface (11, 12).
It is thus possible to inhibit MTORC1 activity by interfering with the function or the interaction of v-ATPase, LAMTOR, and/or Rag as in the case of amino acid deprivation. In addition, v-ATPase inhibitors such as bafilomycin A1 (Baf) and concanomycin A (ConA) could clearly suppress amino acid-triggered MTORC1 activation (10). It was, however, not clear whether other types of lysosome inhibitors, i.e. those not directly targeting v-ATPase, could also cause MTORC1 inhibition. This question was critical to address whether the general lysosome functional status but, not just one of its specific functions, is required for MTORC1 function. Furthermore, whether lysosome inhibitors could in general activate autophagy because of their potential negative effects on MTORC1 would also need to be determined.
This study examined these issues. Our data indicate that there is a general connection between lysosome function and signaling through the Rag GTPase. Inhibition of lysosome function will negatively affect this signaling axis, leading to MTORC1 inhibition and autophagy activation. Thus, our data support the notion that lysosome can be mechanistically involved in autophagy activation, in addition to its traditional role in autophagic degradation.
EXPERIMENTAL PROCEDURES
Antibodies and Chemicals
Niclosamide (N3510), ammonia chloride (AC) (A9434), chloroquine (CQ) (C6628), ConA (C9705), acridine orange (AO) (A6014), and 3-methyladenine (3-MA) (M9281) were from Sigma-Aldrich. Baf (B1080) was from LC Laboratories. Rapamycin (R-1018), E64D (E-2030), and pepstatin A (P-1519) were from A. G. Scientific. Torin 1 (4247) was from Tocris Bioscience. Protease and phosphotase inhibitor mixture tablets (04693116001 and 04906845001) were from Roche Applied Science/Roche Diagnostics. Self-quenched bodipy-conjugated BSA (DQ-BSA Red) (D-12051) and LysoTracker Red (LTR) (L-7528) were from Invitrogen (Molecular Probes). Lipofectamine 2000 (11668019) was from Invitrogen. All chemicals were dissolved in PBS (CQ and AC) or in dimethyl sulfoxide (niclosamide, rapamycin, Torin1, E64D, and pepstatin A). The final concentrations of dimethyl sulfoxide in culture were between 0.05–0.2%, which had no effects on autophagy induction (data not shown).
Anti-cathepsin B/CSTB antibody (sc-13985) was from Santa Cruz Biotechnology; antibodies to cathepsin D/CSTD (2284), ATG12 (2010), MTOR (2983), phospho-MTOR (Ser-2481) (2974), p70 S6K1/RPS6KB1/(9202), phospho-p70 S6K1 (Thr-389) (9234), S6/RPS6 (2217), phospho-S6 (Ser-235/236) (2211), 4E-BP1/EIF4EBP1/(9644), and phospho-4E-BP1 (Thr-37/46) (9459) were from Cell Signaling Technology; antibodies to p62/SQSTM1 (PM045), LC3B/MAP1LC3B (PM036), and ATG16L1 (M150) were from MBL Intl.; antibodies to LAMP2 (ABL-93) was from the Developmental Studies Hybridoma Bank (University of Iowa). Secondary antibodies conjugated with Alexa Fluor 488 (A11001), Cy3 (111-225-144) or horseradish peroxidase (111-006-045 and 111-006-062) were from Invitrogen and Jackson ImmunoResearch Laboratories, respectively.
Cell Culture
Human embryonic kidney cells (293A), human cervical carcinoma cells (HeLa), human lung carcinoma cells (A549), and mouse embryonic fibroblast cells (MEFs) deficient in Atg5 (13) or expressing a knock-in RagA mutant (RagAQ66L) (14) were cultured in DMEM (Thermo Scientific, SH-3024301) supplemented with 10% (v/v) fetal bovine serum (Invitrogen, 10099-141) and standard supplements at 37 °C in a humidified air atmosphere with 5% (v/v) CO2.
Immunoassay
For immunoblot assay, 15 to 20 μg of lysates were used. Proteins were separated by SDS-PAGE and transferred to PVDF membranes (Millipore, ISEQ00010). Primary antibodies were used, followed by horseradish peroxidase-conjugated secondary antibodies. Specific proteins were detected using enhanced chemiluminescence Western blotting agent (Millipore, WBKLS0500), and the images were digitally acquired with a Kodak Image Station 4000 and the companion software (Carestream Health, Inc.).
For immunofluorescence staining, cells were grown on glass coverslips in 24-well plates and were fixed in 4% formaldehyde for 15 min. Cells were washed twice in PBS, permeabilized with 0.1% Triton X-100 for 15 min, followed by another wash in PBS, and blocked in PBS containing 2% BSA for 1 h. Primary and secondary antibodies in PBS containing 2% BSA were applied for overnight and 1 h, respectively. Cells were co-stained with Hoechst 33342 for the nucleus. Samples were washed in PBS containing 0.1% Tween 20 and mounted on glass slides. Fluorescence images were taken using a Nikon Eclipse TE200 epi-fluorescence microscope with NIS-Elements AR3.2 software. For manual quantification of the puncta formation at least three optical fields with at least 50 cells per experimental condition were analyzed. Data from repeated experiments are subjected to statistical analysis.
Long-lived Protein Degradation Assay
Long-lived protein degradation assay was carried out as described previously (15). Briefly, MEFs were cultured in DMEM in 24-well plates, l-[14C]-valine (PerkinElmer, NEC291EU050UC) was added to a final concentration of 0.2 μCi/ml to label intracellular proteins. Cells were incubated for 18 h before changing to fresh medium for another hour with 10% cold l-valine to deplete labeled short-lived proteins. The cells were then incubated in Earle's balanced salt solution or DMEM (plus 0.1% of BSA and 10 mm valine) with or without testing chemicals for an additional 6 or 16 h. The culture medium was recovered, from which the degraded long-lived proteins were measured via liquid scintillation.
Subcellular Fractionation
Following the indicated treatment, cells were suspended in hypotonic buffer (40 mm KCl, 5 mm MgCl2, 2 mm EGTA, 10 mm HEPES, pH 7.5) for 30 min on ice. Cells were homogenized by shearing through a 28.5-gauge needle 30 times. After a centrifugation at 1,000 × g for 10 min, the supernatant was further centrifuged at 12,000 × g for 10 min. The pellet, enriched for the lysosome, was further washed in an isotonic buffer (150 mm NaCl, 5 mm MgCl2, 2 mm EGTA, 10 mm HEPES, pH 7.5) and dissolved in a lysis buffer (1% Triton X-100, 150 mm NaCl, 50 mm Tris-HCl, pH 7.5) for further analysis.
Analysis of Lysosomal Acidity, Enzyme Activity, and Degradation Capacity
To examine the cellular acidic compartment, cells were incubated with LTR (50 nm) or AO (1 μg/ml) during the culture for 20 to 30 min before being examined by fluorescence microscopy.
To measure CSTB activity, 5 μg of fresh cell lysate and 5 μm substrate Z-Phe-Arg-7-amino-4-methylcoumarin (Enzo Life Sciences, BML-P139-0050) were mixed in assay buffer A (100 mm NaCl, 100 mm NaOAc, pH 5.5). Reactions were monitored with an Infinite M200Pro fluorescence spectrometer (Tecan, Morrisville, NC) at λex = 365 nm and λem = 440 nm. For CSTD/CSTE activity, 5 μg of fresh cell lysate and 5 μm substrate Mca-Gly-Lys-Pro-Ile-Leu-Phe-Arg-Leu-Lys(Dnp)-d-Arg-NH2 (Enzo Life Sciences, BML-P145-0001) were mixed in assay buffer B (100 mm NaCl, 100 mm NaOAc, pH 4.0). Reactions were monitored at λex = 328 nm and λem = 393 nm. The relative fluorescence unit at the first 5–30 min of the reaction, which represented the linear acceleration part, was recorded, which is then standardized to that of untreated samples as an indication of the relative change in enzyme activities.
To determine the lysosomal degradation capacity, cells were incubated with 10 μg/ml of DQ-BSA-Red for 1 h at 37 °C. Following the wash, new medium was added with or without the testing chemicals for another 4 to 6 h. Degradation capacity was measured by the red fluorescence signal released due to the degradation of DQ-BSA-Red. The number of red puncta per cell was quantified.
Statistical Analysis
All data are presented as the mean ± S.D. from at least three separate experiments. The p values were determined by Student's t test. p < 0.05 was considered significant.
RESULTS
Niclosamide Induced Autophagy by Suppressing MTOR Activity
A large number of small molecules have been found to regulate autophagy (3, 16–18). Many of them are known compounds with defined effects on other cellular functions.
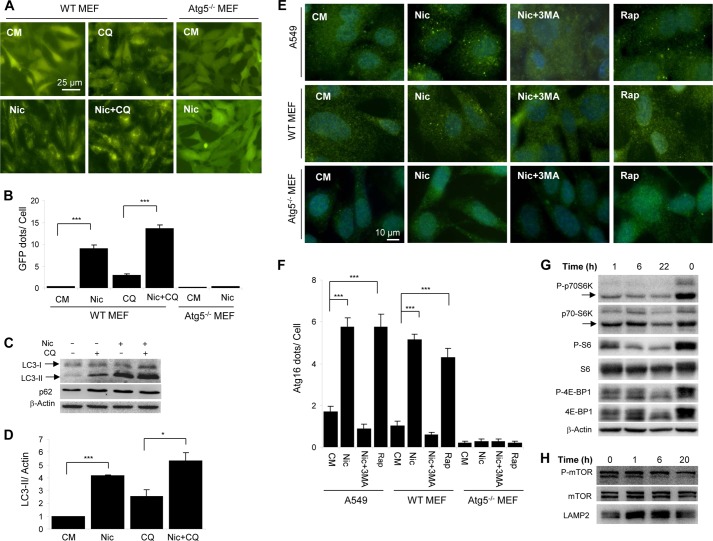
Niclosamide is a salicylanilide derivative that has been used as an Food and Drug Administration-approved anti-helminthic drug (19). Using a GFP-LC3-based high throughput screening we found that niclosamide significantly up-regulated GFP-LC3 puncta following an overnight incubation (20). In the subsequent confirmatory assay, we found that an incubation of 6 h was sufficient to lead to significant GFP-LC3 punctation (Fig. 1, A and B), which was completely abolished in the absence of Atg5, an upstream molecule for LC3 conjugation. The level of the lipidated form of LC3, LC3-II, was also significantly increased in the presence of niclosamide (Fig. 1, C and D). Notably, in both assays, the addition of a well defined lysosome inhibitor, CQ, further enhanced the level of GFP-LC3 puncta and LC3-II, suggesting that niclosamide could promote autophagic flux in a short period of treatment. The level of p62/SQSTM1 was not significantly changed in this short period of treatment by either niclosamide and/or CQ (Fig. 1C).
FIGURE 1.
Niclosamide induces autophagy and inhibition of MTORC1. A and B, wild type and Atg5-deficient MEFs expressing GFP-LC3 were treated with or without niclosamide (Nic; 10 μm) and CQ (40 μm) for 6 h and then assessed for GFP-LC3 puncta formation (A), which was quantified (B). C and D, as treated in A, MEFs were then lysed for immunoblot analysis (C). Densitometry was performed, and the ratio of LC3-II/β-actin was determined (D). E and F, wild type and Atg5-deficient MEFs or A549 cells were treated as indicated (Nic, 10 μm; 3-MA, 10 mm; Rap, 1 μm) for 6 h, followed by immunostaining using an anti-ATG16L1 antibody (E). ATG16L1 puncta were quantified (F). G and H, HeLa cells were treated with niclosamide (10 μm) for different times as indicated. The zero time point represents cells untreated and harvested at the end of the incubation period. Total lysates (G) or the lysosome-enriched heavy membrane fraction (H) was subjected to immunoblot analysis. For B, D, and F, values represent means ± S.D. from three independent experiments. ***, p < 0.001; *, p < 0.05. CM, complete medium.
To further determine that niclosamide initiated autophagy, we examined whether it could promote ATG16L1 punctation. ATG16L1 binds to the Atg5-Atg12 conjugate to form a complex that is necessary for the conjugation of LC3 to phosphatidylethanolamine. The ATG16L1-ATG5-ATG12 complex is present only in the precursor membrane of the autophagosome, i.e. the phagophore (21–23). Vesicular puncta positive for this complex are signs of early autophagosome biogenesis. We found that ATG16L1-positive puncta were significantly increased following a 6-h niclosamide treatment in two different cell lines: A549 lung carcinoma cell line and MEFs (Fig. 1, E and F). As a comparison, rapamycin, a well defined autophagy inducer, induced a similar level of ATG16L1 punctation. ATG16L1 punctation was not observed in the absence of Atg5 or in the presence of 3-MA, an inhibitor of the class III phosphatidylinositol 3-kinase, which is required for the ATG16L1 puncta to form. These findings demonstrated that niclosamide could activate autophagy through the canonical pathway.
Indeed, niclosamide treatment demonstrated a time-dependent dephosphorylation of p70S6 kinase, S6, 4E-BP1 (Fig. 1G), and MTOR (Fig. 1H). This inhibitory effect occurred in 1 h of treatment and reached the plateau by 22 h. Niclosamide could thus induce autophagy by inhibiting MTORC1.
Niclosamide Had a Dual Effect in Suppressing Autophagic Degradation
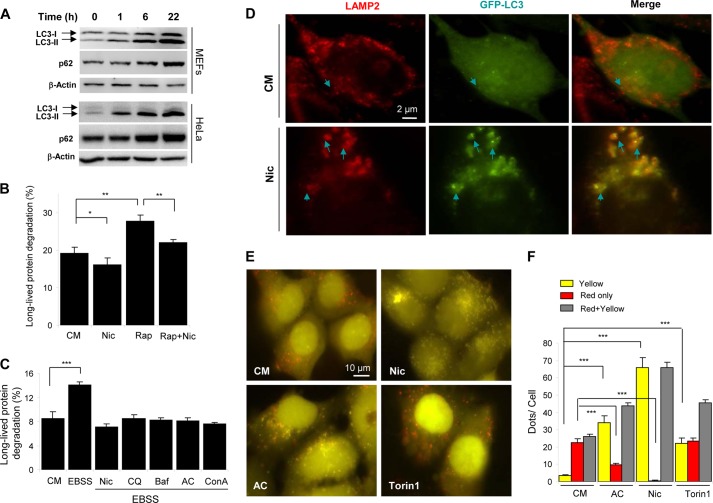
Consistently, the LC3-II level began to elevate in niclosamide-treated cells in 1 h and continued to rise through 22 h (Fig. 2A). Intriguingly, although by 6 h, autophagic flux was elevated (Fig. 1, A–D), the level of ATG16L1 puncta was high (Fig. 1, E and F), and the MTOR activity was suppressed (Fig. 1, G and H), the level of p62 was not decreased at 6 h and paradoxically elevated notably at a later time point of 22 h (Fig. 2A).
FIGURE 2.
Niclosamide inhibits autophagic degradation. A, MEFs or HeLa cells were treated with niclosamide (Nic; 10 μm) for 0–22 h as indicated, followed by immunoblot analysis for p62 and LC3. B and C, MEFs were incubated in complete medium (CM), or in Earle's balanced salt solution (EBSS) with or without niclosamide (10 μm), CQ (40 μm), Baf (0.5 μm), AC (40 mm) or ConA (2.5 μm) for 6 h (C), or incubated in complete medium (CM) with or without niclosamide (10 μm) and rapamycin (Rap; 1 μm) for 16 h (B). The long-lived protein degradation was measured. D, GFP-LC3-expressing MEFs were treated as indicated (Nic, 10 μm) for 6 h, followed by immunostaining with anti-LAMP2. Arrows indicated colocalized GFP-LC3 (green) and LAMP2 (red) puncta. E and F, HEK-293A cells expressing GFP-RFP-LC3 were treated as indicated (Nic, 10 μm; AC, 40 mm; or Torin1, 250 nm) for 6 h (E). Puncta showing both green and red fluorescence (indicated as yellow) or showing only the red fluorescence (indicated as red) were quantified (F). Values represent means ± S.D. from three independent experiments. ***, p < 0.001; **, p < 0.01; *, p < 0.05.
This pattern was different from that of starvation-induced autophagy in which LC3-II level quickly rose in 0.5 to 2 h, followed by a decline, whereas p62 level changed conversely (14, 24, 25). p62 is an adaptor molecule for selective autophagy, but its level is often elevated in autophagy inhibition or deficiency (23, 26). The elevation of p62 and LC3-II during niclosamide treatment at a later time point suggested that this chemical might have a dichotomous effect on autophagic degradation. Indeed, niclosamide treatment did not induce long term protein degradation in a 16-h treatment (Fig. 2B). In contrast, it suppressed long term protein degradation induced by rapamycin or starvation as effectively as some of the well defined lysosome inhibitors, such as CQ, Baf, ConA, and AC (Fig. 2, B and C).
Immunostaining assay further indicated that niclosamide-induced GFP-LC3 puncta were significantly overlapped with the lysosome signals, suggesting that many of the induced autophagosomes could be trapped in the lysosome and were unable to be degraded (Fig. 2D). We confirmed this notion with a RFP-GFP tandem LC3 construct, which has been well used for examining autophagic degradation.(27) If the lysosome successfully degrades autophagic cargos, more red fluorescence (RFP puncta) will be observed because the green fluorescence (green puncta) will be quenched in the acidic compartment. In the suppressive condition, both wavelengths of fluorescence will be retained, resulting in yellow signals in overlapped images.
Cells treated with a known lysosome inhibitor, AC, had a significantly decreased level of red-only puncta and a significantly elevated level of yellow puncta, suggesting that there was an arrest of the transition from the double-colored LC3 puncta to the red-only puncta, a sign of blockage of lysosome degradation. However, cells treated with a well defined autophagy inducer, torin1, exhibited no decrease in the red-only dots despite an increase in yellow puncta, suggesting that the elevation of the total LC3 puncta was not due to inhibition of autophagy degradation but the synthesis of new autophagosomes (double-colored). Niclosamide treatment resulted in a similar pattern as the AC treatment with a decrease of the red-only LC3 puncta. Taken together, these data clearly indicated that niclosamide was able to inhibit autolysosome degradation.
Niclosamide Inhibited Lysosomal Degradation
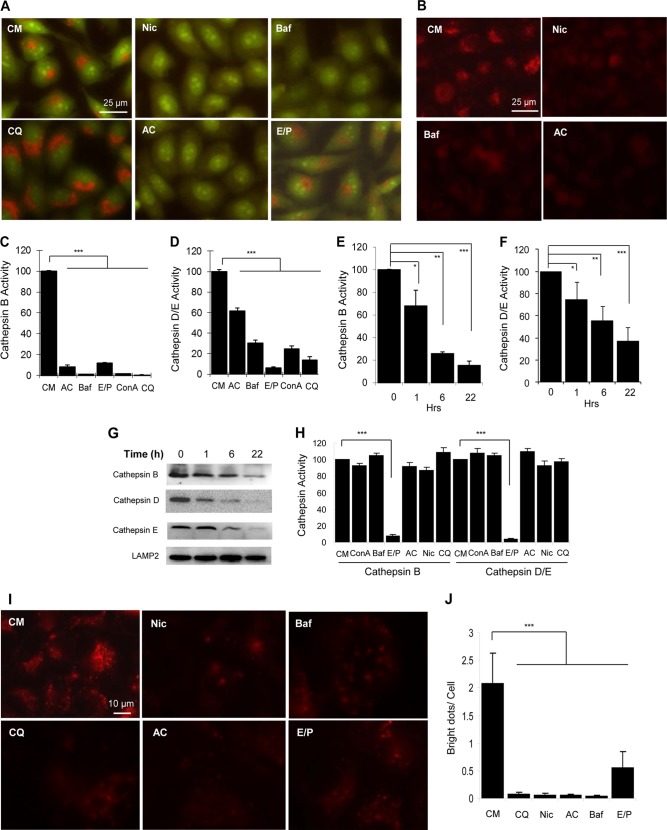
To examine how niclosamide might inhibit autophagic degradation, we treated cells with niclosamide and then stained them with AO (Fig. 3A) or LTR (Fig. 3B) to determine its effects on the acidity of the lysosomal compartment. AO is a lysosomotropic metachromatic fluorochrome, which emits strong red fluorescence inside the lysosomes where it is highly protonated, but weakly green fluorescence in the cytosol due to low protonation (28). Similarly, LTR accumulates in the lysosome in a pH-dependent manner. We found that similar to Baf and AC, niclosamide also elevated lysosomal pH, leading to the loss of both AO and LTR signals in the lysosomal compartment (Fig. 3, A and B). CQ and E64D/pepstatin A did not seem to significantly alter the pH gradient based on the staining of these dyes (Fig. 3A and data not shown).
FIGURE 3.
Niclosamide inhibits lysosomal degradation capacity. A and B, HeLa cells were treated with niclosamide (Nic; 10 μm), CQ (40 μm), Baf (0.5 μm), AC (40 mm), or E64D (25 μm) plus pepstatin A (50 μm) (E/P) for 6 h, followed by staining with AO (1 μg/ml, A) or LTR (50 ng/ml, B) for 30 min. C–G, HeLa cells were treated with various chemicals (40 mm AC; 0.5 μm Baf; 25 μm/50 μm E64D/pepstatin A; 2.5 μm ConA; 40 μm CQ) as indicated (C and D) for 6 h, or with niclosamide (10 μm) for 0 to 20 h (E–G). The lysosome-enriched cytosolic fraction was analyzed for cathepsin B (C and E) and cathepsin D/E (D and F) activities and for immunoblot analysis (G). Cathepsin activities were standardized to that of the untreated sample, which was set at the 100% level. H, whole cell lysates of untreated HeLa cells (4 μg) were mixed with the various chemicals at the concentrations indicated above for 1 h. The activities of cathepsin B and cathepsin D/E were measured. I and J, HeLa cells were preincubated with DQ-BSA(10 μg/ml) for 1 h and then treated as in A (I). The degraded products presented as red puncta, which were quantified (J). For C–F, H, and J, values represent means ± S.D. from three independent experiments. ***, p < 0.001; **, p < 0.01; *, p < 0.05. CM, complete medium.
We then examined the activity of three key lysosome enzymes CSTB, CSTD, and CSTE in the lysosome-enriched cellular fraction. As anticipated, the activities were suppressed by common lysosomal inhibitors, including Baf, ConA, CQ, AC, and E64D/pepstatin A, which work by different mechanisms (Fig. 3, C and D). CSTB seemed to be more sensitive to these inhibitors than CSTD. Notably, niclosamide caused a more rapid inactivation of CSTB than that CSTD and CSTE (Fig. 3, E and F). Immunoblot analysis indicated a reduction of all three cathepsins in the lysosome (Fig. 3G), which correlated with the decreased activity. CSTD and CSTE had the same substrate specificity, and the activity detected with the peptide substrate reflected that of both enzymes (Fig. 3F). This could explain why the detected residual activity was relatively higher than that of CSTB at each time point (Fig. 3E). The reduction in cathepsin level and activity by niclosamide could represent an increased lysosomal membrane permeabilization (28, 29). Niclosamide, as well as Baf, ConA, CQ, and ammonia chloride, had no direct inhibitive effects on CSTB or CSTD/CSTE when incubated directly with cell lysates (Fig. 3H), consistent with the fact that these chemicals affect the acidity of the lysosomes and therefore the enzymatic activities. However, protease inhibitors E64D and pepstatin A can directly bind to and inhibit the enzymes.
Taken together, these findings suggested that niclosamide inhibited lysosomal degradative function not by a direct enzymatic suppression, but likely by altering lysosomal permeability and the pH gradient. These effects can lead to a general reduction of the degradation capacity of the lysosome. Indeed, niclosamide inhibited the degradation of DQ-BSA-Red (Fig. 3, I and J), which was taken to the lysosome by endocytosis.
MTORC1 Activity, Lysosome Function, and Autophagy Induction Were Mechanistically Linked
Activation of MTOR by amino acids seems to depend on a normal lysosome function. Amino acids accumulated in the lumen of lysosome weaken the interaction of v-ATPase with LAMTOR/Regulator, thus activating its guanine nucleotide exchange factor activity toward RagA and RagB (10, 12). GTP-loaded RagA/B can then recruit MTORC1 to the lysosomal surface for activation (11, 12).
Inhibitors of the v-ATPase such as Baf, ConA, and SalA inactivate LAMTOR possibly by locking the interaction of v-ATPase and LAMTOR (12) and, thus, the activation of MTORC1 by amino acids (10, 12). This is true not only when amino acids are added to starved cells to reactivate MTORC1 (10), but also when amino acids are constantly present in the culture where MTORC1 is already activated (Fig. 4). Thus, we found that Baf and ConA inhibited MTORC1 activity even in the continuous presence of amino acids (Fig. 4A). Interestingly, AC, CQ, and E64D/pepstatin A, which suppress the lysosome function via other mechanisms, could also suppress MTORC1 activity in nutrients-replete condition (Fig. 4A), whereasthe dimethyl sulfoxide control had no effects. Together with the finding made with niclosamide (Figs. 1G and 3), our data indicated that the basal MTORC1 activity in the continuous presence of nutrients was also dependent on a normal lysosomal function and that lysosome inhibition by different means could all lead to the inactivation of MTORC1.
FIGURE 4.
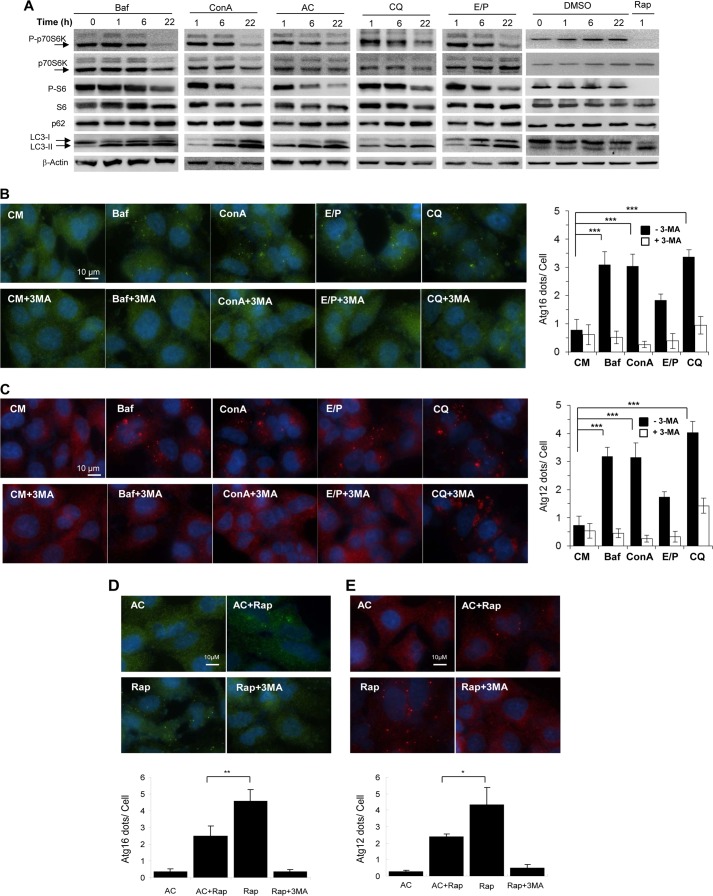
Lysosome inhibitors can inhibit MTORC1 and initiate autophagosome formation. A, MEFs were treated with Baf (0.5 μm), ConA (2.5 μm), AC (40 mm), CQ (40 μm), or E64D (25 μm) plus pepstatin A (50 μm) (E/P) for 0 to 22 h, followed by immunoblot analysis. B and C, A549 cells were treated as indicated with or without 3-MA (10 mm) for 6 h, followed by immunostaining with anti-ATG16L1 (B) or anti-ATG12 (C). D and E, A549 cells were treated as indicated (40 mm AC; 1 μm rapamycin; 10 mm 3-MA) for 6 h, followed by immunostaining with anti-ATG16L1 (D) or anti-ATG12 (E), which were quantified. For panels D and E, values represent means ± S.D. from three independent experiments. **, p < 0.01; *, p < 0.05. DMSO, dimethyl sulfoxide; CM, complete medium; Rap, rapamycin.
As in the case of niclosamide, the inhibition of MTORC1 by these well defined lysosome inhibitors was accompanied with autophagy activation. The elevation of LC3-II in the presence of these inhibitors (Fig. 4A) may not necessarily indicate an increase in autophagosome synthesis because this could be also due to a decrease in autophagosome degradation. We therefore examined two early autophagy activation markers, ATG16L1 and ATG12, by immunostaining assay. Increases in both ATG16L1 and ATG12 puncta formation were observed in cells treated with Baf, ConA, CQ, or E64D/pepstatin A, which were suppressed by 3-MA (Fig. 4, B and C). The induction effect was similar to that of niclosamide (Fig. 1, E and F) and that of rapamycin (Fig. 4, D and E). This observation indicated that lysosomal inhibitors were able to activate autophagy; despite that the overall autophagic function was likely suppressed by these agents at the degradation stage as indicated by the accumulation of p62 (Fig. 4A).
Surprisingly, AC did not induce ATG16L1 or ATG12 puncta formation and in fact suppressed such an induction by rapamycin (Fig. 4, D and E). This observation may suggest that this chemical had a separate inhibitory effect on autophagy induction.
Lysosome Inhibitors Suppressed MTORC1 through the Rag Signaling Module
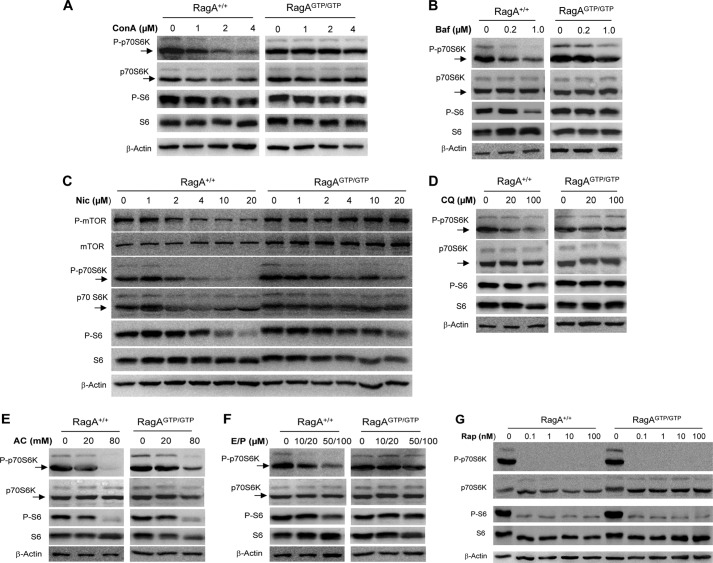
The ability of Rag molecules to recruit MTORC1 to the lysosome surface depends on the GTPase activity (8, 9), which is activated by its guanine nucleotide exchange factor, LAMTOR (10, 12). Notably, LAMTOR activation requires a normal v-ATPase function on the lysosome, which is facilitated by amino acids but suppressed by Baf or ConA. Thus, a constitutively activated RagA mutant (RagAQ66L) can activate MTORC1 independently of the status of LAMTOR or v-ATPase (10, 14). MEFs homozygously expressing this mutant RagA (RagAGTP/GTP) in place of the wild type counterpart have sustained MTORC1 activation regardless the amino acid and glucose levels (10, 14). The MTORC1 activity was rapidly suppressed by amino acid deprivation but was fully recovered following the inclusion of amino acids in the medium (10, 14). Such a recovery of MTORC1 activity was suppressed by v-ATPase inhibitors, ConA or Baf, in wild type MEFs, but not in the RagAGTP/GTP MEFs (Fig. 5, A and B), suggesting that this recovery was driven by v-ATPase-dependent Rag activity so that a constitutively activated RagA was able to resist the effect of ConA or Baf. Notably, we found that niclosamide, CQ, AC, and E64D/pepstatin acted similar to ConA and Baf, and suppressed the recovery of MTOR activity in a dose-dependent manner (Fig. 5, C–F). Furthermore, RagAGTP/GTP conferred a resistance to MTORC1 suppression by these chemicals. Slight reduction of p70S6K phosphorylation was still observed in the RagAGTP/GTP cells for some chemicals at highest doses tested (Fig. 5, C and E). This would be consistent with a similar observation in these cells following a prolonged starvation (14). Thus, it seems that RagAGTP/GTP, while supporting persistent mTORC1 activation, may not sustain such activation in the prolonged presence, or with high doses, of suppressors (starvations or chemicals).
FIGURE 5.
Inhibition of MTORC1 activity by lysosome inhibitors is regulated by Rag GTPases. Wild type (RagA+/+) and RagAGTP/GTP knock-in MEF cells were cultured in Earle's balanced salt solution with ConA (A), Baf (B), niclosamide (Nic; C), CQ (D), AC (E), E64D plus pepstatin A (E/P) (F) or rapamycin (G) at different concentrations as indicated for 1 h. Cells were then cultured in complete medium (CM) containing the same level of chemicals for an additional hour. Cell lysates were prepared and subjected to immunoblot assays with the indicated antibodies.
Notably, in the studies here, this resistance was specific to these lysosomal inhibitors because inhibition of MTORC1 by rapamycin was not protected by the constitutive activation of RagA (Fig. 5G). These observations thus indicated that niclosamide, CQ, AC, and E64D/pepstatin A all eventually inhibited MTORC1 activity through disturbing the lysosomal v-ATPase-LAMTOR-Rag signaling system; despite that their primary targets could be different.
DISCUSSION
Niclosamide Has Dual Effects on Autophagy Induction and Degradation
Niclosamide is an Food and Drug Administration-approved antihelminthic agent, and it has also been reported to possess antitumor activity (30). We (20) and others (5) had found that niclosamide was able to increase GFP-LC3 puncta in high throughput screening assays. This activity was considered to be related to its ability to inhibit MTORC1 but not MTORC2 (5). The present study has confirmed that niclosamide can down-regulate MTORC1 activity. Evidence of increased autophagy activation by niclosamide also includes the dependence of GFP-LC3 puncta on ATG5, and the formation of ATG16L1 puncta, which are also dependent on Atg5. ATG16L1 puncta formation is a well established indicator for autophagosome biogenesis at the early stage (21–23). ATG16L1 binds to the ATG12-ATG5 conjugate to form a high molecular weight complex, guiding the ATG12-ATG5 conjugate to the correct phagophore location where ATG5-ATG12 can act similar to an E3-like enzyme to facilitate LC3 conjugation to phosphatidylethanolamine (31–33).
In contrast to the above observations, we have also found that niclosamide can reduce autophagy degradation capacity. Although autophagy flux was increased in early time points (∼4–6 h, Fig. 1), it was reduced at later time points as indicated by the accumulation of p62 and the reduced long-lived protein degradation (∼16–22 h, Fig. 2). LC3 accumulation through a prolonged stimulation by niclosamide could thus be due to increased autophagy activation as well as reduced autophagy degradation.
Niclosamide does not seem to affect the fusion between the autophagosome and the lysosome because the GFP-LC3 puncta are well colocalized with LAMP2 positive compartments (Fig. 2D). The reduced lysosome degradation capacity is not exclusive to the autophagy process as the degradation of both GFP-RFP-LC3 and DQ-BSA was suppressed. This general lysosomal insufficiency seems to be related to a reduced acidity of the lysosome and a reduced protease activity (Fig. 3). We did not find that niclosamide was able to directly inhibit the enzymatic activity of CSTB or CSTD/CSTE. We found that these proteases were reduced in the lysosome in a time-dependent manner, as suggested by both the enzymatic activity and the protein level (Fig. 3). Notably, a previous study had also shown that niclosamide caused dispersion of protons from the lysosomes to the cytosol, leading to cytosolic acidification (6), which would be consistent with a loss of lysosomal acidity shown here. Changes in both lysosomal pH and levels of cathepsins suggest that niclosamide can promote lysosomal membrane permeabilization, rather than simply acting as a protonophore. Regardless the mechanism, lysosomal functions are likely compromised by niclosamide.
The two effects of niclosamide on autophagy via MTORC1 inhibition and on the lysosome function would likely be mechanistically linked. Niclosamide was able to suppress mTORC1 in TSC2-deficient cells where MTORC1 activity was elevated (5), suggesting that it acted on a different pathway. In addition, it had been proposed that the increased cytosolic acidification was responsible for MTORC1 inhibition although how this could be achieved is not known (6). Our data reported here would support an alternative model that MTORC1 inhibition is caused by lysosomal dysfunction, which affects the v-ATPase-LAMTOR-RAG signaling pathway. This hypothesis is further supported by studies using several other well defined lysosome inhibitors.
Lysosome Function and the v-ATPase-LAMTOR-RAG-MTORC1 Signaling Axis
Inhibitors of lysosomal degradation can reduce lysosomal acidity such as the v-ATPase inhibitors, Baf and ConA, and the alkaline agents, AC and CQ. They may also directly inhibit protease activities such as the cysteine protease inhibitor, E64D, and the aspartic protease inhibitor, pepstatin A. Interestingly CQ does not change the staining pattern of AO as much as the v-ATPase inhibitors or AC does (Fig. 3). This had been noted in another study (34), and we had used up to 100 μm CQ with no obvious differences in AO staining (data not shown). Nevertheless, CQ does alter lysosomal pH (25), which would be responsible for its inhibitory effect on proteolytic activities as it has no direct effect on enzymes (Fig. 3).
Baf and ConA have been found to inhibit amino acid-triggered reactivation of MTORC1 immediately after starvation (10). We found that these two chemicals, similar to niclosamide, also inhibited basal MTORC1 activity under constant nutrient-replete condition. Surprisingly, other types of lysosome inhibitors, CQ, AC, and E64D/pepstatin A, all can inhibit MTORC1 activity at the basal level, despite their different inhibitory mechanisms. This new finding thus suggests that the function of lysosome, in general, has important effects on MTORC1 activity. Disturbance of lysosomal function may ultimately affect v-ATPase or other molecules that are critical to MTORC1 activation.
It is now clear that Baf and ConA inhibit MTORC1 activity via their negative effects directly on v-ATPase (10), which interacts with LAMTOR, the guanine nucleotide exchange factor for the Rag GTPase (11, 12). Activated Rag recruits MTORC1 to the surface of the lysosome, where MTOR could be further activated by RHEB (8, 9). The inhibitory effect of Baf and ConA could thus be antagonized by a constitutively activated RagA mutant (10, 12), whose activity is no longer dependent on the functional v-ATPase and/or LAMTOR, which is suppressible by these two chemicals (8, 9). Remarkably, we found that cells expressing the constitutively activated RagAGTP/GTP were also resistant to the inhibitory effects of niclosamide CQ, AC, and E64D/pepstatin on MTORC1 activity; despite that, these lysosome inhibitors do not seem to possess a direct anti-v-ATPase activity or even to affect the pH of the lysosome. Inhibition of MTORC1 by non-lysosome inhibitors such as rapamycin cannot be reversed by the constitutively active mutant of RagA, indicating that the rescuing effect of this mutant is specific to the lysosome-derived signals.
One plausible explanation is that the normal function of LAMTOR is generally dependent on a normal lysosome function. Disturbance of lysosome integrity may affect the LAMTOR function due to altered v-ATPase activity. Alternatively, the reduced lysosome functionality affects protein degradation and/or amino acids transport into the lysosome, and lysosomal permeabilization could cause amino acid leakage to the cytosol, which all lead to a lower level of intraluminal amino acid level, which would impair the activation of the LAMTOR through v-ATPase. Indeed, a transient lysosomal permeability with Streptolysin O had been shown to suppress the recruitment of MTORC1 to Rag complex (10). Thus, in either scenario, the negative impact can affect MTORC1, as its activation depends on the lysosome through the LAMTOR-RAG complex. Our studies indicate that lysosomal functionality, in general, is closely linked to the LAMTOR-RAG-MTORC1 signaling axis and has significant impact on the MTORC1 activity.
Autophagy Activation Caused by Lysosome Inhibition Is Part of the Lysosome-MTOR-autophagy Dynamics
One of the interesting outcomes of MTORC1 being down-regulated by the lysosome inhibitors is the paradoxical activation of autophagy. Except for ammonia chloride, all of the inhibitors examined in this study could trigger ATG16L1 and ATG12 puncta formation, two of the markers for early autophagosome biogenesis. The fact that the formation of these puncta was blocked by the class III PI3K inhibitor 3-MA indicates that the process was specifically initiated in response to autophagy initiation. Ammonia chloride was not found to be able to induce the early change of ATG16L1 and ATG12. It is possible that this chemical may have additional effects that could prevent these changes despite that it can affect MTORC1 activity. Indeed, ammonia chloride can suppress ATG12 and ATG16L1 punctation induced by rapamycin (Fig. 4, D and E). Additional experiments will be required to further identify the mechanisms involved.
Lysosomes can play a general role in preventing autophagy via supporting MTORC1 activity. Cells expressing constitutively activated RagA (RagAQ66L) are resistant to amino acid or glucose deprivation-induced MTORC1 suppression (10, 14). These cells could not mount an autophagy response upon nutrient deprivation. In addition, mice expressing RagAQ66L died shortly after birth due to failure to activate autophagy as the result of sustained MTORC1 activation (14). Neonatal autophagy is essential to sustain blood glucose level for survival (13, 14, 35).
There is increasing evidence indicating that the relationship among lysosome, MTORC1, and autophagy are intertwined in multiple ways. A successful autophagy process requires functional lysosomes. Although MTORC1 inhibits autophagy activation, its own activation depends on lysosomes both physically and functionally as discussed above. Intracellular lysosome positioning could affect MTORC1 activation and thus autophagosome formation (36). Furthermore, TFEB, a master gene for lysosomal biogenesis, can also promote autophagy (37), but MTORC1 can phosphorylate TFEB to inhibit its translocation to the nucleus, thus preventing it from promoting lysosome biogenesis and autophagy (38–41) Consistently, suppressing MTORC1 could liberate TFEB function for lysosomal activation, which is also facilitated by the concomitantly activated autophagy process and autophagosome-lysosome fusion (42). However, during the process of autophagy, MTORC1 can promote lysosome reformation to prevent lysosome depletion (43). These observations indicate that there are reciprocal regulations involving both negative and positive impacts among these key functions. In fact, it had been hypothesized that lysosome inhibitors might trigger autophagy (44).
Autophagy triggered by reduced lysosome function via MTORC1 down-regulation may constitute a feedback mechanism to raise autophagy capability in response to a reduced degradation output. This mechanism may help to sustain nutrient supplies derived from lysosome degradation. However, it could also pose a pathological concern if autophagosomes are overly accumulated in the absence of sufficient degradation capacity, which would likely cause “autophagy stress” that contributes to disease progression (45). Another implication of the present findings is that caution should be exercised when lysosome inhibitors are used in an autophagy-related research or application. It is prudent that such inhibitors are to be used in transient fashion so that their proautophagy effect would not become apparent to confound the interpretation of the finding. Perhaps more attention should be paid to the use of lysosome inhibitors in vivo so the paradoxical effects on autophagy could be recognized and dissected. Thus, the link from lysosome function to autophagy initiation can bear both a pathophysiological significance and a therapeutic/pharmacological implication.
Acknowledgments
We thank D. M. Sabatini (Whitehead Institute for Biomedical Research, Massachusetts Institute of Technology) for RagAGTP/GTP MEFs and N. Mizushima (Tokyo Medical and Dental University) for Atg5−/− MEFs.
This work was supported by NCI and NIMH, National Institutes of Health Grants R03MH083154, R01CA111456, and R01CA 83817 (to X.-M. Y.). This work used the UPCI Chemical Biology facility that is supported in part by NCI, National Institutes of Health Grant P30CA047904.
- MTORC1
- mammalian target of rapamycin complex 1
- AC
- ammonia chloride
- AO
- acridine orange
- Baf
- bafilomycin A1
- ConA
- concanamycin A
- CQ
- chloroquine
- CSTB
- cathepsin B
- CSTD
- cathepsin D
- CSTE
- cathepsin E
- E/P
- E64D plus pepstatin A
- LTR
- LysoTracker Red
- MEF
- mouse embryonic fibroblast cell
- Nic
- niclosamide
- Rap
- rapamycin
- 3-MA
- 3-methyladenine
- v-ATPase
- vacuolar H+-ATPase.
REFERENCES
- 1. Yang Z., Klionsky D. J. (2010) Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Jung C. H., Ro S. H., Cao J., Otto N. M., Kim D. H. (2010) mTOR regulation of autophagy. FEBS Lett. 584, 1287–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fleming A., Noda T., Yoshimori T., Rubinsztein D. C. (2011) Chemical modulators of autophagy as biological probes and potential therapeutics. Nat. Chem. Biol. 7, 9–17 [DOI] [PubMed] [Google Scholar]
- 4. Yip C. K., Murata K., Walz T., Sabatini D. M., Kang S. A. (2010) Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell 38, 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Balgi A. D., Fonseca B. D., Donohue E., Tsang T. C., Lajoie P., Proud C. G., Nabi I. R., Roberge M. (2009) Screen for chemical modulators of autophagy reveals novel therapeutic inhibitors of mTORC1 signaling. PLoS One 4, e7124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fonseca B. D., Diering G. H., Bidinosti M. A., Dalal K., Alain T., Balgi A. D., Forestieri R., Nodwell M., Rajadurai C. V., Gunaratnam C., Tee A. R., Duong F., Andersen R. J., Orlowski J., Numata M., Sonenberg N., Roberge M. (2012) Structure-activity analysis of niclosamide reveals potential role for cytoplasmic pH in control of mammalian target of rapamycin complex 1 (mTORC1) signaling. J. Biol. Chem. 287, 17530–17545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jewell J. L., Russell R. C., Guan K. L. (2013) Amino acid signalling upstream of mTOR. Nat. Rev. Mol. Cell Biol. 14, 133–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008) The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. L. (2008) Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10, 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zoncu R., Bar-Peled L., Efeyan A., Wang S., Sancak Y., Sabatini D. M. (2011) mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H+-ATPase. Science 334, 678–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bar-Peled L., Schweitzer L. D., Zoncu R., Sabatini D. M. (2012) Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., Mizushima N. (2004) The role of autophagy during the early neonatal starvation period. Nature 432, 1032–1036 [DOI] [PubMed] [Google Scholar]
- 14. Efeyan A., Zoncu R., Chang S., Gumper I., Snitkin H., Wolfson R. L., Kirak O., Sabatini D. D., Sabatini D. M. (2013) Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature 493, 679–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ding W. X., Li M., Chen X., Ni H. M., Lin C. W., Gao W., Lu B., Stolz D. B., Clemens D. L., Yin X. M. (2010) Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology 139, 1740–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cho Y. S., Kwon H. J. (2010) Control of autophagy with small molecules. Arch. Pharm. Res. 33, 1881–1889 [DOI] [PubMed] [Google Scholar]
- 17. Zhang L., Yu J., Pan H., Hu P., Hao Y., Cai W., Zhu H., Yu A. D., Xie X., Ma D., Yuan J. (2007) Small molecule regulators of autophagy identified by an image-based high-throughput screen. Proc. Natl. Acad. Sci. U.S.A. 104, 19023–19028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baek K. H., Park J., Shin I. (2012) Autophagy-regulating small molecules and their therapeutic applications. Chem. Soc. Rev. 41, 3245–3263 [DOI] [PubMed] [Google Scholar]
- 19. Ditzel J., Schwartz M. (1967) Worm cure without tears. The effect of niclosamide on taeniasis saginata in man. Acta Med. Scand. 182, 663–664 [PubMed] [Google Scholar]
- 20. Yin X.-M., Vogt A. (2010) PubChem. BioAssay 463193 [Google Scholar]
- 21. Mizushima N., Kuma A., Kobayashi Y., Yamamoto A., Matsubae M., Takao T., Natsume T., Ohsumi Y., Yoshimori T. (2003) Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J. Cell Sci. 116, 1679–1688 [DOI] [PubMed] [Google Scholar]
- 22. Matsushita M., Suzuki N. N., Obara K., Fujioka Y., Ohsumi Y., Inagaki F. (2007) Structure of Atg5.Atg16, a complex essential for autophagy. J. Biol. Chem. 282, 6763–6772 [DOI] [PubMed] [Google Scholar]
- 23. Itakura E., Mizushima N. (2010) Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 6, 764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mizushima N., Yoshimori T. (2007) How to interpret LC3 immunoblotting. Autophagy 3, 542–545 [DOI] [PubMed] [Google Scholar]
- 25. Ni H. M., Bockus A., Wozniak A. L., Jones K., Weinman S., Yin X. M., Ding W. X. (2011) Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy 7, 188–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirkin V., McEwan D. G., Novak I., Dikic I. (2009) A role for ubiquitin in selective autophagy. Mol. Cell 34, 259–269 [DOI] [PubMed] [Google Scholar]
- 27. Kimura S., Noda T., Yoshimori T. (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3, 452–460 [DOI] [PubMed] [Google Scholar]
- 28. Boya P., Kroemer G. (2008) Lysosomal membrane permeabilization in cell death. Oncogene 27, 6434–6451 [DOI] [PubMed] [Google Scholar]
- 29. Boya P. (2012) Lysosomal function and dysfunction: mechanism and disease. Antioxid. Redox. Signal 17, 766–774 [DOI] [PubMed] [Google Scholar]
- 30. Pan J. X., Ding K., Wang C. Y. (2012) Niclosamide, an old antihelminthic agent, demonstrates antitumor activity by blocking multiple signaling pathways of cancer stem cells. Chin. J. Cancer 31, 178–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hanada T., Noda N. N., Satomi Y., Ichimura Y., Fujioka Y., Takao T., Inagaki F., Ohsumi Y. (2007) The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. J. Biol. Chem. 282, 37298–37302 [DOI] [PubMed] [Google Scholar]
- 32. Fujioka Y., Noda N. N., Nakatogawa H., Ohsumi Y., Inagaki F. (2010) Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J. Biol. Chem. 285, 1508–1515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sakoh-Nakatogawa M., Matoba K., Asai E., Kirisako H., Ishii J., Noda N. N., Inagaki F., Nakatogawa H., Ohsumi Y. (2013) Atg12-Atg5 conjugate enhances E2 activity of Atg3 by rearranging its catalytic site. Nat. Struct. Mol. Biol. 20, 433–439 [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y., Yang N. D., Zhou F., Shen T., Duan T., Zhou J., Shi Y., Zhu X. Q., Shen H. M. (2012) (-)-Epigallocatechin-3-gallate induces non-apoptotic cell death in human cancer cells via ROS-mediated lysosomal membrane permeabilization. PLoS One 7, e46749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., Kominami E., Tanaka K., Chiba T. (2005) Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Korolchuk V. I., Saiki S., Lichtenberg M., Siddiqi F. H., Roberts E. A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F. M., O'Kane C. J., Deretic V., Rubinsztein D. C. (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D. C., Ballabio A. (2011) TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peña-Llopis S., Vega-Rubin-de-Celis S., Schwartz J. C., Wolff N. C., Tran T. A., Zou L., Xie X. J., Corey D. R., Brugarolas J. (2011) Regulation of TFEB and V-ATPases by mTORC1. EMBO J. 30, 3242–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Settembre C., Zoncu R., Medina D. L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M. C., Facchinetti V., Sabatini D. M., Ballabio A. (2012) A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Martina J. A., Chen Y., Gucek M., Puertollano R. (2012) MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy 8, 903–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roczniak-Ferguson A., Petit C. S., Froehlich F., Qian S., Ky J., Angarola B., Walther T. C., Ferguson S. M. (2012) The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci. Signal 5, ra42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou J., Tan S. H., Nicolas V., Bauvy C., Yang N. D., Zhang J., Xue Y., Codogno P., Shen H. M. (2013) Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res. 23, 508–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yu L., McPhee C. K., Zheng L., Mardones G. A., Rong Y., Peng J., Mi N., Zhao Y., Liu Z., Wan F., Hailey D. W., Oorschot V., Klumperman J., Baehrecke E. H., Lenardo M. J. (2010) Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465, 942–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Juhász G. (2012) Interpretation of bafilomycin, pH neutralizing or protease inhibitor treatments in autophagic flux experiments: novel considerations. Autophagy 8, 1875–1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chu C. T. (2006) Autophagic stress in neuronal injury and disease. J. Neuropathol. Exp. Neurol. 65, 423–432 [DOI] [PMC free article] [PubMed] [Google Scholar]