Background: Glucokinase activity is regulated on the posttranslational level in pancreatic beta cells.
Results: Searching a pancreatic islet yeast two-hybrid library, the ubiquitin-like domain of midnolin was identified to bind and inhibit glucokinase at low glucose, thereby reducing insulin secretion.
Conclusion: Midnolin is a new glucokinase interaction partner.
Significance: This is the first report demonstrating midnolin protein expression in pancreatic beta cells.
Keywords: Fluorescence Resonance Energy Transfer (FRET), Glucokinase, Insulin Secretion, Insulin Synthesis, Pancreatic Islets, Parkin, Ubiquitin, Midnolin, Two-hybrid System, Ubiquitin-like Domain
Abstract
Glucokinase acts as a glucose sensor in pancreatic beta cells. Its posttranslational regulation is important but not yet fully understood. Therefore, a pancreatic islet yeast two-hybrid library was produced and searched for glucokinase-binding proteins. A protein sequence containing a full-length ubiquitin-like domain was identified to interact with glucokinase. Mammalian two-hybrid and fluorescence resonance energy transfer analyses confirmed the interaction between glucokinase and the ubiquitin-like domain in insulin-secreting MIN6 cells and revealed the highest binding affinity at low glucose. Overexpression of parkin, an ubiquitin E3 ligase exhibiting an ubiquitin-like domain with high homology to the identified, diminished insulin secretion in MIN6 cells but had only some effect on glucokinase activity. Overexpression of the elucidated ubiquitin-like domain or midnolin, containing exactly this ubiquitin-like domain, significantly reduced both intrinsic glucokinase activity and glucose-induced insulin secretion. Midnolin has been to date classified as a nucleolar protein regulating mouse development. However, we could not confirm localization of midnolin in nucleoli. Fluorescence microscopy analyses revealed localization of midnolin in nucleus and cytoplasm and co-localization with glucokinase in pancreatic beta cells. In addition we could show that midnolin gene expression in pancreatic islets is up-regulated at low glucose and that the midnolin protein is highly expressed in pancreatic beta cells and also in liver, muscle, and brain of the adult mouse and cell lines of human and rat origin. Thus, the results of our study suggest that midnolin plays a role in cellular signaling of adult tissues and regulates glucokinase enzyme activity in pancreatic beta cells.
Introduction
Expression of the glucose phosphorylating enzyme glucokinase in pancreatic beta cells, liver, and neuroendocrine cells is tissue-specifically regulated by two distinct promoters in a single gene (1–3). In pancreatic beta cells glucokinase catalyzes the rate-limiting step for glucose-induced insulin secretion and in liver glucokinase regulates glucose uptake (3–10). Complete knockdown of glucokinase in mice is lethal, and animals heterozygous for the gene knock-out develop diabetes (11). This emphasizes the pivotal role of glucokinase as a glucose sensor maintaining blood glucose homeostasis (3–10).
Hormones stimulate glucokinase regulation on the posttranslational level (12–14). However, in pancreatic beta cells in particular, glucose regulates glucokinase protein content and activity (15, 16). Glucose induces large-scale disorder-order transitions shifting the glucokinase protein to a closed conformation with high phosphorylating activity (17–20). In addition, ubiquitination of glucokinase, and the resulting degradation by the proteasome are relevant quality control steps ensuring cellular glucokinase protein content with high phosphorylating capacity (21–24).
Finally, specific protein interactions modulate the phosphorylating activity of glucokinase in a tissue-specific manner. In liver the glucokinase regulatory protein inhibits glucokinase at low glucose and in addition shuttles the protein to the nucleus (25–28). Both in liver and pancreatic beta cells the bifunctional enzyme phosphofructo-2-kinase/fructose-2,6-bisphosphatase activates glucokinase at high glucose (4, 29–32). Thus, these soluble regulators attenuate or potentiate glucokinase activity, respectively, in a glucose-dependent manner. Although compartmentalization of glucokinase by association with tubulin filaments (33) and binding to insulin granules, a process that is controlled by interaction with nitric-oxide synthase and S-nitrosylation (12, 23, 34) has been described, so far no counterpart to the glucokinase regulatory protein in liver has been identified in pancreatic beta cells.
Thus, the aim of this study was to search for glucokinase binding proteins by a systematic approach. To account for proteins specifically expressed in beta cells, a rat pancreatic islet library was produced, and a yeast two-hybrid library screening was performed. Finally, with a protein sequence containing a fragment of midnolin (35) including its ubiquitin-like domain (ULD),2 a new glucokinase binding partner was identified and further investigated.
EXPERIMENTAL PROCEDURES
Materials
All primers, including random hexamer primers, and chemicals for TaqMan assays were from Invitrogen. The RevertAid™ H Minus M-MuLV reverse transcriptase was from Fermentas (St. Leon-Rot, Germany). The GoTaq® Taq polymerase was from Promega (Mannheim, Germany), and dNTPs were purchased from Genecraft (Münster, Germany). Restriction enzymes for cloning were from New England Biolabs (Beverly, MA).
Yeast Two-hybrid Library Screening
Yeast two-hybrid experiments were performed using pGBKT7 and pGADT7 vectors from the MatchmakerTM GAL4 Two-Hybrid System 3 (Clontech Laboratories, Inc., Palo Alto, CA). The cDNA library was constructed according to the manufacturer's instructions (BD MatchmakerTM Library Construction and Screening kit, Clontech) using pancreatic islets isolated from male Wistar rats. RNA was isolated (36) and used for cDNA first-stranded synthesis. Double-stranded cDNA was produced by Long Distance PCR and purified by a BD Chroma Spin TE-400 column (Clontech) to remove fragments smaller than 200 bp. Finally, recombination of double-stranded cDNA and pGADT7-Rec was performed in Saccharomyces cerevisiae AH109, resulting in the GAL4-AD-library (prey proteins). The cDNA sequence of human beta cell glucokinase was amplified by PCR and subcloned in-frame to the GAL4-DNA-BD (bait protein) into pGBKT7 and transformed into S. cerevisiae Y187. Mating of the two yeast strains was performed according to manufacturer's instructions and selected on SD-Leu, SD-Trp, SD-Leu/Trp, and finally on SD-Leu/Trp/His agar plates. As a control, positive pGADT7-Rec-library clones and pGBKT7-lamin were co-transformed into AH109 and selected on SD-Leu/Trp/His selection agar plates with or without the addition of 3-amino-1,2,4-triazol (Sigma). Plasmid DNA from positive yeast colonies was isolated using the Y-DER® Yeast Extraction Reagent kit (Thermo Scientific, Rockford, IL), expressed in Escherichia coli TOP 10, and analyzed by PCR and sequencing (T7 5′-sequencing primer). For identification of the library inserts, an NCBI-Blast search was performed based on the DNA or protein sequence. For further analysis, the cDNA of sequence17 was removed from pGADT7-Rec-library (NcoI and BamHI restriction sites) and subcloned in-frame into pGBKT7. To quantify β-galactosidase activity in yeast lysates, the Galacto-StarTM reporter assay (Invitrogen) was used with pGBKT7-lamin as control.
Plasmids
Generation of enhanced cyan fluorescence protein (ECFP)-glucokinase has been described previously (25). Both the cDNA of the complete sequence17 and of the ULD were amplified by PCR and subcloned in-frame in EYFP-C1 and mCherry-N1 (37) (SalI and BamHI restriction sites). The cDNA sequence of midnolin was amplified by PCR using the midnolin-tGFP vector (MG208176, Origene) as template and subcloned in-frame to the EYFP-C1 and N1 and mCherry-C1 and N1 vectors (HindIII and KpnI restriction sites). YFP-Parkin (addgene plasmid 23955) and mCherry-Parkin (addgene plasmid 23956) were generated and deposited by Richard Youle (38). The ECFP-Nuc vector was from Clontech.
Cell Culture and Transient Transfection
MIN6, HeLa, and COS cells were grown in Dulbecco's modified Eagle's medium (DMEM, Biochrom AG, Berlin, Germany) supplemented with 25 mmol/liter glucose, 10% (v/v) FCS, 10 units/ml penicillin, 10 μg/ml streptomycin, and 2 mmol/liter glutamine in a humidified atmosphere at 37 °C and 5% CO2. INS1E, RINm5F, MH7777A, and HepG2 cells were grown in RPMI 1640 supplemented with 10 mmol/liter glucose, 10% (v/v) FCS, penicillin, and streptomycin in a humidified atmosphere at 37 °C and 5% CO2. Medium of INS1E cells was additionally supplemented with 50 μmol/liter 2-mercaptoethanol, and medium of MH7777A cells was supplemented with 10 μmol/liter dexamethasone. Cells were transfected with the vector DNA by the use of jetPEI (Qbiogene, Montreal, Canada) or jetPrime (Polyplus-transfection SA, Illkirch, France). Stable MIN6 EYFP, MIN6 EYFP-ULD, or MIN6 EYFP-parkin cell clones were selected through resistance against G418 (1200 μg/ml). Expression of transfected plasmids was checked by quantitative PCR, Western blot, and fluorescence microscopy analyses. mCherry fusion constructs of all proteins served as an additional control.
Pancreatic islets and sections were from NMRI mice. Islets were isolated by collagenase digestion in bicarbonate-buffered Krebs-Ringer solution. Beta cells were obtained using calcium-free Krebs-Ringer solution and kept in RPMI 1640 medium supplemented with 5 mmol/liter glucose for 24 h. Primary hepatocytes were isolated from C57/BL6 mice (25). Isolated cells were suspended in Williams medium E supplemented with 10 mmol/liter glucose, 5% (v/v) fetal calf serum, 1 × 10−4 mmol/liter dexamethasone, and 1 × 10−5 mmol/liter insulin. Hepatocytes were seeded at a density of 4 × 104 cells on glass coverslips and incubated for 24 h in a humidified atmosphere at 37 °C and 5% CO2. Mice were housed at the central animal care facility of the faculties, and cell and tissue isolation were approved by the state's Animal Care Committee.
Mammalian Two-hybrid System (MMTHS)
Mammalian two-hybrid analyses were performed using modified vectors of the CheckMate/Flexi Vector Mammalian Two-Hybrid system (Promega). Generation of pACT-GK, pGL4.EYFP, and pBIND.ECFP were described previously (30). To generate pBIND.ECFP-sequence17 and pBIND.ECFP-ULD, the cDNA of the complete sequence17 and of the ULD were amplified by PCR and subcloned in-frame (SgfI and PmeI restriction sites) to the pBIND.ECFP-vector. MIN6 cells were seeded in six-well microplates at a density of 8 × 104 per well and grown for 3 days. Thereafter, cells were transfected with equimolar amounts of the vectors pGL4.EYFP, pBIND.ECFP-ULD, or pBIND.ECFP-sequence17 and pACT-glucokinase or pACT as the negative control and directly incubated with DMEM containing 3, 10, or 25 mmol/liter glucose. Fluorescence intensities of ECFP and EYFP were determined every 2 h in the nuclei using a semiautomated microscope setup with the scan̂R acquisition software (Olympus, Hamburg, Germany) as described previously (30). The ratio EYFP/ECFP was calculated from single cells to quantify the interaction strength.
Cell Viability Assay
MIN6 cells were seeded in 24-well plates at a density of 100,000 cells and grown for 3 days. Cells were transiently transfected as indicated and grown for 24 h. Incubation with 30 μmol/liter camptothecin (Sigma) was performed in the final 5 h. Cell viability was determined using a microplate-based MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay (39).
Recombinant Glucokinase and ULD
The coding sequence for the ULD of sequence17 (midnolin) was amplified by PCR and subcloned in-frame into the BamHI and SalI sites of pGEX-6P-1 expression vector and expressed in E. coli BL21 using the glutathione S-transferase (GST) Gene Fusion System (GE Healthcare). Recombinant beta cell glucokinase (25) and ULD were expressed and purified as GST-tag protein. Cleavage of the GST tag was achieved by PreScission protease (GE Healthcare).
Glucokinase Enzyme Activity
Glucokinase enzyme activity was measured in an enzyme-coupled photometric assay (31). MIN6, MIN6 EYFP, MIN6 EYFP-ULD, or MIN6 EYFP-parkin cells were seeded in 10-cm dishes at a density of 3 × 106 cells and grown for 3 days. MIN6 cells were transiently transfected with EYFP-midnolin and grown for 48 h. Thereafter, cells were homogenized in phosphate-buffered saline (pH 7.4), and the protein concentration was quantified by a Bio-Rad protein assay. To exclude the cellular hexokinase activity, glucokinase enzyme activities measured at 1 mmol/liter glucose were subtracted from the values obtained at 3.125, 6.25, 12.5, 25, and 50 mmol/liter glucose. Glucose phosphorylating activity of recombinant glucokinase was determined at 1, 3.125, 5, 6.25, 10, 12.5, 25, and 50 mmol/liter glucose after 5 min of incubation with 100 nmol/liter recombinant ULD.
Measurement of Insulin Secretion
MIN6, MIN6 EYFP, MIN6 EYFP-ULD, or MIN6 EYFP-parkin cells were seeded in 6-well plates at a density of 3 × 105 cells and grown for 3 days. MIN6 cells were transiently transfected with EYFP-midnolin and grown for 48 h. Thereafter, the medium was replaced, and cells were incubated in DMEM containing 3 or 25 mmol/liter glucose for 48 h. Finally, cells were incubated for 1 h with bicarbonate-buffered Krebs-Ringer solution without glucose supplemented with 0.1% albumin and thereafter stimulated for 1 h with 3 or 25 mmol/liter glucose. 1 ml of the incubation buffer from each well was carefully harvested and gently centrifuged to remove detached cells. In the final supernatants the secreted insulin was measured. Cells were homogenized by sonication in phosphate-buffered saline (pH 7.4), and insulin content was measured in soluble fractions. Insulin was determined by radioimmunoassay using a rat insulin standard or by ELISA, and the protein concentration was quantified by Bradford protein assay.
Western Blot Analysis
MIN6, INS1E, RINm5F, MH7777A, HepG2, and HeLa cells were seeded in 6-cm dishes at a density of 2.5 × 105 cells and incubated with standard DMEM for 24 h. Cells were homogenized in lysis buffer by sonication, and insoluble material was pelleted by centrifugation. Total cellular protein was fractioned by reducing 10% SDS-PAGE and electroblotted to polyvinylidene difluoride (PVDF) membranes. Nonspecific binding sites of the membranes were blocked with 5% nonfat dried milk for 1 h at room temperature. Blots were incubated with glucokinase antibody (sc-7908, diluted 1:200, Santa Cruz Biotechnology, Santa Cruz, CA), N-terminal midnolin antibody (251273, diluted 1:250, Abbiotec, San Diego, CA), C-terminal midnolin antibody (251274, diluted 1:250, Abbiotec) at 4 °C overnight followed by 2 h of incubation with the appropriate peroxidase-labeled secondary antibody at room temperature. Blots were stripped with Re-blot plus (Millipore, Billerica, MA). The protein bands were visualized by chemiluminescence using the ECL detection system (GE Healthcare). Liver, brain, and muscle tissues were homogenized 2 × 20 s at 6500 rpm in a Precellys 24 system (PEQLAB, Erlangen, Germany) using lysing matrix D (MP Biomedicals, Santa Ana, CA). MIN6 cells were transiently transfected as indicated and grown for 24 h. Cells were homogenized in lysis buffer by sonication, and insoluble material was pelleted by centrifugation. Protein concentration was quantified by a Bio-Rad protein assay. 40 μg of cellular protein were fractioned by reducing 10% SDS-PAGE and electroblotted to PVDF membranes. Nonspecific binding sites of the membranes were blocked with Odyssey Blocking Buffer (Li-Cor Biosciences, Lincoln, NE) for 30 min at room temperature. Blots were incubated with midnolin antibody (251273, diluted 1:250, Abbiotec), parkin antibody (sc-32282, diluted 1:500 Santa Cruz Biotechnology), or glucokinase antibody (sc-7908, diluted 1:200, Santa Cruz Biotechnology), cleaved caspase-3 antibody (Asp 175, 9661, diluted 1:1000, New England Biolabs), caspase-3 antibody (8G10, 9665, diluted 1:1000, New England Biolabs), and GAPDH (sc-137179, diluted 1:2000, Santa Cruz Biotechnology) at 4 °C overnight followed by incubation with the appropriate IRDye secondary antibodies (Li-Cor Biosciences) for 30 min at room temperature. Blots were stripped with Re-blot plus (Millipore). Specific protein bands were visualized in the Li-Cor Infrared Imaging System (Li-Cor Biosciences). Quantification of specific protein bands was performed using Odyssey application software (Li-Cor Biosciences).
Quantitative PCR Analysis
MIN6 cells and NMRI islets were incubated with 3, 10, or 25 mmol/liter glucose for 24 h. Total RNA was isolated from incubated MIN6 cells and NMRI islets and from liver, brain, and muscle using the Qiagen RNeasy Kit (Qiagen, Hilden, Germany) and quality-controlled. Random hexamer primers were used for reverse transcription. The gene expression was measured with TaqMan assays (midnolin, Mm 00491444_m1; Park2, Mm 00450187_m1; Ubac1, Mm00661812_m1; Ubl4a, Mm00455093_g1; Ubl4b, Mm00470977_s1) using the TaqMan Universal PCR Master Mix (Invitrogen) and expressed relative to GAPDH expression. Amplifications were performed in triplicate on a 7900HT real-time PCR system (Invitrogen).
Immunocytochemistry
MIN6, INS1E, and HeLa cells, islets, primary beta cells, and hepatocytes were seeded on cover glasses and grown for 48 h. Thereafter, cells were fixed with 4% paraformaldehyde in PBS at 4 °C overnight. Three-micrometer pancreas sections were deparaffinized in xylene and rehydrated in graded ethanol. High pressure cooking in 0.01 mol/liter citric acid buffer (pH 6.0) for 15 min was used for antigen retrieval. After cooling, the sections were incubated with 5% BSA in PBS (pH 7.4) for 30 min to block unspecific binding sites. Immunostaining was performed as described previously (40) with goat anti-glucokinase antibody (sc-1979, diluted 1:50 or 1:100, Santa Cruz Biotechnology), rabbit N-terminal midnolin antibody (251273, diluted 1:50 or 1:250, Abbiotec), rabbit C-terminal midnolin antibody (251274, diluted 1:50 or 1:250, Abbiotec), goat tubulin antibody (sc-9935, diluted 1:50), or mouse parkin antibody (sc-32282, diluted 1:100, Santa Cruz Biotechnology) and the appropriate secondary antibody Alexa488 donkey anti-goat antibody (diluted 1:200, Dianova, Hamburg Germany), Alexa594 donkey anti-goat antibody (diluted 1:200, Dianova), or Cy5 donkey anti-mouse and anti-rabbit antibodies (diluted 1:200 or 1:500, Dianova). For detection of the mitochondrial network, cells were stained with MitoTracker® Deep Red FM (Invitrogen). Finally, cells were stained with DAPI and fixed onto slides.
Fluorescence Microscopy
Analysis of transient transfected MIN6 cells and fluorescence resonance energy transfer (FRET) experiments were performed with a cellR/Olympus IX81 (Olympus) inverted microscope system equipped with a Cellcubator incubation chamber (Olympus) using 60% humidity, 37 °C, and 5% CO2. Glass-bottom dishes or slides were fixed on the microscope stage, and images were taken with an UPLSAPO 60 × 1.35 NA oil-immersion objective. HQ580/20, ET470/40, S484/15X, HQ620/60, and HC387/11 filter sets (AHF Analysentechnik, Tübingen, Germany) were used for excitation of mCherry, enhanced GFP, Alexa488, Cy5, or MitoTracker® Deep Red FM and for DAPI, respectively. Emission was detected using HQ630/60M, a 510 (30)-630 (100) dual band filter, M517/30, HQ700/75, and 433(50)-517(40)-613(60) filter sets (AHF Analysentechnik). Image processing was done using xcellence software (Olympus) and AutoDeblur Gold WF software (MediaCybernetics, Silver Spring, MD) for image deconvolution. The FRET setup has been described previously (30). Sensitized emission-based FRET efficiency (FRETN) was calculated from the ECFP emission with excitation at 436 nm, EYFP emission with excitation at 436 nm, and EYFP emission with excitation at 500 nm, based on the calculation of Vanderklish (41). Analyses of immunocytochemistry-stained slides were performed with a Fluoview1000 (Olympus) or FV10i (Olympus) confocal microscope system and UPLSAPO 60 × 1.35 NA oil-immersion objective using sequential scanning mode. Image processing was done using FV10-ASW software (Olympus).
Data Analysis
Data are expressed as the means ± S.E. Statistical analyses were performed by ANOVA followed by Bonferroni's test for multiple comparisons or by Student's t test using the Prism analysis program (Graphpad, San Diego, CA).
RESULTS
Identification of Sequence17 as a New Glucokinase Interaction Partner in a Yeast Two-hybrid Screening of a Rat Pancreatic Islet Library
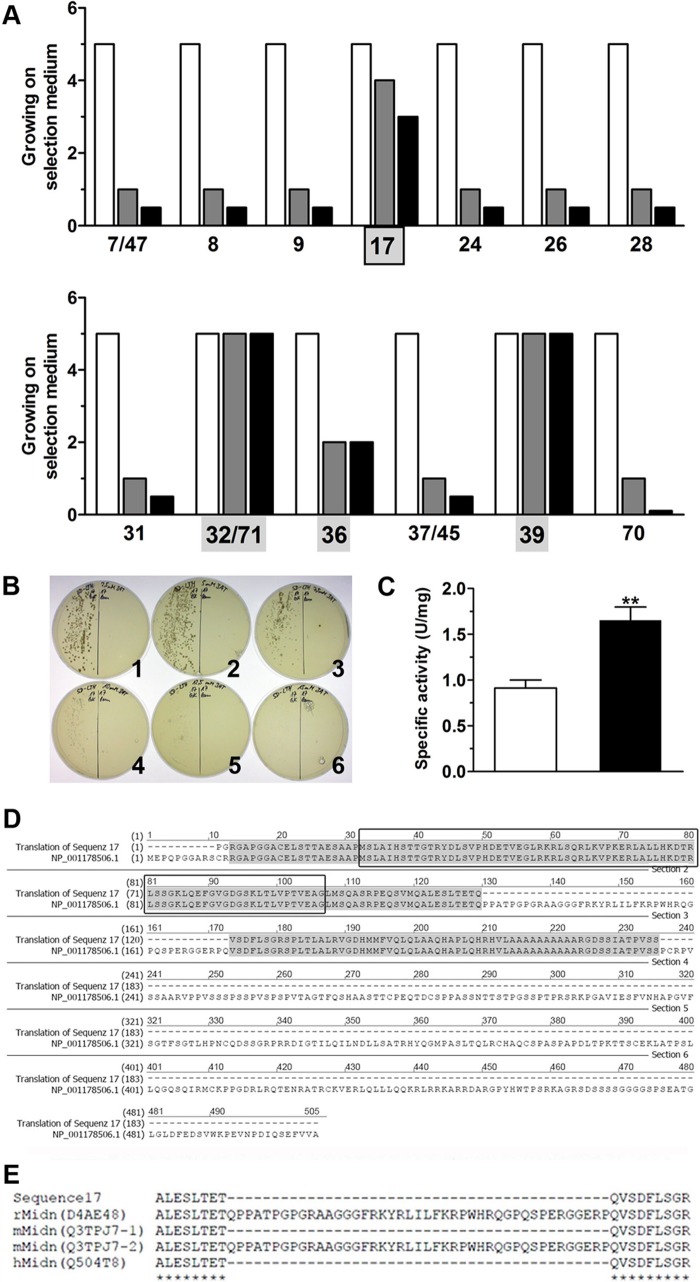
A cDNA library was generated using RNA isolated from rat pancreatic islets with insert sizes ranging from 0.5 to 4 kb. The resulting double-stranded cDNA was cloned into pGADT7-Rec by recombination in yeast AH109 with a high efficiency of 1.2 × 106 transformants. Mating of AH109 pGADT7 library and Y187 pGBKT7-GK was established resulting in 5.5 × 105 library clones in three independent runs. 73 positive yeast clones showing His-reporter gene activity were selected, the plasmid DNA was isolated, and sequence alignments were performed. Because of maximal growth on SD-Leu/Trp/His selection agar plates, 13 yeast clones expressing protein sequences so far unknown to interact with glucokinase were taken for further investigations (Fig. 1A). Four clones of those showed significant growth on SD-Leu/Trp/Ade and SD-Leu/Trp/His/Ade selection agar plates (Fig. 1A). Only yeast expressing glucokinase and sequence17 exhibited a graduated growth on selection agar plates as a function of the selection stringency (Fig. 1A). Yeast expressing glucokinase and sequence 32/71, 36, and 39 showed growth on selection agar plates independent of the stringency (SD-Leu/Trp/His < SD-Leu/Trp/Ade < SD-Leu/Trp/His/Ade) (Fig. 1A), indicating their ability to induce transcription without glucokinase interaction. In fact, His-reporter gene expression could be detected after co-expression of pGADT7-sequence32/71, pGADT7-sequence36, and pGADT7-sequence39 with the negative control pGBKT7-lamin in AH109. In contrast, yeast with pGADT7-sequence17 showed growth on SD-Leu/Trp/His selection agar plates after co-expression with pGBKT7-glucokinase but not with pGBKT7-lamin. This growth was sensitive to the specific inhibitor 3-amino-1,2,4-triazol (Fig. 1B). Finally, a quantitative β-galactosidase assay (Fig. 1C) revealed significantly higher interaction strength for sequence17 with glucokinase compared with the control protein lamin.
FIGURE 1.
Identification of a new glucokinase interaction partner in a yeast two-hybrid library screening. A, 13 different library inserts were identified in a yeast two-hybrid library screening showing different growth on SD-selection agar plates containing Leu/Trp/His (open bars), Leu/Trp/Ade (gray bars), or Leu/Trp/His/Ade (black bars). B, growth of AH109 co-expressing pGADT7-sequence17 and pGBKT7-glucokinase (left side of agar plates) or pGBKT7-lamin (right side of agar plates) on SD-Leu/Trp/His selection agar containing 2.5 (1), 5 (2), 7.7 (3), 10 (4), 12.5 (5), or 15 (6) mmol/liter 3-amino-1,2,4-triazol. C, β-galactosidase reporter gene activity in yeast expressing pGADT7-sequence17 and pGBKT7-glucokinase (black bar) or pGBKT7-lamin (open bar). **, p < 0.05 (Student's t test). U, units. D, sequence alignment of sequence17 with midnolin (NCBI protein accession number NP_001178506.1) with homologies marked in gray. The black box frames the ULD. E, shown is a section of a MAFFT (42) alignment of sequence17 with the rat isoform (D4AE48), mouse isoform1 (Q3TPJ7-1), and isoform2 (Q3TPJ7-2), and human isoform (Q504T8) of midnolin.
Sequence17 Contains a Fragment of Midnolin Including the Complete Sequence of a Ubiquitin-like Domain
Sequence and protein database analyses of sequence17 revealed a 180-amino acid-containing fragment that shows homology to the N-terminal part of midnolin (NCBI Protein Accession Nr. NP_001178506.1) (Fig. 1D). Interestingly, this fragment contains the full-length sequence of a ULD, consisting of 76 amino acids. Sequence alignment by MAFFT (v7.015b) (42) of rat, human, and mouse midnolin isoforms revealed a high homology between the different species. Notably, the 43 amino acids, which are missing in the observed sequence17 in comparison to rat midnolin (Fig. 1D), are also lacking in human and mouse isoform1 of midnolin (Fig. 1E).
Interaction Analyses between 1) Glucokinase and Sequence17 and -2) Glucokinase and the ULD of Sequence17 (midnolin) in a Mammalian Two-hybrid System
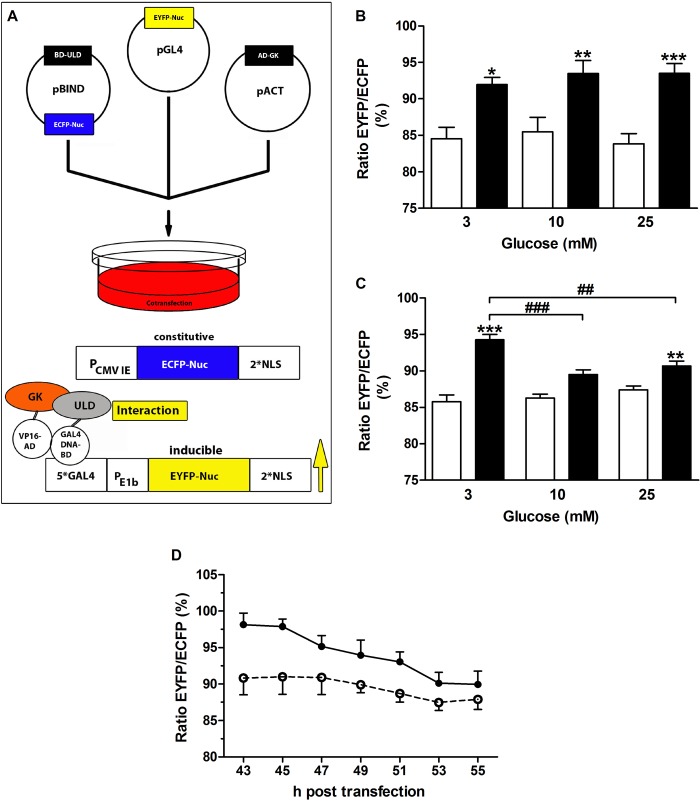
The MMTHS in a fluorescence-based semiautomated microscopy approach has been generated previously to investigate protein-protein interactions in mammalian cells (30). The insulin-secreting MIN6 cells were triple-transfected with the MMTHS vectors (Fig. 2A, top) and incubated at different glucose concentrations. Although ECFP is constitutively expressed to control the transfection efficiency, the expression of EYFP is under control of an inducible promoter (Fig. 2A, bottom). Thus, association of the VP16 activation domain (VP16-AD or VP16-AD-glucokinase) and the GAL4-DNA binding domain (GAL4-DNA-BD-sequence17 or GAL4-DNA-BD-ULD) mediated by the protein-protein interaction will result in EYFP expression. Averaged data from 43–55-h post transfection revealed a significant interaction between sequence17 and glucokinase in comparison to the control independent from the glucose concentration in the culture medium (Fig. 2B). In contrast, the interaction strength between the ULD of sequence17 (midnolin) and glucokinase was glucose-dependent (Fig. 2C) and significantly higher at 3 mmol/liter glucose compared with 10 or 25 mmol/liter glucose (Fig. 2, C and D), suggesting that binding of glucokinase to a ULD-containing protein occurs preferentially at low glucose in insulin-secreting cells.
FIGURE 2.
Glucokinase interacts with the ULD of midnolin preferentially at low glucose. A, the MMTHS plasmids pGL4.EYFP, pBIND.ECFP-ULD or pBIND.ECFP-sequence17 and pACT-glucokinase or pACT were cotransfected, and MIN6 cells were cultured at 3, 10, or 25 mmol/liter glucose. Whereas ECFP is constitutively expressed, EYFP expression indicates protein-protein interaction. The EYFP/ECFP fluorescence ratio was determined every 2 h in the cell nucleus in a semi-automated microscopy approach. B, interaction of BD-sequence17 and AD-glucokinase (black bars) or AD (open bars). C, interaction between BD-ULD and AD-glucokinase (black bars) or AD (open bars). D, time course of interaction between BD-ULD and AD-glucokinase in MIN6 cells cultured at 3 mmol/liter (closed circles, solid line) or 10 mmol/liter (open circles, dashed line). Shown are normalized mean nuclear EYFP/ECFP ratios ± S.E. obtained 43–55 h after transfection of 4–8 individual experiments with a total of 624–6434 nuclei analyzed. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with control; ##, p < 0.01; ###, p < 0.001 compared with 3 mmol/liter glucose (ANOVA/Bonferroni's multiple comparison test).
Glucose-dependent Gene Regulation of ULD-containing Proteins in Beta Cells
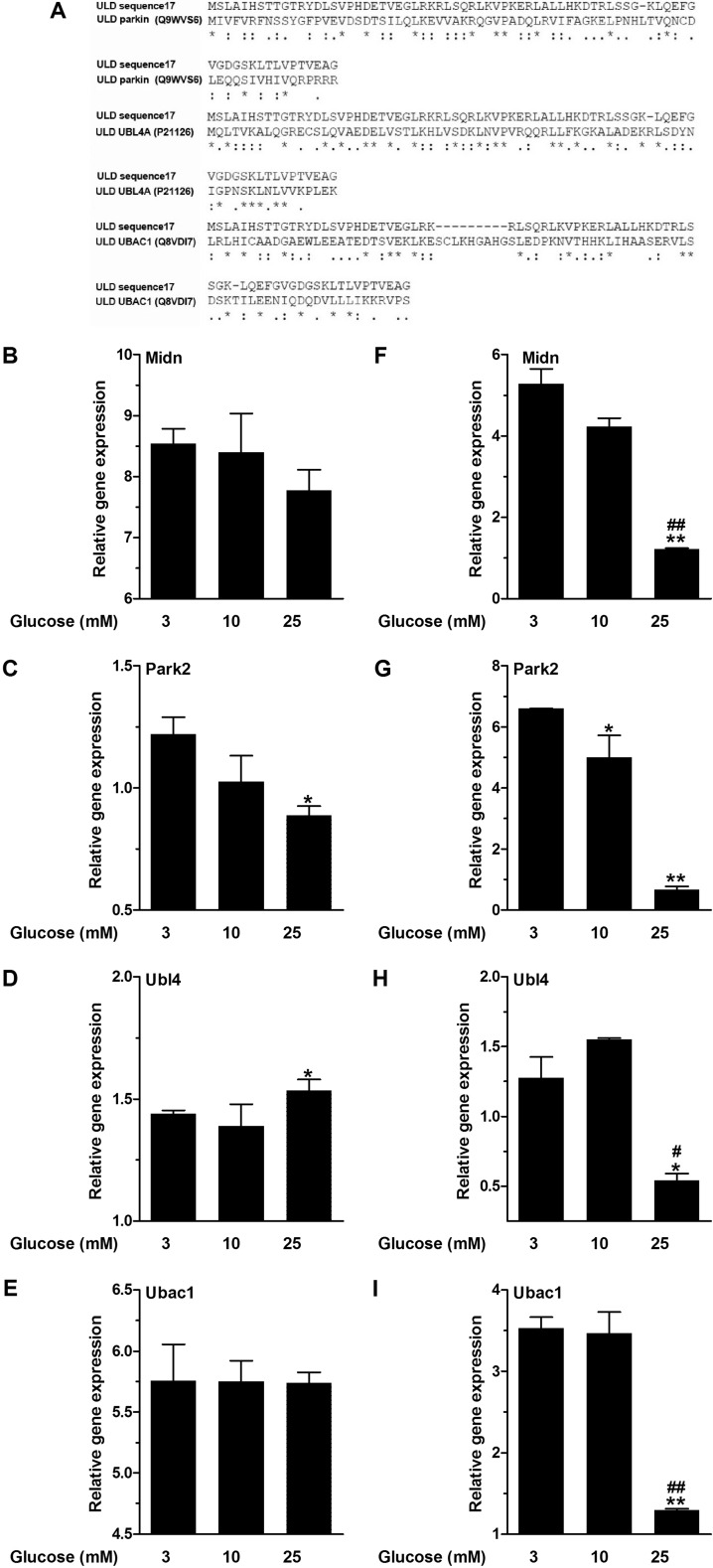
Up-regulation of genes coding for ULD-containing proteins or those involved in the ubiquitin-proteasome pathway at low glucose has been recently shown in insulin-secreting cells and islets (43). To date, ∼30 ULD-containing proteins have been described with at least in-part cytoplasmic localization and, thus, can come into consideration as an interaction partner of glucokinase. Alignments of the ULDs revealed four genes with ∼50% sequence similarity to the ULD of midnolin, namely Park2, Ubl4 (also known as Ubl4a or Gdx), Ubl4b, and Ubac1. Ubl4b could not be detected in beta cells, which is in agreement with previous findings showing Ubl4b expression only in testis (44). Therefore, beyond midnolin, expression of Park2, Ubl4, and Ubac1 (Fig. 3A) has been investigated in MIN6 cells and primary islets after 24 h of incubation at 3, 10, and 25 mmol/liter glucose.
FIGURE 3.
Glucose-dependent gene expression of ULD-containing proteins. A, sequence alignment of the ULD of sequence17 with the ULD of parkin (Q9WVS6), UBL4A (P21126), and UBAC1 (Q8VDI7). Asterisks, fully conserved residues; double dots, strongly similar properties; dots, weakly similar properties. MIN6 cells (B–E) and NMRI islets (F–I) were cultured at 3, 10, or 25 mmol/liter glucose for 24 h. Gene expression levels of midnolin (Midn, B and F), Park2 (C and G), Ubl4 (D and H), and Ubac1 (E and I) were determined. Relative expression levels normalized to housekeeping gene GAPDH are shown. Data are expressed as the means ± S.E. of three individual experiments. *, p < 0.05; **, p < 0.01 compared with 3 mmol/liter glucose; #, p < 0.05; ##. p < 0.01 compared with 10 mmol/liter glucose (ANOVA/Bonferroni's multiple comparison test).
Both in insulin-secreting MIN6 cells and islets midnolin expression levels were highest at 3 mmol/liter glucose (Fig. 3, B and F). Down-regulation was observed at 10 and 25 mmol/liter glucose, which was significant at 25 mmol/liter glucose in islets (Fig. 3F). The same effect, but more pronounced, was observed for Park2 expression. In MIN6 cells and islets a significantly lower Park2 expression was observed at 25 mmol/liter glucose in comparison to 3 mmol/liter glucose (Fig. 3, C and G). Ubl4 and Ubac1 expression levels were comparable at 3 and 10 mmol/liter glucose in MIN6 cells and islets (Fig. 3, D, E, H, and I). At 25 mmol/liter glucose, Ubl4 and Ubac1 expression was down-regulated in islets (Fig. 3, H and I). However, in MIN6 cells, Ubl4 expression significantly increased at 25 mmol/liter glucose (Fig. 3D). Thus, only midnolin and parkin showed distinct glucose-dependent gene expression.
Structural Homologies of the ULDs of Midnolin and Parkin with Ubiquitin
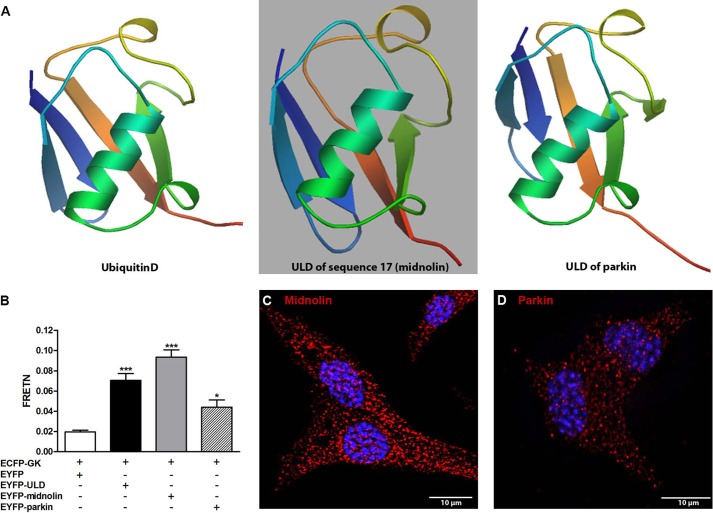
It has been shown that ULDs share strong homologies to ubiquitin (45, 46). The three-dimensional structures of ubiquitinD (Q921A3), the ULD of sequence17 (midnolin), and the ULD of parkin (Q9JK66) were constructed with SWISS-MODEL based on the protein sequences (Fig. 4A) (47–51). The ULD of sequence17 (midnolin) shows distinct homology to the three-dimensional structure of ubiquitin, which adopts a compact, globular form (52). In addition, the three-dimensional structures revealed a strong homology between the ULDs of midnolin and parkin.
FIGURE 4.
Homologies and differences between midnolin and parkin. A, comparison of the three-dimensional structures of ubiquitinD (Q921A3), the ULD of sequence17 (midnolin), and the ULD of parkin (Q9JK66). Images were constructed with SWISS-MODEL (47, 48, 50). B, FRETN efficiencies were calculated in COS cells transfected with ECFP-glucokinase and EYFP (white bar), EYFP-ULD (black bar) EYFP-midnolin (gray bar), or EYFP-parkin (white-striped bar). Shown are the means ± S.E. of three individual experiments. *, p < 0.05; ***, p < 0.001 compared with control (ANOVA/Bonferroni's multiple comparison test). MIN6 cells (C and D) were fixed and stained for midnolin (N-terminal part) (red, C) or parkin (red, D) and DAPI (blue, C and D). Representative images shown were obtained from z-stacks after deconvolution. Scale bars, 10 μm.
Interaction between Glucokinase and Midnolin or Parkin
Using the MMTHS, the strongest interaction between glucokinase and the ULD of midnolin was obtained at 3 mmol/liter glucose. Sensitized emission based fluorescence resonance energy transfer (FRETN) analyses in COS cells confirmed our results. At 3 mmol/liter glucose, FRETN was in ECFP-GK/EYFP-ULD and ECFP-GK/EYFP-midnolin coexpressing cells 3.5 and 4.7 times, respectively, higher than in negative control cells co-expressing ECFP-GK/EYFP (Fig. 4B). Cells co-expressing ECFP-GK/EYFP-parkin showed only 2 times higher interaction strength than the negative control (Fig. 4B). Endogenous midnolin (Fig. 4C) and parkin (Fig. 4D) were detectable in the cytoplasm and nucleus. However, in line with the gene expression analyses (Fig. 3, B and C), the expression level of parkin appears to be lower compared with midnolin.
Effects of the ULD of Midnolin, Full-length Midnolin, and of Parkin on Glucokinase Activity
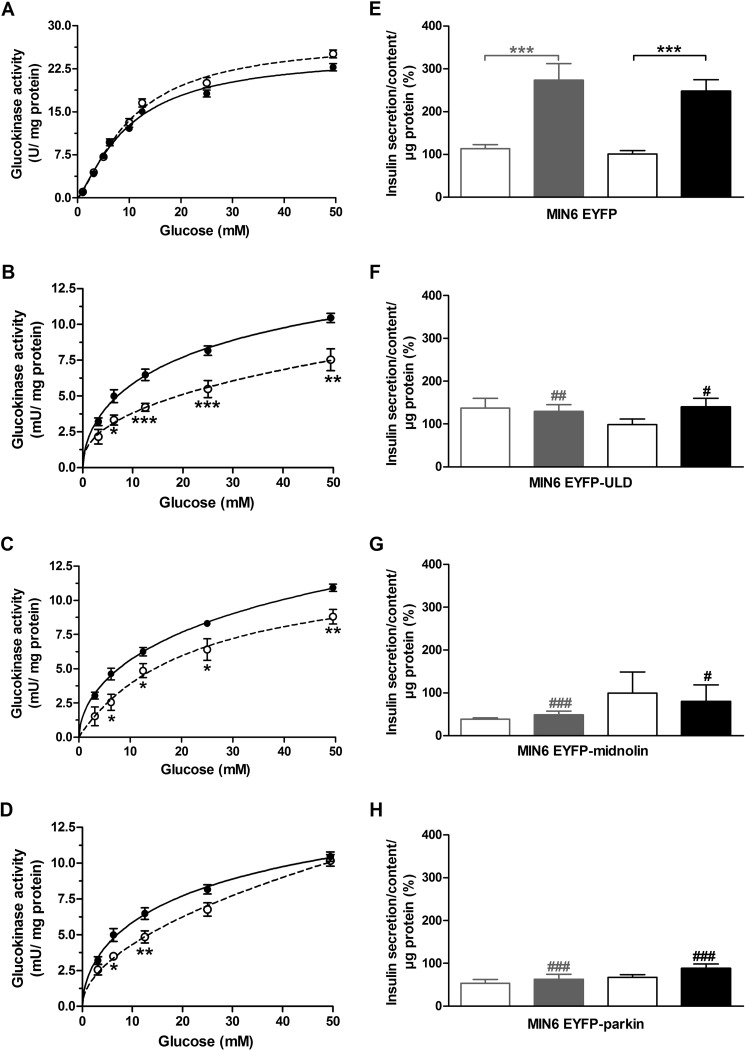
To investigate the effect of the ULD of midnolin on glucokinase activity, glucose phosphorylating activities were determined. In in vitro experiments using recombinant glucokinase and ULD (100 nmol/liter) no influence on glucokinase activity could be detected (Fig. 5A). In contrast, overexpression of EYFP-ULD in insulin-secreting MIN6 cells significantly inhibited endogenous glucokinase activity by reducing both the maximal phosphorylating activity and the affinity to glucose (Fig. 5B). Thus, either the ULD of midnolin was not properly folded in bacteria or the in vitro conditions were not adequate for interaction. The successful use of recombinant ubiquitin (53), which has the same size and high homology with the ULD of midnolin, however, argues against misfolding. Overexpression of full-length EYFP-midnolin significantly inhibited glucokinase enzyme activity and reduced the affinity to glucose (Fig. 5C). Overexpression of EYFP-parkin significantly decreased glucokinase activity in MIN6 cells at 6.25 and 12.5 mmol/liter glucose, thus affecting only the affinity of the enzyme to glucose (Fig. 5D).
FIGURE 5.
Effects of the ULD of midnolin, full-length midnolin, and of parkin on glucokinase activity and glucose-induced insulin secretion in MIN6 cells. A, activity of recombinant beta cell glucokinase was determined at 1, 3.125, 5, 6.25, 10, 12.5, 25, and 50 mmol/liter glucose without (closed circles, solid line) and after incubation with 100 nmol/liter recombinant ULD for 5 min (open circles, dashed line). Shown are the means ± S.E. in units/mg protein of six individual experiments. The activity of endogenous glucokinase was determined in MIN6 EYFP cells (closed circles, solid line; B–D), MIN6 EYFP-ULD cells (open circles, dashed line; B), MIN6 cells transiently transfected with EYFP-midnolin (open circles, dashed line, C), and MIN6 EYFP-parkin cells (open circles, dashed line; D). Glucokinase enzyme activity measured at 1 mmol/liter glucose was subtracted from the values obtained at 3.125, 6.25, 12.5, 25, and 50 mmol/liter glucose to exclude the cellular hexokinase activity. Shown are the means ± S.E. in milliunits/mg of cellular protein of three individual experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared with control at the same glucose concentration (ANOVA/Bonferroni's multiple comparison test). MIN6 EYFP cells (E), MIN6 EYFP-ULD cells (F), MIN6 cells transiently transfected with EYFP-midnolin (G) and MIN6 EYFP-parkin cells (H) were cultured at 3 mmol/liter (gray bars) or 25 mmol/liter (black bars) glucose for 48 h. After starvation for 1 h, cells were incubated at 3 mmol/liter (open bars) or 25 mmol/liter (closed bars) glucose. Data are the means ± S.E. of 4–11 individual experiments. ***, p < 0.001 compared with 3 mmol/liter glucose; #, p < 0.05; ##, p < 0.01; ###, p < 0.001 compared with MIN6 EYFP cells (ANOVA/Bonferroni's multiple comparison test).
Effects of the ULD of Midnolin, Full-length Midnolin, and Parkin on Glucose-induced Insulin Secretion
Insulin-secreting MIN6 cells overexpressing EYFP as a transfection control showed a significant increase in insulin secretion after stimulation with 25 mmol/liter glucose. The glucose responsiveness of MIN6 cells was comparable after 48 h of preincubation at 3 or 25 mmol/liter glucose and a 1-h final incubation period without glucose (Fig. 5E). Glucose-induced insulin secretion was significantly reduced after overexpression of EYFP-ULD (Fig. 5F). Glucose responsiveness was completely lost in MIN6 cells preincubated at 3 mmol/liter glucose, whereas some glucose responsiveness remained after preincubation at 25 mmol/liter glucose. Loss of glucose-induced insulin secretion was also observed after overexpression of EYFP-midnolin, after preincubation with both 3 or 25 mmol/liter glucose (Fig. 5G). Overexpression of EYFP-parkin disturbed insulin secretion in MIN6 cells (Fig. 5H). Not only glucose-induced insulin secretion, but also the basal insulin secretion was significantly reduced (Fig. 5H).
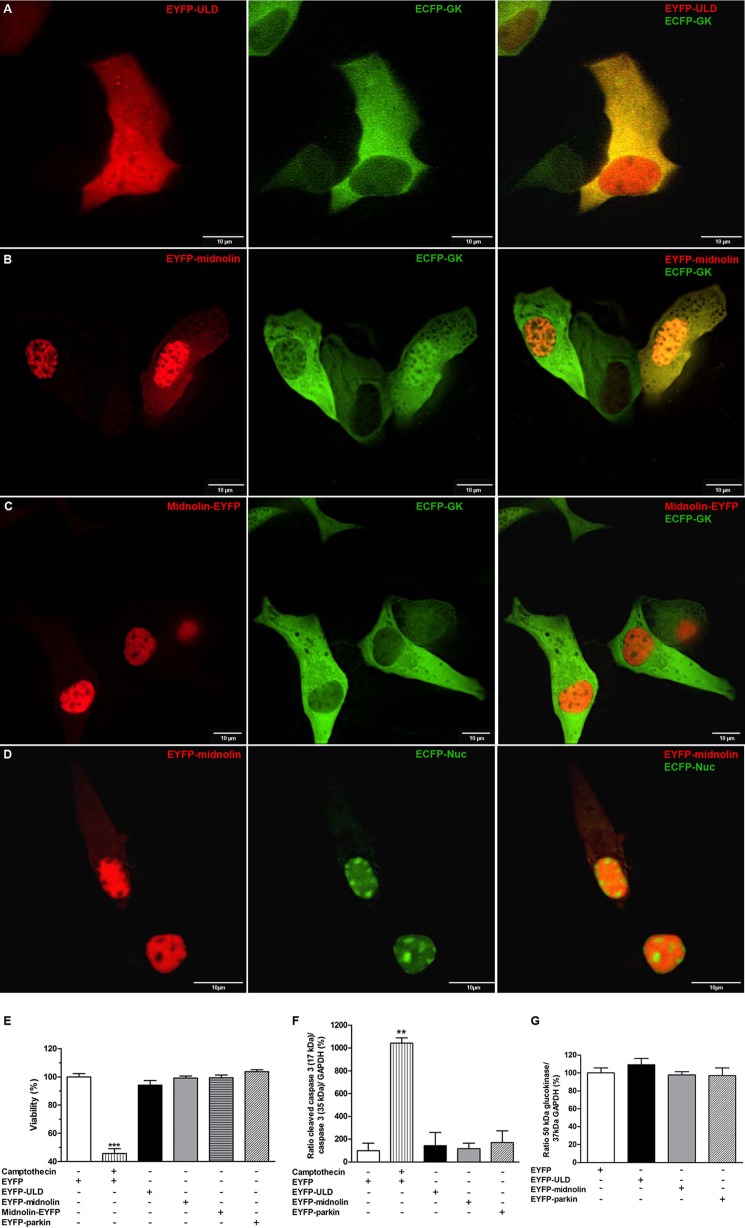
Midnolin Overexpression and Co-localization with Glucokinase in Insulin-secreting MIN6 Cells
Overexpression of EYFP-ULD showed a homogenous distribution in insulin-secreting MIN6 cells with considerable fluorescence in the nucleus (Fig. 6A). In the cytoplasm a distinct co-localization with ECFP-glucokinase was detectable (Fig. 6A). EYFP-midnolin (Fig. 6B) and midnolin-EYFP (Fig. 6C) showed the highest fluorescence in the cell nucleus. However, in ∼50% of the transfected cells both midnolin EYFP fusion constructs were also detectable in the cytoplasm. Co-localization with ECFP glucokinase was observed in the cytoplasm for both midnolin EYFP fusion constructs (Fig. 6, B and C). In some cells, mostly with a high midnolin overexpression level, ECFP-glucokinase was also detectable in the nucleus (Fig. 6B) and co-localized with the overexpressed midnolin EYFP fusion construct. Midnolin contains a nuclear localization signal. Based on overexpression analyses of midnolin GFP fusion constructs in CHO cells, Tsukahara et al. (35) postulated a mainly nucleolar localization of the overexpressed protein. In contrast, co-expression experiments of EYFP-midnolin and a nucleoli-localized fluorescence protein (ECFP-Nuc) in MIN6 cells clearly showed that midnolin is not located in the nucleoli (Fig. 6D).
FIGURE 6.
Subcellular localization of midnolin after overexpression in MIN6 cells and its effect on glucokinase expression and cell viability. A–D, MIN6 cells were transiently co-transfected with EYFP-ULD (A, left panel), EYFP-midnolin (B and D, left panels), midnolin-EYFP (C, left panel), and ECFP-glucokinase (A–C, middle panel) or ECFP-Nuc (D, middle panel). EYFP fusion proteins in red, ECFP fusion proteins in green, and the merge images (A-D, right panels) are shown. Representative images shown were obtained from z-stacks after deconvolution. Scale bars, 10 μm. E–F, MIN6 cells were transiently transfected with EYFP (white bar and white horizontal-striped bar; E and F), EYFP-ULD (black bar; E and F), EYFP-midnolin (gray bar; E and F), midnolin-EYFP (cross-striped gray bar, E), and EYFP-parkin (white striped bar; E and F) and treated with 30 μmol/liter camptothecin (white vertical-striped bar; E and F). Cell viability was determined by MTT assay (E) or quantification of cleaved caspase-3 in relation to caspase-3 and GAPDH expression (F). G, glucokinase expression is shown in relation to GAPDH expression. Data are the means ± S.E. of 3–4 individual experiments. **, p < 0.01; ***, p < 0.001 compared with EYFP transfected MIN6 cells (ANOVA/Bonferroni's multiple comparison test).
Effect of Midnolin Overexpression on Cell Viability and Glucokinase Protein Expression
Overexpression of EYFP-ULD, EYFP-midnolin, midnolin-EYFP, or EYFP-parkin did not reduce cell viability (Fig. 6E) or induce apoptosis (Fig. 6F). The cytotoxic quinoline alkaloid camptothecin served in our experiments as a positive control reducing cell viability by 55% in the MTT assay and induced a 10-fold increase in cleaved caspase-3. The glucokinase protein content determined in MIN6 cells after overexpression of EYFP, EYFP-ULD, EYFP-midnolin, or EYFP-parkin was comparable (Fig. 6G), indicating that neither overexpression of midnolin nor parkin influence glucokinase protein expression.
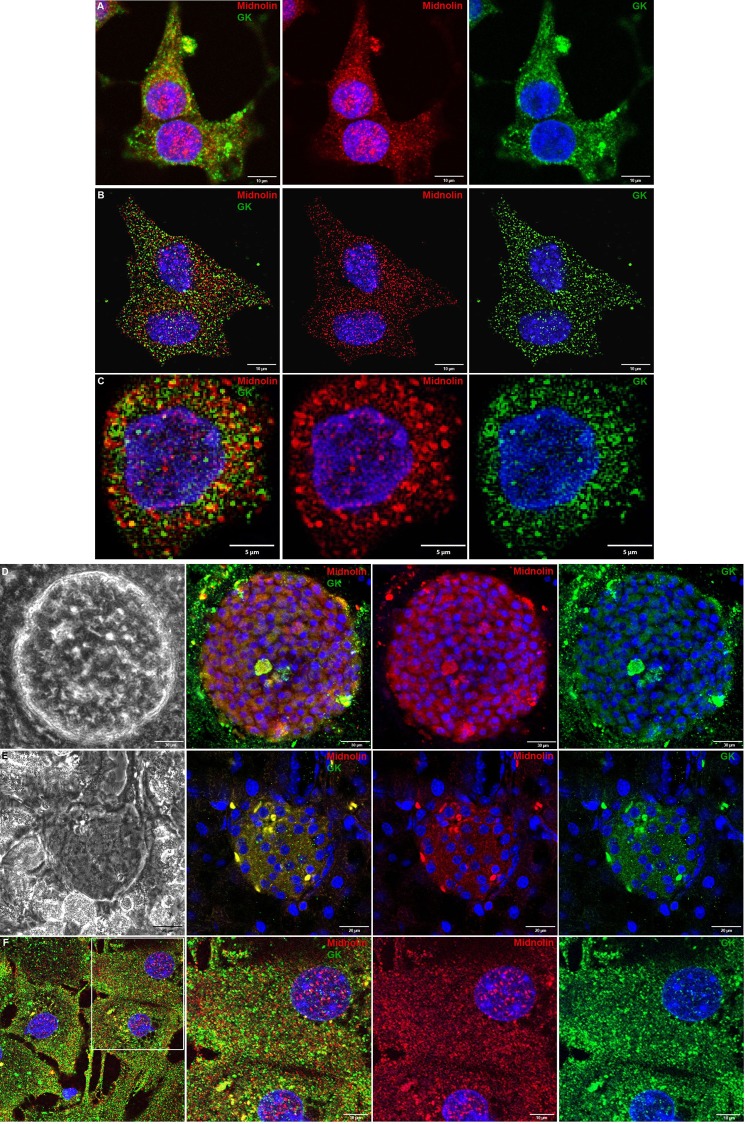
Midnolin Expression and Co-localization with Glucokinase in Insulin-secreting Cells and Primary Cells
In insulin-secreting INS1E cell (Fig. 7A), MIN6 cell (Fig. 7B), and primary mouse beta cell (Fig. 7C) midnolin was detectable in the cytoplasm and the nucleus and showed distinct co-localization with glucokinase in the cytosol. In intact mouse islets, midnolin expression was detected in the whole islet clearly co-localizing with glucokinase (Fig. 7D). Staining of pancreatic sections showed midnolin expression in the islet with only negligible expression in the surrounding exocrine tissue and a distinct co-staining with glucokinase (Fig. 7E). In hepatocytes (Fig. 7F), both midnolin and glucokinase were expressed in the cytosol and the nucleus and showed in part co-localization (Fig. 7F).
FIGURE 7.
Co-localization of midnolin and glucokinase in insulin-secreting cells, primary beta cells, and hepatocytes. INS1E cells (A), MIN6 cells (B), primary mouse beta cells (C), mouse pancreatic islets (D), and hepatocytes (F) were fixed and stained for midnolin (N-terminal part) (red; A-C and F) and (C-terminal part) (red; D), glucokinase (green; A-D and F) and with DAPI (blue; A–D and F). G, pancreatic sections were stained for midnolin (N-terminal part) (red), glucokinase (green), and with DAPI (blue; A–D and F). Representative images shown were obtained from z-stacks after deconvolution (B, C, and F) or processed with FV10-ASW software (A, D, and E). Scale bars, 5 (C), 10 (A, B, and F), 20 (E), or 30 (D) μm.
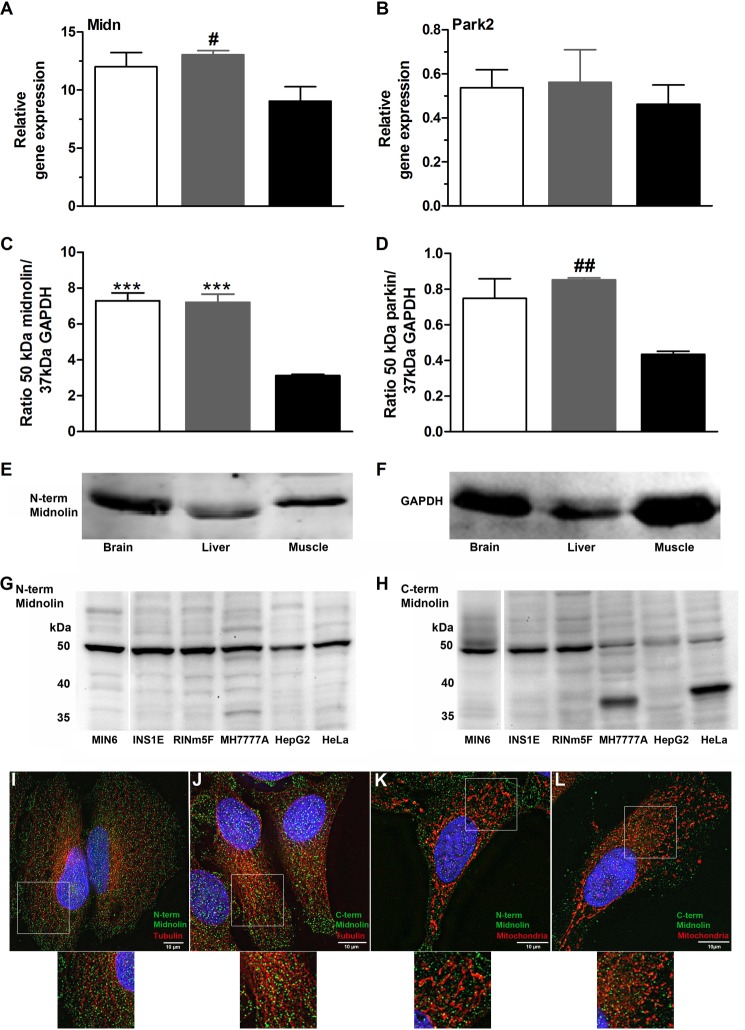
Midnolin Expression in Different Tissues, Species, and Cell Lines
The gene and likewise protein expression levels of midnolin (Fig. 8, A and C) in mouse liver, brain, and muscle appeared to be higher compared with parkin (Fig. 8, B and D). Midnolin expression was comparable in brain and liver but significantly lower in muscle (Fig. 8, A and C). Midnolin was verified as a 50-kDa protein in all the investigated mouse tissues (Fig. 8E), in mouse insulin-secreting MIN6 cells, in rat insulin-secreting INS1E and RINm5F cells, in human MH7777A and rat HepG2 hepatoma cells, and in the human HeLa cell line (Fig. 8, G and H). The specific protein signal at 50 kDa was detectable using the N-terminal (Fig. 8G) and C-terminal (Fig. 8H) midnolin antibody. Interestingly, in cell lines of human origin, a 35-kDa line was additionally detected, particularly using the C-terminal antibody. In HeLa cells (Fig. 8, I–L), comparable to primary mouse beta cells and insulin-secreting cells (Fig. 7, A–C), midnolin was detectable in the nucleus and cytoplasm. The distribution pattern was independent of the antibody used (Fig. 8, I–L). Midnolin showed a punctated cytoplasmic distribution but was marginally co-localized with tubulin (Fig. 8, I and J) and not co-localized with mitochondria (Fig. 8, K and L).
FIGURE 8.
Midnolin expression in different tissues. A and B, brain (open bars), liver (gray bars), and muscle (black bars) were obtained from NMRI mice. Total RNA was isolated, and midnolin (Midn; A) and Park2 (B) gene expression was determined. Shown are relative expression levels normalized to GAPDH. C–F, 40 μg of isolated protein were analyzed by SDS-PAGE and immunoblotted using antibodies against the N-terminal part of midnolin (E), GAPDH (F), and parkin. Quantifications of the 50-kDa midnolin line (C) and the 50-kDa parkin line (D) are shown in relation to the 37-kDa GAPDH line. Shown are the means ± S.E. of three individual experiments. ***, p < 0.001 compared with muscle (ANOVA/Bonferroni's multiple comparison test); #, p < 0.05, ##, p < 0.01 compared with muscle (Student's t test). G and H, 40 μg of protein isolated from MIN6, INS1E, RINm5F, MH7777A, HepG2, and HeLa cells were analyzed by SDS-PAGE and immunoblotted using antibodies against the N-terminal part (G) and C-terminal part (H) of midnolin. I–L, HeLa cells were fixed and stained for midnolin using the N-terminal (green; I and K) or C-terminal (green; J and L) midnolin antibody and for β-tubulin (red; I and J) with MitoTracker® Deep Red FM (red; K and L) and with DAPI (blue; I–L). Representative images shown were obtained from z-stacks after deconvolution. Scale bars, 10 μm.
DISCUSSION
Using yeast two-hybrid screening we elucidated a 180-amino acid protein fragment named sequence17 as a new interaction partner of glucokinase. This fragment is part of midnolin, a nucleolar protein that has been described to be mainly expressed in the mesencephalon during mouse development (35). By the gene trap method and the selection of embryonic stem cells, midnolin has been identified to be more highly expressed during cell differentiation than thereafter (35). Although Tsukahara et al. (35) described midnolin expression in adult tissues, especially in heart, lung, liver, and kidney, they finally concluded that midnolin is involved in regulation of genes related to neurogenesis. Zielak et al. (54) reported that midnolin is important in regulating mRNA transport in developing follicles in cattle. Due to the lack of any further investigations on midnolin, it remains open if the protein could be an interaction partner of glucokinase in pancreatic beta cells.
We confirmed the interaction between glucokinase and sequence17 in insulin-secreting cells by mammalian two-hybrid analyses. Because sequence17 contains the full-length ULD of midnolin, binding of this ULD alone to glucokinase has also been studied. ULDs are domains that strongly resemble ubiquitin (46, 55). The ULD of midnolin was found to have the highest binding affinity to glucokinase if the cells were cultured at 3 mmol/liter glucose. At this glucose concentration, glucokinase exists mainly in its inactive super-open-to-open conformations, and insulin secretion is basal in pancreatic beta cells (4, 17, 19, 20, 30). It is known that proteins containing a ULD are involved in intramolecular interactions and regulation of signal transduction (46, 56, 57). In most cases proteins with a ULD act in close correlation with the ubiquitin-proteasome system (46). In pancreatic islets up-regulation of genes encoding proteins of the ubiquitin-proteasome system at low glucose has been reported (43, 58). Interestingly, RT-PCR analyses revealed the highest midnolin expression at 3 mmol/liter glucose in mouse pancreatic islets and insulin-secreting MIN6 cells. The expression level was stepwise reduced after 24 h incubation at 10 and 25 mmol/liter glucose.
Parkin, containing a ULD with high homology to that of midnolin, showed a comparable glucose-dependent gene expression. This is in agreement with a previous study reporting up-regulation of parkin in rat pancreatic islets at low glucose culture (43). Parkin is an E3 ubiquitin ligase with different expression patterns during development and in adult tissues (59–61). Its function has been best investigated in the dopaminergic system, where loss-of-function mutations in the Park2 gene cause juvenile Parkinson disease (38, 59). Parkin binds via PINK1 (mitochondrial serine/threonine-protein kinase 1)-damaged mitochondria and thereby triggers autophagy (38, 62, 63). Little is known about the role of parkin in pancreatic beta cells. FRETN analyses showed some interaction between glucokinase and parkin. Overexpression of parkin in insulin-secreting MIN6 cells resulted in reduced glucokinase enzyme activity at low glucose, suggesting some relevance of the parkin:glucokinase interaction. However, this cannot explain the loss of glucose-induced and the reduction of basal insulin secretion, which we observed in parkin-overexpressing MIN6 cells. Interestingly, Park2 gene expression in MIN6 cells was much lower compared with mouse pancreatic islets and, thus, is possibly down-regulated in an adaptive process to maintain insulin secretion. Assuming that parkin in pancreatic beta cells is also important to regulate mitochondrial integrity, further work is necessary to characterize this pathway.
Overexpression of the ULD of midnolin and the full-length midnolin in MIN6 cells resulted in reduced glucokinase activity and glucose-induced insulin secretion. This effect was not induced by diminished cell viability or reduced glucokinase protein but was more pronounced in low glucose culture, which strengthens our hypothesis that inactive glucokinase binds to the ULD of midnolin. To our knowledge midnolin expression has not been investigated on the protein level. Quite recently polyclonal antibodies against the N- and C-terminal part of midnolin became available. Both antibodies revealed that midnolin is significantly produced in insulin-secreting cells and localized not only in the nucleus but also in the cytoplasm. Furthermore, we observed distinct co-localization between glucokinase and midnolin in the cytoplasm of isolated primary mouse pancreatic beta cells as well as intact islets and in pancreatic sections. The localization of midnolin in the cytoplasm was not restricted to insulin-secreting cells but was also found in primary hepatocytes and HeLa cells. In our fluorescence microscopy experiments neither the endogenous nor the overexpressed midnolin showed localization in nucleoli. Thus, these results question the exclusively nuclear and nucleolar localization of midnolin, proposed by Tsukahara et al. (35), on the basis of the identified nucleolar localization signal sequence of midnolin and the use of GFP-chimera of the protein. Notably, Tsukahara et al. (35) did not perform immunocytochemistry analyses.
The first protein entry of midnolin (NCBI protein accession number XP_234902) (35) has been somewhat revised recently (NCBI protein accession number NP_001178506.1). Because the identified sequence17 did not contain the 43 amino acids specific for isoform 2 and 3 of midnolin and in all investigated tissues and cell lines of mouse and rat origin a single line of the protein at 50 kDa has been observed, isoform1 is most likely the predominant midnolin expressed in adult mouse and rat tissues. Notably, the midnolin expression level found in liver, brain, and muscle was significant and to a considerable degree higher than the parkin expression. Interestingly, only in human cell lines did we detect in addition another shorter isoform of midnolin of ∼37 kDa. Because this isoform became apparent using the C-terminal antibody, a truncation closer to the N terminus can be assumed.
Together, our findings suggest an important role of midnolin in adult tissues. Because midnolin contains a ULD as an integral component, the protein could play a role in protein quality control or cellular signaling. In pancreatic beta cells, evidence has been obtained that the ULD of midnolin serves as an anchor domain mediating binding to the glucose sensor enzyme glucokinase. This interaction seems to occur preferentially at low glucose and inhibits glucokinase activity. In light of the fact that glucokinase is regulated by the ubiquitin proteasome system (21–24), it has to be the aim of future work to elucidate if midnolin acts for example as known for parkin as an E3 ubiquitin ligase. Characterization of the midnolin isoforms, especially in human tissues, and functional analyses in primary pancreatic beta cells remain another important issue of future work. Furthermore, the results of our study indicate a specific function of parkin in pancreatic beta cells.
Acknowledgments
We thank M. Funk, J. Kresse, B. Leβ, A. Possler, and R. Waterstradt for skillful technical assistance.
The work was supported by Innovative Medicines Initiative Joint Undertaking Grant 155005 (Innovative Medicines Initiative for Diabetes), the resources of which are composed of financial contributions from the European Union Seventh Framework Programme (FP7/2007-2013) and European Federation of Pharmaceutical Industries and Associations (EFPIA) companies in kind contribution.
- ULD
- ubiquitin-like domain
- SD
- synthetic defined
- MMTHS
- mammalian two-hybrid system
- AD
- activation domain
- BD
- binding domain
- Park2
- parkin
- Ubl4 (Gdx)
- ubiquitin-like 4a
- Ubac1
- ubiquitin associated domain containing 1
- FRETN
- fluorescence resonance energy transfer efficiency
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ECFP
- enhanced cyan fluorescence protein
- EYFP
- enhanced yellow fluorescence protein
- GK
- glucokinase
- ANOVA
- analysis of variance.
REFERENCES
- 1. Koranyi L. I., Tanizawa Y., Welling C. M., Rabin D. U., Permutt M. A. (1992) Human islet glucokinase gene. Isolation and sequence analysis of full-length cDNA. Diabetes 41, 807–811 [DOI] [PubMed] [Google Scholar]
- 2. Magnuson M. A., Shelton K. D. (1989) An alternate promoter in the glucokinase gene is active in the pancreatic beta cell. J. Biol. Chem. 264, 15936–15942 [PubMed] [Google Scholar]
- 3. Matschinsky F. M., Magnuson M. A., Zelent D., Jetton T. L., Doliba N., Han Y., Taub R., Grimsby J. (2006) The network of glucokinase-expressing cells in glucose homeostasis and the potential of glucokinase activators for diabetes therapy. Diabetes 55, 1–12 [PubMed] [Google Scholar]
- 4. Baltrusch S., Tiedge M. (2006) Glucokinase regulatory network in pancreatic beta cells and liver. Diabetes 55, S55–S64 [Google Scholar]
- 5. German M. S. (1993) Glucose sensing in pancreatic islet beta cells. The key role of glucokinase and the glycolytic intermediates. Proc. Natl. Acad. Sci. U.S.A. 90, 1781–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Iynedjian P. B., Möbius G., Seitz H. J., Wollheim C. B., Renold A. E. (1986) Tissue-specific expression of glucokinase. Identification of the gene product in liver and pancreatic islets. Proc. Natl. Acad. Sci. U.S.A. 83, 1998–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lenzen S., Panten U. (1988) Signal recognition by pancreatic B-cells. Biochem. Pharmacol. 37, 371–378 [DOI] [PubMed] [Google Scholar]
- 8. Matschinsky F. M. (1990) Glucokinase as glucose sensor and metabolic signal generator in pancreatic beta cells and hepatocytes. Diabetes 39, 647–652 [DOI] [PubMed] [Google Scholar]
- 9. Matschinsky F. M., Ellerman J. E. (1968) Metabolism of glucose in the islets of Langerhans. J. Biol. Chem. 243, 2730–2736 [PubMed] [Google Scholar]
- 10. Wang H., Iynedjian P. B. (1997) Modulation of glucose responsiveness of insulinoma beta cells by graded overexpression of glucokinase. Proc. Natl. Acad. Sci. U.S.A. 94, 4372–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grupe A., Hultgren B., Ryan A., Ma Y. H., Bauer M., Stewart T. A. (1995) Transgenic knockouts reveal a critical requirement for pancreatic beta cell glucokinase in maintaining glucose homeostasis. Cell 83, 69–78 [DOI] [PubMed] [Google Scholar]
- 12. Ding S. Y., Nkobena A., Kraft C. A., Markwardt M. L., Rizzo M. A. (2011) Glucagon-like peptide 1 stimulates post-translational activation of glucokinase in pancreatic beta cells. J. Biol. Chem. 286, 16768–16774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Iynedjian P. B., Jotterand D., Nouspikel T., Asfari M., Pilot P. R. (1989) Transcriptional induction of glucokinase gene by insulin in cultured liver cells and its repression by the glucagon-cAMP system. J. Biol. Chem. 264, 21824–21829 [PubMed] [Google Scholar]
- 14. Sibrowski W., Seitz H. J. (1984) Rapid action of insulin and cyclic AMP in the regulation of functional messenger RNA coding for glucokinase in rat liver. J. Biol. Chem. 259, 343–346 [PubMed] [Google Scholar]
- 15. Liang Y., Najafi H., Smith R. M., Zimmerman E. C., Magnuson M. A., Tal M., Matschinsky F. M. (1992) Concordant glucose induction of glucokinase, glucose usage, and glucose-stimulated insulin release in pancreatic islets maintained in organ culture. Diabetes 41, 792–806 [DOI] [PubMed] [Google Scholar]
- 16. Tiedge M., Lenzen S. (1995) Effects of glucose refeeding and glibenclamide treatment on glucokinase and GLUT2 gene expression in pancreatic B-cells and liver from rats. Biochem. J. 308, 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Antoine M., Boutin J. A., Ferry G. (2009) Binding kinetics of glucose and allosteric activators to human glucokinase reveal multiple conformational states. Biochemistry 48, 5466–5482 [DOI] [PubMed] [Google Scholar]
- 18. Cornish-Bowden A., Storer A. C. (1986) Mechanistic origin of the sigmoidal rate behaviour of rat liver hexokinase D (“glucokinase”). Biochem. J. 240, 293–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kamata K., Mitsuya M., Nishimura T., Eiki J., Nagata Y. (2004) Structural basis for allosteric regulation of the monomeric allosteric enzyme human glucokinase. Structure 12, 429–438 [DOI] [PubMed] [Google Scholar]
- 20. Larion M., Salinas R. K., Bruschweiler-Li L., Miller B. G., Brüschweiler R. (2012) Order-disorder transitions govern kinetic cooperativity and allostery of monomeric human glucokinase. PLoS Biol. 10, e1001452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bjørkhaug L., Molnes J., Søvik O., Njølstad P. R., Flatmark T. (2007) Allosteric activation of human glucokinase by free polyubiquitin chains and its ubiquitin-dependent cotranslational proteasomal degradation. J. Biol. Chem. 282, 22757–22764 [DOI] [PubMed] [Google Scholar]
- 22. Hofmeister-Brix A., Lenzen S., Baltrusch S. (2013) The ubiquitin proteasome system regulates the stability and activity of the glucose sensor glucokinase in pancreatic beta cells. Biochem. J. 456, 173–184 [DOI] [PubMed] [Google Scholar]
- 23. Markwardt M. L., Nkobena A., Ding S. Y., Rizzo M. A. (2012) Association with nitric oxide synthase on insulin secretory granules regulates glucokinase protein levels. Mol. Endocrinol. 26, 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Negahdar M., Aukrust I., Johansson B. B., Molnes J., Molven A., Matschinsky F. M., Søvik O., Kulkarni R. N., Flatmark T., Njølstad P. R., Bjørkhaug L. (2012) GCK-MODY diabetes associated with protein misfolding, cellular self-association, and degradation. Biochim. Biophys. Acta 1822, 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Baltrusch S., Francini F., Lenzen S., Tiedge M. (2005) Interaction of glucokinase with the liver regulatory protein is conferred by leucine-asparagine motifs of the enzyme. Diabetes 54, 2829–2837 [DOI] [PubMed] [Google Scholar]
- 26. de la Iglesia N., Mukhtar M., Seoane J., Guinovart J. J., Agius L. (2000) The role of the regulatory protein of glucokinase in the glucose sensory mechanism of the hepatocyte. J. Biol. Chem. 275, 10597–10603 [DOI] [PubMed] [Google Scholar]
- 27. van Schaftingen E., Vandercammen A., Detheux M., Davies D. R. (1992) The regulatory protein of liver glucokinase. Adv. Enzyme Regul. 32, 133–148 [DOI] [PubMed] [Google Scholar]
- 28. van Schaftingen E., Veiga-da-Cunha M., Niculescu L. (1997) The regulatory protein of glucokinase. Biochem. Soc. Trans. 25, 136–140 [DOI] [PubMed] [Google Scholar]
- 29. Baltrusch S., Lenzen S., Okar D. A., Lange A. J., Tiedge M. (2001) Characterization of glucokinase-binding protein epitopes by a phage-displayed peptide library. Identification of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase as a novel interaction partner. J. Biol. Chem. 276, 43915–43923 [DOI] [PubMed] [Google Scholar]
- 30. Langer S., Kaminski M. T., Lenzen S., Baltrusch S. (2010) Endogenous activation of glucokinase by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase is glucose dependent. Mol. Endocrinol. 24, 1988–1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Massa L., Baltrusch S., Okar D. A., Lange A. J., Lenzen S., Tiedge M. (2004) Interaction of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFK-2/FBPase-2) with glucokinase activates glucose phosphorylation and glucose metabolism in insulin-producing cells. Diabetes 53, 1020–1029 [DOI] [PubMed] [Google Scholar]
- 32. Smith W. E., Langer S., Wu C., Baltrusch S., Okar D. A. (2007) Molecular coordination of hepatic glucose metabolism by the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase:glucokinase complex. Mol. Endocrinol. 21, 1478–1487 [DOI] [PubMed] [Google Scholar]
- 33. Baltrusch S., Lenzen S. (2007) Novel insights into the regulation of the bound and diffusible glucokinase in MIN6 beta cells. Diabetes 56, 1305–1315 [DOI] [PubMed] [Google Scholar]
- 34. Arden C., Harbottle A., Baltrusch S., Tiedge M., Agius L. (2004) Glucokinase is an integral component of the insulin granules in glucose-responsive insulin secretory cells and does not translocate during glucose stimulation. Diabetes 53, 2346–2352 [DOI] [PubMed] [Google Scholar]
- 35. Tsukahara M., Suemori H., Noguchi S., Ji Z. S., Tsunoo H. (2000) Novel nucleolar protein, midnolin, is expressed in the mesencephalon during mouse development. Gene 254, 45–55 [DOI] [PubMed] [Google Scholar]
- 36. Chomczynski P., Sacchi N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 37. Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Improved monomeric red, orange, and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 38. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tiedge M., Lortz S., Drinkgern J., Lenzen S. (1997) Relation between antioxidant enzyme gene expression and antioxidative defense status of insulin-producing cells. Diabetes 46, 1733–1742 [DOI] [PubMed] [Google Scholar]
- 40. Schmitt H., Lenzen S., Baltrusch S. (2011) Glucokinase mediates coupling of glycolysis to mitochondrial metabolism but not to beta cell damage at high glucose exposure levels. Diabetologia 54, 1744–1755 [DOI] [PubMed] [Google Scholar]
- 41. Vanderklish P. W., Krushel L. A., Holst B. H., Gally J. A., Crossin K. L., Edelman G. M. (2000) Marking synaptic activity in dendritic spines with a calpain substrate exhibiting fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 97, 2253–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Katoh K., Misawa K., Kuma K., Miyata T. (2002) MAFFT. A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. López-Avalos M. D., Duvivier-Kali V. F., Xu G., Bonner-Weir S., Sharma A., Weir G. C. (2006) Evidence for a role of the ubiquitin-proteasome pathway in pancreatic islets. Diabetes 55, 1223–1231 [DOI] [PubMed] [Google Scholar]
- 44. Yang F., Skaletsky H., Wang P. J. (2007) Ubl4b, an X-derived retrogene, is specifically expressed in post-meiotic germ cells in mammals. Gene Expr. Patterns 7, 131–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Buchberger A., Howard M. J., Proctor M., Bycroft M. (2001) The UBX domain. A widespread ubiquitin-like module. J. Mol. Biol. 307, 17–24 [DOI] [PubMed] [Google Scholar]
- 46. Grabbe C., Dikic I. (2009) Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem. Rev. 109, 1481–1494 [DOI] [PubMed] [Google Scholar]
- 47. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace. A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 48. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer. An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 49. Kiefer F., Arnold K., Künzli M., Bordoli L., Schwede T. (2009) The SWISS-MODEL Repository and associated resources. Nucleic Acids Res.. 37, D387–D392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) SWISS-MODEL. An automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Peitsch M. C., Wells T. N., Stampf D. R., Sussman J. L. (1995) The Swiss-3DImage collection and PDB-Browser on the World-Wide Web. Trends Biochem. Sci. 20, 82–84 [DOI] [PubMed] [Google Scholar]
- 52. Vijay-Kumar S., Bugg C. E., Cook W. J. (1987) Structure of ubiquitin refined at 1.8 Å resolution. J. Mol. Biol. 194, 531–544 [DOI] [PubMed] [Google Scholar]
- 53. Beers E. P., Callis J. (1993) Utility of polyhistidine-tagged ubiquitin in the purification of ubiquitin-protein conjugates and as an affinity ligand for the purification of ubiquitin-specific hydrolases. J. Biol. Chem. 268, 21645–21649 [PubMed] [Google Scholar]
- 54. Zielak A. E., Canty M. J., Forde N., Coussens P. M., Smith G. W., Lonergan P., Ireland J. J., Evans A. C. (2008) Differential expression of genes for transcription factors in theca and granulosa cells following selection of a dominant follicle in cattle. Mol. Reprod. Dev. 75, 904–914 [DOI] [PubMed] [Google Scholar]
- 55. Kiel C., Serrano L. (2006) The ubiquitin domain superfold. Structure-based sequence alignments and characterization of binding epitopes. J. Mol. Biol. 355, 821–844 [DOI] [PubMed] [Google Scholar]
- 56. Walters K. J., Goh A. M., Wang Q., Wagner G., Howley P. M. (2004) Ubiquitin family proteins and their relationship to the proteasome. A structural perspective. Biochim. Biophys. Acta 1695, 73–87 [DOI] [PubMed] [Google Scholar]
- 57. Welchman R. L., Gordon C., Mayer R. J. (2005) Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6, 599–609 [DOI] [PubMed] [Google Scholar]
- 58. Hartley T., Brumell J., Volchuk A. (2009) Emerging roles for the ubiquitin-proteasome system and autophagy in pancreatic beta cells. Am. J. Physiol. Endocrinol. Metab. 296, E1–E10 [DOI] [PubMed] [Google Scholar]
- 59. Kühn K., Zhu X. R., Lübbert H., Stichel C. C. (2004) Parkin expression in the developing mouse. Brain Res. Dev. Brain Res. 149, 131–142 [DOI] [PubMed] [Google Scholar]
- 60. Shimura H., Hattori N., Kubo S., Yoshikawa M., Kitada T., Matsumine H., Asakawa S., Minoshima S., Yamamura Y., Shimizu N., Mizuno Y. (1999) Immunohistochemical and subcellular localization of Parkin protein. Absence of protein in autosomal recessive juvenile parkinsonism patients. Ann. Neurol. 45, 668–672 [DOI] [PubMed] [Google Scholar]
- 61. Huynh D. P., Dy M., Nguyen D., Kiehl T. R., Pulst S. M. (2001) Differential expression and tissue distribution of parkin isoforms during mouse development. Brain Res. Dev. Brain Res. 130, 173–181 [DOI] [PubMed] [Google Scholar]
- 62. Narendra D. P., Jin S. M., Tanaka A., Suen D. F., Gautier C. A., Shen J., Cookson M. R., Youle R. J. (2010) PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 8, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Narendra D. P., Youle R. J. (2011) Targeting mitochondrial dysfunction. Role for PINK1 and Parkin in mitochondrial quality control. Antioxid. Redox. Signal. 14, 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]