Background: Protons activate acid-sensing ion channels (ASICs).
Results: Proton binding leads to a movement involving the thumb and finger subdomains. Mutation of carboxylates lining the finger leads to loss of activation and loss of this movement.
Conclusion: The carboxylates lining the finger domain are essential for the movement of the thumb and finger domains and in activation.
Significance: This study provides insight into proton-induced conformational changes in ASICs.
Keywords: Acid-sensing Ion Channels (ASICs), Fluorescence Resonance Energy Transfer (FRET), Ligand-binding Protein, Receptor Desensitization, Receptor Structure-Function, Ligand-gated Ion Channels, Receptor Activation
Abstract
Acid-sensing ion channels are cation channels activated by external protons and play roles in nociception, synaptic transmission, and the physiopathology of ischemic stroke. Using luminescence resonance energy transfer (LRET), we show that upon proton binding, there is a conformational change that increases LRET efficiency between the probes at the thumb and finger subdomains in the extracellular domain of acid-sensing ion channels. Additionally, we show that this conformational change is lost upon mutating Asp-238, Glu-239, and Asp-260, which line the finger domains, to alanines. Electrophysiological studies showed that the single mutant D260A shifted the EC50 by 0.2 pH units, the double mutant D238A/E239A shifted the EC50 by 2.5 pH units, and the triple mutant D238A/E239A/D260A exhibited no response to protons despite surface expression. The LRET experiments on D238A/E239A/D260A showed no changes in LRET efficiency upon reduction in pH from 8 to 6. The LRET and electrophysiological studies thus suggest that the three carboxylates, two of which are involved in carboxyl/carboxylate interactions, are essential for proton-induced conformational changes in the extracellular domain, which in turn are necessary for receptor activation.
Introduction
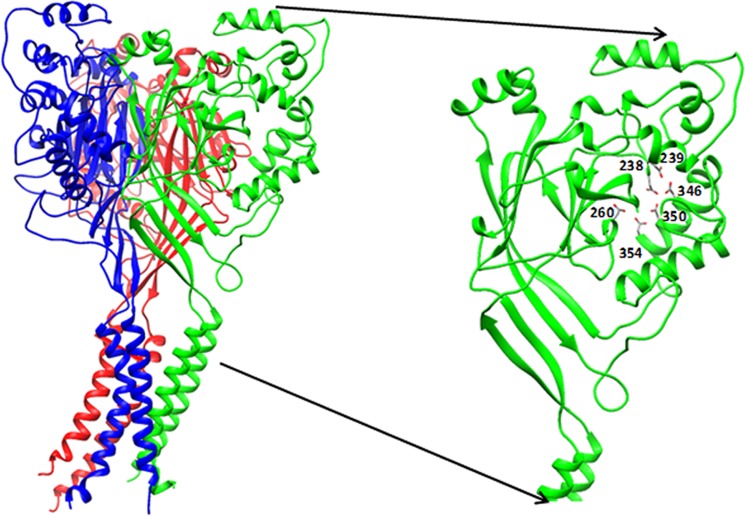
Acid-sensing ion channels (ASICs)2 are cation channels activated by external protons (1, 2). These proteins are important pharmacological targets, as they participate in various sensorial pathways, including nociception, synaptic transmission, and the physiopathology of ischemic stroke (3–5). Functional experiments have identified at least three distinct states: closed resting, open channel, and desensitized. Currently, high resolution structural data are available for chicken ASIC1a (cASIC1a) at pH 5.5 and bound to South American tarantula toxin, psalmotoxin (6–8). These structures are thought to represent the desensitized state and the open channel state of the protein, respectively (6–8). The structures show a trimeric receptor with a large funnel-shaped extracellular domain protruding above the membrane plane, a transmembrane domain, and cytoplasmic N and C termini (Fig. 1) (6). The extracellular domain in each subunit has five subdomains: the palm, finger, thumb, knuckle, and β-ball. Based on the x-ray structure, two pairs of unusually close (<3 Å) carboxyl/carboxylate interactions, Asp-238/Asp-350 and Glu-239/Asp-346, line the finger and thumb domains (Fig. 1, inset) (6–8). This observation led to the hypothesis that at high pH, the negatively charged deprotonated carboxylates repel each other, keeping the thumb and finger domains apart. Reduction in pH and protonation of one of the carboxylates in each pair would allow for the carboxyl/carboxylate interactions, resulting in a movement of the thumb domain toward the finger domain. This initial conformational change is thought to trigger channel activation. Neutralizing Asp-346 or Asp-350 in cASIC1a, as well as pairs 237/350 and 238/346 in rat ASICs, resulted in channels that were still pH-activated, albeit with a right shift in the EC50 for protons (6, 9, 10). In addition to these residues in the thumb and finger domains, several other residues in the extracellular domain have also been implicated in the gating mechanism and proton sensing through mutational studies (9, 11–15) and by cysteine accessibility measurements (16).
FIGURE 1.
Crystal structure of cASIC1a trimer (adapted from Protein Data Bank entry 2QTS) (6). Inset, thumb and finger domains with the three carboxylate pairs highlighted.
Here, we used mutational studies along with luminescence resonance energy transfer (LRET) to investigate the conformational changes in the thumb and finger domains associated with proton gating. We performed these measurements using the wild-type and mutant proteins and neutralizing the carboxylate residues lining the finger domain. These studies show that three carboxylates lining the finger domain, two of which form pairs with the carboxylates in the thumb domain, are essential for proton-mediated activation of the channel, and proton binding induces a movement of the probes in the thumb and finger domains toward each other that is essential for the gating of ASICs.
MATERIALS AND METHODS
Constructs
cASIC1a cloned in a pcDNA vector was used to express the protein in HEK293T cells, and the same was used for in vitro RNA transcription reaction to obtain RNA for injection into oocytes. The receptor was modified appropriately to enable introduction of cysteines by replacement of residues according to the distances being measured. Cysteine residues allow for specific labeling with maleimide-derived fluorophores. Double cysteine mutants with one cysteine in the finger domain (residue 130 or 139) and one in the thumb domain (residue 340) were used for measuring intrasubunit distances with maleimide-derived terbium chelate (Invitrogen) and ATTO 465 (Sigma-Aldrich) as the donor and acceptor fluorophores, respectively. A 1:1 ratio of donor and acceptor fluorophores was used for the labeling. Factor Xa (New England Biolabs) protease cleavage sites (IDGR) were introduced by replacement of residues on either side of one of the cysteines to enable cleavage of the cysteine, after which background LRET was measured. A similar procedure using thrombin as the protease has been used for glutamate receptors, and the detailed procedure has been described previously (17–20).
Site-directed Mutagenesis
Mutants were made using PfuTurbo DNA polymerase (Agilent Technologies). Primers with the mutations incorporated were synthesized by Sigma-Aldrich. PCR was performed using PfuTurbo DNA polymerase. The amplified product was digested with DpnI restriction enzyme (Roche Applied Science) to eliminate template DNA. The resulting amplified plasmid was transformed in Escherichia coli DH5α cells, and plasmid preparations from transformants were sequenced to confirm mutations.
Transfection in HEK293T Cells
HEK293T cells were transfected with pcDNA plasmids expressing the protein of interest. Cells were transfected with Lipofectamine 2000 (Invitrogen) and allowed to express the protein for 24–36 h before being harvested for LRET investigations.
RNA Synthesis and Injections
In addition to HEK293T cells, Xenopus laevis oocytes were used as an expression system. Receptor expression on oocyte membranes was done as described previously (18). Approximately 50 ng of RNA was injected into each oocyte and allowed to recover for 36–48 h at 12 °C. RNA was synthesized in vitro using the Ambion T7 mMESSAGE mMACHINE kit with linearized DNA as the template. Oocytes were then preblocked for exposed cysteines using N-maleoyl-β-alanine (Sigma-Aldrich) for 1 h at 18 °C, washed, and incubated at 18 °C for 24–36 h to allow for expression of the receptor before performing experiments.
LRET
Membrane fractions prepared from oocytes (18) and whole HEK293T cells were used for LRET measurements. A 1:1 ratio of donor (terbium chelate) to acceptor (ATTO 465) was used for all experiments. Three to four 10-cm dishes of HEK293T cells were labeled with the two fluorophores (200 nm each) at room temperature. Cells were washed three to four times with extracellular buffer prior to the LRET measurements. For oocytes, 100–200 oocytes were labeled with the two fluorophores (1 μm each) prior to membrane preparation. All measurements were done at pH 8 and 6. A cuvette-based QuantaMaster model QM3-SS fluorescence spectrometer (Photon Technology International) was used to measure the lifetimes of the fluorophores. A high power pulsed xenon lamp was used for excitation, and the emitted light was passed through a monochromator and passed onto the detector. The temperature was set at 15 °C during the experiments. Fluorescan software (Photon Technology International) was used for acquisition of data, and Origin 4.0 software (OriginLab Corp.) was used for data analysis. The donor-only lifetime measurements were obtained at 545 nm, whereas the sensitized acceptor lifetimes for ATTO 465 were obtained at 508 nm. The LRET measurements were obtained from at least three preparations of protein for each mutant. For each sample, LRET lifetimes were obtained before and after digestion with Factor Xa, and the background LRET after digestion with the protease was subtracted from the initial LRET data to obtain the LRET specific to the ASIC protein (supplemental Fig. 1). The subtracted data were fit to the minimum number of exponentials that best fit the data. The data shown are an average of three samples, with three runs per sample, with each run being an average of 99 scans.
Distance Measurements
Distances between the donor and acceptor fluorophores were calculated using the LRET lifetime (τDA) and donor-only lifetime (τD) using the Förster equation. The R0 value was calculated as described previously (20) and was 36 Å for the terbium chelate/ATTO 465 pair. The largest error in the distances determined by LRET is thought to arise from the orientation factor (κ) included in calculation of R0, although dos Remedios and Moens (21) have argued, using several FRET measurements, that the assumption of 2/3 provides reliable results. For lanthanides that are isotropic, this error is reduced to ±10% at most (21, 22). Additionally, in this study, we were investigating relative changes and not absolute distances; thus, it was expected that the error would be further reduced as the same donor and acceptors were studied under the two pH conditions. Hence, the errors reported Table 1 were calculated based on the error propagation in the fitting of the averaged lifetime data in the Förster equation. The absolute error in the measurements would be ±10% of the distances at most.
TABLE 1.
Sensitized emission lifetimes and corresponding distances measured from receptors tagged with donor and acceptor fluorophores
| Construct/expression system | τD | τDA | R |
|---|---|---|---|
| μs | μs | Å | |
| L139C/Q340C | |||
| Oocytes | |||
| pH 8 | 1648 ± 1.2 | 350 ± 5.4 | 29 ± 0.07 |
| pH 6 | 1672 ± 2.4 | 268 ± 4.3 | 27 ± 0.07 |
| HEK293T cells | |||
| pH 8 | 1555 ± 1.2 | 293 ± 3.0 | 28 ± 0.04 |
| pH 6 | 1575 ± 1.7 | 225 ± 1.3 | 26 ± 0.02 |
| T130C/Q340C | |||
| Oocytes | |||
| pH 8 | 1575 ± 1.1 | 257 ± 2.0, 560 ± 8 | 27 ± 0.03, 32 ± 0.4 |
| pH 6 | 1588 ± 1.3 | 173 ± 1.7, 630 ± 10 | 25 ± 0.06, 33 ± 0.5 |
| HEK293T cells | |||
| pH 8 | 1511 ± 1.2 | 252 ± 8, 520 ± 7 | 28 ± 0.1, 32 ± 0.4 |
| pH 6 | 1589 ± 1.4 | 183 ± 3, 650 ± 9 | 26 ± 0.07, 33 ± 0.5 |
| L139C/Q340C/D238A/E239A | |||
| HEK293T cells | |||
| pH 8 | 1672 ± 0.6 | 304 ± 1.8 | 28 ± 0.02 |
| pH 6 | 1745 ± 0.6 | 312 ± 2.06 | 28 ± 0.03 |
| L139C/Q340C/D238A/E239A/D260A | |||
| HEK293T cells | |||
| pH 8 | 1666 ± 0.6 | 293 ± 1.1 | 28 ± 0.01 |
| pH 6 | 1752 ± 0.6 | 293 ± 1.3 | 28 ± 0.02 |
Electrophysiology
Outside-out patch recordings were performed on HEK293T cells transfected with the indicated ASIC construct and enhanced GFP (7.5:1 μg of ASIC:enhanced GFP cDNA/10 ml of medium) using Lipofectamine 2000. 24–48 h post-transfection, outside-out patches were pulled from enhanced GFP-expressing cells using thick-walled borosilicate glass pipettes of 3–5 megaohms, coated with beeswax, fire-polished, and filled with a solution containing 135 mm CsF, 33 mm CsOH, 11 mm EGTA, 10 mm HEPES, 2 mm MgCl2, and 1 mm CaCl2 (pH 7.4). External solutions were composed of 150 mm NaCl, 10 mm HEPES, 1 mm MgCl2, and 1 mm CaCl2 and adjusted to the indicated pH with 5 n NaOH or 10 n HCl. All recordings were performed with a holding potential of −60 mV using an Axopatch 200B amplifier (Molecular Devices, Sunnyvale, CA), acquired at 30–40 kHz, and filtered at 10 kHz (8-pole Bessel) under the control of pCLAMP 10 software. Series resistances (3–12 megohms) were routinely compensated by >95% where the amplitude exceeded 100 pA. Rapid application was performed using home-built theta (Warner Instruments, Hamden, CT) or multibarrel (VitroCom, Mountain Lakes, NJ) glass application pipettes, pulled to 100–150 μm, and translated using a piezoelectric microstage (Burleigh Instruments). Solution exchange as estimated from open tip potentials was 100–300 μs (10–90% rise time). The extent of apparent desensitization (see Fig. 2) was taken as the percent of the steady-state response during a low pH application compared with the peak response. For dose-response curves (see Fig. 6), various pH values were bracketed by control pH 5.0 responses to assess rundown. Data were normalized to the adjacent control response, averaged across patches, and fit with the standard dose-response logistic equation. Statistical significance was evaluated using Student's two-tailed t test.
FIGURE 2.
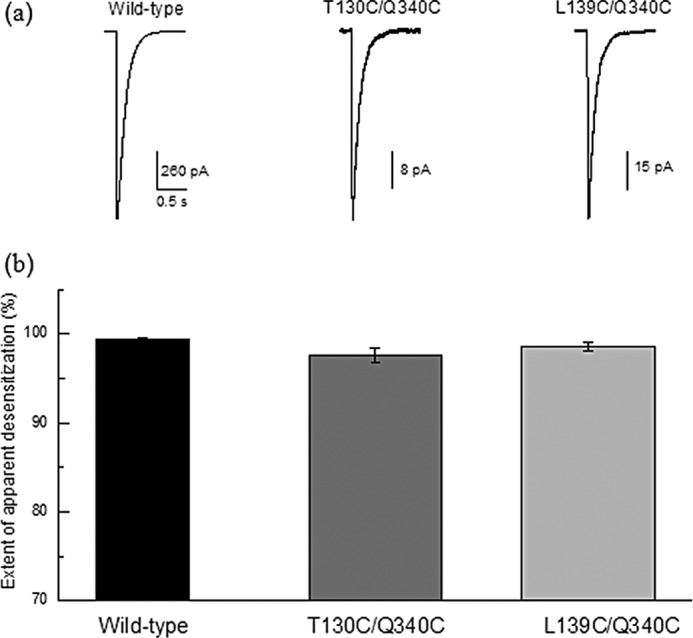
a, currents evoked from outside-out patches expressing wild-type ASIC (left panel) and the 130/340 (middle panel) and 139/340 (right panel) LRET construct channels in response to a pH jump from 8.0 to 6.0 for 1 s. b, extent of desensitization upon reduction in pH from 8.0 to 6.0 for wild-type ASIC and the 130/340 and 139/340 LRET constructs.
FIGURE 6.
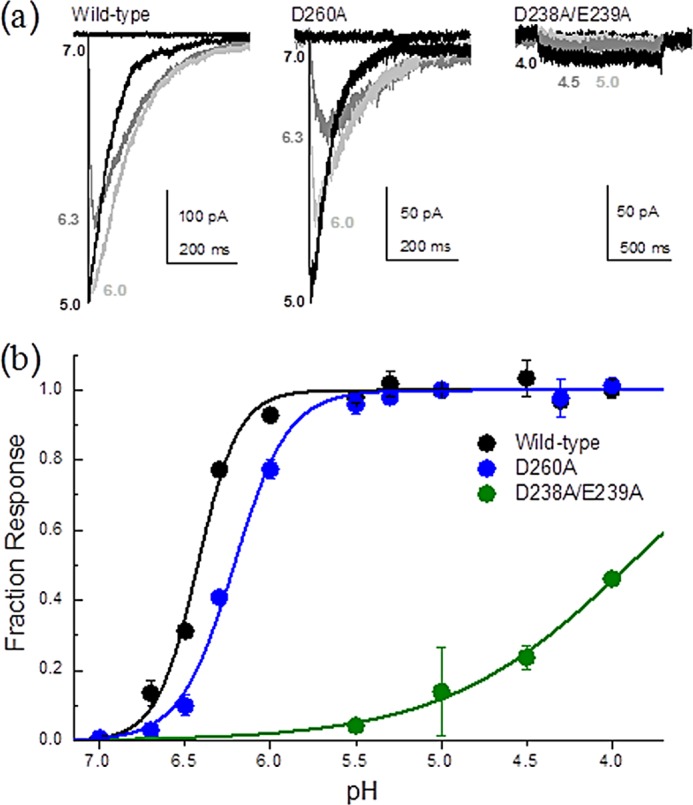
pH dependence of activation in carboxylate mutants. a, example responses from patches containing wild-type ASIC (left panel), D260A (middle panel), and D238A/E239A in the 139/340 LRET background (right panel) at the indicated pH values. b, summary of pH-response curves for wild-type ASIC (black), D260A (blue), and D238A/E239A in the 139/340 LRET background (green). Each point is an average of 3–15 patches.
Surface Biotinylation and NeutrAvidin Pulldown Assay, followed by Western Blotting
Surface expression of the D238A/E239A/D260A triple mutant of cASIC1a was determined using surface biotinylation and NeutrAvidin pulldown, followed by Western blotting as described by Zha et al. (23). The triple mutant with a FLAG tag introduced at the N terminus expressed in HEK293T cells was used for these experiments. Biotinylation was performed 48 h after transfection. Cells were washed with cold PBS solution, and 3 ml of 0.5 mg/ml NHS-biotin (Pierce) in cold PBS was used to tag the surface-expressed proteins with biotin. The reaction was quenched using 0.1 m glycine in ice-cold PBS with calcium chloride and magnesium chloride. The cells were then lysed with lysis buffer containing 30 mm N-ethylmaleimide (Sigma), 1% Nonidet P-40 (Roche Applied Science), 0.5% deoxycholate, and 0.5% SDS in ice-cold PBS+/+, along with protease inhibitor mixture (Roche Applied Science). Cells were then sonicated, and the lysate was spun down. To 300 μl of cleared cell lysate 60 μl of NeutrAvidin slurry was added, followed by overnight incubation at 4 °C. The NeutrAvidin beads were then washed with wash buffer containing 50 mm Tris (pH 7.4) and 1% Triton X-100. After the washes, beads were directly boiled in SDS sample buffer and loaded onto SDS-polyacrylamide gel for Western blot analysis. Western blotting was done following standard procedures, and the blot was probed with anti-FLAG monoclonal antibody (Sigma-Aldrich), followed by secondary antibody (HRP-conjugated anti-mouse antibody, Sigma-Aldrich).
RESULTS
Measuring Conformational Changes in cASIC1a Receptors Expressed in HEK293T Cells and Oocytes
To measure changes in the functional cASIC1a receptors expressed in HEK293T cells and oocytes without purifying the proteins, we introduced the recognition sequence for the protease Factor Xa (IDGR) on either side of one fluorophore-binding site. Measuring the LRET signal before and after digestion with Factor Xa allows for the quantitative subtraction of the background signal (supplemental Fig. 1). Cysteines were introduced at residues 130 and 139 for tagging with donor and acceptor fluorophores in the finger domain. These residues were chosen based on their anticipated ability to reflect conformational changes and because the Factor Xa sequence could be incorporated into the flanking amino acid sequences with minimum perturbation. Electrophysiological measurements from excised patches of HEK293T cells expressing the wild-type and mutant proteins used for the LRET measurements revealed that these constructs were functional and had similar gating kinetics as the wild-type receptor as seen from the rate and extent of desensitization (Fig. 2).
Conformational Changes between the Thumb and Finger Domains upon pH Reduction
To map the distance changes between the thumb and finger domains within a subunit, LRET measurements were performed using the ASIC constructs with cysteines at sites 139 and 340 and with cysteines at sites 130 and 340.
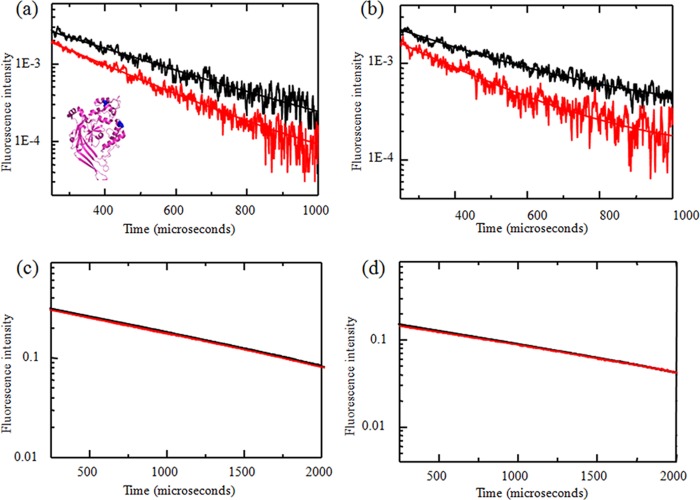
The LRET lifetimes for the 139/340 construct (Fig. 3a, inset) at pH 8 and 6 expressed in HEK293T cells and oocytes (membrane preparations) are shown in Fig. 3 (a and b, respectively), and the corresponding lifetimes from donor only-labeled receptors are shown in Fig. 3 (c and d, respectively). The black and red lines represent pH 8 and 6, respectively, unless indicated otherwise. The LRET lifetimes could be well represented by a single exponential, indicating that the primary energy transfer is within the subunit. This is consistent with the fact that <15% efficiency of LRET is expected for intersubunit LRET at this site based on the x-ray structures. Based on the LRET and donor lifetimes at pH 8, the intrasubunit distance between sites 139 and 340 was calculated to be 28 Å in HEK293T cells and 29 Å in oocytes (Table 1). Reducing the pH from 8 to 6 resulted in a decrease in distance of 2 Å, with the distances being 26 and 27 Å in HEK293T cells and oocytes, respectively. This distance is similar to the distance of 24 Å between the backbone Cα of sites 139 and 340 and the distance of 25 Å between Cδ of the site 139 side chain leucine and Oϵ of the site 340 side chain glutamine observed in the low pH crystal structure of cASIC1a. The pH-dependent decrease in distance between the donor and acceptor fluorophores suggests a conformational change that brings the two fluorophores tagged at the finger and thumb domains closer upon binding protons.
FIGURE 3.
LRET measurements were performed using sensitized emission lifetime measurements from receptors tagged with terbium chelate and ATTO 465, measured using double cysteine mutants at residues 139 and 340 in HEK293T cells (a) and oocyte membranes (b) at pH 8 (black) and pH 6 (red). The corresponding donor lifetime measurements are shown in c and d, respectively. Inset, positions of sites 139 and 340 in the extracellular domain of a single subunit of the ASIC.
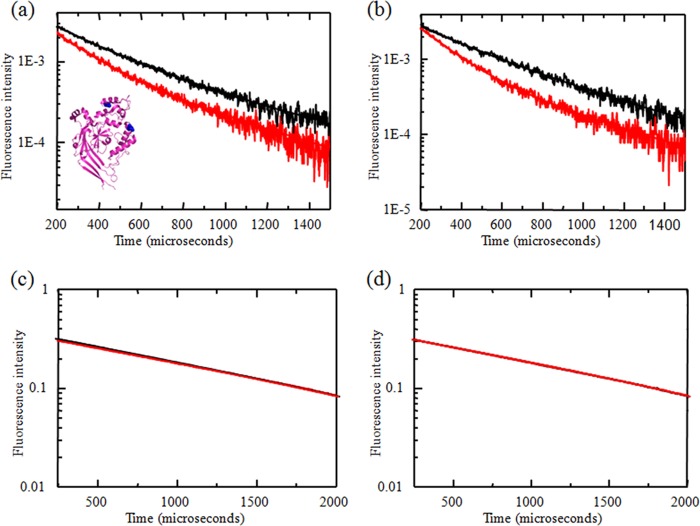
To further test the pH-dependent motion of the thumb and finger domains, we repeated the experiments using the 130/340 construct (Fig. 4). Similar LRET lifetimes were obtained for the 130/340 construct (Fig. 4a, inset) at pH 8 and 6 expressed in HEK293T cells and oocytes (Fig. 4, a and b, respectively). The corresponding lifetimes from donor only-labeled receptors are shown in Fig. 4 (c and d, respectively). The LRET lifetime was fit to two exponentials decays. Based on the shorter lifetimes and the donor-only lifetimes, the distances were calculated to be 28 and 27 Å between sites 130 and 340 in HEK293T cells and oocytes, respectively, at pH 8. Reducing the pH from 8 to 6 resulted in a decrease in the distance of 2 Å between sites 130 and 340 in both HEK293T cells and oocytes. As with the 130/340 mutant, these distances are similar to the distance of 27 Å between the backbone Cα of sites 130 and 340 and the distance of 28 Å between Oγ of the site 130 side chain leucine and Oϵ of the site 340 side chain glutamine observed in the low pH crystal structure of cASIC1a (6). The longer decays were 560 and 630 μs for pH 8 and 6, respectively. These lifetimes correspond to 32 and 33 Å, respectively. The distance of 33 Å at pH 6 is similar to the intersubunit distance of 33 Å at position 130, thus indicating that this component is arising from a small fraction of intersubunit LRET at this site.
FIGURE 4.
LRET measurements were performed using sensitized emission lifetime measurements from receptors tagged with terbium chelate and ATTO 465, measured using double cysteine mutants at residues 130 and 340 in HEK293T cells (a) and oocyte membranes (b) at pH 8 (black) and pH 6 (red). The corresponding donor lifetime measurements are shown in c and d, respectively. Inset, positions of sites 130 and 340 in the extracellular domain of a single subunit of the ASIC.
Because the distance changes upon reduction in pH are similar between residues 130 and 340 and between residues 139 and 340, we can conclude that the change is arising due to a movement of the fluorophore at site 340 toward the fluorophore at sites 139 and 130. Such a motion is consistent with the thumb and finger domains being held together through carboxyl/carboxylate hydrogen bonds as seen in the low pH crystal structure (6). At high pH, these residues are expected to deprotonate and repel each other, leading to the moving apart of the finger and thumb domains.
Effect of Disrupting the Carboxyl/Carboxylate Pairs between the Thumb and Finger Domains
Based on the x-ray structures, the carboxylate pairs Asp-238/Asp-350 and Glu-239/Asp-346 have been hypothesized to be proton sensors and the driving force for the movement of the thumb and finger domains toward each other, ultimately gating/activating the channel (6). To understand the contribution of these carboxyl/carboxylate pairs, we performed electrostatic calculations using the APBS plugin in VMD (24). The results revealed that neutralizing the residues in pairs Asp-238/Asp-350 and Glu-239/Asp-346 significantly reduced the electrostatic potential at these positions. However, there was still a substantial negative electrostatic field deep in the pocket (Fig. 5C). There are two additional carboxylate side chains from Asp-260 and Glu-354 at this location on the finger and thumb domains, respectively. They are 7 Å apart in the crystal structure and have not been suggested as a carboxyl/carboxylate pair in previous studies. However, it is possible that these residues could be closer in the dynamic state of the protein. Additionally, because the electrostatic interactions are expected to be in effect at this distance, they could contribute to proton sensing. We thus performed additional calculations after eliminating this third pair of carboxylate residues (Asp-260 and Glu-354) in addition to the initial two pairs. This resulted in a near loss of the negative electrostatic potential, as indicated by the disappearance of the electric field lines (Fig. 5, C and D). These studies show that in addition to Asp-238, and Glu-239, Asp-260 may also play a role in pH-mediated gating in ASICs. We thus characterized the functional effect of the D260A single, D238A/E239A double, and D238A/E239A/D260A triple mutations.
FIGURE 5.
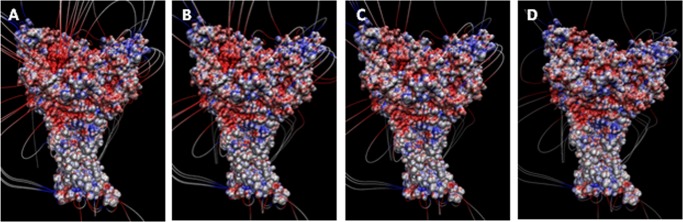
Electrostatic potential and field lines calculated using the adaptive Poisson-Boltzmann solver software (24) for wild-type ASIC protein (A); pair 260/354 mutated to alanines (B); pairs 238/350 and 239/346 mutated to alanines (C); and pairs 238/350, 239/346, and 260/354 mutated to alanines (D).
The electrophysiological studies with the D260A single mutation showed a slight but significant shift in the EC50 by 0.2 pH units relative to the wild-type protein (pH50 for the wild-type protein of 6.42 ± 0.01 (n ≥ 4) and pH50 for D260A of 6.21 ± 0.02 (n ≥ 4); p < 0.0001). This shift in EC50 showed that the electrostatics at this carboxylate site contributed to the proton sensing, consistent with the electrostatic calculations. Analysis of the D238A/E239A ASIC double mutant was complicated by the fact that D238A/E239A showed very small currents (Fig. 6a, right panel) and that extreme proton concentrations (beyond pH 4) compromised our solution delivery system and were inaccessible to LRET measurements (see below). Nonetheless, we did find that the D238A/E239A double mutation substantially shifted the dose-response curves to lower pH (pH50 for D238A/E239A of 3.92 ± 0.04 (n = 3)) (Fig. 6). These studies are consistent with previous work showing that the single mutants at sites 346 (which pairs with site 239) and 350 (which pairs with site 238) in cASIC1a result in a shift in EC50 (6, 9, 10) and that mutating pairs 237/350 and 238/346 in rat ASICs also results in a shift in EC50 (9, 10). The triple mutant D238A/E239A/D260A yielded no electrophysiological responses in 10 patches tested in the pH range of 7 to 4. To ensure that the protein was expressed on the surface of the cell, we performed surface biotinylation and NeutrAvidin pulldown, followed by Western blotting to probe for FLAG-tagged ASIC subunits. The Western blot shows a band corresponding to a molecular mass of 60 kDa, as expected for the single subunit of ASICs, indicating surface expression of the D238A/E239A/D260A ASIC triple mutant (Fig. 7).
FIGURE 7.
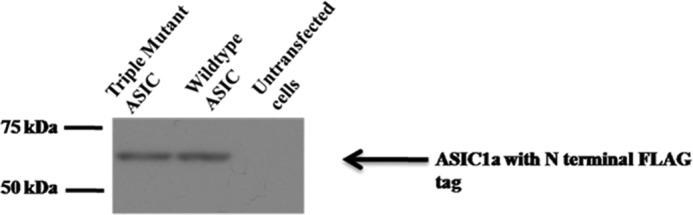
Western blot probing expression of ASICs in cells transfected with the D238A/E239A/D260A triple mutant and wild-type ASIC and untransfected HEK293T cells. ASIC has an N-terminal FLAG tag, and Western blotting was performed using anti-FLAG antibody.
To test the effect of the loss of pH-mediated activation on the conformational changes between the thumb and finger domains, we performed LRET investigations on the D238A/E239A double and D238A/E239A/D260A triple mutants. The LRET lifetimes at pH 8 and 6 (after subtraction of the background signal using Factor Xa cleavage) for the D238A/E239A double mutant in the 139/340 fluorescence construct expressed in HEK293T cells tagged with terbium chelate and ATTO 465 are shown in Fig. 8. The LRET lifetimes could be well represented with a single exponential decay at both pH 8 and 6. The LRET lifetimes for the D238A/E239A double mutant showed no changes upon reduction of the pH from 8 to 6, suggesting no changes between the thumb and finger domains. Additionally, the distances at both pH values for the D238A/E239A double mutant are similar to those for the wild-type 139/340 fluorescence construct at pH 8, suggesting that the distance in the D238A/E239A double mutant is similar to that in the high pH resting state of the corresponding wild-type protein. Because the double mutant protein did not gate at pH 6, the loss in movement in the double mutant confirms that the thumb-to-finger movement is essential for protein-mediated gating. We could not perform experiments at pH <6, as the terbium chelate showed a decrease in intensity possibly due to loss of the terbium from the chelate. These results were further confirmed by the LRET data for the D238A/E239A/D260A triple mutant. The triple mutant in the 139/340 fluorescence construct background expressed in HEK293T cells was also tagged with terbium chelate and ATTO 465 and studied using LRET. The LRET lifetimes at pH 8 and 6 (after subtraction of the background signal using Factor Xa cleavage) are shown in Fig. 8. The LRET intensities observed for the D238A/E239A/D260A triple mutant were similar to those observed for the wild-type LRET construct, showing surface expression, consistent with the biotinylation studies. The LRET lifetimes could be well represented with a single exponential decay at both pH 8 and 6. No significant changes were observed in the lifetimes between pH 8 and 6. Furthermore, the distances based on the LRET lifetimes for the mutant protein are the same as those observed for the wild-type 139/340 LRET construct at pH 8 (Table 1). This shows that the D238A/E239A/D260A triple mutant is similar to the high pH state of the wild-type ASIC protein, with the thumb and finger domains not being able to be held close to each other through the hydrogen bond interactions between the carboxyl/carboxylate pairs.
FIGURE 8.
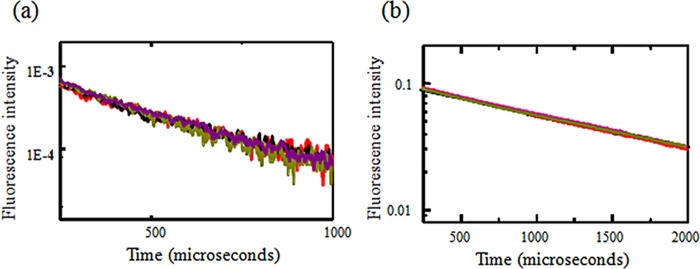
a, LRET measurements using sensitized emission lifetime measurements from receptors tagged with terbium chelate and ATTO 465, measured using double cysteine mutant at residues 139 and 340 with double mutant D238A/E239 (black and red) and triple mutant D238A/E239A/D260A in HEK293T cells at pH 8 and 6 (violet and olive), respectively. b, the corresponding donor lifetime measurements.
DISCUSSION
The x-ray structure of ASIC shows a trimeric structure with a large extracellular domain (6–8). Each subunit has the shape of an upright arm, with the extracellular domain having a structure similar to that of a hand. A large negatively charged electrostatic patch is seen in the extracellular domain of each subunit, which has been suggested as the primary proton sensor. Specifically, within this acidic patch, two pairs of unusually close (<3 Å) carboxyl/carboxylate interactions (Asp-238/Asp-350 and Glu-239/Asp-346) are observed. Based on the proximity of these negatively charged residues, it has been suggested that these residues would have a higher pKa and could get protonated in the range of the gating pH, thus acting as the “proton sensor.” Electrophysiological measurements of receptors with mutations at these two sites showed that the EC50 was shifted for the proton response curves to a more acidic pH (Fig. 6) (6, 9, 10) but did not result in a complete loss of proton gating. This suggests that additional residues are involved in the proton-sensing process. Using electrostatic maps, we identified a third pair of carboxylates deeper in the pocket (Asp-260/Glu-354), which may contribute to proton sensing. Consistent with this hypothesis, eliminating only this carboxylate pair (D260A) resulted in a small but consistent right shift in the pH-response curve (Fig. 6). Moreover, the residual proton sensitivity in the D238A/E239A double mutant was completely eliminated with the addition of the D260A mutation. Despite surface expression, this triple mutation was insensitive to acidic solutions as low as pH 4. Our findings suggest that all three carboxylate pairs help initiate the gating process by acting as proton sensors.
The close proximity of the carboxylate pairs in the thumb and finger domains also suggests that the two domains are likely to be farther apart at higher pH, and upon protonation of one of the carboxylate side chains in each pair, the thumb and finger domains are expected to be locked at a shorter distance, which in turn could be the trigger for activation. Using LRET, we showed that proton binding resulted in the moving closer of fluorophores attached to the thumb and finger domains. In further support of this hypothesis, this movement was lost under the same pH conditions in the D238A/E239A double and D238A/E239A/D260A triple mutants.
The mutational and LRET studies established that there is a conformational change between the thumb and finger domains, and this is essential for channel gating and probably represents the initial step in this process, which, when propagated to other segments such as the palm domain (16) and ultimately to the channel, leads to activation of the receptor.
This work was supported in part by National Science Foundation Grant MCB-1110501 (to V. J), a Harry S. and Isabel C. Cameron Foundation fellowship (to S. S. R.), and an American Heart Association postdoctoral fellowship (to D. M. M.)

This article contains supplemental Fig. 1.
- ASIC
- acid-sensing ion channel
- cASIC1a
- chicken ASIC1a
- LRET
- luminescence resonance energy transfer.
REFERENCES
- 1. Krishtal O. A., Pidoplichko V. I. (1980) A receptor for protons in the nerve cell membrane. Neuroscience 5, 2325–2327 [DOI] [PubMed] [Google Scholar]
- 2. Waldmann R., Champigny G., Bassilana F., Heurteaux C., Lazdunski M. (1997) A proton-gated cation channel involved in acid-sensing. Nature 386, 173–177 [DOI] [PubMed] [Google Scholar]
- 3. Krishtal O. (2003) The ASICs: signaling molecules? Modulators? Trends Neurosci. 26, 477–483 [DOI] [PubMed] [Google Scholar]
- 4. Lingueglia E. (2007) Acid-sensing ion channels in sensory perception. J. Biol. Chem. 282, 17325–17329 [DOI] [PubMed] [Google Scholar]
- 5. Xiong Z.-G., Pignataro G., Li M., Chang S., Simon R. P. (2008) Acid-sensing ion channels (ASICs) as pharmacological targets for neurodegenerative diseases. Curr. Opin. Pharmacol. 8, 25–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jasti J., Furukawa H., Gonzales E. B., Gouaux E. (2007) Structure of acid-sensing ion channel 1 at 1.9 Å resolution and low pH. Nature 449, 316–323 [DOI] [PubMed] [Google Scholar]
- 7. Baconguis I., Gouaux E. (2012) Structural plasticity and dynamic selectivity of acid-sensing ion channel-spider toxin complexes. Nature 489, 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dawson R. J., Benz J., Stohler P., Tetaz T., Joseph C., Huber S., Schmid G., Hügin D., Pflimlin P., Trube G., Rudolph M. G., Hennig M., Ruf A. (2012) Structure of the acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1. Nat. Commun. 3, 936. [DOI] [PubMed] [Google Scholar]
- 9. Li T., Yang Y., Canessa C. M. (2009) Interaction of the aromatics Tyr-72/Trp-288 in the interface of the extracellular and transmembrane domains is essential for proton gating of acid-sensing ion channels. J. Biol. Chem. 284, 4689–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sherwood T., Franke R., Conneely S., Joyner J., Arumugan P., Askwith C. (2009) Identification of protein domains that control proton and calcium sensitivity of ASIC1a. J. Biol. Chem. 284, 27899–27907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li T., Yang Y., Canessa C. M. (2011) Outlines of the pore in open and closed conformations describe the gating mechanism of ASIC1. Nat. Commun. 2, 399. [DOI] [PubMed] [Google Scholar]
- 12. Li T., Yang Y., Canessa C. M. (2011) Asp433 in the closing gate of ASIC1 determines stability of the open state without changing properties of the selectivity filter or Ca2+ block. J. Gen. Physiol. 137, 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li T., Yang Y., Canessa C. M. (2010) Two residues in the extracellular domain convert a nonfunctional ASIC1 into a proton-activated channel. Am. J. Physiol. Cell Physiol. 299, C66–C73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li T., Yang Y., Canessa C. M. (2010) Leu85 in the β1-β2 linker of ASIC1 slows activation and decreases the apparent proton affinity by stabilizing a closed conformation. J. Biol. Chem. 285, 22706–22712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li T., Yang Y., Canessa C. M. (2010) Asn415 in the β11-β12 linker decreases proton-dependent desensitization of ASIC1. J. Biol. Chem. 285, 31285–31291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frey E. N., Pavlovicz R. E., Wegman C. J., Li C., Askwith C. C. (2013) Conformational changes in the lower palm domain of ASIC1a contribute to desensitization and RFamide modulation. PLoS ONE 8, e71733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gonzalez J., Du M., Parameshwaran K., Suppiramaniam V., Jayaraman V. (2010) Role of dimer interface in activation and desensitization in AMPA receptors. Proc. Natl. Acad. Sci. U.S.A. 107, 9891–9896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gonzalez J., Rambhadran A., Du M., Jayaraman V. (2008) LRET investigations of conformational changes in the ligand binding domain of a functional AMPA receptor. Biochemistry 47, 10027–10032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rambhadran A., Gonzalez J., Jayaraman V. (2010) Subunit arrangement in N-methyl-d-aspartate (NMDA) receptors. J. Biol. Chem. 285, 15296–15301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rambhadran A., Gonzalez J., Jayaraman V. (2011) Conformational changes at the agonist binding domain of the N-methyl-d-aspartic acid receptor. J. Biol. Chem. 286, 16953–16957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. dos Remedios C. G., Moens P. D. (1995) Fluorescence resonance energy transfer spectroscopy is a reliable “ruler” for measuring structural changes in proteins. Dispelling the problem of the unknown orientation factor. J. Struct. Biol. 115, 175–185 [DOI] [PubMed] [Google Scholar]
- 22. Selvin S., Abrams B. (1996) Analysing the relationship between maternal weight gain and birth weight: exploration of four statistical issues. Paediatr. Perinat. Epidemiol. 10, 220–234 [DOI] [PubMed] [Google Scholar]
- 23. Zha X. M., Wang R., Collier D. M., Snyder P. M., Wemmie J. A., Welsh M. J. (2009) Oxidant regulated inter-subunit disulfide bond formation between ASIC1a subunits. Proc. Natl. Acad. Sci. U.S.A. 106, 3573–3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]