Background: NEK2 is a mammalian kinase that promotes centrosome separation during the cell cycle.
Results: Agents that demethylate the NEK2 promoter or induce DNA damage repress NEK2 expression in a p53-dependent manner.
Conclusion: p53 represses NEK2 expression and protects its binding region from accumulating DNA methylation.
Significance: Knowledge regarding novel mechanisms of NEK2 regulation may help inform clinical application of NEK2-based anticancer therapeutics.
Keywords: Chromatin Regulation, Chromatin Structure, DNA Methylation, Epigenetics, Nucleosome, p53, MAPit, NEK2
Abstract
Genome-scale mapping suggests that the function of DNA methylation varies with genomic context beyond transcriptional repression. However, the use of DNA-demethylating agents (e.g. 5-aza-2′-deoxycytidine (5aza-dC)) to study epigenetic regulation often focuses on gene activation and ignores repression elicited by 5aza-dC. Here, we show that repression of NEK2, which encodes the never in mitosis A (NIMA)-related kinase, by 5aza-dC is context-specific as NEK2 transcript levels were reduced in HCT116 colon cancer cells but not in isogenic p53−/− cells. Bisulfite sequencing showed that DNA methylation was restricted to the distal region of the NEK2 promoter. Demethylation by 5aza-dC was associated with increased accessibility to micrococcal nuclease, i.e. nucleosome depletion. Conversely, methyltransferase accessibility protocol for individual templates (MAPit) methylation footprinting showed that nucleosome occupancy and DNA methylation at the distal promoter were significantly increased in p53−/− cells, suggesting dynamic regulation of chromatin structure at this region by p53 in HCT116 cells. Stabilization of endogenous p53 by doxorubicin or ectopic expression of p53, but not a p53 DNA-binding mutant, decreased NEK2 expression. Chromatin immunoprecipitation demonstrated direct and specific association of p53 with the distal NEK2 promoter, which was enhanced by doxorubicin. Luciferase reporters confirmed that this region is required for p53-mediated repression of NEK2 promoter activity. Lastly, modulation of p53 abundance altered nucleosome occupancy and DNA methylation at its binding region. These results identify NEK2 as a novel p53-repressed gene, illustrate that its repression by 5aza-dC is specific and associated with nucleosome reorganization, and provide evidence that identification of partially methylated regions can reveal novel p53 target genes.
Introduction
DNA methylation is widely accepted as a transcriptionally repressive epigenetic mark when observed near transcription start sites (TSSs)3 in the core promoters of genes (1). In contrast, the function of DNA methylation in other genomic contexts outside of core promoters, e.g. distal regulatory, insulator, or enhancer regions, remains controversial. For instance, gene body methylation correlates with transcriptionally active rather than repressed genes (2–5). Microarray profiling of transcripts from cells treated with DNA-demethylating agents, such as 5aza-dC (also known as decitabine), is often used to identify epigenetically regulated genes and has identified many derepressed genes. These global studies often also report genes that are repressed by treatment; however, this repression is often regarded as nonspecific. A few groups have shown that gene repression following 5aza-dC treatment results from demethylation of binding sites for transcriptional repressors (6–9).
Maintenance of proper mitotic division during the cell cycle is crucial to the genomic integrity of dividing cells. The serine/threonine kinase NEK2 is the mammalian homolog of never in mitosis A (NIMA), an Aspergillus nidulans protein required for mitosis. Although NEK2 is not required for mitotic entry, it is a well documented regulator of centrosome separation. Loss of NEK2 activity inhibits centrosome separation, whereas overexpression leads to premature centrosome separation. NEK2 overexpression has also been described in a number of human tumor types (10–13), making it an attractive drug target especially for breast cancer treatment (14, 15).
In normal cells, NEK2 transcription is cell cycle-regulated. NEK2 mRNA and protein levels are very low in M and G1, increase in S, and peak in G2 (10, 16–20). Known NEK2 transcriptional regulators include transcription factor E2F4, which represses transcription in G0 and G1 via the retinoblastoma (Rb) family members p107 and p130 (19). By contrast, the transcription factor FoxM1 activates NEK2 expression in G2 (21). Besides its cell cycle regulation, very little is known about modulation of NEK2 expression.
In a previous study, we observed p53-dependent repression of NEK2 following treatment with 5aza-dC (8). Here, we report that NEK2 is repressed by wild-type p53 and that loss of p53 results in local accumulation of DNA methylation. We characterize the relevant p53-repressive region in the NEK2 promoter and show that this region is bound by p53 in vivo. Finally, we show that modulation of p53 levels affects both nucleosome positioning and the DNA methylation status of the distal promoter where the p53-binding site is located. These results not only identify a novel p53-repressed target gene but provide evidence that identification of partially methylated regions can reveal important transcriptional regulatory elements in human cells.
EXPERIMENTAL PROCEDURES
Cell Culture
HCT116, HCT116 p53−/−, and HCT116 p21−/− colon cancer cells (a generous gift from Dr. Bert Vogelstein) were maintained in McCoy's 5A modified growth medium (Invitrogen) supplemented with 10% FBS and 1 unit/ml penicillin plus 1 μg/ml streptomycin. All cells were maintained in a humidified 37 °C incubator with 5% CO2.
Drug Treatments
Cells were grown to 60% confluence and then treated with 0.1, 0.5, or 1 μm doxo (Sigma) or vehicle (sterile water) as indicated for the indicated times. Cells were seeded to 20% confluence and then maintained in 200 nm 5aza-dC (Sigma) or DMSO vehicle for 4 days. 5aza-dC-containing medium was changed daily to provide fresh drug.
RNA Extraction
Cells were homogenized in TriReagent® (Molecular Research Center), and RNA was precipitated with isopropanol, applied to RNeasy spin columns (Qiagen), eluted in nuclease-free water, and treated with RNase-free DNase for 30 min at 37 °C followed by heat inactivation at 75 °C. RNA was stored at −80 °C.
RT-Quantitative PCR (RT-qPCR)
TaqMan real time RT-qPCR assays for NEK2, Ki67, and β-actin mRNAs as well as 18 S rRNA were developed using Primer Express software (Applied Biosystems) based on sequences from GenBankTM. The 18 S rRNA, β-actin, and Ki67 assay details have been published previously (22). Additional RT-qPCR primer sequences are listed in supplemental Table 1. The assays were performed at the Quantitative Genomics Core Laboratory (University of Texas-Houston Medical School) using a 7700 Sequence Detector (Applied Biosystems) as described previously (8). Transcripts were quantified using the ΔΔCt method, normalizing to 18 S rRNA or β-actin mRNA.
Whole Cell Lysate Preparation and Protein Immunoblotting
Cells were harvested in Triton X-100 lysis buffer (1% (v/v) Triton X-100, 150 mm NaCl, 25 mm Tris, pH 7.5 supplemented with 1 mm sodium fluoride, 1 mm glycerol phosphate, and 1× Complete Mini protease inhibitor mixture (Roche Applied Science)) as described previously (8). Membranes were incubated overnight at 4 °C with primary antibodies specific for NEK2 (D-8, Santa Cruz Biotechnology), p53 (D01, Calbiochem), and β-actin (C-4, Millipore) followed by species-specific secondary antibodies and visualization by chemiluminescence.
Cell Cycle Assay
Cells were treated as indicated in each figure, trypsinized, washed in PBS, and resuspended in propidium iodide buffer (50 μg/ml propidium iodide, 0.1% (v/v) Triton X-100, 0.1% (w/v) sodium citrate in PBS). Samples were stored at 4 °C for 2 h, then vortexed, and analyzed for DNA content by flow cytometry on a Guava® Personal Cell Analysis (PCA)-96 flow cytometer. Results were exported into Excel for data analysis.
Plasmids
The construct for overexpression of enhanced GFP (EGFP)-tagged p53 (pp53-EGFP, Clontech; hereafter p53-GFP) was provided by Lawrence Donehower's laboratory at Baylor College of Medicine. Nucleotide 580 in the p53 coding sequence was mutagenized from C to T to generate the L194F DNA-binding mutant using the Stratagene QuikChange mutagenesis kit according to the manufacturer's instructions. Successful mutagenesis was confirmed by sequencing. For long term p53 expression studies, p53-GFP and p53(L194F)-GFP were cloned into tetracycline-inducible pTreDual2 (Clontech). The resulting construct was co-transfected with a linear puromycin resistance marker (Clontech) into p53−/− cells containing a stably integrated, blasticidin (Blast)-resistant construct expressing the reverse tetracycline transactivator (rtTA) from the CMV promoter (pLenti CMV rtTA-3 Blast; w756-1; Addgene plasmid 26429). The p53-inducible luciferase reporter (luciferase expression driven by several repeated p53-binding sites) was provided by Russell Broaddus' laboratory at the M. D. Anderson Cancer Center. The NEK2 promoter-luciferase reporter was purchased from SwitchGear Genomics (Menlo Park, CA). The full-length NEK2 promoter in this reporter construct was digested with XhoI and StuI to excise 102 bp containing the p53-regulatory region. Two other deletions were generated by digesting with XhoI and BglII (removes 399 bp) or AvrII and HindIII (removes 170 bp). All mutants were verified by sequencing.
Luciferase Assays
Cells seeded to 60% confluence in 24-well plates were co-transfected with 150 ng of reporter vector; 10 ng of thymidine kinase (TK) promoter-driven Renilla luciferase (pRL-TK-Renilla, Promega) as an internal control for transfection efficiency and cell viability; and 50 ng of either GFP, p53-GFP, or p53(L194F)-GFP. Cells were harvested 24 h after transfection, and luminescence was measured in triplicate using Promega Dual-Glo luciferase assay reagents according to manufacturer's protocol using an Optima series luminometer.
ChIP
Cells were trypsinized, washed, fixed in 1% (v/v) formaldehyde for 8 min, and quenched with 0.125 m glycine for 5 min. Fixed nuclei were sonicated at 4 °C using a Bioruptor (Diagenode) set to high intensity with alternating 30-s on/off pulses for a total of 40 min. Following verification by agarose gel electrophoresis of chromatin fragments sonicated to an average length of ∼300 bp, chromatin was immunoprecipitated using either anti-p53 clone D01 antibody or IgG control antibody and protein A/G-coated agarose beads. Following several washes, DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and ethanol-precipitated using molecular grade glycogen as carrier. Eluted DNA was analyzed by qPCR using SYBR® Green reagents (Applied Biosystems) according to the manufacturer's recommendations. CDKN1A ChIP primers were published previously (23). Additional ChIP primer sequences are listed in supplemental Table 1. Enrichment levels were quantified using the ΔΔCt method and are presented either as the percentage of input for each amplicon or as enrichment relative to untreated cells, normalized to GAPDH.
EMSA
A dsDNA probe with 28 bp of NEK2 promoter sequence (containing the 23-bp p53-binding site) followed by the T7 promoter was assembled from three oligonucleotides (Integrated DNA Technologies): NEK2 EMSA+ (top strand), GGTTTCGCCATGTTGGCCAGGCTGGTCTTAATACGACTCACTATAGGG; NEK2 EMSA− (3′ bottom strand), AGACCAGCCTGGCCAACATGGCGAAACC; and T7 promoter with a cyanine 5 (Cy5) 5′ fluorescent label (5′ bottom strand), Cy5-CCCTATAGTGAGTCGTATTA. After assembly, the probe product was purified to remove non-annealed oligonucleotides using a PCR purification spin column (Qiagen). Binding reactions (room temperature for 20 min) contained 60 ng of purified p53 protein (Active Motif), buffer (10 mm Tris, pH 7.5, 50 mm KCl, 5 mm MgCl2, 1 mm DTT, 0.05% (v/v) Triton X-100, 2.5% (v/v) glycerol), and 250 fmol of labeled probe. Protein-DNA complexes were resolved by electrophoresis at room temperature on 5% polyacrylamide (w/v) Tris borate-EDTA gels. Competition and supershift reactions were preincubated (before adding Cy5-labeled probe) at 4 °C for 20 min with excess unlabeled competitor DNA and anti-p53 antibody (D01), respectively. For binding competitions, oligonucleotides containing only 28 bp of NEK2 sequence (i.e. no T7 sequence) were synthesized as either unmethylated or C-5 methylated (underlined C residues in the NEK2 EMSA+/− oligonucleotides), annealed, and purified.
Methyltransferase Accessibility Protocol for Individual Templates (MAPit), Single Molecule Footprinting, Bisulfite Genomic Sequencing (BGS), and Pyrosequencing
Nuclei were prepared and probed with 0 or 30 units of M.CviPI (New England Biolabs). Reactions were performed and genomic DNA was extracted and deaminated as described (24). Deaminated DNA was amplified in triplicate using HotStar Taq reagents (Qiagen). Triplicate PCRs with primers specific for bisulfite-converted DNA (supplemental Table 2) were pooled, gel-purified, and TA-cloned using T-easy vector and reagents (Promega). Individual clones were sequenced and analyzed as described (25, 26).
For deep sequencing, primers NEK2 US+/− were synthesized with appended 454 adapters and barcodes according to the manufacturer's recommendations (Roche Applied Science) (supplemental Table 2). After PCR amplification from deaminated DNA, products were gel-purified and submitted for deep sequencing on a 454 Genome Sequencer FLXTM instrument (Roche Applied Science) at the University of Florida Interdisciplinary Center for Biotechnology Research.
For pyrosequencing, deaminated DNA was amplified using primers listed in supplemental Table 2 as described for MAPit but using a 5′-biotinylated reverse primer. Reactions were sequenced as described previously (27) using a PyroMark Q96 MD system (Biotage, Uppsala, Sweden). Methylation frequency was quantified using PyroMark Pyro Q-CpG (version 1.0.9) software.
Quantitative MNase Digestion
Nuclei were prepared and probed with 0–2 units of MNase (Worthington). Reactions were incubated and stopped, and genomic DNA was extracted as described previously (28). DNA was examined by gel electrophoresis and ethidium bromide staining to confirm the presence of a “DNA ladder” indicative of incomplete MNase digestion. Primers for qPCR analysis of NEK2 and GAPDH were the same as used for ChIP. Protection from MNase digestion was quantified using the ΔΔCt method, normalizing to GAPDH.
RESULTS
p53-dependent Repression of NEK2 by 5aza-dC Occurs at a Low Dose and Is Not Due to Cell Line-selective Changes in Cell Cycle or Proliferation
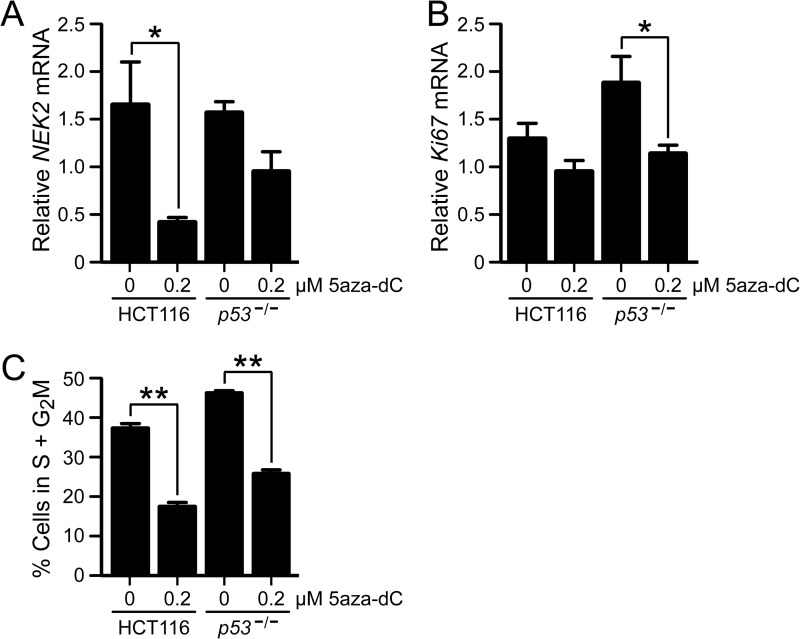
We previously reported microarray and RT-qPCR data that showed that NEK2 was repressed by the DNA demethylation agent 5aza-dC in parental HCT116 cells (hereafter HCT116) but not in isogenic, p53-null HCT116 cells (hereafter p53−/−) (by microarray, 50% repression in HCT116 versus 4% repression in p53−/−, p = 7.36−38; by RT-qPCR, 68% repression in HCT116 versus 10% repression in p53−/−, p = 0.028) (8). Note that treatment with 5aza-dC was previously shown to not induce p53 expression in HCT116 cells (8). To rule out that repression of NEK2 by 5aza-dC treatment was due to one or more off-target effects of the high dose (2 μm) of drug treatment used in that study, experiments were repeated with a lower dose (Fig. 1). Cells treated with 0.2 μm 5aza-dC resulted in a 75% reduction in NEK2 transcript in HCT116 cells (p < 0.05) and a numeric 33% reduction in the p53−/− cells (non-significant) compared with vehicle (Fig. 1A). These results indicate that the repression of NEK2 by 5aza-dC treatment in HCT116 cells is not due to off-target effects of a high dose.
FIGURE 1.
p53-dependent repression of NEK2 by 5aza-dC is not due to nonspecific effects of off-target high dose or on cell cycle and proliferation. RT-qPCR analysis of NEK2 (A) or Ki67 (B) transcript levels following vehicle (distilled water) or low dose (0.2 μm) 5aza-dC treatment of HCT116 or p53−/− cells. Bars indicate the mean ratio of transcript/18 S rRNA (n = 3) following 5aza-dC treatment for each cell type relative to vehicle-treated, control HCT116 cells. C, percentage of cells in S + G2M phases calculated from cell cycle analysis of 5aza-dC- or vehicle-treated cells. Bars indicate the mean percentage of cells in S + G2M (n = 3). *, p < 0.05; **, p < 0.01 by ANOVA followed by Bonferroni ad hoc test. Error bars in each panel represent S.E.
NEK2 expression in many cases is associated with proliferating cells, and 5aza-dC treatment often causes cell growth to slow or stop. Thus, to determine whether the observed p53-dependent repression of NEK2 is due to alteration of the proliferation status of HCT116, but not p53−/−, cells in response to drug treatment, we measured the transcript level of Ki67 by RT-qPCR as described previously (29). Ki67 is a nuclear antigen commonly used as a marker of proliferation. We found that there was a trend of decreased Ki67 transcript levels following 200 nm 5aza-dC treatment (non-significant); however, the decrease was stronger in the p53−/− cells (Fig. 1B; HCT116, 24% decrease; p53−/−, 39% decrease) and therefore could not account for the significant decrease in NEK2 levels in the HCT116 cells. Interestingly, the magnitude of the changes in mRNA levels of NEK2 and Ki67 was similar in the p53−/− cells, suggesting that the modest, non-significant repression of NEK2 observed in the p53−/− cells could be due to decreased cellular proliferation. We conclude that the p53-dependent repression of NEK2 by 5aza-dC was not due to decreased cell proliferation occurring selectively in HCT116 cells.
NEK2 expression is cell cycle-regulated with expression levels peaking during S/G2 of the cell cycle. Therefore, to confirm that p53-dependent NEK2 repression was not due to cell cycle differences between the HCT116 and p53−/− cell lines, we quantified the fraction of cells in S and G2M phases before and after drug treatment by flow cytometry of propidium iodide-stained cells. We observed a similar, significant decrease in the percentage of cells in S and G2M phases following drug treatment in both cell types (Fig. 1C; HCT116, 53% decrease; p53−/−, 44% decrease). These similar decreases in dividing cells are unlikely to explain the significant NEK2 repression that occurs in only the HCT116 cells following treatment. Taken together, these data show that the p53-dependent NEK2 repression observed following 5aza-dC treatment is not likely due to the drug exerting effects on cell cycle distribution selectively in HCT116 cells.
NEK2 Repression by 5aza-dC Correlates with Demethylation of Its Distal Promoter
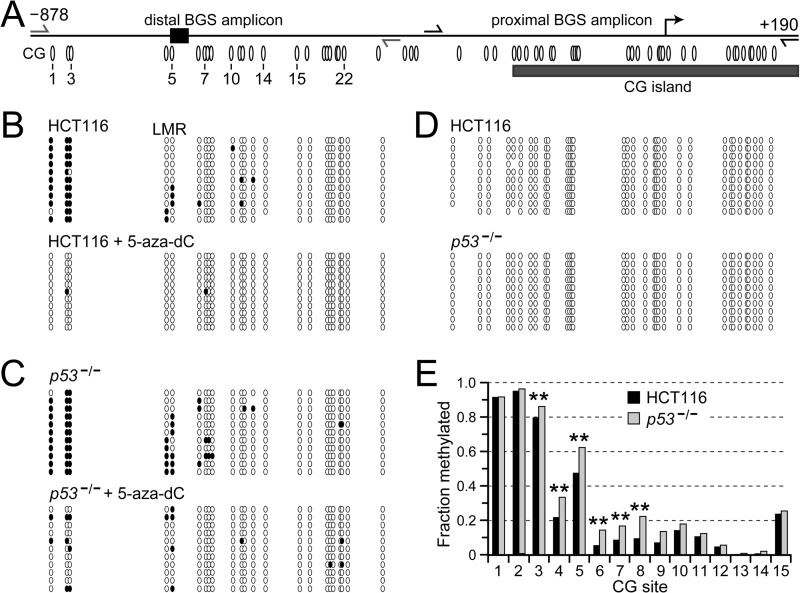
To determine whether 5aza-dC-mediated repression of NEK2 is associated with demethylation of the NEK2 promoter, we conducted BGS (30) (Fig. 2). In the distal promoter region of both HCT116 and p53−/− cells, three CG sites just downstream of −878 were densely methylated in untreated cells (Fig. 2, A; B, top; C, top; and E, CG sites 1–3). By contrast, several CG sites farther downstream exhibited intermediate to low methylation frequencies. Methylation of all these sites was removed in both cell types by 5aza-dC treatment (Fig. 2, B and C, compare top with bottom; p = 0.0001 for both cell lines + 5aza-dC compared with untreated). The proximal promoter was unmethylated in both HCT116 and p53−/− cells (Fig. 2D). Interestingly, DNA methylation was more abundant in the distal promoter region in p53−/− compared with HCT116 cells (Fig. 2, B and C, compare top panels, and E, CG sites 3–8; p = 0.0001).
FIGURE 2.
5aza-dC treatment decreases, whereas depletion of p53 increases, DNA methylation at the distal, but not proximal, NEK2 promoter. A, schematic of 1068 bp of the NEK2 promoter. The bent arrow indicates the TSS. The gray rectangle depicts the 5′ boundary of the annotated CG island that extends further into the gene body. Ovals mark locations to scale of CG sites, numbered to correspond to sites for which quantitative data are reported in E. The gray and black half-arrows, indicate convergent primer-binding sites for BGS of the “distal” and “proximal” promoter regions, respectively. DNA methylation in the distal (B and C) and proximal (D) promoter regions is shown. Each row of ovals represents an individually cloned and sequenced molecule from the indicated cell type. Unfilled ovals represent unmethylated, whereas black ovals represent methylated CG sites. E, for quantitative assessment, barcoded primers were used to generate the amplicons, which were then subjected to deep sequencing. The fraction of molecules methylated at each CG site that attained at least 100-fold coverage is shown. Mean sequencing coverage was 511 ± 147 reads for HCT116 and 950 ± 287 reads for p53−/− cells. Sequencing results are presented starting at the 5′-end of the distal promoter amplicon such that CG site 1 corresponds to the first CG site observed in A–C. **, p < 0.0001 comparing HCT116 with p53−/− using two-tailed Fisher's exact test.
The pattern of intermediate DNA methylation frequencies observed at CG sites 4–12 in the distal NEK2 promoter in HCT116 cells is reminiscent of low-methylated regions (LMRs) recently described by Stadler et al. (31). LMRs were identified in the mouse methylome in embryonic stem cells as CG-poor distal regulatory regions containing intermediate levels (mean 30%) of DNA methylation. Importantly, factor binding to these regions and to promoter-proximal sequences was found to protect them against accumulation of DNA methylation (31, 32). Given the increased accumulation of DNA methylation at the NEK2 LMR in p53−/− cells and the p53-dependent nature of NEK2 repression following 5aza-dC treatment, we hypothesized that NEK2 is directly repressed by p53 and that this LMR harbors a p53-binding site. Note that the NEK2 LMR falls outside the CG island annotated in the UCSC Genome Browser (Fig. 2A).
NEK2 mRNA and Protein Levels Are Repressed by Endogenous p53
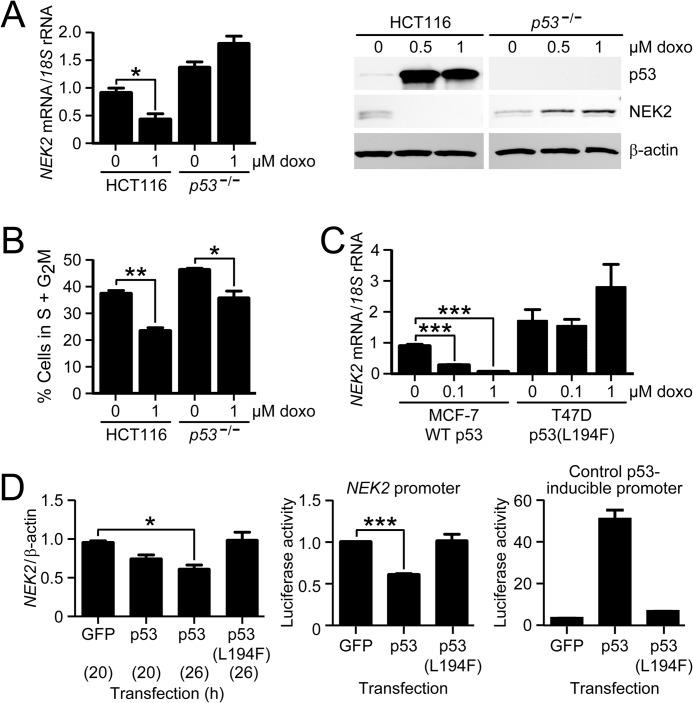
To determine whether NEK2 is repressed by endogenous p53, we treated HCT116 cells with a 1 μm concentration of the p53-inducing drug doxo or vehicle for 48 h and then measured NEK2 transcript levels by RT-qPCR (Fig. 3A, left). We observed that doxo treatment resulted in a significant 62% decrease in NEK2 mRNA levels in HCT116 cells but a numeric 36% increase in p53−/− cells that was not statistically significant. Consistent with the transcript data, NEK2 protein levels were also strongly repressed following 0.5 and 1 μm doxo treatment of HCT116, but not of p53−/−, cells (Fig. 3A, right).
FIGURE 3.
NEK2 transcript and protein levels are repressed by p53 and depend on p53 DNA binding activity. A, RT-qPCR analysis of NEK2 transcript (left) and immunoblot for NEK2 protein (right) after treatment of HCT116 and p53−/− cells with the indicated doses of doxo for 48 h. Bars represent the mean ratio of NEK2/18 S levels (n = 5) relative to vehicle-treated control HCT116 cells. β-Actin serves as the immunoblot loading control. B, quantification of the percentage of HCT116 and p53−/− cells in S plus G2M phases determined by cell cycle analysis after treatment with and without 1 μm doxo. Bars indicate the mean percentage of cells in S + G2M phases (n = 3). C, RT-qPCR analysis of NEK2 transcript in MCF-7 (WT p53) and T47D (L194F mutant p53) cells following doxo treatment at the indicated dosages. Bars represent the mean ratio of NEK2/18 S (n = 3) relative to untreated MCF-7 cells. D, left, RT-qPCR analysis of NEK2 transcript after transfection of HCT116 cells with constructs expressing WT p53 or p53(L194F) mutant for the indicated hours (in parentheses). Bars represent the mean ratio of NEK2/β-actin mRNA levels (n = 3) relative to GFP-transfected control cells. Luciferase activity expressed in HCT116 cells following co-transfection of luciferase reporters under control of either the NEK2 promoter (middle; n = 5) or a control p53-inducible promoter (right; n = 2) plus GFP control, p53-GFP, or p53(L194F)-GFP. Bars represent mean firefly luciferase activity normalized to Renilla luciferase activity, relative to GFP control transfection, measured in triplicate. *, p < 0.05; **, p < 0.01; ***, p < 0.001 by ANOVA and Bonferroni ad hoc test. Error bars in each panel represent S.E.
To again rule out that these results were merely due to differential cell cycle arrest, cells were treated with doxo or vehicle and subjected to propidium iodide staining and cell cycle analysis (Fig. 3B). The percentage of cells in S and G2M phases was quantified pre- and post-drug treatment. The S plus G2M phase fraction decreased similarly following doxo treatment of both HCT116 and p53−/− cells (HCT116, 37% decrease; p53−/−, 23% decrease). Therefore, differential cell cycle regulation cannot explain the HCT116-specific decrease in NEK2 expression.
To determine whether p53-dependent repression of NEK2 by doxo treatment occurs in a different cell type, we measured NEK2 expression levels in MCF-7 and T47D cells (Fig. 3C). Both cell lines are breast cancer-derived; MCF-7 cells express wild-type p53, whereas T47D cells express a mutant p53 protein with an L194F amino acid substitution in the DNA-binding domain that eliminates p53 binding to DNA (International Agency for Research on Cancer TP53 Database). MCF-7 cells treated with doxo exhibited a dose-dependent decrease in NEK2 mRNA. In contrast, T47D cells showed no change in NEK2 levels at 0.1 μm doxo and a non-significant numeric increase at 1 μm doxo. These results suggest that NEK2 expression is repressed by p53 by a mechanism that is dependent on its ability to bind DNA.
NEK2 Repression by Exogenously Expressed p53 Is Dependent on DNA Binding
To determine whether ectopic expression of p53 would repress NEK2 and whether this repression was dependent on DNA binding, HCT116 cells were transiently transfected with fusion protein p53-GFP or p53(L194F)-GFP (Fig. 3D). Transfection of each construct resulted in equivalent levels of expressed protein (data not shown). Wild-type p53-GFP, but not p53(L194F)-GFP, caused a decrease in levels of NEK2 transcript (Fig. 3D, left). Additionally, activity of an NEK2 promoter-luciferase reporter was repressed by co-transfected p53-GFP but not by the p53(L194F)-GFP mutant (Fig. 3D, middle). Transfections using a control p53-inducible luciferase reporter confirmed that p53-GFP, but not p53(L194F)-GFP, activated the reporter (Fig. 3D, right). Together, these data show that p53-dependent transcriptional repression of NEK2 is dependent on the DNA binding activity of p53.
Identification of p53 Regulatory Sites in the NEK2 Promoter
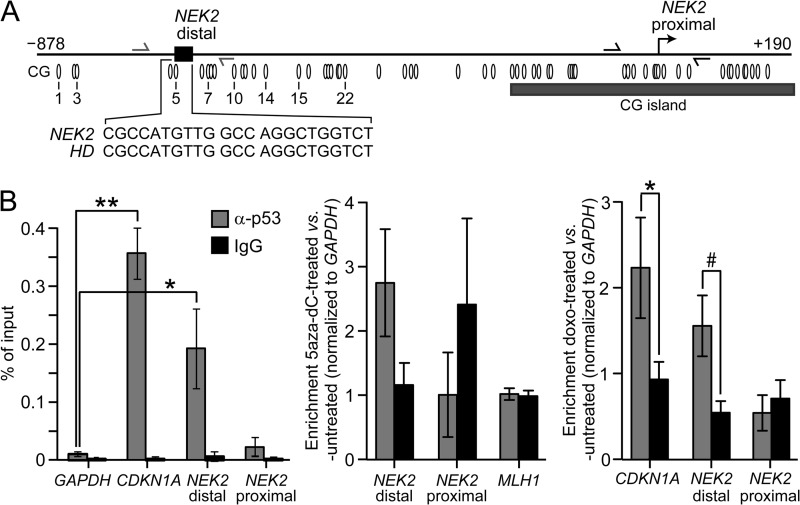
To our knowledge, NEK2 has not previously been shown to be a p53 target gene. Transcription factor analysis using a publically available database (TFSEARCH) did not detect any p53-binding sites in the NEK2 promoter. However, the consensus p53-binding motif is highly degenerate, and many algorithms cannot detect binding sites that substantially diverge from consensus. To further investigate the potential presence of a p53-binding site, we aligned the NEK2 promoter sequence from −1500 to +1 with a list of validated p53-binding sites from 129 known p53 target genes (for a review, see Ref. 33). We identified a putative p53-binding site in the NEK2 promoter 688 bp upstream of exon 1 (Fig. 4A). The binding site is 100% identical to the validated p53-binding site from the huntingtin gene (HD) gene promoter (34). Intriguingly, HD is activated, not repressed, by p53 binding. Note that the identified putative binding site in the NEK2 promoter is located in the region identified as an LMR by BGS in Fig. 2, A and B.
FIGURE 4.
p53 binds to the distal NEK2 promoter in HCT116 cells. A, schematic of the NEK2 promoter. The black rectangle and callout, respectively, indicate the location of the NEK2 p53-binding sequence and its 100% identity to that in the HD promoter. Ovals mark locations to scale of CG sites, numbered the same as in Fig. 2A. The bent arrow demarcates the NEK2 TSS. The gray and black half-arrows indicate ChIP primer binding sites for amplification of distal and proximal NEK2 promoter sequences, respectively. B, left, quantitative ChIP analysis following immunoprecipitation with either anti-p53 or IgG antibodies in untreated HCT116 cells. Data are presented as percentage of input DNA. Each bar represents the mean (n = 4). *, p < 0.05; **, p < 0.01 by ANOVA and Bonferroni ad hoc test versus GAPDH enrichment for the anti-p53 immunoprecipitation. Quantitative ChIP analysis following treatment with 5aza-dC (middle; mean ± error bars that represent 0.5 of the range; n = 2) or doxo (right; mean ± error bars that represent S.E.; n = 3). Data are presented as enrichment relative to untreated control, normalized to GAPDH. *, p < 0.05; #, p = 0.054.
To determine whether this distal region is bound by p53 in vivo, ChIP analysis was conducted using primers flanking the putative binding region (Fig. 4, A and B). We observed enrichment of distal NEK2 promoter sequences following ChIP with an anti-p53 antibody compared with IgG isotype control. PCR amplification of distal NEK2 promoter sequences was enhanced 19-fold compared with the GAPDH promoter (Fig. 4B, left, compare bar 5 with bar 1, respectively, 0.19 versus 0.01% of input; p < 0.05). In contrast, no significant enrichment for proximal NEK2 promoter sequences was obtained (Fig. 4B, left, compare bar 7 with bar 1, 0.02 versus 0.01% of input). The positive control locus, CDKN1A, showed 35-fold enrichment compared with GAPDH (Fig. 4B, left, compare bar 3 with bar 1, 0.35 versus 0.01% of input; p < 0.01). Treatment with 5aza-dC (Fig. 4B, middle) and doxo (Fig. 4B, right) promoted further enrichment of p53 binding to the distal, but not proximal, NEK2 promoter. Note that no enrichment was observed following 5aza-dC treatment at the MLH1 promoter (Fig. 4B, middle). MLH1 is a target gene activated by p53, but it is unmethylated (25, 35), and its expression is not significantly affected by 5aza-dC treatment of HCT116 cells (microarray signals: HCT116 and HCT116 + 5aza-dC, 189 and 152 arbitrary units, respectively).
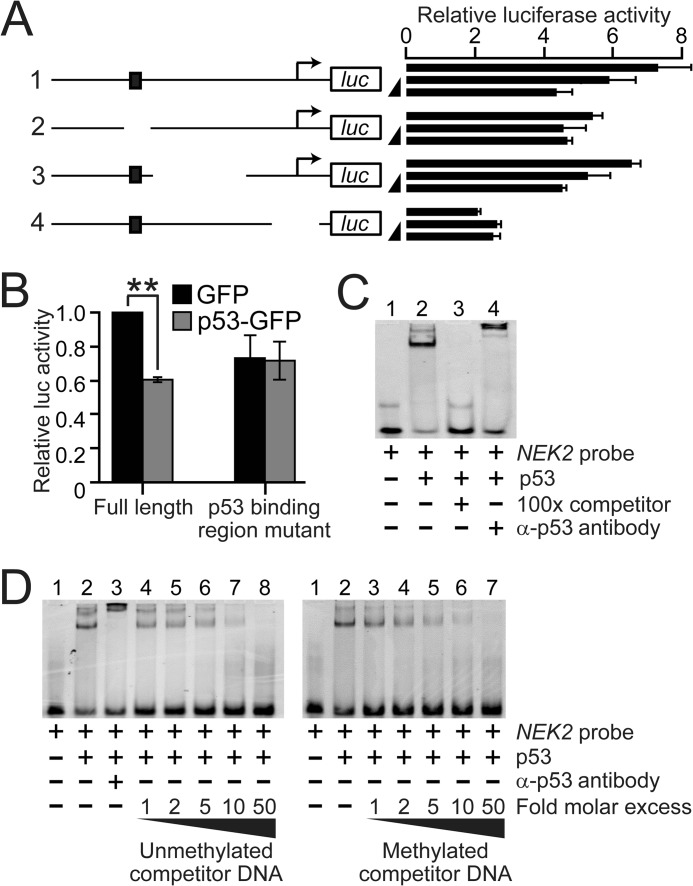
To test whether the distal NEK2 promoter region is required for p53-mediated NEK2 repression, a series of promoter deletion mutants was generated in an NEK2 promoter-luciferase reporter (Fig. 5A). The full-length reporter, containing 1098 bp of the NEK2 promoter from −1018 to +80 relative to the TSS, showed a dose-dependent decrease in promoter activity upon transient transfection of a plasmid expressing p53-GFP (Fig. 5A, construct 1). This repression was significantly dampened in the construct from which the NEK2 p53-binding region (in the LMR) was deleted (Fig. 5, A, construct 2, and B), but relatively unaffected by deletion of downstream sequences (Fig. 5A, construct 3). Luciferase activity was largely reduced, and the p53 response was eliminated in a control construct from which the NEK2 TSS was deleted (Fig. 5A, construct 4).
FIGURE 5.
Characterization of a p53 regulatory region in the NEK2 promoter. A, schematics of NEK2 promoter-luciferase (luc) reporters and associated luciferase activities. The top schematic (labeled 1) represents the full-length reporter containing 1098 bp (from −1018 to +80 relative to the TSS) of NEK2. The black rectangle indicates the p53-binding region. Bent arrows demarcate the NEK2 TSS. Next to each schematic is shown the relative luciferase activity for each reporter in response to transfected GFP (50 ng) or to two doses of transfected p53-GFP (25 or 50 ng; black ramps). Each bar represents the mean of three technical replicates. Error bars represent 1 S.D. B, repeat of the assay in A for the full-length and the second reporter with the p53-binding region deleted using 50 ng of GFP or p53-GFP. Each bar represents the mean ± error bars that represent S.E. (n = 4 independent transfections). **, p < 0.01 by ANOVA and Bonferroni ad hoc test versus GFP control. C, EMSA analysis using Cy5-labeled dsDNA NEK2 probe and purified human p53 protein. Reactions containing the indicated components were analyzed by polyacrylamide gel electrophoresis. D, EMSA analysis comparing the ability of unmethylated (left) versus methylated (right) NEK2 p53-binding sequence to compete for p53 binding. Ramps indicate the increasing -fold molar excesses of unlabeled dsDNA competitor to labeled probe.
To confirm that p53 binds to the identified binding site, EMSA was performed using purified p53 protein and a fluorescently labeled probe containing the 23-bp p53-binding sequence (see Fig. 4A, inset). In the presence of purified human p53 protein, strong formation of a p53-DNA complex of reduced mobility was observed (Fig. 5C, compare lanes 1 and 2). This complex was supershifted by preincubation with anti-p53 antibody (compare lanes 2 and 4) and completely eliminated by preincubation with unlabeled, competitor DNA (compare lanes 2 and 3), indicating a specific p53-DNA interaction.
To determine whether DNA methylation within the p53 site can inhibit binding of p53, binding was competed with unlabeled NEK2 p53-binding site duplex that was either unmethylated (Fig. 5D, left) or methylated on both strands of the single CG site (Fig. 5D, right). We observed that both duplexes competed for p53 binding at equivalent efficiencies (Fig. 5D, left, lanes 2 and 4–8 compared with right, lanes 2–7). This suggests that methylation of the single CG in the NEK2 p53-binding site does not directly impair p53 binding.
Modulation of p53 or Treatment with 5aza-dC Leads to Local Changes in Nucleosome Occupancy at the NEK2 Promoter
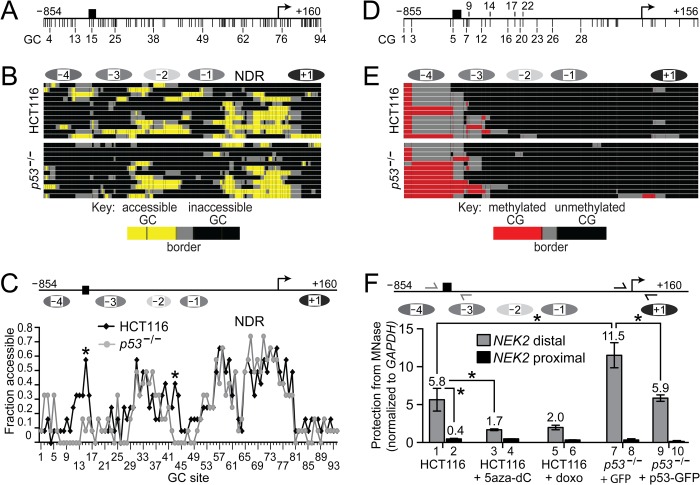
Although often assayed separately, DNA methylation exerts its regulation over gene expression within the context of chromatin. To gain insight into the chromatin structure associated with loss of p53 and how it relates to DNA methylation, we conducted MAPit, a single molecule, high resolution assay for chromatin accessibility (36). MAPit exploits the ability of the M.CviPI enzyme (37) to probe chromatin structure by methylating accessible GC sites that are not protected by histones (i.e. in nucleosomes) or non-histone proteins. The methylation status of sites can then be determined on single molecules by using BGS (30). Thus, probing with M.CviPI enables one to map chromatin accessibility and endogenous CG methylation simultaneously (25).
M.CviPI probing of HCT116 and p53−/− cells showed that the NEK2 TSS co-localizes with a highly accessible or nucleosome-depleted region, a common feature of expressed or expression-competent (poised) genes (Fig. 6, A and B). The NEK2 nucleosome-depleted region is flanked by two inaccessible areas of protection against methylation by M.CviPI that are both of the appropriate size to infer nucleosome occupancy. The nucleosome downstream of the TSS (+1 nucleosome) appears relatively well positioned, whereas the upstream −1 nucleosome occupies more translational positions and is possibly more dynamic. The chromatin structure from the −1 nucleosome through the +1 nucleosome is similar in HCT116 and p53−/− cells. This suggests that neither the DNA methylation status at the LMR nor the absence of p53 affects this “basal,” uninduced chromatin state of the NEK2 proximal promoter.
FIGURE 6.
Modulation of p53 or treatment with 5aza-dC is associated with altered nucleosome occupancy. Schematics of the long NEK2 promoter amplicon indicating GC sites (A) and CG sites (D) marked by vertical ticks. The different upstream and downstream end points reflect the different positions of the first and last sites. Black rectangle, p53-binding site; bent arrow, TSS. MAPit of HCT116 (B and E, top) and p53−/− (B and E, bottom) by treating harvested nuclei with 30 units of M.CviPI activity and then processing purified DNA by BGS. To facilitate pattern recognition, sequencing data were uploaded into a web-based hierarchical clustering program called MethylMapper (26) and are presented as three-color images of GC accessibility in B and DNA methylation in E (see keys at very bottom of B and E). Two or more consecutive accessible sites are connected by color (yellow, GC accessibility; red, CG methylation), whereas two or more consecutive non-methylated sites are connected by black. Gray connects the borders between methylated and non-methylated sites. Each row represents a cloned and sequenced molecule (12 total per group). Also shown in B and E (very top) are the inferred nucleosome positions (ellipses; each the length of a 147-bp nucleosome core particle) based on GC accessibility in HCT116 cells in B. Darker ellipses reflect nucleosomes that have higher apparent occupancy. C, top, schematic of the same region shown in A but with features repositioned to correspond to GC site locations not to scale. C, bottom, quantification of data in B. Each point represents the fraction of molecules accessible at each GC site (i.e. average accessibility across the population of molecules; *, p < 0.05 by two-tailed Fisher's exact test). Note that one GC site at −833 has been omitted from A–C as it overlaps with heavily methylated CG site 2; i.e. both sites are within a GCG. Likewise, all GCG sites have been omitted from D and E as the molecules were probed with M.CviPI. F, top, schematic of the same region shown in A. F, bottom, quantitative MNase assays for the indicated cell lines and treatments. Locations of the distal (gray half-arrows) and proximal (black half-arrows) primer pairs are shown. Data are presented as protection from MNase normalized to protection at GAPDH. Bars indicate the mean ± error bars that represent S.E. (n = 3) for all samples except for HCT116 + doxo (n = 2; mean ± error bars that represent 0.5 of the range). *, p < 0.05 by ANOVA and Bonferroni ad hoc test. NDR, nucleosome-depleted region.
Upstream of the −1 nucleosome is an accessible linker followed by three more variably positioned nucleosomes, −2 to −4 (Fig. 6, A and B). The linkers of nucleosome pairs −1/−2 and −3/−4, the latter harboring the p53-binding site, appear more defined in the parental HCT116 versus p53−/− cells. This conclusion is supported by comparison of the fraction of molecules accessible at each GC site in HCT116 and p53−/− cells (Fig. 6C). This averaged data showed that promoter accessibility differs significantly at only two positions: 1) the linker between the −1 and −2 nucleosomes in HCT116 cells (GC site 43; p = 0.03), whereas protection appears to have shifted upstream in the p53−/− cells, and 2) within the p53-binding site (GC site 15; p = 0.02) where this region is more accessible in the HCT116 cells. These results are consistent with increased nucleosome occupancy in the p53−/− cells; conversely, HCT116 cells with WT p53 have increased GC accessibility at the p53-binding region.
CG methylation of the same NEK2 promoter molecules from HCT116 and p53−/− cells is shown in Fig. 6, D and E. Consistent with data shown in Fig. 2, B and C, the p53−/− cells contained more DNA methylation spanning the p53-binding region than did HCT116 cells (Fig. 6, D and E). Also, DNA methylation was restricted to the distal promoter with negligible methylation observed at the TSS.
Both doxo and 5aza-dC have been shown to alter chromatin structure, including causing eviction or turnover of nucleosomes (35, 38). To determine whether drug treatment causes changes in nucleosome occupancy at the NEK2 promoter, nuclei from HCT116 cells treated with or without 5aza-dC or doxo were digested with MNase and analyzed by qPCR using convergent primers spanning the p53-binding region (Fig. 6F, distal) and within the nucleosome-depleted region encompassing the TSS (Fig. 6F, proximal). In HCT116 cells, as observed by MAPit, the distal promoter exhibited protection against MNase, consistent with nucleosome occupancy, whereas the proximal promoter nucleosome-depleted region exhibited very little protection, consistent with nucleosome depletion (Fig. 6F, bar 1 versus bar 2, all protections relative to GAPDH; p < 0.05). We observed decreased protection against MNase digestion at the distal, but not proximal, NEK2 promoter following 5aza-dC and doxo treatment (Fig. 6F, bar 1 versus bar 3, p < 0.05; bar 1 versus bar 5). Again, consistent with the MAPit data, an increase in protection against MNase, reflective of increased nucleosome occupancy, was observed for the p53−/− cells compared with HCT116 cells at the distal promoter (bar 7 versus bar 1, p < 0.05) but not proximal promoter (bar 9 versus bar 1, non-significant). Notably, ectopic expression of p53-GFP in p53−/− cells for 24 h also caused decreased protection against MNase at the distal promoter (Fig. 6F, bar 9 versus bar 7, p < 0.05). Taken together, these data suggest that the binding of p53 competes with nucleosome occupancy at the distal NEK2 promoter.
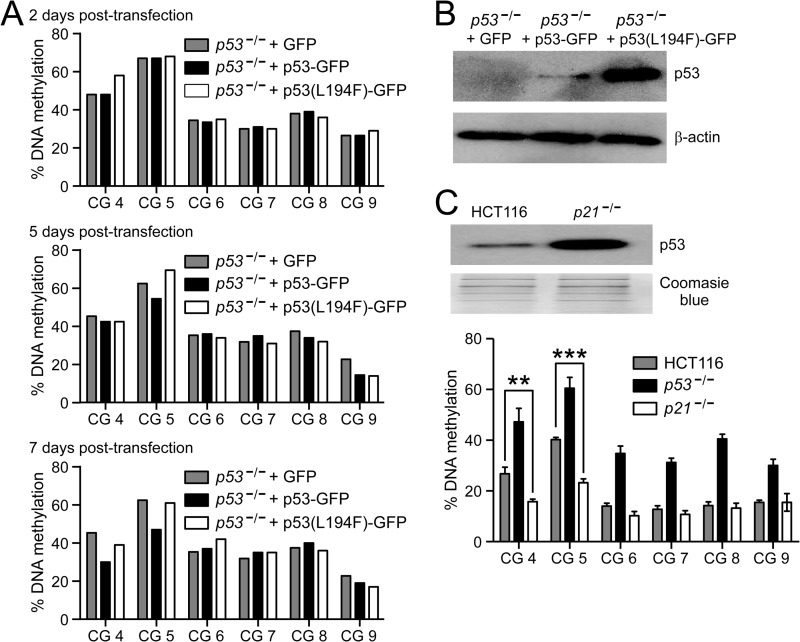
Our MAPit, MNase, and BGS results showed that the loss of p53 is associated with increased nucleosome occupancy and increased DNA methylation at the distal p53-binding region of the NEK2 promoter. To determine whether increased expression of p53 could, in addition to causing nucleosome depletion, lead to demethylation of this region, we took two approaches. First, p53−/− cells were transfected with either GFP control or fusion protein p53-GFP or p53(L194F)-GFP and grown in selective medium. DNA methylation was examined at the NEK2 LMR 2, 5, and 7 days post-transfection. DNA methylation remained largely unchanged at day 2 (Fig. 7A, top). At day 5, some demethylation was observed at CG 5 within the p53-binding site (Fig. 7A, middle; p53−/− + GFP, 62.5%; +p53-GFP, 54.5%; and +p53(L194F-GFP), 69.5%). At day 7, methylation was further decreased by p53 rescue at CG 5 (to 47%). Also, some demethylation was observed at CG 4, located 8 bp upstream of CG 5, between days 5 and 7 (45.3 versus 30%) in cells transfected with p53-GFP but not those transfected with p53(L194F)-GFP (Fig. 7A, bottom). Demethylation was specific to p53-GFP despite higher accumulation of p53(L194F)-GFP in the transfected p53−/− cells (Fig. 7B). Note that nucleosome depletion was observed at 24 h post-transfection (Fig. 6F, bars 9 and 10), indicating an immediate effect of p53 expression on nucleosome occupancy and a delayed effect on DNA methylation.
FIGURE 7.
Increased expression of p53 leads to demethylation of the p53-binding region. A, pyrosequencing analysis comparing NEK2 DNA methylation levels between the indicated cell lines and treatments. CG sites are numbered as in Fig. 2A (CG 5 is located in the p53-binding site, whereas CG 4 is located 8 bp upstream). Data are presented as percentage of methylation at each CG site in p53−/− cells after transfection with the indicated transgenes (+GFP control, p53-GFP, or p53(L194F)-GFP) for 2 (top), 5 (middle), and 7 (bottom) days. B, immunoblot analysis of p53 and β-actin (loading control) in p53−/− cells at 7 days post-transfection with the indicated transgenes. C, top, immunoblot analysis comparing p53 levels in HCT116 and p21−/− cells. The gel stained with Coomassie Blue serves as loading control. C, bottom, pyrosequencing analysis as described in A. Bars indicate the mean ± error bars that represent S.E. (n = 4). **, p < 0.01; ***, p < 0.001 by ANOVA and Bonferroni ad hoc test.
Second, to determine the effects of long term p53 expression on NEK2 CG methylation, we examined the DNA methylation status at the NEK2 LMR in HCT116 p21−/− cells. It has been widely observed that deletion of CDKN1A (encodes p21) in HCT116 cells leads to increased stabilization and accumulation of p53 (39, 40). We confirmed this increase of p53 protein in p21−/− cells compared with HCT116 cells (Fig. 7C, top). DNA methylation was significantly decreased in p21−/− cells compared with HCT116 cells at both CG 5 (p < 0.001) and CG 4 (p < 0.01) (Fig. 7C, bottom). As found by BGS (Fig. 2E), DNA methylation was increased at all six queried CG sites in p53−/− cells (p < 0.01). Taken together, these data support a model whereby binding of p53 causes nucleosome depletion at its binding region and over time inhibits local accumulation of DNA methylation.
DISCUSSION
To our knowledge, NEK2 has not previously been shown to be a p53-repressed target gene. Our data show that NEK2 expression is repressed by WT p53 through binding at the distal NEK2 promoter. Our results are in agreement with several published and unpublished microarray studies that implicated p53-dependent NEK2 repression. We queried the NextBio website, which contains publically available genomics data, and found several microarray data sets that support p53-mediated repression of NEK2. Although data are available from multiple cell types, we have listed in Table 1 relevant results from breast cancer-derived samples in which NEK2 overexpression is particularly common. Notably, although several data sets in addition to ours have shown that NEK2 is repressed by doxo, one in particular showed that this repression is reversed upon treatment with shRNA knockdown of p53 expression (41). Several data sets have also indicated that NEK2 is up-regulated in breast tumors expressing mutant p53 compared with WT. This is of particular importance because NEK2-based anticancer therapies are under preclinical development (42, 43). If NEK2 protein is similarly up-regulated in mutant p53-expressing tumors, then these patients may achieve better benefits from NEK2-based therapies. Also, if they reach the clinic, NEK2 therapies would likely be used in combination with other therapeutic agents, and given the repressive effects of DNA-damaging agents on NEK2 expression, it may be useful to evaluate the efficacy of these combinations.
TABLE 1.
Microarray studies implicate NEK2 as repressed by WT p53
All data were queried from NextBio-curated studies and analyzed using NextBio software. 5-FU; 5-fluorouracil; N.S., not significant; CIT, Cartes d'Identite des Tumeurs project from the French Ligue Nationale Contre le Cancer. BMH compounds are lead novel p53-activating small molecules. Gene Expression Omnibus Series (GSE) accession numbers are provided for unpublished studies.
| Sample | -Fold change | p value | Ref. |
|---|---|---|---|
| NEK2 down-regulated | |||
| Breast cancer fibroblasts + 50 nm doxo (1 day) | −13 | 0.002 | GSE23399 |
| Breast cancer fibroblasts + 50 nm doxo (5 days) | −17.1 | 0.0017 | |
| Breast cancer fibroblasts + 50 nm doxo (7 days) | −11.2 | 0.0008 | |
| MCF7 cells + 1.5 μm doxo (10 h) | −3.5 | 2 × 10−5 | GSE24065 |
| MCF7 cells + 375 μm 5-FU (10 h) | −4.84 | 0.0001 | GSE24065 |
| ZR-75–1 cells + doxo | −2.11 | 0.0023 | 41 |
| ZR-75–1 cells + doxo + p53 shRNA | None | N.S. | |
| MCF7 cells | |||
| +BMH-7 | −1.38 | 0.0017 | 54 |
| +BMH-9 | −1.46 | 0.0002 | |
| +BMH-15 | −1.23 | 0.0056 | |
| +BMH-21 | −1.38 | 0.0013 | |
| +BMH-22 | −1.35 | 0.0005 | |
| +BMH-23 | −1.51 | 0.0012 | |
| NEK2 up-regulated | |||
| Breast cancer biopsies (Uppsala cohort) with TP53 mutation vs. wild type | +2.03 | 3.9 × 10−13 | 55 |
| Human breast cancer after chemotherapy (epirubicin/cyclophosphamide) TP53 mutated vs. wild type | +2.06 | 0.0001 | 56 |
| Human breast tumors (CIT cohort) with TP53 mutation vs. wild type | +1.44 | 0.0003 | 57 |
| Primary breast stroma Li-Fraumeni syndrome patients with TP53 mutation vs. normal | +8.75 | 5.7 × 10−5 | 58 |
We and others have shown that NEK2 transcript and protein expression is repressed upon stimulation of p53 with DNA-damaging agents (Table 1). The functional consequence of p53-mediated repression of NEK2 activity could be inhibition of centrosome separation in response to DNA damage. It has been reported that such an inhibition is a cellular response to DNA damage, independent of growth inhibition (for a review, see Ref. 44). In addition, it was shown that cells exposed to ionizing radiation exhibit both decreased NEK2 expression and kinase activity, and exposure to ionizing radiation inhibits NEK2-mediated separation of the centrosome (45). Thus, p53-mediated repression of NEK2 could link centrosome inactivation to the DNA damage response network.
We have also identified a relevant binding region required for p53-mediated NEK2 repression, although we cannot exclude the possibility that additional p53 regulatory sites are utilized in vivo. This region contains a p53-binding site sequence identical to that associated with transcriptional activation of the HD gene (34). An unanswered question regarding p53 activity is how it can affect activation of some genes while repressing other targets. A recent study suggested that the consensus binding motifs differ between p53-activated and p53-repressed targets (46). That study also suggested that different chromatin structures determine activation versus repression. Comparison of the NEK2 and HD p53-binding regions may provide a novel model system to study how identical sequence motifs can elicit opposing regulatory outcomes.
We have shown that NEK2 repression in HCT116 colon cancer cells by 5aza-dC is p53-dependent and associated with decreased nucleosome occupancy and demethylation of the distal promoter. However, our EMSA studies indicate that DNA methylation does not affect p53 binding to the NEK2-binding site. Thus, the mechanism by which 5aza-dC elicits p53-dependent repression of NEK2 is likely through increased p53-binding site exposure via nucleosome remodeling rather than DNA demethylation. Although nucleosome eviction has been shown in association with gene reactivation (35), to our knowledge, this is the first demonstration that nucleosome eviction may support gene repression by 5aza-dC. It should be noted, however, that the NEK2 p53-binding site contains only one CG site. Fluorescence anisotropy titration has shown that methylation of single CG sites does not directly affect p53 binding (47). However, a genome-wide ChIP study found that, in vivo, p53-bound loci are enriched for hypomethylated DNA compared with adjacent sequences (48). Thus, our in vitro results do not exclude the possibility that DNA methylation in the context of chromatin may affect binding of p53 in vivo. Conversely, DNA methylation could drive preferential binding of a transcriptional activator at this locus. For example, it was recently published that DNA methylation enhances the binding activity of the CCAAT/enhancer-binding protein α transcription factor (49). It has also been shown that p53-mediated repression of some target genes is achieved through competition for binding in place of a transcriptional activator (50, 51). Consistent with this, there does seem to be a small decrease in luciferase reporter activity in the NEK2 promoter-luciferase reporter containing the p53-binding region deletion compared with the full-length reporter (Fig. 5C). This suggests that this region could support binding of a transcriptional activator; however, additional studies are needed to test this model.
Rather than DNA methylation inhibiting binding of p53, we observed that increased binding of p53 inhibits DNA methylation at this locus. This is supported by the increased accumulation of DNA methylation observed at the distal LMR in the p53−/− cells (Figs. 2, B and C, and 6E). Furthermore, DNA methylation was conversely decreased in both p21−/− cells, which hyperaccumulate p53 (Fig. 7C), and p53−/− cells transfected with p53-GFP (Fig. 7A). These changes in DNA methylation were specific as they did not occur in the proximal NEK2 core promoter or in the presence of a p53 protein lacking DNA binding activity (Figs. 2, 6E, and 7A). Several studies have documented factor binding as a mechanism for excluding DNA methylation (31, 32, 52, 53). Further studies are required to elucidate the functional consequences, if any, of DNA methylation at the NEK2 LMR. It would also be of interest to determine how commonly LMRs are present at p53-binding regions and whether there is any difference between DNA methylation patterns surrounding activating versus repressive p53-binding sites.
Acknowledgments
We thank Dr. Lei Deng and Dr. Russell Broaddus for providing the p53-luciferase reporter and RNA from doxo-treated breast cancer cells and Dr. Lawrence Donehower for the p53-EGFP construct. We also thank Dr. Carolina Pardo for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants CA098258 (to D. S. L.) and CA155390 (to M. P. K.). This work was also supported by Bankhead-Coley Florida Cancer Research Program Postdoctoral Fellowship BD-03 (to N. H. N.).

This article contains supplemental Tables 1 and 2.
- TSS
- transcription start site
- 5aza-dC
- 5-aza-2′-deoxycytidine
- MNase
- micrococcal nuclease
- doxo
- doxorubicin
- qPCR
- quantitative PCR
- EGFP
- enhanced GFP
- Cy5
- cyanine 5 fluorescent dye
- MAPit
- methyltransferase accessibility protocol for individual templates
- BGS
- bisulfite genomic sequencing
- LMR
- low-methylated region
- HD
- huntingtin gene
- ANOVA
- analysis of variance.
REFERENCES
- 1. Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21 [DOI] [PubMed] [Google Scholar]
- 2. Hon G. C., Hawkins R. D., Caballero O. L., Lo C., Lister R., Pelizzola M., Valsesia A., Ye Z., Kuan S., Edsall L. E., Camargo A. A, Stevenson B. J., Ecker J. R., Bafna V., Strausberg R. L., Simpson A. J., Ren B. (2012) Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 22, 246–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hellman A., Chess A. (2007) Gene body-specific methylation on the active X chromosome. Science 315, 1141–1143 [DOI] [PubMed] [Google Scholar]
- 4. Ball M. P., Li J. B., Gao Y., Lee J. H., LeProust E. M., Park I. H., Xie B., Daley G. Q., Church G. M. (2009) Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 27, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu H., Coskun V., Tao J., Xie W., Ge W., Yoshikawa K., Li E., Zhang Y., Sun Y. E. (2010) Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329, 444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barletta J. M., Rainier S., Feinberg A. P. (1997) Reversal of loss of imprinting in tumor cells by 5-aza-2′-deoxycytidine. Cancer Res. 57, 48–50 [PubMed] [Google Scholar]
- 7. Renaud S., Loukinov D., Abdullaev Z., Guilleret I., Bosman F. T., Lobanenkov V., Benhattar J. (2007) Dual role of DNA methylation inside and outside of CTCF-binding regions in the transcriptional regulation of the telomerase hTERT gene. Nucleic Acids Res. 35, 1245–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nabilsi N. H., Broaddus R. R., Loose D. S. (2009) DNA methylation inhibits p53-mediated survivin repression. Oncogene 28, 2046–2050 [DOI] [PubMed] [Google Scholar]
- 9. Lai A. Y., Fatemi M., Dhasarathy A., Malone C., Sobol S. E., Geigerman C., Jaye D. L., Mav D., Shah R., Li L., Wade P. A. (2010) DNA methylation prevents CTCF-mediated silencing of the oncogene BCL6 in B cell lymphomas. J. Exp. Med. 207, 1939–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hayward D. G., Fry A. M. (2006) NEK2 kinase in chromosome instability and cancer. Cancer Lett. 237, 155–166 [DOI] [PubMed] [Google Scholar]
- 11. Kokuryo T., Senga T., Yokoyama Y., Nagino M., Nimura Y., Hamaguchi M. (2007) NEK2 as an effective target for inhibition of tumorigenic growth and peritoneal dissemination of cholangiocarcinoma. Cancer Res. 67, 9637–9642 [DOI] [PubMed] [Google Scholar]
- 12. Barbagallo F., Paronetto M. P., Franco R., Chieffi P., Dolci S., Fry A. M., Geremia R., Sette C. (2009) Increased expression and nuclear localization of the centrosomal kinase NEK2 in human testicular seminomas. J. Pathol. 217, 431–441 [DOI] [PubMed] [Google Scholar]
- 13. Brendle A., Brandt A., Johansson R., Enquist K., Hallmans G., Hemminki K., Lenner P., Försti A. (2009) Single nucleotide polymorphisms in chromosomal instability genes and risk and clinical outcome of breast cancer: a Swedish prospective case-control study. Eur. J. Cancer 45, 435–442 [DOI] [PubMed] [Google Scholar]
- 14. Wu G., Qiu X. L., Zhou L., Zhu J., Chamberlin R., Lau J., Chen P. L., Lee W. H. (2008) Small molecule targeting the Hec1/NEK2 mitotic pathway suppresses tumor cell growth in culture and in animal. Cancer Res. 68, 8393–8399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Qiu X. L., Li G., Wu G., Zhu J., Zhou L., Chen P. L., Chamberlin A. R., Lee W. H. (2009) Synthesis and biological evaluation of a series of novel inhibitor of NEK2/Hec1 analogues. J. Med. Chem. 52, 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schultz S. J., Fry A. M., Sütterlin C., Ried T., Nigg E. A. (1994) Cell cycle-dependent expression of NEK2, a novel human protein kinase related to the NIMA mitotic regulator of Aspergillus nidulans. Cell Growth Differ. 5, 625–635 [PubMed] [Google Scholar]
- 17. Iyer V. R., Eisen M. B., Ross D. T., Schuler G., Moore T., Lee J. C., Trent J. M., Staudt L. M., Hudson J., Jr., Boguski M. S., Lashkari D., Shalon D., Botstein D., Brown P. O. (1999) The transcriptional program in the response of human fibroblasts to serum. Science 283, 83–87 [DOI] [PubMed] [Google Scholar]
- 18. Fry A. M. (2002) The NEK2 protein kinase: a novel regulator of centrosome structure. Oncogene 21, 6184–6194 [DOI] [PubMed] [Google Scholar]
- 19. Ren B., Cam H., Takahashi Y., Volkert T., Terragni J., Young R. A., Dynlacht B. D. (2002) E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Whitfield M. L., Sherlock G., Saldanha A. J., Murray J. I., Ball C. A., Alexander K. E., Matese J. C., Perou C. M., Hurt M. M., Brown P. O., Botstein D. (2002) Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13, 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laoukili J., Kooistra M. R., Brás A., Kauw J., Kerkhoven R. M., Morrison A., Clevers H., Medema R. H. (2005) FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat. Cell Biol. 7, 126–136 [DOI] [PubMed] [Google Scholar]
- 22. Xie R., Loose D. S., Shipley G. L., Xie S., Bassett R. L., Jr., Broaddus R. R. (2007) Hypomethylation-induced expression of S100A4 in endometrial carcinoma. Mod. Pathol. 20, 1045–1054 [DOI] [PubMed] [Google Scholar]
- 23. Jain A. K., Allton K., Iacovino M., Mahen E., Milczarek R. J., Zwaka T. P., Kyba M., Barton M. C. (2012) p53 regulates cell cycle and microRNAs to promote differentiation of human embryonic stem cells. PLoS Biol. 10, e1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pardo C. E., Darst R. P., Nabilsi N. H., Delmas A. L., Kladde M. P. (2011) Simultaneous single-molecule mapping of protein-DNA interactions and DNA methylation by MAPit. Curr. Protoc. Mol. Biol. Chapter 21, Unit 21.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pardo C. E., Carr I. M., Hoffman C. J., Darst R. P., Markham A. F., Bonthron D. T., Kladde M. P. (2011) MethylViewer: computational analysis and editing for bisulfite sequencing and methyltransferase accessibility protocol for individual templates (MAPit) projects. Nucleic Acids Res. 39, e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Darst R. P., Nabilsi N. H., Pardo C. E., Riva A., Kladde M. P. (2012) DNA methyltransferase accessibility protocol for individual templates by deep sequencing. Methods Enzymol. 513, 185–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Demircan B., Dyer L. M., Gerace M., Lobenhofer E. K., Robertson K. D., Brown K. D. (2009) Comparative epigenomics of human and mouse mammary tumors. Genes Chromosomes Cancer 48, 83–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cui K., Zhao K. (2012) Genome-wide approaches to determining nucleosome occupancy in metazoans using MNase-Seq. Methods Mol. Biol. 833, 413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nabilsi N. H., Broaddus R. R., McCampbell A. S., Lu K. H., Lynch H. T., Chen L. M., Loose D. S. (2010) Sex hormone regulation of survivin gene expression. J. Endocrinol. 207, 237–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Frommer M., McDonald L. E., Millar D. S., Collis C. M., Watt F., Grigg G. W., Molloy P. L., Paul C. L. (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A. 89, 1827–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stadler M. B., Murr R., Burger L., Ivanek R., Lienert F., Schöler A., van Nimwegen E., Wirbelauer C., Oakeley E. J., Gaidatzis D., Tiwari V. K, Schübeler D. (2011) DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, 490–495 [DOI] [PubMed] [Google Scholar]
- 32. Lienert F., Wirbelauer C., Som I., Dean A., Mohn F., Schübeler D. (2011) Identification of genetic elements that autonomously determine DNA methylation states. Nat. Genet. 43, 1091–1097 [DOI] [PubMed] [Google Scholar]
- 33. Riley T., Sontag E., Chen P., Levine A. (2008) Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 [DOI] [PubMed] [Google Scholar]
- 34. Feng Z., Jin S., Zupnick A., Hoh J., de Stanchina E., Lowe S., Prives C., Levine A. J. (2006) p53 tumor suppressor protein regulates the levels of huntingtin gene expression. Oncogene 25, 1–7 [DOI] [PubMed] [Google Scholar]
- 35. Lin J. C., Jeong S., Liang G., Takai D., Fatemi M., Tsai Y. C., Egger G., Gal-Yam E. N., Jones P. A. (2007) Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell 12, 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kilgore J. A., Hoose S. A., Gustafson T. L., Porter W., Kladde M. P. (2007) Single-molecule and population probing of chromatin structure using DNA methyltransferases. Methods 41, 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu M., Kladde M. P., Van Etten J. L., Simpson R. T. (1998) Cloning, characterization and expression of the gene coding for a cytosine-5-DNA methyltransferase recognizing GpC. Nucleic Acids Res. 26, 3961–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang F., Kemp C. J., Henikoff S. (2013) Doxorubicin enhances nucleosome turnover around promoters. Curr. Biol. 23, 782–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pang L. Y., Scott M., Hayward R. L., Mohammed H., Whitelaw C. B., Smith G. C., Hupp T. R. (2011) p21WAF1 is component of a positive feedback loop that maintains the p53 transcriptional program. Cell Cycle 10, 932–950 [DOI] [PubMed] [Google Scholar]
- 40. Broude E. V., Demidenko Z. N., Vivo C., Swift M. E., Davis B. M., Blagosklonny M. V., Roninson I. B. (2007) p21 (CDKN1A) is a negative regulator of p53 stability. Cell Cycle 6, 1468–1471 [PubMed] [Google Scholar]
- 41. Troester M. A., Herschkowitz J. I., Oh D. S., He X., Hoadley K. A., Barbier C. S., Perou C. M. (2006) Gene expression patterns associated with p53 status in breast cancer. BMC Cancer 6, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tsunoda N., Kokuryo T., Oda K., Senga T., Yokoyama Y., Nagino M., Nimura Y., Hamaguchi M. (2009) NEK2 as a novel molecular target for the treatment of breast carcinoma. Cancer Sci. 100, 111–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suzuki K., Kokuryo T., Senga T., Yokoyama Y., Nagino M., Hamaguchi M. (2010) Novel combination treatment for colorectal cancer using NEK2 siRNA and cisplatin. Cancer Sci. 101, 1163–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Löffler H., Lukas J., Bartek J., Krämer A. (2006) Structure meets function—centrosomes, genome maintenance and the DNA damage response. Exp. Cell Res. 312, 2633–2640 [DOI] [PubMed] [Google Scholar]
- 45. Fletcher L., Cerniglia G. J., Nigg E. A., Yend T. J., Muschel R. J. (2004) Inhibition of centrosome separation after DNA damage: a role for NEK2. Radiat. Res. 162, 128–135 [DOI] [PubMed] [Google Scholar]
- 46. Li M., He Y., Dubois W., Wu X., Shi J., Huang J. (2012) Distinct regulatory mechanisms and functions for p53-activated and p53-repressed DNA damage response genes in embryonic stem cells. Mol. Cell 46, 30–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Petrovich M., Veprintsev D. B. (2009) Effects of CpG methylation on recognition of DNA by the tumour suppressor p53. J. Mol. Biol. 386, 72–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Botcheva K., McCorkle S. R., McCombie W. R., Dunn J. J., Anderson C. W. (2011) Distinct p53 genomic binding patterns in normal and cancer-derived human cells. Cell Cycle 10, 4237–4249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rishi V., Bhattacharya P., Chatterjee R., Rozenberg J., Zhao J., Glass K., Fitzgerald P., Vinson C. (2010) CpG methylation of half-CRE sequences creates C/EBPα binding sites that activate some tissue-specific genes. Proc. Natl. Acad. Sci. U.S.A. 107, 20311–20316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Budhram-Mahadeo V., Morris P. J., Smith M. D., Midgley C. A., Boxer L. M., Latchman D. S. (1999) p53 suppresses the activation of the Bcl-2 promoter by the Brn-3a POU family transcription factor. J. Biol. Chem. 274, 15237–15244 [DOI] [PubMed] [Google Scholar]
- 51. Hoffman W. H., Biade S., Zilfou J. T., Chen J., Murphy M. (2002) Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J. Biol. Chem. 277, 3247–3257 [DOI] [PubMed] [Google Scholar]
- 52. Hsieh C. L. (1999) Evidence that protein binding specifies sites of DNA demethylation. Mol. Cell. Biol. 19, 46–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lin I. G., Tomzynski T. J., Ou Q., Hsieh C. L. (2000) Modulation of DNA binding protein affinity directly affects target site demethylation. Mol. Cell. Biol. 20, 2343–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Peltonen K., Colis L., Liu H., Jäämaa S., Moore H. M., Enbäck J., Laakkonen P., Vaahtokari A., Jones R. J., af Hällström T. M., Laiho M. (2010) Identification of novel p53 pathway activating small-molecule compounds reveals unexpected similarities with known therapeutic agents. PLoS One 5, e12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miller L. D., Smeds J., George J., Vega V. B., Vergara L., Ploner A., Pawitan Y., Hall P., Klaar S., Liu E. T., Bergh J. (2005) An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. U.S.A. 102, 13550–13555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bertheau P., Turpin E., Rickman D. S., Espié M., de Reyniès A., Feugeas J. P., Plassa L. F., Soliman H., Varna M., de Roquancourt A., Lehmann-Che J., Beuzard Y., Marty M., Misset J. L., Janin A., de Thé H. (2007) Exquisite sensitivity of TP53 mutant and basal breast cancers to a dose-dense epirubicin-cyclophosphamide regimen. PLoS Med. 4, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Guedj M., Marisa L., de Reynies A., Orsetti B., Schiappa R., Bibeau F., MacGrogan G., Lerebours F., Finetti P., Longy M., Bertheau P., Bertrand F., Bonnet F., Martin A. L., Feugeas J. P., Bièche I., Lehmann-Che J., Lidereau R., Birnbaum D., Bertucci F., de Thé H., Theillet C. (2012) A refined molecular taxonomy of breast cancer. Oncogene 31, 1196–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Herbert B. S., Chanoux R. A., Liu Y., Baenziger P. H., Goswami C. P., McClintick J. N., Edenberg H. J., Pennington R. E., Lipkin S. M., Kopelovich L. (2010) A molecular signature of normal breast epithelial and stromal cells from Li-Fraumeni syndrome mutation carriers. Oncotarget 1, 405–422 [DOI] [PMC free article] [PubMed] [Google Scholar]