Background: Triacylglycerols are stored in lipid droplets and can be mobilized by lipases.
Results: Ayr1p was identified as a novel triacylglycerol lipase.
Conclusion: In addition to the known lipases Tgl3p, Tgl4p, and Tgl5p, further hydrolytic enzymes contribute to the mobilization of non-polar lipids in yeast.
Significance: This study opens the view for a broader network of lipolytic enzymes in yeast.
Keywords: Lipase, Lipids, Lipid Droplets, Peroxisomes, Triacylglycerol, Yeast
Abstract
Saccharomyces cerevisiae, as well as other eukaryotes, preserves fatty acids and sterols in a biologically inert form, as triacylglycerols and steryl esters. The major triacylglycerol lipases of the yeast S. cerevisiae identified so far are Tgl3p, Tgl4p, and Tgl5p (Athenstaedt, K., and Daum, G. (2003) YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J. Biol. Chem. 278, 23317–23323; Athenstaedt, K., and Daum, G. (2005) Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae, are localized to lipid particles. J. Biol. Chem. 280, 37301–37309). We observed that upon cultivation on oleic acid, triacylglycerol mobilization did not come to a halt in a yeast strain deficient in all currently known triacylglycerol lipases, indicating the presence of additional not yet characterized lipases/esterases. Functional proteome analysis using lipase and esterase inhibitors revealed a subset of candidate genes for yet unknown hydrolytic enzymes on peroxisomes and lipid droplets. Based on the conserved GXSXG lipase motif, putative functions, and subcellular localizations, a selected number of candidates were characterized by enzyme assays in vitro, gene expression analysis, non-polar lipid analysis, and in vivo triacylglycerol mobilization assays. These investigations led to the identification of Ayr1p as a novel triacylglycerol lipase of yeast lipid droplets and confirmed the hydrolytic potential of the peroxisomal Lpx1p in vivo. Based on these results, we discuss a possible link between lipid storage, lipid mobilization, and peroxisomal utilization of fatty acids as a carbon source.
Introduction
In Saccharomyces cerevisiae as well as in other eukaryotes, an excess of fatty acids is stored as triacylglycerols (TG)2 and steryl esters (SE), often referred to as non-polar lipids. Both lipids are stored in organelle-like structures called lipid droplets (LD), which are about 400 nm in diameter and consist of a highly hydrophobic core of TG, surrounded by shells of SE and a phospholipid monolayer containing a distinct set of proteins (1–4). TG are synthesized by the acyltransferases Dga1p and Lro1p, and SE are synthesized by the SE synthases Are1p and Are2p (5–10). All TG- and SE-synthesizing enzymes are located in the endoplasmic reticulum, but Dga1p is also found on LD. TG serves as the main energy storage, and both TG and SE are depots of membrane lipid components. Upon requirement (i.e. during growth or starvation), TG and SE can be mobilized by lipases or hydrolases. Currently, three major TG lipases are known, namely Tgl3p, Tgl4p, and Tgl5p, which are located on LD (11, 12). The hydrolysis of SE is conducted by Tgl1p and Yeh1p localized to LD and Yeh2p, which was found to be associated with the plasma membrane (13–16).
Tgl3p, Tgl4p, and Tgl5p share a common consensus sequence GXSXG, where serine is the essential residue of the catalytic triad aspartic acid/glutamic acid and histidine (17). They also contain a patatin domain, named after a plant storage protein that possesses lipid acylhydrolase activity (18). In vitro, all three proteins exhibit lipolytic activity, whereas in vivo only Tgl3p and Tgl4p mobilize TG efficiently. Previous studies from our laboratory have described functions for Tgl3p, Tgl4p, and Tgl5p in addition to their lipase activities. Tgl3p, Tgl4p, and Tgl5p harbor an acyltransferase motif (HX4D), and in vitro enzyme assays showed that both Tgl3p and Tgl5p act as lysophospholipid acyltransferases. Besides the conserved lipase motif, Tgl4p contains a (G/A)XGXXG calcium-independent phospholipase A2 domain. Phospholipase as well as SE hydrolase activity of Tgl4p was also established in vitro (19). A Δtgl3Δtgl4Δtgl5 (TM) yeast strain lacking all three TG lipases does not reveal any growth defect under standard growth conditions, although mutations in TGL3 or TLG4 lead to fat yeast cells that accumulate TG (19–21). Interestingly, we observed that upon cultivation on oleic acid, TG mobilization did not come to a halt in the TM deficient in all currently known TG lipases, suggesting the presence of novel not yet characterized hydrolases.
S. cerevisiae grown in the presence of oleic acid proliferates peroxisomes (Px) and at the same time accumulates large LD (22). Px are small ubiquitous organelles involved in the decomposition of toxic substances like H2O2 as well as degradation of fatty acids via β-oxidation. The mechanism of fatty acid transport to its site of degradation is not yet completely understood. In contrast to mammalian cells, the degradation of fatty acids in the yeast exclusively takes place in Px (23–25). Thus, functional Px are crucial for growth of yeast cells on fatty acids as a carbon source. Binns et al. (26) proposed a direct link between Px and LD, indicating a putative pathway for lipid supply to Px. It was suggested that Px can even penetrate LD, forming a structure called pexopodia, and that this contact may stimulate non-polar lipid turnover.
The aim of the present study was to identify novel hydrolytic enzymes possibly involved in non-polar lipid metabolism in the yeast S. cerevisiae. Our data suggest that in addition to Tgl3p, Tgl4p, and Tgl5p, further hydrolases are responsible for the mobilization of TG. Our approach identified a novel TG lipase and shed more light on a possible link between lipid storage, lipid mobilization, and peroxisomal utilization of fatty acids as a carbon source.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Conditions
Table 1 gives an overview of strains used in this study. Yeast cells were either grown in YPD medium containing 1% yeast extract, 2% glucose, and 2% peptone or on oleic acid-supplemented medium (YPO) containing 0.3% yeast extract, 0.5% peptone, 0.1% glucose, 0.5% KH2PO4, and 0.1% oleic acid. For solubilizing oleic acid in YPO, 0.2% Tween 80 was added to the medium. Yeast strains bearing plasmids were cultivated in synthetic minimal medium (SD) containing 0.67% yeast nitrogen base (U. S. Biochemical Corp.), 2% glucose, and the respective amino acid supplements. If not stated otherwise, all cells were cultivated in liquid media at 30 °C under vigorous shaking until early stationary phase. For gene expression studies, yeast cells were grown to the midexponential phase. Growth was monitored by measuring optical density at 600 nm (A600). Expression of CUP1 promoter-controlled genes was induced after growth for 12 h by adding CuSO4 at a final concentration of 0.5 mm to the medium.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| WT | BY4741 Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Euroscarf |
| TM | WT; tgl3Δ::kanMX4; tgl4Δ::kanMX4; tgl5Δ::kanMX4 | Ref. 12 |
| QiM | tgl3Δ::kanMX4;tgl4Δ::kanMX4;tgl5Δ::kanMX4; are1Δ::LEU2;are2Δ::kanMX4 | Kindly provided by A. Wagner |
| LPX1+ | TM + pYEX4T-1-LPX1 | This study |
| LDH1+ | TM + pYEX4T-1-LDH1 | This study |
| YJU3+ | TM + pYEX4T-1-YJU3 | This study |
| AYR1+ | TM + pYEX4T-1-AYR1 | This study |
| EHT1+ | TM + pYEX4T-1-EHT1 | This study |
| TSC10+ | TM + pYEX4T-1-TSC10 | This study |
| YBR056w+ | TM + pYEX4T-1-YBR056w | This study |
| YKL050c+ | TM + pYEX4T-1-YKL050c | This study |
| AYR1S18A+ | TM + pYEX4T-1-AYR1S18A | This study |
| VC | TM + pYEX4T-1 (vector control) | This study |
Genetic Techniques
A list of primers used for creating deletion cassettes is shown in Table 2. Gene deletions were performed following the PCR-mediated method described by Longtine et al. (27). Deletion cassettes were transformed employing the high efficiency lithium acetate transformation protocol (28). Correct integration of the knock-out cassettes was verified by growth auxotrophy as well as colony PCR. For the expression of candidate hydrolases/lipases, the open reading frames of the respective genes were amplified from BY4741 chromosomal DNA using primers listed in Table 2. Restricted PCR fragments of LPX1, LDH1, YJU3, AYR1, and YBR056w were inserted into the BamHI and EcoRI sites, whereas EHT1, TSC10, and YKL050c were inserted into the BamHI and SalI sites of the plasmid pYEX4T-1, a vector for high level expression of glutathione S-transferase (GST) fusion proteins in yeast under the control of a copper-inducible CUP1 promoter.
TABLE 2.
Primers used in this study
| Name | Sequence 5′ → 3′ |
|---|---|
| Primers used for construction of deletion mutants | |
| Delare1fw | GTTCAGCACGGCTTGCAGCAAGAGCGCCAAAACAGATTGCAAGACAGCTGAAGCTTCGTACGC |
| Delare1rev | TATATCTATCAAGGGCTTGCGAGGGACACACGTGGTATGGTGGCAGTGCATAGGCCACTAGTGGATCTG |
| Delare2fw | TATATCTATCAAGGGCTTGCGAGGGACACACGTGGTATGGTGGCAGTGCATAGGCCACTAGTGGATCTG |
| Delare2ev | AACAGACACATTACGTTAGCAAAAGCAACAATAACAAACACAACCCAGCTGAAGCTTCGTACGC |
| Are1fwctrl | GAAAAATGTGAGATGGTGTAGAGTG |
| Are1revctrl | ATGGTTCTGCCCCAGATTTACC |
| Are2fwctrl | CTTTCATCAATACATCTATATATTCG |
| Are2revctrl | GTAATTGTGGTAGCTGTGTGTTCAT |
| Primers for construction of overexpressing constructs | |
| Lpx1fw | GCGTGGATCCATGGAACAGAACAGGTTCAAGAAAG |
| Lpx1rev | CGGGAATTCTTACAGTTTTTGTTTAGTCGTTTTAACC |
| Ldh1fw | GCGTGGATCCATGAATATGGCAGAACGTGCAG |
| Ldh1rev | CGGGAATTCCTACAATTTGGAATTATCAATCACCTCTCG |
| Yju3fw | GCGTGGATCCATGGCTCCGTATCCATACAAAG |
| Yju3rev | CGGGAATTCTTATGGTTTAGCTTCGGTCGTG |
| Ayr1fw | GCGTGGATCCATGTCGGAGTTACAGTCACAAC |
| Ayr1rev | CGGGAATTCCTAATCGTCCTTATTCTTCTGTTTCGAC |
| Eht1fw | GCGTGGATCCATGTCAGAAGTTTCCAAATGGCC |
| Eht1rev | CGAGTCGACTCATACGACTAATTCATCAAACTTAGTG |
| Tsc10fw | GCGTGGATCCATGAAGTTTACGTTAGAAGACCAAGTTG |
| Tsc10rev | CGAGTCGACTCAGTTGGCCTTCTTGCCGTC |
| YBR056wfw | GCGTGGATCCATGATTGGCTCACTTAGAAACAAATTTGAG |
| YBR056wrev | CGGGAATTCTTAATACTTATTAAACTCATCTAACCCACGTTG |
| YKL050cfw | GCGTGGATCCATGTCACTAATATCTGCGTTGCAAAC |
| YKL050crev | CGAGTCGACTTAGATAACCTCTTTGAAAAAACTTTCCTTAGGAG |
| Primers for site-directed mutagenesis | |
| Ayr1_S18Afw | GTTGTTACAGGCGCCGCCGGTGGTATTGGATATG |
| Ayr1_S18Arev | CATATCCAATACCACCGGCGGCGGCTGTAACAACG |
Point mutations in AYR1 were introduced by site-directed mutagenesis. Plasmid pYEX 4T-1_AYR1S18A was constructed by overlap extension PCR using the primers listed in Table 2.
Isolation of Organelles
Isolation of highly purified LD and Px was performed as previously described (29–31) and will only be described in brief here. For the preparation of Px, cells were grown in YPO to the late exponential phase. After harvesting, washing, and determining of the cell wet weight, cells were incubated with 0.5 g/ml SP-A (0.1 m Tris/SO4, pH 9.4) and 1.54 mg of DTT/ml of SP-A for at least 10 min at 30 °C with shaking. Cells were then washed and resuspended in prewarmed SP-B (1.2 m sorbitol, 20 mm KH2PO4, pH 7.4), and spheroplasts were generated by using Zymolyase-20 T (Seikaguku Corp.) at a concentration of 2 mg/g cell wet weight in 6 ml of SP-B/g of cell wet weight for 1 h at 30 °C with shaking. The resulting spheroplasts were then washed with cold SP-B and resuspended in breaking buffer (5 mm MES/KOH, pH 6.0, 0.6 m sorbitol, 1 mm KCl, and 0.5 mm EDTA) in twice the cell volume. The cell suspension was homogenized on ice using a Dounce homogenizer with a tight fitting pestle. Nuclei, unbroken cells, and cell debris were removed by centrifugation at 5,000 × g for 5 min in a Sorvall SLC3000 rotor. The resulting pellet was resuspended twice, rehomogenized, and centrifuged again to enhance the yield. The combined supernatants were centrifuged at 15,000 × g for 15 min in an SS34 rotor. The organelle pellet, consisting of mitochondria and Px, was again suspended in breaking buffer and centrifuged at 5,000 × g for 5 min to clean it from larger aggregates. The centrifugation step at 15,000 × g was repeated, and after suspension of the pellet in breaking buffer, it was loaded onto a Nycodenz gradient (17–35%; w/v) in 5 mm MES/KOH, pH 6.0, 1 mm KCl, 0.24 m sucrose. The loaded gradient tubes were then centrifuged at 26,000 × g for 90 min in a swing out rotor (Sorvall AH-629). The white Px layer was taken with a syringe, diluted with 4 volumes of breaking buffer, and sedimented at 15,000 × g in an SS34 rotor for 15 min at 4 °C.
For the isolation of LD, cells were grown in minimal medium (SD) to the early stationary phase. After preparation of spheroplasts as described above, they were resuspended in 1 ml/g cell wet weight LD-A (12% Ficoll 400 in 10 mm MES/Tris, pH 6.9, 0.2 mm Na2EDTA·2H2O), followed by mechanical disintegration using a Dounce homogenizer with a loose fitting pestle. The resulting homogenate was diluted with a half-volume of LD-A and centrifuged at 7,000 × g for 5 min at 4 °C. The supernatant was collected, and the pellet was resuspended in LD-A. Spheroplast disintegration and centrifugation were repeated, and the combined supernatants were transferred into Ultra-Clear centrifuge tubes (Beckman), up to one-third of height overlaid with LD-A to the top of the tube. Ultracentrifugation at 28,000 × g for 45 min at 4 °C using a swing out rotor yielded a white layer on top (crude LD) that was lifted with a spatula and transferred into a 15-ml Dounce homogenizer. The crude LD were homogenized with a loose fitting pestle. Then the sample was transferred to a new ultracentrifuge tube (one-quarter of the total tube volume) and overlaid with LD-B (8% Ficoll 400 in 10 mm MES/Tris, pH 6.9, 0.2 mm Na2EDTA·2H2O). Ultracentrifugation at 28,000 × g for 30 min at 4 °C resulted in a top layer containing LD, which was again transferred to a 15-ml Dounce homogenizer, where the LD were rehomogenized. Prior to the last ultracentrifugation step, buffer LD-C (0.25 m sorbitol in 10 mm MES/Tris, pH 6.9, 0.2 mm Na2EDTA·2H2O) was filled into a fresh ultracentrifuge tube up to three-quarters of the tube volume. The homogenized LD were loaded to the bottom of the tube with the aid of a syringe. The last ultracentrifuge step at 28,000 × g for 30 min at 4 °C yielded a top layer containing highly purified LD. The top layer was collected with a pipette and transferred into a 7-ml Dounce homogenizer, and LD were homogenized for further analysis.
Protein Determination
The protein concentration of isolated fractions and whole cell extracts was determined by the method of Lowry et al. (32) using bovine serum albumin as a standard. Samples of LD fractions were delipidated with 2–3 volumes of diethyl ether prior to protein analysis. The organic phase was withdrawn, and residual diethyl ether was removed under a stream of nitrogen. Proteins were precipitated with trichloroacetic acid at a final concentration of 10% and solubilized in 100 μl of 0.1% SDS, 0.1 m NaOH. Purified proteins were quantified by using the Bio-Rad protein assay based on the method of Bradford (33) and bovine serum albumin as a standard. 10 μg of each fraction were loaded onto SDS gels for Western blot analysis. SDS-PAGE was performed by the method of Laemmli (34) using 12.5% separation gels. Proteins on gels were detected by staining with Coomassie Blue. Western blot analysis was performed according to Haid and Suissa (35). Samples were denatured at 37 °C to avoid hydrolysis of polypeptides. Proteins were detected by a primary antibody directed against the GST tag and a peroxidase-conjugated secondary antibody. Enhanced chemiluminescent signal detection reagents (SuperSignalTM, Pierce) were used to visualize immunoreactive bands.
Lipid Analysis
For lipid analysis, crude homogenates were prepared by harvesting cells in the early stationary phase, resuspending the cell pellet in breaking buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl) and disintegrating by vigorous shaking in the presence of glass beads for 10 min at 4 °C. After disruption, cell debris were removed at 5,000 × g for 5 min. The supernatant was further used for protein determination and lipid extraction by the method of Folch et al. (36), using chloroform/methanol (2:1; v/v) as solvent. For the quantification of non-polar lipids, extracts were applied to Silica Gel 60 plates, and chromatograms were developed by a two-step developing system using first the solvent system light petroleum/diethyl ether/acetic acid (35:15:1) for two-thirds of the plate. Plates were dried briefly and further developed to the top of the plate using the second solvent system light petroleum/diethyl ether (49:1; v/v). Non-polar lipids were visualized by postchromatographic charring after dipping the plates in a solution consisting of 0.63 g of MnCl2 × 4H2O, 60 ml of water, 60 ml of methanol, and 4 ml of concentrated sulfuric acid. The plates were then heated at 105 °C for 40 min. Visualized bands were quantified by densitometric scanning (CAMAG TLC SCANNER 3) at 400 nm with authentic standards containing defined amounts of the respective lipids.
Radioactive in Vivo TG Mobilization Assay
Precultures from wild type BY4741 (WT) and tgl3Δtgl4Δtgl5Δare1Δare2Δ (QiM) were used to inoculate a main culture to an A600 of 0.1 in 100 ml of liquid YPO containing 10 μl of [3H]oleic acid (50 μCi = 110,000,000 dpm). Cells were grown for 20 h at 30 °C with shaking. Then cells were harvested and washed twice with a sterile solution containing 0.5% bovine serum albumin (essentially fatty acid-free; Sigma) and resuspended in 100 ml of non-labeled YPO. Cells were incubated at 30 °C, and aliquots were taken at the time points indicated. Cells were washed with 0.5% bovine serum albumin prior to cell density measurement at 600 nm. Aliquots of 10 ml were harvested at 4,500 × g for 5 min in a Hettich table top centrifuge. Cells were washed with 0.5% bovine serum albumin, and cell pellets were frozen at −20 °C and then used for lipid extraction as described above. Dried lipid extracts were separated by TLC as described above. Bands of TG were scraped off and, after the addition of 7 ml of scintillation mixture, subjected to scintillation counting using LSC Safety (Baker, Deventer, The Netherlands) with 5% water as a scintillation mixture.
Lipase/Esterase Inhibitor Assays
Specific inhibitors were used to screen for the occurrence of TG lipases (NBD-sn1/3-TGP) and hydrolases (NBD-HE-HP) (37–40). The reactive groups of the inhibitors bind specifically to the respective enzymes. The covalently linked enzyme-inhibitor complex can be detected through the NBD tag. For the inhibitor assay, purified LD or Px samples were incubated with 0.1% (final concentration) of the respective inhibitors and 0.6% Triton X-100 in 50 mm Tris-Cl, pH 7.4, at 37 °C on a thermomix shaker at 600 × g overnight. Proteins were precipitated with 10% (final concentration) TCA and washed in cold acetone to remove all unbound inhibitor. The remaining sample was dissolved in 10 μl of SDS-sample buffer (63 mm Tris/HCl, pH 6.8, 2% SDS, 10% glycerol, 0.0025% bromphenol blue, 2% β-mercaptoethanol) and separated by SDS-PAGE as described above. To identify fluorescent protein-inhibitor complexes in gel, gels were scanned at 530 nm and at an excitation wavelength of 488 nm using an Imager FXProPlus (Bio-Rad). For total protein staining, gels were incubated with Sypro Ruby at room temperature for 1 h and washed in acetic acid/ethanol for 2 h. The fluorescence was scanned at 605 nm (excitation wavelength of 488 nm). LC-MS/MS analyses were performed as described by Birner-Gruenberger et al. (41). In addition, a detailed description of the method is provided in the supplemental material. Protein quantification was carried out as described above.
Protein Expression, Solubilization, and Purification
All candidate proteins were expressed from plasmid pYEX4T-1 in S. cerevisiae TM. GST fusion proteins were purified batch-wise by affinity chromatography using reduced glutathione agarose. Ayr1p and Eht1p were purified from LD fractions after solubilization by Zwittergent® (Sigma) at a final concentration of 20 mm; Lpx1p was solubilized from Px fractions with Zwittergent® at 10 mm; Ldh1p and YBR056wp were obtained from homogenates after solubilization with 2 mm Zwittergent. Reduced glutathione-agarose beads (Invitrogen) were washed with PBS to remove azide and equilibrated with PBS containing 1% Triton X-100 at 4 °C just prior to use. Solubilized fractions were applied to GST-beads and incubated for 2 h with moderate shaking at 4 °C. After washing with PBS, fusion proteins were eluted with 20 mm reduced glutathione (Sigma) in PBS, pH 8, at 4 °C for 30 min.
Esterase Activity Assays Using p-Nitrophenyl Esters as Substrates
p-Nitrophenylacetate (pNPA), p-nitrophenylbutyrate (pNPB), and p-nitrophenypalmitate were used as substrates to determine esterase activity of purified proteins. The total reaction volume was 1 ml, containing 890 μl of 1× PBS buffer, 100 μl of substrate solution (ranging from 25 μm to 10 mm final concentrations), and 10 μl of purified protein, which was added directly prior to measurement. Absorbance was measured at 405 nm for 3 min at 30 °C. Enzyme activity was determined using different substrate concentrations. Michaelis-Menten kinetics was analyzed using GraphPad Prism version 5.
RNA Isolation and Real-time PCR
For the isolation of RNA, cells were grown to the midlogarithmic phase on YPD and YPO at 30 °C. RNA was isolated using the RNeasy kit from Qiagen as described by the manufacturer. After DNase I digestion, real-time PCR was performed using the SuperScript III Platinum SYBR Green One-Step qRT-PCR kit (Invitrogen) by following the manufacturer's instructions. Reactions were performed in sealed MicroAmp optical 96-well reaction plates, and amplification was measured using an ABI 7500 instrument (Applied Biosystems). Samples were quantified using the ΔΔCt method described by Livak and Schmittgen (42). Differences in mRNA expression after ACT1 normalization relative to the control can be calculated with this method. Primers used for real-time PCR are listed in Table 3.
TABLE 3.
Primers for qRT-PCR
| Primer | Sequence (5′ → 3′) |
|---|---|
| ACT1_RTfw | CCAGCCTTCTACGTTTCCATCCAAG |
| ACT1_RTrev | GACGTGAGTAACACCATCACCGGA |
| LPX1_RTfw | AACTCCCTACGTTTAAAGACGTGCGACCATTTC |
| LPX1_RTrev | CAGGATTTGGCTGTGCGCATTGGT |
| LDH1_RTfw | TCATAAGATAGTGCTTGTAGGGCATTCTATGGGTT |
| LDH1_RTrev | GCGTCAACAAAACTAGCGTTTGTACCGCT |
| YJU3_RTfw | AGGAATCACATCTGATAAAGCCTATC |
| YJU3_RTrev | TCGTGTATTTGCCTAAACGACCCA |
| AYR1_RTfw | GCTGATAAAAGGCCCTTGCCTGAAACCTCAA |
| AYR1_RTrev | CAGCTGGCATGGGCTTATTGTCCTTAGC |
| EHT1_RTfw | TTCCAAATGGCTCTCTCCCCGATCA |
| EHT1_RTrev | TTCCCGGGTCGATTTAAAGCTCTTTG |
| TSC10_RTfw | CAAGTCGCTGGCCAGAGGTGATGA |
| TSC10_RTrev | AGCGGCTTTTCTTTGCGGTGAGC |
| YBR056w_RTfw | GGACTTTGCAGTTTGAATACGGTG |
| YBR056w_RTfw | AAGTCATCACCGTGAGGA |
| YKL050c_RTfw | GCGTGTTGGATGATGGGCCTAA |
| YKL050c_RTrev | CGGTGCCATCTACTGCTTGGTGCAC |
In Vivo TG Mobilization Assays
TG mobilization of strains overexpressing putative lipases/esterases was measured in vivo by letting cells grow until the early stationary phase in synthetic minimal medium. Then fresh medium was inoculated with the pregrown cultures to an A600 of 3. For the mobilization of TG, the fatty acid synthesis inhibitor cerulenin was added to a final concentration of 10 μg/ml. At the time points indicated, 10-ml aliquots were taken, cells were harvested, and lipids were extracted and analyzed as described above.
Triacylglycerol Lipase Activity Assays Using [9,10-3H]Triolein as Substrate
TG lipase activity of isolated LD and purified enzymes was determined using [9,10-3H]triolein (PerkinElmer Life Sciences) as substrate. TG lipase activity was measured in a final volume of 200 μl. Samples were incubated in a mixture containing 100 mm potassium phosphate buffer, pH 7.5, 125 μm [9,10-3H]triolein (specific activity 50 μCi/ml), 45 μm phosphatidylcholine/phosphatidylinositol (3:1; mol/mol), 25 mm MgCl2, and 0.2% fatty acid-free bovine serum albumin at 30 °C for 1 h in a water bath. The reaction was stopped by adding 2 ml of chloroform/methanol (2:1, v/v), and lipids were extracted as described above. Lipids were dried under a stream of nitrogen, dissolved in 50 μl of chloroform/methanol (2:1, v/v), and separated by TLC. Chromatograms were developed using chloroform/acetone/acetic acid (45:4:0.4, v/v/v) as a solvent system. Lipids were visualized by staining with iodine vapor, and bands corresponding to fatty acids were scraped off the plate. Radioactivity was measured by liquid scintillation counting using LSC Safety (Baker) with 5% water as a scintillation mixture. Enzyme activity was further determined, depending on the triolein substrate concentration (0.2–250 μm) and time (1–200 min). Michaelis-Menten kinetics was analyzed using GraphPad Prism version 5.
RESULTS
A Yeast Strain Lacking All Currently Known TG Lipases Is Able to Mobilize TG in Vivo
The mobilization of TG from LD requires the catalytic activities of TG lipases. Previous research from our laboratory identified Tgl3p as the major TG lipase and the two enzymes Tgl4p and Tgl5p as showing minor lipolytic activities (11, 12). In the course of these studies, however, it became evident that the turnover of TG in TM did not come to a complete halt when cells were forced to mobilize TG in the presence of cerulenin, an inhibitor of fatty acid synthesis, suggesting the presence of additional TG lipases. To test this hypothesis, we constructed a mutant strain lacking both acyl-CoA:sterol acyltransferases, Are1p and Are2p, as well as the major known TG lipases Tgl3p, Tgl4p, and Tgl5p and performed a pulse-chase assay monitoring the fate of TG in vivo. This QiM accumulated high levels of TG but did not produce any SE, ensuring that incorporation of [3H]oleate occurred primarily into TG and that the loss of label due to incorporation into other compounds was minimized. Cells were pulse-labeled with [3H]oleic acid, and levels of 3H in TG were followed during the chase on unlabeled YPO. Both wild type and QiM grew normally under the given conditions (Fig. 1A). As expected, in wild type cells, the label in TG decreased over the time, indicating that TG was subject to turnover. Interestingly, in the QiM, lacking all known TG lipases, TG mobilization was observed with a slight delay but at a similar rate as wild type (Fig. 1B). Thus, we concluded that in cells grown on oleic acid, one or more additional lipases might become active, accounting for TG mobilization.
FIGURE 1.
In vivo triacylglycerol mobilization. Cells from either WT (●) or the QiM (■) were grown in the presence of [3H]oleate for 5 h and then shifted to fresh YPO medium. At the time points indicated, aliquots were withdrawn and analyzed for cellular density (A) and for the amount of [3H]TG (B). Data were obtained from two independent experiments with S.E. values (error bars) as indicated.
Screening for Novel Lipases and Hydrolases in LD and Px
The findings described above led us to search for novel TG lipases/hydrolases in the yeast. To test whether such enzymes became active in yeast grown on oleic acid, we employed a functional proteome assay making use of fluorescently labeled inhibitors of serine lipid hydrolases as probes (37, 38). Isolated organelles from the yeast were used to screen for such novel enzymes. LD from cells grown on glucose or oleic acid, respectively, as well as Px from YPO-grown cells were incubated with fluorescent phosphonic acid esters (NBD-sn1/3-TGP and NBD-HE-HP), which covalently bind to the active sites of serine lipid hydrolases. After incubation, proteins were separated by SDS-PAGE, and labeled polypeptides were identified by the NBD-reporter tag. In LD from wild type grown on YPD and YPO, protein bands of the size of the three lipases Tgl3p (∼73 kDa), Tgl4p (∼102 kDa), and Tgl5p (∼85 kDa) were detected (Fig. 2A, asterisks). Moreover, bands representing possible new lipases were identified as well (Fig. 2A, arrows). A comparison of the two growth conditions revealed that the protein pattern of cells grown on YPO was different from YPD. We concluded from these data that indeed a different subset of proteins existed when cells were grown in the presence of oleic acid compared with glucose. To follow up this idea, we analyzed LD from a TM grown on YPD or YPO, respectively. Again, we were able to detect additional protein bands that did not correspond to the already known TG lipases (Fig. 2B, arrows).
FIGURE 2.
In vitro inhibitor assay screening for novel hydrolases from lipid droplets and peroxisomes. LD and Px from WT and TM were isolated from cells grown on YPD or YPO as described under “Experimental Procedures.” Aliquots were incubated with inhibitors as described and incubated at 37 °C overnight. Proteins were then precipitated and separated by SDS-PAGE. Gels were scanned at a wavelength characteristic for NBD. A, LD protein pattern from WT cells after incubation with inhibitors for TG lipases or hydrolases from YPD- and YPO-grown cells. Asterisks indicate the occurrence of the three known TG lipases Tgl3p (73 kDa), Tgl4p (102 kDa), and Tgl5p (84 kDa). Arrows show the formation of additional bands indicating the presence of additional lipases or hydrolases. blank, background of proteins incubated with no inhibitor. B, LD protein pattern from TM cells grown on either YPD or YPO. Arrows indicate the occurrence of new hydrolases/lipases. C, Px from either WT or TM grown on YPO were incubated with a subset of inhibitors and analyzed for formation of protein bands indicating lipases or esterases (arrows).
Similar results were obtained when we analyzed Px from wild type and TM with the same subset of inhibitors (Fig. 2C). Arrows indicate the occurrence of novel proteins exhibiting an esterase (NBD-HE-HP) or even lipase (NBD-sn1/3-TGP) activity. Proteins detected were of different molecular mass compared with those from LD. Thus, we speculated that on LD and Px, distinct hydrolases were expressed under oleic acid-induced conditions. Altogether, our screening apparently identified new and not yet characterized candidates for novel hydrolases/lipases on LD and Px. To identify these proteins reacting with NBD-HE-HP, they were separated by SDS-PAGE (Fig. 3A), tryptically digested, and subjected to MS analysis. A complete list of proteins identified by this approach is shown in supplemental Table 1. Proteins are listed according to their respective molecular weight as well as by peptide length and score value. All known TG lipases were identified with high abundance in LD samples. One exception was Tgl5p, which was found only on LD from cells grown on YPD. Interestingly, only a few gene products were detected in this screening in more than one organelle sample (see supplemental Table 1).
FIGURE 3.
Mass spectrometric analysis of novel lipase/esterase candidates. LD and Px were prepared and incubated with NBD-HE-HP, which resembles a single chain carboxylic acid ester as described under “Experimental Procedures.” A, proteins were separated by SDS-PAGE, and fluorescent bands were cut from the gel, subjected to tryptic digestion, and analyzed by MS/MS. B, Venn diagram showing the number of proteins found in either LD from YPD, LD from YPO, or Px from YPO, indicating the overlapping hits.
As expected, both LD samples showed the highest overlap in protein candidates (Fig. 3B). Eighteen candidate proteins were found on LD from cells grown on both carbon sources. These proteins represented putative new candidate hydrolases or lipases under oleic acid-induced conditions. The most promising candidates, containing a TG lipase motif (GXSXG) and showing high abundance in the performed screening, were selected for further investigations. Further analysis was therefore restricted to Lpx1p, Ldh1p, Yju3p, Ayr1p, Eht1p, Tsc10p, Ybr056wp, and Ykl050cp (Table 4).
TABLE 4.
Short list of candidate hydrolases
Listed are putative new lipases or hydrolases on LD from yeast cells grown on either YPD or YPO. The selection was based on motif, localization, abundance in the inhibitor assay screening, and already known functions. TGL, triacylglycerol lipase motif (GXSXG); HYD, hydrolase motif (DXDX(T/V)); LD, lipid droplets; Mt, mitochondrion; Cyt, cytosol; ER, endoplasmic reticulum; N, nucleus.
| ORF | Gene name | Localization | Motif |
|---|---|---|---|
| YOR084W | LPX1 | Px | TGL |
| YBR204C | LDH1 | LD, Mt, Px | TGL, HYD |
| YKL094W | YJU3 | LD | TGL (2×) |
| YIL124W | AYR1 | ER, LD, Mt, Cyt | TGL |
| YBR177C | EHT1 | LD, Mt | TGL |
| YBR265W | TSC10 | Cyt, ER, Mt | TGL |
| YBR056W | Cyt | TGL | |
| YKL050C | Unknown | TGL |
Because previous studies from our laboratory described functions of Tgl3p, Tgl4p, and Tgl5p in addition to their lipase activities, we speculated about dual functions and localizations of other proteins involved in non-polar lipid metabolism (19, 20). Lpx1p is an oleic acid-inducible lipase that localizes to the peroxisomal matrix and is required for normal Px morphology. However, thus far, it has been impossible to identify the reaction catalyzed by Lpx1p. In previous studies (43), Lpx1p was heterologously expressed and purified from Escherichia coli. The purified protein was tested for esterase, TG lipase, and phospholipase C and D activities, but only weak lipolytic activity was measured in vitro. Therefore, Lpx1p was chosen as a positive control for enzymes with minor TG lipase activity in vivo.
Ldh1p was reported as a hydrolase with a GXSXG type motif, primarily localized to LD exerting esterase and weak TG lipase activities in vitro (44, 45). Yju3p was identified as a functional orthologue of mammalian monoacylglycerol lipase specifically hydrolyzing monoacylglycerols (46). Because TG lipase activity of Yju3p was unlikely, it served as our negative control. Ayr1p has been described as NADPH-dependent 1-acyldihydroxyacetone phosphate reductase found on LD, in the endoplasmic reticulum, and on the mitochondrial outer membrane involved in phosphatidic acid biosynthesis (47). In addition, Ayr1p was shown to display 17β-hydroxysteroid dehydrogenase activity (48). Eht1p localizes to LD and the mitochondrial outer membrane and has been annotated as acyl-coenzyme A:ethanol O-acyltransferase (2, 49, 50). It plays a minor role in medium-chain fatty acid ethyl ester biosynthesis and possesses esterase activity on short-chain substrates. Tsc10p is a ketosphinganine reductase that catalyzes the second step in phytosphingosine synthesis (51). Ybr056wp and Ykl050cp are two proteins of currently unknown functions.
Gene Expression of Candidate Genes in Cells Grown on YPO
The different protein patterns of LD from cells grown on YPD or YPO, respectively, suggested that different proteins may be active on the different carbon sources. To address this question, the expression levels of the candidate proteins in wild type cultivated on YPD and YPO were compared. All candidate proteins showed an increased expression level on YPO (Table 5). This effect, which we called here the “YPO induction factor,” became most obvious for LPX1. This result can be explained by the fact that Lpx1p is a peroxisomal protein. Because Px strongly proliferate in cells grown on YPO, these increased mRNA levels of peroxisomal proteins were not surprising. Interestingly, the unannotated genes YBR056w and YKL050c also showed a very high expression in cells grown on YPO. AYR1 became especially interesting because it showed a 5-fold increase in the relative expression level on YPO. In a second set of experiments, we compared expression levels of candidate genes in the TM cultivated on YPD or YPO, respectively (Table 5). The increase of mRNA levels of LPX1 could again be observed but was less pronounced than in wild type. LDH1, EHT1, YKL050c, and AYR1 also showed significantly elevated levels of mRNA in cells grown on YPO.
TABLE 5.
“YPO induction factor” of putative hydrolases in wild type and TM
Relative gene expression of LPX1, LDH1, YJU3, AYR1, EHT1, TSC10, YBR056w, and YKL050c was measured by qRT-PCR from isolated RNA of WT and TM, respectively, cultivated on YPD and YPO. Expression of the respective genes in YPD-grown cells was set at 1, and values obtained with cells grown on YPO were set in relation (“YPO induction factor”). Data are mean values from three independent experiments with the respective deviations <5%. WT, wild type; TM, triple mutant (Δtgl3Δtgl4Δtgl5).
| Gene | YPO induction factor |
|
|---|---|---|
| WT | TM | |
| LPX1 | 140 | 63 |
| LDH1 | 4 | 2.5 |
| YJU3 | 4 | 1.3 |
| AYR1 | 5 | 3 |
| EHT1 | 3 | 2.5 |
| TSC10 | 2 | 1.3 |
| YBR056w | 20 | 1.5 |
| YKL050c | 40 | 3 |
In Vitro Esterase Assays Confirm Hydrolytic Activities of Candidate Proteins
The eight candidate genes described above were overexpressed in a Δtgl3/4/5 background (TM) and purified to perform enzyme analyses. For this purpose, full-length proteins were expressed as fusion proteins with an N-terminal GST tag in S. cerevisiae (Fig. 4A) and purified using affinity chromatography (Fig. 4B). Lpx1p was successfully purified from peroxisomal fractions, Ayr1p and Eht1p from LD samples, and Ldh1p and Ybr056wp were isolated from whole cell extracts. Yju3p, Tsc10p, and Ykl050cp could not be purified at sufficient amounts and purity.
FIGURE 4.
Protein expression and purification of novel triacylglycerol lipase candidates. A, Western blot analysis from total cell proteins from overexpressing strains grown to stationary phase after induction with CuSO4 for 4 h. Putative hydrolases were expressed as fusion proteins with a GST tag and purified after induction by affinity chromatography. Western blot analyses are representative for at least two independent experiments. B, Coomassie Blue-stained SDS gel of purified proteins. Lpx1p was purified from Px, Ayr1p and Eht1p were isolated from LD, and Ybr056wp and the tag itself as a control were purified from homogenates. All other proteins could not be purified at sufficient amounts or purity.
Purified enzymes were analyzed for esterase activities using pNPB as well as pNPA as substrates (Table 6). Lpx1p hydrolyzed pNPB with a Km of 0.89 mm and a Vmax of 3.76 μmol/min/mg, which resembled values reported by Thoms et al. (43). Higher activities were found with pNPA as substrate. The highest activities were found for Ayr1p, which cleaved pNPA with a Km of 3.50 mm and a Vmax of 18.50 μmol/min/mg, and pNPB, with a Km of 1.52 mm and a Vmax of 14.06 μmol/min/mg. All other enzymes tested showed only minor hydrolytic activities with these substrates. p-Nitrophenyl palmitate was not converted by any of the enzymes (data not shown). Control assays with the GST tag showed that the tag alone did not affect our results.
TABLE 6.
In vitro esterase activities
Esterase activity was measured from purified enzymes using pNPB and pNPA as test substrates. Km and Vmax values were calculated using Michaelis-Menten approximations. Data are mean values from three independent experiments with the respective S.D. values.
| Enzyme | Substrate | Vmax | Km |
|---|---|---|---|
| μmol/min/mg | mm | ||
| Lpx1p | pNPB | 3.76 ± 0.54 | 0.89 ± 0.13 |
| pNPA | 10.94 ± 0.98 | 2.50 ± 0.22 | |
| Ldh1p | pNPB | 0.90 ± 0.15 | 0.89 ± 0.15 |
| pNPA | 1.26 ± 0.18 | 1.78 ± 0.25 | |
| Ayr1p | pNPB | 14.06 ± 2.23 | 1.52 ± 0.24 |
| pNPA | 18.50 ± 2.76 | 3.50 ± 0.52 | |
| Eht1p | pNPB | 1.01 ± 0.14 | 0.88 ± 0.12 |
| pNPA | 3.44 ± 0.48 | 2.06 ± 0.29 | |
| Ybr056wp | pNPB | 1.439 ± 0.22 | 0.81 ± 0.12 |
| pNPA | 7.02 ± 1.26 | 3.51 ± 0.63 | |
| Control: GST tag | pNPB | 0.35 ± 0.05 | 0.37 ± 0.05 |
Overexpression of Candidate Genes Affects TG Content of Yeast Cells in the Stationary Phase
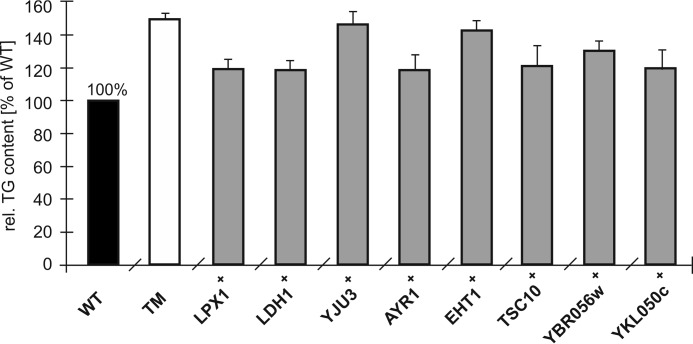
Because TG mobilization is already strongly impaired in a TM strain, additional major lipolytic activities were not expected. To make possible effects of our candidates visible, we chose overexpression of these genes in the TM background rather than additional deletions. Analysis of non-polar lipids from whole cell extracts was performed in cells grown to the stationary phase under these conditions. The TG content in the TM was increased to ∼150% of wild type (Fig. 5). Strains overexpressing LPX1+, LDH1+, AYR1+, TSC10+, YBR056w+, and YKL050c+ showed a reduction of TG of about 30% compared with the TM. In contrast, overexpression of YJU3+ and EHT1+ did not significantly change the level of TG (Fig. 5). Lpx1p and Ldh1p had already been described as new TG lipases (43–45). As expected, deletion of the respective genes in addition to TGL3, TGL4, and TGL5 did not show an effect on growth in the presence of glucose or oleic acid as a carbon source; nor did it show an effect on the TG content compared with the TM (data not shown).
FIGURE 5.
Triacylglycerol levels of strains overexpressing candidate lipases. Cells from WT, TM, and strains overexpressing putative novel TG lipases in the TM background were grown in minimal medium until early stationary phase. Total cell and lipid extracts were obtained as described under “Experimental Procedures.” Lipids were separated by TLC and quantified densitometrically. Results shown are the average from at least three independent experiments with the S.D. values (error bars) indicated.
In Vivo Mobilization of TG Identifies Ayr1p as Novel TG Lipase
Changes in the TG content of strains overexpressing putative TG lipases suggested that these proteins also exhibit lipolytic activities in vivo. Therefore, we examined the mobilization of TG in the presence of the fatty acid synthase inhibitor cerulenin with all overexpressing strains described above. Because synthesis of fatty acids is inhibited under these conditions, cells are forced to mobilize TG to ensure sufficient supply for the synthesis of membrane lipids. At the same time, the synthesis of TG is blocked. Degradation of TG was compared both with the wild type and with the TM. Wild type mobilized TG very quickly and showed a nearly complete depletion of TG within 7 h, whereas the TM showed poor mobilization, which leveled off at 80% of the initial value after this time period. This result confirmed previous studies from our laboratory (12). Furthermore, we observed that LPX1+ and AYR1+ showed a significantly altered mobilization rate compared with the TM. The TG level of the AYR1-overexpressing strain dropped to 53%, and the strain overexpressing LPX1 even showed enhanced mobilization to 40% of the starting value (Fig. 6A). All other overexpression strains did not show more mobilization of TG than the TM (Table 7). These results demonstrated that Lpx1p acts as a TG lipase in vivo, confirming previous results obtained in vitro (43). Additionally, the role of Ayr1p as a novel TG lipase of the yeast was fostered. In parallel to the determination of the cellular TG content, growth experiments were performed to exhibit sensitivity to cerulenin. All overexpression strains showed a growth behavior similar to that of TM, which was more sensitive to cerulenin than the wild type (Fig. 6B). In the absence of cerulenin, all strains grew like wild type (data not shown).
FIGURE 6.
In vivo triacylglycerol mobilization in the presence of cerulenin. Cellular TG levels (A) and growth curves (B) of WT (♦), TM (●), and the two overexpressing strains TM + AYR1+ (+) and TM + LPX1+ (▴) in the presence of cerulenin (inhibitor of fatty acid synthesis) are shown. Data are expressed as relative amounts of TG per culture aliquot with the value at the time point 0 set at 100%. Analyses are representative of at least four independent experiments. Data are mean values with respective S.D. values of about 10%.
TABLE 7.
Triacylglycerol mobilization of overexpressing strains in the presence of cerulenin
Strains overexpressing putative new lipases as well as the TM were grown in the presence of cerulenin. Culture aliquots were taken between 0 and 7 h and analyzed for TG content as described under “Experimental Procedures.” TG content at the starting point was set as 100% and followed over time. Results shown are the average from at least three independent experiments. S.D. values were <10%.
| Strain | TG content after 7 h |
|---|---|
| % of starting point | |
| Δtgl3/4/5 (VC) | 80 |
| TM + LPX1+ | 40 |
| TM + LDH1+ | 70 |
| TM + YJU3+ | 75 |
| TM + AYR1+ | 53 |
| TM + EHT1+ | 70 |
| TM + TSC10+ | 80 |
| TM + YBR056w+ | 70 |
| TM + YKL050c+ | 70 |
In Vitro Enzyme Assays Confirm Ayr1p as Novel TG Lipase
Having shown that Lpx1p and Ayr1p exhibit lipolytic activities in vivo, we also tested them for in vitro TG lipase activity. First, we tested lipase activity of purified Ayr1p and Lpx1p. It had previously been reported that Lpx1p exhibits lipase activity using 1,2-dioleoyl-3-(pyren-1-yl)decanoyl-rac-glycerol as substrate (43). Therefore, Lpx1p served as a positive control. Purified Ayr1p showed an even higher TG lipase activity than Lpx1p (Fig. 7A). To confirm lipase activity of Ayr1p, LD fractions from the AYR1-overexpressing strain were compared with wild type and TM (Fig. 7B). In agreement with our data of the in vivo TG mobilization assays as well as with results obtained with purified enzymes, LD from AYR1+ showed increased TG mobilization activity compared with the TM, reaching the same level as wild type. Michalis-Menten enzyme analysis revealed Ayr1p to be an active TG lipase hydrolyzing [9,10-3H]triolein with a Vmax of 10.93 pmol/h/mg and a Km of 61.21 μm. Linear increase of product formation was observed up to 100 min.
FIGURE 7.
In vitro triacylglycerol lipase assays. A, analysis of TG lipase activity of purified Ayr1p and Lpx1p compared with the tag control. B, analysis of TG lipase activity of LD fractions from WT, TM, and TM + AYR1+. C, analysis of TG lipase activity of mutated Ayr1p (S18A) compared with wild type Ayr1p. Experiments were performed in triplicate and are representative of at least two independent biological experiments. Data are mean values with the respective deviation (error bars). n.d., not detectable.
Ayr1p possesses the characteristic GXSXG motif. To determine the functional significance of this conserved sequence, a point mutation was introduced (S18A), and the purified mutated protein was tested for lipase activity as described above. Replacement of serine by alanine in the lipase motif of Ayr1p completely abolished lipase activity (Fig. 7C).
Altogether, we provide evidence for additional hydrolytic enzymes in the yeast S. cerevisiae involved in non-polar lipid metabolism. Moreover, we show that Lpx1p exhibits lipolytic activity not only in vitro as described by Thoms et al. (43) but also in vivo. Identification of Ayr1p as novel TG lipase of the yeast S. cerevisiae provides an additional piece of evidence in the puzzle of non-polar lipid metabolism.
DISCUSSION
Previous studies from our laboratory identified Tgl3p, Tgl4p, and Tgl5p as major TG lipases of the yeast S. cerevisiae being localized to LD (11, 12). In triple deletion strains lacking these major lipolytic enzymes, TG mobilization was strongly impaired but not completely abolished when cells were grown on glucose. Here, we show that the QiM lacking the three TG lipases and the sterol acyltransferases Are1p and Are2p grown on oleic acid is not only able to hydrolyze TG in vivo but also able to mobilize TG close to wild type level (see Fig. 1). This result tempted us to speculate about the presence of further hydrolytic enzymes.
A major goal of the present study was to identify enzymes that might play a role in the mobilization of TG in the absence of the three major lipases. To address this question, we designed a functional proteome approach, making use of specific fluorescent inhibitors of esterases and lipases as probes. This study led to the detection of a subset of proteins that showed TG hydrolase activity in vitro on isolated LD but also with Px (see Fig. 2). Although these probes specifically recognized catalytic serine residues, detection of false positives could not be excluded. We were also aware that non-catalytic proteins were detected by chance due to the overlap with reactive enzyme bands on the gel. However, the sensitivity and selectivity of the assay was proven to be good enough because all known TG lipases were identified, although Yeh1p, a steryl ester hydrolase of LD, was not found. Detection of all prominent LD and peroxisomal proteins in the respective samples served as a proof of purity of the isolated organelles. However, our screening detected additional, so far unannotated, proteins on LD or in Px. Our findings complement a previous study from our laboratory aimed at the completion of the LD proteome (3). Data provided in the present study also support the view that many proteins may have multiple localizations and functions in the cell. Regulatory mechanisms governing localization and function of proteins in different compartments have to be taken into account. One recent example for such aspects is the stability, gene expression, and localization of Tgl3p, depending on the availability and the presence of its substrate TG (52).
The list of proteins found in our screening was narrowed to a subset of proteins with putative hydrolase or lipase function (see Table 4) based on computational sequence analysis, assigned functions, and subcellular localizations. Nevertheless, proteins of already annotated localization and function different from non-polar lipid metabolism were included. The reason for this strategy was the discovery of a dual role for the known TG lipases Tgl3p, Tgl4p, and Tgl5p, which also possess high acyltransferase activities (19, 20). Consequently, other proteins might have multiple functions, too, maybe not only on LD but also in other organelles. The short list of putative novel TG lipases included Lpx1p, Ldh1p, Yju3p, Ayr1p, Eht1p, Tsc10p, Ybr056wp, and Ykl050cp. All of these proteins contain the consensus sequence for a classical lipase (GXSXG motif) with a classical catalytic triad containing a nucleophile serine. Lpx1p and Ldh1p had been described previously as lipases based on in vitro analysis, and Lpx1p was localized on Px, whereas Ldh1p was identified as an LD component (43–45).
One major question, of course, was the physiological role or relevance of the newly detected hydrolytic/lipolytic enzymes. The TG content analysis of whole cell extracts from overexpression strains indeed proposed a role for Lpx1p, Ldh1p, Ayr1p, Tsc10p, Ybr056wp, and Ykl050cp in TG mobilization in the absence of the major lipases, at least in the stationary growth phase (see Fig. 5). In vivo mobilization of TG by Lpx1p and Ayr1p confirmed the role of these two proteins as lipases in living cells, whereas all other enzymes did not exhibit lipolytic activities in vivo (see Fig. 6 and Table 7). This result was perfectly in line with in vitro enzyme assays (see Fig. 7). Moreover, these two proteins showed the highest activities when esterase substrates were used (Table 6). Interestingly, a yeast strain lacking not only in the major lipases Tgl3p, Tgl4p, and Tgl5p but also Ayr1p, Lpx1p, or Ldh1p, respectively, still showed mobilization of TG. This finding supported the view that other, most likely minor lipolytic activities set fatty acids free as supply for energy production and/or synthesis of membrane lipid components. This view was supported by gene expression studies. In these experiments, Lpx1p and Ayr1p were among the most strongly induced genes in the TM grown on oleic acid compared with cells grown on glucose-containing medium (see Table 5). The question remains why TG are mobilized in cells suffering already from an excess of free fatty acids by supply from the medium. Currently, we can only speculate that different pools of lipids exist, which are mobilized upon different requirements. One possibility might be that fatty acids mobilized from TG may serve primarily for phospholipid synthesis or β-oxidation. However, proof for this model is still missing. In a previous study, it had already been reported that growth on oleate led to the accumulation of large amounts of free fatty acids (53). From this finding, it can be assumed that free fatty acids do not play an essential role in regulating the non-polar lipid turnover because TG mobilization occurred normally under these conditions.
In summary, our results suggest that TG turnover is not only catalyzed by the main lipases Tgl3p, Tgl4p, and Tgl5p but is also performed by a subset of lipases and hydrolases with lower activities. Our data demonstrate that the action of such enzymes is strongly dependent on growth conditions and that they may come to the fore when the major enzymes are inactive. Finally, our investigations identified Ayr1p as a novel TG lipase and confirmed the role of Lpx1p as lipase in vivo. These data contribute to a more detailed knowledge of non-polar lipid turnover and set the stage to investigate regulatory aspects of non-polar lipid metabolism in more detail.
Acknowledgments
We thank the Austrian Centre of Industrial Biotechnology Graz for providing the ABI 7500 instrument and Karin Athenstaedt for fruitful discussions.
This work was supported by Austrian Science Fund (FWF) Projects W901 DK Molecular Enzymology and P23029 (to G. D.), the Austrian Federal Ministry of Science and Research (bmwf) (GOLD project in the framework of GEN-AU/Genome Research in Austria), and the Austrian Science Funds, FWF (Project F3005 SFB LIPOTOX).

This article contains supplemental Table 1.
- TG
- triacylglycerol(s)
- LD
- lipid droplet(s)
- Px
- peroxisome(s)
- QiM
- quintuple mutant
- SE
- steryl ester(s)
- TM
- triple mutant
- NBD
- 12-(N-methyl-N-(7-nitrobenz-2-oxa-1,3-diazol-4-yl))
- NBD-sn1/3-TGP
- nitrobenz-2-oxa-1,3-diazole-sn1/3-triacylglyceride phosphonate
- NBD-HE-HP
- O-((6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino)hexanoyl)-aminoethyl-P-(n-hexyl)phosphonic acid p-nitrophenyl ester
- pNPB
- p-nitrophenylacetate
- pNPA
- p-nitrophenylacetate.
REFERENCES
- 1. Czabany T., Wagner A., Zweytick D., Lohner K., Leitner E., Ingolic E., Daum G. (2008) Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 283, 17065–17074 [DOI] [PubMed] [Google Scholar]
- 2. Athenstaedt K., Zweytick D., Jandrositz A., Kohlwein S. D., Daum G. (1999) Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181, 6441–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Grillitsch K., Connerth M., Köfeler H., Arrey T. N., Rietschel B., Wagner B., Karas M., Daum G. (2011) Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited. Lipidome meets proteome. Biochim. Biophys. Acta 1811, 1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kohlwein S. D., Veenhuis M., van der Klei I. J. (2013) Lipid droplets and peroxisomes. Key players in cellular lipid homeostasis or a matter of fat. Store 'em up or burn 'em down. Genetics 193, 1–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oelkers P., Tinkelenberg A., Erdeniz N., Cromley D., Billheimer J. T., Sturley S. L. (2000) A lecithin cholesterol acyltransferase-like gene mediates diacylglycerol esterification in yeast. J. Biol. Chem. 275, 15609–15612 [DOI] [PubMed] [Google Scholar]
- 6. Dahlqvist A., Stahl U., Lenman M., Banas A., Lee M., Sandager L., Ronne H., Stymne S. (2000) Phospholipid:diacylglycerol acyltransferase. An enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants. Proc. Natl. Acad. Sci. U.S.A. 97, 6487–6492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sorger D., Daum G. (2002) Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J. Bacteriol. 184, 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Oelkers P., Cromley D., Padamsee M., Billheimer J. T., Sturley S. L. (2002) The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277, 8877–8881 [DOI] [PubMed] [Google Scholar]
- 9. Yang H., Bard M., Bruner D. A., Gleeson A., Deckelbaum R. J., Aljinovic G., Pohl T. M., Rothstein R., Sturley S. L. (1996) Sterol esterification in yeast. A two-gene process. Science 272, 1353–1356 [DOI] [PubMed] [Google Scholar]
- 10. Zweytick D., Leitner E., Kohlwein S. D., Yu C., Rothblatt J., Daum G. (2000) Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 267, 1075–1082 [DOI] [PubMed] [Google Scholar]
- 11. Athenstaedt K., Daum G. (2003) YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J. Biol. Chem. 278, 23317–23323 [DOI] [PubMed] [Google Scholar]
- 12. Athenstaedt K., Daum G. (2005) Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae, are localized to lipid particles. J. Biol. Chem. 280, 37301–37309 [DOI] [PubMed] [Google Scholar]
- 13. Rajakumari S., Grillitsch K., Daum G. (2008) Synthesis and turnover of non-polar lipids in yeast. Prog. Lipid Res. 47, 157–171 [DOI] [PubMed] [Google Scholar]
- 14. Köffel R., Tiwari R., Falquet L., Schneiter R. (2005) The Saccharomyces cerevisiae YLL012/YEH1, YLR020/YEH2, and TGL1 genes encode a novel family of membrane-anchored lipases that are required for steryl ester hydrolysis. Mol. Cell. Biol. 25, 1655–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jandrositz A., Petschnigg J., Zimmermann R., Natter K., Scholze H., Hermetter A., Kohlwein S. D., Leber R. (2005) The lipid droplet enzyme Tgl1p hydrolyzes both steryl esters and triglycerides in the yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta 1735, 50–58 [DOI] [PubMed] [Google Scholar]
- 16. Müllner H., Deutsch G., Leitner E., Ingolic E., Daum G. (2005) YEH2/YLR020c encodes a novel steryl ester hydrolase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 280, 13321–13328 [DOI] [PubMed] [Google Scholar]
- 17. Schrag J. D., Cygler M. (1997) Lipases and α/β hydrolase fold. Methods Enzymol. 284, 85–107 [DOI] [PubMed] [Google Scholar]
- 18. Mignery G. A., Pikaard C. S., Park W. D. (1988) Molecular characterization of the patatin multigene family of potato. Gene 62, 27–44 [DOI] [PubMed] [Google Scholar]
- 19. Rajakumari S., Daum G. (2010) Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 285, 15769–15776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rajakumari S., Daum G. (2010) Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol. Biol. Cell 21, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kurat C. F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., Scholz H., Zimmermann R., Leber R., Zechner R., Kohlwein S. D. (2006) Obese yeast. Triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 281, 491–500 [DOI] [PubMed] [Google Scholar]
- 22. Rosenberger S., Connerth M., Zellnig G., Daum G. (2009) Phosphatidylethanolamine synthesized by three different pathways is supplied to peroxisomes of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1791, 379–387 [DOI] [PubMed] [Google Scholar]
- 23. Lazarow P. B., De Duve C. (1976) A fatty acyl-CoA oxidizing system in rat liver peroxisomes. Enhancement by clofibrate, a hypolipidemic drug. Proc. Natl. Acad. Sci. U.S.A. 73, 2043–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kunau W. H., Bühne S., de la Garza M., Kionka C., Mateblowski M., Schultz-Borchard U., Thieringer R. (1988) Comparative enzymology of β-oxidation. Biochem. Soc. Trans. 16, 418–420 [DOI] [PubMed] [Google Scholar]
- 25. Hiltunen J. K., Mursula A. M., Rottensteiner H., Wierenga R. K., Kastaniotis A. J., Gurvitz A. (2003) The biochemistry of peroxisomal β-oxidation in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 27, 35–64 [DOI] [PubMed] [Google Scholar]
- 26. Binns D., Januszewski T., Chen Y., Hill J., Markin V. S., Zhao Y., Gilpin C., Chapman K. D., Anderson R. G., Goodman J. M. (2006) An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173, 719–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 28. Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360 [DOI] [PubMed] [Google Scholar]
- 29. Zinser E., Daum G. (1995) Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast 11, 493–536 [DOI] [PubMed] [Google Scholar]
- 30. Zinser E., Sperka-Gottlieb C. D., Fasch E. V., Kohlwein S. D., Paltauf F., Daum G. (1991) Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173, 2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Connerth M., Grillitsch K., Köfeler H., Daum G. (2009) Analysis of lipid particles from yeast. Methods Mol. Biol. 579, 359–374 [DOI] [PubMed] [Google Scholar]
- 32. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) Protein measurement with the folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 33. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 34. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 35. Haid A., Suissa M. (1983) Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96, 192–205 [DOI] [PubMed] [Google Scholar]
- 36. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 37. Schmidinger H., Birner-Gruenberger R., Riesenhuber G., Saf R., Susani-Etzerodt H., Hermetter A. (2005) Novel fluorescent phosphonic acid esters for discrimination of lipases and esterases. Chembiochem 6, 1776–1781 [DOI] [PubMed] [Google Scholar]
- 38. Schmidinger H., Hermetter A., Birner-Gruenberger R. (2006) Activity-based proteomics. Enzymatic activity profiling in complex proteomes. Amino Acids 30, 333–350 [DOI] [PubMed] [Google Scholar]
- 39. Schmidinger H., Susani-Etzerodt H., Birner-Gruenberger R., Hermetter A. (2006) Inhibitor and protein microarrays for activity-based recognition of lipolytic enzymes. Chembiochem 7, 527–534 [DOI] [PubMed] [Google Scholar]
- 40. Oskolkova O. V., Saf R., Zenzmaier E., Hermetter A. (2003) Fluorescent organophosphonates as inhibitors of microbial lipases. Chem. Phys. Lipids 125, 103–114 [DOI] [PubMed] [Google Scholar]
- 41. Birner-Gruenberger R., Susani-Etzerodt H., Waldhuber M., Riesenhuber G., Schmidinger H., Rechberger G., Kollroser M., Strauss J. G., Lass A., Zimmermann R., Haemmerle G., Zechner R., Hermetter A. (2005) The lipolytic proteome of mouse adipose tissue. Mol. Cell. Proteomics 4, 1710–1717 [DOI] [PubMed] [Google Scholar]
- 42. Livak K. J., Schmittgen T. D. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 43. Thoms S., Debelyy M. O., Nau K., Meyer H. E., Erdmann R. (2008) Lpx1p is a peroxisomal lipase required for normal peroxisome morphology. FEBS J. 275, 504–514 [DOI] [PubMed] [Google Scholar]
- 44. Thoms S., Debelyy M. O., Connerth M., Daum G., Erdmann R. (2011) The putative Saccharomyces cerevisiae hydrolase Ldh1p is localized to lipid droplets. Eukaryot. Cell 10, 770–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Debelyy M. O., Thoms S., Connerth M., Daum G., Erdmann R. (2011) Involvement of the Saccharomyces cerevisiae hydrolase Ldh1p in lipid homeostasis. Eukaryot. Cell 10, 776–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Heier C., Taschler U., Rengachari S., Oberer M., Wolinski H., Natter K., Kohlwein S. D., Leber R., Zimmermann R. (2010) Identification of Yju3p as functional orthologue of mammalian monoglyceride lipase in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1801, 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Athenstaedt K., Daum G. (2000) 1-Acyldihydroxyacetone-phosphate reductase (Ayr1p) of the yeast Saccharomyces cerevisiae encoded by the open reading frame YIL124w is a major component of lipid particles. J. Biol. Chem. 275, 235–240 [DOI] [PubMed] [Google Scholar]
- 48. Vico P., Cauet G., Rose K., Lathe R., Degryse E. (2002) Dehydroepiandrosterone (DHEA) metabolism in Saccharomyces cerevisiae expressing mammalian steroid hydroxylase CYP7B. Ayr1p and Fox2p display 17β-hydroxysteroid dehydrogenase activity. Yeast 19, 873–886 [DOI] [PubMed] [Google Scholar]
- 49. Saerens S. M., Verstrepen K. J., Van Laere S. D., Voet A. R., Van Dijck P., Delvaux F. R., Thevelein J. M. (2006) The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J. Biol. Chem. 281, 4446–4456 [DOI] [PubMed] [Google Scholar]
- 50. Zahedi R. P., Sickmann A., Boehm A. M., Winkler C., Zufall N., Schönfisch B., Guiard B., Pfanner N., Meisinger C. (2006) Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 17, 1436–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Beeler T., Bacikova D., Gable K., Hopkins L., Johnson C., Slife H., Dunn T. (1998) The Saccharomyces cerevisiae TSC10/YBR265w gene encoding 3-ketosphinganine reductase is identified in a screen for temperature-sensitive suppressors of the Ca2+-sensitive csg2Delta mutant. J. Biol. Chem. 273, 30688–30694 [DOI] [PubMed] [Google Scholar]
- 52. Schmidt C., Athenstaedt K., Koch B., Ploier B., Daum G. (2013) Regulation of the yeast triacylglycerol lipase tgl3p by formation of nonpolar lipids. J. Biol. Chem. 288, 19939–19948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Connerth M., Czabany T., Wagner A., Zellnig G., Leitner E., Steyrer E., Daum G. (2010) Oleate inhibits steryl ester synthesis and causes liposensitivity in yeast. J. Biol. Chem. 285, 26832–26841 [DOI] [PMC free article] [PubMed] [Google Scholar]