Background: CD154 is released from T-cells in a manner that remains elusive.
Results: CD154 is released from Jurkat E6.1 T-cells by ADAM10 and ADAM17 upon its interaction with CD40 exclusively.
Conclusion: The cleavage of CD154 is mediated through specific signaling events that bypass many other CD154 signaling events.
Significance: These data may lead to the generation of novel targets for the treatment of CD154-associated disorders.
Keywords: Cell Signaling, Inflammation, Integrins, Molecular Biology, Receptors, CD154, CD40, T-cells
Abstract
CD154 (CD40 ligand) is a type II transmembrane protein that belongs to the tumor necrosis factor superfamily. The soluble form of CD154 (sCD154), which results from the shedding of membrane-bound CD154, plays a key role in the production of proinflammatory cytokines and has been linked to various autoimmune and vascular disorders. Therefore, elucidating the mechanisms by which CD154 is released from the cell surface following its interaction with its various receptors is of primordial importance. Using co-culture experiments, we show that CD154 is shed predominantly upon its engagement with CD40. Indeed, only CD40 (both membrane-bound and soluble) and not α5β1 or αMβ2 is involved in the cleavage and release of CD154 from Jurkat E6.1 T-cells. Interestingly, CD154 is cleaved independently of the formation of cell surface CD40 homodimers and independently of its association into lipid rafts. In contrast, we found that the protein kinase C (PKC) signaling family and the matrix metalloproteinases ADAM10 and ADAM17 are intimately involved in this process. In conclusion, our data indicate that CD154 is released from T-cells by ADAM10 and ADAM17 upon CD40 ligation. These findings add significant insights into the mechanisms by which CD154 is down-regulated and may lead to the generation of novel therapeutic targets for the treatment of CD154-associated disorders.
Introduction
CD154, also known as CD40 ligand (CD40L), is a type II transmembrane protein belonging to the TNF superfamily. CD154 was shown to play a crucial role in humoral and cellular immunity as well as in numerous inflammatory events. This immunoinflammatory molecule is expressed on a wide variety of cells of hematopoetic and nonhematopoetic origin. Indeed, CD154 expression is exhibited on activated T-cells, activated platelets, monocytes, dendritic cells, mast cells, fibroblasts, endothelial cells, and others. In addition to the membrane-bound molecule, a soluble form of CD154 (sCD154)3 was also reported, and increased circulating levels of sCD154 have been associated with many pathological conditions including autoimmune diseases and inflammatory conditions. Interestingly, sCD154 was shown to be biologically active, given its ability to interact with its classical CD40 receptor (1). In addition to CD40, CD154 can also bind to active and inactive αIIbβ3, inactive α5β1, and active αMβ2 (2–4).
Many membrane-bound members of the TNF family as well as their receptors are cleaved from the cell surface and released in the circulation and biological fluids as soluble molecules (5). It has been shown that CD154 is cleaved from the cell surface of activated platelets in a metalloproteinase-dependent manner. Soluble CD154 was reported to be biologically active and to induce the activation of B-lymphocytes and endothelial cells (6). Interestingly, the shedding of CD154 from platelets is triggered following its interaction with CD40, which is also expressed on platelets (7). Recent evidence supports the role of matrix metalloproteinase-2 (MMP-2) (8), in association with the αIIbβ3 integrin (9), in mediating the proteolytic cleavage of CD154 from activated platelets. Moreover, MMP-9 was also reported to be involved in the shedding of CD154 from platelets with Crohn disease. Indeed, these platelets exhibit high contents of CD154, and MMP-9 inhibition significantly down-regulates the cleavage of CD154 (10).
Soluble CD154 was first identified in supernatants from activated T-cells (11). The authors demonstrated that the soluble form of CD154 is released following a proteolytic cleavage at methionine 113 of the full-length molecule. The 18-kDa fragment consists only of part of the extracellular domain of membrane-bound CD154, albeit it remains capable of CD40 binding. Subsequent studies have also described sCD154 and outlined its biological activity (1, 12).
Even though sCD154 has been fairly well characterized, the underlying mechanisms mediating its production remain elusive. Recently, Matthies et al. demonstrated that the release of sCD154 by T-cells is enhanced in the presence of PKC agonists. In addition, the authors highlight the importance of ADAM10 as the proteinase controlling the production of sCD154 (13). In the present study, we investigated the underlying mechanisms involved in the production of sCD154 upon its ligation with its different receptors. Our results demonstrate that CD154 is shed predominantly from Jurkat E6.1 T-cells upon its engagement with CD40, as ligation with α5β1 or αMβ2 showed no effect on this process. Interestingly, the formation of cell surface CD40 homodimers does not appear essential for CD154 cleavage. Moreover, we show herein that CD154 cleavage is independent of its association into lipid rafts, but requires the PKC signaling family and the metalloproteinases ADAM10 and ADAM17.
MATERIALS AND METHODS
Antibodies and Reagents
The monoclonal antibody C4.14 raised against human CD154 was produced in our laboratory as described previously (clone C4.14 does not interfere with the binding of CD154 with its receptors) (14). The anti-α5β1 antibody (clone JBS5) came from Santa Cruz Biotechnology, whereas the anti-αMβ2 antibody (clone ICRF44) was procured from BD Biosciences. The anti-CD40 monoclonal antibody (clone G28.5) was purified from hybridoma cell lines as outlined previously (15). Human soluble CD40-Fc was generated in our laboratory as described previously (16). Mouse and human IgGs (isotype controls) were purchased from Santa Cruz Biotechnology. Polyclonal antibodies against p38 and ERK1/2 (phosphorylated and total forms) were from Cell Signaling. Antibodies directed against ADAM17 and ADAM10 came from Calbiochem. The p38 (SB203580), ERK1/2 (U0126), PKC (chelerythrine), and MMP (TAPI-1) inhibitors were all from Calbiochem.
Cell Lines and Culture Conditions
The human Jurkat E6.1 T-cell line, as well as HEK293, BJAB, and U937 cells were obtained from ATCC. Jurkat E6.1 cells were stably transfected with human wild-type CD154 (CD154WT), CD154 lacking its cytoplasmic domain (CD154-ΔCyto), or CD154 chimeric molecules containing the transmembrane domain of transferrin receptor 1 (CD154-RTF) as described previously (17). Cells were cultured at 37 °C under a humidified 5% CO2 atmosphere in RPMI 1640 medium containing 10% fetal bovine serum (FBS), l-glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin (Wisent, Montreal, QC, Canada), and 100 μg/ml Zeocin (InvivoGen). HEK293 cells were stably transfected with human CD40 (HEK-CD40), human αMβ2 (HEK-αMβ2), human CD40 mutated at position 238 to an alanine (HEK293-CD40C238A), or control vector (HEK-Vector), as outlined previously (15). All HEK293 cells were maintained in DMEM supplemented with 5% FBS and 400 μg/ml hygromycin B (Wisent).
Mutagenesis and Oligonucleotide Synthesis
The antisense oligonucleotides (ASO) directed against both ADAM17 (5′-CCTAGTCAGTGCTGTTATCA-3′) and ADAM10 (5′-GGTCTGAGGATATGATCTCT-3′) containing five 2′-O-methoxyethyl modifications at the 5′-end and 3′-end were obtained from Operon (Huntsville, AL). A scramble antisense oligonucleotide (GGATGTTCGTCCTCCTCACA) was used as negative control. To down-express ADAM17 and ADAM10, Jurkat E6.1-CD154-transfected cells were incubated with 15 μm ASO or scramble oligonucleotide in Opti-MEM without serum for 6 h.
CD154 Shedding
Jurkat E6.1-CD154-transfected T-cells (5 × 106/ml) were either co-cultured with HEK293 cells (5 × 106/ml) (HEK-Vector, HEK-αMβ2, or HEK-CD40) or stimulated with sCD40-Fc (1.25 μg/ml) for different time points at 37 °C. CD154 shedding was then measured by residual cell surface expression by FACS analysis or by CD154 expression within cell lysates (full-length CD154) and supernatants (sCD154) by Western blot assays.
CD154 cleavage was also assessed on T-cells from human peripheral blood mononuclear cells (PBMC) samples. Briefly, blood was withdrawn in acid/citrate/dextrose-anticoagulated tubes from donors who gave their written consent according to the guidelines established by the CR-CHUM ethical committee. PBMCs were obtained from Ficoll-Paque (GE Healthcare) according to the manufacturer's instructions. Washed PBMCs (2 × 106) were then stimulated with staphylococcal enterotoxin B (SEB) (1 μg/ml) in 96-well round bottom plates for 4, 6, and 24 h at 37 °C under a humidified 5% CO2 atmosphere. CD154 shedding was then measured by residual cell surface expression by FACS analysis.
FACS Analysis
Jurkat E6.1-CD154-transfected T-cells (5 × 106/ml) were stained with the anti-CD154 mAb C4.14 for 1 h at 4 °C, washed, and incubated with a secondary anti-mouse Alexa Fluor 488-coupled antibody for 30 min at 4 °C. Cells were then washed and analyzed (10,000 events) on a FACS-Calibur cytometer (BD Biosciences) by their characteristic forward and side scatter properties.
Western Blot Analysis
Whole cell lysates and supernatants were resolved on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, blocked in 5% nonfat dry milk in TBST (20 mm Tris, 150 mm NaCl (pH 7.5), 0.1% Tween 20) for 1 h at room temperature, and immunoblotted overnight at 4 °C with the appropriate primary antibody. The membranes were then washed three times in TBST, incubated with secondary antibody-HRP for 1 h at room temperature, and developed by enhanced chemiluminescence (PerkinElmer Life Sciences).
Statistical Analysis
Data are presented as means ± S.E. Data were analyzed using the Prism Software (GraphPad Software) by a Student's t test, with *p < 0.05 considered significant.
RESULTS
Membrane-bound CD154 Is Cleaved upon Its Interaction with CD40
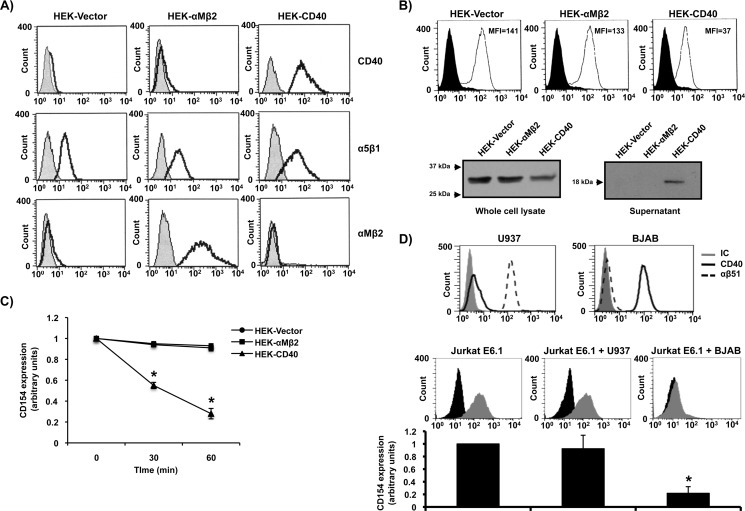
Although the importance of CD40 in CD154 shedding from activated platelets has already been established (7), its contribution to CD154 cleavage from T-cells remains controversial (12, 18). Moreover, the involvement of the other CD154 receptors (α5β1, αMβ2, and αIIbβ3) in this process has yet to be investigated. To address this issue, Jurkat E6.1 T-cells were stably transfected with CD154 and co-cultured with HEK293 cells transfected with control vector alone (HEK-Vector), HEK293 cells transfected with CD40 (HEK-CD40) or HEK293 cells transfected with αMβ2 (HEK-αMβ2) for different time points. As shown in Fig. 1, B and C, only HEK293-CD40-transfected cells induced a time-dependent CD154 cleavage from Jurkat E6.1 cells, whereas HEK293-Vector cells, which constitutively express α5β1 (Fig. 1A), or HEK293-αMβ2-transfected cells showed no effect. To confirm that CD154 is cleaved and not internalized upon CD40 engagement, total cell lysates and supernatants were analyzed for CD154 expression. Results show that the total amount of cellular CD154 decreases with time as T-cells are co-cultured with CD40-expressing HEK293 cells (Fig. 1B). This decrease was associated with an up-regulation of sCD154 within the respective supernatants, confirming sCD154 release upon CD40 engagement. In contrast, co-culturing of Jurkat E6.1 cells with HEK293-Vector or HEK293-αMβ2 failed to induce sCD154 release. Given that cell surface levels of α5β1 (HEK293-Vector) are lower than αMβ2 (HEK293-αMβ2) and CD40 (HEK293-CD40), we performed additional co-culturing experiments to rule out the implication of α5β1 in CD154 cleavage. Fig. 1D shows that co-culturing of Jurkat E6.1 cells with U937 cells, which constitutively express high levels of the α5β1 integrin receptor, failed to induce CD154 shedding, compared with CD40-positive BJAB cells, thereby confirming data from our HEK293 co-culture system. Taken together, these results indicate that the interaction of CD154 with membrane-bound CD40 plays a predominant role in CD154 cleavage and release, whereas its interaction with α5β1 or αMβ2 does not appear to be involved in this process.
FIGURE 1.
Membrane-bound CD154 is cleaved upon its interaction with CD40. A, expression profiles of CD40, α5β1, and αMβ2 on HEK293-Vector, HEK293-αMβ2, and HEK293-CD40 cells are shown. Cells (5 × 106/ml) were stained with the appropriate antibody (CD40, clone G28.5; α5β1, clone JBS5; and αMβ2, clone ICRF44) or its isotype control (IC) for 1 h at 4 °C, washed, and incubated with an anti-mouse IgG Alexa Fluor 488-coupled secondary antibody for 30 min at 4 °C. Sample were then washed and analyzed by FACS analysis. Histogram plots are representative of four independent experiments. B, Jurkat E6.1-CD154 cells (10 × 106/ml) were co-cultured with HEK-Vector (10 × 106/ml), HEK-αMβ2 (10 × 106/ml), or HEK-CD40 (10 × 106ml) cells for 1 h at 37 °C. Total cell lysates and supernatants were then analyzed by Western blotting for CD154 expression (clone C4.14). Full-length membrane-bound CD154 (≈33 kDa) is shown in whole cell lysates, whereas sCD154 (≈18 kDa) is found within the supernatants from co-cultures with HEK-CD40 cells only. Upper histogram plots show representative CD154 shedding at 60 min of co-culturing experiments as described in C. C, Jurkat E6.1 cells stably transfected with CD154 (5 × 106/ml) were co-cultured with HEK293 cells (5 × 106/ml) stably transfected with vector alone (HEK-Vector), with αMβ2 (HEK-αMβ2), or with CD40 (HEK-CD40) for the indicated time at 37 °C. Cells were then stained with an anti-CD154 antibody (clone C4.14) or its isotype control for 1 h at 4 °C, washed, and incubated with an anti-mouse IgG Alexa Fluor 488-coupled secondary antibody. CD154 shedding was then measured by FACS analysis as arbitrary units of residual membrane expression over time. D, upper representative histogram plots show the expression of α5β1 (clone JBS5) and CD40 (clone G28.5) on both U937 and BJAB cells. Cells (5 × 106/ml) were stained as described in A and analyzed by FACS analysis. Lower plots and histogram bars of mean of data (n = 3; *p < 0.05 versus Jurkat E6.1 alone) show the impact of U937 and BJAB cells on CD154 cleavage from Jurkat E6.1-CD154 cells. Jurkat E6.1 cells stably transfected with CD154 (5 × 106/ml) were co-cultured with U937 (5 × 106/ml) or BJAB (5 × 106/ml) cells or left alone for 1 h at 37 °C. Cells were then double stained with an anti-CD3-FITC antibody (clone UCHT1) and an anti-CD154-biotin-conjugated antibody (clone C4.14) or their respective isotype controls for 1 h at 4 °C. Samples were then washed and incubated with streptavidin-phycoerythrin for 30 min at 4 °C for CD154 fluorescence. Cells were washed, and double fluorescence was detected by FACS analysis using the appropriate compensation parameters. Histogram plots are representative of CD154 fluorescence within the CD3-positive population (Jurkat E6.1 T-cells).
CD154 Is Cleaved from Activated Human T-cells in a CD40-dependent Manner
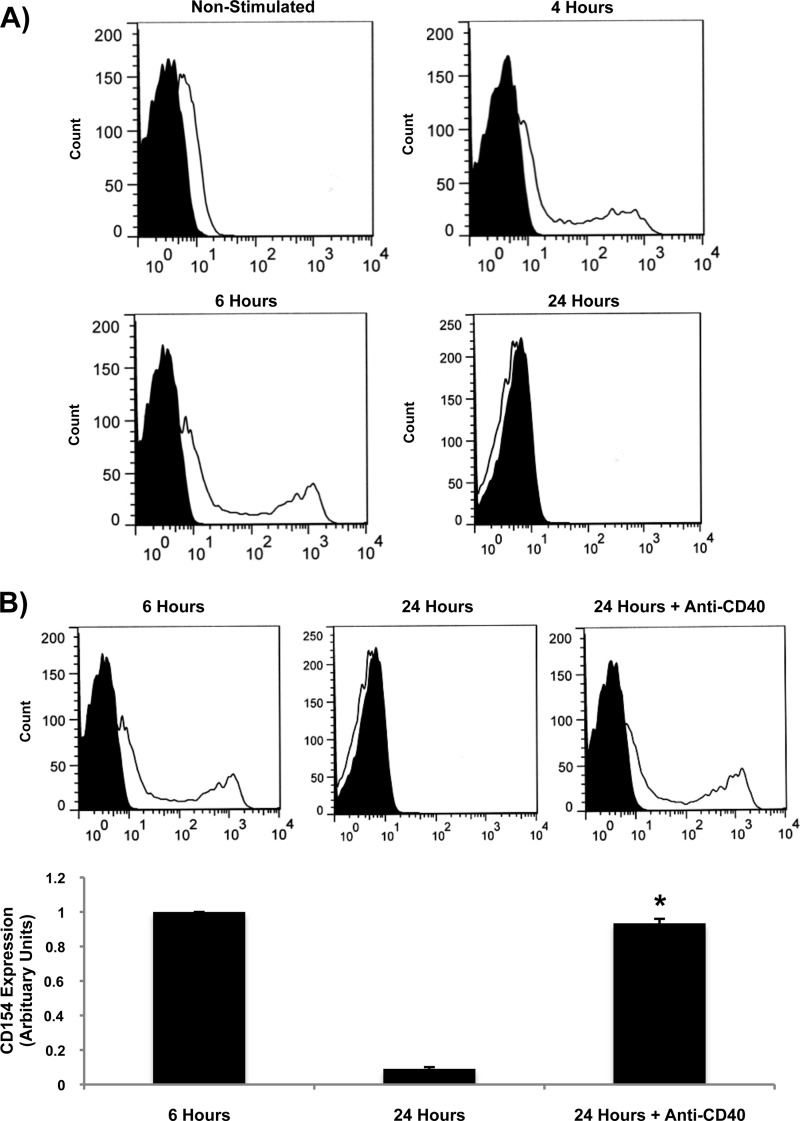
To confirm the importance of CD40 in CD154 release from T-cells, PBMC samples were isolated from human peripheral blood and stimulated with SEB, a bacterial superantigen that activates T-cells in a Vβ-restricted fashion. As illustrated in Fig. 2A, SEB stimulation led to maximal CD154 up-regulation at 4–6 h, after which levels returned to base line at 24 h poststimulation as a result of cleavage of the membrane-bound form. More importantly, pretreatment of PBMCs with a specific blocking anti-CD40 mAb (clone 82102) completely prevented SEB-induced CD154 cleavage at 24 h after stimulation (Fig. 2B). These results thus confirm the requirement of CD40 in this process in a physiological setting involving primary human T-cells.
FIGURE 2.
CD154 is cleaved from activated human T-cells in a CD40-dependent manner. A, freshly isolated human PBMCs were stimulated with SEB (1 μg/ml) for 4, 6, and 24 h. Cells were then stained with a phycoerythrin (PE)-coupled anti-CD154 antibody or its isotype control (IgG-PE) for 1 h at 4 °C. Samples were then washed and analyzed by FACS analysis for total CD154 fluorescence within the PBMC population. Histogram plots are representative of three independent experiments. B, PBMCs were left untreated or preincubated with a blocking anti-CD40 mAb (clone 82102; 5 μg/ml) for 1 h at 37 °C. Cells were then stimulated with SEB (1 μg/ml) for different time points, and CD154 fluorescence was determined as described in A. Lower histogram represents the mean of data of CD154 membrane expression (n = 3; *, p < 0.05 versus 24 h; error bars, S.E.).
Homodimerization of CD40 Is Not Required for CD154 Shedding
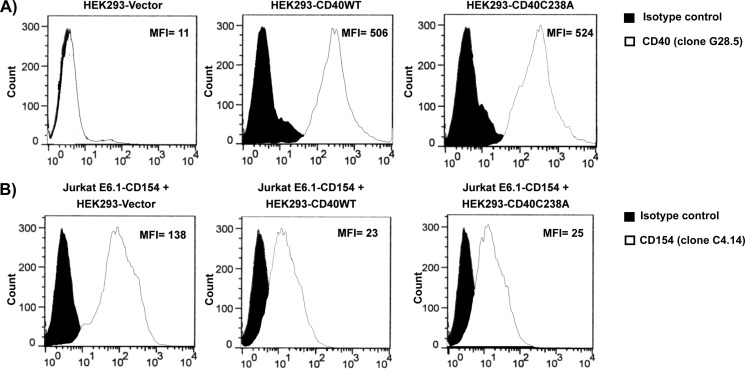
The intracellular domain of CD40 contains cysteine-rich domains, allowing disulfide bridging and dimerization of two CD40 molecules. Indeed, crystallographic analyses indicate that CD40 complexes with CD154 in a 2:3 ratio, suggesting that CD40 may form dimers in solution (19). Moreover, we have previously reported that CD40 forms homodimers through cysteine 238 of the cytoplasmic domain of CD40 and that this process is involved in IL-2 secretion by T-cells (15). Based on these data, we sought to investigate the importance of CD40 homodimers in CD154 cleavage from Jurkat E6.1 T-cells. For this purpose, HEK293 cells were stably transfected with CD40 molecules to which the cysteine 238 residue was replaced by an alanine (HEK293-CD40C238A), resulting in an inability to produce CD40 dimers on the cell surface (data not shown). These cells, which express comparable levels of cell surface CD40 (Fig. 3A) and similarly bind CD154 (data not shown) compared with their CD40WT counterparts, showed no effect on CD154 shedding from Jurkat E6.1 T-cells. Indeed, both HEK293-CD40WT and HEK293-CD40C238A induced similar CD154 cleavage upon co-culture with Jurkat E6.1-CD154-transfected cells (Fig. 3B). These data indicate that although CD40 homodimers are required for specific elements of CD154-induced signaling, CD154 is shed from the cell surface independently of this process.
FIGURE 3.
Homodimerization of CD40 is not required for CD154 shedding. A, expression profile of CD40 on HEK293-Vector, HEK293-CD40WT, and HEK293-CD40C238A cells are shown. Cells (5 × 106/ml) were stained an anti-CD40 antibody (clone G28.5) or its isotype control for 1 h at 4 °C, washed, and incubated with an anti-mouse IgG Alexa Fluor 488-coupled secondary antibody for 30 min at 4 °C. Sample were then washed and analyzed by FACS analysis. Histogram plots are representative of four independent experiments (MFI, mean fluorescence intensity). B, Jurkat E6.1 cells stably transfected with CD154 (5 × 106/ml) were co-cultured with HEK293 cells (0.5 × 106/ml) stably transfected with vector alone (HEK293-Vector), with CD40 wild type (HEK293-CD40WT), or with CD40 C238A mutants (HEK293-CD40C238A) for 1 h at 37 °C. Cells were then stained with an anti-CD154 antibody (clone C4.14) or its isotype control for 1 h at 4 °C, washed, and incubated with an anti-mouse IgG Alexa Fluor 488-coupled secondary antibody. CD154 cell surface expression was then measured by FACS analysis. Histogram plots are representative of four independent experiments.
Soluble CD40 Induces CD154 Shedding
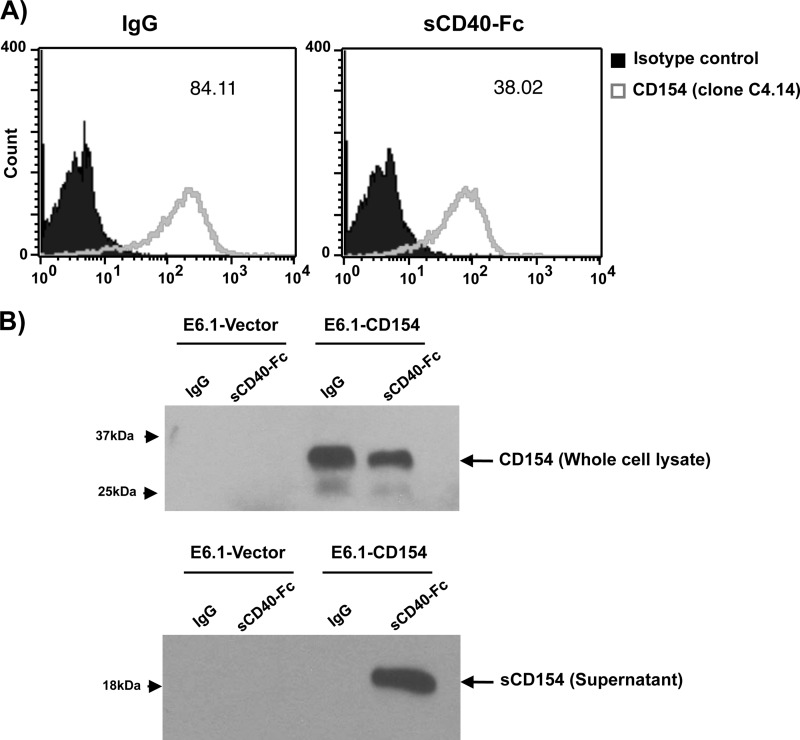
Next, we sought to investigate whether sCD40 could also induce CD154 cleavage from T-cells, as shown above with membrane-bound CD40. Fig. 4A indicates that stimulation of CD154-transfected Jurkat E6.1 T-cells with sCD40-Fc significantly decreases CD154 expression on the surface of T-cells, compared with negative control IgG. This response was also associated with a decreased CD154 expression in total cell lysates and an up-regulation of sCD154 in the supernatant from these cells (Fig. 4B). Moreover, neither soluble α5β1 nor soluble αMβ2 was able to induce CD154 shedding from the surface of these cells (data not shown), confirming results from our co-culturing assays that CD40 represents the predominant CD154 receptor involved in its cleavage and release from the cell surface. These data show that CD40 can exclusively trigger CD154 release and that signaling through CD154 alone (without the implication of CD40 signaling) is sufficient to induce this process.
FIGURE 4.
Soluble CD40 induces CD154 shedding. A, Jurkat E6.1-CD154-transfected cells (5 × 106/ml) were stimulated with soluble CD40-Fc (1.25 μg/ml) or negative control IgG (1.25 μg/ml) for 1 h at 37 °C. Cells were then washed and stained with an anti-CD154 antibody (clone C4.14) for 1 h at 4 °C. Samples were thereafter incubated with an anti-mouse IgG Alexa Fluor 488-coupled secondary antibody for 30 min at 4 °C, washed, and analyzed by FACS for membrane-bound CD154 expression. Histogram plots are representative of five independent experiments (values within plots represent the mean fluorescence intensity for each sample). B, Jurkat E6.1-Vector-transfected (10 × 106/ml) and Jurkat E6.1-CD154-transfected (10 × 106/ml) cells were stimulated with sCD40-Fc (1.25 μg/ml) or negative control IgG (1.25 μg/ml) for 1 h at 37 °C. Total cell lysates and supernatants were then analyzed by Western blotting for CD154 expression (clone C4.14). Full-length membrane-bound CD154 (≈33 kDa) is shown in whole cell lysates, whereas sCD154 (≈18 kDa) is found within the supernatants of E6.1-CD154 cells stimulated with sCD40-Fc only. Blots are representative of four independent experiments.
CD154 Shedding Is Independent of Lipid Raft Integrity
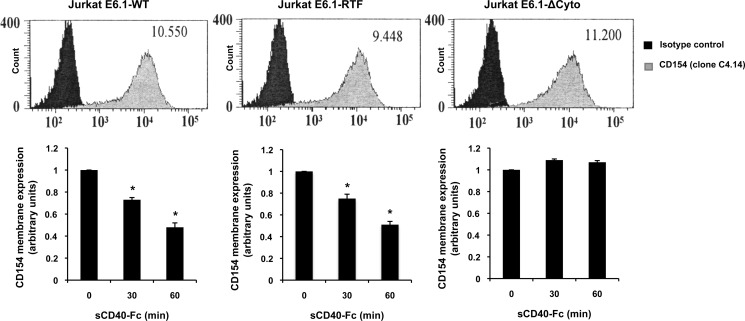
In search of the underlying cellular mechanisms involved in CD154 shedding, we first sought to evaluate the role of lipid rafts in this process. Because we have previously reported that the translocation of CD154 into lipid rafts is required for the induction of important signaling pathways in response to CD40 ligation (17, 20), these critical cellular microdomains might serve as key focal points for the assembly of specific signaling pathways involved in CD154 cleavage. Also, it has been proposed that lipid rafts regulate the shedding activity of specific ADAMs, which are responsible for the cleavage of several transmembrane proteins (20–22). To study the influence of lipid rafts in CD154 shedding from T-cells, Jurkat E6.1 cells were transfected with chimeric CD154 molecules (E6.1-Chim) containing the transmembrane domain of transferrin receptor 1, a type II protein excluded from lipid rafts. Fig. 5 shows that stimulation of both CD154WT-transfected and CD154-Chim-transfected Jurkat E6.1 cells with sCD40-Fc induced comparable levels of CD154 cleavage. In contrast, Jurkat E6.1 cells which have been transfected with CD154 molecules lacking the cytoplasmic domain (E6.1-ΔCyto) failed to trigger CD154 shedding upon sCD40-Fc ligation, thus demonstrating the importance of CD154 signaling in this process. Of note, all three Jurkat E6.1 transfectants (WT, Chim, and ΔCyto) expressed similar levels of CD154 (Fig. 5), thereby excluding the possibility that these differential responses were a result of variable CD154 membrane expressions among the three constructions. These results were also confirmed with the cholesterol-chelating agent, methyl β-cyclodextrin (data not shown). Taken together, these data indicate that lipid rafts are not required for the CD154 signaling pathways involved in its cleavage and release by T-cells.
FIGURE 5.
CD154 shedding is independent of lipid raft integrity. Jurkat E6.1-CD154 wild-type transfected (Jurkat E6.1-WT), Jurkat E6.1-CD154 transferrin receptor I mutant (Jurkat E6.1-RTF), and Jurkat E6.1-CD154 cytoplasmic domain deletion mutant (Jurkat E6.1-ΔCyto) cells were assessed for CD154 cell surface expression by FACS analysis (upper histogram panels). Cells (5 × 106/ml) were stained with an anti-CD154 antibody (clone C4.14) or its isotype control for 1 h at 4 °C, washed, and incubated with a secondary anti-mouse IgG Alexa Fluor 488-coupled antibody for an additional 30 min at 4 °C. All histogram plots are representative of four independent experiments (values within plots represent the mean fluorescence intensity). Lower histogram bars represent the mean of data (n = 4; *p < 0.05; error bars, S.E.) of CD154 cell surface cleavage for all three populations. Cells (5 × 106/ml) were stimulated with sCD40-Fc (1.25 μg/ml) for the indicated time, washed, and stained with an anti-CD154 antibody (clone C4.14) or its isotype control for 1 h at 4 °C. Samples were then washed and incubated with a secondary anti-mouse IgG Alexa Fluor 488-coupled antibody for an additional 30 min at 4 °C. Cleavage was assessed by FACS analysis, and results are expressed as arbitrary units of CD154 membrane expression in the absence (time 0) or presence of sCD40-Fc (30- and 60-min stimulation).
CD154 Shedding Is Modulated by PKC Activation
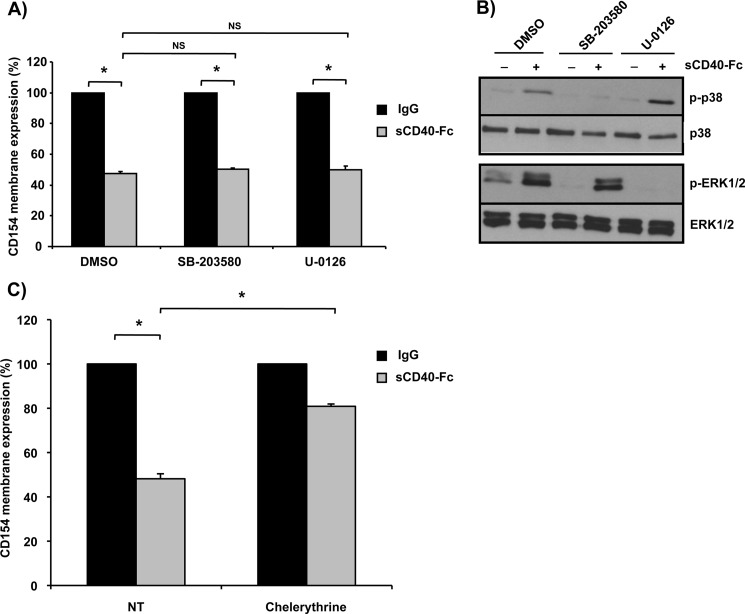
Several studies have reported that the ectodomain shedding of many transmembrane proteins is dependent upon the activation of MAP kinases such as p38 and ERK1/2 (23–25). To determine whether these members of the MAP kinase superfamily are also involved in CD154 cleavage, CD154-transfected Jurkat E6.1 cells were stimulated with sCD40-Fc in the presence or absence of SB203580 or U0126, p38 and ERK1/2 inhibitors, respectively. As shown in Fig. 6, A and B, neither inhibitor showed an effect on CD40-Fc-mediated CD154 shedding, indicating that the cleavage of CD154 from the cell surface is independent of MAPK activation. Due to its implication in membrane-bound protease activation (26), the role of PKCs in CD154 shedding was also determined. Pretreatment of CD154-transfected E6.1 cells with the broad spectrum PKC inhibitor chelerythrine significantly prevented CD154 cleavage in response to sCD40-Fc stimulation (Fig. 6C). Taken together, these results indicate that the cleavage of CD154 from T-cells is dependent on the activation of PKCs but not MAPKs.
FIGURE 6.
CD154 shedding is modulated by PKC activation. A, Jurkat E6.1-CD154-transfected cells (5 × 106/ml) were preincubated with the p38 SB203580 inhibitor (20 μm), the ERK1/2 U0126 inhibitor (20 μm), or control dimethyl sulfoxide (DMSO) for 15 min at 37 °C. Cells were then stimulated with sCD40-Fc (1.25 μg/ml) or negative control IgG (1.25 μg/ml) for an additional hour at 37 °C. CD154 shedding was assessed by residual cell surface expression by FACS analysis as outlined previously (n = 3; *p < 0.05 and NS = nonsignificant; error bars, S.E.). B, Jurkat E6.1-CD154 transfected cells (5 × 106/ml) cells were preincubated with inhibitors as described in A and subsequently stimulated with sCD40-Fc (1.25 μg/ml) or not for 5 min at 37 °C. Samples were then lysed and phospho-p38 (p-p38), phospho-EKR1/2 (p-ERK1/2), as well as total p38 (T-p38) and total ERK1/2 (T-ERK1/2) were analyzed by Western blotting. Blots are representative of four independent experiments. C, Jurkat E6.1-CD154-transfected cells (5 × 106/ml) were stimulated with sCD40-Fc (1.25 μg/ml) or negative control IgG (1.25 μg/ml) for 1 h at 37 °C in the absence (control dimethyl sulfoxide) or presence of the PKC inhibitor, chelerythrine (1 μm). CD154 shedding was thereafter assessed by FACS analysis as outlined previously (n = 4; *, p < 0.05).
CD154 Shedding Is Mediated by ADAM10 and ADAM17
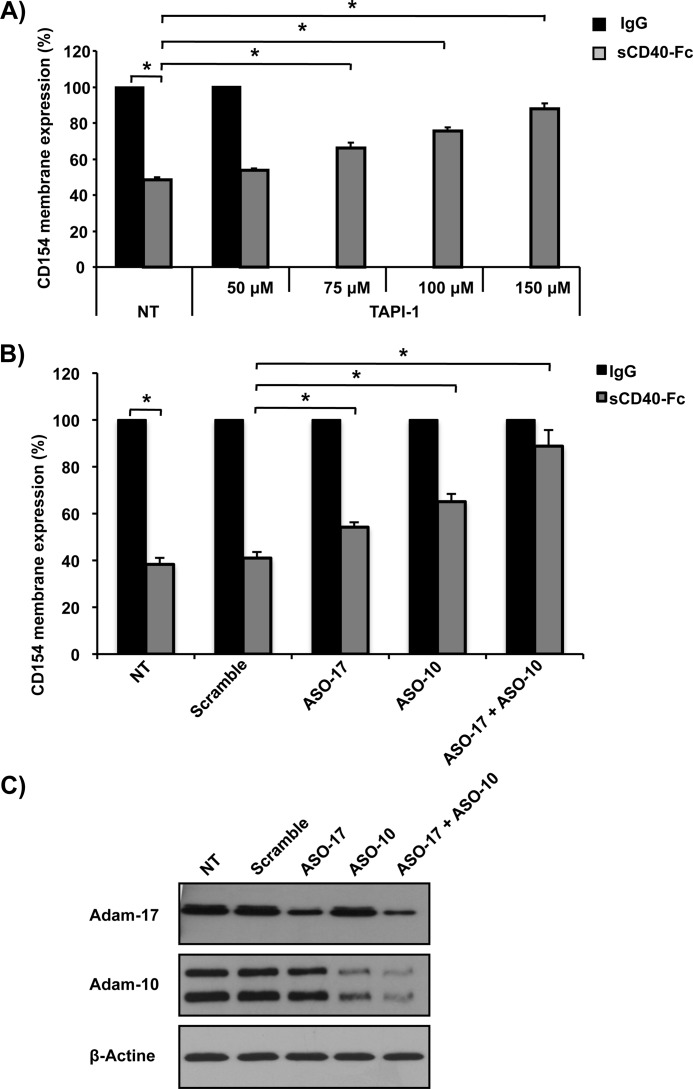
The enzyme responsible for CD154 shedding on the surface of activated platelets has been demonstrated previously (8), but little is known about this process regarding T-cells. The metalloproteinase ADAM10 has been highlighted as a possible candidate (13), albeit no direct demonstration of its implication in CD154 cleavage has been provided. Because ADAM proteins are part of the metalloproteinase family, we first evaluated the impact of a broad spectrum matrix metalloproteinase inhibitor (TAPI-2) on sCD40-Fc-mediated CD154 cleavage. Fig. 7A shows that preincubation with TAPI-2 dose-dependently reverses CD154 release from the surface of Jurkat E6.1 T-cells. To delineate the metalloproteinases responsible for CD154 shedding, knockdowns of ADAM10 and ADAM17 through antisense targeting were conducted (Fig. 7, B and 7C). Our results show that specific down-regulation of either ADAM10 or ADAM17 significantly prevents CD154 shedding from Jurkat E6.1 cells in response to sCD40-Fc stimulation, whereas a negative control scramble oligonucleotide antisense had no effect. In addition, specific targeting of both emzymes (ADAM10 and ADAM17) almost entirely reversed CD154 cleavage. These data show for the first time a direct implication of the metalloproteinases ADAM10 and ADAM17 in CD154 cleavage from T-cells.
FIGURE 7.
CD154 shedding is mediated by ADAM10 and ADAM17. A, Jurkat E6.1-CD154 (5 × 106/ml)-transfected cells were preincubated with the broad spectrum MMP inhibitor TAPI-2 at the indicated concentration for 15 min at 37 °C or left untreated (NT), stimulated with sCD40-Fc (1.25 μg/ml) or negative control IgG (1.25 μg/ml) for 1 h at 37 °C, and CD154 cell surface expression was determined by FACS analysis as outlined previously (n = 4; *, p < 0.05; error bars, S.E.). B, Jurkat E6.1 cells stably expressing CD154 (0.5 × 106/ml) were transiently transfected with 15 μm antisense oligonucleotide against ADAM17 (ASO17), ADAM10 (ASO10) or both ADAM17 and ADAM10 (ASO17 + ASO10). A negative control scramble antisense oligonucleotide was also employed (15 μm). Cells were then stimulated with sCD40-Fc (1.25 μg/ml) or control IgG (1.25 μg/ml) for 1 h at 37 °C, and CD154 shedding was assessed by FACS analysis as the relative percentage of residual CD154 membrane expression over nontreated cells (n = 4; *, p < 0.05 and NS = nonsignificant). C, effect of oligonucleotide antisense targeting on the level of expression of ADAM10 and ADAM17 in Jurkat E6.1-CD154-transfected cells. Cells were treated as in B, lysed 72 h after transfection, and analyzed for ADAM10 and ADAM17 by Western blotting. Blots are representative of three independent experiments.
DISCUSSION
As a member of the TNF family, CD154 exists in membrane-bound and soluble forms. Indeed, many studies have described sCD154 with respect to its structure, production, and biological significance. Despite evidence outlining the release of soluble CD154 from activated T-cells, the underlying mechanisms remained unclear. Here, we demonstrated that among the different CD154 receptors, only CD40 is involved in CD154 cleavage and release from the surface of T-cells. However, homodimerization of cell surface CD40 does not appear essential for this process. Moreover, signaling through CD154 alone is sufficient to induce this process as stimulation with recombinant purified sCD40-Fc led to significant sCD154 release from CD154-positive T-cells. We were also able to show that cleavage of membrane-bound CD154 does not require translocation of the molecule into lipid rafts. This process was nevertheless shown to be dependent upon PKC activation and the subsequent enzymatic activity of the metalloproteinases ADAM17 and ADAM10.
Since sCD154 was first described in 1995 by Graf et al., much attention has been focused on elucidating the mechanisms by which CD154 is produced and released. As T-cells and platelets are the main reservoirs of CD154, studies were drawn toward these two types of cells. Jin et al. demonstrated that sCD154 is released within 30 min of platelet activation in the presence of agonists such as thrombin and collagen. This release was shown to result from cleavage of cell surface CD154 (6). Interestingly, the binding of membrane-bound CD154 to its receptor CD40, which is constitutively expressed on platelets, leads to a decrease of cell surface CD154 and a concomitant increase of sCD154 within the supernatant of agonist-activated platelets. Indeed, specific inhibition of the CD40 receptor prevents release of sCD154 from activated platelets, indicating that the CD154/CD40 interaction is required for platelet CD154 shedding (7). Here, we were able to demonstrate that CD154 is shed predominantly from Jurkat E6.1 T-cells upon its engagement with CD40, as ligation with α5β1 or αMβ2 (both membrane-bound or soluble) showed no effect on this process. These data imply that although the other CD154 receptors can trigger intracellular signaling and modulate inflammatory reactions (2, 4, 27), only the CD154/CD40 interaction is responsible for CD154 shedding. This could be explained by the fact that the CD154 residues involved in CD40 binding are different from those implicated in binding to its other integrin receptors (α5β1, αMβ2 and αIIbβ3) (14). Thus, the CD154/CD40 receptor-ligand pair may regulate unique signaling elements required for the secretion of the main metalloproteinase(s) responsible for CD154 cleavage. Interestingly, stimulation of CD154-transfected Jurkat E6.1 cells with sCD40-Fc induced significant CD154 cleavage, indicating that signaling through CD154 alone is sufficient to trigger this process. However, these data do not exclude the potential importance of CD40 signaling in CD154 shedding and release from the cell surface. In fact, deletion of the cytoplasmic domain of CD154 or CD40 alone significantly reverses CD154 cleavage from the cell surface in co-culture assays, whereas deletion of the cytoplasmic domains of both entirely prevent this process (data not shown). Hence, both CD154 and CD40 signaling share important roles in CD154 shedding from T-cells.
We have previously documented that CD40 forms homodimers on the cell surface and that this process is involved in certain elements of CD154-induced signaling such as IL-2 secretion by T-cells (15). Indeed, crystallographic and mutagenesis analyses have confirmed the presence of CD40 dimers, while highlighting the importance of the cysteine 238 residue in this process (15, 19). In the present study, we have established that CD40 homodimers are not essential for CD154 cleavage from T-cells, given that HEK293-CD40C238A mutant cells, which fail to produce CD40 dimers on the cell surface (data not shown), induced similar CD154 shedding compared with HEK293-CD40WT cells. Receptor oligomerization is a phenomenon that normally enhances cell signaling and facilitates subsequent biological responses (28). This process often occurs in Triton X-100 insoluble fractions such as lipid raft microdomains. Indeed, we have determined previously that CD40 homodimers are preferentially distributed into lipid rafts, as disruption of these domains through methyl-β-cyclodextrin treatment abolishes their formation. Interestingly, we have also shown herein that lipid rafts are not required for CD154 cleavage and release from T-cells (Fig. 5), thus supporting our CD40 dimerization data. Hence, CD154 is shed from the cell surface independently of lipid raft integrity and CD40 oligomerization, both of which appear interconnected. However, these results do not undermine the importance of CD40 dimerization in CD154-induced signaling, but rather support the concept that CD154/CD40 activation leads to diverging signaling pathways involved in distinct cellular responses.
Lipid rafts are critical microdomains required for the assembly of numerous signaling molecules. We have recently demonstrated that the translocation of CD154 into lipid rafts mediates several CD40-driven biological responses such as Akt and p38 activation, as well as IL-2 production (17). Herein, our data indicate that cells expressing chimeric CD154 molecules that are unable to translocate into lipid raft compartments released similar levels of sCD154 as did CD154-transfected Jurkat E6.1 cells, thereby indicating that these cellular domains are not essential for CD154 cleavage from T-cells. These results also imply that CD40 ligation leads to diverging CD154 signaling pathways that ultimately lead to distinct biological responses. As the signaling pathways involved in CD154 cleavage bypass lipid rafts, those implicated in MAPKs activation and cytokine production such as IL-2 depend on them. Interestingly, our results also indicate that MAPKs (p38 and ERK1/2) are not required for CD154 cleavage (Fig. 6), further supporting the rationale of diverging signaling pathways through CD154 in response to CD40 ligation.
Because the ectodomain shedding of many transmembrane proteins is dependent upon the activation of MAPKs such as p38 and ERK1/2, (29–31) it was imperative that we determine the importance of these kinases in CD154 cleavage from T-cells. Although CD40 ligation induced activation of both p38 and ERK1/2, their specific inhibition showed no impact of CD154 shedding. These results indicate that signaling through CD154 is not specific or exclusive to CD154 cleavage and that this process requires an alternate pathway. In contrast, and as outlined previously, shedding requires activation of the PKC family (13). The PKC family plays a pivotal role in various cellular functions such as differentiation, proliferation, transcription regulation, and cell death (32). It has been shown previously using PKC agonists and antagonists that this pathway regulates the generation of sCD154 (13). In the present study, we were able to confirm these data through a pharmacological approach. Indeed, cells pretreated with a broad spectrum PKC inhibitor (chelerythrine) failed to produce sCD154 upon sCD40-Fc stimulation. These results imply that the PKC family controls the activation the main enzymes involved in CD154 cleavage and release from the cell surface. However, it would be pertinent to delineate further the specific PKC isoform(s) implicated in this process, as it has become fairly well established that specific PKC isoforms regulate numerous yet distinct cellular functions. For instance, cleavage of the amyloid-β precursor protein into β-amyloid, the primary component of amyloid plaques found in the brains of patients with Alzheimer disease, is modulated by PKCα and PKCϵ (26). Interestingly, this proteolysis is also mediated by the metalloproteinase ADAM17, indicating that these PKC isoforms could perhaps regulate the cleavage and release of CD154 from T-cells. However, additional studies will be needed to specifically address this issue.
The MMP family mediates the proteolytic cleavage of numerous cell surface proteins. Indeed, CD154 shedding from the surface of activated platelets is modulated by MMP-2 (8, 9). However, prior to this study, little information regarding the enzyme implicated in CD154 cleavage and release from T-cell had been provided. Matthies et al. have shown that ADAM10 could perhaps regulate this process, as recombinant ADAM10 can cleave CD154 at the surface of peripheral CD4+ T-cells (13). Unfortunately, these data did not provide clear demonstration that ADAM10 is involved in CD154 shedding form the surface of T-cells. Here, we were first able to confirm that cleavage is dependent upon the MMP family, as a broad spectrum MMP inhibitor significantly reverses sCD154 production. Using specific antisense targeting we were thereafter able to demonstrate that ADAM10 and ADAM17 are critically involved in CD154 cleavage from Jurkat E6.1 T-cells. Because inhibition of both ADAM10 and ADAM17 nearly completely inhibits CD154 shedding from Jurkat E6.1 T-cells, other ADAM members expressed within T-cells are presumably not involved in this process. These results also indicate that the enzymatic regulation of CD154 shedding in cell-dependent, because platelets and T-cells employ different proteases to release sCD154 from the cell surface. Interestingly, both platelets and T-cells also contain ADAM 10/17 and MMP-2, respectively (33, 34). The expression level of these enzymes may vary between these two cell types, thereby explaining this differential regulation. Hence, all three proteases (MMP-2, ADAM10, and ADAM17) have the ability to cleave CD154, albeit their relative contribution to this process may differ depending on the importance of the cytoplasmic reservoir found within one particular cell type. Moreover, the signaling pathways triggered upon CD154/CD40 activation may also differ between these two cell types, further explaining this phenomenon. However, CD154 signaling in platelets remains uncharacterized and additional experiments will be needed to validate these hypotheses.
The biological significance of CD154 shedding from the cell surface has yet to be fully demonstrated. Although it has been documented that cleaved sCD154, in contrast to membrane-bound CD154, can no longer induce an inflammatory response on endothelial cells (7), it has also been shown that sCD154 remains a biologically active trimer that can induce B-cell proliferation (1). Even though released sCD154 can retain biological properties and bind CD40, it may be relatively unstable in biological fluids and decidedly not as active as transmembrane CD154. This would therefore indicate that shedding helps terminate the effects of CD154 signaling through a critical negative feedback loop required to temper cellular activation. Nevertheless, further studies are required to fully address this issue, and a more in depth comparison between the biological activities of both CD154 forms would support this hypothesis.
In conclusion, this study adds significant insights into the mechanisms by which sCD154 is released by T-cells. Our results indicate mainly that CD154 is cleaved from the surface of T-cells by ADAM10 and ADAM17 upon CD40 engagement in a manner that bypasses many important CD154 signaling events such as MAPK activation, lipid raft association, and CD40 dimerization. These data may lead to the generation of novel therapeutic targets for the treatment of CD154-associated inflammatory diseases, as circulating levels of sCD154 have been linked to autoimmune and cardiovascular diseases.
Acknowledgments
We are grateful to Mr. Haydar Alturaihi for the generation of recombinant sCD40-Fc. We would also like to acknowledge Dr. Amal Nadiri for her critical insights and revision of the manuscript
This work was supported by the Canadian Institutes of Health Research (IRSC) and the Canadian Arthritis Network (CAN).
- sCD154
- soluble CD154
- ADAM
- a disintegin and metalloproteinase domain-containing protein
- ASO
- antisense oligonucleotide
- MMP
- matrix metalloproteinase
- PBMC
- peripheral blood mononuclear cell
- SEB
- staphylococcal enterotoxin B.
REFERENCES
- 1. Pietravalle F., Lecoanet-Henchoz S., Blasey H., Aubry J. P., Elson G., Edgerton M. D., Bonnefoy J. Y., Gauchat J. F. (1996) Human native soluble CD40L is a biologically active trimer, processed inside microsomes. J. Biol. Chem. 271, 5965–5967 [DOI] [PubMed] [Google Scholar]
- 2. Léveillé C., Bouillon M., Guo W., Bolduc J., Sharif-Askari E., El-Fakhry Y., Reyes-Moreno C., Lapointe R., Merhi Y., Wilkins J. A., Mourad W. (2007) CD40 ligand binds to α5β1 integrin and triggers cell signaling. J. Biol. Chem. 282, 5143–5151 [DOI] [PubMed] [Google Scholar]
- 3. Zirlik A., Maier C., Gerdes N., MacFarlane L., Soosairajah J., Bavendiek U., Ahrens I., Ernst S., Bassler N., Missiou A., Patko Z., Aikawa M., Schönbeck U., Bode C., Libby P., Peter K. (2007) CD40 ligand mediates inflammation independently of CD40 by interaction with Mac-1. Circulation 115, 1571–1580 [DOI] [PubMed] [Google Scholar]
- 4. André P., Prasad K. S., Denis C. V., He M., Papalia J. M., Hynes R. O., Phillips D. R., Wagner D. D. (2002) CD40L stabilizes arterial thrombi by a β3 integrin-dependent mechanism. Nat. Med. 8, 247–252 [DOI] [PubMed] [Google Scholar]
- 5. Jones S. A., Rose-John S. (2002) The role of soluble receptors in cytokine biology: the agonistic properties of the sIL-6R/IL-6 complex. Biochim. Biophys. Acta 1592, 251–263 [DOI] [PubMed] [Google Scholar]
- 6. Jin Y., Nonoyama S., Morio T., Imai K., Ochs H. D., Mizutani S. (2001) Characterization of soluble CD40 ligand released from human activated platelets. J. Med. Dent. Sci. 48, 23–27 [PubMed] [Google Scholar]
- 7. Henn V., Steinbach S., Büchner K., Presek P., Kroczek R. A. (2001) The inflammatory action of CD40 ligand (CD154) expressed on activated human platelets is temporally limited by coexpressed CD40. Blood 98, 1047–1054 [DOI] [PubMed] [Google Scholar]
- 8. Reinboldt S., Wenzel F., Rauch B. H., Hohlfeld T., Grandoch M., Fischer J. W., Weber A. A. (2009) Preliminary evidence for a matrix metalloproteinase-2 (MMP-2)-dependent shedding of soluble CD40 ligand (sCD40L) from activated platelets. Platelets 20, 441–444 [DOI] [PubMed] [Google Scholar]
- 9. Choi W. S., Jeon O. H., Kim D. S. (2010) CD40 ligand shedding is regulated by interaction between matrix metalloproteinase-2 and platelet integrin αIIbβ3. J. Thromb. Haemost. 8, 1364–1371 [DOI] [PubMed] [Google Scholar]
- 10. Menchén L., Marín-Jiménez I., Arias-Salgado E. G., Fontela T., Hernández-Sampelayo P., Rodríguez M. C., Butta N. V. (2009) Matrix metalloproteinase 9 is involved in Crohn's disease-associated platelet hyperactivation through the release of soluble CD40 ligand. Gut 58, 920–928 [DOI] [PubMed] [Google Scholar]
- 11. Graf D., Müller S., Korthäuer U., van Kooten C., Weise C., Kroczek R. A. (1995) A soluble form of TRAP (CD40 ligand) is rapidly released after T cell activation. Eur. J. Immunol. 25, 1749–1754 [DOI] [PubMed] [Google Scholar]
- 12. Ludewig B., Henn V., Schröder J. M., Graf D., Kroczek R. A. (1996) Induction, regulation, and function of soluble TRAP (CD40 ligand) during interaction of primary CD4+ CD45RA+ T cells with dendritic cells. Eur. J. Immunol. 26, 3137–3143 [DOI] [PubMed] [Google Scholar]
- 13. Matthies K. M., Newman J. L., Hodzic A., Wingett D. G. (2006) Differential regulation of soluble and membrane CD40L proteins in T cells. Cell Immunol. 241, 47–58 [DOI] [PubMed] [Google Scholar]
- 14. El Fakhry Y., Alturaihi H., Yacoub D., Liu L., Guo W., Leveillé C., Jung D., Khzam L. B., Merhi Y., Wilkins J. A., Li H., Mourad W. (2012) Functional interaction of CD154 protein with α5β1 integrin is totally independent from its binding to αIIbβ3 integrin and CD40 molecules. J. Biol. Chem. 287, 18055–18066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reyes-Moreno C., Sharif-Askari E., Girouard J., Léveillé C., Jundi M., Akoum A., Lapointe R., Darveau A., Mourad W. (2007) Requirement of oxidation-dependent CD40 homodimers for CD154/CD40 bidirectional signaling. J. Biol. Chem. 282, 19473–19480 [DOI] [PubMed] [Google Scholar]
- 16. El Fakhry Y., Alturaihi H., Diallo D., Merhi Y., Mourad W. (2010) Critical role of lipid rafts in CD154-mediated T cell signaling. Eur. J. Immunol. 40, 770–779 [DOI] [PubMed] [Google Scholar]
- 17. Benslimane N., Hassan G. S., Yacoub D., Mourad W. (2012) Requirement of transmembrane domain for CD154 association to lipid rafts and subsequent biological events. PLoS One 7, e43070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yellin M. J., Sinning J., Covey L. R., Sherman W., Lee J. J., Glickman-Nir E., Sippel K. C., Rogers J., Cleary A. M., Parker M. (1994) T lymphocyte T cell-B cell-activating molecule/CD40-L molecules induce normal B cells or chronic lymphocytic leukemia B cells to express CD80 (B7/BB-1) and enhance their costimulatory activity. J. Immunol. 153, 666–674 [PubMed] [Google Scholar]
- 19. An H. J., Kim Y. J., Song D. H., Park B. S., Kim H. M., Lee J. D., Paik S. G., Lee J. O., Lee H. (2011) Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation. J. Biol. Chem. 286, 11226–11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murai T., Maruyama Y., Mio K., Nishiyama H., Suga M., Sato C. (2011) Low cholesterol triggers membrane microdomain-dependent CD44 shedding and suppresses tumor cell migration. J. Biol. Chem. 286, 1999–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tellier E., Canault M., Rebsomen L., Bonardo B., Juhan-Vague I., Nalbone G., Peiretti F. (2006) The shedding activity of ADAM17 is sequestered in lipid rafts. Exp. Cell Res. 312, 3969–3980 [DOI] [PubMed] [Google Scholar]
- 22. Gil C., Cubí R., Aguilera J. (2007) Shedding of the p75NTR neurotrophin receptor is modulated by lipid rafts. FEBS Lett. 581, 1851–1858 [DOI] [PubMed] [Google Scholar]
- 23. Thorp E., Vaisar T., Subramanian M., Mautner L., Blobel C., Tabas I. (2011) Shedding of the Mer tyrosine kinase receptor is mediated by ADAM17 protein through a pathway involving reactive oxygen species, protein kinase Cδ, and p38 mitogen-activated protein kinase (MAPK). J. Biol. Chem. 286, 33335–33344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Killock D. J., Ivetić A. (2010) The cytoplasmic domains of TNFα-converting enzyme (TACE/ADAM17) and L-selectin are regulated differently by p38 MAPK and PKC to promote ectodomain shedding. Biochem. J. 428, 293–304 [DOI] [PubMed] [Google Scholar]
- 25. Xu P., Derynck R. (2010) Direct activation of TACE-mediated ectodomain shedding by p38 MAP kinase regulates EGF receptor-dependent cell proliferation. Mol. Cell 37, 551–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cisse M., Braun U., Leitges M., Fisher A., Pages G., Checler F., Vincent B. (2011) Mol. Cell. Neurosci. 47, 223–232 [DOI] [PubMed] [Google Scholar]
- 27. Wolf D., Hohmann J. D., Wiedemann A., Bledzka K., Blankenbach H., Marchini T., Gutte K., Zeschky K., Bassler N., Hoppe N., Rodriguez A. O., Herr N., Hilgendorf I., Stachon P., Willecke F., Duerschmied D., von zur Muhlen C., Soloviev D. A., Zhang L., Bode C., Plow E. F., Libby P., Peter K., Zirlik A. (2011) Binding of CD40L to Mac-1 I-domain involves the EQLKKSKTL motif and mediates leukocyte recruitment and atherosclerosis but does not affect immunity and thrombosis in mice. Circ. Res. 109, 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lingwood D., Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 29. Wang Y., Robertson J. D., Walcheck B. (2011) Different signaling pathways stimulate a disintegrin and metalloprotease-17 (ADAM17) in neutrophils during apoptosis and activation. J. Biol. Chem. 286, 38980–38988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luo X., Prior M., He W., Hu X., Tang X., Shen W., Yadav S., Kiryu-Seo S., Miller R., Trapp B. D., Yan R. (2011) Cleavage of neuregulin-1 by BACE1 or ADAM10 protein produces differential effects on myelination. J. Biol. Chem. 286, 23967–23974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kohutek Z. A., diPierro C. G., Redpath G. T., Hussaini I. M. (2009) ADAM10-mediated N-cadherin cleavage is protein kinase C-α-dependent and promotes glioblastoma cell migration. J. Neurosci. 29, 4605–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rosse C., Linch M., Kermorgant S., Cameron A. J., Boeckeler K., Parker P. J. (2010) PKC and the control of localized signal dynamics. Nat. Rev. Mol. Cell Biol. 11, 103–112 [DOI] [PubMed] [Google Scholar]
- 33. Nagase H., Woessner J. F., Jr. (1999) Matrix metalloproteinases. J. Biol. Chem. 274, 21491–21494 [DOI] [PubMed] [Google Scholar]
- 34. Santos-Martínez M. J., Medina C., Jurasz P., Radomski M. W. (2008) Role of metalloproteinases in platelet function. Thromb. Res. 121, 535–542 [DOI] [PubMed] [Google Scholar]