Background: Yeast cells grown in choline exhibit a reduction in the level of the choline transporter Hnm1.
Results: Hnm1 transporter activity triggers its endocytosis by a mechanism dependent on the ubiquitin ligase Rsp5 and the casein kinases Yck1/2.
Conclusion: Choline transport activity affects the stability of the Hnm1 choline transporter.
Significance: Regulation of choline transporter activity constitutes an important regulatory node for phosphatidylcholine homeostasis.
Keywords: Glycerophospholipid, Membrane Transport, Phosphatidylcholine, Phospholipid Metabolism, Yeast Metabolism
Abstract
Choline is a precursor for the synthesis of phosphatidylcholine through the CDP-choline pathway. Saccharomyces cerevisiae expresses a single high affinity choline transporter at the plasma membrane, encoded by the HNM1 gene. We show that exposing cells to increasing levels of choline results in two different regulatory mechanisms impacting Hnm1 activity. Initial exposure to choline results in a rapid decrease in Hnm1-mediated transport at the level of transporter activity, whereas chronic exposure results in Hnm1 degradation through an endocytic mechanism that depends on the ubiquitin ligase Rsp5 and the casein kinase 1 redundant pair Yck1/Yck2. We present details of how the choline transporter is a major regulator of phosphatidylcholine synthesis.
Introduction
Phosphatidylcholine (PC)2 is the most abundant cellular phospholipid in eukaryotic cells. PC fulfills an essential role in maintaining membrane permeability barriers while at the same time serving as a substrate for the generation of numerous second messenger signaling molecules (1, 2). Cellular PC homeostasis is controlled by balancing rates of PC synthesis with degradation. Regulatory pathways impinge upon these two processes to coordinate PC level with other cellular processes, such as cell proliferation and vesicular trafficking (3–7). PC synthesis in eukaryotic cells occurs through the CDP-choline and phosphatidylethanolamine (PE) methylation pathways, whereas PC turnover is via multiple lipases.
For PC synthesis through the CDP-choline pathway, choline is activated sequentially to phosphocholine and then CDP-choline, which is condensed with diacylglyerol (DAG), to form PC (Fig. 1). In the yeast Saccharomyces cerevisiae, net synthesis of PC through this pathway depends on exogenous choline uptake, a process mediated by a single high affinity choline transporter localized at the plasma membrane encoded by the HNM1 gene (8–11). The PE methylation pathway catalyzes the addition of three methyl groups to PE to synthesize PC and contributes greatly to total PC synthesis in S. cerevisiae when exogenous choline is scarce (12).
FIGURE 1.
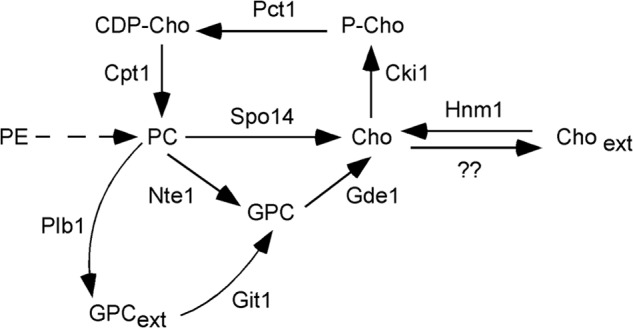
Metabolism of choline in the yeast S. cerevisiae. A scheme illustrates the transformations involving choline-containing metabolites. Solid arrows indicate direct enzyme conversions, and a dashed arrow indicates a conversion that requires more than one enzymatic step.
Degradation of PC in yeast takes place through the activity of three phospholipases. The phospholipase B Plb1 is localized at the plasma membrane and within the periplasmic space. Plb1 deacylates PC with the glycerophosphocholine produced released extracellularly (13). Nte1, a second phospholipase B, is an integral membrane protein localized at the endoplasmic reticulum and is responsible for intracellular glycerophosphocholine formation. Accumulation of intracellular glycerophosphocholine due to increased Nte1 activity correlates with increased PC synthesis (14). The glycerophosphocholine produced by Nte1 can be further catabolized to choline by the action of the glycerophosphodiesterase Gde1 (15, 16). Intracellular choline can also be generated by the PC phospholipase D Spo14. Spo14 is a soluble enzyme that, upon activation, becomes membrane-bound, localizing at the endosomal compartment and plasma membrane. Spo14 is essential for spore formation and for Sec14-independent vesicular trafficking (17). Choline generated intracellularly by Nte1/Gde1 or by Spo14 is either recycled back into PC through the CDP-choline pathway or excreted out of the cell (9, 17, 18).
The availability of the lipid precursors choline and inositol has a profound effect on yeast phospholipid synthesis. Inositol abundance sustains a high rate of phosphatidylinositol synthesis and allosterically inhibits phosphatidylserine synthase, diverting the common precursor CDP-DAG toward the synthesis of phosphatidylinositol. Simultaneously, a sustained high rate of phosphatidylinositol synthesis drags upon the phosphatidic acid pool via CDP-DAG consumption, leading to transcriptional attenuation of genes encoding enzymes for phospholipid biosynthesis (3, 12, 19). The presence of choline in the growth media enhances the repressive effect of inositol on gene expression, presumably as it affects the same pool of phosphatidic acid via DAG consumption for PC synthesis (3, 12, 20). However, choline on its own lacks such an effect on the transcriptional regulation of phospholipid synthesis genes (20), suggesting the existence of regulatory mechanisms that control PC metabolism to prevent a skewed phospholipid composition. Indeed, increased turnover of PC through Nte1 takes place when yeast cells grow in inositol-free medium, and this turnover is necessary to prevent transcriptional attenuation triggered by exogenous choline (8).
From these observations a basic question arises. How do yeast cells control PC synthesis when choline is abundant? The ultimate regulation would be limiting the amount of choline channeled into the CDP-choline pathway. Supporting this notion, a pronounced reduction in choline transporter steady state level and consequent choline uptake was observed for yeast cells grown in choline-rich medium independent of inositol content (8, 10, 21). In this paper, we explore the underlying mechanism providing this regulation.
Cells are equipped with several mechanisms to acquire nutrients for growth, coordinating substrate abundance with cellular demand. We show here that upon choline exposure, the sole high affinity choline transporter in S. cerevisiae, Hnm1, is subject to endocytosis and vacuolar degradation. Our data are consistent with the major driver of this event being the choline transporter rate. We go on to determine that Hnm1 endocytosis requires the ubiquitin ligase Rsp5 and is contingent on the casein kinase 1 pair Yck1/Yck2.
MATERIALS AND METHODS
Reagents
[methyl-14C]Choline chloride and [methyl-14C]methionine were from American Radiolabeled Chemicals. Antibodies used were anti-mouse horseradish peroxidase (HRP)-linked IgG and anti-rabbit HRP-linked IgG (Cell Signaling), anti-TAP (Open Biosystems), anti-ubiquitin (P4D1, Cell Signaling), anti-GFP (JL-8, Clontech), and anti-Pgk1 (Molecular Probes). Growth media were from Difco. Choline, choline oxidase from Arthrobacter globiformis, horseradish peroxidase, cycloheximide, homovanillic acid, and hemicholinium were from Sigma.
Media and Culture Conditions
Yeast cells were maintained in YEPD (1% bacto-yeast extract, 2% bacto-peptone, 2% dextrose) medium or SD (0.67% bacto-yeast nitrogen base without amino acids, 2% dextrose) medium supplemented as required for plasmid maintenance and nutrient auxotrophies. Yeast cells were routinely grown at 25 °C.
Yeast Strains and Plasmids
The yeast strains used in this study are shown in Table 1. Single deletion mutants in the BY4741 background were obtained from Euroscarf. When necessary, the genetic KanMX4 marker was replaced with the nourseothricin acetyltransferase gene cassette NatMX4 using plasmid pAG25 or with the Candida albicans URA3 gene (URA3MX cassette) using plasmid pAG60 as described (22). Double mutant strains or single mutant strains carrying a tagged allele were constructed by standard genetic crosses and sporulation. The HNM1 gene tagged at its 3′-end in frame with the GFP coding sequence was amplified by PCR from genomic DNA of the strain PMY764 using the following primers: 5′-HNM1, CAAATTGGTTAAGTCCATGAA; 3′-HNM1, TGCTTTTTATCCTTCTTTTCC. A 5-kb DNA fragment was generated extending from 0.5 kb upstream to 0.5 kb downstream of the HNM1 open reading frame and encompassing the complete insertion encoding GFP plus the HIS3 gene marker. This PCR product was subcloned into a pCR2.1 vector (Invitrogen), and its identity was verified by DNA sequencing. A fragment of 3.2 kb was released using SpeI (352 nt upstream of the HNM1 start codon) and PstI (446 nt downstream of GFP encoding the sequence stop codon) and ligated into the pRS415 low copy plasmid.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source/Laboratory |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 | Euroscarf |
| PMY756 | BY4741 HNM1-TAP::HIS3MX6 | Open Biosystems |
| PMY764 | BY4741 HNM1-GFP::HIS3MX6 | Invitrogen |
| PMY915 | BY4741 HNM1-TAP::HIS3MX6 cki1Δ::KanMX4 | This study |
| PMY742 | BY4741 HNM1-TAP::HIS3MX6 pct1Δ::KanMX4 | This study |
| PMY892 | BY4741 HNM1-TAP::HIS3MX6 cpt1Δ::KanMX4 | This study |
| PMY762 | BY4741 HNM1-GFP::HIS3MX6 cki1Δ::KanMX4 | This study |
| PMY867 | BY4741 HNM1-GFP::HIS3MX6 pct1Δ::KanMX4 | This study |
| PMY869 | BY4741 HNM1-GFP::HIS3MX6 cpt1Δ::KanMX4 | This study |
| PMY888 | BY4741 HNM1-TAP::HIS3MX6 eki1Δ::URA3MX | This study |
| PMY908 | BY4741 cki1Δ::KanMX4 hnm1Δ::NatMX4 | This study |
| RH448 | MATa leu2 ura3 his4 lys2 bar1 | H. Riezman |
| RH1597 | MATa leu2 ura3 his4 bar1 end4-1 | H. Riezman |
| SUB62 | MATa his3 leu2 trp1 ura3 (DF5 background) | D. Finley |
| Y0753 | his3 leu2 lys2 trp1 ura3 rsp5::HIS3 pSPT23#5 (DF5 background) | S. Jentsch |
| LRB341 | MATa his3 leu2 ura3 | L. Robinson |
| LRB362 | MATa his3 leu2 ura3 yck1-1::URA3 yck2-2ts | L. Robinson |
Protein Extraction and Western Blot Analysis
Logarithmically growing cells from a 5-ml culture (A600 nm of 0.5) were harvested, washed, and taken up in 200 μl of 50 mm Tris-HCl buffer, pH 8, 20% glycerol-containing Complete protease inhibitor mixture (Roche Applied Science) plus 1 μg/ml pepstatin A. Cells were broken with glass beads using a bead beater for two periods at 4 °C of 1 min intercalated with 1 min on ice. Cell debris was removed by a 3-min centrifugation at 500 × g, and supernatants were saved. Proteins were fractionated by 10% SDS-PAGE, transferred to PVDF membrane, and detected using appropriate primary and HRP-conjugated secondary antibodies followed by enhanced chemiluminescence.
For detection of ubiquitinated Hnm1-GFP adducts, logarithmically growing cells from a 50-ml culture (A600 nm of 0.6) were treated with 10 mm NaN3, 10 mm NaF, and 5 mm N-ethylmaleimide and placed in an ice-water bath. Cells were harvested, washed, and resuspended in 0.5 ml of lysis buffer composed of 50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Triton X-100, 10 mm N-ethylmaleimide, 1 μg/ml pepstatin A, and Complete protease inhibitor mixture (Roche Applied Science). Cells were broken with glass beads using a bead beater at 4 °C for three periods of 1 min intercalated with 3 min on ice. Extracts were recovered by pinching a hole at the bottom of the tube and collection by low speed centrifugation into a new tube. After the addition of 0.5 ml of fresh lysis buffer, the extracts were incubated for 30 min at 4 °C on a spinning wheel. Cell debris were eliminated by centrifugation at 500 × g for 3 min. Supernatants (1 ml) were incubated with 10 μl of anti-GFP monoclonal antibody (JL-8) and 40 μl of a 1:1 slurry of protein A-Sepharose (Amersham Biosciences) in lysis buffer overnight at 4 °C on a spinning wheel. Beads were washed four times with 10 ml of lysis buffer for 15 min on a spinning wheel at 4 °C. Proteins were eluted from the beads with 50 μl of SDS-PAGE loading buffer for 5 min at 95 °C. Proteins were fractionated by 7.5% SDS-PAGE, transferred to PVDF membrane, and probed with anti-ubiquitin monoclonal antibody and HRP-conjugated secondary antibodies followed by enhanced chemiluminescence.
Microscopy
Yeast cells were grown to mid-log phase at 25 °C in synthetic medium and treated with choline as indicated in the corresponding figure legends. Aliquots (0.5 ml) were withdrawn, and cells were spun down at 500 × g for 3 min and resuspended in a small amount of the same medium warmed at 25 °C for visualization. The live cells were mounted on slides and observed using a Zeiss Axiovert 200 M microscope fitted with a plan-neofluor ×100 oil immersion lens. Images were captured using a Zeiss Axio Cam HR using Axiovision version 4.5 software. Staining of vacuolar membranes with FM4-64 or of the cell periphery with calcofluor white was performed as described (23).
[methyl-14C]Methionine Labeling
Yeast cells were grown in synthetic defined medium containing [methyl-14C]methionine (2500 dpm/nmol) for at least seven generations for steady-state labeling of metabolites derived from the PE methylation pathway. Analyses of radioactivity distribution in aqueous cellular fractions were performed as described previously (9). Most, if not all, of the radioactivity associated with the aqueous fraction was distributed among choline, phosphocholine, glycerophosphocholine, and methionine.
Choline Transport Assay
Cells were harvested, thoroughly washed three times with ice-cold choline-free fresh medium, and resuspended in fresh medium prewarmed at 25 °C. A half-milliliter of 30 μm [14C]choline (∼25,000 dpm/nmol) prepared in prewarmed fresh medium was added to 0.5 ml of cell suspension. Uptake was allowed to proceed at 25 °C for 1.5 and 3 min and then stopped by the addition of 5 ml of ice-cold 10 mm choline, 10 mm NaN3, 10 mm NaF. Cells were filtered under vacuum through Whatman GF/C glass fiber filters and rinsed twice with 25 ml of ice-cold Tris-buffered saline containing 2 mm choline. The filters were allowed to dry, and the associated radiolabel was determined by liquid scintillation counting. Under the experimental conditions described above, the amount of choline taken up by cells never exceeded 5% of input.
Choline Mass Assay
Intracellular choline content was estimated enzymatically using choline oxidase coupled to a peroxidase-homovanillic acid detection array. Briefly, cells were grown to mid-log phase and immediately placed on ice. Lipids were extracted using the Folch method (21). The aqueous phase was chromatographed through a column of Dowex AG1-X8 (OH form) developed with water as previously described (21). Choline-containing eluates were concentrated by evaporation under vacuum and resuspended in 10 mm Tris-Cl, pH 8.0, containing 1% (w/v) KCl. To this, 0.2 unit of choline oxidase, 2 units of horseradish peroxidase, and 1 mm homovanillic acid were added. Samples were incubated at 37 °C for 1 h protected from light. Samples were analyzed upon excitation at 315 nm and detected at an emission of 425 nm.
RESULTS
Active Choline Transport Triggers Endocytosis of the Choline Transporter Hnm1
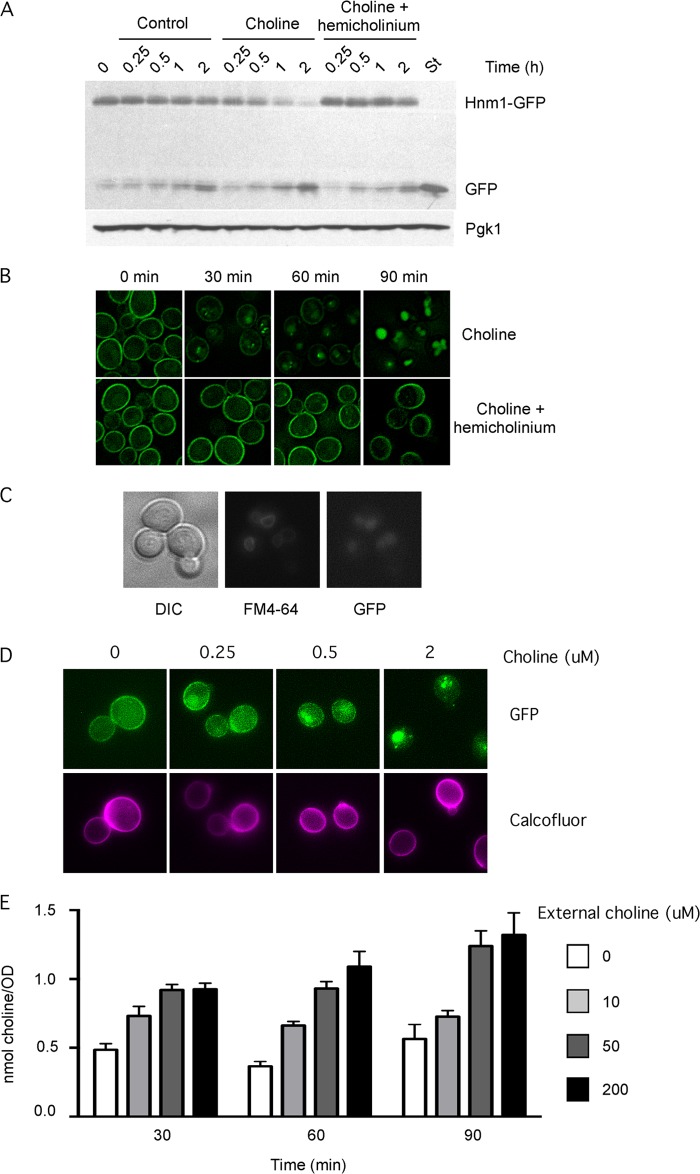
A functional C-terminal GFP-tagged variant of Hnm1 encoded at the HNM1 locus (Table 2) was used to analyze the stability of Hnm1 protein upon choline exposure by Western blot of protein extracts from cycloheximide-arrested cells. The addition of 200 μm choline substantially reduced the stability of Hnm1-GFP fusion and led to GFP accumulation. The simultaneous addition of 5 mm hemicholinium (a choline analog that blocks choline transport) protected Hnm1 from choline-induced degradation (Fig. 2A) to a level comparable with Hnm1 decay in control cells. Active transport of choline by Hnm1 into the cell appears to be required to decrease Hnm1 level.
TABLE 2.
Choline uptake rates for cells expressing tagged variants of Hnm1
Cells of the indicated genotypes were grown into log phase in synthetic defined media.
| Relevant genotype | Uptake ratea |
|---|---|
| pmol/min 106 cells | |
| HNM1 | 10.1 ± 0.2 |
| HNM1-TAP | 9.5 ± 0.3 |
| HNM1-GFP | 11.1 ± 0.2 |
a Data represent the mean ± S.E. of triplicate samples.
FIGURE 2.
Choline uptake induces choline transporter Hnm1 degradation. A, logarithmically growing wild type cells, expressing a functional Hnm1-GFP variant were treated with 100 μg/ml cycloheximide. After 15 min, choline (0.2 mm) or choline (0.2 mm) plus hemicholinium (5 mm) were added to the cells as indicated. Aliquots were withdrawn from the cultures at the indicated times and processed for immunoblot analysis using antibodies against GFP and against Pgk1. Stationary phase cells, characterized by intense vacuolar and no plasma membrane GFP staining, were included (lane St). B, wild type cells expressing a functional Hnm1-GFP variant were treated or not with choline (0.2 mm) or choline (0.2 mm) plus hemicholinium (5 mm), and at the indicated times, they were observed by fluorescence microscopy. C, wild type cells expressing Hnm1-GFP were exposed to 0.2 mm choline for 2 h. Vacuolar membranes were stained with FM4-64. Cells were observed by fluorescence microscopy. D, wild type cells expressing Hnm1-GFP were exposed to the indicated choline concentrations for 30 min. Cell walls were stained with Calcofluor. Cells were observed by fluorescence microscopy. E, wild type cells were grown to mid-log phase and exposed to different choline concentrations for the indicated times. Choline concentrations are listed on the right and range from 0 to 200 μm. Cells were processed for choline mass determination. Data represent the mean ± S.E. (error bars) of duplicate determination of two independent experiments.
We monitored the effect of choline addition on Hnm1-GFP localization by fluorescence microscopy. Intense plasma membrane staining was observed, consistent with the cellular role of Hnm1 as a high affinity transporter for choline (Fig. 2B). After 30 min in the presence of 200 μm choline, a decrease in the intensity at the plasma membrane and the appearance of a few bright spots per cell were evident. At later time points (60 and 90 min), plasma membrane staining was further reduced, whereas internal fluorescence increased. It is known that the GFP fold is relatively resistant to vacuolar degradation. The shift of the fluorescence signal from plasma membrane to round internal compartments is consistent with internalization and vacuolar degradation of Hnm1. Indeed, vacuolar membrane staining with FM4-64 confirmed that Hnm1-GFP was trafficked to the vacuole upon choline exposure (Fig. 2C). Hemicholinium prevented choline-induced delocalization of Hnm1-GFP from the plasma membrane (Fig. 2B), consistent with uptake of choline eliciting a mechanism leading to the internalization and vacuolar degradation of Hnm1.
We went on to determine the concentration of exogenous choline required for Hnm1-GFP internalization. When yeast cells were exposed for 30 min to external choline concentrations from 5 to 200 μm, a strong reduction on Hnm1-GFP signal at plasma membrane and formation of few bright foci were evident (Fig. 2B) (data not shown). At external choline concentrations of 0, 0.25, 0.5, and 2 μm, a transition from mostly plasma membrane Hnm1-GFP distribution to a diminution at this location alongside the formation of internal bright foci after a 30-min exposure was evident (Fig. 2D). Considering that Km of Hnm1 for choline transport is 0.5 μm (24), these data revealed a correlation between choline transport activity and signaling Hnm1 for targeted endocytosis.
It is known that choline kinase activity becomes a limiting factor for PC synthesis when yeast cells are exposed for 30 min to choline concentrations above 100 μm. As a consequence, the rate of choline uptake exceeds the rate of phosphocholine formation, leading to the accumulation of intracellular choline (21). Yeast cells were exposed to 10, 50, and 200 μm choline, and intracellular choline mass was determined after 30-, 60-, and 90-min exposure. From 50 to 200 μm external choline, increasing concentrations of intracellular choline were detected over time, whereas for cells exposed to 10 μm choline, intracellular choline levels did not increase during that period and did not differ significantly from endogenous levels (Fig. 2E).
Together, these data indicate that intracellular choline build-up is probably not the signal for targeted degradation of Hnm1. The data suggest that once the external choline concentration exceeds the Km for choline transport, Hnm1 is internalized from the plasma membrane for degradation by the vacuole. A conformational state of Hnm1 acquired during choline transport might be susceptible to targeted degradation.
Level and Localization of Hnm1 in PC Synthesis Mutants
To elucidate if a metabolite of the CDP-choline pathway for PC synthesis regulates Hnm1 stability, we assessed the level and localization of Hnm1 in mutant strains defective in each step of the CDP-choline pathway under both choline-deprived and -supplemented regimes. For cpt1Δ mutant cells, defective in the last step of PC synthesis, the level and localization of Hnm1 under choline-free growth conditions, as well as its susceptibility to choline-induced turnover, were similar to those in wild type cells (Fig. 3, A and B). Cells deficient in the Pct1 enzyme displayed a similar pattern, although the level and plasma membrane staining of Hnm1 under choline-free conditions were slightly reduced compared with wild type or cpt1Δ cells under the same conditions. Remarkably, cki1Δ cells grown in choline-free medium exhibited an extremely reduced level of Hnm1 and very faint plasma membrane localization, a pattern also observed in choline-containing medium (Fig. 3, A and B).
FIGURE 3.
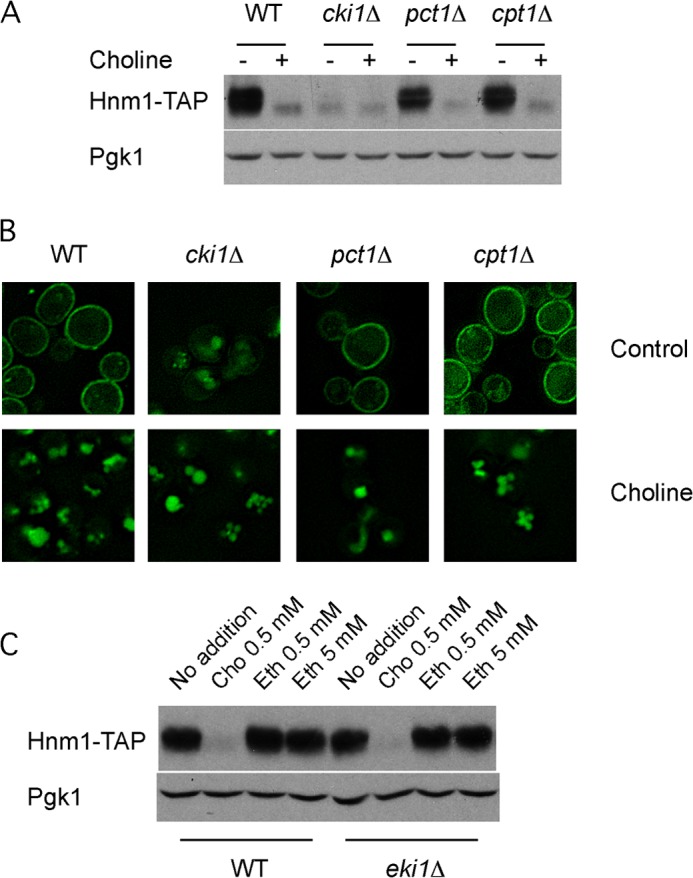
Phosphatidylcholine synthesis is not required for choline-induced Hnm1 endocytosis. A, yeast strains of the indicated genotypes expressing Hnm1-TAP protein were grown into log phase in the absence or the presence of 1 mm choline. Cells were processed for immunoblot analysis using antibodies against TAP and against Pgk1. B, a similar experiment was conducted with the indicated strains expressing Hnm1-GFP and grown in the absence or presence of 1 mm choline. Live cells were observed by fluorescence microscopy. C, wild type and eki1Δ cells expressing Hnm1-TAP were grown into log phase in the presence of the indicated concentration of choline or ethanolamine. Cells were processed for immunoblot analysis using antibodies against TAP and against Pgk1.
Choline uptake rate measurements (Table 3) corroborated these observations. Cells deficient for the Cki1 enzyme took up choline at a low rate under both choline-deficient and -replete conditions. Wild type and cpt1Δ cells exhibited a ∼10-fold decrease in the rate of choline uptake in cells grown in the presence of choline compared with those grown in choline-free medium, mirroring their choline-induced change on Hnm1 level and localization. The pct1Δ cells grown in choline-free medium took up choline at 40% of the wild type rate, a drop in transport activity suggested by the reduced level of Hnm1 at the plasma membrane on this mutant strain (Fig. 3, A and B) under the same conditions.
TABLE 3.
Choline uptake rates for mutants of the CDP-choline pathway
Cells were grown to mid-log phase in synthetic complete medium containing or not 1 mm choline.
| Relevant genotype | Choline uptake ratea |
|
|---|---|---|
| Choline-free medium | Choline-rich medium | |
| pmol/min 106 cells | ||
| Wild type | 10.5 ± 0.2 | 0.9 ± 0.05 |
| cki1Δ | 0.9 ± 0.05 | 0.5 ± 0.05 |
| pct1Δ | 2.6 ± 0.1 | 0.8 ± 0.04 |
| cpt1Δ | 10.2 ± 0.3 | 0.8 ± 0.05 |
a Data represent the mean ± S.E. of two independent experiments performed in triplicate.
These results taken together indicate that choline-induced Hnm1 degradation is not a response to increased PC synthesis because a block in the CDP-choline pathway did not prevent it. The reduced level of Hnm1 observed in cki1Δ cells grown in choline-free media is consistent with intracellular choline build-up signaling Hnm1 for degradation. A block at the choline kinase not only prevents choline taken up from the medium from being incorporated into PC but also prevents choline generated intracellularly by PC turnover from recycling back into PC (Fig. 1).
In addition to choline, Hnm1 also transports ethanolamine (10, 11). A separate set of enzymes constitutes the CDP-ethanolamine pathway for PE synthesis. We analyzed by Western blot the level of Hnm1-TAP for cells growing in ethanolamine-containing medium (Fig. 3C). Contrary to choline, ethanolamine did not trigger Hnm1 degradation. Similar results were observed for mutant cells deficient in the ethanolamine kinase Eki1, the first enzyme of this pathway. These results highlight the specificity of choline as a metabolite triggering Hnm1 degradation.
Metabolic Labeling of PC Biosynthesis Mutants
We measured the content of choline-containing metabolites in wild type, cki1Δ, pct1Δ, and cpt1Δ cells as well as cki1Δ eki1Δ and cki1Δ spo14Δ nte1Δ strains by labeling them at steady state with [14C-methyl]methionine. The radiolabeled methyl group is transferred from S-adenosylmethionine, a downstream metabolite of methionine, to PE during PC synthesis through the PE methylation pathway. PC turnover, in turn, generates choline that is either reused for PC synthesis through the CDP-choline pathway or excreted from the cell (Fig. 1). Strains were grown for 7–8 generations in medium containing radiolabeled methionine to steady state so that metabolite labeling profiles were reflective of metabolite distribution. The absence of Pct1 and Cpt1 elicited the expected metabolic phenotypes: phosphocholine and CDP-choline accumulation for pct1Δ and cpt1Δ cells, respectively (Fig. 4A). Surprisingly, different strains carrying a deletion of the CKI1 gene alone or in combination with eki1Δ or spo14Δ nte1Δ inactivated genes did not accumulate an amount of choline comparable with phosphocholine or CDP-choline observed in pct1Δ and cpt1Δ cells, respectively. Indeed, the choline level in cki1Δ cells was not different from choline levels measured in wild type cells.
FIGURE 4.
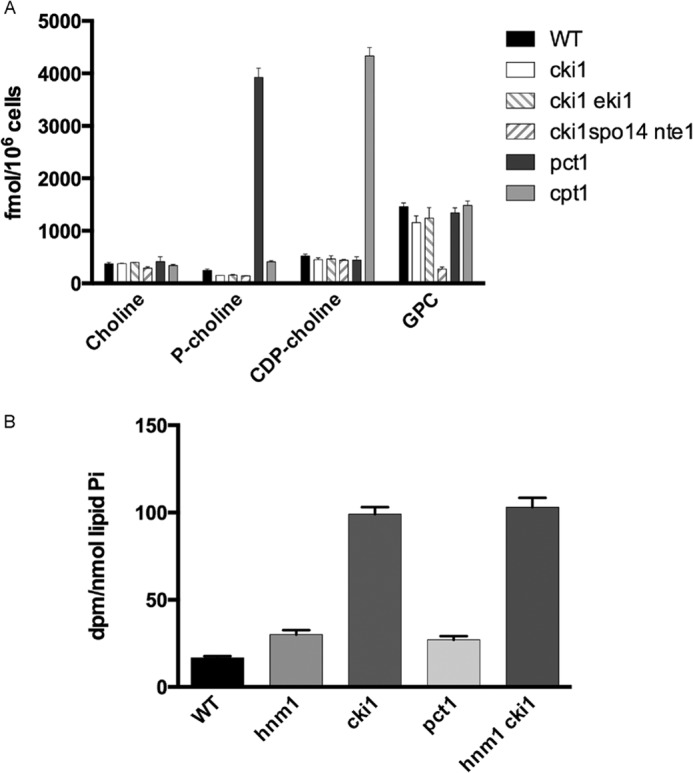
A block in the choline kinase step promotes choline excretion. A, cells of the indicated genotypes were labeled to steady state with [methyl-14C]methionine. Harvested cells were processed for separation and quantification of radioactive metabolites present in aqueous fractions. Data represent the mean ± S.E. (error bars) of single determination of two independent strains for each genotype. This experiment was repeated three times with qualitatively similar results. B, cells of the indicated genotypes were labeled to steady state with [methyl-14C]methionine. Cells were transferred into fresh non-radioactive medium and grown for an additional 1.5 generations. Aliquots of cell-free media were separated by thin layer chromatography, and radioactivity associated with choline was determined by liquid scintillation counting. Data represent the mean ± S.E. of duplicate determination of two independent strains for each genotype.
The excretion of choline under blockage of the CDP-choline pathway has been reported previously (25). We assessed excretion of choline in wild type cells and PC synthesis mutants by chasing cells previously labeled to steady state with [methyl-14C] methionine in fresh medium for another 1.5 generations and then quantified radiolabeled choline excreted into the medium. A higher amount of radiolabeled choline was detected in the culture medium in cki1Δ cells compared with wild type, hnm1Δ, and pct1Δ cells (Fig. 4B). In cells defective in the metabolism of choline, there is increased excretion of choline out of the cell. When the same analysis was performed in a double cki1Δ hnm1Δ mutant strain, choline excretion was very similar to that of cki1Δ cells, indicating that choline is excreted out of the cell independent of Hnm1.
Choline-induced Down-regulation of Hnm1 Involves Endocytosis through a Ubiquitin Ligase Rsp5-mediated Mechanism
The vacuolar GFP staining observed under choline-rich conditions suggests that Hnm1 plasma membrane level is regulated by endocytic delivery for vacuolar turnover (26–29). We assessed the dependence on endocytosis by looking at Hnm1-GFP trafficking in the endocytic mutant end4-1 strain. END4 encodes a transmembrane actin-binding protein involved in endocytosis, and the product of the allele end4-1 is inactive at 37 °C (30, 31). Upon a shift to the restrictive temperature for the end4-1 allele, the addition of choline did not trigger Hnm1-GFP internalization in the end4-1 strain, whereas in an End4-proficient strain, choline supplementation resulted in Hnm1-GFP trafficking out of the plasma membrane (Fig. 5A).
FIGURE 5.
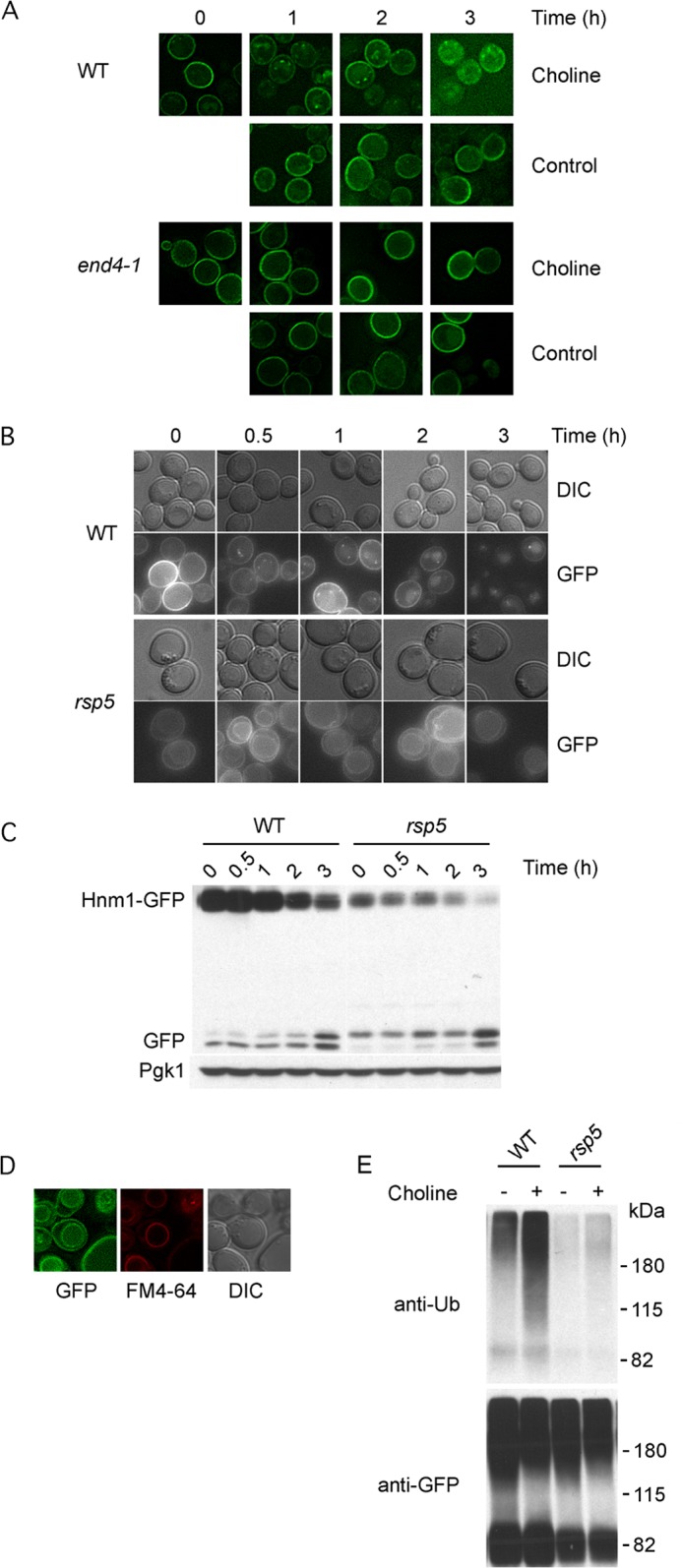
Rsp5 mediates choline-induced Hnm1 endocytosis and degradation. A, end4-1 and wild type cells were transformed with a plasmid bearing a GFP-tagged allele of HNM1. Logarithmically growing cells were shifted to 37 °C, and after 15 min, choline (200 μm) was added. At the indicated times, cells were observed by fluorescence microscopy. B and C, rsp5Δ and wild type cells expressing Hnm1-GFP from a plasmid were exposed to 200 μm choline. At the indicated times, cells were processed for fluorescence microscopy (B) and Western blotting (C). D, vacuolar membrane staining with FM4–64 for rsp5Δ cells expressing Hnm1-GFP. E, choline exposure promotes Hnm1 ubiquitination. Yeast cells of the indicated genotypes expressing Hnm1-GFP from a plasmid were exposed or not to 200 μm choline for 20 min. Hnm1-GFP was immunoprecipitated from cell lysates, separated by 7.5% SDS-PAGE, and probed with the indicated antibodies. Molecular weight markers are indicated on the left. DIC, differential interference contrast.
The ubiquitin ligase Rsp5, among its various functions, regulates the stability of many permeases, targeting them by ubiquitination for endocytosis and vacuolar degradation. The essential character of Rsp5 is associated with the ubiquitin-proteasome-dependent processing of the transcription factor Spt23. Rsp5 is dispensable when yeast cells express a C-terminal truncation of Spt23 (32). We monitored changes in Hnm1-GFP level and localization upon the addition of choline in rsp5Δ cells kept alive by the expression of a C-terminally truncated version of Spt23. Over 3 h of choline exposure, there was a slight decrease in plasma membrane staining and increase in vacuolar lumen staining for rsp5Δ cells, whereas in wild type cells, internal bright spots formed upon choline exposure and plasma membrane localization of Hnm1-GFP declined alongside increasing diffuse vacuolar lumen staining (Fig. 5B). Western blot analysis revealed a strong stabilization of Hnm1-GFP in choline-treated rsp5Δ cells compared with its rapid decrease in wild type cells (Fig. 5C). These results taken together point toward the ubiquitin ligase Rsp5 being responsible for targeting Hnm1 for choline-induced endocytosis and degradation. It is worth highlighting that in rsp5Δ cells, vacuolar membrane staining was evident in all conditions examined (Fig. 5, C and D). It is known that Rsp5 has a role for cargo sorting from late endosomes into multivesicular bodies, and defects in sorting at this step lead to delivery of cargo proteins to the vacuolar membrane (33–35).
We analyzed by Western blotting the ubiquitination of Hnm1 by exposing wild type and rsp5Δ cells expressing Hnm1-GFP to 200 μm choline for 20 min. GFP adducts were immunoprecipitated, separated by SDS-PAGE, and probed for ubiquitin (Fig. 5E). An increased amount of ubiquitination of Hnm1-GFP was evident upon choline exposure for wild type cells, whereas a minor increase was detected for rsp5Δ cells.
The casein kinase 1 redundant protein pair Yck1/Yck2 has been involved in targeting transporters and other integral membrane proteins for trafficking from the plasma membrane (36–38). We explored the participation of these protein kinases in Hnm1 regulation by monitoring choline-induced Hnm1-GFP internalization in a yck1-1 yck2-1ts strain whereby protein kinase activity can be inactivated by shifting the cells to the restrictive temperature. After a temperature shift to inactivate casein kinase activity, choline was added, and Hnm1-GFP internalization was examined (Fig. 6A). Reduction of casein kinase activity attenuated choline-induced Hnm1-GFP turnover. These results suggest that casein kinase 1 pair Yck1/Yck2 participates in the cascade of events that result in Hnm1 transport from the plasma membrane to the vacuole upon choline exposure.
FIGURE 6.
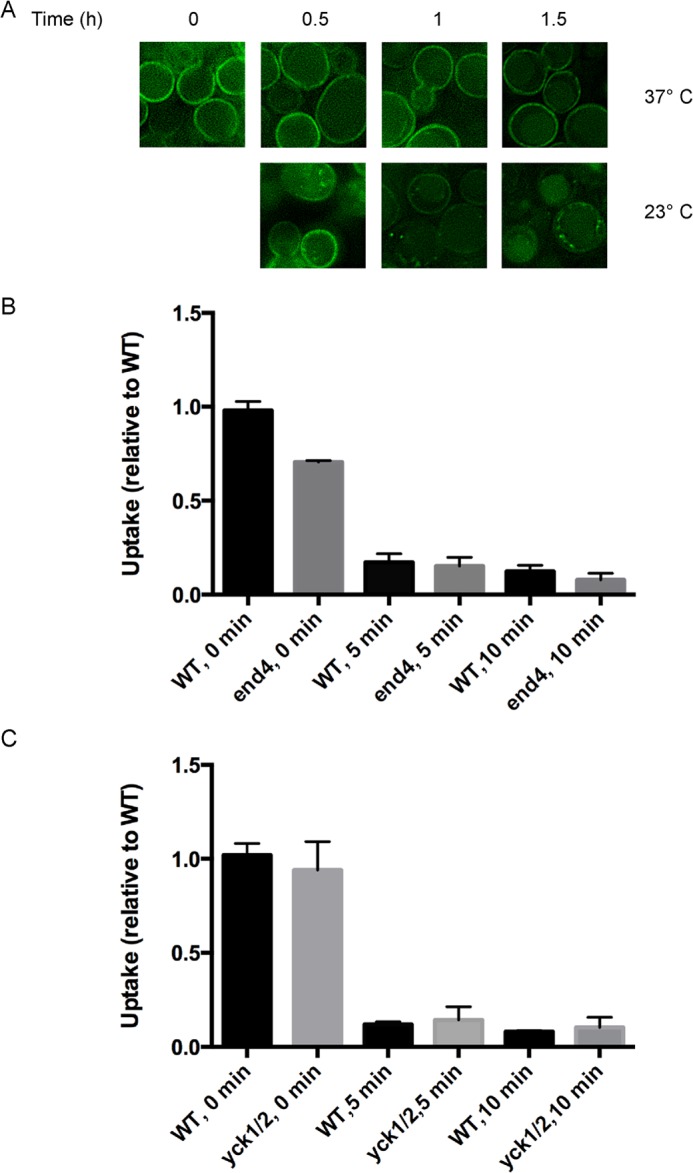
Choline exposure triggers a rapid Hnm1 inactivation independent of endocytosis. A, yck1-1 yck2-1ts and wild type cells expressing Hnm1-GFP from a plasmid were grown at 23 °C and shifted at 37 °C for 30 min. Choline (200 μm) was added to the cells, and they were observed by fluorescence microscopy at the indicated times. B, end4-1 and wild type cells grown at 25 °C were shifted at 37 °C. After 15 min, the cells were exposed to 200 μm choline for the indicated periods, and the choline uptake rate was measured. C, yck1-1 yck2-1ts and wild type cells grown at 23 °C were shifted at 37 °C for 30 min. The cells were then exposed to 200 μm choline for the indicated periods, and choline uptake was measured. For C and D, plotted data are mean ± S.E. (error bars) for quadruplicates and relative to their wild type growing in choline-free medium. Choline uptake rate for wild type strain RH448 (parental of end4-1) was 3.8 ± 0.3 pmol min−1 106 cells. Choline uptake rate for wild type strain LRB341 (parental of yck1-1 yck2-2ts) was 16.0 ± 0.8 pmol min−1 106 cells.
Rapid Decrease of Choline Transport upon Choline Exposure Can Occur Independent of Hnm1 Internalization
A defect in endocytosis prevents choline-induced Hnm1 internalization. We assessed choline transport activity upon choline exposure when Hnm1 internalization was impeded. Cells carrying inactive End4 protein were exposed to 200 μm choline for different periods, and choline uptake was measured (Fig. 6B). A rapid decrease in Hnm1 activity was observed in end4-1 cells after choline exposure, occurring as fast as the wild type parental strain. A rapid decrease of choline transport appears to precede endocytosis of the Hnm1 transporter.
We also examined choline transport in the strain defective in casein kinase 1 activity. Inactivation of the casein kinase 1 pair Yck1/Yck2 did not prevent the fast reduction of choline transport upon choline exposure (Fig. 6C). In addition, these results support the notion that a rapid decrease in choline transport occurs independent of targeted endocytosis.
DISCUSSION
In this paper, we describe new regulatory mechanisms that control choline uptake. Upon choline exposure, the sole choline transporter in yeast, Hnm1, is endocytosed from the plasma membrane to the vacuole in an Rsp5- and Yck1/2-dependent manner. We also observed a rapid inactivation of Hnm1 transporter activity upon choline exposure that occurred prior to endocytosis.
We show that choline-induced down-regulation of Hnm1 was not a response to high rates of PC synthesis upon choline supplementation. The integrity of the CDP-choline pathway was not necessary for choline to elicit both inactivation and degradation of Hnm1, suggesting that the rate of PC synthesis itself does not regulate choline uptake. Inactivation of the final step in the PC synthesis pathway resulted in an accumulation of CDP-choline and decreased PC synthesis, but the level of Hnm1 at the plasma membrane and its susceptibility to endocytosis were unaffected. Inactivation of the penultimate step resulted in a modest decrease in Hnm1 activity and endocytosis.
The absence of the choline kinase Cki1 decreased choline transport activity. This phenotype arose from the very reduced plasma membrane content of Hnm1. In addition, cki1Δ cells did not accumulate choline intracellularly but excreted choline into the medium through an Hnm1-independent mechanism. Regulation of intracellular choline concentration seems to be stringent because the choline levels in cki1Δ cells unexpectedly were not different from those measured for choline kinase-proficient cells. How is Hnm1 targeted for degradation in cki1Δ if choline levels are not increased? The data suggest that intracellular choline accumulation is not the signal triggering Hnm1 down-regulation. Intracellular choline content did not increase when yeast cells were exposed to 10 μm choline. However, this extracellular choline concentration triggered Hnm1 down-regulation. We show a correlation of concentration around the Km of Hnm1 for choline transport (0.5 μm), implying that the rate of choline transport can modulate Hnm1 endocytosis. Consistent with the notion that the choline transporter becomes susceptible to degradation upon choline transport is our observation that hemicholinium prevented choline-dependent Hnm1 down-regulation. These observations support a model where Hnm1, when delivering choline into the cytoplasm, acquires a “degradable” conformational state.
Choline has a pivotal role in PC metabolism; it is a precursor for PC synthesis as well as an end product of PC catabolism (Fig. 1). Its dual character contributes to understanding the observation that degradation of Hnm1 is not a response to high rates of PC synthesis through the CDP-choline pathway but merely to choline transport activity. Choline content is the result of a balance between uptake, consumption, generation by PC degradation, and excretion. The existence of an Hnm1-independent mechanism of choline excretion working alongside Hnm1 down-regulation highlights the cellular need to regulate choline content. Unrestricted flow through the CDP-choline pathway would perturb PC metabolism and impact its intercalation with key cellular processes, including vesicle trafficking and regulation of nucleotide levels, because DAG and CTP are both consumed by the CDP-choline pathway (3, 39–42).
We show that choline affects Hnm1 endocytosis through the ubiquitin ligase Rsp5. It is highly probable that Rsp5 catalyzes the ubiquitination of Hnm1. First, Hnm1 is resistant to choline-induced degradation in an rsp5Δ mutant strain, consistent with the observed reduction on Hnm1 ubiquitination upon choline exposure. Second, the down-regulation that Hnm1 undergoes upon choline exposure closely resembles the regulation of other yeast transporters by their substrates, for which it has been demonstrated that Rsp5-mediated ubiquitination is necessary to trigger their endocytosis and vacuolar degradation (26, 29).
We also show that casein kinase 1 essential pair Yck1/Yck2 is required for choline-induced Hnm1 endocytosis. This protein kinase is known to be involved in targeting transporters and receptors at the plasma membrane for ubiquitin-mediated endocytosis. Based on the overall conservation of mechanisms and factors involved in the regulated endocytosis of yeast transporters and receptors, we hypothesize that Hnm1 becomes phosphorylated directly or indirectly by Yck1/Yck2, and this event is upstream of Rsp5-mediated ubiquitination of Hnm1.
Acknowledgments
We thank Dan Finley, Howard Riezman, Stefan Jentsch, and Lucy Robinson for strains used in this study. We also thank Drs. Vanina Zaremberg and Pia Elustondo for helpful comments on the manuscript.
This study was supported by a Canadian Institutes of Health Research grant (to C. R. M.).
- PC
- phosphatidylcholine
- DAG
- diacylglycerol
- PE
- phosphatidylethanolamine.
REFERENCES
- 1. Boumann H. A., Gubbens J., Koorengevel M. C., Oh C. S., Martin C. E., Heck A. J., Patton-Vogt J., Henry S. A., de Kruijff B., de Kroon A. I. (2006) Depletion of phosphatidylcholine in yeast induces shortening and increased saturation of the lipid acyl chains. Evidence for regulation of intrinsic membrane curvature in a eukaryote. Mol. Biol. Cell 17, 1006–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Howe A. G., Zaremberg V., McMaster C. R. (2002) Cessation of growth to prevent cell death due to inhibition of phosphatidylcholine synthesis is impaired at 37 degrees C in Saccharomyces cerevisiae. J. Biol. Chem. 277, 44100–44107 [DOI] [PubMed] [Google Scholar]
- 3. Carman G. M., Han G. S. (2011) Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu. Rev. Biochem. 80, 859–883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carman G. M., Kersting M. C. (2004) Phospholipid synthesis in yeast. Regulation by phosphorylation. Biochem. Cell Biol. 82, 62–70 [DOI] [PubMed] [Google Scholar]
- 5. Skinner H. B., McGee T. P., McMaster C. R., Fry M. R., Bell R. M., Bankaitis V. A. (1995) The Saccharomyces cerevisiae phosphatidylinositol-transfer protein effects a ligand-dependent inhibition of choline-phosphate cytidylyltransferase activity. Proc. Natl. Acad. Sci. U.S.A. 92, 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cottrell S. F., Getz G. S., Rabinowitz M. (1981) Phospholipid accumulation during the cell cycle in synchronous cultures of the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 256, 10973–10978 [PubMed] [Google Scholar]
- 7. Lykidis A., Jackowski S. (2001) Regulation of mammalian cell membrane biosynthesis. Prog. Nucleic Acid Res. Mol. Biol. 65, 361–393 [DOI] [PubMed] [Google Scholar]
- 8. Fernández-Murray J. P., Gaspard G. J., Jesch S. A., McMaster C. R. (2009) NTE1-encoded phosphatidylcholine phospholipase b regulates transcription of phospholipid biosynthetic genes. J. Biol. Chem. 284, 36034–36046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murray J. P., McMaster C. R. (2005) Nte1p-mediated deacylation of phosphatidylcholine functionally interacts with Sec14p. J. Biol. Chem. 280, 8544–8552 [DOI] [PubMed] [Google Scholar]
- 10. Nikawa J., Hosaka K., Tsukagoshi Y., Yamashita S. (1990) Primary structure of the yeast choline transport gene and regulation of its expression. J. Biol. Chem. 265, 15996–16003 [PubMed] [Google Scholar]
- 11. Nikawa J., Tsukagoshi Y., Yamashita S. (1986) Cloning of a gene encoding choline transport in Saccharomyces cerevisiae. J. Bacteriol. 166, 328–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carman G. M., Henry S. A. (1999) Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog. Lipid. Res. 38, 361–399 [DOI] [PubMed] [Google Scholar]
- 13. Lee K. S., Patton J. L., Fido M., Hines L. K., Kohlwein S. D., Paltauf F., Henry S. A., Levin D. E. (1994) The Saccharomyces cerevisiae PLB1 gene encodes a protein required for lysophospholipase and phospholipase B activity. J. Biol. Chem. 269, 19725–19730 [PubMed] [Google Scholar]
- 14. Fernández-Murray J. P., McMaster C. R. (2007) Phosphatidylcholine synthesis and its catabolism by yeast neuropathy target esterase 1. Biochim. Biophys. Acta 1771, 331–336 [DOI] [PubMed] [Google Scholar]
- 15. Fernández-Murray J. P., McMaster C. R. (2005) Glycerophosphocholine catabolism as a new route for choline formation for phosphatidylcholine synthesis by the Kennedy pathway. J. Biol. Chem. 280, 38290–38296 [DOI] [PubMed] [Google Scholar]
- 16. Fisher E., Almaguer C., Holic R., Griac P., Patton-Vogt J. (2005) Glycerophosphocholine-dependent growth requires Gde1p (YPL110c) and Git1p in Saccharomyces cerevisiae. J. Biol. Chem. 280, 36110–36117 [DOI] [PubMed] [Google Scholar]
- 17. Rudge S. A., Zhou C., Engebrecht J. (2002) Differential regulation of Saccharomyces cerevisiae phospholipase D in sporulation and Sec14-independent secretion. Genetics 160, 1353–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xie Z., Fang M., Rivas M. P., Faulkner A. J., Sternweis P. C., Engebrecht J. A., Bankaitis V. A. (1998) Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc. Natl. Acad. Sci. U.S.A. 95, 12346–12351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Carman G. M., Henry S. A. (2007) Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282, 37293–37297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jesch S. A., Zhao X., Wells M. T., Henry S. A. (2005) Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J. Biol. Chem. 280, 9106–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McMaster C. R., Bell R. M. (1994) Phosphatidylcholine biosynthesis via the CDP-choline pathway in Saccharomyces cerevisiae. Multiple mechanisms of regulation. J. Biol. Chem. 269, 14776–14783 [PubMed] [Google Scholar]
- 22. Voth W. P., Jiang Y. W., Stillman D. J. (2003) New “marker swap” plasmids for converting selectable markers on budding yeast gene disruptions and plasmids. Yeast 20, 985–993 [DOI] [PubMed] [Google Scholar]
- 23. Vida T. A., Emr S. D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hosaka K., Yamashita S. (1980) Choline transport in Saccharomyces cerevisiae. J. Bacteriol. 143, 176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patton-Vogt J. L., Griac P., Sreenivas A., Bruno V., Dowd S., Swede M. J., Henry S. A. (1997) Role of the yeast phosphatidylinositol/phosphatidylcholine transfer protein (Sec14p) in phosphatidylcholine turnover and INO1 regulation. J. Biol. Chem. 272, 20873–20883 [DOI] [PubMed] [Google Scholar]
- 26. Lauwers E., Erpapazoglou Z., Haguenauer-Tsapis R., André B. (2010) The ubiquitin code of yeast permease trafficking. Trends Cell Biol. 20, 196–204 [DOI] [PubMed] [Google Scholar]
- 27. Lin C. H., MacGurn J. A., Chu T., Stefan C. J., Emr S. D. (2008) Arrestin-related ubiquitin-ligase adaptors regulate endocytosis and protein turnover at the cell surface. Cell 135, 714–725 [DOI] [PubMed] [Google Scholar]
- 28. Nikko E., Pelham H. R. (2009) Arrestin-mediated endocytosis of yeast plasma membrane transporters. Traffic 10, 1856–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rotin D., Staub O., Haguenauer-Tsapis R. (2000) Ubiquitination and endocytosis of plasma membrane proteins. Role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176, 1–17 [DOI] [PubMed] [Google Scholar]
- 30. Raths S., Rohrer J., Crausaz F., Riezman H. (1993) end3 and end4. Two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J. Cell Biol. 120, 55–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wesp A., Hicke L., Palecek J., Lombardi R., Aust T., Munn A. L., Riezman H. (1997) End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell 8, 2291–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H. D., Jentsch S. (2000) Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102, 577–586 [DOI] [PubMed] [Google Scholar]
- 33. Blondel M. O., Morvan J., Dupré S., Urban-Grimal D., Haguenauer-Tsapis R., Volland C. (2004) Direct sorting of the yeast uracil permease to the endosomal system is controlled by uracil binding and Rsp5p-dependent ubiquitylation. Mol. Biol. Cell 15, 883–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Erpapazoglou Z., Froissard M., Nondier I., Lesuisse E., Haguenauer-Tsapis R., Belgareh-Touzé N. (2008) Substrate- and ubiquitin-dependent trafficking of the yeast siderophore transporter Sit1. Traffic 9, 1372–1391 [DOI] [PubMed] [Google Scholar]
- 35. Kim Y., Deng Y., Philpott C. C. (2007) GGA2- and ubiquitin-dependent trafficking of Arn1, the ferrichrome transporter of Saccharomyces cerevisiae. Mol. Biol. Cell 18, 1790–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hicke L., Zanolari B., Riezman H. (1998) Cytoplasmic tail phosphorylation of the α-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 141, 349–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Marchal C., Haguenauer-Tsapis R., Urban-Grimal D. (2000) Casein kinase I-dependent phosphorylation within a PEST sequence and ubiquitination at nearby lysines signal endocytosis of yeast uracil permease. J. Biol. Chem. 275, 23608–23614 [DOI] [PubMed] [Google Scholar]
- 38. Paiva S., Vieira N., Nondier I., Haguenauer-Tsapis R., Casal M., Urban-Grimal D. (2009) Glucose-induced ubiquitylation and endocytosis of the yeast Jen1 transporter. Role of lysine 63-linked ubiquitin chains. J. Biol. Chem. 284, 19228–19236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chang Y. F., Carman G. M. (2008) CTP synthetase and its role in phospholipid synthesis in the yeast Saccharomyces cerevisiae. Prog. Lipid Res. 47, 333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cleves A. E., McGee T. P., Whitters E. A., Champion K. M., Aitken J. R., Dowhan W., Goebl M., Bankaitis V. A. (1991) Mutations in the CDP-choline pathway for phospholipid biosynthesis bypass the requirement for an essential phospholipid transfer protein. Cell 64, 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Litvak V., Dahan N., Ramachandran S., Sabanay H., Lev S. (2005) Maintenance of the diacylglycerol level in the Golgi apparatus by the Nir2 protein is critical for Golgi secretory function. Nat. Cell Biol. 7, 225–234 [DOI] [PubMed] [Google Scholar]
- 42. Sarri E., Sicart A., Lazaro-Diéguez F., Egea G. (2011) Phospholipid synthesis participates in the regulation of diacylglycerol required for membrane trafficking at the Golgi complex. J. Biol. Chem. 286, 28632–28643 [DOI] [PMC free article] [PubMed] [Google Scholar]