Recent genome sequencing studies have identified several classes of complex genomic rearrangements that appear to be derived from a single catastrophic event rather than numerous incremental steps. In this review, Zhang et al. discuss these phenomena, summarize current models, and consider the impact of massive chromosomal change on the development of diseases such as cancer.
Keywords: chromothripsis, chromoplexy, chromoanasynthesis, genome evolution, cancer, chromosomal translocation, copy number alteration
Abstract
Recent genome sequencing studies have identified several classes of complex genomic rearrangements that appear to be derived from a single catastrophic event. These discoveries identify ways that genomes can be altered in single large jumps rather than by many incremental steps. Here we compare and contrast these phenomena and examine the evidence that they arise “all at once.” We consider the impact of massive chromosomal change for the development of diseases such as cancer and for evolution more generally. Finally, we summarize current models for underlying mechanisms and discuss strategies for testing these models.
The evolution of a normal cell to a cancer cell is generally thought of as a sequential accumulation of many independent lesions to the genome (Nowell 1976; Greaves and Maley 2012; Yates and Campbell 2012). In addition to somatic point mutations, these lesions include copy number alterations, such as chromosomal deletions or duplications (Fig. 1A,B), and balanced structural rearrangements, such as translocations and inversions (Mitelman et al. 2007; Rabbitts 2009; Stephens et al. 2009; Stratton et al. 2009). By chance, these somatic alterations in the genome will hit cancer genes—disrupting tumor suppressors or activating proto-oncogenes—affecting cells and causing their progeny to gradually accumulate the necessary “hallmarks of cancer” (Hanahan and Weinberg 2011). Although there is ample evidence that cancer can develop through this type of gradual mutational accrual, there are reasons to think that there could be advantages if the oncogenic changes could be acquired all at once (at least from the vantage point of the aspiring cancer cell). One obvious advantage is speed, with the possibility of rapidly accruing a large phenotypic effect from combinatorial acquisition of smaller-effect genetic changes. Another potential advantage of all-at-once or “punctuated equilibrium” (Gould and Eldredge 1977) mechanisms is that intermediate steps in tumor development can actually be deleterious. For example, oncogene activation can trigger stress responses, such as p53 activation, that can inhibit proliferation or promote cellular senescence (Lowe et al. 1994; Fearnhead et al. 1997; Kastan and Bartek 2004; Halazonetis et al. 2008; Jackson and Bartek 2009). In principle, an all-at-once mutational mechanism could circumvent this problem by simultaneously generating an active oncogene together with changes that eliminate the stress response.
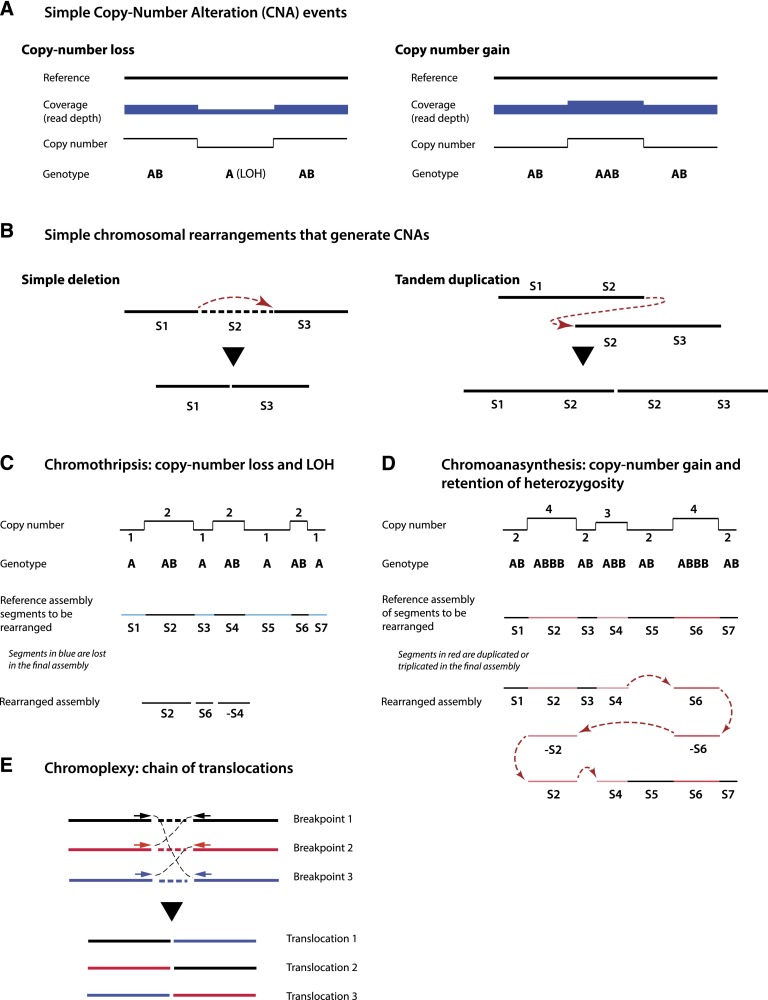
Figure 1.
Characteristics of simple and complex chromosomal rearrangements. (A) Copy number losses or gains can be detected by array-based or sequencing-based technologies from abrupt changes in the read depth signal (sequencing) or the intensity of fluorescent probes (array). Loss or gain generally affects only one homolog; therefore, a copy number loss also results in LOH. (B) Copy number alterations are often generated by simple chromosomal rearrangements, such as deletions or tandem duplications. (C) Chromothripsis is characterized by an alternating copy number profile with loss and retention of heterozygosity. Minus signs in C and D denote chromosomal segments that are inverted. Fluorescent in situ hybridization (FISH) characterization confirms that only one chromatid (allele B) is affected. (D) Chromoanasynthesis is characterized by a copy number profile that alternates between euploid and higher ploidy. In contrast to chromothripsis, chromoanasynthesis does not necessarily exhibit LOH but reflects resynthesis of segments from one chromatid. Chromoanasynthesis also contains frequent insertions of short sequences between the rearrangement junctions (red arrows) that are copied from the rearranged segments (templated insertions). Both DNA resynthesis and templated insertions are suggestive of replication-based mechanisms. (E) Chromoplexy is characterized by a closed chain of translocations, with little or no copy number alteration.
Although all-at-once massive genomic alterations might accelerate the acquisition of growth-promoting mutations, they have an obvious downside: They could also potentially generate massive deleterious effects that could overwhelm any growth-promoting mutations. One way to minimize this collateral damage would be to have the massive genomic change be restricted to a portion of the genome. Although it is common to think of mutation rates as numbers that apply evenly across the genome (Durrett 2008; Bozic et al. 2010; Tomasetti et al. 2013), there is a growing appreciation of localized variability in these rates (Hodgkinson and Eyre-Walker 2011; Hodgkinson et al. 2012; Roberts et al. 2012; Schuster-Böckler and Lehner 2012). Because of transcription-coupled DNA repair, transcriptionally silent portions of the genome have approximately threefold higher mutation rates (Fousteri and Mullenders 2008; Lawrence et al. 2013). Likewise, late-replicating regions of the genome exhibit a similar fold increase in mutation rates (Stamatoyannopoulos et al. 2009; Chen et al. 2010; Koren et al. 2012; Donley and Thayer 2013). Localized hypermutation has also been associated with chromosomal rearrangements. In budding yeast, DNA breaks repaired by mitotic gene conversion are accompanied by surprisingly high mutation rates (>1000-fold higher than spontaneous mutations) in the vicinity of the break (Hicks et al. 2010; Malkova and Haber 2012). Similarly, localized regions of hypermutation (“kataegis”) that often occurred at the site of somatic rearrangements were discovered in breast cancer genomes (Nik-Zainal et al. 2012; Alexandrov et al. 2013). Even more dramatic local genomic change can be generated from critical telomere shortening, resulting in breakage–fusion–bridge (BFB) cycles (McClintock 1941) that occur over several cell division cycles (and hence are not “all-at-once” events). Analysis of genome copy number alterations in cancer also identified “firestorms” of local amplifications and deletions whose origin is uncertain but might originate from BFB cycles (Hicks et al. 2006).
Recently, whole-genome sequencing has led to the discovery of three new classes of complex catastrophic chromosomal rearrangement: chromothripsis, chromoanasynthesis, and chromoplexy (Fig. 1C–E; Liu et al. 2011; Stephens et al. 2011; Rausch et al. 2012; Baca et al. 2013). Although the underlying mechanisms for these phenomena are unknown and likely to be different, all of these patterns of rearrangements appear to have originated from a single event, simultaneously generating tens or even hundreds of translocations and/or DNA copy number changes (Meyerson and Pellman 2011; Tubio and Estivill 2011; Forment et al. 2012; Jones and Jallepalli 2012; Maher and Wilson 2012). To encompass the potentially all-at-once origin of these mutational mechanisms, a unifying term, chromoanagenesis, has been proposed (Holland and Cleveland 2012). Two of these classes of chromosomal rearrangement, chromothripsis and chromoanasynthesis, affect limited portions of the genome (often a single chromosome or chromosome arm), raising the mechanistic question of how extensive chromosomal rearrangements can be so highly localized. The frequency of these types of events in cancer remains unclear, but many recent studies suggest that they may be more common than initially suspected (Malhotra et al. 2013; Yang et al. 2013; Zack et al. 2013). These new classes of mutagenesis are not restricted to human cancer and have turned up in such exotic places as the Tasmanian devil's transmissible facial tumor (Deakin et al. 2012) and human congenital diseases (Kloosterman et al. 2011a, 2012; Liu et al. 2011; Chiang et al. 2012; Nazaryan et al. 2013). Although these new classes of chromosome rearrangement have mostly been found in the context of disease, it seems likely that they reflect very general mechanisms for genome alteration that could be important for organismal evolution.
A new game at the genome casino
Genome instability is not only a hallmark but also an enabling characteristic of cancer (Negrini et al. 2010; Hanahan and Weinberg 2011; Gordon et al. 2012). Disruption of genome maintenance and elevated rates of somatic mutations (e.g., due to disruption of DNA repair machineries or increased damage) accelerate the transformation from precancerous tissue into malignant tumors (Gorgoulis et al. 2005; Halazonetis et al. 2008; Vogelstein et al. 2013). Despite accelerated tumor evolution, individual mutation events are usually regarded as independent of each other, acquired randomly and gradually. In addition to site-specific mutation events (point mutations, chromosomal translocations, and copy number variations), doubling of the genome content due to cytokinesis failure is a one-off event that can generate aneuploidy and structural alterations of chromosomes (Fujiwara et al. 2005; Holland and Cleveland 2009; Carter et al. 2012; Ganem and Pellman 2012; Zack et al. 2013).
In studying the tumor genome of a patient with chronic lymphocytic leukemia (CLL), Stephens et al. (2011) observed a striking pattern of chromosomal rearrangements that was at odds with all known models of genomic alteration. The tumor from this patient contained 45 tumor-specific translocations, 42 of which were concentrated on the long arm of chromosome 4. Furthermore, the rearrangement events were clustered within multiple focal regions in chromosome 4, raising the question of how the damage could be so highly localized. Did this result from a genuinely new mechanism for genome damage, or was the genome center playing the conventional game of random chance but had been dealt a rare hand?
Several lines of evidence suggested this was, in fact, a novel phenomenon. First, despite the large number of rearrangements, SNP array copy number analysis showed that the altered region of chromosome 4 existed in only two copy number states, with many transitions between these two states. (As in the study by Stephens et al. (2011), we use the term copy number states because of the highly aneuploid nature of cancer genomes, where each of the parental haplotypes can be present in multiple copies.) This pattern of having only two copy number states is in sharp contrast to conventional clusters of complex rearrangements (Hicks et al. 2006; Bignell et al. 2007; Zhang et al. 2009a). Known causes of complex rearrangements such as BFB cycles characteristically result in multiple amplified copy number states that are assumed to derive from events occurring over many rounds of cell division (McClintock 1941; Stark et al. 1989; Gisselsson et al. 2000; Bignell et al. 2007; Greenman et al. 2012b).
The alternation between two copy number states was accompanied by loss and preservation of heterozygosity (Fig. 1A,C). Whole-genome sequencing further revealed a very complex pattern of segmental rearrangements at the boundaries of high copy number states, with events that were head to tail (deletion type), head to head/tail to tail (inversion type), and tail to head (tandem duplication type) (Fig. 2A). However, despite this chaotic pattern of rearrangement, loci that preserved parental copy number also preserved heterozygosity of the parental genotypes (Fig. 1C). As discussed below, this maintenance of islands of heterozygosity among many large regions with loss of heterozygosity (LOH) is difficult to explain by multiple independent deletions.
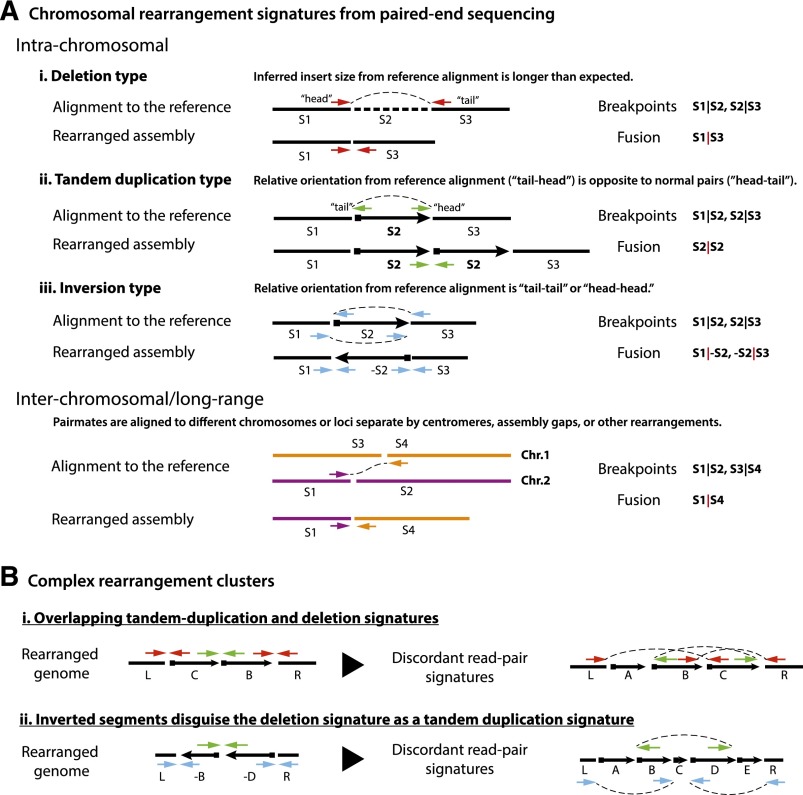
Figure 2.
Signatures of chromosomal rearrangements from paired-end sequencing. (A) Four major types of chromosomal rearrangement signatures deduced from paired-end sequencing: deletion type, tandem duplication type, inversion type, and long-range type (including interchromosomal). Each type of read pair shows aberrations from proper pairs when aligned to the reference assembly, which implies a new junction in the test chromosome. A deletion (read pairs shown as red arrows) or tandem duplication (shown as green arrows) only introduce one fusion and hence can be unambiguously inferred from a single type of signature (in the absence of other overlapping events). Inversion events, however, introduce at least two rearranged junctions (shown here is a simple inversion); hence, the rearranged genome can only be constructed by considering all related events. The same is true for long-range events with pair mates residing in different chromosomes or separated by centromeres, assembly gaps, or other rearrangements. In general, it is necessary to incorporate copy number information together with rearrangement signatures to derive the structure of the rearranged genome (for a computational framework, see Greenman et al. 2012a). (B) Examples of complex chromosomal rearrangements exhibiting multiple overlapping rearrangement signatures that do not reflect the same events as the “typing.” The discordant read pair signatures (color arrows) are shown both in the rearranged genome and according to their alignment to the reference genome. (i) A complex rearrangement resulting in the deletion of segment A and swapping of segments B and C generates two “tandem duplication” and one “deletion” read pair signatures, none of which corresponds to a simple event. (ii) Two inversions alter a deletion signature into a tandem duplication signature (green arrows).
A third feature of the observation was that the pattern of end-joining strongly suggested an origin from a DNA double-strand break (DSB). As highlighted in later studies (Kloosterman et al. 2011a, 2012; Chiang et al. 2012), the preponderance of rearrangements involved ligation of both ends of a single DNA break to other genomic segments (ends being operationally defined as sequences within ∼1 kb of each other; hereafter referred to as a pairing of ends). This contrasted with complex genomic rearrangements that arise by replication-based mechanisms (Lee et al. 2007; Carvalho et al. 2009, 2011) where the rearrangement is often seen only on one side of the initial lesion (Haber and Debatisse 2006; see below for more details).
The final defining feature of this new mutagenesis event was the nonrandom, highly localized clustering of chromosomal breakpoints. These four hallmarks—alternating copy number profiles, loss and preservation of heterozygosity, pairing of the breakpoint ends, and clustering of rearrangements—define the class of chromosomal alteration termed chromothripsis (Greek for shattering of chromosomes).
All at once?
The defining hallmarks of chromothripsis cannot be explained by a gradual accumulation of independent events. First, truly independent chromosomal rearrangements are expected to be randomly distributed across the genome rather than clustered in focal regions (Beroukhim et al. 2007, 2010). Second, the gradual accumulation of such a large number of overlapping rearrangements, both copy number neutral and copy number changing, would almost certainly produce a final outcome with more than two copy number states. Finally, maintenance of islands of heterozygosity interspersed between regions that have lost heterozygosity is hard to explain by multiple sequential events. This is illustrated by the fact that a deletion early in the process will remove sequences that cannot be regained. Thus, if the observed pattern resulted from sequential mutation events, deletions would need to be restricted to a later time period, when they could not interrupt the acquisition of balanced rearrangements—a complex “programmed” event for which it is hard to envision a mechanism (one example is shown in Fig. 3).
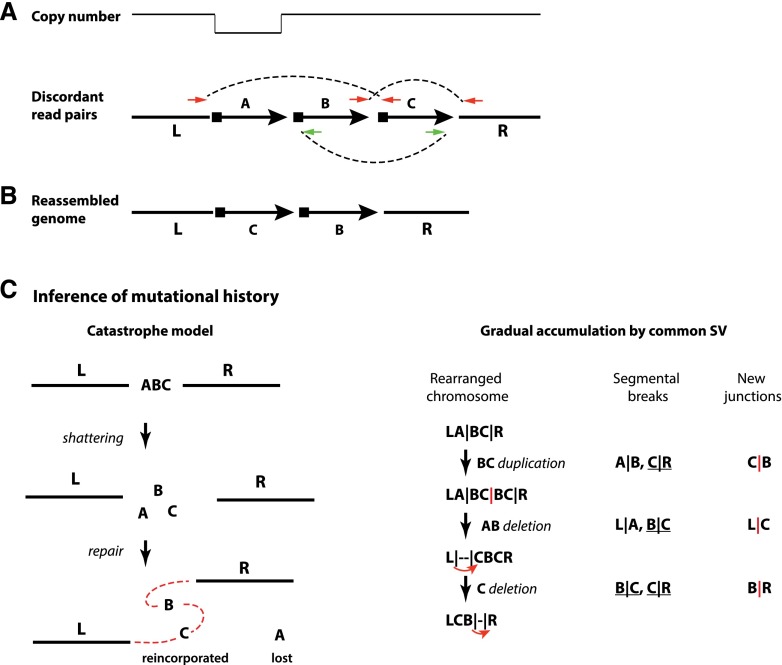
Figure 3.
Reconstruction of a local complex rearrangement and its mutational history from read pair signatures and copy number information. (A) Copy number alterations can be detected by array methods or read depths in sequencing data; rearrangement signatures can be detected from discordant read pairs in sequencing data. (B) By combining copy number information and rearrangement signatures, one can construct the configuration of the rearranged segments in the chromothriptic chromosome. (C) The history of mutation events that could have generated the outcome in B is often not unique. However, all solutions have to break four junctions in the reference assembly (L|A, A|B, B|C, and C|R) and generate three new junctions (L|C, C|B, and B|R). A catastrophe model can easily solve this puzzle because all of the breaks and translocations occur all at once. By comparison, a solution from the gradual model of simple events requires at least three simple events to generate the final configuration (one for each new junction). As each event causes two breakpoints, three events need six breakpoints, which requires two additional breaks in addition to the four original junctions to be broken. These two additional breaks arise from either within the lost A segment (lost history) or hitting the same junction multiple times (recurrent breaks). Shown here is one possible solution that involves recurrent breaks at both B|C and C|R junctions. Both lost history and recurrent breaks are highly unlikely. As the number of rearranged fragments increases, the need to include these types of unlikely events further increases, strongly arguing against the gradual model.
In contrast to the gradual model, Stephens et al. (2011) proposed a much more compelling catastrophic model of chromothripsis that offers a parsimonious explanation of the observation: One chromosome (or regions from one or more chromosome arms) is shattered into segments of different lengths in a single catastrophic event; the segments are then stitched together, most likely by nonhomologous end-joining, resulting in a derivative chromosome containing a subset of the shattered segments in random orientations. The retained segments maintain the parental copy number and heterozygosity, yielding the high copy number state. The segments that are not incorporated into the derivative chromosome are either lost, yielding the low copy number state, or incorporated into a double-minute chromosome, as discussed below.
Historiography of the cancer genome
These general arguments make intuitive sense but ideally would be backed up by a detailed molecular assembly of the evolutionary path taken to generate the chromothriptic chromosome. Stephens et al. (2011) faced the obvious challenge of inferring prior history from a terminal snapshot of the present, which is already difficult for cancer genomes with simple rearrangement patterns (Greenman et al. 2012a) and quickly becomes impractical for a complex case such as chromothripsis (Yates and Campbell 2012).
The first complication for inferring an evolutionary trajectory is purifying selection: The mature tumor is biased to preserve genomic changes that are beneficial to tumor development and to have lost deleterious changes (Beroukhim et al. 2007; Greenman et al. 2007; Bignell et al. 2010). By viewing only the endpoint of selection, we could miss intermediate steps in the development of the rearranged chromosome whose signatures were lost during tumor evolution. An additional technical complication for solving the genomic Rubik's cube for the chromothriptic chromosome is that one cannot interpret paired-end sequences in a simple straightforward way when they occur in the context of other overlapping rearrangements. For example, a discordant pair of sequencing reads that is inferred to be a “tandem duplication type” event when directly mapped onto the reference genome sequence (Fig. 2A; Raphael et al. 2003; Tuzun et al. 2005; Korbel et al. 2007; Campbell et al. 2008; Medvedev et al. 2009) has a completely different interpretation under a rearranged genome when additional rearrangement events have occurred between the pair mates (Fig. 2B). This technical issue is less problematic for simple genomes where rearrangements seldom overlap and can be decoupled and interpreted unambiguously (Greenman et al. 2012a,b; Quinlan and Hall 2012). However, it becomes prohibitively complex in chromothripsis, where tens or hundreds of rearrangements overlap, resulting in an enormous number of possible permutations. This provides an important cautionary note for aspiring genome analysts: An accurate interpretation of a highly rearranged cancer genome from paired-end sequencing data requires an integrative analysis of rearrangement signatures (such as deletion type or tandem duplication type) (Fig. 2A) and copy number data. In other words, context matters.
The following back-of-the-envelope calculations illustrate the magnitude of the problem. The number of different outcomes created by random permutations of 10 overlapping rearrangements is 10! = 3,628,800 and the number grows to ∼1064 for 50 rearrangements. Typical chromothripsis harbors tens or even hundreds of overlapping rearrangements, and it is clearly impossible to enumerate all possible outcomes. Additional complications are caused by intermediate events being obscured by later events; e.g., a later deletion event that removes the signature of an earlier translocation. This estimate also does not account for rearrangements that are not detectable due to technical issues such as uncertainties in the reference genome assembly or inaccuracies in short-read alignment.
The most difficult challenge stemming from the vast number of outcomes due to permutation complexity, however, is finding not just one solution, but the most likely solution based on a certain background statistical model of tumor evolution. Ultimately, we want to know the likelihood of the multievent solution to chromothripsis based on the background model of tumor evolution to dismiss the gradual model in favor of a single catastrophe. (Fig. 3 illustrates this problem by a simple example.) To answer this last question without enumerating all possible outcomes of a gradual model, Stephens et al. (2011) focused on the number of copy number states during genome evolution, essentially reducing the multidimensional space of chromosome configurations to a single dimension of “copy number.” They set up simulations in which individual rearrangement events were applied to the genome sequentially, starting from the reference sequence. Instead of brute force enumeration of all genomic configurations, the eventual number of copy number states was scored, making a testable prediction about the structure of the chromothriptic chromosome without the need for a complete molecular reconstruction of the rearranged chromosome. The gradual model rapidly generated multiple high copy number states because gradual accumulation allows for overlapping tandem duplications to affect the same region multiple times. The discrepancy between the large copy number complexity implied by the gradual model and the hallmark two copy number states observed in chromothripsis provided a strong statistical argument against a gradual model for chromothripsis. It is worth noting that chromothriptic chromosomes can sometimes exhibit multiple copy number states (Stephens et al. 2011; Rausch et al. 2012). One explanation is the overlap between chromothripsis and other independently generated copy number gains; another possibility is that chromothripsis that occurred during or after DNA replication can result in a shattering and assembly of both sister chromatids to generate copy number gains. In either scenario, however, the low copy number states (three or four) are still in stark contrast to the rapid accumulation of multiple copy number states, as the gradual model suggests.
The focal pattern of breakpoints in chromothripsis provides yet another argument against the gradual model. Independent rearrangements are generally assumed to be randomly distributed across the genome. The large size of the human genome implies that it is extremely unlikely to see two events that are very close to each other (∼1 kb apart). The likelihood of seeing tens or hundreds of breakpoints within one chromosome (or chromosome arm) with few events elsewhere in the genome is vanishingly small if these events were independent and not due to specific targeted mechanisms. This statistical argument suggests that clustered rearrangements in chromothripsis very likely occurred simultaneously. A generalization of this statistical argument into a systematic way of inferring co-occurring translocations (“chained” translocations) was recently proposed (Baca et al. 2013). This new approach, discussed further below, potentially provides a complementary method to validate the idea that chromothriptic lesions occurred at the same time.
In summary, chromothripsis is best explained by a catastrophic all-at-once model and cannot be reconciled with the gradual model. The catastrophic model yields a simple explanation of the low number of observed copy number states. It also provides an explanation for the many overlapping tandem duplication type events that do not increase copy number (see Fig. 2B, i, for an example): These duplication type events are instead explained by the random orientations at which the segments are stitched together. Similar explanations can apply to deletion and other “type” events (Fig. 2B).
When is it chromothripsis?
The discovery of chromothripsis in cancer immediately raises the question of how frequently it occurs. After the initial discovery, there have been a plethora of studies confirming that chromothripsis is indeed a widespread and previously unappreciated phenomenon in both primary tumors (Kloosterman et al. 2011b; Magrangeas et al. 2011; Berger et al. 2012; Rausch et al. 2012) and cancer cell lines such as HeLa (Adey et al. 2013; Landry et al. 2013). Many cancer genomes reported in earlier studies also exhibited features that resemble chromothripsis (Stephens et al. 2009; Campbell et al. 2010). So how are we to recognize chromothripsis? How many rearrangements “make” the definition, and how focal is focal? Because copy number data are the most available, a common sense approach was to adopt operational definitions that would require, say, 10, 20, or 50 oscillating copy number changes (Magrangeas et al. 2011). In some cases, corrections were applied to normalize the number of focal events to the general frequency of events on that chromosome (Kim et al. 2013).
Although an operational criterion is reasonable for a first-pass screening and many instances with multiple oscillations between two copy number states likely are bona fide chromothripsis, the field is moving away from operational definitions and adopting criteria that incorporate information from paired-end sequencing. Through more precise analysis of the structure of the derivative chromosome, one can better distinguish chromothripsis from a large number of progressively acquired rearrangements in tumors with high levels of genetic instability. Because an excellent review by Korbel and Campbell (2013) has recently covered this topic in depth, we only briefly review the main elements of these criteria here.
As discussed above, one hallmark of chromothripsis is the clustering of rearrangements. The degree of clustering can be assessed by simple statistical methods because the null hypothesis of random breakpoints predicts that the distance between breakpoints should be distributed exponentially (Beroukhim et al. 2010; Malhotra et al. 2013). However, it is important to note that clustering of breakpoints can also be generated by other processes involving multiple steps over several cell division cycles. For example, clustering can result from BFB cycles (Gisselsson et al. 2000), chromosome fragile sites (Debatisse et al. 2012), and strong positive selection for local oncogene amplifications (Bignell et al. 2007). Thus, clustering by itself is a necessary but not sufficient criterion for chromothripsis. An additional criterion is provided by the fact that chromothripsis is characterized by regions with interspersed retention and LOH, which can be verified from genotype information from either SNP array or sequencing. Finally, if restricted to a single chromosome, the regions of rearrangement and segment loss should be confined to a single parental haplotype because the shattering and stitching back together occurs (mostly) on a single chromatid. This can be directly verified by cytogenetic methods, such as fluorescence in situ hybridization (FISH), or inferred from phased haplotypes in trio family studies (Browning and Browning 2011).
Additional information from sequencing can evaluate the model that a chromosome has been shattered and randomly stitched together in more detail. First, the model predicts that the order of the fragments on the derivative chromosome would be random or near random with respect to their positions on the reference genome. Second, if reassembly of the derivative chromosome is random, the orientations of fragments on the derivative chromosome should also be random. This means that there should be no bias for fragments to be joined head to tail (deletion type), tail to head (tandem duplication type), head to head, or tail to tail (inversion type). Such randomness in the orientation of fragments contrasts with, for example, BFB cycles, which are characterized by inverted segments biased to be head to head or tail to tail (Bignell et al. 2007). Finally, if the shattering and restitching indeed happen all at once, then each rearranged fragment will have a unique head and tail sequence (with respect to the reference sequence). Thus, when “walking” down a chromothriptic chromosome, there will be an invariant alternation between head and tail (or tail and head) sequences, reflecting a linear connection (Korbel and Campbell 2013). This contrasts with the prediction for a mechanism involving sequential acquisition of rearrangements, particularly those involving amplifications. In this latter case, rearranged segments could be “reused” in ways that would generate heads followed by heads and tails followed by tails when amplifications are nested.
Clearly, within a short period of time, the field has advanced far beyond “know it when you see it” operational definitions for chromothripsis. The above-delineated criteria each address a different hallmark feature of chromothripsis and can be subjected to rigorous statistical evaluation. Nevertheless, challenges remain. How exactly to combine multiple criteria and weigh them properly to make the most probable case for or against chromothripsis needs further development. There are also numerous technical challenges. Short sequence reads can only be aligned to the mappable regions of the genome, and the interesting question of what happens within highly repetitive sequences remains to be resolved. Complications also come from other factors, such as inaccuracies in short-read alignments, the obscuring effect of other structural rearrangements (e.g., simple deletions/amplifications/translocations) layered on top of chromothripsis, tumor heterogeneity, and aneuploidy. All of these technical factors may lead to an underestimate of the frequency of chromothripsis.
How frequently does chromothripsis happen?
So what, then, is our best guess for the frequency of chromothripsis? Unfortunately, the largest volume of cancer genomic data (such as in The Cancer Genome Atlas [TCGA] project) is array-based and does not have the power to provide an accurate estimate of the overall frequency of chromothripsis. However, it is becoming evident that the frequency of chromothripsis tends to vary between different tumor tissue types. In a survey of whole-genome sequencing data for 64 samples of seven tumor types from TCGA, Malhotra et al. (2013) found no chromothriptic rearrangements in basal-like breast cancers, colon adenocarcinomas, ovarian cancers, or renal adenomas but found rearrangements consistent with chromothripsis in two of six (33%) lung adenocarcinomas, two of 13 (15%) lung squamous cell carcinomas, and 39% of glioblastomas (n = 18). In a recent survey of 4934 cancers from TCGA, Zack et al. (2013) employed multiple criteria to model chromothripsis by SNP array copy number alone and suggested that chromothripsis occurred in 5% of all samples, with frequencies ranging from 0% in head and neck squamous carcinoma to a maximum of 16% in glioblastoma. In addition, chromothripsis has been observed in colorectal cancer (Kloosterman et al. 2011b), melanoma (Berger et al. 2012; Hammond et al. 2013; Hirsch et al. 2013), small-cell lung cancer (Iwakawa et al. 2013), prostate cancer (Wu et al. 2012), and a variety of brain tumors (Molenaar et al. 2012; Northcott et al. 2012; Brastianos et al. 2013) and blood malignancies (MacKinnon and Campbell 2013; Morin et al. 2013; Nagel et al. 2013). It is interesting to point out that the rate of chromothripsis seems to be unrelated to the overall rate of somatic copy number alterations (SCNAs) (Zack et al. 2013).
The high frequency of chromothripsis in certain tumor types suggests that chromothripsis depends on the genetic and environmental background of cancers. Among the most frequently mutated genes in human cancer is TP53 (Olivier et al. 2010), which plays central roles in maintaining genome stability (Zhu et al. 2002; Meek 2004; Thompson and Compton 2010). The correlation between TP53 mutation and chromothripsis was first demonstrated in Sonic Hedgehog (SHH) medulloblastoma. Rausch et al. (2012) applied high-throughput whole-genome sequencing and long-fragment mate-pair sequencing to characterize genomic rearrangements in four patients of SHH medulloblastoma who also had germline-inactivating TP53 mutations (Li-Fraumeni syndrome). Sequencing of each of these tumors not only confirmed clustering of chromosomal breaks indicative of chromothripsis but also revealed high-level amplifications due to double-minute chromosomes that originated from the shattered fragments. Thus, TP53 mutations are permissive or conducive for either the initiating chromothriptic event, the selective advantage of cells bearing the rearranged chromosome, or both. Rausch et al. (2012) further identified strong correlations between TP53 mutation and chromothripsis in acute myeloid leukemia, demonstrating that the effect of TP53 mutation can manifest in multiple tissue types.
How accurate are the estimates of the frequency of chromothripsis? Although there could be some overestimation due to ambiguities in the interpretation of genomic data, we must also consider underestimation due to the fact that the overwhelming majority of chromothriptic events are almost certainly lost because of deleterious fitness effects. Negatively selected events in normal or premalignant tissues could be detected only at a single-cell level immediately after the chromothriptic rearrangement occurs, which is currently not feasible on the appropriate scale. Ultimately, understanding the mechanisms that generate chromothripsis will be integral to estimating its frequency. If the underlying cellular defects could lead to, but not cause, chromothripsis every time they occur, then the observed cases of chromothripsis only represent the tip of the iceberg for less dramatic but possibly more common mutational events.
Does chromothripsis cause cancer?
In general, we expect that the disruption of chromosomal integrity in a catastrophe such as chromothripsis will be negatively selected or even trigger apoptosis. However, this does not exclude the possibility that rare catastrophes could promote cellular transformation, which could happen in at least two ways through copy number alterations. First, the massive DNA loss can include one or more tumor suppressor genes (Stephens et al. 2011; Yang et al. 2013). In their survey of the TCGA SNP array data, Zack et al. (2013) found that 72% of chromothripsis events resulted in SCNAs in regions that are frequently disrupted in cancer, with the most dramatic case in glioblastoma showing that 20 out of 22 samples with chromothripsis had one event disrupting the CDKN2A locus. Second, and perhaps most importantly, shattered chromosomal segments not incorporated into the derivative chromosome can be linked to form a double-minute (circular) chromosome. These double minutes are readily amplified, which commonly occurs if they harbor an oncogene. An illustration of this comes from the original reports on chromothripsis (Stephens et al. 2011; Rausch et al. 2012). Stephens et al. (2011) described a small-cell lung cancer cell line where a chromothriptic event generated a derivative chromosome 8 and a double minute containing MYC and other segments of chromosome 8. Most notably, the sequences on the derivative chromosome and the double minute were exclusive, providing compelling evidence that early in the development of this tumor, chromosome 8 was shattered, with some bits being contemporaneously incorporated into the derivative and others going into the double minute.
Having recognized the association between chromothripsis, double minutes, and cancer-causing mutations, one can infer chromothripsis as the most likely underlying mechanism in the formation of high-level focal amplifications that incorporate multiple disconnected segments (Iwakawa et al. 2013; Sanborn et al. 2013). The multiple focal amplifications can be either extrachromosomal (double minutes) or intrachromosomal (homogeneously staining regions [HSRs]) due to chromosomal integration of double minutes (Storlazzi et al. 2010; Gibaud et al. 2013). Indeed, data linking the formation of double minutes to the formation of derivative chromosomes containing complex rearrangements were known prior to the first description of chromothripsis. Gibaud et al. (2010) characterized five different extrachromosomal amplifications of double-minute chromosomes in a glioma sample: One double-minute chromosome consisted of multiple segments from chromosome 5 and chromosome 9, which, in retrospect, was likely to have resulted from chromothripsis. FISH analysis verified that the extrachromosomal amplicon sequences were not present in the derivative chromosomes derived from chromosome 5 and chromosome 9. We further note that copy number profiles of both chromosomes showed the hallmark copy number patterns suggestive of chromothripsis. In addition, the rearrangements also created oncogenic lesions. The double minute contained the TERT (telomere reverse transcriptase) gene from a fragment in chromosome 5. TERT is amplified or overexpressed in many cancer types (Beroukhim et al. 2010). The functional significance of TERT overexpression is remarkably underscored by the recent discovery that ∼80% of glioblastomas harbored mutations in the 5′ flanking region of TERT that activate its expression (Horn et al. 2013; Huang et al. 2013; Killela et al. 2013). Interestingly, the glioma sample studied by Gibaud et al. (2010) also had deletions encompassing the CDKN2A locus on the chromosome 9 derivatives. Thus, one episode of chromothripsis appears to have generated two critical oncogenic lesions (TERT amplification and CDKN2A deletion).
Besides copy number alterations, translocations in chromothripsis can also lead to gene fusions or dysregulated gene expression. Mehine et al. (2013) suggested that chromothripsis is a frequent driver event in uterine leiomyomas, resulting in increased expression of the translocated HMGA1 and HMGA2 genes. However, the probability of generating in-frame gene fusions—the major class of oncogenic translocations—through random chromosomal translocations is generally small (Stephens et al. 2009), and the observed frequency of in-frame fusions due to chromothripsis is comparable with those resulting from simple rearrangement events (Stephens et al. 2011). Nonetheless, although each rearranged segment in chromothripsis has a small probability of generating a functionally important fusion gene (Teles Alves et al. 2013), it is reasonable to speculate that chromothripsis, because it generates many rearrangements, will increase the frequency of such fusions. This is not because chromothripsis has any unique ability to generate gene fusions but simply reflects the fact that in chromothripsis, more shots are taken on goal.
Although there is good reason to think that chromothripsis can generate oncogenic alterations and potentially multiple mutations at one fell swoop, we are just starting to understand the impact of chromothripsis on cancer pathogenesis or prognosis. Recent studies have shown that chromothripsis can independently arise in either the primary tumor or its metastasis (Kloosterman et al. 2011b) and does not necessarily predict resistance to drug treatment (Bassaganyas et al. 2013). Even though chromothripsis has been suggested as a poor prognostic factor (Molenaar et al. 2012; Hirsch et al. 2013), direct experimental characterization of chromothripsis-derived mutations is lacking.
One important question is when chromothripsis occurs during tumor development: Can it be an early driving event, or is it just another consequence of genomic chaos in late stage cancers? The latter possibility is intuitive, but there is also interesting evidence for chromothripsis occurring in the absence of massive genetic instability. First, allele frequencies of chromothriptic rearrangements suggest that chromothripsis generally occurs earlier than simple rearrangements (Malhotra et al. 2013). Moreover, the high frequency of chromothripsis identified in tumors with low to moderate mutation rates (neuroblastoma, medulloblastoma, glioblastoma, and blood and bone marrow malignancies) suggests that chromothripsis does not, in any obligatory way, require a background of elevated genomic instability. On the other hand, tumor types with frequent copy number alterations, such as ovarian or lung cancers, exhibit a larger number of chromosomal alterations, but they are often independently acquired rather than correlated as in chromothripsis (Malhotra et al. 2013; Zack et al. 2013). Additionally, as described in more detail below, de novo complex rearrangements that resemble chromothripsis have also been observed in patients with congenital disorders. So far, there has been no evidence that these patients have a cancer predisposition or defects in the maintenance of genome stability like patients with germline mutations in the TP53 or BRCA genes. Together, these findings imply that chromothripsis can originate in cells with normal or near-normal genetic stability.
Chromothripsis in congenital disorders
Strong selection during embryonic development ensures that only mild genomic alterations can be tolerated through development. It was therefore very surprising when recent studies reported massive constitutional chromosomal rearrangements similar to those of chromothripsis in cancer in some patients with congenital developmental disorders (Kloosterman et al. 2011a, 2012; Chiang et al. 2012; Nazaryan et al. 2013). In comparison with chromothripsis in cancer, these instances of apparent germline chromothripsis showed minimal DNA loss, presumably reflecting the selection against haploinsufficiency or complete loss of gene function due to monoallelic gene expression. Array-based copy number analysis, such as array comparative genome hybridization (aCGH) or SNP arrays, cannot, as expected, detect these copy-neutral chromosomal alterations, necessitating whole-genome sequencing.
Chromoanasynthesis
A different pattern of constitutional complex genome rearrangements was reported by Liu et al. (2011) in their investigation of patients with congenital developmental defects. The investigators combined aCGH and FISH to characterize copy number alterations and chromosomal rearrangements in these patients and sequenced the PCR products at rearrangement junctions to resolve breakpoint sequences. In contrast to chromothripsis, Liu et al. (2011) observed frequent copy number gains with interspersed regions containing duplication or triplication of one parental allele but no LOH (Fig. 1D). Breakpoint junction sequences exhibited microhomology but also included templated sequence insertions (100–1000 base pairs [bp]) on the rearranged chromosome. The short template insertions suggested a replication-based mechanism for incorporating these segments during the repair of DNA DSBs (Lee et al. 2007; Hastings et al. 2009). The observation of microhomology further led Liu et al. (2011) to propose microhomology-mediated breakage-induced repair (MMBIR) as the major mechanism for generating the observed translocations (Carvalho et al. 2009; Zhang et al. 2009b; Liu et al. 2012). In this model, the extra allele copies are incorporated due to template-switching events in DNA replication instead of ligation (via end-joining) of shattered fragments. To emphasize the difference in mechanisms, Liu et al. (2011) termed this phenomenon chromoanasynthesis (synthesis of new chromosomes). They also suggested that the same replicative mechanism may play a role in chromothripsis due to similar features in both phenomena, such as localized clusters of rearrangements and copy number alternations.
One of the hallmark features of chromothripsis is interspersed loss and preservation of heterozygosity, which suggests a “shatter-and-stitch” process of one chromatid. This is in sharp contrast to chromoanasynthesis, which reflects extra synthesis of DNA segments. Although extra synthesis can lead to alternating DNA copy numbers, it does not necessarily result in LOH: LOH occurs by DNA deletion or gene conversion. Another difference between chromoanasynthesis and chromothripsis is that adjacent chromosomal ends in chromothripsis (especially in the germline) often result from one DNA DSB, whereas segmental breaks in chromoanasynthesis are separated by longer distances (0.01∼1 Mb) and appear to result from template-switching events. Although replication-based DNA repair can contribute to chromothripsis, as evidenced from examples of templated insertion sequences (Chiang et al. 2012), it is difficult to explain the proximity of adjacent chromosomal breaks based purely on template switching without DNA DSBs. Finally, chromoanasynthesis results in one aberrant chromosome with duplicated segments due to extra DNA synthesis; in contrast, chromothripsis generally gives rise to one or multiple derivative chromosomes with either massive DNA loss (in cancer) or no apparent DNA copy number alterations (in the germline). Based on these differential characteristics, we agree with the conclusion that chromoanasynthesis is a distinct phenomenon from chromothripsis (Holland and Cleveland 2012; Liu et al. 2012; Kloosterman and Cuppen 2013).
The distinction between chromothripsis and chromoanasynthesis is also evident in a more recent report of germline genomic rearrangements. Kloosterman et al. (2012) identified eight cases that resemble chromothripsis, exhibiting random stitching of segments from multiple chromosomes with little DNA loss, and two cases with apparent gain or deletion of DNA segments that are suggestive of replication errors. One interesting observation is that the rearranged alleles in both chromothripsis and chromoanasynthesis are preferentially of paternal origin; this was attributed to errors that result from the higher number of mitotic divisions in gametogenesis in males relative to females (Kloosterman and Cuppen 2013). In contrast, most human germline aneuploidies are maternally derived (Handyside et al. 2012; Nagaoka et al. 2012), and de novo chromosomal instability in early embryonic development does not show a preferred origin (Vanneste et al. 2009; Voet et al. 2011).
Chromoplexy
Chromothripsis and chromoanasynthesis are two examples of complex genomic rearrangement triggered or generated by a single event. A single catastrophic event was also invoked to explain a recently described pattern of rearrangement found in prostate cancers (Fig. 1E; Berger et al. 2011; Baca et al. 2013). Termed chromoplexy (Greek for braid of chromosomes), this event is characterized by “chains” of translocations involving multiple chromosomes—up to eight so far described. The sequence signatures of chromoplexy suggest that all of the partner segments involved in the translocations originate from DNA DSBs, with both ends of each break contributing to the daisy chain cycle of translocations (Fig. 4A). This organization of breakpoint ends bears some similarity to the pairing of the ends described above in chromothripsis. As with chromothripsis, the pairing pattern strongly implies that chromoplexy is another example of an all-at-once mechanism for chromosomal rearrangement: If the breakpoint ends originate from DSBs and are joined into an integrated pattern of translocations, then the breaks must have occurred simultaneously.
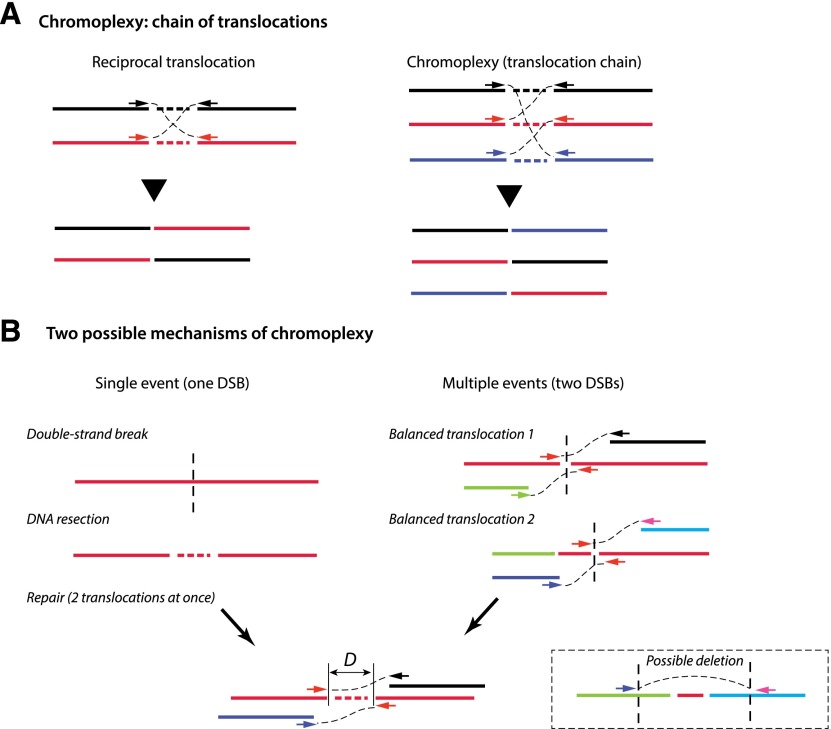
Figure 4.
Chromoplexy and ChainFinder. (A) Chromoplexy is an extension of balanced rearrangements to multiple partners. The different colors of linked discordant pair mates reflect different chromosomes or loci to which each mate is aligned. DNA resection during the repair of DSBs often results in loss of a small segment between the two ends. (B) Adjacent translocations of two breakpoint ends can arise in two different ways. (Left) In a single-event model, a single DNA DSB generates two broken ends with a small deletion due to DNA resection; joining of the two broken ends with ends from other DNA breaks produces the observed translocations. (Right) In a multiple-event model, two independent balanced translocations occur independently at different times; the inner DNA segment could have been lost in a subsequent deletion event (shown in the dashed box) or due to DNA resection. The distance between the two adjacent translocation ends, D, provides a statistical measure of the likelihood of either scenario. The two-event model implies that two independent breaks must have occurred within distance D. This likelihood is approximately given by 1 − exp(−μD), where μ is the average density of chromosomal breaks that can be estimated for each sample at any given locus (≲10−6 per base pair). When the ends are very close (100–1000 bp), this probability is vanishingly small, favoring the single-event model and rejecting the multievent model. The central idea of ChainFinder is to find “chained” translocations that form a closed cycle and where the multiple-event model can be rejected for any two pairs of adjacent translocations (i.e., no reciprocal translocation can be postulated as an intermediate state, connecting two smaller chains).
To put the intuitive argument into a more rigorous framework, Baca et al. (2013) developed an algorithm called ChainFinder to identify the chained translocations of chromoplexy. The assumption is that the probability that the observed translocations of two adjacent breakpoint ends occurred simultaneously is related to the probability that two breakpoint ends originate from a single DSB. The inference of a DSB is not straightforward because the breakpoint ends originating from a single DSB due to resection and other processing are not precisely connected in the reference sequence (see Fig. 4B for an illustration); a null model for independently generated breaks is therefore necessary for a statistical inference. To create the null model of independently generated chromosomal breaks, ChainFinder first estimates the density of chromosomal breaks at any locus by averaging over multiple tumor samples and then multiplying this by the total number of chromosomal breaks detected in a particular sample. This generates a sample-specific, locus-dependent frequency of breaks. Based on the null model of local breakage frequency, ChainFinder calculates the probability that two adjacent translocation ends have originated from independent breaks (Fig. 4B). This probability monotonically decreases with the separation between the two ends and becomes vanishingly small when the two ends are very close (100∼1000 bp), enabling a rejection of the null model that the two translocations occurred independently. ChainFinder then searches for all “cycles” of rearrangements (closed chains of rearrangements) and tests the null model for every two pairs of adjacent breakpoints: If both pairs of ends have a reasonable chance of having originated from independent breaks, then these two pairs of ends can arise from two independent events, and the original cycle is reduced to two smaller cycles. Chromoplexy events are therefore defined as chains of translocations that cannot be further reduced.
The ChainFinder algorithm is conceptually different from and potentially complementary to the Monte Carlo simulations used by Stephens et al. (2011) to infer that complex rearrangements have resulted from a single catastrophic event. The Monte Carlo simulations monitor DNA segment copy number as a surrogate, enabling rejection of the gradual model without directly testing the all-at-once model. In contrast, ChainFinder directly examines the interdependency of rearrangements and thus gets closer to a direct test of the all-at-once hypothesis. The complex patterns of rearrangements in chromothripsis are almost certainly “chained”; it is thus possible to apply ChainFinder to deconvolute chromothripsis and chromoplexy from other independent rearrangements that occur on the same chromosome or its homologous counterpart. Malhotra et al. (2013) adopted a similar heuristic idea to search for clusters of translocations to infer chromothripsis. A more rigorous generalization of the idea of “two simultaneous translocations due to one DSB” as judged from the spatial proximity of breakpoint ends could be a powerful method to deconvolute the evolution history of rearranged genomes.
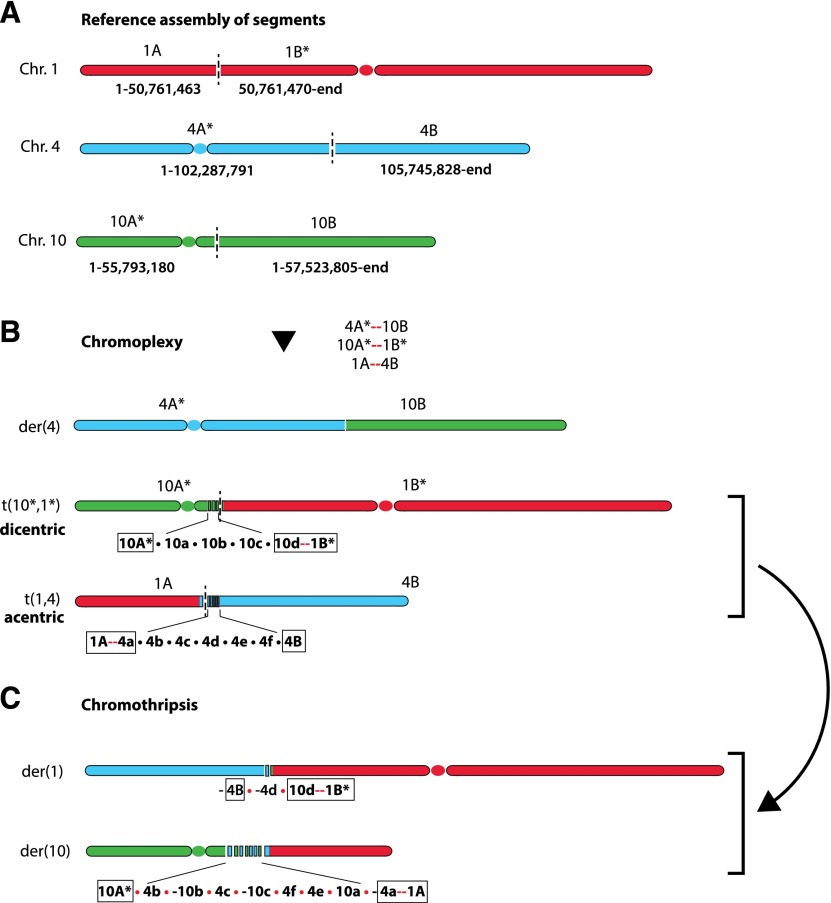
The following example illustrates the potential for a more general application of this type of approach to resolve complex rearrangements into smaller chains and decipher the history of events during tumor evolution. In an early report of germline chromothripsis (Kloosterman et al. 2011a), we noticed that the complex rearrangements of multiple chromosomal segments in three chromosomes (1, 4, and 10) can possibly be reduced to two cycles: the first cycle containing three DSBs originating from each chromosome (Fig. 5A,B) and the second containing only DSBs in chromosome 4 and chromosome 10 (Fig. 5B,C). Because the pairs of adjacent breakpoint ends are close to each other (<1000 bp), the translocations are almost certainly chained together in each cycle. Interestingly, if the first chain of three translocations occurred all at once, the result would have been a functional centromere-containing derivative chromosome, t(4*,10q), which is retained in the final chromosomal configuration, plus two mitotically unstable chromosomes: a dicentric chromosome, t(10*,1*), and an acentric chromosome fragment, t(1p,4q) (Fig. 5B). The two mitotically unstable derivatives would either be lost or undergo additional rearrangement. It is therefore possible that unstable chromosomes generated by the original chain of translocations (chromoplexy) could result in subsequent catastrophes that add chromothripsis on top of the original translocations. This speculative model, which we term translocation-induced chromothripsis, is one potential mechanism for chromothripsis involving multiple chromosomes.
Figure 5.
Possible decomposition of rearrangements reported by Kloosterman et al. (2011a) into two chains. The first chain is plausibly a chromoplexy event that swaps partners between six chromosomal arms, shown in A. Asterisks denote chromosomal segments containing a centromere. Newly formed junctions are shown as red links. This chromoplexy episode would create one functional (single centromere-containing) derivative chromosome, t(4p*, 10q), plus two aberrant ones, t(10p*,1q*) (dicentric) and t(1p,4q) (acentric), as shown in B. The unstable dicentric and acentric chromosomes could then have triggered the remaining rearrangements shown in C, which are interlinked and resemble a chromothripsis. The outcome of this second chain is the swap of partners between the two nonfunctional chromosomes 10A*–1B* and 1A–4B, which results in two new derivative chromosomes, 10A*–(-1A) and (-4B)–1B*. This event is accompanied by fragmentation and repair of multiple segments near the junctions from the first chromoplexy event (10a, 10b, and 10c; and 4b, 4c, 4d, 4e, and 4f).
Mechanisms for complex genome rearrangements
The discovery of chromothripsis, chromoanasynthesis, and chromoplexy suggests the existence of novel processes impacting genome architecture. The underlying mechanisms are likely to be different for each phenomenon. However, any mechanisms need to explain the common theme of single-event-mediated extensive damage and, for chromothripsis and chromoanasynthesis, the fact that the damage is highly localized. Little is known about these underlying mechanisms, but we briefly review current thinking below.
For chromoanasynthesis, the sequence signatures at translocation junctions strongly implicate replication-based mechanisms (Liu et al. 2011, 2012). The fact that chromoanasynthesis is characterized by apparent template insertions from nearby genomic regions, often contains microhomology at the breakpoint junctions, and lacks the sequence signatures of DSBs strongly implicates replication template-switching mechanisms. These mechanisms have been termed fork stalling and template switching (FoSTeS) and MMBIR (Slack et al. 2006; Lee et al. 2007; Carvalho et al. 2009; Hastings et al. 2009; Zhang et al. 2009a,b). Best characterized in yeast (for recent reviews, see Anand et al. 2013; Lambert and Carr 2013), these mechanisms can account for duplications (Narayanan et al. 2006; Hicks et al. 2010) and fold-back inversions (Mizuno et al. 2013). The template switches can arise by recombination involving invasion of homologous sequences or the use of microhomologies to prime ectopic DNA synthesis. Also consistent with a role for replication defects in this type of errors, replication stress induced by hydroxyurea or aphidicolin can induce copy number variation in human cells (Arlt et al. 2009, 2012). Replication defects also can explain the regional bias for the abnormalities of chromoanasynthesis: There is evidence that breakpoint ends of copy number abnormalities tend to be in physical proximity (Lieberman-Aiden et al. 2009; De and Michor 2011; Fudenberg et al. 2011); hence, different template-switching events could have occurred in loosely defined nuclear territories. Nevertheless, without direct experimental recapitulation of chromoanasynthesis, the inferred models for the mechanism, although supported by strong indirect evidence, remain provisional (Currall et al. 2013; Kloosterman and Cuppen 2013).
For chromoplexy, no mechanistic information is currently available. It is clear that many breaks must be present around the same time for chromoplexy to occur. Thus any event that can induce multiple breaks, such as DNA replication stress (Donley and Thayer 2013) or active transcription (for review, see Kim and Jinks-Robertson 2012), could lead to concurrent DSBs in multiple chromosomes. Chained translocations to repair these breaks could then occur by chance, with selection acting to preserve copy number balance and oncogene activation at the breakpoint junctions. It is also possible that chained translocations are promoted by physical clustering of the breakpoints in the nucleus (Roix et al. 2003; Meaburn et al. 2007; Misteli 2010; Alt et al. 2013). This could occur by clustering of commonly regulated transcriptional sites or other clustering events, such as the assembly of DNA replication factories. In the case of prostate cancer, colocalization of transcriptional sites could be mediated by colocalization of androgen-responsive elements (Lin et al. 2009), which could cluster topoisomerase II-mediated DSBs (Haffner et al. 2010).
The origin of chromothripsis is also unclear, but plausible hypotheses have been recently advanced (Forment et al. 2012; Holland and Cleveland 2012; Jones and Jallepalli 2012; Kloosterman and Cuppen 2013). The major puzzle is how a large number of DSBs can be so highly localized. One proposal is that shattering occurs during mitosis when the chromosomes are highly compacted. At this point in the cell cycle, it is conceivable that external damage such as from ionizing radiation could slice through multiple segments of a single chromosome (Stephens et al. 2011). Critical telomere shortening leading to BFB cycles has also been considered because the induced rearrangements are local and chromothripsis is often observed near the telomeres (Stephens et al. 2011). However, disfavoring this explanation is the fact that BFB cycles can result in gene amplification and should produce a bias for inverted rearrangements.
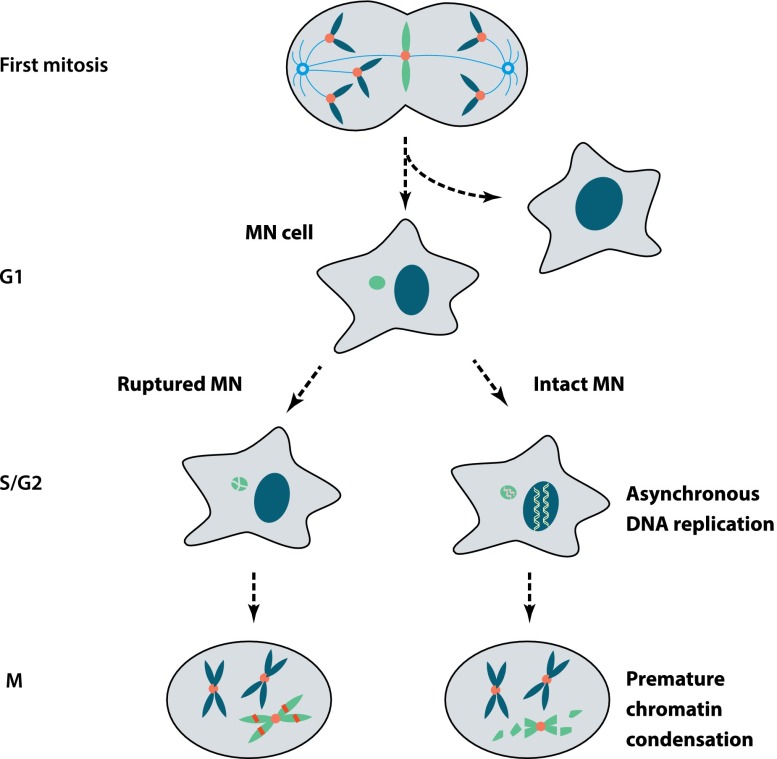
One simple solution to generate massive local mutagenesis is when one or two chromosomes (or an isolated acentric chromosome arm) are physically isolated from the other chromosomes. A simple and appealing hypothesis for how this could occur with fairly high frequency in human cancers is the partitioning of whole chromosomes or acentric chromosome fragments into micronuclei (Fig. 6; Crasta et al. 2012). It has become apparent that chromosomes in micronuclei can be subject to massive spontaneous DNA damage (Terradas et al. 2009; Forment et al. 2012; Holland and Cleveland 2012), and this damage occurs in the first cell cycle after micronuclei are formed (Crasta et al. 2012). Importantly, damaged micronuclei can be reincorporated into the main nucleus during subsequent cell division (Crasta et al. 2012; Hatch et al. 2013). This means that damaged micronuclei can, at least in principle, generate mutations that can be integrated back into the genome.
Figure 6.
Extensive DNA damage of chromosomes in micronuclei (MN). Micronuclei can form by missegregation of whole chromosomes after mitotic errors. Pictured here is a “merotelic” kinetochore attachment where a single intact chromatid is caught in a tug-of-war between microtubules from opposite poles. Micronuclei can also originate from chromosome breaks, generating acentric chromosome fragments, or be generated from chromosome bridges by a mechanism that has not been determined (Hoffelder et al. 2004). (Left) Newly generated micronuclei can undergo irreversible loss of nuclear membrane integrity (Hatch et al. 2013). If this occurs during S phase, chromosomes in micronuclei will acquire massive DNA damage. (Right) Alternatively, late-replicating chromosomes in micronuclei may undergo DNA damage if the cell enters mitosis before micronuclear replication is complete (Crasta et al. 2012), subjecting the chromosome to premature chromosome compaction (PCC). The extent and mechanism of DNA damage during PCC remains to be established. During the subsequent cell division cycle, the micronuclear genome can be reincorporated into the primary nucleus, remain isolated in a micronucleus, or possibly be lost due to autophagy or extrusion.
Micronuclei are structurally similar to regular nuclei in that they have a double-membrane structure and nuclear pores but contain only one or a few chromosomes or chromosomal segments. Spatial isolation of individual chromosomes in micronuclei provides a natural setup for localized chromosomal damage, but how does damage happen?
One possible mechanism is that newly generated micronuclei acquire their DNA damage as they progress into the S phase and G2 phase of the subsequent cell cycle (Crasta et al. 2012; Hatch et al. 2013). Micronuclei replicate their DNA asynchronously with the primary nucleus, with many micronuclei showing DNA replication in G2 (Fig. 6; Crasta et al. 2012). This delayed replication means that some micronuclei might enter mitosis and undergo premature chromosome compaction (PCC) (Obe and Beek 1975) before they have completed DNA replication (Fig. 6; Donley and Thayer 2013). PCC is believed to cause DNA damage, as evidenced by gaps observed on chromosome spreads (Kato and Sandberg 1967; Johnson and Rao 1970) and the fact that PCC can induce breakage at chromosome fragile sites (Smith et al. 2001; El Achkar et al. 2005; Ravi et al. 2013). Indeed, our group has found that in chromosome spreads, late-replicating micronuclear DNA develops the pulverized appearance that is characteristic of PCC (Crasta et al. 2012).
A recent study from Hetzer's laboratory (Hatch et al. 2013) has provided important new insight into additional defects in micronuclei that could cause DNA damage. Using live-cell imaging, the group had previously shown that cancer cell nuclei occasionally undergo spontaneous loss of nuclear integrity (as assayed by cytoplasmic spilling of a nuclear green fluorescent protein) that is reversible (Vargas et al. 2012). They have now reported that many micronuclei undergo irreversible loss of nuclear envelope (NE) integrity, as evidenced by fluorescent imaging and electron microscopy. The basis for the defect in NE integrity is still not clear, but it is correlated with abnormal and asymmetric nuclear lamina assembly. Most strikingly, the rupture of micronuclei is tightly correlated with the appearance of massive damage (Fig. 6). Although it has not been possible to directly image the timing of damage acquisition relative to the timing of rupture, fixed cell experiments strongly suggest that damage occurs close to the time of rupture.
How NE rupture contributes to DNA damage is not known. We found that micronuclei in serum-starved cells readily undergo rupture but acquire little or no damage (A Spektor and D Pellman, unpubl.). Thus, rupture seems to be required for damage but not sufficient to generate damage. One possibility would be that NE rupture during DNA replication would abruptly dilute replication factors and cause DNA replication fork stalling and thus generate damage. Alternatively, rupture during S phase might enable enzymes such as endonucleases or topoisomerases to act aberrantly on micronuclear chromosomes.
Conclusion
The discovery of chromothripsis by Stephens et al. (2011) opened a floodgate of genome sequencing studies of complex, potentially all-at-once genome rearrangement. This has led to the identification of chromothripsis in many cancer types and in some congenital diseases. The flurry of genome sequencing has also uncovered other classes of complex rearrangements (chromoanasynthesis and chromoplexy), which also appear to originate from single catastrophic events. These discoveries have expanded our view of the mechanisms by which genomes can evolve. Although we have no evidence at present that these mechanisms contribute to speciation, this is certainly plausible.
These unexpected discoveries were the fruits of new technical advances, particularly next-generation sequencing and the accompanying novel bioinformatics tools. The recent studies illustrate the fact that these events are more common than could have been expected, and further sequencing efforts will clarify their true frequency. However, as discussed above, the phenomena identified to date could be the extreme examples of even more frequent but less dramatic events. Thus, understanding the underlying mechanism is not only interesting in its own right but may impact our understanding of how common these events really are.
In this review, we tried to illustrate the importance, power, and limitations of bioinformatic and computational approaches to reconstruct the history of complex chromosomal rearrangements (Stephens et al. 2011; Greenman et al. 2012a; Baca et al. 2013). Although reconstructing history from a snapshot of the genome is a challenging task, we hope that we conveyed the tremendous progress toward solving this open problem.
The understanding of catastrophic chromosomal rearrangements is certainly incomplete until the biological mechanisms underlying these phenomena are defined. Indeed, one exciting fallout from the genome sequencing efforts is the potential to discover new biological mechanisms. For example, if it can be shown that damage occurring in micronuclei can lead to chromothripsis, this could provide an important example of the emerging role of nuclear architecture and subnuclear organization in maintaining the stability of the genome (Meaburn et al. 2007; Misteli and Soutoglou 2009; Misteli 2010; Alt et al. 2013). Ultimately, linking postulated biological mechanisms to these new classes of genome rearrangement will require an experimental recapitulation of the phenomena in the laboratory. As the physicist Richard Feynman famously said, “What I cannot create, I do not understand.” Combining experimental live-cell imaging approaches with emerging single-cell genome sequencing technologies (Shapiro et al. 2013) may be one way to recreate and thus understand complex chromosomal rearrangements. Thus, the use of genome sequencing as a hypothesis-generating discovery tool may be ushering in a new era where genome sequencing is harnessed for hypothesis-testing experimental biology.
Acknowledgments
We thank I.M. Hall, D.M. Weinstock, S. Baca, H. Cornils, J. Francis, P.J. Campbell, and P.J. Stephens for discussions. C.Z.Z. was supported by the National Cancer Institute (U24CA143867). M.L.L was supported by the National Science Foundation (DGE1144152). D.P. was supported by the Howard Hughes Medical Institute, the National Institute of Health (GM083299), and the G. Harold and Leila Y. Mathers Charitable Foundation.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.229559.113.
References
- Adey A, Burton JN, Kitzman JO, Hiatt JB, Lewis AP, Martin BK, Qiu R, Lee C, Shendure J 2013. The haplotype-resolved genome and epigenome of the aneuploidy HeLa cancer cell line. Nature 500: 207–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borrensen-Dale AL, et al. 2013. Signatures of mutational processes in human cancer. Nature 500: 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt FW, Zhang Y, Meng FL, Guo C, Schwer B 2013. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell 152: 417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand RP, Lovett ST, Haber JE 2013. Break-induced DNA replication. Cold Spring Harb Perspect Biol doi: 10.1101/cshperspect.a010397 [DOI] [PMC free article] [PubMed]
- Arlt MF, Mulle JG, Schaibley VM, Ragland RL, Durkin SG, Warren ST, Glover TW 2009. Replication stress induces genome-wide copy number changes in human cells that resemble polymorphic and pathogenic variants. Am J Hum Genet 84: 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt MF, Rajendran S, Birkeland SR, Wilson TE, Glover TW 2012. De novo CNV formation in mouse embryonic stem cells occurs in the absence of Xrcc4-dependent nonhomologous end-joining. PLoS Genet 8: e1002981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, et al. 2013. Punctuated evolution of prostate cancer genomes. Cell 153: 666–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassaganyas L, Bea S, Escaramis G, Tornador C, Salaverria I, Zapata L, Drechsel O, Ferreira PG, Rodriguez-Santiago B, Tubio JM, et al. 2013. Sporadic and reversible chromothripsis in chronic lymphocytic leukemia revealed by longitudinal genomic analysis. Leukemia (in press). doi: 10.1038/leu.2013.127 [DOI] [PMC free article] [PubMed]
- Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. 2011. The genomic complexity of primary human prostate cancer. Nature 470: 214–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. 2012. Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485: 502–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. 2007. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci 104: 20007–20012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Santarius T, Pole JC, Butler AP, Perry J, Pleasance E, Greenman C, Menzies A, Taylor S, Edkins S, et al. 2007. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res 17: 1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell GR, Greenman CD, Davies H, Butler AP, Edkins S, Andrews JM, Buck G, Chen L, Beare D, Latimer C, et al. 2010. Signatures of mutation and selection in the cancer genome. Nature 463: 893–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic I, Antal T, Ohtsuki H, Carter H, Kim D, Chen S, Karchin R, Kinzler KW, Vogelstein B, Nowak MA 2010. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci 107: 18545–18550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brastianos PK, Horowitz PM, Santagata S, Jones RT, McKenna A, Getz G, Ligon KL, Palescandolo E, Van Hummelen P, Ducar MD, et al. 2013. Genomic sequencing of meningiomas identifies oncogenic SMO and AKT1 mutations. Nat Genet 45: 285–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL 2011. Haplotype phasing: Existing methods and new developments. Nat Rev Genet 12: 703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. 2008. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet 40: 722–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell PJ, Yachida S, Mudie LJ, Stephens PJ, Pleasance ED, Stebbings LA, Morsberger LA, Latimer C, McLaren S, Lin ML, et al. 2010. The patterns and dynamics of genomic instability in metastatic pancreatic cancer. Nature 467: 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. 2012. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 30: 413–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Zhang F, Liu P, Patel A, Sahoo T, Bacino CA, Shaw C, Peacock S, Pursley A, Tavyev YJ, et al. 2009. Complex rearrangements in patients with duplication of MECP2 can occur by fork stalling and template switching. Hum Mol Genet 18: 2188–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho CM, Ramocki MB, Pehlivan D, Franco LM, Gonzaga-Jauregui C, Fang P, McCall A, Pivnick EK, Hines-Dowell S, Seaver LH, et al. 2011. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nat Genet 43: 1074–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Rappailles A, Duquenne L, Huvet M, Guilbaud G, Farinelli L, Audit B, d'Aubenton-Carafa Y, Arneodo A, Hyrien O, et al. 2010. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res 20: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C, Jacobsen JC, Ernst C, Hanscom C, Heilbut A, Blumenthal I, Mills RE, Kirby A, Lindgren AM, Rudiger SR, et al. 2012. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat Genet 44: 390–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currall BB, Chiang C, Talkowski ME, Morton CC 2013. Mechanisms for structural variation in the human genome. Curr Genet Med Rep 1: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De S, Michor F 2011. DNA replication timing and long-range DNA interactions predict mutational landscapes of cancer genomes. Nat Biotechnol 29: 1103–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deakin JE, Bender HS, Pearse AM, Rens W, O'Brien PC, Ferguson-Smith MA, Cheng Y, Morris K, Taylor R, Stuart A, et al. 2012. Genomic restructuring in the Tasmanian devil facial tumor: Chromosome painting and gene mapping provide clues to evolution of a transmissible tumor. PLoS Genet 8: e1002483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debatisse M, Le Tallec B, Letessier A, Dutrillaux B, Brison O 2012. Common fragile sites: Mechanisms of instability revisited. Trends Genet 28: 22–32 [DOI] [PubMed] [Google Scholar]
- Donley N, Thayer MJ 2013. DNA replication timing, genome stability and cancer: Late and/or delayed DNA replication timing is associated with increased genomic instability. Semin Cancer Biol 23: 80–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrett R. 2008. Probability models for DNA sequence evolution. Springer, New York. [Google Scholar]
- El Achkar E, Gerbault-Seureau M, Muleris M, Dutrillaux B, Debatisse M 2005. Premature condensation induces breaks at the interface of early and late replicating chromosome bands bearing common fragile sites. Proc Natl Acad Sci 102: 18069–18074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnhead HO, McCurrach ME, O'Neill J, Zhang K, Lowe SW, Lazebnik YA 1997. Oncogene-dependent apoptosis in extracts from drug-resistant cells. Genes & Dev 11: 1266–1276 [DOI] [PubMed] [Google Scholar]
- Forment JV, Kaidi A, Jackson SP 2012. Chromothripsis and cancer: Causes and consequences of chromosome shattering. Nat Rev Cancer 12: 663–670 [DOI] [PubMed] [Google Scholar]
- Fousteri M, Mullenders LH 2008. Transcription-coupled nucleotide excision repair in mammalian cells: Molecular mechanisms and biological effects. Cell Res 18: 73–84 [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Getz G, Meyerson M, Mirny LA 2011. High order chromatin architecture shapes the landscape of chromosomal alterations in cancer. Nat Biotechnol 29: 1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437: 1043–1047 [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D 2012. Linking abnormal mitosis to the acquisition of DNA damage. J Cell Biol 199: 871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibaud A, Vogt N, Hadj-Hamou NS, Meyniel JP, Hupe P, Debatisse M, Malfoy B 2010. Extrachromosomal amplification mechanisms in a glioma with amplified sequences from multiple chromosome loci. Hum Mol Genet 19: 1276–1285 [DOI] [PubMed] [Google Scholar]
- Gibaud A, Vogt N, Brison O, Debatisse M, Malfoy B 2013. Characterization at nucleotide resolution of the homogeneously staining region sites of insertion in two cancer cell lines. Nucleic Acids Res 41: 8210–8219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Pettersson L, Hoglund M, Heidenblad M, Gorunova L, Wiegant J, Mertens F, Dal Cin P, Mitelman F, Mandahl N 2000. Chromosomal breakage–fusion–bridge events cause genetic intratumor heterogeneity. Proc Natl Acad Sci 97: 5357–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D 2012. Causes and consequences of aneuploidy in cancer. Nat Rev Genet 13: 189–203 [DOI] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA Jr, Kastrinakis NG, Levy B, et al. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434: 907–913 [DOI] [PubMed] [Google Scholar]
- Gould SJ, Eldredge N 1977. Punctuated equilibria: The tempo and mode of evolution reconsidered. Paleobiology 3: 115–151 [Google Scholar]
- Greaves M, Maley CC 2012. Clonal evolution in cancer. Nature 481: 306–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman CD, Pleasance ED, Newman S, Yang F, Fu B, Nik-Zainal S, Jones D, Lau KW, Carter N, Edwards PA 2012a. Estimation of rearrangement phylogeny for cancer genomes. Genome Res 22: 346–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman CD, Cooke SL, Marshall J, Stratton MR, Campbell PJ 2012b. Modeling breakage–fusion–bridge cycles as a stochastic paper folding process.http://arxiv.org/abs/1211.2356 [DOI] [PMC free article] [PubMed]
- Haber JE, Debatisse M 2006. Gene amplification: Yeast takes a turn. Cell 125: 1237–1240 [DOI] [PubMed] [Google Scholar]
- Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, et al. 2010. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet 42: 668–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J 2008. An oncogene-induced DNA damage model for cancer development. Science 5868: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Hammond D, Zeng K, Espert A, Bastos RN, Baron RD, Gruneberg U, Barr FA 2013. Melanoma-associated mutations in protein phosphatase 6 cause chromosome instability and DNA damage owing to dysregulated Aurora-A. J Cell Sci 126: 3429–3440 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2011. Hallmarks of cancer: The next generation. Cell 144: 646–674 [DOI] [PubMed] [Google Scholar]
- Handyside AH, Montag M, Magli MC, Repping S, Harper J, Schmutzler A, Vesela K, Gianaroli L, Geraedts J 2012. Multiple meiotic errors caused by predivision of chromatids in women of advanced maternal age undergoing in vitro fertilization. Eur J Hum Genet 20: 742–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G 2009. Mechanisms of change in gene copy number. Nat Rev Genet 10: 551–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW 2013. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154: 47–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks J, Krasnitz A, Lakshmi B, Navin NE, Riggs M, Leibu E, Esposito D, Alexander J, Troge J, Grubor V, et al. 2006. Novel patterns of genome rearrangement and their association with survival in breast cancer. Genome Res 16: 1465–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks WM, Kim M, Haber JE 2010. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science 329: 82–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch D, Kemmerling R, Davis S, Camps J, Meltzer PS, Ried T, Gaiser T 2013. Chromothripsis and focal copy number alterations determine poor outcome in malignant melanoma. Cancer Res 73: 1454–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson A, Eyre-Walker A 2011. Variation in the mutation rate across mammalian genomes. Nat Rev Genet 12: 756–766 [DOI] [PubMed] [Google Scholar]
- Hodgkinson A, Chen Y, Eyre-Walker A 2012. The large-scale distribution of somatic mutations in cancer genome. Hum Mutat 33: 136–143 [DOI] [PubMed] [Google Scholar]
- Hoffelder DR, Luo L, Burke NA, Watkins SC, Gollin SM, Saunders WS 2004. Resolution of anaphase bridges in cancer cells. Chromosoma 112: 389–397 [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW 2009. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW 2012. Chromoanagenesis and cancer: Mechanisms and consequences of localized, complex chromosomal rearrangements. Nat Med 18: 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn S, Figl A, Rachakonda PS, Fischer C, Sucker A, Gast A, Kadel S, Moll I, Nagore E, Hemminki K, et al. 2013. TERT promoter mutations in familial and sporadic melanoma. Science 339: 959–961 [DOI] [PubMed] [Google Scholar]
- Huang FW, Hodis E, Xu MJ, Kryukov GV, Chin L, Garraway LA 2013. Highly recurrent TERT promoter mutations in human melanoma. Science 339: 957–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa R, Takenaka M, Kohno T, Shimada Y, Totoki Y, Shibata T, Tsuta K, Nishikawa R, Noguchi M, Sato-Otsubo A, et al. 2013. Genome-wide identification of genes with amplification and/or fusion in small cell lung cancer. Genes Chromosomes Cancer 52: 802–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J 2009. The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RT, Rao PN 1970. Mammalian cell fusion: Induction of premature chromosome condensation in interphase nuclei. Nature 226: 717–722 [DOI] [PubMed] [Google Scholar]
- Jones MJ, Jallepalli PV 2012. Chromothripsis: Chromosomes in crisis. Dev Cell 23: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Bartek J 2004. Cell-cycle checkpoints and cancer. Nature 432: 316–323 [DOI] [PubMed] [Google Scholar]
- Kato H, Sandberg AA 1967. Chromosome pulverization in human binucleate cells following colcemid treatment. J Cell Biol 34: 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. 2013. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci 110: 6021–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Jinks-Robertson S 2012. Transcription as a source of genome instability. Nat Rev Genet 13: 204–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TM, Xi R, Luquette LJ, Park RW, Johnson MD, Park PJ 2013. Functional genomic analysis of chromosomal aberrations in a compendium of 8000 cancer genomes. Genome Res 23: 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Cuppen E 2013. Chromothripsis in congenital disorders and cancer: Similarities and differences. Curr Opin Cell Biol 25: 341–348 [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Guryev V, van Roosmalen M, Duran KJ, de Bruijn E, Bakker SC, Letteboer T, van Nesselrooij B, Hochstenbach R, Poot M, et al. 2011a. Chromothripsis as a mechanism driving complex de novo structural rearrangements in the germline. Hum Mol Genet 20: 1916–1924 [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Hoogstraat M, Paling O, Tavakoli-Yaraki M, Renkens I, Vermaat JS, van Roosmalen MJ, van Lieshout S, Nijman IJ, Roessingh W 2011b. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol 12: R103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Tavakoli-Yaraki M, van Roosmalen MJ, van Binsbergen E, Renkens I, Duran K, Ballarati L, Vergult S, Giardino D, Hansson K, et al. 2012. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep 1: 648–655 [DOI] [PubMed] [Google Scholar]
- Korbel JO, Campbell PJ 2013. Criteria for inference of chromothripsis in cancer genomes. Cell 152: 1226–1236 [DOI] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren A, Polak P, Nemesh J, Michaelson JJ, Sebat J, Sunyaev SR, McCarroll SA 2012. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am J Hum Genet 91: 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Carr AM 2013. Replication stress and genome rearrangements: Lessons from yeast models. Curr Opin Genet Dev 23: 132–139 [DOI] [PubMed] [Google Scholar]
- Landry JJ, Pyl PT, Rausch T, Zichner T, Tekkedil MM, Stutz AM, Jauch A, Aiyar RS, Pau G, Delhomme N, et al. 2013. The genomic and transcriptomic landscape of a HeLa cell line. G3 (Bethesda) 3: 1213–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Carvalho CM, Lupski JR 2007. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131: 1235–1247 [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang L, Tanasa B, Hutt K, Ju BG, Ohgi K, Zhang J, Rose DW, Fu XD, Glass CK, et al. 2009. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell 139: 1069–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. 2011. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Carvalho CM, Hasings PJ, Lupski JR 2012. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev 22: 211–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SW, Jacks T, Housman DE, Ruley HE 1994. Abrogation of oncogene-associated apoptosis allows transformation of p53-deficient cells. Proc Natl Acad Sci 91: 2026–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKinnon RN, Campbell LJ 2013. Chromothripsis under the microscope: A cytogenetic perspective of two cases of AML with catastrophic chromosome rearrangement. Cancer Genet 206: 238–251 [DOI] [PubMed] [Google Scholar]
- Magrangeas F, Avet-Loiseau H, Munshi NC, Minvielle S 2011. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood 118: 675–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CA, Wilson RK 2012. Chromothripsis and human disease: Piecing together the shattering process. Cell 148: 29–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Lindberg M, Faust GG, Leibowitz ML, Clark RA, Layer RM, Quinlan AR, Hall IM 2013. Breakpoint profiling of 64 cancer genomes reveals numerous complex rearrangements spawned by homology-independent mechanisms. Genome Res 23: 762–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkova A, Haber JE 2012. Mutations arising during repair of chromosome breaks. Annu Rev Genet 46: 455–473 [DOI] [PubMed] [Google Scholar]
- McClintock B 1941. The stability of broken ends of chromosomes in Zea Mays. Genetics 26: 234–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T, Soutoglou E 2007. Spatial genome organization in the formation of chromosomal translocations. Semin Cancer Biol 17: 80–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev P, Stanciu M, Brudno M 2009. Computational methods for discovering structural variation with next-generation sequencing. Nat Methods 6: S13–S20 [DOI] [PubMed] [Google Scholar]
- Meek DW 2004. The p53 response to DNA damage. DNA Repair (Amst) 3: 1049–1056 [DOI] [PubMed] [Google Scholar]
- Mehine M, Kaasinen E, Makinen N, Katainen R, Kampjarvi K, Pitkanen E, Heinonen HR, Butzow R, Kilpivaara O, Kuosmanen A, et al. 2013. Characterization of uterine leiomyomas by whole-genome sequencing. N Engl J Med 369: 43–53 [DOI] [PubMed] [Google Scholar]
- Meyerson M, Pellman D 2011. Cancer genomes evolve by pulverizing single chromosomes. Cell 144: 9–10 [DOI] [PubMed] [Google Scholar]
- Misteli T 2010. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol 2: a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Soutoglou E 2009. The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 10: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F 2007. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 7: 233–245 [DOI] [PubMed] [Google Scholar]
- Mizuno K, Miyabe I, Schalbetter SA, Carr AM, Murray JM 2013. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature 493: 246–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J 2012. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature 483: 589–593 [DOI] [PubMed] [Google Scholar]
- Morin RD, Mungall K, Pleasance E, Mungall AJ, Goya R, Huff RD, Scott DW, Ding J, Roth A, Chiu R, et al. 2013. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 122: 1256–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, Hunt PA 2012. Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat Rev Genet 13: 493–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel S, Meyer C, Quentmeier H, Kaufmann M, Drexler HG, MacLeod RA 2013. Chromothripsis in Hodgkin lymphoma. Genes Chromosomes Cancer 52: 741–747 [DOI] [PubMed] [Google Scholar]
- Narayanan V, Mieczkowski PA, Kim HM, Petes TD, Lobachev KS 2006. The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125: 1283–1296 [DOI] [PubMed] [Google Scholar]
- Nazaryan L, Stefanou EG, Hansen C, Kosyakova N, Bak M, Sharkey FH, Mantziou T, Papanastasiou AD, Velissariou V, Liehr T, et al. 2013. The strength of combined cytogenetic and mate-pair sequencing techniques illustrated by a germline chromothripsis rearrangement involving FOXP2. Eur J Hum Genet doi: 10.1038/ejhg.2013.147 [DOI] [PMC free article] [PubMed]
- Negrini S, Gorgoulis VG, Halazonetis TD 2010. Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol 11: 220–228 [DOI] [PubMed] [Google Scholar]
- Nik-Zainal S, Alexandrov LB, Wedge DC, Van Loo P, Greenman CD, Raine K, Jones D, Hinton J, Marshall J, Stebbings LA, et al. 2012. Mutational processes molding the genomes of 21 breast cancers. Cell 149: 979–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northcott PA, Shih DJ, Peacock J, Garzia L, Morrissy AS, Zichner T, Stutz AM, Korshunov A, Reimand J, Schumacher SE, et al. 2012. Subgroup-specific structural variation across 1000 medulloblastoma genomes. Nature 488: 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowell PC 1976. The clonal evolution of tumor cell populations. Science 194: 23–28 [DOI] [PubMed] [Google Scholar]
- Obe G, Beek B 1975. The human leukocyte test system. VII. Further investigations concerning micronucleus-derived premature chromosome condensation. Humangenetik 30: 143–154 [DOI] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P 2010. TP53 mutations in human cancers: Origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2: a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2012. Characterizing complex structural variation in germline and somatic genomes. Trends Genet 28: 43–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabbitts TH 2009. Commonality but diversity in cancer gene fusions. Cell 137: 391–395 [DOI] [PubMed] [Google Scholar]
- Raphael BJ, Volik S, Collins C, Pevzner PA 2003. Reconstructing tumor genome architectures. Bioinformatics 19: ii162–ii171 [DOI] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. 2012. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148: 59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi M, Nivedita K, Pai GM 2013. Chromatin condensation dynamics and implications of induced premature chromosome condensation. Biochimie 95: 124–133 [DOI] [PubMed] [Google Scholar]
- Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, et al. 2012. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell 46: 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T 2003. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet 34: 287–291 [DOI] [PubMed] [Google Scholar]
- Sanborn JZ, Salama SR, Grifford M, Brennan CW, Mikkelsen T, Jhanwar S, Katzman S, Chin L, Haussler D 2013. Double-minute chromosomes in glioblastoma multiforme are revealed by precise reconstruction of oncogenic amplicons. Cancer Res 73: 6036–6045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster-Böckler B, Lehner B 2012. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 488: 504–507 [DOI] [PubMed] [Google Scholar]
- Shapiro E, Biezuner T, Linnarsson S 2013. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat Rev Genet 14: 618–630 [DOI] [PubMed] [Google Scholar]
- Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ 2006. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet 4: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L, Plug A, Thayer M 2001. Delayed replication timing leads to delayed mitotic chromosome condensation and chromosomal instability of chromosome translocations. Proc Natl Acad Sci 98: 13300–13305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos JA, Adzhubei I, Thurman RE, Kryukov GV, Mirkin SM, Sunyaev SR 2009. Human mutation rate associated with DNA replication timing. Nat Genet 41: 393–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark GR, Debatisse M, Giulotto E, Wahl GM 1989. Recent progress in understanding mechanisms of mammalian DNA amplification. Cell 57: 901–908 [DOI] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. 2009. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi CT, Lonoce A, Guastadisegni MC, Trombetta D, D'Addabbo P, Daniele G, L'Abbate A, Macchia G, Surace C, Kok K, et al. 2010. Gene amplification as double minutes or homogeneously staining regions in solid tumors: Origin and structure. Genome Res 20: 1198–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA 2009. The cancer genome. Nature 458: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles Alves I, Hiltemann S, Hartjes T, van der Spek P, Stubbs A, Trapman J, Jenster G 2013. Gene fusions by chromothripsis of chromosome 5q in the VCaP prostate cancer cell line. Hum Genet 132: 709–713 [DOI] [PubMed] [Google Scholar]
- Terradas M, Martin M, Tusell L, Genesca A 2009. DNA lesions sequestered in micronuclei induce a local defective-damage response. DNA Repair (Amst) 8: 1225–1234 [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA 2010. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol 188: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasetti C, Vogelstein B, Parmigiani G 2013. Half or more of the somatic mutations in cancers of self-renewing tissues originate prior to tumor initiation. Proc Natl Acad Sci 110: 1999–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubio JM, Estivill X 2011. Cancer: When catastrophe strikes a cell. Nature 470: 476–477 [DOI] [PubMed] [Google Scholar]
- Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. 2005. Fine-scale structural variation of the human genome. Nat Genet 37: 727–732 [DOI] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F 2009. Chromosome instability is common in human cleavage-stage embryos. Nat Med 15: 577–583 [DOI] [PubMed] [Google Scholar]
- Vargas JD, Hatch EM, Anderson DJ, Hetzer MW 2012. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus 3: 88–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voet T, Vanneste E, Vermeesch JR 2011. The human cleavage stage embryo is a cradle of chromosomal rearrangements. Cytogenet Genome Res 133: 160–168 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW 2013. Cancer genome landscapes. Science 339: 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Wyatt AW, McPherson A, Lin D, McConeghy BJ, Mo F, Shukin R, Lapuk AV, Jones SJ, Zhao Y, et al. 2012. Poly-gene fusion transcripts and chromothripsis in prostate cancer. Genes Chromosomes Cancer 51: 1144–1153 [DOI] [PubMed] [Google Scholar]
- Yang L, Luquette LJ, Gehlenborg N, Xi R, Haseley PS, Hsieh CH, Zhang C, Ren X, Protopopov A, Chin L, et al. 2013. Diverse mechanisms of somatic structural variations in human cancer genomes. Cell 153: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LR, Campbell PJ 2012. Evolution of the cancer genome. Nat Rev Genet 13: 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. 2013. Pan-cancer patterns of somatic copy-number alteration. Nat Genet 45: 1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Carvalho CM, Lupski JR 2009a. Complex human chromosomal and genomic rearrangements. Trends Genet 25: 298–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR 2009b. The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet 41: 849–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW 2002. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell 109: 811–821 [DOI] [PubMed] [Google Scholar]