The Fbw7 tumor suppressor targets proteins for ubiquitylation. Welcker et al. found that dimerization enables Fbw7 to target substrates through concerted binding to two suboptimal and independent recognition sites. Dimerization increases Fbw7's robustness and regulates Fbw7 stability, and this likely involves Fbw7 trans-autoubiquitylation. This study reveals an unanticipated complexity in substrate degradation.
Keywords: Fbw7, SCF, phosphodegron, cyclin E
Abstract
The Fbw7 tumor suppressor targets a broad network of proteins for ubiquitylation. Here we show critical functions for Fbw7 dimerization in regulating the specificity and robustness of degradation. Dimerization enables Fbw7 to target substrates through concerted binding to two suboptimal and independent recognition sites. Accordingly, an endogenous dimerization-deficient Fbw7 mutation stabilizes suboptimal substrates. Dimerization increases Fbw7's robustness by preserving its function in the setting of mutations that disable Fbw7 monomers, thereby buffering against pathogenic mutations. Finally, dimerization regulates Fbw7 stability, and this likely involves Fbw7 trans-autoubiquitylation. Our study reveals novel functions of Fbw7 dimerization and an unanticipated complexity in substrate degradation.
Fbw7 is an evolutionarily conserved substrate receptor of an SCF (Skp1, Cul1, and F-box protein) ubiquitin ligase that targets proteins for degradation (Welcker and Clurman 2008; Skaar et al. 2013). Approximately 20 Fbw7 substrates are known, including critical oncoproteins (e.g., cyclin E, Myc, Jun, and Notch), and Fbw7 is mutated in 8%–10% of human cancers (Akhoondi et al. 2007; Welcker and Clurman 2008; Crusio et al. 2010). Fbw7 and its orthologs (Cdc4, Ago, Sel-10, and Pop1/Pop2) contain three functional domains: a β propeller formed by WD40 repeats that binds to substrates, an F-box that recruits SCF components, and a dimerization domain.
Fbw7 binds substrates upon their phosphorylation within Cdc4 phosphodegrons (CPDs) (Koepp et al. 2001; Nash et al. 2001; Strohmaier et al. 2001; Welcker and Clurman 2008). All CPDs contain a central phosphothreonine/serine and other conserved residues, whereas optimal CPDs also contain a P+4 phosphorylation that increases binding affinity (Nash et al. 2001; Welcker et al. 2003; Hao et al. 2007). Two Fbw7 β-propeller pockets interact with both degron phosphorylations, while upstream CPD residues fit into a hydrophobic groove (Orlicky et al. 2003; Hao et al. 2007). Fbw7 substrates have either a single CPD (e.g., Myc, SREBP, and TGIF1) (Welcker et al. 2004; Yada et al. 2004; Sundqvist et al. 2005; Bengoechea-Alonso and Ericsson 2010) or two identified degrons (e.g., cyclin E, PGC1, MCL1, and KLF2) (Koepp et al. 2001; Strohmaier et al. 2001; Welcker et al. 2003; Olson et al. 2008; Inuzuka et al. 2011; Wertz et al. 2011; Wang et al. 2013). The significance of multiple degrons is poorly understood. While each CPD could provide an independent degradation signal, multiple degrons might also cooperatively bind to Fbw7, as speculated for cyclin E (Hao et al. 2007; Welcker and Clurman 2007).
Previous studies have explored the consequences of Fbw7/Cdc4 dimerization (Zhang and Koepp 2006; Hao et al. 2007; Tang et al. 2007; Welcker and Clurman 2007). SCFCdc4 dimers target diverse substrate lysines and hyperextend polyubiquitin chains compared with monomers, indicating that dimer asymmetry regulates catalysis (Tang et al. 2007). Fbw7 dimerization was also required to degrade a suboptimal CPD cyclin E mutant, suggesting that dimerization impacts substrate binding (Hao et al. 2007; Welcker and Clurman 2007). However, because structural studies have used Fbw7 monomers bound to degron peptides, the relationships between Fbw7 dimers and multiple degrons are poorly characterized.
We show that Fbw7 is exclusively a dimer and that dimerization enables binding modes that establish the specificity and robustness of Fbw7–substrate interactions. By studying full-length substrates with optimal and suboptimal degrons, we show that dimerization facilitates their degradation through the concerted interactions of two suboptimal degrons. Accordingly, an endogenous Fbw7 mutation that prevents dimerization selectively stabilizes suboptimal substrates. An unanticipated consequence of Fbw7 dimerization involves the robustness of degradation. Dimers tolerate mutations in the Fbw7–substrate interface that disable monomers, and dimerization may “buffer” against deleterious mutations. Finally, in contrast with a recent study (Min et al. 2012), we found that Fbw7 monomers are stable and that disruption of dimerization is unlikely to destabilize Fbw7 in cancers.
Results and Discussion
Cyclin E degrons cooperate for efficient Fbw7 binding
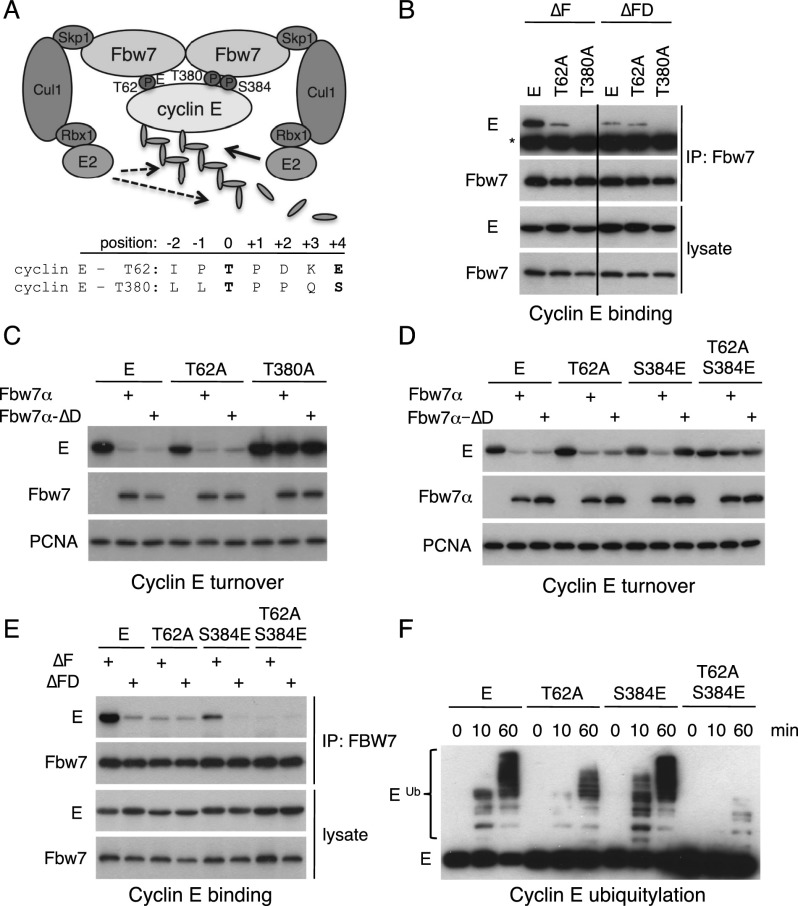
We tested the hypothesis that an Fbw7 dimer interacts simultaneously with both cyclin E degrons and found that dimerization and both CPDs contributed to binding and that the degrons acted synergistically. Cyclin E contains a suboptimal N-terminal T62 CPD and a high-affinity C-terminal T380 CPD (Fig. 1A). We cotransfected mutants that inactivate each degron (cyclin ET62A and cyclin ET380A) or prevent Fbw7 dimerization to determine how each degron and Fbw7 dimerization contribute to cyclin E regulation. We used Fbw7 or Fbw7ΔD (a deletion of residues FQSWS in the dimerization domain) constructs with F-box deletions (Fbw7ΔF) that prevent SCF recruitment and thus uncouple cyclin E binding from its turnover. Cyclin E bound readily to dimers (Fbw7ΔF), which was greatly diminished with cyclin ET62A and eliminated with cyclin ET380A (Fig. 1B). In contrast, monomers (Fbw7ΔFD) bound to cyclin E and cyclin ET62A equally but at the low amount seen with cyclin ET62A and dimers (Fig. 1B). These data suggest that Fbw7 monomers bind exclusively via the high-affinity T380 CPD and that the concerted interactions of both degrons with Fbw7 dimers produce maximal binding. Dimers that cannot engage the T62 CPD act essentially like monomers. Thus, the amount of cyclin ET62A binding (Fig. 1B), turnover (Supplemental Fig. SF1A), and ubiquitylation (Supplemental Fig. SF1B) is indistinguishable with either Fbw7 dimers or monomers. Although the cyclin ET62A and Fbw7ΔD mutations reduced binding, their residual affinity still supported cyclin E degradation in cotransfection assays (Fig. 1C). However, whereas the T380 degron was sufficient for cyclin E degradation (Fig. 1C, cyclin ET62A), the T62 degron was not (Fig. 1C, cyclin ET380A). Because cyclin E abundance solely reflects its turnover when Fbw7 is cotransfected, we examined abundance as a surrogate for degradation in this and related experiments.
Figure 1.
Degrons cooperate to bind Fbw7 dimers. (A) Model of cyclin E bound to dimeric Fbw7 through both degrons. (B) 293A cells were cotransfected with Flag-Fbw7 and MYC-cyclin E as indicated, and lysates were immunoprecipitated with Flag antibody and blotted with tag antibodies. (*) Heavy chain. The vertical line indicates cropped lanes. (C) Cyclin E turnover by dimers (Fbw7α) or monomers (Fbw7α-ΔD). 293A cells were cotransfected as indicated and analyzed by Western blotting (PCNA-loading control). (D,E) A weakened T380 degron requires both the T62 degron and Fbw7 dimers for cyclin E turnover (D) and binding to Fbw7 (E). The assays are as in B and C. (F) Cyclin E's degrons cooperate in a ubiquitylation assay. Lysates expressing the indicated MYC-tagged cyclin E constructs were subjected to in vitro ubiquitylation and immunoblotted for cyclin E (MYC tag).
Two suboptimal degrons bind to Fbw7 dimers
The T380 degron suffices for cyclin E turnover and prevented us from determining whether the combined interactions of two suboptimal degrons with Fbw7 dimers can drive degradation. However, because previous work found that cyclin E with a weakened T380 degron is degraded by dimers but not monomers (Hao et al. 2007; Welcker and Clurman 2007), we speculated that this context might unmask degron cooperativity. In fact, many Fbw7 substrates have suboptimal CPDs with unfavorable residues in various positions, including a P+4 glutamate instead of a phospho-S/T. We thus used cyclin E with a weakened T380 degron (cyclin ES384E) as a model substrate to determine how two suboptimal CPDs interact with Fbw7.
Cyclin ES384E was readily degraded by transfected Fbw7 dimers but not monomers, and this required the normally dispensable T62 degron, as shown by Fbw7's inability to degrade cyclin ET62A/S384E (Fig. 1D). Thus, two suboptimal CPDs cooperatively drove cyclin E degradation by Fbw7 dimers. The two suboptimal degrons also cooperatively bound Fbw7, since T62 was required for coimmunoprecipitation of cyclin ES384E with Fbw7ΔF (Fig. 1E). Finally, we showed that cyclin ES384E ubiquitylation by Fbw7 dimers in vitro also required the T62 CPD (Fig. 1F). Thus, productive interactions between Fbw7 dimers and cyclin ES384E required an intact T62 degron, indicating functional cooperation between two suboptimal CPDs, presumably by increased avidity.
We used two additional Fbw7 dimerization mutants to rule out possible artifacts of the Fbw7ΔD deletion. Both Fbw7-M240 (a truncation mutant) (Welcker and Clurman 2007) and Fbw7-LI (a double point mutant) (see the Materials and Methods; Tang et al. 2007) required an intact T380 degron for cyclin E turnover, confirming that monomers require a high-affinity CPD to degrade cyclin E (Supplemental Fig. SF2). Of note, we previously reported that cyclin ES384E was degraded by Fbw7 monomers (Welcker and Clurman 2007). This discrepancy resulted from increased Fbw7 affinity caused by an inadvertent Fbw7 mutation (A626T) of a residue that binds to the T380 CPD (data not shown).
Defining dimer-dependent degron positions
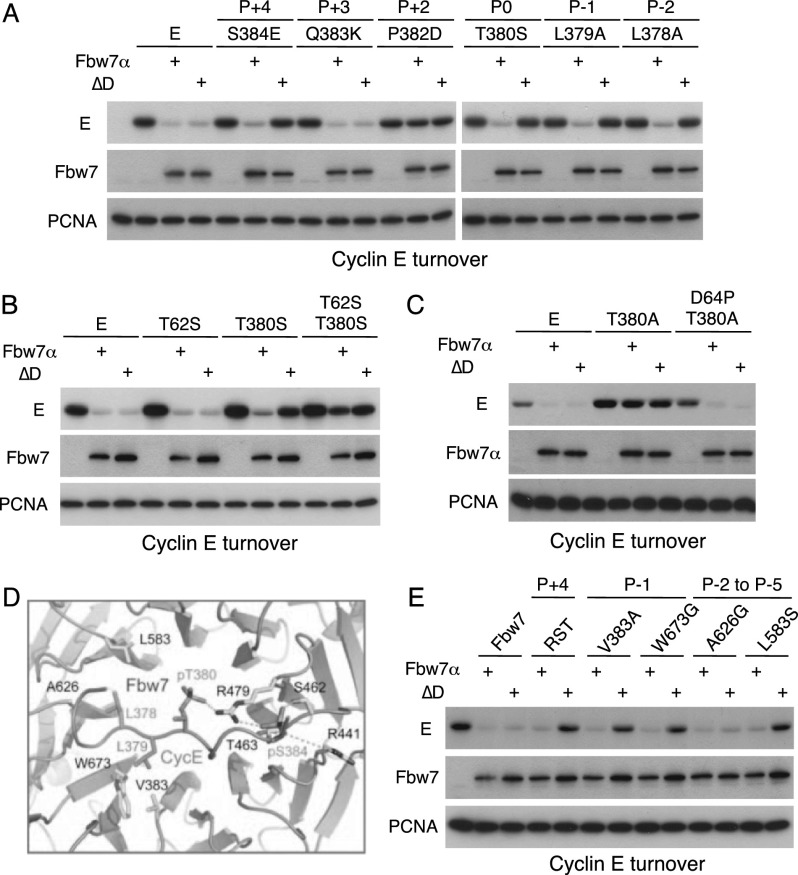
Because substrate CPDs differ in various positions, we examined which residues determine dimer dependence. Cyclin E selectively resisted degradation by Fbw7 monomers in transfection assays when the hydrophobic P−1 or P−2 positions were unfavorable, or when the P0 threonine is converted to serine (Fig. 2A; Supplemental Fig. SF3A). Phosphoserine in P0 was previously shown to reduce CPD affinity (Nash et al. 2001), and changing both central threonines to serines (cyclin ET62S/T380S) rendered cyclin E nearly resistant to Fbw7 (Fig. 2B, degradation; Supplemental Fig. SF3B, binding). Notably, some Fbw7 substrates naturally contain serine in P0 (SREBP2 and KLF5). While the P+3 position did not impact cyclin E turnover, the P+2 proline proved essential (Fig. 2A, cyclin EP382D). We attempted to increase T62 degron affinity by mimicking the T380 CPD and introduced a P+2 proline. Remarkably, cyclin ED64P was degraded and bound by transfected Fbw7 dimers and monomers even with a disabled T380 degron (Fig. 2C; Supplemental Fig. SF3C, cyclin ED64P/T380A).
Figure 2.
Degron determinants of dimer dependence. (A) 293A cells were cotransfected with MYC-cyclin E (mutations and their CPD positions, as indicated) and Flag-Fbw7 and analyzed as in Figure 1. (B) Weakened degrons containing serine in the central positions render cyclin E nearly resistant to degradation by Fbw7. The assay is as in A. (C) Introducing proline into P+2 of the T62 degron (D64P) results in a strong degron that overcomes the need for the T380 degron (D64P/T380A). The assay is as in A. (D) Schematic of the interaction of a cyclin E degron peptide with the Fbw7 propeller. (E) Fbw7 propeller mutants reveal mutational buffering by Fbw7 dimerization. Cells were cotransfected with Flag-Fbw7 dimer and monomer (ΔD) constructs containing mutations of degron-contacting residues as indicated and tested for MYC-cyclin E turnover by Western blotting as above.
In summary, a single high-affinity degron is sufficient for cyclin E turnover, even by Fbw7 monomers. However, many substrates naturally contain suboptimal degrons, suggesting that their degradation requires compensatory binding affinity elsewhere. For cyclin E, this is achieved by the accessory T62 degron, which can be engaged through Fbw7 dimerization. In other cases, we suggest that additional interaction signals remain to be discovered within either substrates themselves or perhaps their binding partners.
Fbw7 dimerization as a mutational buffer
We next tested the relationship between binding affinity and the requirement for Fbw7 dimerization by mutating Fbw7 propeller residues that make direct contact with the P−1, P−2, or P+4 positions of the T380 degron (Fig. 2D; Hao et al. 2007). Four β-propeller surface residues contact phospho-S384: R441, S462, T463, and R479, the latter of which also interacts with phospho-T380. We combined three mutations (R441M, S462A, and T463V; Fbw7RST) to weaken the interactions with phospho-S384 and used transfection to show that cyclin E degradation (Fig. 2E) and binding (Supplemental Fig. SF3E) by Fbw7RST required dimers. Similar results were achieved with mutations of residues that contact the cyclin E P−1 and P−2 positions (Fbw7V383/W673 and Fbw7L583, respectively) (Fig. 2E; Supplemental Fig. SF3E). Only one Fbw7 residue that is predicted to bind the CPD did not impact cyclin E degradation when mutated (A626). Fbw7 dimers thus tolerate mutations that weaken the Fbw7–substrate interface and disable substrate degradation by monomers. Dimerization may thus have evolved in part to buffer against a large set of mutations in substrates and Fbw7 that would impair monomer function, thereby restricting truly detrimental mutations to a few critical degron and β-propeller positions, such as the Fbw7 arginine missense mutations observed in cancers (Welcker and Clurman 2008). Moreover, these data suggest that Fbw7 arginine missense mutations might preferentially impact substrates that are dependent on functional dimers for their degradation.
Engineered human cell lines prevent dimerization of endogenous Fbw7
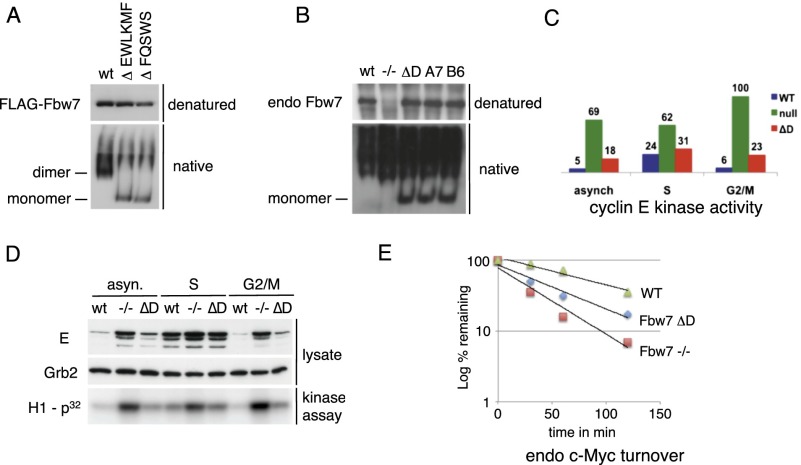
In order to study the role of endogenous Fbw7 dimerization, we used gene targeting to create multiple and independent HCT116 cell lines with homozygous endogenous Fbw7ΔD mutations (Supplemental Fig. SF4). The Fbw7ΔD deletion described above spans two exons and is not easily created by gene targeting. We thus generated another D-domain deletion (EWLKMF) upstream of the exon border that also eliminated dimerization (Supplemental Fig. SF4). To confirm that endogenous Fbw7 in our engineered cell lines is monomeric, we analyzed Fbw7 migration in nondenaturing gels. Recombinant Fbw7 dimers and monomers migrate with the expected molecular weights in sizing columns (data not shown) and are easily distinguished in native gels (Supplemental Fig. SF5). Both dimer-deficient Fbw7 mutants (FQSWS and EWLKMF) migrated as monomers when ectopically expressed (Fig. 3A). Importantly, endogenous Fbw7 monomers were detected in all three dimer-deficient cell lines but not in parental cells (Fig. 3B). The absence of ectopic or endogenous wild-type Fbw7 monomers strongly suggests that Fbw7 is exclusively a dimer in vivo. Of note, the nonspecific background signal that migrates slower than Fbw7 dimers in the native gels results from the antibodies used for immunoprecipitation and is seen even in Fbw7-null cells (Fig. 3B, lane 2). Thus, while dimers appeared absent from ectopic Flag-Fbw7ΔD mutants (Fig. 3A), the high background prevented us from showing this for endogenous Fbw7ΔD (Fig. 3B).
Figure 3.
Differential regulation of endogenous substrates by endogenous Fbw7 monomers. (A) Ectopic wild-type or dimerization mutants of Flag-Fbw7 expressed in 293A cells were subjected to denaturing and nondenaturing gel analysis and Western-blotted. The positions of Fbw7 monomers and dimers are indicated. (B) Engineered HCT116 cells express monomeric Fbw7. Three independent clones were generated (ΔD, A7, and B6) using two different gene targeting strategies (see the Materials and Methods). Analysis was as in A using immunoprecipitation-Western with Fbw7-specific antibodies. The position of monomers is indicated. (C,D). HCT116 cells expressing Fbw7 dimers (wild type [wt]) or monomers (ΔD) and Fbw7-null cells (−/−) were arrested by aphidicolin in S or G2/M phase or nocodazole in prometaphase or left unsynchronized (asyn) and analyzed for cyclin E abundance and activity. The bar graph depicts the relative amount of cyclin E activity. (E) HCT116 lines (wild type, null, and monomer) were analyzed by quantitative Western blot for c-Myc half-life after cycloheximide chase. The plot averages two independent experiments.
Differential substrate regulation in Fbw7ΔD cells
We examined three substrates that are stabilized in Fbw7-null cells: cyclin E, c-Myc, and SREBP. Cyclin E degradation is triggered by autophosphorylation and is most severely impacted by Fbw7 mutations in mitosis (Grim et al. 2008). We examined cyclin E in asynchronous cells and cells arrested in S phase or prometaphase. Cyclin E abundance and activity were constitutively high in Fbw7-null cells but only modestly increased in Fbw7ΔD cells, which exhibited nearly normal cyclin E periodicity (Fig. 3C,D). Thus, consistent with the finding that cyclin E's high-affinity T380 CPD supports its degradation by transfected Fbw7 monomers (Fig. 1C), endogenous cyclin E turnover does not strictly require endogenous Fbw7 dimerization, although dimerization does contribute somewhat to normal cyclin E regulation.
Fbw7 mediates c-Myc phospho-T58-dependent degradation (Welcker et al. 2004; Yada et al. 2004). Endogenous Myc turnover was somewhat more dimer-dependent than cyclin E, and its stability in Fbw7ΔD cells was intermediate to that of wild-type or Fbw7-null cells (Fig. 3E). Interestingly, the Myc CPD-Fbw7 contact area is predicted to be smaller than the cyclin E's T380 CPD (1002 Å vs. 1304 Å), although it is difficult to infer affinity differences from these predictions (Schumann 2007).
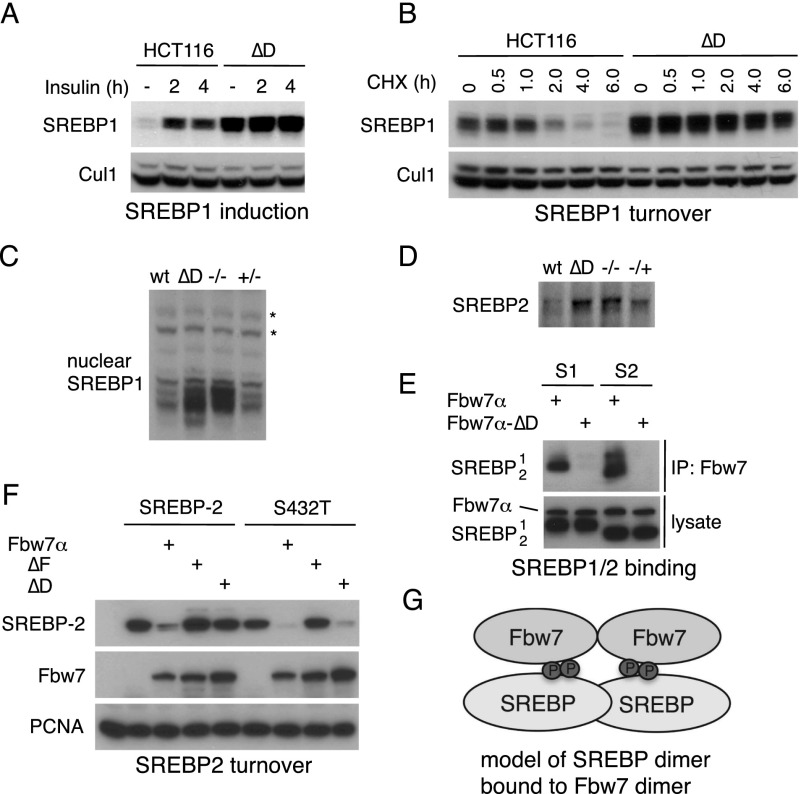
Finally, we examined SREBP to study an endogenous substrate with a suboptimal degron (Sundqvist et al. 2005). The SREBP1 CPD contains various unfavorable residues, most notably a threonine in P−2. SREBP1 abundance was greatly increased in resting and insulin-stimulated Fbw7ΔD cells (Fig. 4A), and this reflected impaired degradation (Fig. 4B). Strikingly, SREBP1 was nearly maximally stabilized in Fbw7ΔD cells when compared with Fbw7-null cells (Fig. 4C). Thus, endogenous SREBP1 turnover is almost completely dependent on Fbw7 dimerization. SREBP2 levels were also maximally increased in Fbw7ΔD cells when compared with Fbw7-null cells (Fig. 4D).
Figure 4.
SREBP regulation requires Fbw7 dimers. (A) Cells in serum-free medium were treated with insulin as indicated and immunoblotted for nuclear SREBP1 and Cul1 (loading control). (B) SREBP1 half-life is prolonged in HCT116 ΔD cells. Cells were treated with cycloheximide and analyzed by Western blotting. (C) Active nuclear SREBP1 is nearly maximally stabilized in cells expressing Fbw7 monomers compared with Fbw7-null cells. (*) Background band/loading control. (D) Nuclear SREBP2 abundance in HCT116 cell lines was determined by immunoprecipitation-Western analysis. (E) Dimer-dependent SREBP binding. 293A cells were transfected as indicated, immunoprecipitated with Flag antibody (Fbw7), and Western-blotted. (S1) SREBP1; (S2) SREBP2. dnCul1 was coexpressed to prevent SREBP turnover by coexpressed Fbw7. (F) 293A cells were transfected and blotted as indicated (PCNA-loading control). Converting the central serine to a threonine in the SREBP2 degron (LMSPPAS and S432T) allows SREBP2 degradation by Fbw7 monomers. (G) Model for the SREBP–Fbw7 interaction.
Transfected SREBP1 and SREBP2 exhibited dimer-dependent Fbw7 binding either when coexpressed with dnCul1 to inhibit turnover (Fig. 4E) or when lysates from transfected cells were mixed (Supplemental Fig. SF6A). The SREBP2 CPD is weakened by a P0 phosphoserine. Remarkably, conversion of this serine to threonine (SREBP2S432T) enabled degradation by Fbw7 monomers, again demonstrating that a high-affinity CPD allows substrate degradation by monomers, whereas only dimers productively interact with suboptimal degrons (Fig. 4F). Because SREBP1/2 form homodimers and heterodimers (Osborne 2000), we speculate that Fbw7 dimers interact with the CPDs of each protomer within an SREBP dimer (Fig. 4G). Supporting this idea, a dimer-deficient SREBP mutant bound Fbw7 poorly in vitro (Supplemental Fig. SF6B,C).
Fbw7 monomers are stable
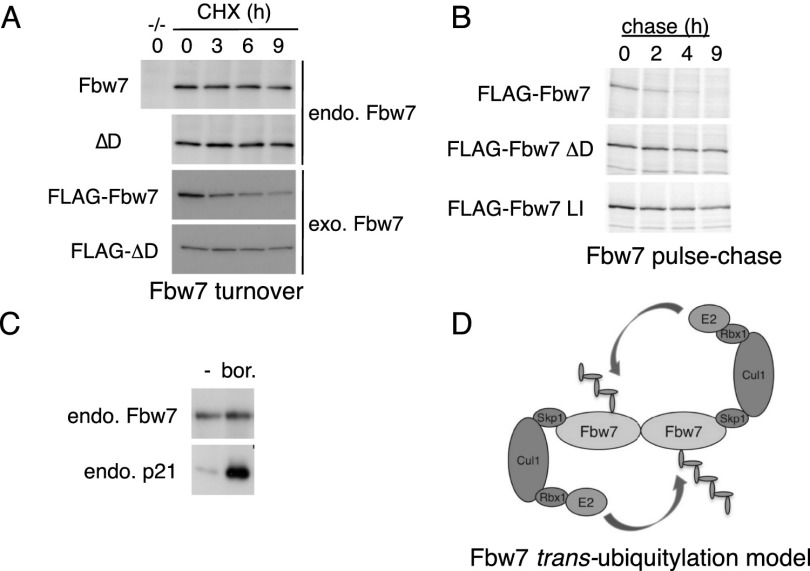
A recent report found that transiently transfected Fbw7 monomers are unstable due to increased autoubiquitylation (Min et al. 2012). In contrast, endogenous Fbw7 was equally abundant in wild-type and Fbw7ΔD cell lines (Fig. 3B). Moreover, transfected Fbw7ΔD is expressed at much higher levels from equal amounts of plasmid DNA (Supplemental Fig. SF3A). We thus examined the stability of Fbw7 monomers and dimers. Unexpectedly, Flag-Fbw7ΔD was much more stable than Flag-Fbw7 (Fig. 5A). We used 35S-methionine pulse chase to confirm that two different Fbw7 dimerization mutants (Fbw7ΔD and Fbw7LI) were stable compared with wild-type Fbw7 (Fig. 5B). Thus, ectopic dimers are labile, whereas monomers are stable. In contrast, endogenous dimers and monomers are both stable because neither appreciably decayed during a 9-h cycloheximide chase (Fig. 5A) or accumulated upon proteasome inhibition (Fig. 5C).
Figure 5.
Dimerization regulates Fbw7 stability and autoubiquitylation. (A) Cycloheximide chase of endogenous Fbw7 dimers and monomers (immunoprecipitated from HCT116 cells; shown in the top panels) and exogenous Flag-Fbw7 (transfected 293A cells; less Fbw7ΔD was loaded to allow comparable exposures). (B) Pulse chase of ectopic Flag-Fbw7 and two Flag-Fbw7 dimerization mutants in 293A cells. (LI) L256E/I257E. (C) Endogenous Fbw7 is insensitive to proteasome inhibition. HCT116 cells were treated with bortezomib, and Fbw7 was visualized by immunoprecipitation-Western. p21Cip1 serves as positive control. (D) Model of Fbw7 autoubiquitylation in trans.
These data contradict a study showing that ectopic Fbw7 monomers are unstable and that conversion of ectopic Fbw7 dimers to monomers by the Pin1 prolyl isomerase causes Fbw7 turnover (Min et al. 2012). Indeed, native gel analyses revealed that cotransfection of Pin1 with Fbw7 did not instigate the formation of Fbw7 monomers (Supplemental Fig. SF7). It is unclear why ectopic Fbw7 is unstable compared with endogenous Fbw7, although we confirmed that its stability depends on the proteasome and its F-box (Supplemental Fig. SF8A,B). One possibility is that a limiting factor that prevents Fbw7 degradation, such as a deubiquitylating enzyme, is overwhelmed by Fbw7 overexpression.
Dimerization regulates Fbw7 autoubiquitylation
The stability of ectopic Fbw7 monomers suggested a trans-ubiquitylation model in which one Fbw7 protomer is ubiquitylated by its associated protomer (Fig. 5D). To test this idea, we purified differentially tagged Fbw7 heterodimers from cotransfected cells by sequential immunoprecipitation and subjected the complexes to autoubiquitylation. The target Fbw7 protomer (MYC-Fbw7ΔF) cannot bind SCF, and its autoubiquitylation depends on the associated protomer, which we expressed as Flag-Fbw7 or Flag-Fbw7ΔF. Supplemental Figure SF8C shows that MYC-Fbw7ΔF is ubiquitylated in trans by an associated Flag-Fbw7 protomer but not by Flag-Fbw7ΔF. Fbw7 dimers can thus trans-ubiquitylate in vitro. We also observed cis-ubiquitylation of recombinant Fbw7 monomers in vitro (Supplemental Fig. SF8D), and both cis and trans mechanisms may contribute to Fbw7 autoubiquitylation.
If trans-ubiquitylation regulates Fbw7 stability in vivo, overexpressed Fbw7ΔF might dominantly stabilize Fbw7 by forming heterodimers in which the wild-type protomer cannot be trans-ubiquitylated. Indeed, MYC-tagged Fbw7 abundance was increased by coexpressed Flag-Fbw7ΔF but not Flag-Fbw7 (Supplemental Fig. SF8E); stabilization required dimerization and did not occur with Flag-Fbw7ΔFD. Fbw7 truncation mutants further supported the idea that dominant Fbw7 stabilization requires dimerization (Supplemental Fig. SF8F). These data support a model in which dimerization regulates Fbw7 stability via trans-autoubiquitylation.
Dimerization and multiple substrate recognition motifs in Cullin ring ligases (CRLs)
An important implication of our study is that multiple degrons allow complex signaling pathways to impact Fbw7 pathway activity. In addition to cyclin E, MCL1 and PGC1α may also be examples of Fbw7 substrates that are regulated by multiple degrons (Olson et al. 2008; Inuzuka et al. 2011; Wertz et al. 2011).
The concept that dimerization allows multiple degron interactions is not mutually exclusive with the idea that dimerization also regulates SCF catalysis. Indeed, we used Fbw7 dimers or monomers to ubiquitylate cyclin E in the presence of a ubiquitin mutant (K48R) that reveals the number of substrate conjugation sites because it cannot be chain-extended (Supplemental Fig. SF9). As predicted, dimers conjugated Ub-K48R to multiple cyclin E sites, whereas monomers targeted a single site. Dimerization thus impacts SCF function through both substrate binding and catalysis.
Sic1 is a multi-CPD Cdc4 substrate that has been heavily studied (Feldman et al. 1997; Nash et al. 2001; Hao et al. 2007; Orlicky et al. 2010). However, there are differences between the interactions of an Fbw7 dimer with cyclin E and those of Sic1 with Cdc4. Although individual diphosphorylated high-affinity Sic1 peptides may bind Cdc4 similarly to cyclin E/Fbw7, systematic mutational analyses revealed that numerous low-affinity CPDs dynamically engage a single Cdc4 substrate-binding domain such that its affinity appears independent of Cdc4 dimerization (Mittag et al. 2008; Tang et al. 2012). Nonetheless, Cdc4 dimerization is needed for catalytic activity in vitro and function in vivo. The different types of substrate interactions found among these orthologs highlight the complexity of protein degradation by this critical ligase complex.
In addition to other SCF substrate receptors (e.g., β-TrCP) (Suzuki et al. 2000) dimerization is also found in other CRLs; notably, Cul3-BTB ligases. For example, the flexibility of a Cul3-SPOP dimer may allow it to engage multiple binding sites (Zhuang et al. 2009), and the interaction of a Cul3-Keap1 dimer with Nrf2 involves two Nrf2 recognition sites that are engaged by a Keap1 dimer (Tong et al. 2007). Thus, the interactions of dimeric substrate receptors with multiple substrate-binding motifs may broadly regulate CRL function.
Materials and methods
Reagents
Plasmids and mutagenesis
MYC-Cyclin E and Flag-Fbw7 constructs were described previously (Welcker et al. 2003). Human SREBP1 and SREBP2 were cloned from cDNA as processed forms (truncated at amino acids 490 and 484, respectively). Site-directed mutants were made by QuikChange (Stratagene). Fbw7ΔD and truncation mutants were previously described (Welcker and Clurman 2007). Fbw7LI mutates residues 256 and 257 into aspartates. Fbw7ΔF deletes the entire F-box (281–321).
Antibodies
The antibodies used were as follows: Flag tag (M2, Sigma), HA tag (Y-11, Sigma), MYC tag (9E10, in-house), Fbw7 (A301-720 and A301-721, Bethyl Laboratories), SREBP1 (H-160, Sigma), SREBP2 (Cayman Chemical Company and R&D Systems), PIN1 (G8, Sigma), PCNA (PC-10, in-house), and Cul1 (71-8700, Zymed).
Native gel analysis
Cells were lysed in Tween-20 buffer (50 mM Tris, 150 mM NaCl, 10% glycerol, 0.1% Tween-20, 1 mM EDTA, plus protease and phosphatase inhibitors), mixed in native loading buffer (50 mM Tris at pH 6.8, 10% glycerol, bromophenol blue), and run in gels excluding SDS.
Acknowledgments
We thank Dr. Tanel Punga for SREBP1ΔD. This work was supported by National Institutes of Health grants CA084069 and CA102742 (to B.E.C.), CA107134 (to N.Z.), and 2T32 GM007270 (to E.A.L.); Howard Hughes Medical Institute (N.Z.); Science Foundation Ireland grants 07/SK/B1242b and 10/IN.1/B2986 (to J.E.); and the American Cancer Society (J.E.G.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.229195.113.
References
- Akhoondi S, Sun D, von der Lehr N, Apostolidou S, Klotz K, Maljukova A, Cepeda D, Fiegl H, Dafou D, Marth C, et al. 2007. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res 67: 9006–9012 [DOI] [PubMed] [Google Scholar]
- Bengoechea-Alonso MT, Ericsson J 2010. Tumor suppressor Fbxw7 regulates TGFβ signaling by targeting TGIF1 for degradation. Oncogene 29: 5322–5328 [DOI] [PubMed] [Google Scholar]
- Crusio KM, King B, Reavie LB, Aifantis I 2010. The ubiquitous nature of cancer: The role of the SCF(Fbw7) complex in development and transformation. Oncogene 29: 4865–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ 1997. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell 91: 221–230 [DOI] [PubMed] [Google Scholar]
- Grim JE, Gustafson MP, Hirata RK, Hagar AC, Swanger J, Welcker M, Hwang HC, Ericsson J, Russell DW, Clurman BE 2008. Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. J Cell Biol 181: 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP 2007. Structure of a Fbw7–Skp1–cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell 26: 131–143 [DOI] [PubMed] [Google Scholar]
- Inuzuka H, Shaik S, Onoyama I, Gao D, Tseng A, Maser RS, Zhai B, Wan L, Gutierrez A, Lau AW, et al. 2011. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471: 104–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp DM, Schaefer LK, Ye X, Keyomarsi K, Chu C, Harper JW, Elledge SJ 2001. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294: 173–177 [DOI] [PubMed] [Google Scholar]
- Min SH, Lau AW, Lee TH, Inuzuka H, Wei S, Huang P, Shaik S, Lee DY, Finn G, Balastik M, et al. 2012. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol Cell 46: 771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittag T, Orlicky S, Choy WY, Tang X, Lin H, Sicheri F, Kay LE, Tyers M, Forman-Kay JD 2008. Dynamic equilibrium engagement of a polyvalent ligand with a single-site receptor. Proc Natl Acad Sci 105: 17772–17777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P, Tang X, Orlicky S, Chen Q, Gertler FB, Mendenhall MD, Sicheri F, Pawson T, Tyers M 2001. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414: 514–521 [DOI] [PubMed] [Google Scholar]
- Olson BL, Hock MB, Ekholm-Reed S, Wohlschlegel JA, Dev KK, Kralli A, Reed SI 2008. SCFCdc4 acts antagonistically to the PGC-1α transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev 22: 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Willems A, Tyers M, Sicheri F 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112: 243–256 [DOI] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M 2010. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol 28: 733–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne TF 2000. Sterol regulatory element-binding proteins (SREBPs): Key regulators of nutritional homeostasis and insulin action. J Biol Chem 275: 32379–32382 [DOI] [PubMed] [Google Scholar]
- Schumann S. 2007. “Structure determination and biochemical characterization of the protein ubiquitin ligases SCFFbw7and APC.” PhD thesis, Martin Luther University of Halle-Wittenberg, Halle-Wittenberg, Germany. [Google Scholar]
- Skaar JR, Pagan JK, Pagano M 2013. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol 14: 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strohmaier H, Spruck CH, Kaiser P, Won KA, Sangfelt O, Reed SI 2001. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413: 316–322 [DOI] [PubMed] [Google Scholar]
- Sundqvist A, Bengoechea-Alonso M, Ye X, Lukiyanchuk V, Jin J, Harper JW, Ericsson J 2005. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCFFbw7. Cell Metab 1: 379–391 [DOI] [PubMed] [Google Scholar]
- Suzuki H, Chiba T, Suzuki T, Fujita T, Ikenoue T, Omata M, Furuichi K, Shikama H, Tanaka K 2000. Homodimer of two F-box proteins βTrCP1 or βTrCP2 binds to IκBα for signal-dependent ubiquitination. J Biol Chem 275: 2877–2884 [DOI] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M 2007. Suprafacial orientation of the SCF(Cdc4) dimer accommodates multiple geometries for substrate ubiquitination. Cell 129: 1165–1176 [DOI] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Mittag T, Csizmok V, Pawson T, Forman-Kay JD, Sicheri F, Tyers M 2012. Composite low affinity interactions dictate recognition of the cyclin-dependent kinase inhibitor Sic1 by the SCFCdc4 ubiquitin ligase. Proc Natl Acad Sci 109: 3287–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong KI, Padmanabhan B, Kobayashi A, Shang C, Hirotsu Y, Yokoyama S, Yamamoto M 2007. Different electrostatic potentials define ETGE and DLG motifs as hinge and latch in oxidative stress response. Mol Cell Biol 27: 7511–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Wang Y, Liu N, Ren C, Jiang C, Zhang K, Yu S, Chen Y, Tang H, Deng Q, et al. 2013. FBW7 regulates endothelial functions by targeting KLF2 for ubiquitination and degradation. Cell Res 23: 803–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE 2007. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div 2: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE 2008. FBW7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer 8: 83–93 [DOI] [PubMed] [Google Scholar]
- Welcker M, Singer J, Loeb KR, Grim J, Bloecher A, Gurien-West M, Clurman BE, Roberts JM 2003. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol Cell 12: 381–392 [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JA, Harper JW, Eisenman RN, Clurman BE 2004. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci 101: 9085–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, Kusam S, Lam C, Okamoto T, Sandoval W, Anderson DJ, Helgason E, Ernst JA, Eby M, Liu J, et al. 2011. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 471: 110–114 [DOI] [PubMed] [Google Scholar]
- Yada M, Hatakeyama S, Kamura T, Nishiyama M, Tsunematsu R, Imaki H, Ishida N, Okumura F, Nakayama K, Nakayama KI 2004. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J 23: 2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Koepp DM 2006. Fbw7 isoform interaction contributes to cyclin E proteolysis. Mol Cancer Res 4: 935–943 [DOI] [PubMed] [Google Scholar]
- Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, et al. 2009. Structures of SPOP–substrate complexes: Insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell 36: 39–50 [DOI] [PMC free article] [PubMed] [Google Scholar]