Deregulation of DNA replication origin firing and rereplication promote genome instability and tumorigenesis by largely elusive mechanisms. Neelsen et al. investigate the consequences of Emi1 depletion in human cells, previously associated with rereplication. DNA fiber labeling shows that origin reactivation occurs at the earliest stages of Emi1 depletion and is associated with checkpoint-blind ssDNA gaps and replication fork reversal. The authors propose a new model for rereplication-induced chromosomal breakage, which may contribute to cancer-relevant genome rearrangements.
Keywords: DNA replication, genome integrity, origin licensing, rereplication, tumorigenesis
Abstract
Deregulated origin licensing and rereplication promote genome instability and tumorigenesis by largely elusive mechanisms. Investigating the consequences of Early mitotic inhibitor 1 (Emi1) depletion in human cells, previously associated with rereplication, we show by DNA fiber labeling that origin reactivation occurs rapidly, well before accumulation of cells with >4N DNA, and is associated with checkpoint-blind ssDNA gaps and replication fork reversal. Massive RPA chromatin loading, formation of small chromosomal fragments, and checkpoint activation occur only later, once cells complete bulk DNA replication. We propose that deregulated origin firing leads to undetected discontinuities on newly replicated DNA, which ultimately cause breakage of rereplicating forks.
The activation of DNA replication origins is a tightly regulated mechanism, entailing two main steps: (1) “origin licensing,” restricted to late mitosis and early G1, when essential replication initiation proteins (ORC1, Cdc6, Cdt1, and Mcm2–7) are sequentially loaded on origin DNA sequences, forming the “prereplicative complex” (preRC), and (2) “origin firing,” occurring throughout S phase, when additional proteins are recruited to the preRC and start unwinding and DNA synthesis (Arias and Walter 2007). As relicensing and thus rereplication are detrimental to genome stability, several cyclin-dependent kinase (CDK)-dependent and -independent mechanisms have evolved to coordinate these steps with cell cycle progression (Blow and Dutta 2005; Arias and Walter 2007).
Although several preRC components are targets of regulation, the major mechanism by which metazoans prevent origin licensing during S phase is inactivation of Cdt1 by ubiquitin-mediated degradation or binding to its inhibitor Geminin. Cdt1 proteolysis is tightly linked to the cell cycle, as ubiquitylation requires CDK-dependent phosphorylation (Li et al. 2003; Sugimoto et al. 2004; Nishitani et al. 2006). Moreover, CUL4/DDB1-mediated ubiquitylation of Cdt1 occurs in S phase or in response to DNA damage (Arias and Walter 2006; Nishitani et al. 2006; Senga et al. 2006). Geminin exerts its inhibitory function on Cdt1 in S, G2, and early M phase and is inactivated in late M phase by anaphase-promoting complex (APC/C)-dependent polyubiquitylation, leading to reactivation of origin licensing (McGarry and Kirschner 1998; Wohlschlegel et al. 2000; Tada et al. 2001; Li and Blow 2004). Accordingly, Geminin depletion induces rereplication and activation of the DNA damage response (DDR) (Melixetian et al. 2004; Zhu et al. 2004).
By direct control of Geminin and indirect control of Cdt1 proteolysis via regulation of CycA–CDK activity, APC/C plays a pivotal role coordinating origin licensing with cell cycle progression (Hook et al. 2007). APC/C activity is inhibited by Early mitotic inhibitor 1 (Emi1) (Wang and Kirschner 2013), which thereby stabilizes APC/C substrates like Geminin and Cyclin A (Di Fiore and Pines 2007). Thus, inactivation of Emi1 leads to degradation of both inhibitors of Cdt1 activity, resulting in massive rereplication and DDR activation (Machida and Dutta 2007).
As many origin licensing genes are overexpressed in cancer cells and several oncogenes are known to affect origin licensing, it is suspected that deregulated licensing contributes to genome instability and tumorigenesis (Hook et al. 2007; Blow and Gillespie 2008). However, our understanding of how rereplication challenges genome stability is very limited. Studies with Xenopus laevis egg extracts provided the first insight into the effects of rereplication. Addition of recombinant Cdt1 to G2-arrested egg extracts was shown to trigger DNA breaks, proposed to arise from head-to-tail collision of rereplicating forks (Davidson et al. 2006). However, little information is available on the mechanisms leading to DNA damage and DDR activation in rereplicating human cells.
We combined cell/molecular biology and in vivo single-molecule approaches to investigate how deregulated origin licensing by Emi1 depletion affects replicating chromosomes. We show that cells experience mild DNA replication stress and ssDNA accumulation during the first replication round upon licensing deregulation, which may act as precursor for DNA breaks, when rereplicating forks approach ssDNA gaps on the template. Extending the analysis to other experimental systems of deregulated licensing (Geminin depletion and Cdt1 addition in X. laevis extracts), we propose a new model for rereplication-induced chromosomal breakage, which may contribute to cancer-relevant genome rearrangements.
Results and Discussion
Emi1 depletion affects DNA synthesis prior to accumulation of >4N DNA, chromosomal breakage, and checkpoint activation
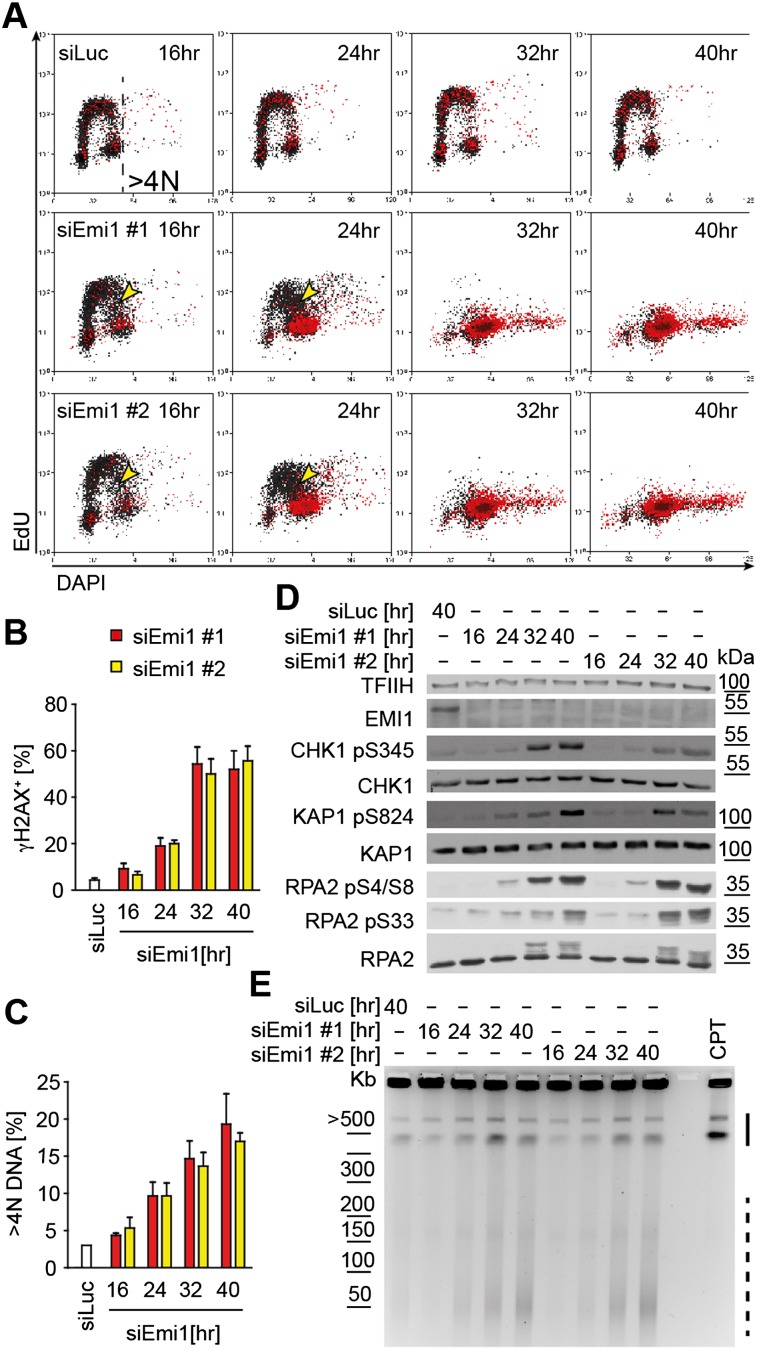
To gain mechanistic insight into how deregulated origin licensing affects the replication process, leading to DNA breaks and DDR activation, we depleted Emi1 in U2OS cells, a condition previously associated with rereplication and DNA damage (Machida and Dutta 2007). Using flow cytometry, we monitored cell cycle progression (DNA content), DNA synthesis (EdU incorporation), and DDR activation (phosphorylation of H2AX [γH2AX]) (Supplemental Fig. S1) after Emi1 depletion. Sixteen hours to 24 h after siEmi1 transfection, we noticed accumulation of cells in S phase and a reduced incorporation rate in mid–late S phase (Fig. 1A). In synchronized cells, the impact of deregulated origin licensing on DNA synthesis was detected from the onset of the first S phase (Supplemental Fig. S2). At these time points, γH2AX was only detected in cells close to having completed a first round of bulk DNA replication (Fig. 1A,B). Later (32–40 h), γH2AX and a markedly reduced rate of DNA synthesis were detected in cells displaying >4N DNA, a commonly used readout for rereplication (Fig. 1A,C). Only at 32–40 h did cells accumulate detectable levels of double-strand breaks (DSB) and display activation of ATM and ATR pathways (phosphorylation of KAP1/RPA2-S4/S8 and CHK1/RPA2-S33, respectively), as expected for DSB-induced DDR (Fig. 1D,E). A relevant fraction of chromosomal fragments induced by Emi1 depletion is significantly smaller (20–100 kb) than camptothecin-induced DSB (0.5–2 Mb) (Fig. 1E; Supplemental Fig. S1B; Hanada et al. 2007), suggesting that rereplication-induced DSBs are clustered. DNA breakage at 32–40 h was also confirmed by colocalization of γH2AX and 53BP1, particularly evident in cells with “giant nuclei,” a sign of extensive rereplication (Supplemental Fig. S1C–E; Zhu et al. 2004). Altogether, these data indicate that mild replication stress during the first S phase after Emi1 depletion precedes cell cycle arrest, DNA breakage, and DDR activation, which are coupled to overt rereplication (DNA content >4N). Similar observations were made in untransformed human epithelial cells (RPE-1) (Supplemental Fig. S3), showing that the stepwise impact on DNA replication and genome stability is a general consequence of Emi1 depletion.
Figure 1.
Emi1 depletion causes DNA replication stress in S phase and DDR activation and DNA breakage in cells with ≥4 DNA. (A) FACS analysis of DNA synthesis (EdU), DNA content (DAPI), and DDR activation (γH2AX) after mock (siLUC) or Emi1 depletion in U2OS cells using two different siRNAs. γH2AX+ cells are in red (see also Supplemental Fig. S1A). Yellow arrowheads indicate cells with compromised DNA synthesis. γH2AX+ cells (B) and cells with >4N DNA (C) after mock (siLUC) or Emi1 depletion quantified by FACS. Mean + SEM; n = 3. (D) ATR (pCHK1), ATM (pKAP1) activation, RPA phosphorylation (RPA2 pS4/S8 and pS33), and total DDR proteins (CHK1, KAP1, and RPA2) assessed by Western blot upon Emi1 depletion. (TFIIH) Loading control. (E) DNA breakage after mock (siLUC) or Emi1 depletion monitored by pulse-field gel electrophoresis. The solid and dashed lines indicate large (0.5- to 2-Mb) and smaller (20- to 100-kb) chromosomal fragments, respectively. The molecular size markers are based on data in Supplemental Figure S1B. Four-hour treatment with 1 μM camptothecin (CPT) served as a positive control for DSB.
Progressive RPA accumulation on chromatin precedes rereplication-associated DNA damage
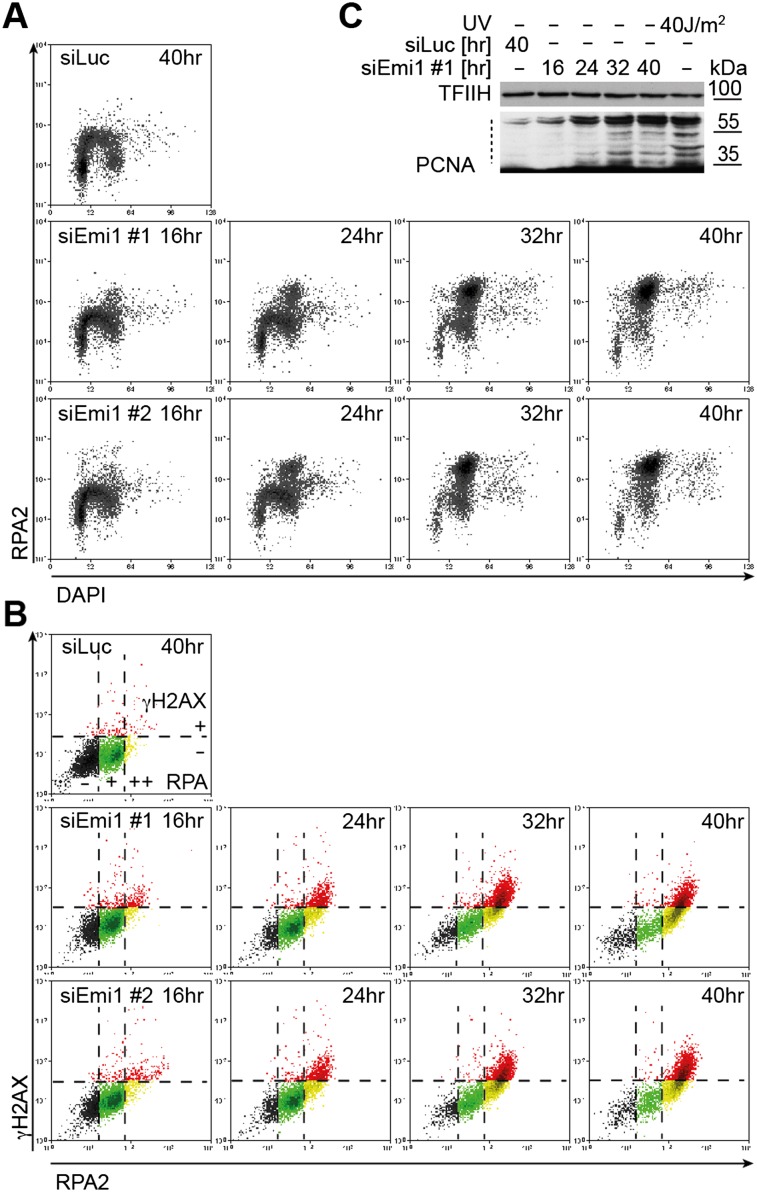
To further characterize DNA replication stress early after Emi1 depletion, we monitored chromatin loading of the human ssDNA-binding protein (RPA) (Forment et al. 2012). Limited amounts of ssDNA are present during DNA replication, leading to RPA chromatin loading in S phase (Fig. 2A; Supplemental Fig. S4A; Forment et al. 2012). While this signal is rapidly lost as control cells complete S phase, Emi1 depletion leads to progressive accumulation of RPA on chromatin and unusually high RPA levels in mid–late S-phase cells (Fig. 2A). RPA foci colocalized with γH2AX foci at late time points (32–40 h) (Supplemental Fig. S4A), presumably marking processing of the detected DSBs (Fig. 1E; Supplemental Fig. S1C,D). However, in both U2OS and RPE-1 cells, some RPA accumulation was already observed at earlier time points (16–24 h), when it was largely uncoupled from DDR activation, extensive rereplication (measured by flow cytometry), and DNA breaks (Figs. 1, 2A; Supplemental Fig. S3D). Furthermore, even at later time points (≥24 h) after Emi1 depletion, when γH2AX is clearly detectable in the population, our FACS experiments identified a cell population with unusually high RPA content in the absence of γH2AX (Fig. 2B, yellow dots, RPA++ γH2AX−). These data suggest that Emi1 depletion progressively induces ssDNA accumulation, which goes undetected by the DDR and precedes rereplication-associated DNA breaks. Importantly, PCNA ubiquitylation, a sensitive marker of replication-associated ssDNA gaps (for review, see Chen et al. 2011), was detectable within 24 h (Fig. 2C) and thus earlier than other DDR markers (Fig. 1D).
Figure 2.
siEmi1-induced deregulation of origin licensing promotes RPA chromatin binding and ubiquitylation of PCNA from the first S phase. (A) FACS analysis of chromatin-bound RPA and DNA content (DAPI) after mock (siLuc) or Emi1 depletion in U2OS cells. (B) γH2AX/RPA levels in samples in A. Black, green, and yellow regions identify RPA negative cells (−), cells with S-phase RPA levels (+), and cells with elevated RPA (++), respectively. The red region identifies γH2AX+ cells. See Supplemental Figure S4B for Emi1 levels. (C) Analysis of PCNA ubiquitylation in mock-transfected cells (siLuc) and at the indicated time points after Emi1 depletion (siEmi1 #1). The dotted line indicates ubiquitylated PCNA. UV-irradiated cells served as positive control. (TFIIH) Loading control.
Emi1 depletion does not detectably impair fork progression but induces refiring of clustered origins before accumulation of >4N DNA content
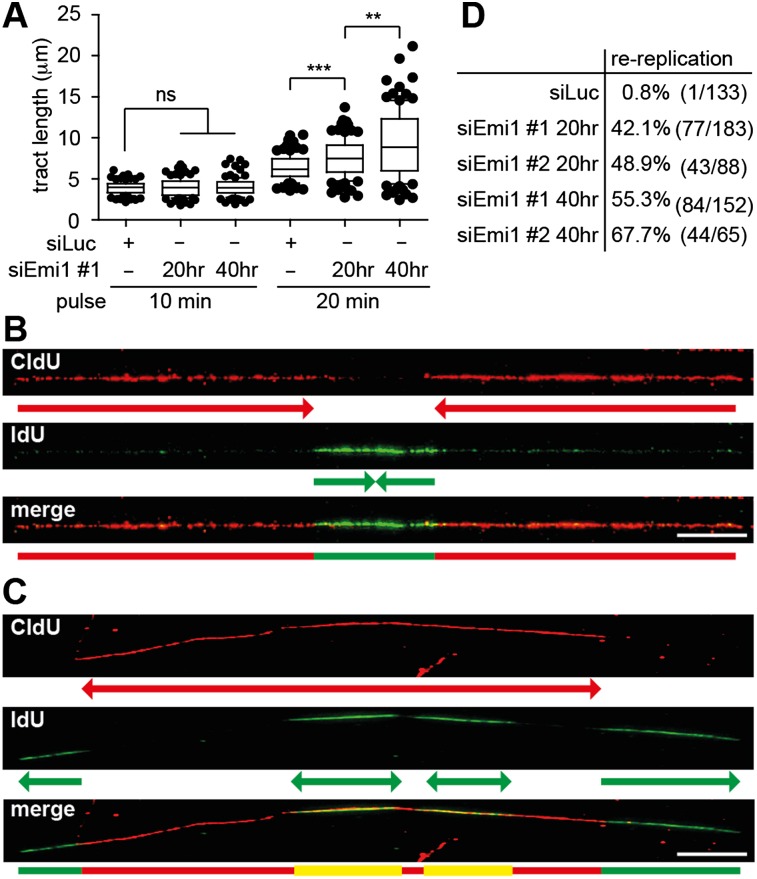
We next monitored the effect of Emi1 depletion on fork progression by DNA fiber spreading (Jackson and Pombo 1998). Ongoing forks were identified by a red–green pattern (Supplemental Fig. S5A). Fork progression appeared unaffected by Emi1 depletion when we used a 10-min labeling time (Fig. 3A). However, in Emi1-depleted cells, tract length increased more markedly than in control cells with longer labeling times (Fig. 3A; Supplemental Fig. S5C,D). This suggests that Emi1 depletion does not affect progression of individual forks but that deregulated activation of clustered replication origins leads to more frequent fork fusion and thus longer tracts. We then adapted the labeling protocol to detect DNA rereplication events, modifying a published protocol (Dorn et al. 2009). A 120-min CldU pulse followed by a 30-min IdU pulse allowed us to follow fork progression and reactivation of replication origins in previously replicated tracts (Supplemental Fig. S5A). “Rereplication” events during the second labeling should appear as green signals embedded in a longer red tract. A similar pattern is expected for physiological fork fusion events during the second label (“termination”), but rereplication events can be distinguished by the substantial overlap of red and green signals (Fig. 3B,C). As expected, in control cells, virtually all green signals identified within red tracts displayed a termination pattern (“red–green–red”) (Fig. 3B,D). In contrast, Emi1-depleted cells showed rereplication events (“red–yellow–red”), often coupled to further progression of the first set of forks during the second labeling period (Fig. 3C, note the distal green tracts). Surprisingly, these “rereplication” events were almost as frequent as “termination” events already 20 h after siRNA transfection (Fig. 3D). At 40 h, when rereplication has led to >4N DNA content (Fig. 1A,C), rereplication events were more frequent than fork fusions and were occasionally clustered on the same DNA fiber (Fig. 3C,D). These data demonstrate that origin reactivation can be detected by DNA fiber spreading before it is detectable by flow cytometry and that refiring of clustered origins occurs already during a first round of replication with deregulated origin licensing.
Figure 3.
Rereplication is detectable by a DNA fiber-spreading assay before completion of bulk DNA synthesis. (A) Length of newly replicated tracts (IdU; green) in mock-depleted U2OS cells (siLuc) and after Emi1 depletion (siEmi1 #1), using 10-min or 20-min labeling pulses. (B,C) Representative DNA tracts labeled with CldU for 2 h and IdU for 30 min to identify termination and rereplication events. (B) A replication “termination” event. (C) Two “rereplication” events in close proximity. (D) Quantification of rereplication/termination events as shown in B and C after mock (siLuc) or Emi1 depletion. The percentage indicated represents the fraction of rereplication events in the total population of “red–green–red” tracts analyzed. (Whiskers) 10–90 percentile; (***) P < 0.0001; (**) P < 0.005; (ns) not significant, Mann-Whitney test; n = 100 in A. Bar, 10 μm. See Supplemental Figure S5, A and B, for Emi1 levels and labeling protocols to study fork progression (A) and rereplication events (B,C).
Deregulation of origin licensing induces ssDNA gaps on replicated duplexes, which can be template for rereplication
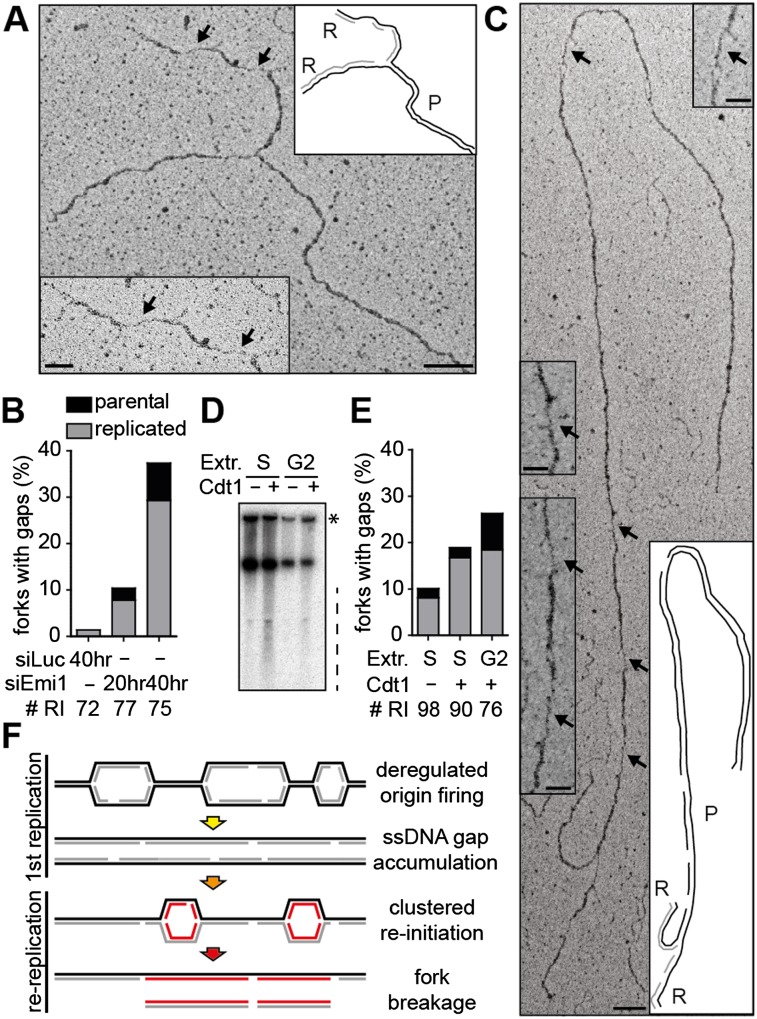
To gain additional insight into the molecular consequences of deregulated origin licensing, we investigated in vivo replication fork structure by electron microscopy (EM) (Neelsen et al. 2014). Already 20 h after siRNA transfection, several marks of replication stress were detectable (Fig. 4). Small (<1-kb) replication bubbles were overrepresented upon Emi1 depletion (9%–10% compared with 1%–2% in control U2OS cells), suggesting that deregulated firing is accompanied by reduced fork progression from the origin. These replicated tracts would be too small for detection in DNA fiber assays, which may explain why the reduced EdU incorporation after Emi1 depletion (Fig. 1A) is not accompanied by detectable reduction in fork progression (Fig. 3A). As implied by RPA chromatin loading and PCNA ubiquitylation (Fig. 2A,C), 11% of forks exposed ssDNA gaps 20 h after siEmi1 transfection compared with 1% of forks in control cells (Fig. 4A,B; Supplemental Fig. S6A). Furthermore, ∼13% of the replication forks had undergone reversal (Supplemental Fig. S6B,C). The latter two features closely resemble the effects of oncogene activation (Neelsen et al. 2013) and thus most likely reflect the licensing defects common to these genetic conditions (Hook et al. 2007; Blow and Gillespie 2008) and their consequences in terms of nucleotide depletion (Bester et al. 2011) and/or interference with transcription (Jones et al. 2013). Although ssDNA sensors (e.g., RPA chromatin loading and PCNA ubiquitylation) (Fig. 2A,C) detected these changes in the architecture of replication intermediates, they are “checkpoint-blind” (i.e., not associated per se with DDR activation) (Fig. 1A,D), as already shown for CycE overexpression (Neelsen et al. 2013). The proportion of forks displaying ssDNA gaps increases from 11% to 37% between 20 and 40 h, when rereplication and DSB become detectable by flow cytometry and PFGE, respectively (Figs. 1A,C,E, 4B). Intriguingly, a significant proportion of the observed ssDNA gaps were located on template DNA ahead of the replication forks (Fig. 4B,C; Supplemental Fig. S6D). At 40 h, when all cells completed a first round of replication and the relative proportion of rereplicating forks in our EM samples is expected to increase, 20% of the ssDNA gaps (n = 6 of 22) were detected on template DNA ahead of the replication forks (Fig. 4B). These data strongly suggest that gaps accumulating during the first round of replication after Emi1 depletion persist and present a damaged template for new replication rounds.
Figure 4.
Emi1 depletion leads to ssDNA gaps on the replicated duplex, which persist as a template for rereplicating forks. (A,C) Electron micrographs of representative replication forks from U2OS cells 40 h after transfection with siEmi1. Black arrows indicate ssDNA gaps. The insets show magnified ssDNA gaps and schemes of fork structure, indicating parental (P) and replicated (R) duplexes. Gaps are on a replicated duplex in A and on the parental duplex in C. Black and gray lines describe parental and newly synthesized DNA strands in the replicated duplexes, respectively. Bars: 100 nm (250 base pairs [bp]); inset, 50 nm. (B) Frequency of replication forks with ssDNA gaps in mock-depleted cells (siLuc) and after Emi1 depletion (siEmi1 #1). #RI is the number of analyzed replication intermediates. (D) Sperm nuclei replication assays in Xenopus interphase extracts. For S-phase experiments, extracts were optionally supplemented with 10 ng/μL Cdt1 at the time of sperm and [α-32P]dATP addition and incubated for 60 min. For G2 experiments, Cdt1 was optionally added with [α-32P]dATP 90 min after sperm addition and incubated for a further 60 min. After incubation, DNA was isolated, separated by neutral agarose gel electrophoresis, and autoradiographed. The dashed line indicates sperm DNA fragmentation. The asterisk indicates branched replicating DNA molecules retained in the well. (E) Frequency of replication forks with ssDNA gaps recovered after sperm nuclei incubation in S-phase or G2-phase extracts (see D), with optional addition of Cdt1. # RI is the number of analyzed replication intermediates. (F) Model for the formation of chromosomal breaks upon deregulation of origin licensing by Emi1 depletion. Excessive firing of clustered origins leads to replication stress during the first S phase and accumulation of ssDNA gaps. Uncontrolled reactivation of replication origins in this context triggers chromosomal breakage by replication of a discontinuous template.
To further test this hypothesis, we analyzed by EM a different experimental system associated with rereplication and DNA breakage; i.e., addition of Cdt1 to replication sperm nuclei in X. laevis egg extracts (Davidson et al. 2006). In line with published results, addition of Cdt1 (Ferenbach et al. 2005) induced [α-32P]dATP incorporation in G2 extracts, particularly visible on branched DNA molecules retained in the well (Fig. 4D; Supplemental Fig. S6E). Moreover, rereplication was associated with DNA breakage when Cdt1 was added to S-phase and G2 extracts (Fig. 4D). The analysis of replication intermediates confirmed an accumulation of ssDNA gaps upon Cdt1 addition, particularly marked in G2 extracts (Fig. 4E) where multiple rounds of rereplication have been reported (Davidson et al. 2006). Similar to Emi1 depletion, ssDNA gaps were also observed ahead of the replication forks, showing that rereplication was impaired by template discontinuities (Fig. 4E).
A new model for chromosomal breakage associated with deregulated origin licensing and rereplication
Our data strongly suggest that deregulated origin firing rapidly induces ssDNA gaps during DNA replication and that these persist in the template, where they cause stalling and eventually breakage of rereplicating forks (Fig. 4F). Rereplicating forks could break by simply impacting ssDNA gaps on the template (“runoff”). Alternatively, they could stall upstream of the gap and later be resolved into DSBs by slow “runoff,” nucleolytic processing, or head-to-tail collision with forks generated in following rounds of rereplication. In support of transient stalling and remodeling, small replication bubbles accumulated upon Emi1 depletion, indicative of early fork stalling from reactivated origins. Moreover, the frequency of reversed forks remained high 40 h after Emi1 depletion, when rereplicating forks are overrepresented in our EM samples (Fig. 3D; Supplemental Fig. S6C). Fork reversal was associated with fork slowing during replication of a nicked template, thereby protecting forks from breakage (Ray Chaudhuri et al. 2012). Thus, rereplicating forks could transiently arrest and reverse at ssDNA gaps before eventually undergoing breakage.
A prediction of our model is that the genotoxicity of rereplication correlates with the extent of origin firing deregulation in the previous replication round, as this creates the template discontinuities for rereplication. We tested this hypothesis by comparing the described effects for Emi1 depletion with Geminin depletion, a genetic condition associated with mild overreplication (Melixetian et al. 2004; Zhu et al. 2004), in which CDK-dependent Cdt1 inactivation partially restrains deregulated origin firing (Hook et al. 2007; Machida and Dutta 2007). In line with a recent study (Klotz-Noack et al. 2012), Geminin-depleted cells showed unperturbed EdU incorporation and progression in the first S phase, indicating marginal replication stress (Supplemental Fig. S7A). Accordingly, Geminin-depleted cells undergoing overt rereplication (>4N DNA content) displayed γH2AX only after completion of the first S phase and transition into a mitotic state (H3 phosphorylation) (Supplemental Fig. S7B). In line with our model, rereplication induced by Geminin depletion is associated with higher EdU incorporation and less DNA damage than Emi1 depletion, indicated by reduced γH2AX in cells with >4N DNA content (40 h) (Supplemental Fig. S7A,C). Intriguingly, if deregulation of origin licensing is induced after S-phase completion, DNA breakage requires at least two rounds of rereplication, as observed with Cdt1 addition to sperm nuclei in G2-arrested X. laevis extracts (Davidson et al. 2006), reinforcing the conclusions of this study. In this view, fork breakage during rereplication would not require head-to-tail fork collision (Davidson et al. 2006) but would rather occur as forks approach ssDNA gaps in close proximity to the origin, resulting in the observed release of small DNA fragments (Figs. 1E, 4F; Davidson et al. 2006). Accordingly, even in experimental conditions where rereplicating forks should represent a substantial fraction of total replication intermediates (siEmi1 40 h, Cdt1 addition in G2 extracts), we could never identify by EM a replicating and a rereplicating fork on the same DNA fragment.
The molecular mechanisms characterized here under conditions of severe rereplication could also be relevant for milder deregulation of origin licensing, associated with genome evolution and tumorigenesis (Hook et al. 2007; Green et al. 2010). As mild oncogene-induced replication stress can go undetected by cell cycle checkpoints (Fig. 1D; Neelsen et al. 2013), reactivation of specific replication origins in the presence of unrepaired ssDNA gaps may compromise chromosome integrity. Intriguingly, complex rearrangements in tumors have been recently associated with replication errors and copy number changes (Liu et al. 2011), which could result from breakage and repair of overreplicating chromosomes by mechanisms similar to those described here.
Materials and methods
Cell culture and transfections
U2OS and hTERT RPE-1 retinal pigmented epithelial cells were grown in DMEM + 10% FCS. Cells were transfected with the indicated siRNAs using RNAiMAX (Invitrogen) according to the manufacturer's instructions: siLuc (10 nM; 5′-GGUACGCGGAAUACUUCGAdTdT-3′), siEmi1 #1 (10 nM; 5′-GAUUGUGAUCUCUUAUUAAdTdT-3′), siEmi1 #2 (10 nM; 5′-GAGAAUUUCGGUGACAGUCUAdTdT-3′), and siGeminin (20 nM; 5′- UGCCAACUCUGGAAUCAAAdTdT-3′).
Methods
Flow cytometry was essentially performed as described previously for γH2AX/EdU/DAPI in Neelsen et al. (2013) and for γH2AX/RPA/DAPI in Forment et al. (2012). DNA fiber spreadings were performed according to Ray Chaudhuri et al. (2012) with the modifications outlined in the text. Pulse-field gel electrophoresis, immunofluorescence, and sample preparation for EM have been described in Neelsen et al. (2013, 2014). Protocols for Cdt1 purification, replication assays in X. laevis egg extracts, and isolation of genomic DNA for electron microscopic analysis can be found in the Supplemental Material. Detailed protocols for all other methods and a list of antibodies are included in the Supplemental Material.
Acknowledgments
We are grateful to all members of the Lopes laboratory for helpful discussions. We thank the Center for Microscopy and Image Analysis of the University of Zurich for technical assistance with the EM experiments, and D. Hühn, A. Sartori, S. Bregenhorn, and J. Jiricny (Institute of Molecular Cancer Research, Zurich) for sharing reagents and technical assistance. This work was supported by the Swiss National Science Foundation (grants PP0033-114922 and PP00P3_135292), Krebsliga Zurich, Stiftung zur Krebsbekämpfung, Research Priority Program on Systems Biology of the University of Zurich, and Wellcome Trust (grant WT097945). K.C. was supported by a Biotechnology and Biological Sciences Research Council/Doctoral Training Account studentship.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.226373.113.
References
- Arias EE, Walter JC 2006. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat Cell Biol 8: 84–90 [DOI] [PubMed] [Google Scholar]
- Arias EE, Walter JC 2007. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes Dev 21: 497–518 [DOI] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B 2011. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell 145: 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Dutta A 2005. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol 6: 476–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Gillespie PJ 2008. Replication licensing and cancer—a fatal entanglement? Nat Rev Cancer 8: 799–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Bozza W, Zhuang Z 2011. Ubiquitination of PCNA and its essential role in eukaryotic translesion synthesis. Cell Biochem Biophys 60: 47–60 [DOI] [PubMed] [Google Scholar]
- Davidson IF, Li A, Blow JJ 2006. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol Cell 24: 433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore B, Pines J 2007. Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J Cell Biol 177: 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn ES, Chastain PD II, Hall JR, Cook JG 2009. Analysis of re-replication from deregulated origin licensing by DNA fiber spreading. Nucleic Acids Res 37: 60–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenbach A, Li A, Brito-Martins M, Blow JJ 2005. Functional domains of the Xenopus replication licensing factor Cdt1. Nucleic Acids Res 33: 316–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forment JV, Walker RV, Jackson SP 2012. A high-throughput, flow cytometry-based method to quantify DNA-end resection in mammalian cells. Cytometry A 81: 922–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green BM, Finn KJ, Li JJ 2010. Loss of DNA replication control is a potent inducer of gene amplification. Science 329: 943–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 14: 1096–1104 [DOI] [PubMed] [Google Scholar]
- Hook SS, Lin JJ, Dutta A 2007. Mechanisms to control rereplication and implications for cancer. Curr Opin Cell Biol 19: 663–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DA, Pombo A 1998. Replicon clusters are stable units of chromosome structure: Evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140: 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Mortusewicz O, Afzal I, Lorvellec M, Garcia P, Helleday T, Petermann E 2013. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene 32: 3744–3753 [DOI] [PubMed] [Google Scholar]
- Klotz-Noack K, McIntosh D, Schurch N, Pratt N, Blow JJ 2012. Re-replication induced by geminin depletion occurs from G2 and is enhanced by checkpoint activation. J Cell Sci 125: 2436–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Blow JJ 2004. Non-proteolytic inactivation of geminin requires CDK-dependent ubiquitination. Nat Cell Biol 6: 260–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhao Q, Liao R, Sun P, Wu X 2003. The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J Biol Chem 278: 30854–30858 [DOI] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. 2011. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146: 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida YJ, Dutta A 2007. The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev 21: 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarry TJ, Kirschner MW 1998. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell 93: 1043–1053 [DOI] [PubMed] [Google Scholar]
- Melixetian M, Ballabeni A, Masiero L, Gasparini P, Zamponi R, Bartek J, Lukas J, Helin K 2004. Loss of Geminin induces rereplication in the presence of functional p53. J Cell Biol 165: 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen KJ, Zanini IM, Herrador R, Lopes M 2013. Oncogenes induce genotoxic stress by mitotic processing of unusual replication intermediates. J Cell Biol 200: 699–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen KJ, Chaudhuri AR, Follonier C, Herrador R, Lopes M 2014. Visualization and interpretation of eukaryotic DNA replication intermediates in vivo by electron microscopy. Methods Mol Biol 1094: 177–208 [DOI] [PubMed] [Google Scholar]
- Nishitani H, Sugimoto N, Roukos V, Nakanishi Y, Saijo M, Obuse C, Tsurimoto T, Nakayama KI, Nakayama K, Fujita M, et al. 2006. Two E3 ubiquitin ligases, SCF–Skp2 and DDB1–Cul4, target human Cdt1 for proteolysis. EMBO J 25: 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, Bermejo R, Cocito A, Costanzo V, Lopes M 2012. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat Struct Mol Biol 19: 417–423 [DOI] [PubMed] [Google Scholar]
- Senga T, Sivaprasad U, Zhu W, Park JH, Arias EE, Walter JC, Dutta A 2006. PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J Biol Chem 281: 6246–6252 [DOI] [PubMed] [Google Scholar]
- Sugimoto N, Tatsumi Y, Tsurumi T, Matsukage A, Kiyono T, Nishitani H, Fujita M 2004. Cdt1 phosphorylation by cyclin A-dependent kinases negatively regulates its function without affecting geminin binding. J Biol Chem 279: 19691–19697 [DOI] [PubMed] [Google Scholar]
- Tada S, Li A, Maiorano D, Mechali M, Blow JJ 2001. Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat Cell Biol 3: 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kirschner MW 2013. Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase-promoting complex. Nat Cell Biol 15: 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dwyer BT, Dhar SK, Cvetic C, Walter JC, Dutta A 2000. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290: 2309–2312 [DOI] [PubMed] [Google Scholar]
- Zhu W, Chen Y, Dutta A 2004. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol Cell Biol 24: 7140–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]