Insulin-like growth factor 2 (IGF2) is frequently overexpressed in pediatric cancers. In a miRNA screen of primary Wilms' tumors, Haber and colleagues identify specific overexpression of miR-483-5p, which is embedded in the IGF2 gene. Interestingly, the IGF2 mRNA itself is transcriptionally up-regulated by miR-483-5p. A nuclear pool of miR-483-5p binds the IGF2 mRNA, enhancing the association of RNA helicase and promoting IGF2 transcription. This unexpected nuclear function for a miRNA points to the complexity of mechanisms regulating IGF2 expression, implicated in both normal development and cancer.
Keywords: IGF2, fetal promoters, nuclear miRNA, transcription
Abstract
Insulin-like growth factor 2 (IGF2), a developmentally regulated and maternally imprinted gene, is frequently overexpressed in pediatric cancers. Although loss of imprinting (LOI) at fetal promoters contributes to increased IGF2 in tumors, the magnitude of IGF2 expression suggests the involvement of additional regulatory mechanisms. A microRNA (miRNA) screen of primary Wilms' tumors identified specific overexpression of miR-483-5p, which is embedded within the IGF2 gene. Unexpectedly, the IGF2 mRNA itself is transcriptionally up-regulated by miR-483-5p. A nuclear pool of miR-483-5p binds directly to the 5′ untranslated region (UTR) of fetal IGF2 mRNA, enhancing the association of the RNA helicase DHX9 to the IGF2 transcript and promoting IGF2 transcription. Ectopic expression of miR-483-5p in IGF2-dependent sarcoma cells is correlated with increased tumorigenesis in vivo. Together, these observations suggest a functional positive feedback loop of an intronic miRNA on transcription of its host gene.
Human insulin-like growth factor 2 (IGF2), a maternally imprinted gene, is transcribed from five promoters whose activity is both tissue-specific and developmentally regulated. In the fetus, IGF2 mRNA is highly transcribed primarily from the paternal promoters P2, P3, and P4, which are largely silenced in most tissues after birth. In the adult liver, IGF2 expression is driven by biallelic activation of the promoter P1 (Supplemental Fig. 1; Monk et al. 2006). Expression of IGF2 is frequently elevated in malignancies, including Wilms' tumor (a pediatric kidney cancer) (Huff 2011), sarcomas, and some colorectal cancers. In these cancers, the fetal promoters P2, P3, and P4 are often preferentially used, and loss of imprinting (LOI) commonly leads to their biallelic expression (Pollak 2008). LOI at fetal promoters has been widely considered the cause of the elevation of IGF2 expression in tumors (Jelinic and Shaw 2007), but additional mechanisms are likely to contribute to the very high levels of IGF2 expression seen in many cancers.
The global expression pattern of microRNAs (miRNAs) is typically much higher in differentiated tissues than in cancers, consistent with a broad role for miRNAs in suppressing cellular proliferation and enhancing differentiation (Chang et al. 2008). However, in comparing miRNA expression patterns in the pediatric kidney cancer Wilms' tumor versus normal renal tissues, we observed selective up-regulation of miR-483-5p in the cancer cells. We demonstrate that miR-483-5p, which resides in an intron of the IGF2 gene, enhances transcription of fetal but not adult IGF2 transcripts by binding to its 5′ untranslated region (UTR), thereby up-regulating expression of its host gene. This unexpected nuclear function for a miRNA points to the complexity of mechanisms regulating the expression of the IGF2 gene, implicated in both normal development and cancer.
Results and Discussion
The IGF2 intronic miR-483 is overexpressed in Wilms' tumor
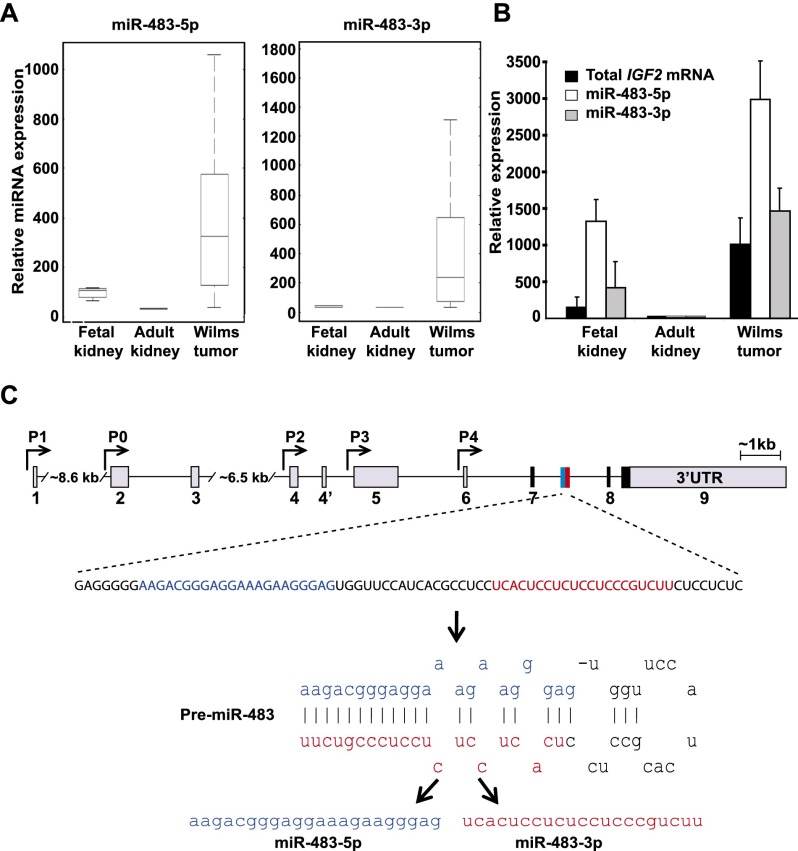
In screening 431 miRNAs for differential expression in Wilms' tumor compared with normal adult and fetal kidneys, we identified miR-483 (comprising two distinct miRNAs: miR-483-5p and miR-483-3p) as being the most dramatically and specifically up-regulated miRNA (Fig. 1A; Supplemental Tables 1, 2). Although most miRNAs are more abundantly expressed in mature nonproliferating tissues than in embryonic tissues or cancers (Chang et al. 2008), both miR-483s were expressed >10-fold higher in the highly proliferative Wilms' tumor (N = 20) compared with adult differentiated kidneys (N = 3). miR-483 expression was also more than threefold higher in the tumor compared with fetal kidney tissue (N = 5), which contains proliferative renal precursors thought to be the cells of origin for this embryonic malignancy (Fig. 1B). The pri-miR-483 lies in intron 2 of the IGF2 transcript and encodes two miRNAs on opposite strands of the pre-miR-483 hairpin (Fig. 1C). The levels of both miRNAs parallel the expression of the IGF2 host transcript in Wilms' tumor, fetal kidneys, and adult kidneys (Fig. 1B; Veronese et al. 2010; Ma et al. 2011). miR-483-5p has also recently been reported among miRNAs, conferring an adverse prognosis in both Wilms' tumor and adrenal cortical carcinoma (Patterson et al. 2011; Watson et al. 2013).
Figure 1.
The IGF2 intronic miR-483s are highly expressed in Wilms' tumor along with the host IGF2 mRNA. (A) Box plots showing higher miR-483 expression in Wilms' tumors compared with normal adult and fetal kidney tissues. In each box, the central mark (red) indicates the median, the edges of the box represent the 25th and 75th percentiles, and the deviations extend to the most extreme data points. The Y-axis is in units of fluorescence intensity. (B) qRT–PCR measurement of coexpression of IGF2 mRNA and the miR-483s in fetal kidneys, adult kidneys, and Wilms' tumors. Expression is relative to levels in adult kidney tissues. (C) Localization of miR-483s within the human IGF2 gene. Arrows denote promoters P0–P4, gray boxes indicate promoter-specific 5′ UTRs or 3′ UTRs, black boxes indicate coding regions, and exons are numbered. The miR-483 gene encodes two miRNAs: miR-483-5p (blue) and miR-483-3p (red).
miR-483-5p enhances IGF2 transcription
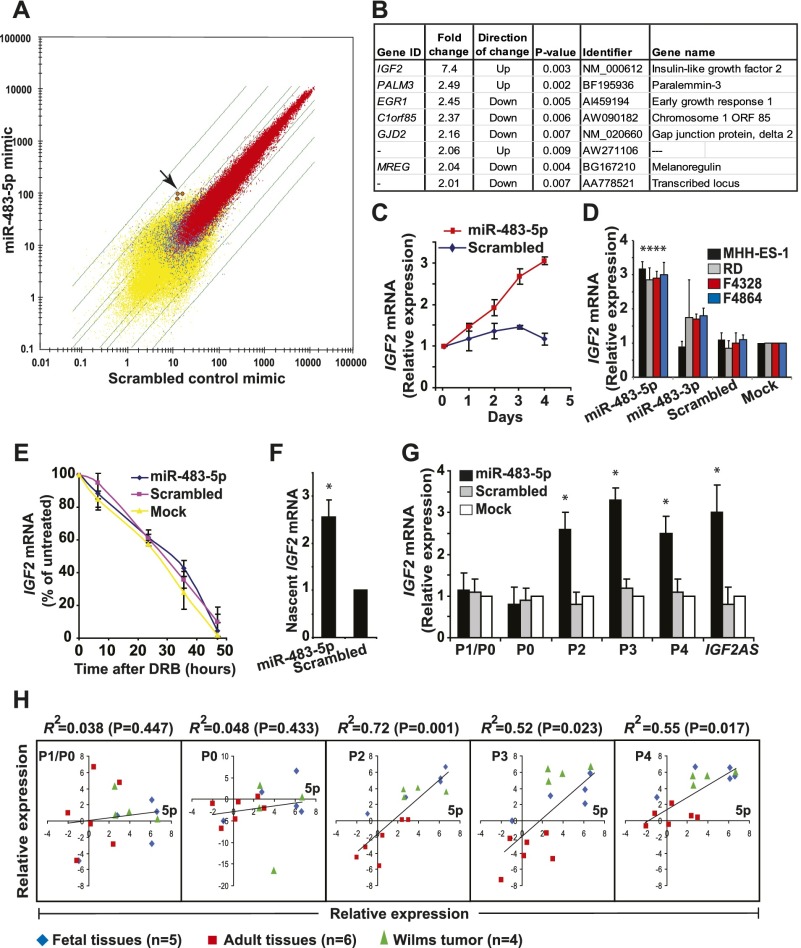
Although both miR-483s are highly conserved among placental mammals (Supplemental Fig. 2), we were unable to validate 3′ UTR target sequences that had been predicted by available computational algorithms using traditional luciferase reporter experiments. We therefore tested the effect of ectopic miR-483-5p or miR-483-3p expression on endogenous transcripts by microarray profiling. Given the scarcity of Wilms' tumor cell lines, we selected MHH-ES-1 Ewing's sarcoma cells (Wisniewski et al. 2002), derived from a highly aggressive pediatric tumor expressing endogenous IGF2, to ensure a cellular context in which this developmental pathway is active. Remarkably, an increase in the IGF2 transcript itself was the most significant gene expression change following transfection of a miR-483-5p mimic, with up to sevenfold induction of IGF2 mRNA (Fig. 2A,B). This observation was validated using quantitative RT–PCR (qRT–PCR) of control and miR-483-5p transfected cells, showing a progressive increase in IGF2 mRNA over time (Fig. 2C) and a consistent change over a wide range of concentration of ectopic miR-483-5p (Supplemental Fig. 3). miR-483-5p also induces IGF2 mRNA in the human rhabdomyosarcoma cell line RD as well as in mouse sarcoma cell lines (Fig. 2D). In contrast, no significant changes were evident following ectopic expression of a miR-483-3p mimic (data not shown). We therefore pursued miR-483-5p as a candidate regulator of IGF2 mRNA expression.
Figure 2.
miR-483-5p induces promoter-specific IGF2 transcription. (A) Scatter plot (log) of mRNA profiles of MHH-ES-1 cells transfected with miR-483-5p (Y-axis) versus scrambled control (X-axis). Transcripts expressed in both samples (red), one sample (blue), or neither sample (yellow) are shown (as determined by default settings of the Affymetrix GCOS software), with green lines indicating twofold, threefold, 10-fold, and 30-fold differential expression between the two samples. The arrow points to signals from three independent IGF2 mRNAs probes. (B) Genes that are changed (either up-regulated or down-regulated) more than twofold in MHH-ES-1 cells transfected with a miR-483-5p mimic compared with scrambled control. (C) Time course of IGF2 mRNA expression after transfection of MHH-ES-1 with a miR-483-5p mimic, as measured by qRT–PCR. Expression is relative to levels at day 0. (D) Induction of IGF2 mRNA by miR-483-5p in two human sarcoma cell lines (MMH-ES-1 and RD) and two mouse sarcoma lines (F4328 and F4864), as measured by qRT–PCR. The asterisk denotes a significant difference (P < 0.01) compared with mock treatment, whose measurement is set to 1. (E) Stability of IGF2 mRNA in miR-483-5p transfected MHH-ES-1 cells treated with the transcriptional inhibitor DRB. Signal is normalized to total RNA and expressed as percentage of IGF2 mRNA in untreated cells. (F) Nascent IGF2 mRNA transcripts, measured by nuclear run-on assays in MHH-ES-1 cells transfected with a miR-483-5p mimic. Expression is relative to levels of nascent GAPDH mRNA. The asterisk denotes a significant difference (P < 0.01) compared with scrambled control. (G) Expression of promoter-specific IGF2 transcripts in MHH-ES-1 cells transfected with a miR-483-5p mimic. P1/P0 refers to the common transcript from these promoters (see the Supplemental Material for promoter information). The asterisk denotes a significant difference (P < 0.01) compared with mock treatment, whose expression is set to 1. (H) Linear regression of expression of each IGF2 promoter-specific transcript (Y-axis; expression relative to that of ACTB mRNA) with miR-483-5p (5p for short) expression (X-axis; expression relative to that of RNU48 small RNA) in human primary tissues (adult or fetal livers, hearts, kidneys, muscles, and brains; umbilical) and Wilms' tumors, as measured by qRT–PCR. The coefficient of determination (R2) and associated P-value are shown.
We first tested whether IGF2 mRNA is increased through stabilization by miR-483-5p. IGF2 mRNA in MHH-ES-1 cells treated with the transcriptional inhibitor 5,6-dichloro-1-β-D ribofuranosylbenzimidazole (DRB) or actinomycin D showed no difference following transfection with either a miR-483-5p mimic, a scrambled mimic, or mock transfected controls (Fig. 2E), indicating that mRNA stability is not altered by the miRNA. In contrast, nuclear run-on assays showed increased nascent IGF2 transcripts in MHH-ES-1 nuclei overexpressing miR-483-5p compared with a scrambled mimic, suggesting that miR-483-5p expression enhances transcription of the IGF2 mRNA (Fig. 2F).
To test which of the five IGF2 promoters are affected by miR-483-5p, we used specific qRT–PCR primers that distinguish between individual transcripts. miR-483-5p specifically increased IGF2 mRNA from the grouped fetal promoters P2, P3, and P4 but not from the far upstream P0 and P1 promoters that regulate IGF2 expression in adult tissues (Fig. 2G). IGF2AS, a 2-kb antisense RNA whose promoter lies adjacent to P3 (Supplemental Fig. 1), was also up-regulated by miR-483-5p. Consistent with the promoter-specific effect of miR-483-5p, the expression of this miRNA in a panel of human fetal and adult tissues and Wilms' tumors correlated with P2-, P3-, and P4-derived IGF2 transcripts (R2 > 0.5) but not with the P0- and P1-initiated mRNAs (Fig. 2H). Enhanced expression of IGF2 mRNAs driven from the fetal P2–P4 promoters by miR-483-5p is also accompanied by a slight relaxation of gene imprinting (Supplemental Fig. 4).
Nuclear miR-483-5p interacts with the 5′ UTR of the major fetal IGF2 transcript
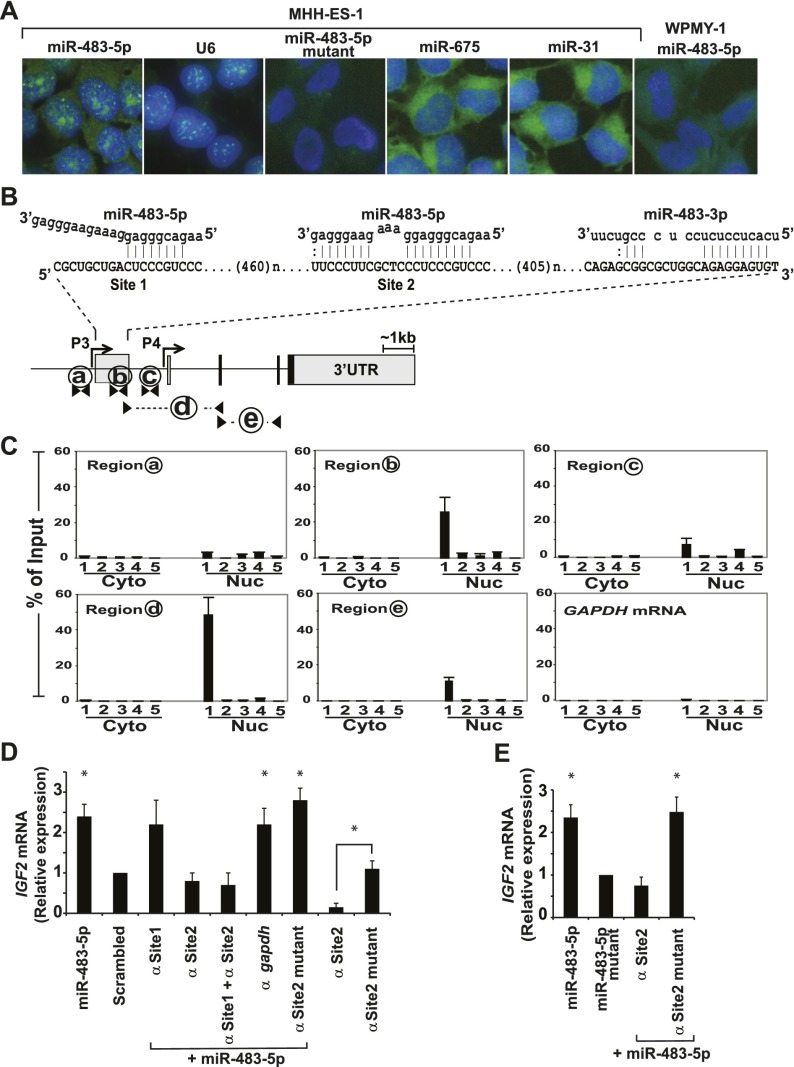
Given the positive regulatory effect of miR-483-5p on IGF2 mRNA transcription, rather than the characteristic role of miRNAs in regulating the stability of mature mRNA, we tested whether the miRNA localizes to the nucleus. Using fluorescence in situ hybridization (FISH) with digoxygenin (DIG)-conjugated locked nucleic acid (LNA) probes and confocal microscopy, we observed multiple punctate foci of endogenous miR-483-5p within the nuclei of MHH-ES-1 cells, along with a diffuse cytoplasmic signal (Fig. 3A). The nuclear miR-483-5p signal was comparable with that of the small nuclear RNA U6. Mutant miR-483-5p probes with three mismatched nucleotides in the seed region (see Supplemental Table 5 for sequences) failed to detect the endogenous miR-483-5p in MHH-ES-1 cells. The wild-type probes failed to detect the signal in a fibroblast cell line that lacks IGF2 mRNA expression, and no nuclear staining was evident with the control miRNAs miR-675 and miR-31, which have well-defined cytoplasmic functions (Valastyan et al. 2009; Keniry et al. 2012). Further analysis of nuclear and cytoplasmic RNA fractions from three different cell lines confirmed the presence of miR-483-5p in both compartments (Supplemental Table 3).
Figure 3.
miR-483-5p localizes to the nucleus and binds the 5′ UTR of nuclear IGF2 P3 mRNA to induce gene activation. (A) FISH showing nuclear localization of endogenous miR-483-5p (FITC; green) using a specific LNA probe against the miRNA. The nucleus was stained with DAPI (blue). The small nuclear RNA U6 is shown as positive control for nuclear signal. LNA probes detecting the miRNA-483-5p mutant and the WPMY-1 fibroblast cell line that lacks IGF2 expression were used as negative controls. Two unrelated miRNAs (miR-675 and miR-31) lack apparent nuclear localization. (B) Schematic representation of potential miR-483-binding sites within the 5′ UTR region of IGF2 P3 transcript, including miR-483-5p (site 1 and site 2) and miR-483-3p. Fragments subjected to pull-down assays are shown with letters (a–e), with amplification primers denoted by arrowheads. (C) Biotinylated miRNA affinity pull-down was conducted using streptavidin beads with nuclear and cytosolic extracts of cells transfected with different biotinylated miRNA baits followed by qRT–PCR analysis for the presence of specific regions of IGF2 mRNA. The miRNA baits are as follows: biotinylated miR-483-5p (1), biotinylated miR-483-5p mutant (2), sense strand biotinylated miR-483-5p (3), biotinylated miR-483-3p (4), and nonbiotinylated miR-483-5p (5). PCR-amplified fragments are as follows: P3 promoter (region a), P3 5′ UTR (region b), and the intronic region between the P3 5′ UTR and the P4 5′ UTR (region c) that represents the spliced intron sequence. Region d identifies the spliced mature P3 mRNA, region e identifies total IGF2 mRNAs, and region f identifies GAPDH mRNA, which serves as a control. Cyto and Nuc represent pull-down from cytoplasmic and nuclear fractions, respectively. (D) Suppression of IGF2 mRNA induction in MHH-ES-1 cells by miR-483-5p (vs. scrambled control) following transfection of a 2′-O-methyl antisense protector RNA that specifically targets site 2. The 2′-O-methyl antisense RNAs target site 1 alone (α Site1), site 2 alone (α Site2), site 1 plus site 2 (α Site1 + α Site2), GAPDH 5′ UTR (α GAPDH), or mutant site 2 (α Site2 mutant). The asterisk denotes a significant difference (P < 0.01) compared with scrambled control. Two transfections with site 2 alone or mutant site 2 alone in the absence of ectopic miR-483-5p are also shown to demonstrate the endogenous effect of miR-483-5p on IGF2 mRNAs. (E) Nascent IGF2 mRNA transcripts, measured by nuclear run-on assays in MHH-ES-1 cells transfected with miR-483-5p, miR-483-5p mutant, or miR-483-5p along with site 2 alone or mutant site 2. Expression is relative to levels of nascent GAPDH mRNA. The asterisk denotes a significant difference (P < 0.05) compared with miR-483-5p mutant samples, whose measurement is set to 1.
To search for potential miR-483-5p target-binding sites, we scanned the entire 30-kb human IGF2 gene, from the P1 promoter to the 3′ UTR region, identifying two strong potential binding sites (site 1 and site 2) within the 5′ UTR of the major P3 fetal transcript (Fig. 3B). A single such site is also present in the 5′ UTR of the mouse and rat IGF2 mRNA encoding the comparable transcript (variant 1) (Supplemental Fig. 5A). Although miR-483-3p does not modulate IGF2 levels, a conserved miR-483-3p-binding site exists within the P3 5′ UTR region. Since these candidate target sequences are present in potential promoter regulatory DNA elements as well as in the 5′ UTR of the mRNA transcript, we used biotinylated miRNA affinity pull-down assays (Lal et al. 2011; Kang et al. 2012; Yoon et al. 2012) to test for binding of the miRNAs to genomic DNA versus RNA. The assays were conducted separately, with nuclear versus cytosolic extracts derived from cells that had been transfected with the various biotinylated miRNA baits, followed by qRT–PCR analysis of specific regions of IGF2 DNA or mRNA. Neither biotinylated miR-483-5p nor miR-483-3p showed specific enrichment for the relevant genomic DNA sequences (Supplemental Fig. 5B,C). However, specific IGF2 mRNA sequences were coprecipitated with the miRNA baits: miR-483-5p pull-down experiments revealed >50-fold enrichment of the P3 mRNA transcript containing the P3-driven 5′ UTR region compared with the pull-down using a mutant miR-483-5p with a three-nucleotide change in the seed sequence or a miR-483-5p biotinylated on the opposite strand (Fig. 3C; Supplemental Fig. 6). The P3 IGF2 mRNA pull-down by miR-483-5p was observed from the nuclear but not the cytosolic fraction. Biotinylated miR-483-3p failed to pull down the IGF2 transcript. Thus, miR-483-5p selectively binds to the nuclear IGF2 P3 transcript.
To determine whether miR-483-5p binding to its predicted target sites within the 5′ UTR is critical for up-regulation of IGF2 mRNA expression, we cotransfected 2′-O-methyl-modified antisense oligonucleotide protectors (Giraldez et al. 2006) complementary to either site 1 or site 2 to block binding by the 5p miRNA. Cotransfection of the antisense oligonucleotide targeting site 2 abolished miR-483-5p-mediated induction of IGF2 (Fig. 3D). A mutant antisense oligonucleotide against site 2 (17 out of 34 sequence mismatches) or an antisense targeting the GAPDH 5′ UTR had no effect on IGF2 mRNA induction. In contrast to antisense protectors targeting site 2, targeting site 1 had no effect on IGF2 mRNA expression. Thus, only one of the two miR-483-5p-binding sites within the 5′ UTR of the IGF2 mRNA appears to mediate a positive feedback loop, leading to increased transcription. Moreover, the antisense oligonucleotide targeting site 2 alone, but not its mutant version, significantly lowers the baseline endogenous IGF2 mRNA level (Fig. 3D). This suggests that the effect that we observed is not restricted to ectopic overexpression of miR-483-5p but that the native miRNA positively regulates the steady-state levels of its host mRNA.
To determine whether induction of IGF2 transcription occurs through nuclear miR-483-5p directly interacting with IGF2 mRNA, we preformed nuclear run-on assays where either the miR-483-5p sequence is mutated or its target sequence is blocked. Expression of miR-483-5p increased the levels of nascent IGF2 transcripts in MHH-ES-1 cells. This effect was suppressed by antisense protectors targeting site 2 but was not affected by mutant protectors or observed following expression of mutant miR-483-5p (Fig. 3E). These results suggest that the interaction of miR-483-5p with IGF2 mRNA P3 site 2 alters IGF2 mRNA transcription.
miR-483-5p promotes the association of IGF2 mRNA with the RNA helicase DHX9
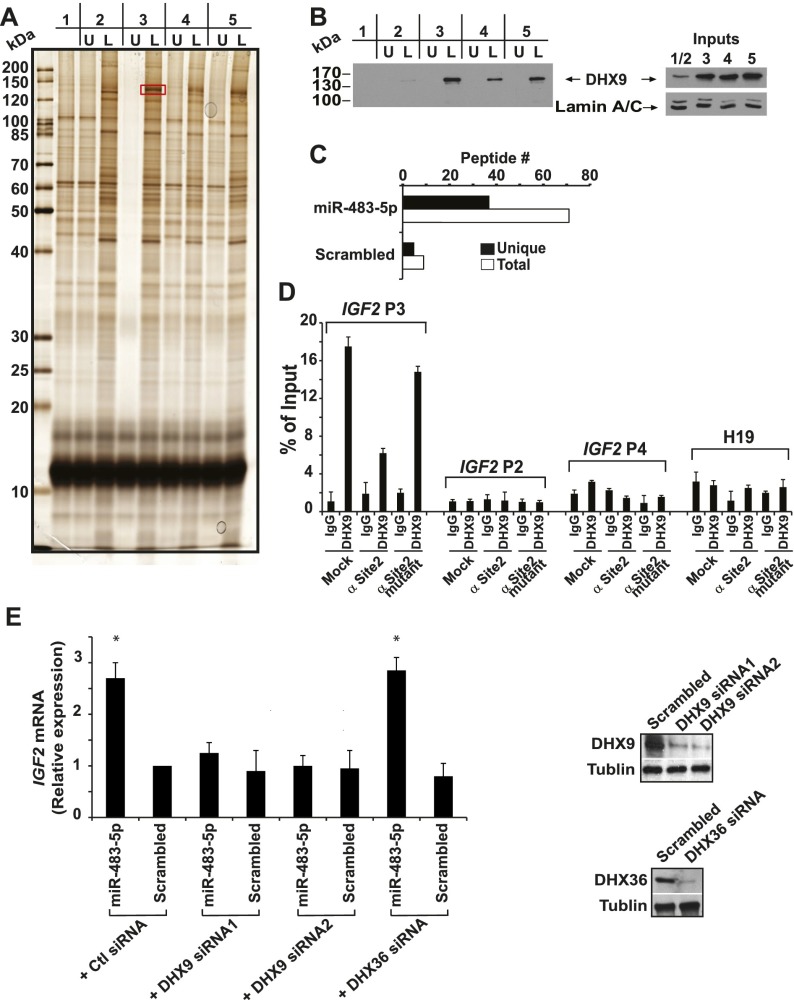
To better understand the mechanisms involved in miR-483-5p-mediated regulation of IGF2 expression, we searched for associated proteins using complementary strategies. First, we used biotinylated 5′ UTR of the IGF2 P3 transcript as bait to isolate interacting proteins from the nuclear extracts of MHES-ES-1 cells transfected with miR-483-5p or a scrambled control. As an additional control, we also transfected cells simultaneously with miR-483-5p together with an antisense protector targeting site 2 or a mutated antisense protector. Electrophoresis of affinity-purified proteins followed by silver staining identified a 140-kDa protein that was enriched in cells transfected with miR-483-5p compared with cells transfected with either control sequences or antisense sequences targeting site 2 (Fig. 4A; Supplemental Fig. 7). Targeted sequencing of this protein identified it to be ATP-dependent RNA helicase A (DHX9). Western blot analysis confirmed the coprecipitation of DHX9, and we note that DHX9 protein expression increases upon overexpression of miR-483-5p (Fig. 4B). The interaction between miR-483-5p, IGF2 mRNA, and DHX9 was confirmed using a miRNA–protein affinity purification assay as a second approach (Newman et al. 2008). Biotinylated miR-483-5p or a scrambled control was used as bait to pull down associated proteins from MHH-ES-1 nuclear extracts, followed by mass spectrometric analysis. In this analysis, DHX9 was one of the predominant proteins specifically associated with miR-483-5p (Fig. 4C; Supplemental Table 4).
Figure 4.
miR-483-5p enhances the interaction of IGF2 P3 mRNA with the RNA helicase A (DHX9). (A) Silver-stained gel of electrophoresed proteins pulled down using the biotinylated 5′ UTR of IGF2 P3 mRNA following transfection of cells with scrambled miRNA control (lanes 1,2), miR-483-5p (lane 3), miR-483-5p + antisense protector targeting site 2 (lane 4), and miR-483-5p + antisense mutated protector (lane 5). The pull-down experiments were performed using biotin-labeled IGF2 P3 mRNA 5′ UTR (L), with the identical, unlabeled IGF2 mRNA fragment serving as negative control (U). The protein that is differentially bound to IGF2 P3 mRNA 5′ UTR in the presence of miR-483-5p and in the absence of the inhibitory protector (red box in lane 3, L) was sequenced and found to be DHX9. (B) Western blot confirmation of DHX9 pull-down by IGF2 P3 5′ UTR RNA. DHX9 content for each input sample is shown. Lamin A/C serves as the loading control. (C) Number of DHX9 peptides associated with miR-483-5p or control miRNA baits, measured by mass spectrometry. Black and white bars represent unique peptide numbers and total peptide numbers identified, respectively. (D) Immunoprecipitation of endogenous DHX9 in MHH-ES-1 cells and those transfected with antisense protector targeting site 2 (α Site2) or antisense mutated protector (α Site2 mutant). Four mRNAs—IGF2 P2, P3, P4 and H19—were assayed by qRT–PCR. (E) siRNA-mediated knockdown of DHX9 abrogates the induction of IGF2 by miR-483-5p and reduces endogenous IGF2 mRNA in the absence of ectopic miR-483-5p. The asterisk denotes a significant difference (P < 0.01) compared with scrambled control, which is set to 1. DHX9 knockdown is shown by Western blot.
To further test the potential interaction between DHX9, the IGF2 mRNA, and miR-483-5p, we immunoprecipitated endogenous DHX9 to see whether it directly binds to the RNAs. We observed that DHX9 is bound specifically to IGF2 mRNA P3 but not to P2 or P4, the two IGF2 mRNA variants that lack the miR-483-5p-binding site, or H19, a mRNA transcribed from the same genomic locus (Fig. 4D). Blocking site 2 using antisense sequences but not mutant sequences against the site significantly reduced but did not completely abolish DHX9 binding to IGF2 P3, suggesting that miR-483-5p increases the association between DHX9 and IGF2 mRNA, and DHX9 may also interact with other regions of the mRNA besides the miR-483-5p-binding site. Cross-linking assays to determine whether DHX9 directly interacts with miR-483-5p were inconclusive due to high levels of background (data not shown). Together, these results suggest that DHX9 specifically associates with IGF2 P3 mRNA and that miR-483-5p greatly enhances this interaction.
While its precise functions remain to be elucidated, DHX9 has previously been reported to regulate transcription (Padmanabhan et al. 2012) as well as miRNA-dependent pathways (Robb and Rana 2007). To test the functional significance of the interaction between DHX9, miR-483-5p, and IGF2 mRNA, we knocked down expression of the helicase in MHH-ES-1 cells and analyzed the consequences on miR-483-5p-mediated induction of IGF2 mRNA. Suppression of DHX9 not only abrogated the induction of IGF2 mRNA by ectopic miR-483-5p but also reduced endogenous IGF2 mRNA levels (Fig. 4E), suggesting that it contributes to the miRNA-mediated up-regulation of IGF2. As a control for specificity, we also tested another ATP-dependent RNA helicase, DHX36, that was enriched in the miR-483-5p pull-down (Supplemental Table 4). Knockdown of the DHX36, however, did not affect miR-483-5p-mediated IGF2 mRNA induction (Fig. 4E).
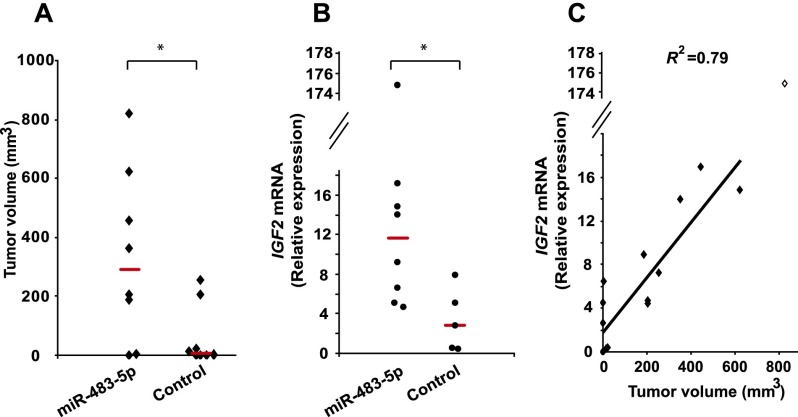
miR-483-5p expression is correlated with enhanced tumorigenesis in vivo
To test whether the miR-483-5p effect on IGF2 expression has physiological consequences in modulating tumorigenicity, MHH-ES-1 cells infected with lentiviral constructs encoding miR-483-5p were inoculated subcutaneously into immunodeficient mice. MHH-ES-1 cells expressing miR-483-5p generated significantly larger tumors in a higher fraction of animals compared with cells transfected with empty vector (Fig. 5A; Supplemental Fig. 8). IGF2 mRNA expression in tumors derived from miR-483-5p-infected cells was higher (Fig. 5B) and correlated with tumor size (Fig. 5C).
Figure 5.
Ectopic expression of miR-483-5p in sarcoma cells enhances tumorigenesis. (A) Tumor volume of xenografts in 16 mice generated by MHH-ES-1 cells stably expressing miR-483-5p or empty control vector. The bar represents the median tumor size. The asterisk denotes a significant difference (P < 0.01) compared with the control group. (B) IGF2 mRNA expression in mouse tumors generated from miR-483-5p versus control vector transfected MHH-ES-1 cells. The bar represents the median expression. The asterisk denotes a significant difference (P < 0.05) compared with the control group. (C) Linear regression of tumor volume (X-axis) versus IGF2 expression (Y-axis) for 15 mice, excluding one outlier (diamond) with a very large tumor expressing extraordinarily high levels of IGF2. The coefficient of determination (R2) (P = 0.0004) is shown.
Concluding remarks
We showed that miR-483-5p, a miRNA embedded in the IGF2 gene, mediates a function in the nucleus, targeting the 5′ UTR of the major fetal IGF2 P3 transcript, leading to its up-regulation. The unexpected functional properties of miR-483-5p are particularly interesting in view of the complex IGF2 physiology, where imprinted fetal IGF2 promoters enforce high IGF2 production during rapid placental and fetal development (Moore and Haig 1991). Interestingly, miR-483 is present only in placental mammals (Supplemental Fig. 2) despite the broad evolutionary conservation of IGF2 signaling, raising the possibility that the miRNA may have coevolved with this complex gene regulatory mechanism. In addition to this physiological role, positive feedback regulation by miR-483-5p may further contribute to increased fetal IGF2 promoter expression during tumorigenesis. More traditional 3′ UTR mRNAs targeted by miR483-5p have been reported, leading to reduced expression of some potential candidates, including ERK1, SRF, Socs3, and MeCP2 (Qiao et al. 2011; Ma et al. 2012; Wang et al. 2012; Han et al. 2013). miR-483-5p may thus exhibit distinct nuclear and cytoplasmic properties, both contributing to its physiological effects.
Further studies will be required to dissect the mechanisms by which miR-483-5p mediates its effect on IGF2 transcription, but the apparent requirement for RNA helicase DHX9 raises the possibility that small RNA-mediated RNA processing is involved, as proposed for splicing-coupled transcriptional silencing promoted by siRNAs (Allo et al. 2009). The 5′ UTR of IGF2 P3 mRNA has a 72% GC content and is therefore highly structured. DHX9 may play a role in modulating this structure to facilitate immediate processing of nascent transcripts, thereby enhancing the overall transcription efficiency. Indeed, mRNA processing and transcription are tightly coupled in mammalian cells, with protein factors modulating transcriptional initiation, elongation, and termination (Dahan et al. 2011). Whether other affected mRNAs that reside in close genomic proximity (IGF2 P2 and P3 and IGF2AS) are induced by miR-483-5p through localized changes in chromosomal structure and thus facilitated transcription remains to be determined. Taken all together, the self-regulation of IGF2 transcription by the intronic and coexpressed miR-483-5p points to a novel miRNA functional property modulating a complex developmental pathway in mammalian cells.
Material and methods
Tumor and tissue samples and cell lines
Frozen Wilms' tumor, normal adult kidney, or fetal kidney tissue samples used for miRNA screen were obtained from Children's Hospital Boston and Brigham and Women's Hospital. All samples were collected with institutional review board approval. Human primary tissues used to assess the expression of IGF2 promoter–specific mRNAs and miR-483-5p were obtained from Stratagene and Zyagen, as described in the Supplemental Material. Cell lines were grown under standard culture conditions, as described in the Supplemental Material.
miRNA, RNA, and protein analysis and mouse xenografts
miRNA screen, qRT–PCR, RNA stability assay, miRNA FISH, nuclear RUN-ON, RIP, and immunoprecipitation experiments are described in the Supplemental Material.
Acknowledgments
We thank J. Avruch, N. Dai, K. Morris, and J. Whetstine for reagents and helpful discussions. We thank Dr. Martina Schnölzer and Dr. Tore Kempf at the DKFZ Genomics and Proteomics Core facility for mass spectrometry analysis. This work was supported by funding from the Howard Hughes Medical Institute (to M.L., M.Y., F.B., M.N.R., and D.A.H), the Burroughs Wellcome Fund (to M.N.R.), National Cancer Institute 5R37CA058596-19 (to D.A.H.), and a Helmholtz-University-Junior Research Group (VH-NG-504; to S.D.).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.224170.113.
References
- Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, Elela SA, et al. 2009. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struct Mol Biol 16: 717–724 [DOI] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT 2008. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet 40: 43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan O, Gingold H, Pilpel Y 2011. Regulatory mechanisms and networks couple the different phases of gene expression. Trends Genet 27: 316–322 [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Han K, Gennarino VA, Lee Y, Pang K, Hashimoto-Torii K, Choufani S, Raju CS, Oldham MC, Weksberg R, Rakic P, et al. 2013. Human-specific regulation of MeCP2 levels in fetal brains by microRNA miR-483-5p. Genes Dev 27: 485–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff V 2011. Wilms' tumours: About tumour suppressor genes, an oncogene and a chameleon gene. Nat Rev Cancer 11: 111–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinic P, Shaw P 2007. Loss of imprinting and cancer. J Pathol 211: 261–268 [DOI] [PubMed] [Google Scholar]
- Kang H, Davis-Dusenbery BN, Nguyen PH, Lal A, Lieberman J, Van Aelst L, Lagna G, Hata A 2012. Bone morphogenetic protein 4 promotes vascular smooth muscle contractility by activating microRNA-21 (miR-21), which down-regulates expression of family of dedicator of cytokinesis (DOCK) proteins. J Biol Chem 287: 3976–3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keniry A, Oxley D, Monnier P, Kyba M, Dandolo L, Smits G, Reik W 2012. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 14: 659–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal A, Thomas MP, Altschuler G, Navarro F, O'Day E, Li XL, Concepcion C, Han YC, Thiery J, Rajani DK, et al. 2011. Capture of microRNA-bound mRNAs identifies the tumor suppressor miR-34a as a regulator of growth factor signaling. PLoS Genet 7: e1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N, Wang X, Qiao Y, Li F, Hui Y, Zou C, Jin J, Lv G, Peng Y, Wang L, et al. 2011. Coexpression of an intronic microRNA and its host gene reveals a potential role for miR-483-5p as an IGF2 partner. Mol Cell Endocrinol 333: 96–101 [DOI] [PubMed] [Google Scholar]
- Ma N, Li F, Li D, Hui Y, Wang X, Qiao Y, Zhang Y, Xiang Y, Zhou J, Zhou L, et al. 2012. Igf2-derived intronic miR-483 promotes mouse hepatocellular carcinoma cell proliferation. Mol Cell Biochem 361: 337–343 [DOI] [PubMed] [Google Scholar]
- Monk D, Sanches R, Arnaud P, Apostolidou S, Hills FA, Abu-Amero S, Murrell A, Friess H, Reik W, Stanier P, et al. 2006. Imprinting of IGF2 P0 transcript and novel alternatively spliced INS-IGF2 isoforms show differences between mouse and human. Hum Mol Genet 15: 1259–1269 [DOI] [PubMed] [Google Scholar]
- Moore T, Haig D 1991. Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet 7: 45–49 [DOI] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM 2008. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA 14: 1539–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan K, Robles MS, Westerling T, Weitz CJ 2012. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 337: 599–602 [DOI] [PubMed] [Google Scholar]
- Patterson EE, Holloway AK, Weng J, Fojo T, Kebebew E 2011. MicroRNA profiling of adrenocortical tumors reveals miR-483 as a marker of malignancy. Cancer 117: 1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak M 2008. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer 8: 915–928 [DOI] [PubMed] [Google Scholar]
- Qiao Y, Ma N, Wang X, Hui Y, Li F, Xiang Y, Zhou J, Zou C, Jin J, Lv G, et al. 2011. miR-483-5p controls angiogenesis in vitro and targets serum response factor. FEBS Lett 585: 3095–3100 [DOI] [PubMed] [Google Scholar]
- Robb GB, Rana TM 2007. RNA helicase A interacts with RISC in human cells and functions in RISC loading. Mol Cell 26: 523–537 [DOI] [PubMed] [Google Scholar]
- Valastyan S, Reinhardt F, Benaich N, Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL, Weinberg RA 2009. A pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell 137: 1032–1046 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Veronese A, Lupini L, Consiglio J, Visone R, Ferracin M, Fornari F, Zanesi N, Alder H, D'Elia G, Gramantieri L, et al. 2010. Oncogenic role of miR-483-3p at the IGF2/483 locus. Cancer Res 70: 3140–3149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Shi M, Hou S, Ding B, Liu L, Ji X, Zhang J, Deng Y 2012. miR-483-5p suppresses the proliferation of glioma cells via directly targeting ERK1. FEBS Lett 586: 1312–1317 [DOI] [PubMed] [Google Scholar]
- Watson JA, Bryan K, Williams R, Popov S, Vujanic G, Coulomb A, Boccon-Gibod L, Graf N, Pritchard-Jones K, O'Sullivan M 2013. miRNA profiles as a predictor of chemoresponsiveness in Wilms' tumor blastema. PLoS ONE 8: e53417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewski D, Lambek CL, Liu C, Strife A, Veach DR, Nagar B, Young MA, Schindler T, Bornmann WG, Bertino JR, et al. 2002. Characterization of potent inhibitors of the Bcr-Abl and the c-kit receptor tyrosine kinases. Cancer Res 62: 4244–4255 [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M 2012. LincRNA-p21 suppresses target mRNA translation. Mol Cell 47: 648–655 [DOI] [PMC free article] [PubMed] [Google Scholar]