The Mediator complex is crucial for eukaryotic transcription. In this study, Eyboulet et al. define a previously unsuspected role for Mediator in connecting transcription with DNA repair. The authors identify a functional interaction between the Mediator subunit Med17 and the DNA repair 3′ endonuclease Rad2/XPG. Rad2 occupancy of RNA Pol II-transcribed genes is transcription-dependent. Moreover, med17 mutants defective in Mediator–Rad2 interaction are UV-sensitive. Thus, Mediator functions in DNA repair by facilitating Rad2/XPG recruitment to transcribed genes.
Keywords: Mediator, transcription, DNA repair, Rad2/XPG, RNA polymerase II, Saccharomyces cerevisiae
Abstract
Mediator is a large multiprotein complex conserved in all eukaryotes. The crucial function of Mediator in transcription is now largely established. However, we found that this complex also plays an important role by connecting transcription with DNA repair. We identified a functional contact between the Med17 Mediator subunit and Rad2/XPG, the 3′ endonuclease involved in nucleotide excision DNA repair. Genome-wide location analyses revealed that Rad2 is associated with RNA polymerase II (Pol II)- and Pol III-transcribed genes and telomeric regions in the absence of exogenous genotoxic stress. Rad2 occupancy of Pol II-transcribed genes is transcription-dependent. Genome-wide Rad2 occupancy of class II gene promoters is well correlated with that of Mediator. Furthermore, UV sensitivity of med17 mutants is correlated with reduced Rad2 occupancy of class II genes and concomitant decrease of Mediator interaction with Rad2 protein. Our results suggest that Mediator is involved in DNA repair by facilitating Rad2 recruitment to transcribed genes.
Mediator of transcription regulation is a large multiprotein complex conserved in all eukaryotes. It interacts with RNA polymerase II (Pol II) to transmit signals from specific regulators to the Pol II transcription machinery. Together with the general transcription factors (GTFs; TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH), this coactivator stimulates the assembly of the preinitiation complex (PIC) and, ultimately, transcription initiation (Kornberg 2005; Ries and Meisterernst 2011). Mediator is generally required at Pol II-transcribed gene promoters for regulated transcription (Holstege et al. 1998; Andrau et al. 2006; Zhu et al. 2006). As expected given its central role in transcription activation, it has been implicated in numerous developmental processes. Mutations that affect Mediator subunits lead to a number of human pathologies (Risheg et al. 2007; Schwartz et al. 2007). Recently, mutations in the Med17 Mediator subunit have been associated with infantile cerebral atrophy (Kaufmann et al. 2010), and a mutation in the Med23 subunit cosegregated with intellectual disability (Hashimoto et al. 2011). Since oncogenesis results from gene disregulation, it is not unexpected that Mediator is involved in several cancers (Zhang et al. 2005; Vijayvargia et al. 2007; Firestein et al. 2008; Gade et al. 2009; Li et al. 2010; Kuuselo et al. 2011). Mediator has at least 25 subunits organized into four modules: head, middle, tail, and Cdk8 kinase modules. The complete crystallographic structure of the Mediator complex has not been determined. However, recent structural data have been obtained for the Mediator head module (Lariviere et al. 2012; Robinson et al. 2012). Mediator subunits are engaged in numerous contacts within the complex (Guglielmi et al. 2004) and probably with partners belonging to other components of the transcription machinery. For example, a cooperation of Mediator with TFIID has been reported in yeast and humans (Johnson et al. 2002; Lariviere et al. 2006; Cai et al. 2010; Takahashi et al. 2011). Recently, we identified a direct interaction between the Med17 Mediator subunit and the Rpb3 Pol II subunit required for global Pol II transcription in vivo (Soutourina et al. 2011). Previously, we showed that a direct contact between the Med11 subunit of Mediator and the Rad3 subunit of TFIIH is essential for the recruitment of the GTF to the PIC independently of Pol II (Esnault et al. 2008). In addition to the crucial role of Mediator as a link between activators and the basal transcription machinery, recent studies suggest broader Mediator transcriptional functions, notably in the post-recruitment steps of transcription (Wang et al. 2005; Takahashi et al. 2011) and in DNA looping between the enhancers and promoters through a functional connection with cohesin (Kagey et al. 2010).
Transcription is coupled with DNA repair, ensuring the continuity of Pol II progression. DNA lesions impede the elongating transcription machinery and can potentially affect gene expression, leading to problems in development, cell growth, and survival. Proteins first identified as components of transcription or DNA repair machineries may be involved in both processes, operating beyond their initially described pathway. The TFIIH complex is the best-characterized dual factor, playing an essential role in Pol II transcription as a GTF and in nucleotide excision DNA repair (NER) (Compe and Egly 2012). NER is a major and evolutionarily conserved DNA repair pathway that removes DNA lesions such as cyclobutane pyrimidine dimers (CPD), the most abundant photoproduct arising upon UV irradiation. During NER, >30 proteins cooperate to recognize, incise, and excise a damaged oligonucleotide from the genomic DNA to protect the cell from the deleterious effects of DNA damages (Lagerwerf et al. 2011). There are two distinct NER pathways: global genome repair (GGR), which removes DNA lesions in the genome overall, and transcription-coupled repair (TCR), which removes DNA lesions that interfere with the progression of RNA polymerase through actively transcribed genes (Svejstrup 2002, 2007; Hanawalt and Spivak 2008). A single irreversibly stalled Pol II in an essential gene could be enough to lead to cell death. This situation explains why a large number of mechanisms and factors have evolved to protect the cell from stalled Pol II. While the NER reaction and the components required for efficient DNA lesion recognition and repair are fairly well understood, many questions on the active coupling of DNA repair with transcription remain unanswered.
Recent reports suggest that an interplay between transcription and DNA repair is likely to be more complex than previously thought. PAF and Ccr4–Not complexes, known for their role in transcription, have been proposed to be required for efficient TCR in yeast (Gaillard et al. 2009). In addition to TFIIH, other NER factors have been suggested to play a dual role in DNA repair and transcription. Rad2/XPG, the 3′ endonuclease implicated in NER, is one of such factor. Mutations in the human XPG gene give rise to a xeroderma pigmentosum (XP) sometimes associated with Cockayne syndrome (CS) (Clarkson 2003; Scharer 2008). Rad2/XPG, together with the Rad1–Rad10/XPF–ERCC1 5′ endonuclease, cleaves the damaged DNA strand in both NER pathways, GGR and TCR, resulting in the release of a DNA fragment containing the DNA lesion. Moreover, a specific nonenzymatic function of XPG in recognition of stalled transcription in TCR initiation has been also proposed (Sarker et al. 2005). In addition, a transcriptional role for the NER factor Rad2/XPG has been suggested in the yeast Saccharomyces cerevisiae (Lee et al. 2002). In humans, mutations in XPG result in impaired transactivation (Ito et al. 2007), and XPG binds to the promoters and distal regions of several nuclear receptor (NR)-dependent genes in the absence of any genotoxic stress (Le May et al. 2010). Recently, XPG and XPF endonucleases have been found to be involved in chromatin looping between the promoter and the terminator of the activated RARβ2 human gene (Le May et al. 2012).
In this study, we identified a functionally important interaction between the essential Med17 Mediator subunit and the Rad2/XPG DNA repair protein, suggesting a functional link between Mediator of transcription regulation and DNA repair. Genome-wide location analyses revealed that Rad2 was associated with Pol II-transcribed genes but also with Pol III-transcribed genes and telomeric regions in the absence of exogenous genotoxic stress. We showed that Rad2 occupancy of Pol II-transcribed genes was transcription-dependent. Moreover, Rad2 occupancy of class II gene promoters was highly correlated with that of Mediator. Several Mediator med17 mutants were UV-sensitive in a GGR-deficient background, and their UV sensitivity was correlated with reduced Rad2 occupancy of class II genes and a concomitant decrease of the interaction between Mediator and Rad2 protein. We thus uncovered a previously unsuspected role of Mediator in DNA repair via Rad2/XPG recruitment.
Results
Physical interaction between the Med17 Mediator subunit and the Rad2/XPG protein
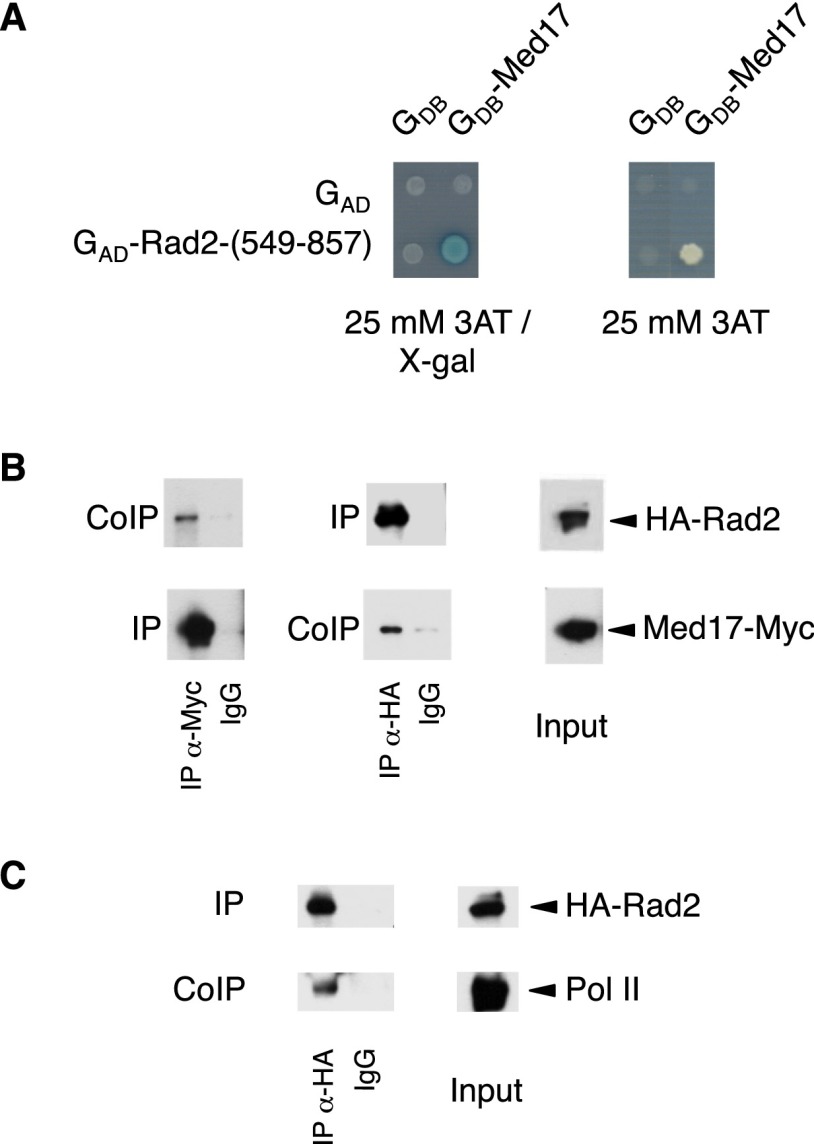
Previously, we characterized Mediator complex organization by screening the Mediator subunits with a yeast genomic library in a two-hybrid system. Protein–protein interactions within the Mediator complex have been published (Guglielmi et al. 2004). Many interactions that we uncovered in the screen were confirmed crystallographically and served as a basis for other Mediator studies (for example, Lariviere et al. 2012; Robinson et al. 2012). In addition, our two-hybrid screening revealed a large number of interactions between S. cerevisiae Mediator and other nuclear proteins (B Guglielmi, NL van Berkum, C Boschiero, FCP Holstege, and M Werner, unpubl.). One particularly interesting interaction is the contact between the Med17 Mediator head module subunit and the Rad2 endonuclease, the yeast homolog of human XPG protein involved in DNA repair. Med17 fused to the Gal4 DNA-binding domain interacted in the two-hybrid assay with the Rad2 fragment (549–857) fused to the Gal4 activation domain (Fig. 1A). The contact between Mediator and Rad2 was confirmed by coimmunoprecipitation (co-IP) experiments with crude extracts of a yeast strain expressing Med17-Myc and HA-Rad2 from their native promoters (Fig. 1B; Supplemental Fig. S1). Our results show that Mediator coimmunoprecipitates with Rad2 in crude extracts and vice versa. Rad2 was detected by Western blotting when Med17-Myc was used to immunoprecipitate Mediator complex, and the Med17-Myc Mediator subunit was coimmunoprecipitated when Rad2 was immunoprecipitated via the HA tag.
Figure 1.
The Med17 Mediator subunit interacts with the Rad2/XPG DNA repair protein. (A) Two-hybrid interaction between the Med17 Mediator subunit and Rad2. Two-hybrid assays were performed in the Y190 strain, and the activation of the lacZ or HIS3 reporters was tested using an X-Gal overlay plate assay or 3AT plate assay (Werner et al. 1993). Med17 was fused to the Gal4 DNA-binding domain (GDB-Med17), and the Rad2-(549–857) fragment was fused to the Gal4 activation domain [GAD-Rad2-(549–857)]. (B) Co-IP of Mediator and Rad2. Mediator was immunoprecipitated through the Med17-Myc subunit with α-Myc antibody from crude yeast extracts (Input; right panel) and analyzed by Western blotting with α-HA antibody (co-IP) against Rad2 (left panel). (Middle panel) Vice versa, Rad2 was immunoprecipitated with α-HA antibody from crude extracts and analyzed by Western blotting with α-Myc antibody (co-IP) against Med17. (IgG) Control immunoprecipitation with IgG magnetic beads only. (C) Co-IP of Rad2 and Pol II. Rad2 was immunoprecipitated with α-HA antibody from crude extracts (Input). Coimmunoprecipitated Pol II was detected by Western blotting with α-Rpb1 antibody (co-IP). (IgG) Control immunoprecipitation with IgG magnetic beads only.
Pol II is the main component of the transcription machinery and the first complex in TCR that recognizes the DNA damage (Svejstrup 2002, 2007; Hanawalt and Spivak 2008). The XPG protein was reported to coimmunoprecipitate with Pol II in crude extracts from undamaged HeLa cells (Sarker et al. 2005). In line with these results, our co-IP experiments also indicated that Rad2 coimmunoprecipitated with Pol II in yeast crude extracts (Fig. 1C). Coimmunoprecipitated Pol II forms phosphorylated on Ser5 and Ser2 of the Rpb1 C-terminal repeat domain (CTD) were also detected by Western blotting (data not shown).
Genome-wide location analysis of Rad2
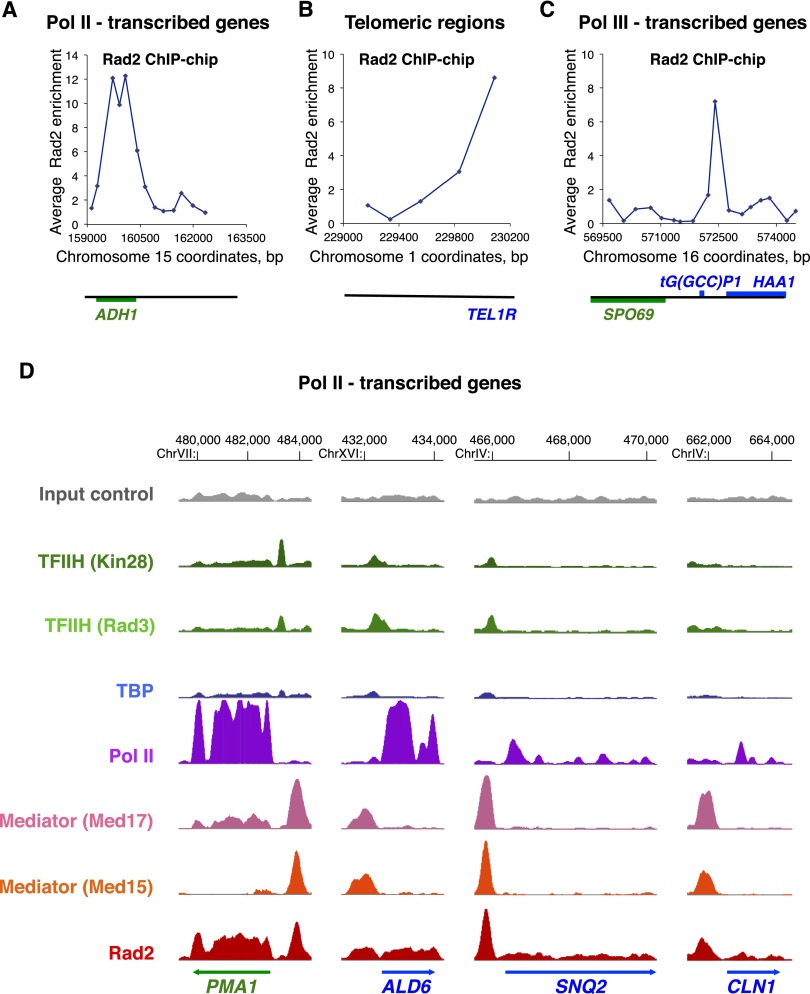
Recently, an association of XPG, the human homolog of Rad2, with several NR-dependent genes was shown in the absence of genotoxic stress (Le May et al. 2010, 2012). However, to date, no genomic data are available on the Rad2/XPG occupancy in humans or yeast. To determine the targets of Rad2 in the yeast genome in the absence of exogenous genotoxic stress, we performed Rad2 chromatin immunoprecipitation (ChIP)–chip experiments. Two yeast strains carrying N-terminal or C-terminal HA-tagged version of Rad2 were constructed and used in conventional ChIP experiments. No increase in the UV sensitivity of these strains was observed, suggesting that both tagged versions of Rad2 were functional (data not shown). We found that N-terminally and C-terminally tagged Rad2 were significantly enriched inside three constitutively expressed Pol II-transcribed genes tested: ADH1, PYK1, and PMA1 (Supplemental Fig. S2A). The strain carrying an N-terminal tagged version of Rad2 was used for ChIP–chip analysis, since the ChIP signal to noise ratios were higher. We found 2471 oligonucleotides significantly bound by Rad2 (P-value < 0.005; out of 40,000 oligonucleotide probes present on the DNA arrays). The genomic profile of Rad2 occupancy was complex, spanning from Pol II-transcribed genes (promoter and transcribed regions) to, unexpectedly, Pol III-transcribed genes and telomeric/subtelomeric regions. Examples of Rad2 ChIP–chip enrichment profiles illustrate these different genomic region categories (Fig. 2A–C).
Figure 2.
Genome-wide analysis of Rad2, Mediator, Pol II, TBP, and TFIIH. (A–C) Examples of Rad2 ChIP–chip enrichment profiles on Pol II-transcribed genes (A), on a telomeric region (B), and on a Pol III-transcribed gene (C). Rad2 enrichment profiles were assessed from ChIP–chip experiments with HA-Rad2 strain grown at 30°C in YPD medium. Watson strand-transcribed genes are colored in blue, and Crick strand-transcribed genes are colored in green. The genomic positions of probe regions on corresponding chromosomes are indicated along the X-axis. The enrichment ratio is indicated along the Y-axis. (D) Examples of Rad2, Mediator, Pol II, TBP, and TFIIH ChIP-seq enrichment profiles on selected Pol II-transcribed genes. Densities of sequence tags were assessed from ChIP-seq experiments performed with HA-Rad2-tagged, Med15-HA-tagged (Mediator), Med17-HA-tagged (Mediator), HA-TBP-tagged, Rad3-HA-tagged (core TFIIH), and Kin28-HA-tagged (TFIIH kinase module) strains with α-HA antibody. Pol II was immunoprecipitated using α-Rpb1 antibody. ChIP-seq density profiles are displayed using IGB yeast genome browser. Watson strand-transcribed genes are colored in blue, and Crick strand-transcribed genes are colored in green. Input DNA and DNA from ChIP with an untagged strain were used as negative controls. Densities of sequence tags were displayed after subtraction of normalized control of untagged strain as described in the Materials and Methods.
To gain in resolution and determine precisely the yeast genomic regions enriched by Rad2 protein, we performed ChIP-seq experiments. In agreement with our Rad2 ChIP–chip results, we confirmed the presence of this protein inside and on the promoter regions of Pol II-transcribed genes as well as on Pol III-transcribed genes. Telomeric regions are highly repetitive and therefore are poorly represented in ChIP-seq analysis. We identified 2600 significantly enriched Rad2 peaks using MACS2 peak calling software; 1203 of these peaks were located on promoter regions of Pol II-transcribed genes, 33 were on snRNA genes, and 268 were on Pol III-transcribed genes. This bioinformatics tool, MACS2, also allowed us to identify some Rad2 enrichment peaks inside the transcribed regions of class II genes but was not particularly adapted for detection of large Rad2 distributions in these regions, missing many Rad2-enriched ORFs. To identify precisely the Rad2 enrichment signals inside ORFs and thus complete the Rad2 ChIP-seq distribution analysis, we compared Rad2 mean densities for each ORF with density distributions of two negative controls: untagged strain and input DNA samples. We observed that 818 regions inside the protein-coding genes were significantly enriched by Rad2.
The results obtained by our genome-wide analyses were confirmed by conventional ChIP on a set of selected regions representing the categories of Rad2 genomic targets that we identified. Supplemental Figure S2B illustrates significant enrichment of Rad2 on several promoter and transcribed regions of selected class II genes compared with background levels measured on the coding region of the nontranscribed GAL1 gene. All selected Pol III-transcribed genes displayed significant Rad2 enrichment (Supplemental Fig. S2C). The spatial Rad2 distribution on the longest class III gene, SCR1, was similar to that of Pol III (Supplemental Fig. S2D; Ghavi-Helm et al. 2008). We also confirmed the presence of Rad2 on telomeric regions (Supplemental Fig. S2E). In addition, a significant Rad2 enrichment was detected on the Pol I-transcribed 35S rDNA region compared with the intergenic NTS2 region (Supplemental Fig. S2F). Taken together, these results indicated a complex Rad2 distribution on the yeast genome in the absence of exogenous genotoxic stress, spanning the genes of all three transcription classes and telomeric/subtelomeric regions. For further study, we focused particularly on Pol II-transcribed genes.
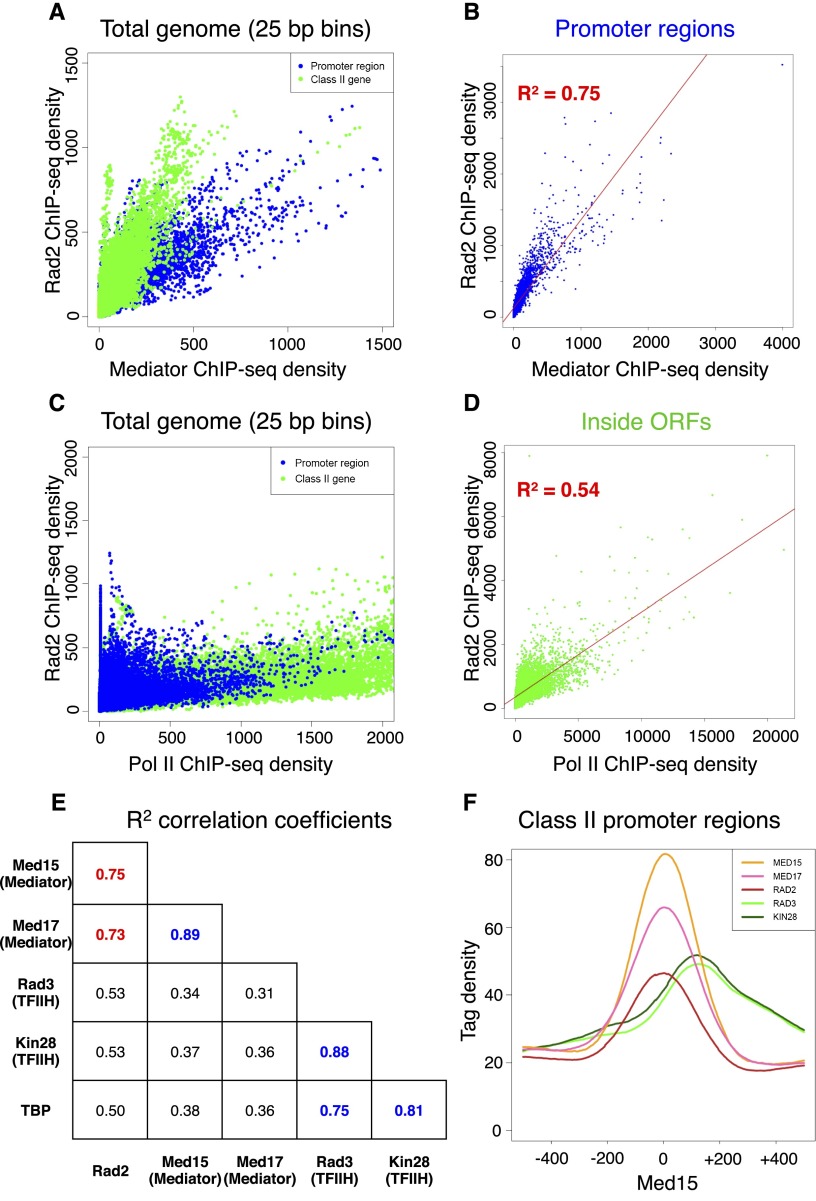
Genome-wide Rad2 occupancy correlates with Mediator occupancy on promoter regions of Pol II-transcribed genes
To compare Rad2 ChIP-seq profiles with those of Mediator, Pol II, and GTFs, we performed ChIP-seq experiments for Mediator (Med15 and Med17 subunits), Pol II, and GTFs (TBP and two TFIIH subunits: the Rad3/XPD subunit of core TFIIH and the Kin28/Cdk7 kinase subunit of TFIIK). Input DNA and DNA from ChIP with an untagged strain were used as negative controls. Figure 2D and Supplemental Figure S3 show examples of ChIP-seq density distributions using the Integrated Genome Browser (IGB) for all of the proteins on selected class II (Fig. 2D) or class III (Supplemental Fig. S3) genes. Transcription by all three RNA polymerases requires TBP protein. As expected, density peaks of this protein were also detected on the Pol III-transcribed genes. These TBP peaks on class III genes coincided with Rad2 peaks (Supplemental Fig. S3). On the selected promoter regions of Pol II-transcribed genes, there was a particular correlation between Rad2 and Mediator for the location and intensity of peaks (Fig. 2D). Furthermore, genome-wide Mediator and Rad2 occupancies were well correlated. Rad2 and Mediator ChIP-seq densities (25-base-pair [bp] bin) on the yeast genome were computed and compared (Fig. 3A; Supplemental Fig. S4A). As expected, the Rad2 enrichment of several genomic categories, including class II promoter and transcribed regions, made the correlation between Rad2 and Mediator densities complex. Considering only class II gene promoter regions, regression analysis showed a linear correlation with a correlation coefficient of 0.75 for the Med15 Mediator subunit (Fig. 3B) or 0.73 for the Med17 Mediator subunit (Supplemental Fig. S4B). Intergenic regions encompassing Pol III-transcribed genes were excluded from this analysis. The global correlation between Rad2 and Mediator occupancies on promoter regions is consistent with a functional link between Rad2 and the Mediator complex. We determined whether Rad2 association with Pol II-transcribed regions was correlated with Pol II occupancy (Fig. 3C). Considering only Pol II-transcribed regions inside ORFs, encompassing most of the yeast Pol II ChIP signals, no strong correlation could be observed between Rad2 and Pol II ChIP-seq densities (Fig. 3D). Regression analysis showed a lower correlation coefficient of 0.54. In comparison, regression analysis indicated, as expected, a linear correlation between two subunits of the Mediator complex (Med15 and Med17) or TFIIH complex (Rad3 and Kin28), with a high correlation coefficient of 0.89 (Supplemental Fig. S4C,D) or 0.88 (Supplemental Fig. S5A,B), respectively, and between TBP and Rad3 or Kin28 TFIIH subunits with a high correlation coefficient of 0.75 or 0.81, respectively (Fig. 3E). Pairwise correlations between ChIP-seq densities of Rad2, Mediator, Kin28, Rad3, and TBP showed that the strongest correlation for Rad2 protein occupancy on promoter regions was with Mediator (Fig. 3E). On the other hand, no strong correlation was observed between Rad2 and Kin28 ChIP-seq occupancies, with a correlation coefficient of 0.53 (Supplemental Fig. S5C,D).
Figure 3.
Correlation of Rad2 ChIP-seq density with those of Mediator and Pol II. (A) Rad2 ChIP-seq density versus Mediator (Med15 subunit) ChIP-seq density on total yeast genome. Density of sequence tags in Mediator and Rad2 ChIP-seq experiments has been calculated for each 25-bp bin of yeast genome. Each point on the plot corresponds to one genomic bin. The genomic bins located inside ORFs or class II promoter regions are indicated in green and blue, respectively. Pol III-transcribed genes were excluded from this analysis. (B) Rad2 ChIP-seq density versus Mediator (Med15 subunit) ChIP-seq density on class II promoter regions. The density of sequence tags in Mediator and Rad2 ChIP-seq experiments has been calculated for promoter region of each Pol II-transcribed gene. Each point on the plot corresponds to one promoter region. Promoter regions correspond to intergenic regions for Pol II-transcribed genes in tandem or in divergent orientation, excluding intergenic regions encompassing Pol III-transcribed genes. A linear regression for Rad2 ChIP-seq density versus Mediator ChIP-seq density (red line) and a R2 correlation coefficient are indicated. (C) Rad2 ChIP-seq density versus Pol II ChIP-seq density on total yeast genome, as described in A. (D) Rad2 ChIP-seq density versus Pol II ChIP-seq density inside ORFs. The density of sequence tags in Rad2 and Pol II ChIP-seq experiments has been calculated for each ORF. Each point on the plot corresponds to one ORF. A linear regression for Rad2 ChIP-seq density versus Pol II ChIP-seq density (red line) and a R2 correlation coefficient are indicated. (E) Pairwise correlation coefficients between ChIP-seq densities of Rad2, Med15 (Mediator), Med17 (Mediator), Rad3 (TFIIH core module), Kin28 (TFIIH kinase module), and TBP on promoter regions. R2 correlation coefficients for each pairwise combination are shown. A correlation coefficient for Mediator versus Rad2 is highlighted in red, and pairwise correlation coefficients for different GTF (Rad3, Kin28, and TBP) or Mediator subunit combinations (Med15 and Med17) are shown in blue. (F) Distribution of Mediator, Rad2, and TFIIH ChIP-seq densities on Pol II-transcribed gene promoters. Tag densities are shown relative to the localization of the maximum of Mediator enrichment peaks. Intergenic regions encompassing Pol III-transcribed genes were excluded. The tag density was determined for each protein in a 1000-bp window centered on the maximum of Mediator enrichment peaks per each nucleotide position. Mean tag density was then calculated and plotted over the window.
The location of Rad2 peaks on the promoter regions of Pol II-transcribed genes was compared with those of Mediator and GTFs. Mean ChIP-seq read densities were calculated for each protein on promoter regions of Pol II-transcribed genes centered on Mediator enrichment peaks (Fig. 3F). To consider only Pol II-transcribed gene promoters, intergenic regions encompassing Pol III-transcribed genes were excluded. These analyses showed that Rad2 peak distribution coincides with that of Mediator (Med15 and Med17 subunits) but not with those of TFIIH core (Rad3) or kinase (Kin28) modules. Taken together, our results revealed that genome-wide Rad2 occupancy was highly correlated with that of Mediator on promoter regions of Pol II-transcribed genes and that Rad2 enrichment distribution in these regions superposed with that of Mediator.
Rad2 occupancy on Pol II-transcribed genes is transcription-dependent
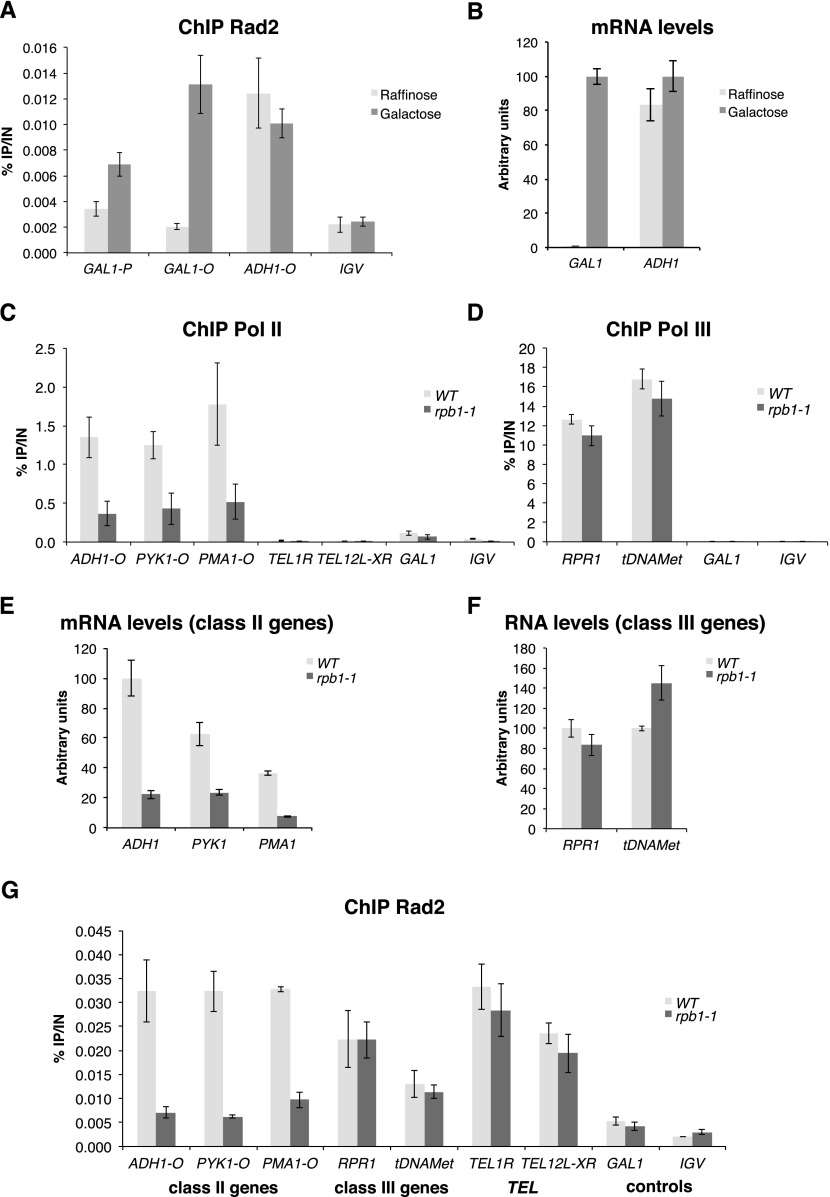
An essential question arising from Rad2 recruitment in the absence of exogenous genotoxic stress is whether Rad2 occupancy on class II genes depends on Pol II transcription. To address this question, we examined Rad2 occupancy on the GAL1 gene by ChIP experiments upon galactose induction. When yeast cells were grown in raffinose-supplemented medium, Rad2 was absent from the GAL1 promoter and ORF (Fig. 4A). After 60 min of galactose induction, Rad2 was recruited to the promoter and transcribed region of the GAL1 gene. The constitutively expressed ADH1 gene was always bound by the Rad2 protein. As expected, the GAL1 mRNA level was highly induced after galactose induction, while the ADH1 mRNA level was comparable in raffinose- and galactose-supplemented medium (Fig. 4B). These results indicated that Rad2 recruitment to GAL1 gene was transcription-dependent.
Figure 4.
Transcription dependency of Rad2 occupancy on Pol II-transcribed genes. (A) Quantitative ChIP analysis of Rad2 occupancy on the GAL1 gene before and after galactose induction. The HA-Rad2 tagged strain was grown in a raffinose-supplemented medium, and then galactose was added for 60 min. Quantitative ChIP experiments were performed with α-HA antibody against HA-Rad2. Mean values and standard deviation (indicated by error bars) of three independent experiments are shown. Immunoprecipitated fragments from ChIP experiments were amplified with primers corresponding to the GAL1 gene promoter (GAL1 P) or ORF (GAL1 O). ADH1 ORF represents a constitutively expressed gene control. A nontranscribed region on chromosome V (IGV) was used as a negative control. (B) mRNA level of GAL1 and ADH1 genes before and after galactose induction. Yeast cells were grown in a raffinose-supplemented medium, and then galactose was added for 60 min. RNA levels for GAL1 and ADH1 genes were quantified by RT-qPCR and normalized to 25S rRNA, 18S rRNA, and SCR1 levels. Values are expressed in arbitrary units representing a percentage of the value in galactose-supplemented medium. Mean values and standard deviation (indicated by error bars) of three independent experiments are shown. (C,D,G) Effect of rpb1-1 mutation on Rad2, Pol II, and Pol III occupancies on selected regions. Quantitative ChIP assays were performed using α-Rpb1 antibody (Pol II) (C), α-Myc antibody against Rpc160-Myc (Pol III) (D), and α-HA antibody against HA-Rad2 (G). Cells were grown in selective SD medium complemented with amino acids at 25°C and then shifted for 90 min at 37°C. A GAL1 ORF and a nontranscribed region on chromosome V (IGV) were used as negative controls. (E,F) Effect of rpb1-1 mutation on RNA level of class II (E) or class III (F) genes. Cells were grown in selective SD medium complemented with amino acids at 25°C and then shifted for 90 min at 37°C. RNA levels for selected genes were quantified by RT-qPCR and normalized to 25S rRNA, 18S rRNA, and SCR1 levels. Values are expressed in arbitrary units representing a percentage of the value in the wild-type strain for the ADH1 gene (E) or RPR1 or tDNAMet genes (F). Mean values and standard deviation (indicated by error bars) of three independent experiments are shown.
To address more generally the question of the transcriptional dependency of Rad2 occupancy on Pol II-transcribed genes, we used a Pol II mutant, rpb1-1, that rapidly stops transcription after a shift to nonpermissive temperature (Nonet et al. 1987). When cells were incubated for 90 min at 37°C, as expected, Pol II occupancy on Pol II-transcribed genes was strongly reduced in the rpb1-1 mutant compared with the wild-type strain (Fig. 4C), whereas Pol III occupancy on tested class III genes remained unchanged (Fig. 4D). We also checked that, as expected, RNA levels were strongly decreased in the rpb1-1 mutant for class II genes (Fig. 4E) but not class III genes (Fig. 4F). Rad2 occupancy on Pol II-transcribed genes followed that of Pol II and was also greatly reduced after incubation at 37°C (Fig. 4G; Supplemental S6A,B). Rad2 occupancy decrease was observed for Pol II-transcribed gene promoters and ORFs. This decrease of Rad2 occupancy was not related to Mediator occupancy, since it did not change in the rpb1-1 mutant (Supplemental Fig. S6C–E). Conversely, Rad2 occupancy on class III genes or telomeric regions remained unchanged under the same conditions (Fig. 4G). Thus, the presence of Rad2 on class II genes is specifically dependent on Pol II transcription.
UV irradiation sensitivity is the only observed phenotype of rad2 deletion
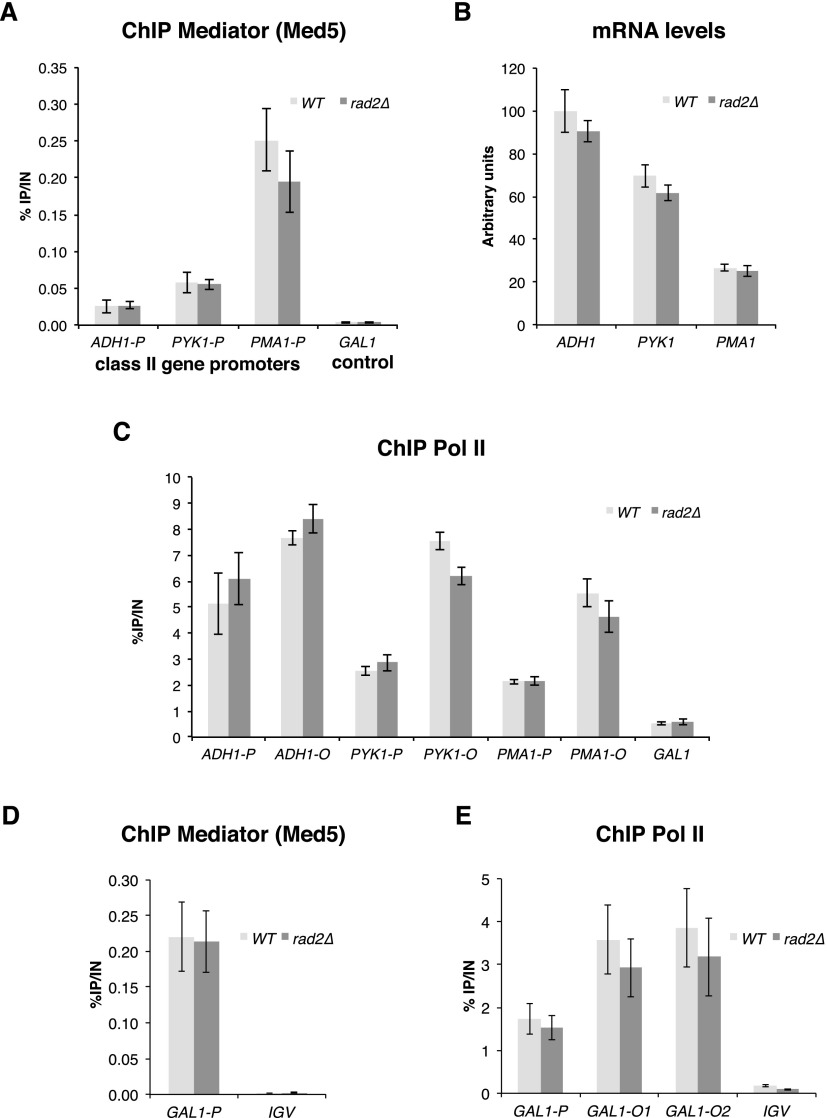
One hypothesis for the functional interplay between Mediator and Rad2 consists of a possible implication of Rad2/XPG in transcription, since such a role has been previously suggested (Lee et al. 2002; Le May et al. 2010, 2012). In S. cerevisiae, the RAD2 gene is not essential for viability. We started by examining whether, under standard growth conditions (rich glucose medium, YPD), any effect of rad2 deletion on Mediator or Pol II occupancy could be identified. We did not observe any difference between Mediator occupancy of the rad2Δ strain compared with the wild-type strain on constitutively expressed gene promoters (Fig. 5A; Supplemental Fig. S7A,B). In addition, Pol II occupancy on selected genes and the corresponding mRNA level in the rad2Δ strain were similar to those of the wild-type strain (Fig. 5B,C; Supplemental Fig. S7C). Similarly, no effect of rad2 deletion on Mediator or Pol II occupancy on the GAL1 gene was observed in galactose-inducing medium (Fig. 5D,E). Previously, Lee et al. (2002) have stated that rad2 deletion slowed down the kinetics of galactose-inducible genes. To check this possibility, the Pol II recruitment to the GAL1 gene promoter and transcribed region and the GAL1 mRNA level were examined at different times after galactose induction by ChIP and quantitative RT–PCR (qRT–PCR) experiments. However, there was no difference between the rad2Δ mutant and the wild-type strain (Supplemental Fig. S8).
Figure 5.
Mediator and Pol II occupancies in the rad2 deletion strain on selected regions. (A,C) Quantitative ChIP analysis of Mediator and Pol II occupancies under standard growth conditions. rad2Δ and wild-type strains were grown in YPD medium at 30°C. Immunoprecipitations were performed using α-HA antibody against Med5-HA (Mediator) (A) and α-Rpb1 antibody (Pol II) (C). (A) Immunoprecipitated fragments from Mediator ChIP experiments were amplified with primers corresponding to selected class II gene promoters. (C) Immunoprecipitated fragments from Pol II ChIP experiments were amplified with primers corresponding to selected class II gene promoters (P) or ORFs (O). Mean values and standard deviation (indicated by error bars) of three independent experiments are shown. A GAL1 ORF was used as a negative control. (B) Effect of rad2Δ mutation on mRNA levels of class II genes. rad2Δ and wild-type strains were grown in YPD medium at 30°C. RNA levels for selected genes were quantified by RT-qPCR and normalized to 25S rRNA, 18S rRNA, and SCR1 levels. Values are expressed in arbitrary units representing a percentage of the value in the wild-type strain for ADH1 gene. Mean values and standard deviation (indicated by error bars) of three independent experiments are shown. (D,E) Quantitative ChIP analysis of Mediator and Pol II occupancies upon galactose induction. rad2Δ and wild-type strains were grown in a raffinose-supplemented medium, and then galactose was added for 60 min. Quantitative ChIP experiments were performed with α-HA antibody against Med5-HA (Mediator) (D) and α-Rpb1 antibody (Pol II) (E). Mean values and standard deviation (indicated by error bars) of three independent experiments are shown. Immunoprecipitated fragments from ChIP experiments were amplified with primers corresponding to GAL1 gene promoter (GAL1-P) or ORF (GAL1-O1 and GAL1-O2). A nontranscribed region on chromosome V (IGV) was used as a negative control.
To identify growth phenotypes of rad2 deletion strains and therefore a possible transcriptional role for Rad2, we tested different growth conditions in two backgrounds: BY4741 and YPH499. No growth difference between rad2Δ and wild-type strains was observed on YPD medium under different temperature conditions (16°C, 25°C, 30°C, or 37°C), on different carbon sources (YP medium supplemented with galactose, lactate, ethanol, or glycerol), and on drugs to test for multidrug resistance transcriptional response (cycloheximide and ketoconazole) or for NTP depletion conditions (6-azauracil and mycophenolic acid) (Supplemental Fig. S9; data not shown). The only phenotype of the rad2 deletion strains that we observed is a high UV sensitivity, as expected for a DNA repair protein. Taken together, our results strongly suggest that Rad2 does not play a major role in the transcriptional process in yeast, at least in the two backgrounds examined here.
Physiological significance of Mediator–Rad2 contact in DNA repair
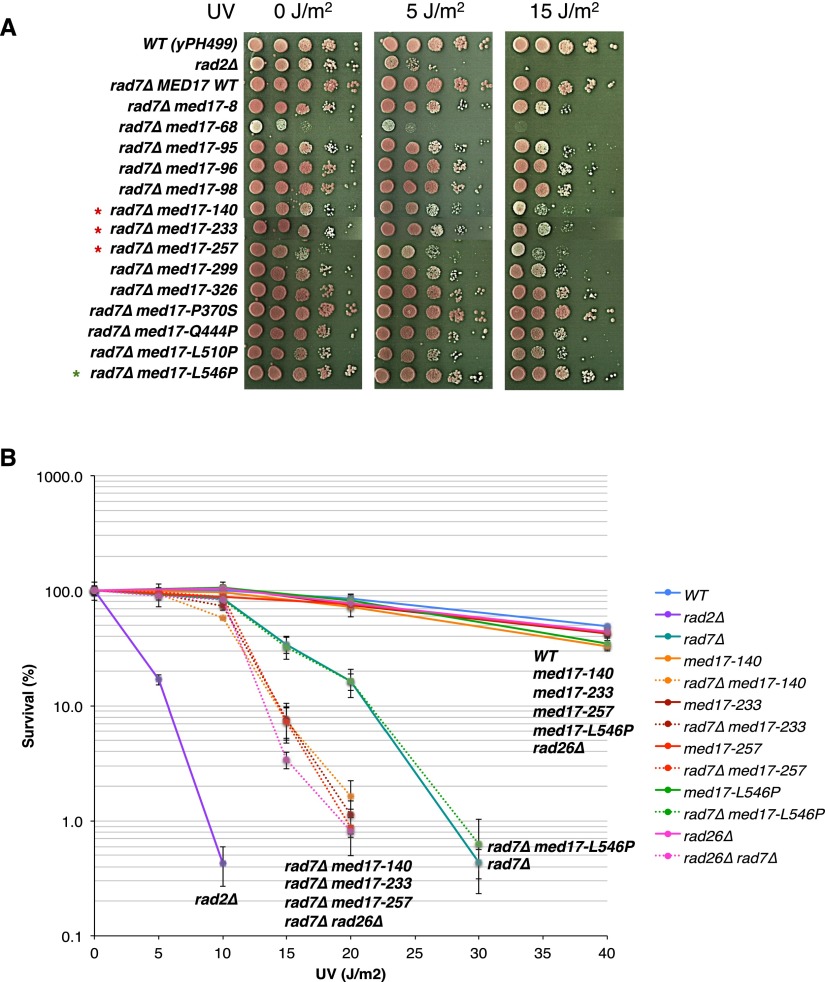
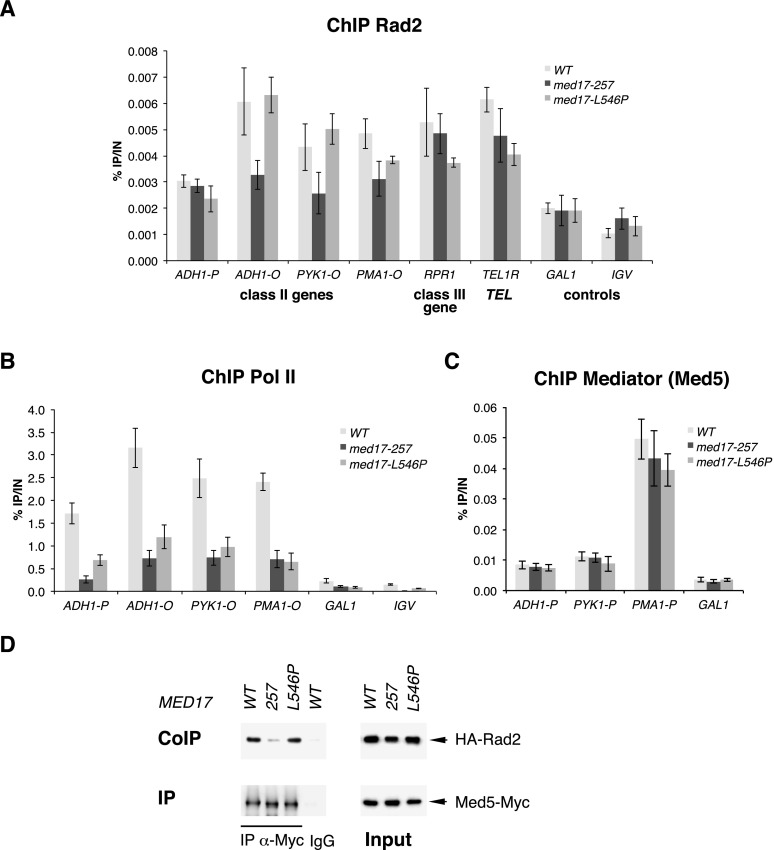
We could not identify a major implication of Rad2 in yeast Pol II transcription. This led to the hypothesis that Mediator might play a role in DNA repair via its interaction with the Rad2 protein. To investigate the functional relationship between Mediator and Rad2, we used our collection of 29 conditional mutants for the Med17 essential Mediator subunit (Soutourina et al. 2011). The integrity of Mediator in med17 mutants has been verified by co-IP experiments (Soutourina et al. 2011). To directly address a possible role of Mediator in DNA repair, a UV sensitivity phenotype was systematically investigated for med17 mutants. No UV sensitivity was found for med17 mutants in the context where both NER pathways, GGR and TCR, were functional (Supplemental Fig. S10). Next, we used a GGR-deficient context (rad7Δ strains) to analyze specifically the potential implication of Med17 in the TCR pathway by UV sensitivity spot assays. Several Mediator mutants among our collection of med17-ts were UV-sensitive in a rad7Δ GGR-deficient context (Fig. 6A). To refine the UV sensitivity analysis, we determined UV survival curves for several med17 mutants (three UV-sensitive med17-140, med17-233, and med17-257 mutants and one UV-insensitive med17-L546P mutant) alone or in combination with rad7Δ mutation (Fig. 6B). The UV dose response analysis further confirmed UV sensitivity spot assays and clearly indicated that the combination of rad7 deletion with several med17 mutations leads to an increased UV sensitivity. An extremely UV-sensitive rad2 deletion mutant was also included for comparison. Interestingly, the UV sensitivity of med17-ts rad7Δ mutants was comparable with that of the rad26Δ rad7Δ double mutant deficient in both GGR (rad7Δ) and TCR (rad26Δ) NER pathways (Fig. 6B). Rad26 is the yeast homolog of the TCR-specific human CSB protein. In yeast, RAD26 deletion does not lead to UV sensitivity but significantly increases the UV sensitivity of GGR-deficient mutants. Since med17 mutants were UV-sensitive in a GGR-deficient context, we wondered whether these mutants could be epistatic with the TCR-deficient rad26 deletion mutant. No UV sensitivity increase was found for med17 mutants in the rad26Δ context (Supplemental Fig. S11), demonstrating that, indeed, med17 mutants are epistatic with the rad26Δ mutant. Taken together, these results are consistent with a specific implication of Mediator in the TCR pathway. Our results also indicated that the UV sensitivity of med17-ts mutants was not a consequence of a transcriptional effect, since this phenotype was observed only in a GGR-deficient context and not in a context where both NER pathways, GGR and TCR, were functional or in a TCR-deficient context. Three of the UV-sensitive med17-ts mutants were selected for further molecular study (med17-257, med17-140, and med17-233) (the results on the two latter mutants are shown in Supplemental Fig. S12). Rad2 occupancy in these mutants was determined by ChIP experiments for selected Pol II-transcribed genes. Figure 7A and Supplemental Figure S12A show a significant decrease in Rad2 association in UV-sensitive Mediator mutants compared with the wild-type strain and med17-L546P mutant, one of the UV-insensitive med17-ts mutants. Rad2 occupancy to a class III gene and a telomeric region was not affected in these mutants. All tested med17 mutations, including med17-L546P, which is UV-insensitive, led to a marked decrease in Pol II association with Pol II-transcribed genes but did not modify the Mediator association to corresponding gene promoters (Fig. 7B,C; Supplemental Fig. S12B,C). A decrease in Pol II occupancy was not responsible for Rad2 occupancy defects of UV-sensitive mutants (med17-257, med17-140, and med17-233) or their UV sensitivity, since a reduction of Pol II occupancy in the med17-L546P mutant, similar to that of the UV-sensitive med17-ts mutants, was not accompanied by Rad2 occupancy decrease and UV sensitivity. To determine the effects of med17-ts mutations on Mediator contact with the Rad2 protein, we performed co-IP experiments and showed that Mediator–Rad2 interaction was reduced in UV-sensitive mutants med17-257, med17-140 , and med17-233 compared with the wild-type strain and the med17-L546P UV-insensitive mutant (Fig. 7D; Supplemental Fig. S12D). These results indicated that Mediator is involved in Rad2 recruitment and therefore suggest that it is involved in DNA repair.
Figure 6.
UV sensitivity of med17 mutants. (A) UV sensitivity of med17 mutants in a GGR-deficient rad7Δ context by spot assays. Cells were serially diluted, spotted on YPD agar plates, treated or not with the indicated doses of UV (UV Stratalinker 1800), and incubated for 3 d at 30°C. Red asterisks mark selected UV-sensitive med17 mutants, and a green asterisk marks a selected UV-insensitive med17 mutant. (B) UV survival curves of med17 and rad26Δ mutants alone and in a GGR-deficient rad7Δ context. Cells were grown in YPD medium to a mid-exponential phase, diluted, plated on YPD agar, and irradiated with the different doses of UV (UV Stratalinker 1800). Plates were incubated for 3 d at 30°C in the dark before colonies were counted and survival was calculated. Mean values and standard deviation (indicated by error bars) of at least three independent experiments are shown.
Figure 7.
Effects of med17 mutants on Rad2, Pol II, and Mediator occupancies and on Rad2–Mediator interaction. (A–C) Quantitative ChIP analysis of Rad2, Pol II, and Mediator occupancies on selected regions. ChIP assays were performed using α-HA antibody against HA-Rad2 (A), α-Rpb1 antibody (Pol II) (B), and α-HA antibody against Med5-HA (Mediator) (C). Cells were grown in YPD medium at 30°C and then shifted for 45 min at 37°C. Immunoprecipitated fragments from ChIP experiments were amplified with primers corresponding to selected class II gene promoters (P) or ORFs (O), a selected class III gene, and a telomeric region. Mean values and standard deviation (indicated by error bars) of three independent experiments are shown. A GAL1 ORF and a nontranscribed region on chromosome V (IGV) were used as negative controls. (D) Co-IP of Mediator and Rad2. Mediator was immunoprecipitated through a Med5-Myc subunit with a α-Myc antibody from crude extracts (Input; right panel) and analyzed by Western blotting with α-HA antibody (co-IP) against Rad2 (left panel). (IgG) Control immunoprecipitation with IgG magnetic beads only.
Discussion
Substantial evidence has been obtained on the essential function of the Mediator complex in transcription activation. However, our experiments indicate that this complex plays an important role in linking transcription with other nuclear processes. In this study, we identified a functionally important contact between the Med17 Mediator subunit and the Rad2 DNA repair protein, the S. cerevisiae homolog of human XPG. Mediator might play a role in DNA repair by loading Rad2/XPG to active genes, or Rad2/XPG could be implicated in transcription, influencing PIC assembly or stability on several gene promoters or both. Our results suggest that Mediator is involved in DNA repair through a functional link with the Rad2/XPG protein. (1) The essential Med17 Mediator subunit interacts with Rad2/XPG DNA repair protein. (2) We showed that Rad2 is located on Pol II-transcribed genes (promoter and transcribed regions) in the absence of exogenous genotoxic stress. (3) Genome-wide Rad2 occupancy of class II gene promoters is well correlated with that of Mediator. (4) Moreover, Rad2 distribution on these regions overlapped with that of Mediator but not of TFIIH, further highlighting a functional cooperation between Mediator and Rad2/XPG. It should be noted that Mediator and Rad2 ChIP-seq peaks on class II gene promoters are generally located on the upstream regulating regions, whereas TFIIH and other GTFs peaks are located on core promoter regions close to transcription start sites. (5) Rad2 occupancy of Pol II-transcribed genes is transcription-dependent, since rpb1-1 Pol II mutation reduces Rad2 occupancy after a shift to nonpermissive temperature when transcription is rapidly inhibited, and since Rad2 is recruited to GAL1 gene upon galactose induction of this gene. (6) However, no growth phenotypes (except UV sensitivity) or transcriptional effects were observed in the rad2Δ context, suggesting that Rad2 does not play a major role in the transcriptional process in yeast, at least in our two backgrounds. (7) On the contrary, Mediator med17-ts mutants are UV-sensitive in a GGR-deficient background (rad7Δ) and are epistatic with a TCR-deficient mutant (rad26Δ). (8) This UV sensitivity of several med17 mutants is correlated with reduced Rad2 occupancy of class II genes and concomitant decrease of interaction with Rad2 protein.
We suggest that the contact between Med17 and Rad2 is direct, since our two-hybrid screening permitted us to reveal only specific interactions between different subunits within the Mediator complex (Guglielmi et al. 2004) that were further confirmed as direct by structural analysis (for example, Lariviere et al. 2012; Robinson et al. 2012). Moreover, we demonstrated that several mutations in the Med17 Mediator subunit resulted in a decreased interaction between Mediator and Rad2, an observation in favor of a direct Mediator–Rad2 contact.
Therefore, Mediator is likely to facilitate Rad2 recruitment to transcribed genes, setting the stage for rapid DNA lesion removal. We propose that Mediator could function in transcription-coupled DNA repair, revealing a previously unknown role of the Mediator complex as a link between transcription and DNA repair. Whether Rad2/XPG is the only NER factor that is recruited through an interaction with Mediator or assembles with other NER factors to remove lesions on transcribed genes remains to be addressed. Further investigations will determine whether Mediator is engaged in functional interplay with other NER factors.
In the light of Mediator and Rad2/XPG conservation from yeast to humans, the molecular events governing the Mediator link with DNA repair are likely to exist in all eukaryotes. In support of this idea, we observed the Mediator–XPG contact by co-IP experiments with crude extracts from HeLa cells (data not shown). The conservation of Mediator connection to DNA repair might give insights into our understanding of human diseases like XP/CS.
Interestingly, both Mediator and Rad2/XPG interact with Pol II, the main component of transcription machinery and the first complex in TCR that recognizes the DNA damage. The mechanisms of functional orchestration of these contacts remain to be investigated. Our genome-wide analyses show that Mediator and Rad2 co-occupy class II gene promoter regions and, moreover, that a high correlation is observed between the two profiles on promoter regions. Rad2 is also bound to Pol II-transcribed regions that are occupied by Pol II. However, a correlation between Rad2 and Pol II on these regions was modest, suggesting a complex relationship between Rad2 and Pol II. At this stage, we propose that Mediator facilitates Rad2 recruitment on class II genes promoters. While we do not know what the precise mechanisms of Rad2 loading on ORFs are, our results suggest that they could entail both the loading of Rad2 on Pol II and another independent mechanism. A modest genome-wide correlation between Rad2 and Pol II contrasts with the good genome-wide correlation between TFIIS and Pol II or Pol III occupancies on Pol II- or Pol III-transcribed genes, respectively (Ghavi-Helm et al. 2008). These observations suggested that TFIIS associates at the 5′ end of genes and accompanies Pol II and Pol III but cannot be loaded on traveling enzymes. It should be noted that one of the med17 mutants used in this study (med17-257) that showed a UV-sensitive phenotype was lethal in combination with the rpb3-2 Pol II mutation (Soutourina et al. 2011) and was affected in Pol II–Mediator interaction (data not shown). However, other med17 mutations with UV-sensitive phenotypes did not lead to rpb3-2 colethality phenotypes. Our results showed that Pol II occupancy was reduced in UV-sensitive and -insensitive med17 mutants. Thus, the lower occupancy of Pol II cannot explain the UV sensitivity of the med17 mutants that are defective in Rad2 interaction. We also showed that the UV sensitivity of med17 mutants was not a consequence of a transcriptional defect, since this phenotype was observed only in a GGR-deficient context and not in a context where both NER pathways were functional or a TCR-deficient context. Taken together, our results suggest that UV sensitivity of Mediator mutants was not an indirect consequence of their transcriptional defects.
A transcriptional role for the NER factor Rad2/XPG was first proposed in the yeast S. cerevisiae (Lee et al. 2002). Rad2 was suggested to promote efficient Pol II transcription. However, we did not observe the mild growth phenotypes of rad2 deletion strains or slower galactose induction kinetics in the absence of the RAD2 gene reported by Lee et al. (2002). Moreover, we did not find any genetic interaction between rad2Δ and rad26Δ. Rad26 is the yeast homolog of the TCR-specific human CSB protein. This result might be due to the different genetic backgrounds used by us and Lee et al. (2002). In spite of our considerable efforts, we did not obtain any evidence for a direct implication of Rad2 in transcription. In human cells, it has been demonstrated that mutations in XPG prevent its association with TFIIH, resulting in the dissociation of CAK and XPD/Rad3 from the core TFIIH and in impaired transactivation (Ito et al. 2007). Recently, XPG and other NER factors were shown to be associated with the promoters and distal regions of several NR-dependent genes in the absence of any exogenous genotoxic stress (Le May et al. 2010). The presence of these repair proteins at the promoters of activated genes is sensitive to transcription inhibitors and necessary for optimal DNA demethylation and histone post-translational modifications (H3K4/H3K9 methylation and H3K9/14 acetylation) and thus for efficient transcription. Deficiencies in some NER factors impede the recruitment of others and affect NR transactivation. Recently, XPG and XPF endonucleases were found to be involved in chromatin looping between the promoter and the terminator of the activated RARβ2 human gene (Le May et al. 2012). It should be noted that NRs do not exist in S. cerevisiae, which moreover has no DNA methylation system. It remains to be determined whether XPG and other NER factors are generally important for transcription activation in mammalian cells or whether their action is limited to specific pathways such as NR-mediated transcription.
We showed that in addition to Pol II-transcribed genes, Rad2 is also located on Pol III-transcribed genes and telomeric regions in the absence of exogenous genotoxic stress, indicating a complex Rad2 distribution on the yeast genome and opening interesting perspectives of possible novel Rad2 functions. Recently, a role of Mediator on telomeres was proposed, since this complex influences telomeric silencing, cellular life span, and telomere heterochromatin maintenance (Zhu et al. 2011; Peng and Zhou 2012). In yeast, Mediator is bound to the telomeric regions. Our results revealed that Rad2 is also enriched in these regions. The colocalization of Mediator and Rad2 on telomeres raises the possibility that Mediator function on the telomeres could be related to Rad2.
In conclusion, our results suggest an intriguing model that Mediator might play more roles in nuclear processes than previously assumed by serving as an assembly platform or a regulatory element linking transcription with DNA repair and possibly other chromatin-related processes.
Materials and methods
Strains and plasmids
All S. cerevisiae strains are described in Supplemental Table S1. All plasmids are listed in Supplemental Table S2. The oligonucleotides used in this study can be found in Supplemental Table S3.
ChIP, ChIP–chip, and ChIP-seq
ChIP and ChIP–chip experiments were performed as described (Ghavi-Helm et al. 2008). In ChIP experiments, we considered a significant enrichment compared with negative controls or a significant difference between ChIP occupancies when P-value was <0.05 in a Student's t-test. Chromatin preparation for ChIP-seq experiments was performed as described for conventional ChIP, except that an additional sonication step with Bioruptor (Diagenode; six cycles of 30 sec with medium intensity setting) was included to generate DNA fragments of ∼200-bp mean size. DNA sequencing of 40-nucleotide (nt) tags was performed on Solexa genome analyzer GA-IIx using the procedures recommended by the manufacturer (Illumina). Input DNA and DNA from ChIP with an untagged strain were used as negative controls. The ChIP–chip and ChIP-seq data have been deposited to the Array Express under accession numbers E-MEXP-3875 and E-MTAB-1595.
Co-IP experiments
Whole-yeast extract preparation and immunoprecipitation in immunoprecipitation buffer (50 mM HEPES at pH 7.5, 100 mM NaCl, 20% glycerol, 1 mM dithiothreitol, 0.5 mM EDTA, 0.05% NP-40) supplemented with a protease inhibitor cocktail (Complete, Roche) and 1 mM PMSF and Western blotting were performed as described previously (Soutourina et al. 2006). The 12CA5 anti-HA and 9E10 anti-Myc antibodies were used against HA- or Myc-tagged proteins, and the 8WG16 anti-Rpb1 antibody was used against Pol II.
qRT–PCR analysis
RNA was extracted with hot acidic phenol following a protocol derived from Schmitt et al. (1990). Reverse transcription of 0.5-μg RNA samples was performed using iScript cDNA synthesis kit (Bio-Rad) with a mix of oligo(dT) and random hexamers for priming. qPCR results were normalized using 25S rRNA, 18S rRNA, and SCR1 RNA as internal controls. Values represent the average of three independent experiments, and error bars indicate standard deviations.
Data analysis
ChIP-seq data were analyzed using the following bioinformatics tools. The sequences were aligned on S. cerevisiae genome (University of California at Santa Cruz [UCSC] version sacCer3) with Bowtie version 0.12.7 (Langmead et al. 2009). Conversions to different file formats were performed using Samtools version 0.1.16. Only uniquely mapped tags were used, and a maximum of two mismatches was allowed. To avoid possible sequencing artifacts, reads beyond five repetitions at the same position of the genome were removed, and reads with the best-quality scores were conserved for each position. The number of mappable tags for each ChIP-seq experiment is indicated in Supplemental Table S4. To calculate ChIP-seq density, reads were extended to an assumed fragment length of 180 nt, and a count of reads was determined per 1-bp bin using Bedtools version 2.15.0. ChIP-seq density profiles were displayed using UCSC or IGB yeast genome browsers.
We used input DNA and DNA from ChIP with untagged strain as negative controls. The ChIP sample from the untagged strain generally shows very low tag density on the genome, except some regions representing <1% of the genome that display an apparent enrichment in this negative control. It should be noted that the majority of these regions were located inside the highly transcribed class II genes. To correct for this nonrandom background distribution, we subtracted the normalized signal of untagged strain sample from the ChIP samples for each protein. The ChIP signals of untagged strain have different impacts on the ChIP signals for different proteins. For this reason, the subtraction step was preceded by a normalization of the ChIP signal of an untagged strain compared with the corresponding ChIP sample based on the qPCR analyses of nontranscribed control regions.
The significantly enriched regions were identified using peak calling MACS2 software version 2.0.10.20120703 (Zhang et al. 2008) with a minimum false discovery rate (q-value) cutoff of 0.01 for peak detection followed by PeakSplitter step of PeakAnalyzer software for subdivision of ChIP-seq regions into discrete peaks (Salmon-Divon et al. 2010). Significantly enriched Rad2 peaks were annotated on the yeast genome using ChIPpeakAnno R script from the Bioconductor project (Zhu et al. 2010). To identify the Rad2 enrichment signals inside protein-coding genes, Rad2 mean densities were computed for each ORF and compared with mean densities of two negative controls: the untagged strain sample, normalized as described above, and the input DNA sample, normalized using NCIS R script version 1.0.1 (Liang and Keles 2012). We required that enriched ORFs have the mean density in Rad2 sample higher than that in the untagged strain control plus two standard deviations and, at the same time, in the input DNA control plus two standard deviations.
Acknowledgments
We thank N. van Berkum for two-hybrid screening with the Med17 Mediator subunit, S. Wydau for yeast strains, the SPI (CEA/Saclay) for monoclonal antibodies, A. Polesskaya for advice in human cell culture, and C. Mann and S. Un for critical reading of the manuscript. This work has benefited from the facilities and expertise of the high-throughput sequencing platform of IMAGIF (Centre de Recherche de Gif). This work was supported by the Agence Nationale de la Recherche (grant no. ANR_11_BSV8_020_01). F.E. was supported by a grant from the Fondation ARC (grant no. DOC20130606697).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.225813.113.
References
- Andrau JC, van de Pasch L, Lijnzaad P, Bijma T, Koerkamp MG, van de Peppel J, Werner M, Holstege FC 2006. Genome-wide location of the coactivator mediator: Binding without activation and transient Cdk8 interaction on DNA. Mol Cell 22: 179–192 [DOI] [PubMed] [Google Scholar]
- Cai G, Imasaki T, Yamada K, Cardelli F, Takagi Y, Asturias FJ 2010. Mediator head module structure and functional interactions. Nat Struct Mol Biol 17: 273–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson SG 2003. The XPG story. Biochimie 85: 1113–1121 [DOI] [PubMed] [Google Scholar]
- Compe E, Egly JM 2012. TFIIH: When transcription met DNA repair. Nat Rev Mol Cell Biol 13: 343–354 [DOI] [PubMed] [Google Scholar]
- Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M 2008. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell 31: 337–346 [DOI] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, et al. 2008. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature 455: 547–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gade P, Singh AK, Roy SK, Reddy SP, Kalvakolanu DV 2009. Down-regulation of the transcriptional mediator subunit Med1 contributes to the loss of expression of metastasis-associated dapk1 in human cancers and cancer cells. Int J Cancer 125: 1566–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard H, Tous C, Botet J, Gonzalez-Aguilera C, Quintero MJ, Viladevall L, Garcia-Rubio ML, Rodriguez-Gil A, Marin A, Arino J, et al. 2009. Genome-wide analysis of factors affecting transcription elongation and DNA repair: A new role for PAF and Ccr4–not in transcription-coupled repair. PLoS Genet 5: e1000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J 2008. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev 22: 1934–1947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi B, van Berkum NL, Klapholz B, Bijma T, Boube M, Boschiero C, Bourbon HM, Holstege FC, Werner M 2004. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res 32: 5379–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt PC, Spivak G 2008. Transcription-coupled DNA repair: Two decades of progress and surprises. Nat Rev Mol Cell Biol 9: 958–970 [DOI] [PubMed] [Google Scholar]
- Hashimoto S, Boissel S, Zarhrate M, Rio M, Munnich A, Egly JM, Colleaux L 2011. MED23 mutation links intellectual disability to dysregulation of immediate early gene expression. Science 333: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Ito S, Kuraoka I, Chymkowitch P, Compe E, Takedachi A, Ishigami C, Coin F, Egly JM, Tanaka K 2007. XPG stabilizes TFIIH, allowing transactivation of nuclear receptors: Implications for Cockayne syndrome in XP-G/CS patients. Mol Cell 26: 231–243 [DOI] [PubMed] [Google Scholar]
- Johnson KM, Wang J, Smallwood A, Arayata C, Carey M 2002. TFIID and human mediator coactivator complexes assemble cooperatively on promoter DNA. Genes Dev 16: 1852–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann R, Straussberg R, Mandel H, Fattal-Valevski A, Ben-Zeev B, Naamati A, Shaag A, Zenvirt S, Konen O, Mimouni-Bloch A, et al. 2010. Infantile cerebral and cerebellar atrophy is associated with a mutation in the MED17 subunit of the transcription preinitiation mediator complex. Am J Hum Genet 87: 667–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30: 235–239 [DOI] [PubMed] [Google Scholar]
- Kuuselo R, Savinainen K, Sandstrom S, Autio R, Kallioniemi A 2011. MED29, a component of the mediator complex, possesses both oncogenic and tumor suppressive characteristics in pancreatic cancer. Int J Cancer 129: 2553–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagerwerf S, Vrouwe MG, Overmeer RM, Fousteri MI, Mullenders LH 2011. DNA damage response and transcription. DNA Repair (Amst) 10: 743–750 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lariviere L, Geiger S, Hoeppner S, Rother S, Strasser K, Cramer P 2006. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat Struct Mol Biol 13: 895–901 [DOI] [PubMed] [Google Scholar]
- Lariviere L, Plaschka C, Seizl M, Wenzeck L, Kurth F, Cramer P 2012. Structure of the Mediator head module. Nature 492: 448–451 [DOI] [PubMed] [Google Scholar]
- Lee SK, Yu SL, Prakash L, Prakash S 2002. Requirement of yeast RAD2, a homolog of human XPG gene, for efficient RNA polymerase II transcription: Implications for Cockayne syndrome. Cell 109: 823–834 [DOI] [PubMed] [Google Scholar]
- Le May N, Mota-Fernandes D, Velez-Cruz R, Iltis I, Biard D, Egly JM 2010. NER factors are recruited to active promoters and facilitate chromatin modification for transcription in the absence of exogenous genotoxic attack. Mol Cell 38: 54–66 [DOI] [PubMed] [Google Scholar]
- Le May N, Fradin D, Iltis I, Bougneres P, Egly JM 2012. XPG and XPF endonucleases trigger chromatin looping and DNA demethylation for accurate expression of activated genes. Mol Cell 47: 622–632 [DOI] [PubMed] [Google Scholar]
- Li LH, He J, Hua D, Guo ZJ, Gao Q 2010. Lentivirus-mediated inhibition of Med19 suppresses growth of breast cancer cells in vitro. Cancer Chemother Pharmacol 68: 207–215 [DOI] [PubMed] [Google Scholar]
- Liang K, Keles S 2012. Normalization of ChIP-seq data with control. BMC Bioinformatics 13: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet M, Scafe C, Sexton J, Young R 1987. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol Cell Biol 7: 1602–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Zhou JQ 2012. The tail-module of yeast Mediator complex is required for telomere heterochromatin maintenance. Nucleic Acids Res 40: 581–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries D, Meisterernst M 2011. Control of gene transcription by Mediator in chromatin. Semin Cell Dev Biol 22: 735–740 [DOI] [PubMed] [Google Scholar]
- Risheg H, Graham JM Jr, Clark RD, Rogers RC, Opitz JM, Moeschler JB, Peiffer AP, May M, Joseph SM, Jones JR, et al. 2007. A recurrent mutation in MED12 leading to R961W causes Opitz-Kaveggia syndrome. Nat Genet 39: 451–453 [DOI] [PubMed] [Google Scholar]
- Robinson PJ, Bushnell DA, Trnka MJ, Burlingame AL, Kornberg RD 2012. Structure of the Mediator Head module bound to the carboxy-terminal domain of RNA polymerase II. Proc Natl Acad Sci 109: 17931–17935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon-Divon M, Dvinge H, Tammoja K, Bertone P 2010. PeakAnalyzer: Genome-wide annotation of chromatin binding and modification loci. BMC Bioinformatics 11: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker AH, Tsutakawa SE, Kostek S, Ng C, Shin DS, Peris M, Campeau E, Tainer JA, Nogales E, Cooper PK 2005. Recognition of RNA polymerase II and transcription bubbles by XPG, CSB, and TFIIH: Insights for transcription-coupled repair and Cockayne Syndrome. Mol Cell 20: 187–198 [DOI] [PubMed] [Google Scholar]
- Scharer OD 2008. XPG: Its products and biological roles. Adv Exp Med Biol 637: 83–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res 18: 3091–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CE, Tarpey PS, Lubs HA, Verloes A, May MM, Risheg H, Friez MJ, Futreal PA, Edkins S, Teague J, et al. 2007. The original Lujan syndrome family has a novel missense mutation (p.N1007S) in the MED12 gene. J Med Genet 44: 472–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J, Bordas-Le Floch V, Gendrel G, Flores A, Ducrot C, Dumay-Odelot H, Soularue P, Navarro F, Cairns BR, Lefebvre O, et al. 2006. Rsc4 connects the chromatin remodeler RSC to RNA polymerases. Mol Cell Biol 26: 4920–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina J, Wydau S, Ambroise Y, Boschiero C, Werner M 2011. Direct interaction of RNA polymerase II and mediator required for transcription in vivo. Science 331: 1451–1454 [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ 2002. Mechanisms of transcription-coupled DNA repair. Nat Rev Mol Cell Biol 3: 21–29 [DOI] [PubMed] [Google Scholar]
- Svejstrup JQ 2007. Contending with transcriptional arrest during RNAPII transcript elongation. Trends Biochem Sci 32: 165–171 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. 2011. Human mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayvargia R, May MS, Fondell JD 2007. A coregulatory role for the mediator complex in prostate cancer cell proliferation and gene expression. Cancer Res 67: 4034–4041 [DOI] [PubMed] [Google Scholar]
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ 2005. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell 17: 683–694 [DOI] [PubMed] [Google Scholar]
- Werner M, Chaussivert N, Willis IM, Sentenac A 1993. Interaction between a complex of RNA polymerase III subunits and the 70-kDa component of transcription factor IIIB. J Biol Chem 268: 20721–20724 [PubMed] [Google Scholar]
- Zhang X, Krutchinsky A, Fukuda A, Chen W, Yamamura S, Chait BT, Roeder RG 2005. MED1/TRAP220 exists predominantly in a TRAP/Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol Cell 19: 89–100 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Wiren M, Sinha I, Rasmussen NN, Linder T, Holmberg S, Ekwall K, Gustafsson CM 2006. Genome-wide occupancy profile of mediator and the Srb8-11 module reveals interactions with coding regions. Mol Cell 22: 169–178 [DOI] [PubMed] [Google Scholar]
- Zhu LJ, Gazin C, Lawson ND, Pages H, Lin SM, Lapointe DS, Green MR 2010. ChIPpeakAnno: A Bioconductor package to annotate ChIP-seq and ChIP–chip data. BMC Bioinformatics 11: 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Liu B, Carlsten JO, Beve J, Nystrom T, Myers LC, Gustafsson CM 2011. Mediator influences telomeric silencing and cellular life span. Mol Cell Biol 31: 2413–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]