Abstract
Cellular function can be controlled through the gene expression program, but often protein post-translational modifications (PTMs) provide a more precise and elegant mechanism. Key functional roles of specific modification events—for instance, during the cell cycle—have been known for decades, but only in the past 10 years has mass-spectrometry-(MS)-based proteomics begun to reveal the true extent of the PTM universe. In this overview for the special PTM issue of Molecular and Cellular Proteomics, we take stock of where MS-based proteomics stands in the large-scale analysis of protein modifications. For many PTMs, including phosphorylation, ubiquitination, glycosylation, and acetylation, tens of thousands of sites can now be confidently identified and localized in the sequence of the protein. The quantification of PTM levels between different cellular states is likewise established, with label-free methods showing particular promise. It is also becoming possible to determine the absolute occupancy or stoichiometry of PTM sites on a large scale. Powerful software for the bioinformatic analysis of thousands of PTM sites has been developed. However, a complete inventory of sites has not been established for any PTM, and this situation will persist into the foreseeable future. Furthermore, although PTM coverage by MS-based methods is impressive, it still needs to be improved, especially in tissues and in clinically relevant systems. The central challenge for the field is to develop streamlined methods for determining biological functions for the myriad of modifications now known to exist.
The human genome project has revealed a surprisingly small number of protein-coding genes: around 20,000—not many more than those of the lowly worm (1). In contrast, it has turned out that there is a bewildering number and diversity of RNA species, and biological functions for many of them are still not known. Proteins constitute the next level of the gene expression program, and here diversity comes in two forms: variants affecting the primary sequence of amino acids, and post-translational modifications (PTMs).1 Today primary sequence variation is mainly studied indirectly by means of deep RNA sequencing, although top-down and shotgun proteomics could provide more direct and quantitative results in the future (2). In contrast, the unbiased discovery and analysis of large numbers of PTM sites is the exclusive domain of MS-based proteomics, and great strides are being made in identifying the myriad possible protein modifications. The study of PTMs via MS now constitutes a large and vibrant field of research, as exemplified in the original and review papers in this special issue of Molecular and Cellular Proteomics. Here we provide a snapshot of where the field stands, what progress has already been made, and where we see the key challenges for the future.
PTM Analysis via MS
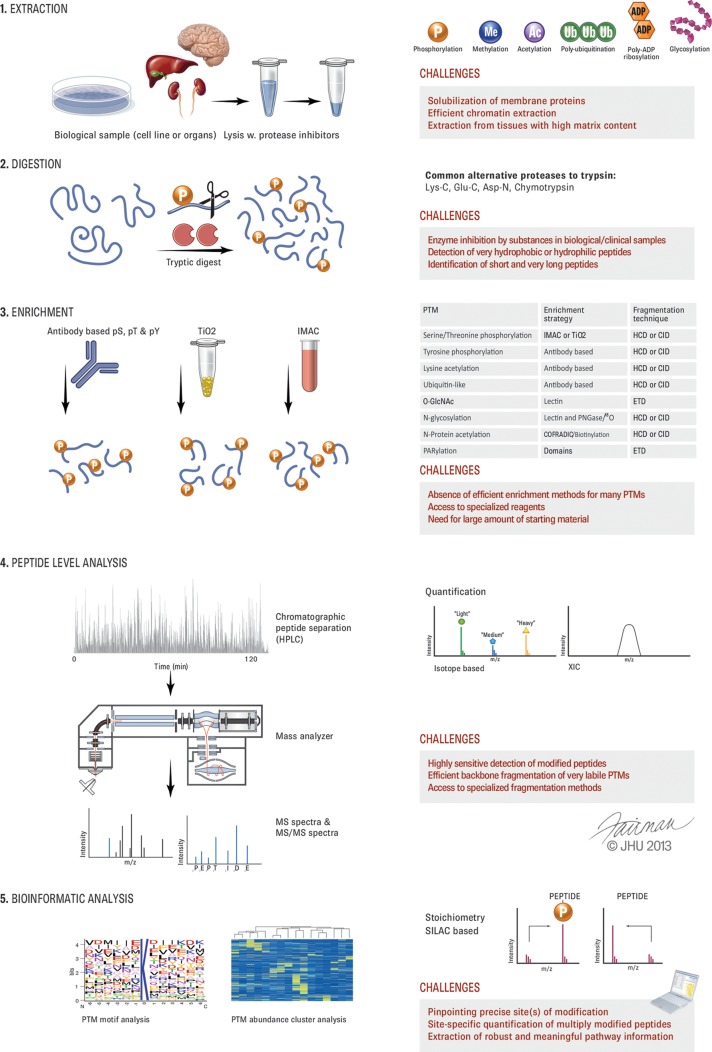
Mass spectrometry has long been used to map modifications on purified proteins; in fact, this was one of the first applications of MS in protein research (3). In contrast, proteomic investigations of PTMs—meaning their analysis on a large number of proteins, such as those in an organelle or in an entire cell—have taken off only since the beginning of the past decade (4–6). Although the obstacles to complete proteome analysis are finally being overcome (7), this is not the case for PTMs, because their analysis is more difficult both technologically and conceptually. There are several principal differences between proteome and PTM analysis. In proteome measurements, several peptides are generally available to characterize each protein, which makes the measurement much more robust, whereas in PTM measurements each peptide representing a modification site of interest needs to stand on its own. Modified peptides can be of lower abundance and more difficult to identify from their fragmentation spectra than nonmodified peptides. Furthermore, these tandem mass spectra should contain sufficient information to localize the modification with single amino acid resolution. Below we address the challenges of the PTM workflow in turn (Fig. 1).
Fig. 1.
Overview of PTM analysis and challenges. The principal steps in the workflow for PTM analysis are depicted on the left. On the right, the main types of modifications, alternative proteases, PTM enrichment methods, and quantification types are symbolized. For each step, some of the main challenges are listed.
Extraction, Solubilization, and Digestion
The sample preparation methods used for PTM analysis are generally the same as for standard shotgun proteomics. Exceptions are the PTM enrichment step and the strict requirement to inactivate enzymes responsible for adding or removing the PTM of interest. The extraction and solubilization of proteins from cell culture lysates is relatively straightforward, and a number of robust protocols exist. In contrast, more effort should be directed toward the efficient retrieval of proteins from tissues with high contents of extracellular matrix. As an example, cardiac muscle tissue is very rigid, and special homogenization technologies are needed to efficiently extract proteins for enrichment of their phosphopeptides (8). Chromatin-bound and transmembrane proteins are easily analyzed in proteome studies, but they are still challenging in PTM analysis, as the enrichment strategies might not be compatible with the specific buffers applied (9) and peptides from all parts of the proteins need to be generated.
As in expression proteomics, trypsin is the protease of choice for PTM analysis because of its high cleavage specificity (10) and the MS/MS-friendly properties of the resulting peptides. In some instances tryptic peptides might be too small or too large for LC-MS/MS analysis, a fact that needs to be taken into account when certain critical PTM sites need to be quantified in planned experiments. Likewise, some PTMs modify the solubility properties of the peptides significantly. For example, glycosylated or phosphorylated tryptic peptides can be too hydrophilic to be captured by conventional reversed-phase C18 material. Using proteases with different cleavage specificities such as chymotrypsin, Lys-N, or endoproteinase Glu-C (11, 12) in addition to trypsin can help one overcome some of these issues and should be more broadly considered. Developments in these areas are often underappreciated, but they are critical to the overall success of PTM-based proteomics.
Enrichment and Sensitivity in PTM Analysis
Regulatory PTMs, by definition, have substoichiometric occupancy of their target proteins, making it a major challenge to achieve sensitivity in MS analysis (Fig. 1). Although proteomics workflows have become much more sensitive overall, they still require a specific step in which the PTM of interest is enriched with respect to the unmodified proteins or peptides. There are now protocols with very high specificity for phosphorylated peptides, often leading to a phosphopeptide proportion of more than 90%. Enrichment efficiency can be around a factor of 100, making it possible to start with much more protein material than in proteome analysis and thereby boosting sensitivity (13). However, the fact that these starting amounts are much greater than what is needed for proteome analysis needs to be considered in PTM-based projects. Metal affinity chromatography—for instance, using titanium dioxide (14)—and/or anti-phosphotyrosine antibodies (15) are frequently employed. It is also possible to enrich with antibodies raised against specific phosphorylation motifs, but generally it is more attractive to analyze the entire phosphoproteome instead. With a few exceptions such as N-linked glycopeptide enrichment via lectins (16), the specificity and enrichment for other PTMs are much lower.
The difference that a good enrichment strategy can make is exemplified by the recent introduction of diglycine-specific antibodies to detect the remnant modification after tryptic digestion of ubiquitinated proteins (17, 18). Before the development of these antibodies, target proteins of interest had to be tagged or the ubiquitin itself had to be used as a purification handle, whereas now tens of thousands of ubiquitination sites can be specifically enriched and analyzed in a generic workflow (19, 20). For most PTMs, however, efficient enrichment strategies do not exist at all, making them very difficult to study other than on a protein-by-protein basis. The development of specific and highly enriching protocols for more classes of PTMs therefore remains an urgent task for the community. We recommend that these tasks be recognized as important scientific endeavors in their own right that should be supported by the academic funding system.
Efficiency of Fragmentation, Identification, and Localization
The MS community has worked on peptide fragmentation mechanisms for several decades, and the basic mechanisms are well understood and robustly implemented. One major development over the past few years has been the wide adoption of the “high-high” strategy (21)—high resolution at both MS and MS/MS levels—and this has been especially beneficial for PTMs.
In contrast to the identification of proteins, PTM analysis inherently has to rely on single peptide species, making it much more challenging. The same fragmentation methods, scoring schemes, and stringent false discovery rate controls can generally be used. However, apart from the unambiguous identification of the modified peptides themselves, the modified amino acids need to be localized in the sequence, which requires a separate “localization score.” This is unproblematic for stable modifications, and it is our experience that the mean localization probability is typically greater than 99% in large data sets (22–28).
Guidelines in the proteomics literature state that the annotated spectra of PTMs bearing peptides should be provided (29). We fully support these requirements and note that they are particularly needed in the general biological literature, where novel modifications are still reported without any or with only low-quality MS/MS evidence.
In MS-based PTM analysis, it is important to generate sufficient peptide fragmentation information for peptide identification and site localization. Among the different strategies that have been developed for this purpose, multistage activation in ion trap instruments (30), “beam-type” fragmentation on time-of-flight instruments (14, 31), or the equivalent higher-collisional dissociation (HCD) implementation on Orbitrap instruments (32, 33) provide a high chance that the relevant modification-specific fragment events will be generated (Fig. 1).
Although the techniques described above are almost universal, it can be difficult to localize phospho-serine in multiphosphorylated peptides, and very labile modifications such as O-GlcNAc might require the use of electron transfer dissociation (ETD) (34, 35). Furthermore, the quantification of regulated changes in multiphosphorylated peptides remains challenging. Some PTMs can be very heterogeneous, as is the case for N-linked glycosylation. For these PTMs, the detailed determination of the structure of the PTM can be even more difficult than defining the modification site (36, 37). Nevertheless, even the accurately measured fold changes of the modified peptides themselves are already very useful for planning biological follow-up experiments. Some protein modifications, such as intermediates in redox reactions, occur very transiently in the cell and can currently be studied only when they are trapped by stabilizing chemical reagents (38). We see a great opportunity for PTM proteomics in the large-scale analysis of these processes, which have so far been studied almost exclusively in in vitro reactions.
Completeness of the PTM Catalog: How Much Is Enough?
MS-based proteomics can now determine complete model proteomes, and this will soon be possible in mammalian systems as well (7). In contrast, there are no complete PTM catalogs so far. Not only are the MS-based efforts far from saturation for most PTMs, but it is difficult to even define what a complete PTM proteome would be. For instance, a given serine/threonine kinase may have some phosphorylation propensity toward a very large set of linear sequences, but this does not necessarily imply a biological function of these substrates in vivo. Conversely, biologically relevant substrates might become modified only under a very restricted set of circumstances (39) that were not part of the set of conditions used in the global PTM analysis. It appears that almost all proteins can be post-translationally modified. Extrapolating from current data, at least 70% of proteins are phosphorylated at some point (40, 41), with similar proportions for ubiquitin (42) and lysine acetylation (43). The PTMs of several proteins or protein classes have been studied in very great depth, with histones being a prime example. Nearly all common modifications have been reported to occur on histones, and with the use of new enrichment strategies, fragmentation methods, and computational approaches, more are still being discovered. Histone PTMs also illustrate challenges to complete PTM analysis even on individual proteins, such as the difficulty of generating peptides of suitable length for MS, the detection of histone modifications that are restricted to few sites in the genome, and the interplay of modifications—in this case termed the histone code. Some of the proteins that are intensely studied, such as p53, have likewise been reported to have a plethora of modifications (44). However, much of these data were acquired via low-resolution and statistically uncontrolled methods and should therefore be treated with caution. The same caveats apply to some large-scale PTM studies. Researchers should not be satisfied with the mere presence of a PTM in a database; they should also critically evaluate the associated data. Fortunately, there is increasing awareness of the need for software tools to evaluate data quality within and between studies.
In our opinion, a useful minimum depth in large-scale PTM analysis should cover the key functional sites in the process under investigation. A phosphoproteome of 10,000 sites will include the majority of the sites classically studied using phospho-specific antibodies. Reaching such a depth would have been a daunting prospect just a few years ago. Today this can be done in single LC-MS/MS runs, making it possible to follow multiple conditions and replicates.
Relative Quantification and Stoichiometry of PTMs
The quantification of PTMs is highly desirable because regulation in a biological context makes it much more likely that a site will in fact be functional. The PTM-bearing peptides are quantified in the same way as unmodified peptides. However, it is more challenging because it is based on single peptides, often of very low abundance. Peptide quantification by means of stable isotope labeling with amino acids in cell culture (SILAC) is the gold standard because it eliminates many workflow-induced sources of error (45, 46). Nevertheless, many chemical labeling strategies have been successfully used as well, and they can be applied to any sample type (47, 48). For PTM analysis, large amounts of starting materials typically need to be labeled. With reporter-ion-based quantification, such as tandem mass tags and iTRAQ (49, 50), care needs to be taken that the observed regulation is due to the PTM-bearing peptide of interest, rather than co-fragmented precursors (51).
Recent improvements in data quality and in algorithms now make label-free quantification very attractive in the PTM field. In addition to being the most straightforward method operationally, label-free analysis allows direct comparison of MS signals between any number of samples, is applicable to any source material, and avoids reagent costs.
In addition to relative quantification of the modification site of interest, it is very useful to determine the site's occupancy or stoichiometry (i.e. the fraction of proteins that are modified). A large fractional stoichiometry, especially when combined with dynamic regulation, is a very good indication that the site may be functional. The converse does not necessarily hold true, because a modification may be located on a small pool of proteins that are temporally or spatially distinct. The cell cycle and DNA damage response are examples of this. In phosphoproteome analysis, stoichiometry has been determined for thousands of sites from the data themselves (40) or after deliberate dephosphorylation of half of the sample (52, 53). In the future, this will be possible not only with stable isotope labeling with amino acids in cell culture, but also with label-free methods.
The value of stoichiometry information has been illustrated in the case of acetylation. Although this modification can readily be found on many abundant proteins, it appears to frequently occur at very low stoichiometries and to be of nonenzymatic origin (54). In contrast, certain classes of PTMs, such as N-linked glycosites, are mainly generated co-translationally and generally occur at very high stoichiometries.
Less Studied or Unknown Modifications
Almost all of the efforts in large-scale PTM mapping have been directed at the most prominent modifications, especially phosphorylation, ubiquitylation, glycosylation, and acetylation. However, more than 200 in vivo modifications are known, and there are even more peptide modifications that can be induced chemically—for instance, during sample preparation. Although PTM identification normally requires specifying the modified amino acid in search software, unbiased methods for PTM analysis have also been developed (55, 56). These call for the in silico addition of the mass difference between the modified and the unmodified peptide to each amino acid in turn. Although this works very well even on a proteomic scale, the lack of an enrichment step means that only the most abundant of such unknown modifications are likely to be found.
Novel modifications are still being described regularly, especially on lysines (57–59). In addition to demonstrating biologically relevant regulation of such novel modifications, it is very useful to determine their stoichiometry to provide a functional context.
Modifications that are large relative to the peptide can be difficult to study via conventional LC-MS/MS and might require the development of special methods. Poly(ADP-ribosyl)ation, a prominent cellular response to genotoxic stress, is an example of such a PTM, with the additional challenge that it is very labile in the fragmentation step (61–62). Other understudied PTMs include those on extracellular matrix proteins, which can be large, labile, and insoluble. Analyzing and biologically characterizing these classes of PTMs will occupy researchers for many years to come.
Bioinformatic Analysis
This was until recently seen as perhaps the largest bottleneck in MS-based proteomics (63). Fortunately, this area is now well developed in both expression proteomics and large-scale PTM analysis, and comprehensive and statistically rigorous software tools are readily available. Typical steps include motif analysis (64), Gene Ontology enrichment (65), pathway analysis (66), and analysis of protein–protein interactions (67). In the MaxQuant environment, the Perseus software allows extensive statistical and functional analysis of proteome and PTM data (68). The NetWorKIN software (69, 70) combines protein–protein information from STRING (67) with information on linear kinase motifs (62) to provide likely kinase–substrate relationships in large-scale data.
Bioinformatic analysis also typically includes comparison to one of the PTM databases. These are still in a state of flux with regard to the data that they accept and retain. Curators of the widely used UniProt database, for instance, have largely stopped the indiscriminate incorporation of all data from large-scale PTM projects. This is in contrast to the PhosphoSite database (71), which even includes unpublished data. An advantage of the latter approach is the higher degree of confidence in sites identified in multiple groups; however, it needs to be kept in mind that common misidentifications will also accumulate. Clearly, the community needs to establish more refined ways of integrating and presenting the PTM data that it generates. We envision that the usefulness of the PTM information in specialized and general databases will continue to improve as a result of the exponential increase in the data generated, as well as better data quality and reporting standards.
Throughput in PTM Analysis
So far global PTM analysis has been very challenging and resource demanding, with some of the large-scale studies taking months to complete. The proteomics community needs to develop ways to make PTM analysis much faster, just like the genomics community has done with next-generation sequencing technologies. Fortunately, current trends are very encouraging. Deep phosphoanalysis can now be done in a single day of measurement time. Single-shot approaches (72–74) already allow the analysis of more than 10,000 sites in a few hours (75), making it realistic to perform an entire project in a few days. Added advantages of single-shot approaches are that they tend to use less input material and are more robust because of the absence of fractionation steps.
Targeted approaches aim to identify and quantify a small number of peptides of interest in a short analysis time (76, 77). In the context of PTMs, the targeted approach allows the measurement of key PTM sites in many conditions (78–80). However, this comes at the expense of the unbiased nature of shotgun proteomics, as new and unexpected sites cannot be discovered. Targeted analysis is conceptually straightforward and has therefore proven very attractive to nonspecialists. However, extensive method optimization and process control are required in order to achieve and maintain acceptable levels of specificity. This is especially true if more than a few sites are targeted and when low-resolution instrumentation is used.
The high throughput promised by the methods described above is a precondition in order for PTM analysis to make an impact in the clinic. This is an especially intriguing area of PTM-based proteomics, because many diseases deregulate signaling pathways, and because this may be more directly evidenced in the dynamic modifications of pathway members than in the expression levels of transcripts or proteins. For instance, cancer genome projects have revealed large numbers of mutations in the same pathway, each occurring in small numbers of cells or patients (81). PTM analysis on clinical samples would determine, in a straightforward way, whether a pathway is critically modulated in a given tumor and how this activity is affected by treatment. Currently the community is still at the stage of proof of principle investigations of these concepts. Much development of the PTM workflow and instrumentation will be required in order to reach the level of robustness and validation required for clinical application.
Functional Assignment of PTMs
The signaling community has grown up with the concept of very few but biologically highly important PTMs—for example, key sites controlling the cell cycle or a growth factor pathway. In contrast, MS-based proteomics provides tens of thousands of sites, raising the question of their biological relevance. Clearly, if properly controlled, these large-scale data sets can directly point to key regulatory sites (82, 83, 85). In these instances, proteomics functions as an initial screen in a cell-signaling project. More often, however, researchers are faced with the challenge of how to select a very small number of sites from large-scale data and how to perform functional follow-up on these (86). Obvious prioritization criteria include high identification and quantification accuracy, regulation of the site in the process of interest, and a reasonable stoichiometry. Often, MS-based proteomics itself can be used in follow-up, through measurement of additional conditions or with additional proteomics data such as interactions of the PTM in question. However, classical in vitro enzyme assays might not correspond to the situation in vivo, a fact that should be kept in mind when validating sites via these methods. Likewise, point mutations might not be sufficiently informative, especially in the case of lysine, which can be the target of several PTM types. Finally, classical approaches typically do not take account of the cooperative nature of PTM sites, which is becoming more and more appreciated. We believe that it is unrealistic to hope that established signaling assays can validate more than a small fraction of the PTM sites now being identified. Instead we envisage proteomics approaches themselves providing the answer to the current dilemma. The throughput and accuracy of MS-based proteomics now lead to the possibility of quantifying a PTM on a global scale while systematically perturbing it. As an early example, each of the kinases and phosphatases in the yeast proteome has been deleted, followed by measurement of the phosphoproteome (87).
Enzyme–Substrate Relationships
Although advanced workflows can now routinely be used to analyze tens of thousands of phosphorylation sites in single experiments, it remains difficult to assign the specific kinase or kinases that directly catalyze the modification of a phosphorylation site of interest. This is an important question that can partially be addressed bioinformatically via linear kinase motif analyses (69, 88, 89) or experimentally via pharmacological inhibition or knockdown of kinases involved in the cellular response under investigation. In the latter case, global quantitative phosphoproteomics can efficiently identify phosphorylation sites that are dependent on or downstream of specific kinases (24, 90, 91). Chemical proteomics strategies using analog-sensitive kinases (in which single site-directed mutagenesis of the so-called gatekeeper residue in the kinase domain renders the mutated kinase specific to a bulky ATP analog) can more directly determine kinase–substrate relationships (92–94). With the recent breakthroughs in genome editing technologies (95), we expect that analog-sensitive kinase approaches will in the future be applied widely to all susceptible kinases and analyzed via quantitative phosphoproteomics. Streamlined genome editing also holds great promise for the functional study of PTM networks and even individual PTM sites that are key determinants in cell fate decisions.
PTM Analysis in the Future
As discussed above, there are now robust MS-based proteomic workflows for the rigorous identification and quantification of very large numbers of PTMs. Enrichment protocols exist for the PTMs of greatest biological interest. Novel capabilities not generally available in classical approaches have been developed, with the quantification of site occupancy providing a prime example. Bioinformatic analysis of PTM data has become rigorous and streamlined, and it now constitutes an important foundation of systems biology.
The major challenges of the field lie in functional interpretation and follow-up. However, with increasing availability of the technology to signaling biologists, large-scale PTM quantification will often simply serve as an initial screen, followed by detailed study of key sites. How all these PTM data should be integrated in PTM databases and made available to the community is still an unsolved problem.
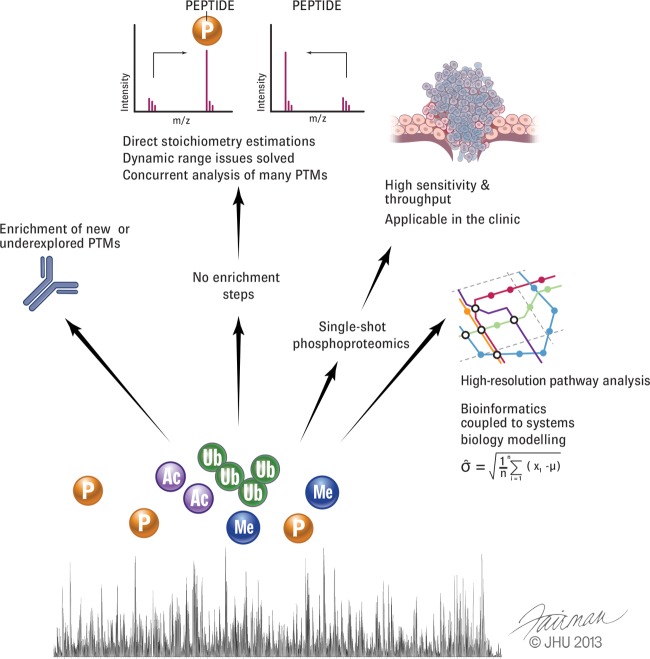
Many technological challenges remain (Fig. 2). PTM screens still need to be made faster, more sensitive, and more reproducible, and they need to cover an even greater dynamic range. Further development of the single-shot approach would combine the advantages of targeted approaches with those of shotgun approaches. With such advances, large-scale PTM analysis will be ready to make a direct impact in the clinic. The potential impact of such a development cannot be overemphasized, given the need for more precise therapies targeted at the molecular nature of the individual disease.
Fig. 2.
A vision for PTM analysis. Proteomics of PTMs would greatly benefit from a more complete repertoire of enrichment tools and reagents. With further development of mass spectrometric capabilities, however, it would be desirable to detect modified peptides without specific enrichment. This would make it possible to study many PTMs simultaneously and estimate their stoichiometry directly. Robust PTM analysis with high throughput could deliver patient classification and unique information on treatment efficacy. Finally, very deep and quantitatively accurate PTM analysis will provide a crucial basis for systems biology.
In the longer term, it is conceivable that dramatically improved mass spectrometric performance could allow the analysis of PTM-bearing peptides without prior enrichment (Fig. 2). This would have many benefits, including the ability to work with much less biological material and the comprehensive analysis of different PTMs at once, including the stoichiometry of each.
Acknowledgments
We thank all the members of the involved labs for helpful discussion and input. We are thankful to Dorte Bekker-Jensen for help with the figures.
Footnotes
Author contributions: J.V.O. and M.M. analyzed data; J.V.O. and M.M. wrote the paper.
* The NNF Center for Protein Research is supported by a generous donation from the Novo Nordisk Foundation. This work was supported by the 7th Framework Program of the European Union (262067-PRIME-XS) and the research career program FSS Sapere Aude (J.V.O.).
1 The abbreviations used are:
- ETD
- electron transfer dissociation; stable isotope labeling with amino acids in cell culture (SILAC)
- PTM
- post-translational modification.
REFERENCES
- 1. Clamp M., Fry B., Kamal M., Xie X., Cuff J., Lin M. F., Kellis M., Lindblad-Toh K., Lander E. S. (2007) Distinguishing protein-coding and noncoding genes in the human genome. Proc. Natl. Acad. Sci. U.S.A. 104, 19428–19433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tran J. C., Zamdborg L., Ahlf D. R., Lee J. E., Catherman A. D., Durbin K. R., Tipton J. D., Vellaichamy A., Kellie J. F., Li M., Wu C., Sweet S. M., Early B. P., Siuti N., LeDuc R. D., Compton P. D., Thomas P. M., Kelleher N. L. (2011) Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 480, 254–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stenflo J., Fernlund P., Egan W., Roepstorff P. (1974) Vitamin K dependent modifications of glutamic acid residues in prothrombin. Proc. Natl. Acad. Sci. U.S.A. 71, 2730–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ficarro S. B., McCleland M. L., Stukenberg P. T., Burke D. J., Ross M. M., Shabanowitz J., Hunt D. F., White F. M. (2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305 [DOI] [PubMed] [Google Scholar]
- 5. Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villen J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 7. Mann M., Kulak N. A., Nagaraj N., Cox J. (2013) The coming age of complete, accurate, and ubiquitous proteomes. Mol. Cell. 49, 583–590 [DOI] [PubMed] [Google Scholar]
- 8. Lundby A., Andersen M. N., Steffensen A. B., Horn H., Kelstrup C. D., Francavilla C., Jensen L. J., Schmitt N., Thomsen M. B., Olsen J. V. (2013) In vivo phosphoproteomics analysis reveals the cardiac targets of beta-adrenergic receptor signaling. Sci. Signal. 6, rs11. [DOI] [PubMed] [Google Scholar]
- 9. Jensen S. S., Larsen M. R. (2007) Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun. Mass Spectrom. 21, 3635–3645 [DOI] [PubMed] [Google Scholar]
- 10. Olsen J. V., Ong S. E., Mann M. (2004) Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol. Cell. Proteomics 3, 608–614 [DOI] [PubMed] [Google Scholar]
- 11. Taouatas N., Drugan M. M., Heck A. J., Mohammed S. (2008) Straightforward ladder sequencing of peptides using a Lys-N metalloendopeptidase. Nat. Methods 5, 405–407 [DOI] [PubMed] [Google Scholar]
- 12. Gilmore J. M., Kettenbach A. N., Gerber S. A. (2012) Increasing phosphoproteomic coverage through sequential digestion by complementary proteases. Anal. Bioanal. Chem. 402, 711–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou H., Low T. Y., Hennrich M. L., van der Toorn H., Schwend T., Zou H., Mohammed S., Heck A. J. (2011) Enhancing the identification of phosphopeptides from putative basophilic kinase substrates using Ti (IV) based IMAC enrichment. Mol. Cell. Proteomics 10, M110.006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-nanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 15. Rush J., Moritz A., Lee K. A., Guo A., Goss V. L., Spek E. J., Zhang H., Zha X. M., Polakiewicz R. D., Comb M. J. (2005) Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 23, 94–101 [DOI] [PubMed] [Google Scholar]
- 16. Zielinska D. F., Gnad F., Wisniewski J. R., Mann M. (2010) Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 [DOI] [PubMed] [Google Scholar]
- 17. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Udeshi N. D., Svinkina T., Mertins P., Kuhn E., Mani D. R., Qiao J. W., Carr S. A. (2013) Refined preparation and use of anti-diglycine remnant (K-epsilon-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12, 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wagner S. A., Beli P., Weinert B. T., Scholz C., Kelstrup C. D., Young C., Nielsen M. L., Olsen J. V., Brakebusch C., Choudhary C. (2012) Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteomics 11, 1578–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mann M., Kelleher N. L. (2008) Precision proteomics: the case for high resolution and high mass accuracy. Proc. Natl. Acad. Sci. U.S.A. 105, 18132–18138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 23. Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., Mann M. (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 24. Pan C., Olsen J. V., Daub H., Mann M. (2009) Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol. Cell. Proteomics 8, 2796–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Daub H., Olsen J. V., Bairlein M., Gnad F., Oppermann F. S., Korner R., Greff Z., Keri G., Stemmann O., Mann M. (2008) Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell. 31, 438–448 [DOI] [PubMed] [Google Scholar]
- 26. Gnad F., de Godoy L. M., Cox J., Neuhauser N., Ren S., Olsen J. V., Mann M. (2009) High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics 9, 4642–4652 [DOI] [PubMed] [Google Scholar]
- 27. Zanivan S., Meves A., Behrendt K., Schoof E. M., Neilson L. J., Cox J., Tang H. R., Kalna G., van Ree J. H., van Deursen J. M., Trempus C. S., Machesky L. M., Linding R., Wickstrom S. A., Fassler R., Mann M. (2013) In vivo SILAC-based proteomics reveals phosphoproteome changes during mouse skin carcinogenesis. Cell Rep. 3, 552–566 [DOI] [PubMed] [Google Scholar]
- 28. Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3. [DOI] [PubMed] [Google Scholar]
- 29. Bradshaw R. A., Burlingame A. L., Carr S., Aebersold R. (2006) Reporting protein identification data: the next generation of guidelines. Mol. Cell. Proteomics 5, 787–788 [DOI] [PubMed] [Google Scholar]
- 30. Schroeder M. J., Shabanowitz J., Schwartz J. C., Hunt D. F., Coon J. J. (2004) A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76, 3590–3598 [DOI] [PubMed] [Google Scholar]
- 31. Nuhse T. S., Stensballe A., Jensen O. N., Peck S. C. (2003) Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 2, 1234–1243 [DOI] [PubMed] [Google Scholar]
- 32. Olsen J. V., Macek B., Lange O., Makarov A., Horning S., Mann M. (2007) Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4, 709–712 [DOI] [PubMed] [Google Scholar]
- 33. Nagaraj N., D'Souza R. C., Cox J., Olsen J. V., Mann M. (2010) Feasibility of large-scale phosphoproteomics with higher energy collisional dissociation fragmentation. J. Proteome Res. 9, 6786–6794 [DOI] [PubMed] [Google Scholar]
- 34. Syka J. E., Coon J. J., Schroeder M. J., Shabanowitz J., Hunt D. F. (2004) Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wells L., Vosseller K., Hart G. W. (2001) Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science 291, 2376–2378 [DOI] [PubMed] [Google Scholar]
- 36. Dell A., Morris H. R. (2001) Glycoprotein structure determination by mass spectrometry. Science 291, 2351–2356 [DOI] [PubMed] [Google Scholar]
- 37. Reinhold V. N., Reinhold B. B., Costello C. E. (1995) Carbohydrate molecular weight profiling, sequence, linkage, and branching data: ES-MS and CID. Anal. Chem. 67, 1772–1784 [DOI] [PubMed] [Google Scholar]
- 38. Meissner F., Molawi K., Zychlinsky A. (2008) Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nat. Immunol. 9, 866–872 [DOI] [PubMed] [Google Scholar]
- 39. Dix M. M., Simon G. M., Cravatt B. F. (2008) Global mapping of the topography and magnitude of proteolytic events in apoptosis. Cell 134, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 41. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villen J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mertins P., Qiao J. W., Patel J., Udeshi N. D., Clauser K. R., Mani D. R., Burgess M. W., Gillette M. A., Jaffe J. D., Carr S. A. (2013) Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat. Methods 10, 634–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lundby A., Lage K., Weinert B. T., Bekker-Jensen D. B., Secher A., Skovgaard T., Kelstrup C. D., Dmytriyev A., Choudhary C., Lundby C., Olsen J. V. (2012) Proteomic analysis of lysine acetylation sites in rat tissues reveals organ specificity and subcellular patterns. Cell Rep. 2, 419–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vousden K. H., Prives C. (2009) Blinded by the light: the growing complexity of p53. Cell 137, 413–431 [DOI] [PubMed] [Google Scholar]
- 45. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 46. Blagoev B., Kratchmarova I., Ong S. E., Nielsen M., Foster L. J., Mann M. (2003) A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat. Biotechnol. 21, 315–318 [DOI] [PubMed] [Google Scholar]
- 47. Mallick P., Kuster B. (2010) Proteomics: a pragmatic perspective. Nat. Biotechnol. 28, 695–709 [DOI] [PubMed] [Google Scholar]
- 48. Bantscheff M., Kuster B. (2012) Quantitative mass spectrometry in proteomics. Anal. Bioanal. Chem. 404, 937–938 [DOI] [PubMed] [Google Scholar]
- 49. Thompson A., Schafer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 50. Ross P. L., Huang Y. L. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 51. Mertins P., Udeshi N. D., Clauser K. R., Mani D. R., Patel J., Ong S. E., Jaffe J. D., Carr S. A. (2012) iTRAQ labeling is superior to mTRAQ for quantitative global proteomics and phosphoproteomics. Mol. Cell. Proteomics 11, M111.014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamagata A., Kristensen D. B., Takeda Y., Miyamoto Y., Okada K., Inamatsu M., Yoshizato K. (2002) Mapping of phosphorylated proteins on two-dimensional polyacrylamide gels using protein phosphatase. Proteomics 2, 1267–1276 [DOI] [PubMed] [Google Scholar]
- 53. Wu R., Haas W., Dephoure N., Huttlin E. L., Zhai B., Sowa M. E., Gygi S. P. (2011) A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods 8, 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Weinert B. T., Iesmantavicius V., Wagner S. A., Scholz C., Gummesson B., Beli P., Nystrom T., Choudhary C. (2013) Acetyl-phosphate is a critical determinant of lysine acetylation in E. coli. Mol. Cell. 51, 265–272 [DOI] [PubMed] [Google Scholar]
- 55. Savitski M. M., Nielsen M. L., Zubarev R. A. (2006) ModifiComb, a new proteomic tool for mapping substoichiometric post-translational modifications, finding novel types of modifications, and fingerprinting complex protein mixtures. Mol. Cell. Proteomics 5, 935–948 [DOI] [PubMed] [Google Scholar]
- 56. Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005) InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626–4639 [DOI] [PubMed] [Google Scholar]
- 57. Tan M., Luo H., Lee S., Jin F., Yang J. S., Montellier E., Buchou T., Cheng Z., Rousseaux S., Rajagopal N., Lu Z., Ye Z., Zhu Q., Wysocka J., Ye Y., Khochbin S., Ren B., Zhao Y. (2011) Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang Z., Tan M., Xie Z., Dai L., Chen Y., Zhao Y. (2011) Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Moellering R. E., Cravatt B. F. (2013) Functional lysine modification by an intrinsically reactive primary glycolytic metabolite. Science 341, 549–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Deleted in proof.
- 61. Zhang Y., Wang J., Ding M., Yu Y. (2013) Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat. Methods 10, 981–984 [DOI] [PubMed] [Google Scholar]
- 62. Jungmichel S., Rosenthal F., Altmeyer M., Lukas J., Hottiger M. O., Nielsen M. L. (2013) Proteome-wide identification of poly(ADP-ribosyl)ation targets in different genotoxic stress responses. Mol. Cell 52, 272–285 [DOI] [PubMed] [Google Scholar]
- 63. Aebersold R. (2009) A stress test for mass spectrometry-based proteomics. Nat. Methods 6, 411–412 [DOI] [PubMed] [Google Scholar]
- 64. Schwartz D., Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 65. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Haw R., Hermjakob H., D'Eustachio P., Stein L. (2011) Reactome pathway analysis to enrich biological discovery in proteomics datasets. Proteomics 11, 3598–3613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Franceschini A., Szklarczyk D., Frankild S., Kuhn M., Simonovic M., Roth A., Lin J., Minguez P., Bork P., von Mering C., Jensen L. J. (2013) STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41, D808–D815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cox J., Mann M. (2012) 1D and 2D annotation enrichment: a statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinformatics 13, S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Miller M. L., Jensen L. J., Diella F., Jorgensen C., Tinti M., Li L., Hsiung M., Parker S. A., Bordeaux J., Sicheritz-Ponten T., Olhovsky M., Pasculescu A., Alexander J., Knapp S., Blom N., Bork P., Li S., Cesareni G., Pawson T., Turk B. E., Yaffe M. B., Brunak S., Linding R. (2008) Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal. 1, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Linding R., Jensen L. J., Ostheimer G. J., van Vugt M. A., Jorgensen C., Miron I. M., Diella F., Colwill K., Taylor L., Elder K., Metalnikov P., Nguyen V., Pasculescu A., Jin J., Park J. G., Samson L. D., Woodgett J. R., Russell R. B., Bork P., Yaffe M. B., Pawson T. (2007) Systematic discovery of in vivo phosphorylation networks. Cell 129, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hornbeck P. V., Chabra I., Kornhauser J. M., Skrzypek E., Zhang B. (2004) PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics 4, 1551–1561 [DOI] [PubMed] [Google Scholar]
- 72. Thakur S. S., Geiger T., Chatterjee B., Bandilla P., Frohlich F., Cox J., Mann M. (2011) Deep and highly sensitive proteome coverage by LC-MS/MS without prefractionation. Mol. Cell. Proteomics 10, M110.003699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kocher T., Pichler P., Swart R., Mechtler K. (2012) Analysis of protein mixtures from whole-cell extracts by single-run nanoLC-MS/MS using ultralong gradients. Nat. Protoc. 7, 882–890 [DOI] [PubMed] [Google Scholar]
- 74. Pirmoradian M., Budamgunta H., Chingin K., Zhang B., Astorga-Wells J., Zubarev R. A. (2013) Rapid and deep human proteome analysis by single-dimension shotgun proteomics. Mol. Cell. Proteomics 12, 3330–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lundby A., Secher A., Lage K., Nordsborg N. B., Dmytriyev A., Lundby C., Olsen J. V. (2012) Quantitative maps of protein phosphorylation sites across 14 different rat organs and tissues. Nat. Commun. 3, 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lange V., Picotti P., Domon B., Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Oliveira A. P., Ludwig C., Picotti P., Kogadeeva M., Aebersold R., Sauer U. (2012) Regulation of yeast central metabolism by enzyme phosphorylation. Mol. Syst. Biol. 8, 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bisson N., James D. A., Ivosev G., Tate S. A., Bonner R., Taylor L., Pawson T. (2011) Selected reaction monitoring mass spectrometry reveals the dynamics of signaling through the GRB2 adaptor. Nat. Biotechnol. 29, 653–658 [DOI] [PubMed] [Google Scholar]
- 80. Zheng Y., Zhang C., Croucher D. R., Soliman M. A., St-Denis N., Pasculescu A., Taylor L., Tate S. A., Hardy W. R., Colwill K., Dai A. Y., Bagshaw R., Dennis J. W., Gingras A. C., Daly R. J., Pawson T. (2013) Temporal regulation of EGF signalling networks by the scaffold protein Shc1. Nature 499, 166–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Garraway L. A., Lander E. S. (2013) Lessons from the cancer genome. Cell 153, 17–37 [DOI] [PubMed] [Google Scholar]
- 82. Hsu P. P., Kang S. A., Rameseder J., Zhang Y., Ottina K. A., Lim D., Peterson T. R., Choi Y., Gray N. S., Yaffe M. B., Marto J. A., Sabatini D. M. (2011) The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332, 1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Yu Y., Yoon S. O., Poulogiannis G., Yang Q., Ma X. M., Villen J., Kubica N., Hoffman G. R., Cantley L. C., Gygi S. P., Blenis J. (2011) Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 332, 1322–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Deleted in proof.
- 85. Francavilla C., Rigbolt K. T., Emdal K. B., Carraro G., Vernet E., Bekker-Jensen D. B., Streicher W., Wikstrom M., Sundstrom M., Bellusci S., Cavallaro U., Blagoev B., Olsen J. V. (2013) Functional proteomics defines the molecular switch underlying FGF receptor trafficking and cellular outputs. Mol. Cell. 51, 707–722 [DOI] [PubMed] [Google Scholar]
- 86. Roux P. P., Thibault P. (2013) The coming of age of phosphoproteomics; from large data sets to inference of protein functions. Mol. Cell. Proteomics 12, 3453–3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., Pedrioli P. G., Gerrits B., Picotti P., Lam H., Vitek O., Brusniak M. Y., Roschitzki B., Zhang C., Shokat K. M., Schlapbach R., Colman-Lerner A., Nolan G. P., Nesvizhskii A. I., Peter M., Loewith R., von Mering C., Aebersold R. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 3, rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Gnad F., Ren S., Cox J., Olsen J. V., Macek B., Oroshi M., Mann M. (2007) PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 8, R250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Linding R., Jensen L. J., Pasculescu A., Olhovsky M., Colwill K., Bork P., Yaffe M. B., Pawson T. (2008) NetworKIN: a resource for exploring cellular phosphorylation networks. Nucleic Acids Res. 36, D695–D699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Koch A., Krug K., Pengelley S., Macek B., Hauf S. (2011) Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci. Signal. 4, rs6. [DOI] [PubMed] [Google Scholar]
- 91. Kettenbach A. N., Schweppe D. K., Faherty B. K., Pechenick D., Pletnev A. A., Gerber S. A. (2011) Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4, rs5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., Blethrow J., Shimizu E., Tsien J. Z., Schultz P. G., Rose M. D., Wood J. L., Morgan D. O., Shokat K. M. (2000) A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature 407, 395–401 [DOI] [PubMed] [Google Scholar]
- 93. Holt L. J., Tuch B. B., Villen J., Johnson A. D., Gygi S. P., Morgan D. O. (2009) Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Oppermann F. S., Grundner-Culemann K., Kumar C., Gruss O. J., Jallepalli P. V., Daub H. (2012) Combination of chemical genetics and phosphoproteomics for kinase signaling analysis enables confident identification of cellular downstream targets. Mol. Cell. Proteomics 11, O111.012351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Esvelt K. M., Wang H. H. (2013) Genome-scale engineering for systems and synthetic biology. Mol. Syst. Biol. 9, 641. [DOI] [PMC free article] [PubMed] [Google Scholar]