Abstract
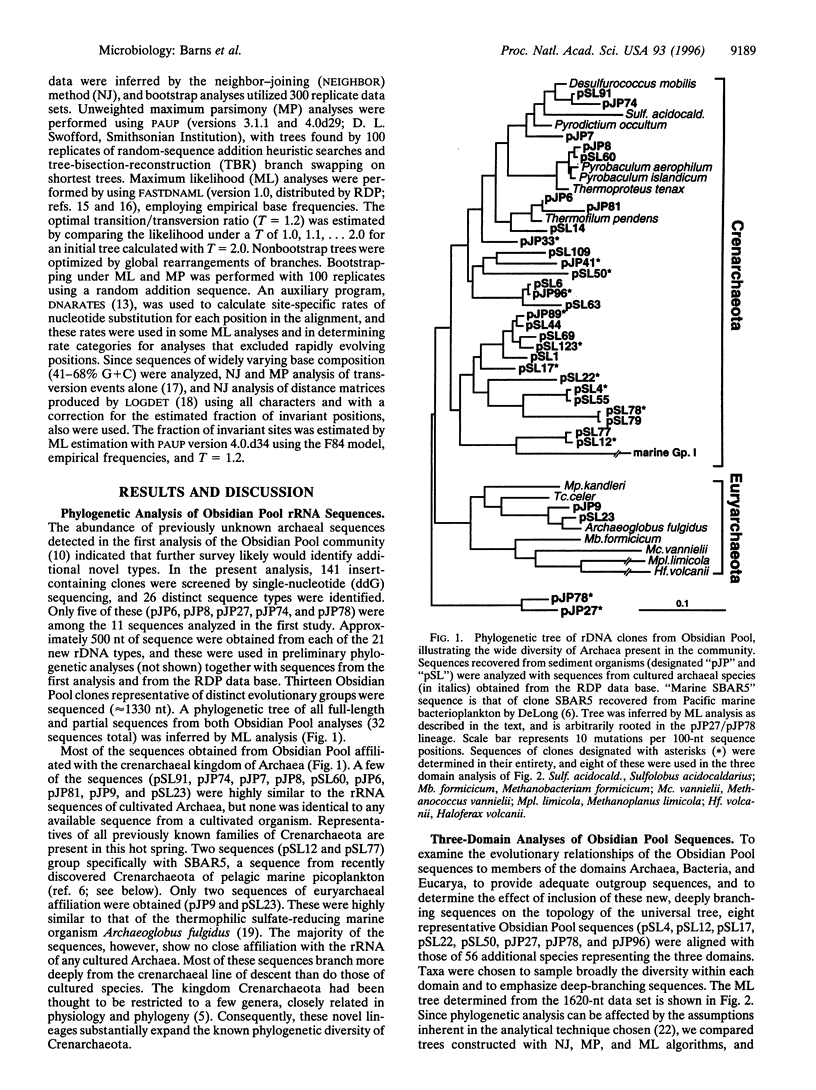
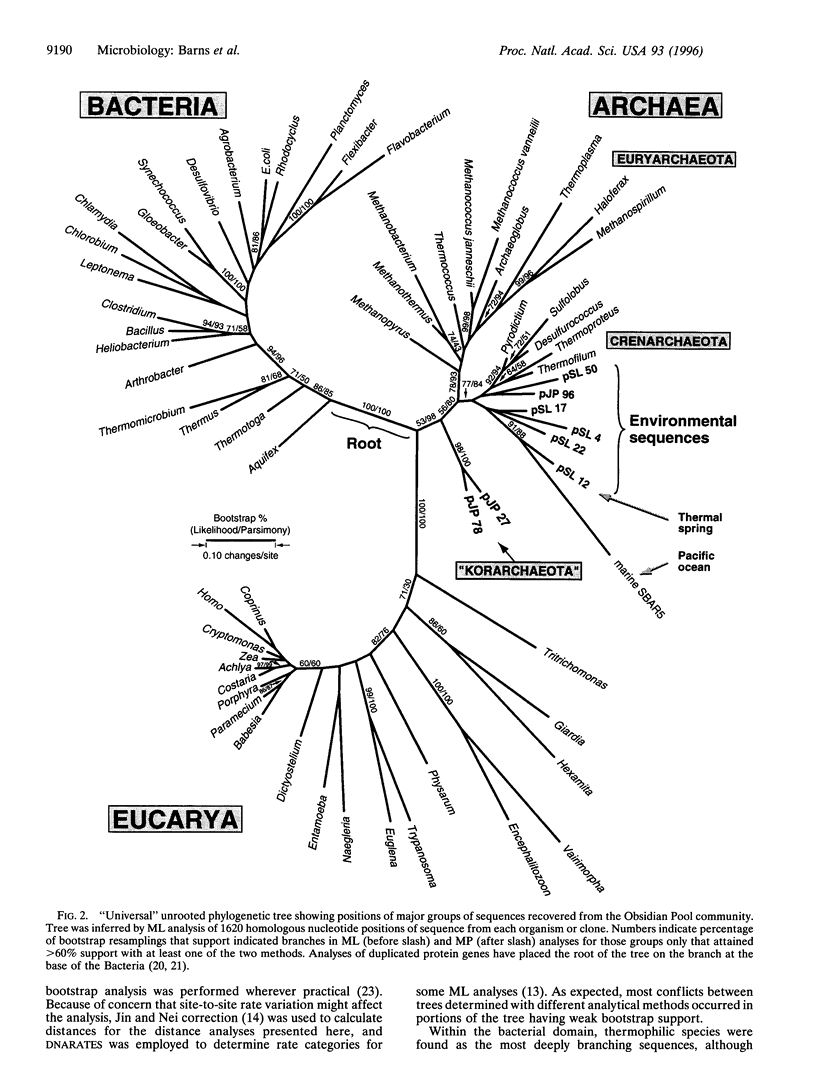
Phylogenetic analysis of ribosomal RNA sequences obtained from uncultivated organisms of a hot spring in Yellowstone National Park reveals several novel groups of Archaea, many of which diverged from the crenarchaeal line of descent prior to previously characterized members of that kingdom. Universal phylogenetic trees constructed with the addition of these sequences indicate monophyly of Archaea, with modest bootstrap support. The data also show a specific relationship between low-temperature marine Archaea and some hot spring Archaea. Two of the environmental sequences are enigmatic: depending upon the data set and analytical method used, these sequences branch deeply within the Crenarchaeota, below the bifurcation between Crenarchaeota and Euryarchaeota, or even as the sister group to Eukaryotes. If additional data confirm either of the latter two placements, then the organisms represented by these ribosomal RNA sequences would merit recognition as a new kingdom, provisionally named "Korarchaeota."
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann R. I., Ludwig W., Schleifer K. H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995 Mar;59(1):143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldauf S. L., Palmer J. D., Doolittle W. F. The root of the universal tree and the origin of eukaryotes based on elongation factor phylogeny. Proc Natl Acad Sci U S A. 1996 Jul 23;93(15):7749–7754. doi: 10.1073/pnas.93.15.7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barns S. M., Fundyga R. E., Jeffries M. W., Pace N. R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci U S A. 1994 Mar 1;91(5):1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. R., Doolittle W. F. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggraf S., Olsen G. J., Stetter K. O., Woese C. R. A phylogenetic analysis of Aquifex pyrophilus. Syst Appl Microbiol. 1992 Aug;15(3):352–356. doi: 10.1016/S0723-2020(11)80207-9. [DOI] [PubMed] [Google Scholar]
- Burggraf S., Stetter K. O., Rouviere P., Woese C. R. Methanopyrus kandleri: an archaeal methanogen unrelated to all other known methanogens. Syst Appl Microbiol. 1991;14:346–351. doi: 10.1016/s0723-2020(11)80308-5. [DOI] [PubMed] [Google Scholar]
- De Rijk P., Van de Peer Y., Van den Broeck I., De Wachter R. Evolution according to large ribosomal subunit RNA. J Mol Evol. 1995 Sep;41(3):366–375. doi: 10.1007/BF01215184. [DOI] [PubMed] [Google Scholar]
- DeLong E. F. Archaea in coastal marine environments. Proc Natl Acad Sci U S A. 1992 Jun 15;89(12):5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong E. F., Wu K. Y., Prézelin B. B., Jovine R. V. High abundance of Archaea in Antarctic marine picoplankton. Nature. 1994 Oct 20;371(6499):695–697. doi: 10.1038/371695a0. [DOI] [PubMed] [Google Scholar]
- Delwiche C. F., Kuhsel M., Palmer J. D. Phylogenetic analysis of tufA sequences indicates a cyanobacterial origin of all plastids. Mol Phylogenet Evol. 1995 Jun;4(2):110–128. doi: 10.1006/mpev.1995.1012. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol. 1981;17(6):368–376. doi: 10.1007/BF01734359. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- Fuhrman J. A., McCallum K., Davis A. A. Novel major archaebacterial group from marine plankton. Nature. 1992 Mar 12;356(6365):148–149. doi: 10.1038/356148a0. [DOI] [PubMed] [Google Scholar]
- Jin L., Nei M. Limitations of the evolutionary parsimony method of phylogenetic analysis. Mol Biol Evol. 1990 Jan;7(1):82–102. doi: 10.1093/oxfordjournals.molbev.a040588. [DOI] [PubMed] [Google Scholar]
- Lake J. A. Tracing origins with molecular sequences: metazoan and eukaryotic beginnings. Trends Biochem Sci. 1991 Feb;16(2):46–50. doi: 10.1016/0968-0004(91)90020-v. [DOI] [PubMed] [Google Scholar]
- Larsen N., Olsen G. J., Maidak B. L., McCaughey M. J., Overbeek R., Macke T. J., Marsh T. L., Woese C. R. The ribosomal database project. Nucleic Acids Res. 1993 Jul 1;21(13):3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leipe D. D., Gunderson J. H., Nerad T. A., Sogin M. L. Small subunit ribosomal RNA+ of Hexamita inflata and the quest for the first branch in the eukaryotic tree. Mol Biochem Parasitol. 1993 May;59(1):41–48. doi: 10.1016/0166-6851(93)90005-i. [DOI] [PubMed] [Google Scholar]
- Lloyd A. T., Sharp P. M. Evolution of the recA gene and the molecular phylogeny of bacteria. J Mol Evol. 1993 Oct;37(4):399–407. doi: 10.1007/BF00178869. [DOI] [PubMed] [Google Scholar]
- Maidak B. L., Larsen N., McCaughey M. J., Overbeek R., Olsen G. J., Fogel K., Blandy J., Woese C. R. The Ribosomal Database Project. Nucleic Acids Res. 1994 Sep;22(17):3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInerney J. O., Wilkinson M., Patching J. W., Embley T. M., Powell R. Recovery and phylogenetic analysis of novel archaeal rRNA sequences from a deep-sea deposit feeder. Appl Environ Microbiol. 1995 Apr;61(4):1646–1648. doi: 10.1128/aem.61.4.1646-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Matsuda H., Hagstrom R., Overbeek R. fastDNAmL: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994 Feb;10(1):41–48. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- Olsen G. J. Microbial ecology. Archaea, Archaea, everywhere. Nature. 1994 Oct 20;371(6499):657–658. doi: 10.1038/371657a0. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R. A brief note concerning archaebacterial phylogeny. Can J Microbiol. 1989 Jan;35(1):119–123. doi: 10.1139/m89-018. [DOI] [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R. Ribosomal RNA: a key to phylogeny. FASEB J. 1993 Jan;7(1):113–123. doi: 10.1096/fasebj.7.1.8422957. [DOI] [PubMed] [Google Scholar]
- Reysenbach A. L., Wickham G. S., Pace N. R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994 Jun;60(6):2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera M. C., Lake J. A. Evidence that eukaryotes and eocyte prokaryotes are immediate relatives. Science. 1992 Jul 3;257(5066):74–76. doi: 10.1126/science.1621096. [DOI] [PubMed] [Google Scholar]
- Stein J. L., Marsh T. L., Wu K. Y., Shizuya H., DeLong E. F. Characterization of uncultivated prokaryotes: isolation and analysis of a 40-kilobase-pair genome fragment from a planktonic marine archaeon. J Bacteriol. 1996 Feb;178(3):591–599. doi: 10.1128/jb.178.3.591-599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viale A. M., Arakaki A. K., Soncini F. C., Ferreyra R. G. Evolutionary relationships among eubacterial groups as inferred from GroEL (chaperonin) sequence comparisons. Int J Syst Bacteriol. 1994 Jul;44(3):527–533. doi: 10.1099/00207713-44-3-527. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Achenbach L., Rouviere P., Mandelco L. Archaeal phylogeny: reexamination of the phylogenetic position of Archaeoglobus fulgidus in light of certain composition-induced artifacts. Syst Appl Microbiol. 1991;14(4):364–371. doi: 10.1016/s0723-2020(11)80311-5. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Kandler O., Wheelis M. L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]