Abstract
The post-translational modification of intracellular proteins by O-linked N-acetylglucosamine (O-GlcNAc) regulates essential cellular processes such as signal transduction, transcription, translation, and protein degradation. Misfolded, damaged, and unwanted proteins are tagged with a chain of ubiquitin moieties for degradation by the proteasome, which is critical for cellular homeostasis. In this review, we summarize the current knowledge of the interplay between O-GlcNAcylation and ubiquitination in the control of protein degradation. Understanding the mechanisms of action of O-GlcNAcylation in the ubiquitin-proteosome system shall facilitate the development of therapeutics for human diseases such as cancer, metabolic syndrome, and neurodegenerative diseases.
THE UBIQUITIN-PROTEASOME SYSTEM
Ubiquitination is a post-translational process by which ubiquitin is covalently attached to the lysine residues of target proteins. Ubiquitin is an 8.5-kDa protein that exists in all eukaryotic cells (1). It is encoded in mammals by four genes. The UBB and UBC genes encode polyubiquitin precursors, whereas UBA52 and RPS27A genes encode fusion proteins composed of a ubiquitin and the ribosomal proteins L40 and S27a, respectively (2). The ubiquitination process consists a cascade of reactions: (i) activation of ubiquitin by the ubiquitin-activating enzyme (E1), (ii) transfer of ubiquitin from E1 to a ubiquitin-conjugating enzyme (E2), and (iii) recognizing target proteins and mediating transfer of ubiquitin from E2 to the target by a ubiquitin ligase (E3) (3).
Polyubiquitination (at least four subunits) through lysine 48 (K48) of ubiquitin normally marks target proteins for proteasomal degradation. The ubiquitin-proteasome system (UPS)1 is the key machinery by which cells dispose of misfolded and damaged proteins in order to maintain cellular homeostasis. In addition, monoubiquitination through K48 or polyubiquitination through other lysine residues of ubiquitin regulates distinct cellular processes, including subcellular localization, endocytosis, and enzymatic activity (4, 5).
Crosstalk between different types of post-translational modifications (PTMs) encodes a wealth of biological information. It is known that ubiquitination and other forms of PTMs are mutually regulated. A large body of evidence shows that phosphorylation and ubiquitination are connected either positively or negatively (6, 7). Regulatory crosstalk between lysine acetylation and ubiquitination has been shown to control protein stability (8). Crosstalk between histone methylation and ubiquitination is involved in gene expression and protein stability (9).
O-GlcNAc Modification and Its Interplay with Other PTMs
Thousands of cytoplasmic and nuclear proteins are modified by a single O-linked β-N-acetylglucosamine (O-GlcNAc) moiety at serine (S) or threonine (T) residues, termed O-GlcNAcylation (10, 11). O-GlcNAcylation is catalyzed by O-GlcNAc transferase (OGT), whereas the reverse reaction is mediated by O-GlcNAcase (OGA, NCOAT, or MGEA5). UDP-GlcNAc, the donor substrate for O-GlcNAcylation, is derived from extracellular glucose through the hexosamine biosynthetic pathway. Because UDP-GlcNAc and protein O-GlcNAc levels in the cell fluctuate with the availability of glucose, free fatty acids, uridine, and the amino acid glutamine, O-GlcNAc is proposed as a nutrient sensor and metabolic regulator (12, 13). This dynamic and reversible modification is emerging as a key regulator of diverse cellular processes, such as signal transduction, transcription, translation, and cytoskeletal functions (14–16). Aberrant O-GlcNAcylation has been implicated in a spectrum of human diseases, including diabetes, cancer, cardiovascular disease, and Alzheimer disease.
Since its discovery in 1984, O-GlcNAcylation has been extensively studied in relationship with phosphorylation (10, 17). Interplay between O-GlcNAcylation and other PTMs is emerging as an important area of investigation. It has been shown that OGT overexpression alters the acetylation and methylation of histones and the activity of an arginine methyltransferase, CARM1 (18). Allison et al. show that O-GlcNAcylation of RelA at T305 is required in order for p300-mediated acetylation at K310 to fully activate NF-κB transcription (19). O-GlcNAcylation of a histone lysine methyltranferase, MLL5, promotes methylation of H3K4 to facilitate retinoic-acid-induced granulopoiesis (20). Recent studies reveal that the ten-eleven translocation proteins TET2 and TET3 form a complex with OGT that sustains H3K4 methylation through O-GlcNAcylating host cell factor C1 (HCF-1), a component of the H3K4 methyltransferase SET1/COMPASS complex (21). Meanwhile, a growing body of evidence demonstrates that O-GlcNAcylation regulates mono- and polyubiquitination, protein stability, and proteasome function, which is the focus of this review.
O-GlcNAcylation Regulates Protein Ubiquitination via Phosphorylation
Because O-GlcNAcylation can affect phosphorylation (10) and phosphorylation can regulate ubiquitination (6), it is conceivable that O-GlcNAcylation controls protein ubiquitination and stability through interplay with phosphorylation (Table I, Fig. 1A), as exemplified below.
Table I. List of proteins for which stability is regulated by O-GlcNAc signaling.
| Protein | O-GlcNAc site | Expression and stability | Ubiquitination | Mechanism | Function | Reference |
|---|---|---|---|---|---|---|
| p53 | S149 | Increased by O-GlcNAc | Decreased by O-GlcNAc | Reduce phosphorylation at T155 | Cancer | (24) |
| Δ-lactoferrin | S10 | Increased by O-GlcNAc | Decreased by O-GlcNAc | Compete with phosphorylation at S10 | Cell cycle arrest and apoptosis | (27) |
| Snail1 | S112 | Increased by O-GlcNAc | Decreased by O-GlcNAc | Block phosphorylation | Epithelial-mesenchymal transition | (30) |
| ERβ | S16 | Increased by O-GlcNAc | N/D | Compete with phosphorylation at S16 | Transcription | (32) |
| CK2α | S347 | Reduced by O-GlcNAc | N/D | Reduce phosphorylation at T344 | Cell proliferation | (34) |
| CRTC2 | S70, S171 | N/D | N/D | Compete with phosphorylation at S70, S171 | Gluconeogenesis | (36, 37) |
| PGC-1α | S333 | Increased by O-GlcNAc | Decreased by O-GlcNAc | Recruit BAP1 | Gluconeogenesis | (13) |
| Clock | Site N/D | Increased by O-GlcNAc | Decreased by O-GlcNAc | Recruit BAP1 | Circadian rhythm | (46) |
| Bmal1 | S418 | Increased by O-GlcNAc | Decreased by O-GlcNAc | Recruit BAP1 | Circadian rhythm | (45, 46) |
| ChREBP | Site N/D | Increased by O-GlcNAc | Decreased by O-GlcNAc | Lipogenesis | (49, 50) | |
| Keratins 8/18 (K8/18) | K8 (N/D) | Reduced by O-GlcNAc | Increased by O-GlcNAc | Filament architecture | (53) | |
| K18 (S29/S30/S48) | ||||||
| A20 | Site N/D | Reduced by high glucose | Increased by high glucose | Atherosclerosis | (54) | |
| β-catenin | Site N/D | Increased by HBP flux | N/D | Cell proliferation | (55) | |
| p67eIF2 | S60/T62/S63 | Increased by O-GlcNAc | N/D | Protein synthesis | (56) | |
| FoxM1 | Not O-GlcNAcylated | Increased by OGT | N/D | Breast cancer | (57) | |
| Plakoglobin | Site N/D | Increased by OGT | N/D | Cell–cell adhesion | (58) | |
| Sp1 | Site N/D | Reduced upon starvation | N/D | Transcription | (59) | |
| Nkx2.5 | Site N/D | Reduced by OGA inhibitors | N/D | Diabetic cardiomyopathy | (60) | |
| SirT1 | Not known whether O-GlcNAcylated | Cytosolic level decreased by HBP | N/D | β cell apoptosis | (61) |
N/D, not determined.
Fig. 1.
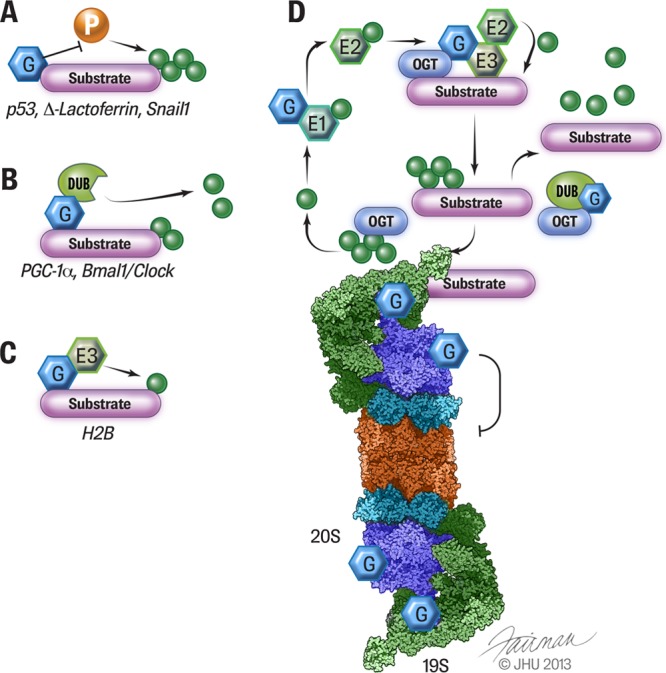
Modes of interaction between O-GlcNAcylation and ubiquitination. A, O-GlcNAcylation of the substrate antagonizes phosphorylation at the same or adjacent site to control ubiquitination. B, O-GlcNAcylation provides a docking site for DUBs that deubiquitinate the substrate. C, O-GlcNAcylation of H2B recruits an E3 complex to monoubiquitinate H2B. D, targeting the ubiquitin-proteasome system. OGT physically interacts with ubiquitin precursors, E3s, and DUBs. O-GlcNAcylation has been found on E1s, E3s, DUBs, and 19S and 20S proteasomes.
p53
The expression of the tumor suppressor p53 is tightly controlled by proteasomal degradation so as to maintain low levels under normal conditions and rapidly accumulate upon DNA damage (22). The fate of p53 is dictated by a variety of PTMs, including phosphorylation, acetylation, methylation, ubiquitination, and O-GlcNAcylation (23). Cho and colleagues demonstrate that the treatment of MCF-7 cells with an OGA inhibitor increases the level of O-GlcNAcylated p53 and decreases cell viability (24). O-GlcNAcylation of p53 at S149 inhibits phosphorylation at T155 by the COP9 signalosome, thereby reducing p53 ubiquitination and degradation.
Δ-lactoferrin
Δ-lactoferrin is a transcription factor that induces cell cycle arrest by up-regulating the expression of genes including Skp1, DcpS, and Bax (25). Δ-lactoferrin expression is down-regulated in cancer cells, whereas its high-level expression is correlated with a good prognosis in human breast cancer (25). It has been shown that Δ-lactoferrin is reciprocally O-GlcNAcylated and phosphorylated at S10 (26, 27). O-GlcNAcylation stabilizes Δ-lactoferrin and retains a basal level of transcriptional activity. Upon activation, Δ-lactoferrin is phosphorylated at S10, which promotes transcription and subsequent degradation through K379 polyubiquitination (27). These studies point to the idea that protein functions can be precisely controlled by dynamic and coordinated changes in O-GlcNAcylation, phosphorylation, and ubiquitination.
Snail1
The zinc-finger protein Snail1 regulates epithelial-mesenchymal transition and tumor progression by repressing the transcription of E-cadherin, a major component of cell adhesion junctions (28). It has been shown that phosphorylation of Snail1 by casein kinase 1 and glycogen synthase kinase-3β promotes the ubiquitination and proteasomal degradation of Snail1 (29). Park et al. show that O-GlcNAcylation of Snail1 at S112 decreases glycogen synthase kinase-3β-mediated phosphorylation and increases the stability of the protein (30). Consistently, the pharmacological inhibition of OGA by PUGNAc increases the half-life of Snail1 by inhibiting ubiquitination. In response to hyperglycemia, O-GlcNAc modification of Snail1 down-regulates E-cadherin transcription and therefore promotes cell migration and invasive programs (30).
Estrogen Receptor β
The nuclear receptor estrogen receptor β (ERβ) mediates many aspects of estrogen action, including reproduction, inflammation, behavior, and energy metabolism (31). Cheng and Hart demonstrate that ERβ is reciprocally modified by O-GlcNAcylation and phosphorylation at S16. The S16A mutant devoid of both modifications reduces ERβ turnover, whereas the S16E mutant mimicking constitutive phosphorylation has an increased turnover rate (32). A simple interpretation of these results is that O-GlcNAcylation protects the S16 site from phosphorylation and Pro-Glu-Ser-Thr region–mediated proteasomal degradation. However, whether this process involves the ubiquitination of ERβ remains unclear.
Casein Kinase 2-α
Casein kinase 2 (CK2) is a serine/threonine protein kinase that has been implicated in cell proliferation, DNA repair, circadian rhythm, and other cellular processes (33). Tarrant et al. demonstrate that phosphorylation of the catalytic subunit of CK2α at T344 increases protein stability by promoting the interaction with Pin1 (34). Moreover, the proximal S347 is modified by O-GlcNAc, which antagonizes T344 phosphorylation and leads to proteasomal degradation of CK2α (34). It will be interesting to explore the role of ubiquitination of CK2α in this context.
cAMP-response Element-binding-protein-regulated Transcription Coactivator 2
cAMP-response element-binding-protein-regulated transcription coactivator 2 (CRTC2) is a transcriptional coactivator for cAMP-response element-binding protein and an important regulator of gluconeogenesis in the liver (35). Under fasting conditions, glucagon induces dephosphorylation of CRTC2 at S171, resulting in CRTC2 translocation into the nucleus to activate the transcription of gluconeogenic genes (35). During feeding, insulin activates the Ser/Thr kinase SIK2 to phosphorylate CRTC2 at S171. Subsequently, phosphorylated CRTC2 translocates to the cytoplasm and undergoes ubiquitination-dependent degradation (36). Dentin et al. reveal that O-GlcNAcylation of CRTC2 at S70 and S171 competes with phosphorylation to suppress cytoplasmic sequestration, thereby contributing to hyperglycemia-induced hepatic gluconeogenesis (37). Whether O-GlcNAcylation has a direct effect on CRTC2 ubiquitination and stability warrants further investigation.
O-GlcNAcylation Stabilizes Proteins by Recruiting Deubiquitinase
Using a proteomic approach, Ruan et al. recently identified a large number of putative OGT-binding proteins. Many proteins in the ubiquitination pathway, including ubiquitin precursors, E3 ubiquitin ligases, and deubiquitinases (DUBs), are enriched (Table II). Although the functions of these interactions have not been determined, these findings raise the possibility that O-GlcNAc signaling directly modulates the ubiquitin system (13).
Table II. Putative OGT-binding proteins involved in ubiquitination.
| Family | Symbol | Name |
|---|---|---|
| Ubiquitin | RPS27A | Ubiquitin and ribosomal protein S27a precursor |
| Ubiquitin | UBA52 | Ubiquitin and ribosomal protein L40 precursor |
| Ubiquitin | UBB | Ubiquitin B precursor |
| Ubiquitin | UBC | UBC ubiquitin C |
| E3a | DDB1 | DNA damage-binding protein 1 |
| E3 | HUWE1 | HECT, UBA, and WWE domain containing 1, E3 ubiquitin-protein ligase |
| E3 | MYCBP2 | MYC binding protein 2, E3 ubiquitin-protein ligase |
| E3 | PRPF19 | Pre-mRNA-processing factor 19 |
| DUBb | BAP1 | BRCA1 associated protein 1 |
| DUB | OTUD4 | OTU domain-containing protein 4 |
| DUB | PRPF8 | Pre-mRNA-processing-splicing factor 8 |
| DUB | USP9X | Ubiquitin specific protease 9 |
Peroxisome Proliferator-activated Receptor Gamma Co-activator 1-α
Peroxisome proliferator-activated receptor gamma co-activator 1-α (PGC-1α) is a key transcriptional cofactor that promotes mitochondrial biogenesis and hepatic gluconeogenesis. It integrates multiple metabolic signals and is extensively regulated by PTMs, including phosphorylation, methylation, acetylation, ubiquitination, and O-GlcNAcylation (38). Ruan et al. demonstrate that OGT forms a glucose-sensitive complex with HCF-1 (13). As a scaffold protein, HCF-1 recruits OGT to O-GlcNAcylate PGC-1α at S333. BAP1 is a DUB known to interact with HCF-1 through an HCF-1 binding motif (39, 40). Ruan et al. further show that O-GlcNAcylation of PGC-1α facilitates the recruitment of BAP1 that deubiquitinates and stabilizes PGC-1α. This is the first demonstration that O-GlcNAcylation regulates protein ubiquitination and stability through a DUB. Diabetic animals have increased levels of HCF-1 and BAP1 in the liver, which is associated with increased PGC-1α levels and gluconeogenesis. Knockdown of OGT and HCF-1 improves glucose metabolism in diabetic db/db mice (13). This study elucidates the role of the antagonism between O-GlcNAcylation and ubiquitination as a regulatory mechanism governing metabolic homeostasis.
BMAL1/CLOCK
The circadian clock functions to align physiological and behavioral processes with daily environmental cycles (41). The molecular clock involves a transcriptional feedback loop in which BMAL1 and CLOCK activate the Period (Per1, -2, and -3) and Cryptochrome (Cry1 and -2) genes. PERs and CRYs accumulate rhythmically and form an inhibitory complex against BMAL1/CLOCK to repress their own transcription (42). The pace of the clock is controlled by various regulatory mechanisms, including PTMs of core clock proteins (43).
The core clock proteins, including BMAL1, CLOCK, PER, and CRY, have been shown to be modified by O-GlcNAc in Drosophila and mammals (44–47). BMAL1 and CLOCK are rhythmically modified and stabilized by O-GlcNAcylation (45, 46). Li et al. further demonstrate that the OGT–BAP1 complex O-GlcNAcylates and deubiquitinates BMAL1 and CLOCK to control the amplitude of circadian oscillation in response to nutrient availability. Disruption of O-GlcNAc signaling in mouse liver perturbs the diurnal rhythm of glucose metabolism (46).
Taken together, the studies by Ruan et al. and Li et al. define the OGT–HCF-1–BAP1 complex as a key modulator of ubiquitination, suggesting a novel mechanism by which O-GlcNAcylation controls protein stability (13, 46) (Fig. 1B).
O-GlcNAcylation Regulates Protein Ubiquitination via Unknown Mechanisms
Carbohydrate-response Element-binding Protein
Carbohydrate-response element-binding protein (ChREBP) is a basic helix-loop-helix leucine zipper transcription factor that regulates glucose and lipid metabolism in a glucose-dependent manner (48). Under low glucose conditions, phosphorylated, inactive ChREBP primarily resides in the cytoplasm. High glucose levels trigger ChREBP dephosphorylation at S196 by PP2A and translocation into the nucleus, followed by dephosphorylation at T666 to induce the transcriptional activity of ChREBP. However, dephosphorylation at these sites is not sufficient for the constitutive activation of ChREBP, suggesting additional layers of regulation (48). Guinez et al. show that ChREBP is modified by O-GlcNAc, which stabilizes ChREBP to increase the transcription of lipogenic genes (49). OGT overexpression induces lipogenesis by increasing ChREBP levels, whereas OGA overexpression prevents hepatic steatosis in db/db mice (49). Recently, another study showed that FoxO1 reduces ChREBP stability by inhibiting O-GlcNAcylation and promoting ubiquitination of the protein (50). How O-GlcNAcylation of ChREBP affects its ubiquitination is not known.
Keratins 8 and 18
Type I and II keratin proteins are expressed in specific pairs in various tissues during development and differentiation (51). Keratin pair 8/18 is widely studied in terms of the regulation of protein interaction, ubiquitination, and filament organization by phosphorylation (52). Keratins 8 and 18 are also highly O-GlcNAcylated (53). O-GlcNAcylation increases keratin 8/18 solubility, ubiquitination, and proteasomal degradation. O-GlcNAc-deficient keratin 18 is more stable and causes changes in the filament architecture. There does not seem to be a reciprocal relationship between O-GlcNAcylation and phosphorylation of keratin 8/18 in vivo. The mechanism by which O-GlcNAcylation increases ubiquitination is still a mystery (53).
A20
The zinc-finger protein A20 is a negative regulator of NF-κB signaling that has been shown to suppress apoptosis and inflammation. Shrikhande et al. reported that hyperglycemia promotes the O-GlcNAcylation, ubiquitination, and degradation of A20, which accelerate atherosclerosis in diabetic mice (54). Interestingly, A20 has both ubiquitin ligase and deubiquitinase activities, suggesting that regulation of the A20 protein level by O-GlcNAcylation is a control point for the ubiquitination of A20 target proteins.
As listed in Table I, the hexosamine/O-GlcNAc pathway can modulate the stability of many other proteins either positively or negatively. For example, O-GlcNAc signaling increases the stability of β-catenin (55), p67eIF2 (56), FoxM1 (57), plakoglobin (58), and Sp1 (59) but decreases the stability of Nkx2.5 (60) and cytosolic SirT1 (61). However, whether the UPS is involved in the regulation of these proteins has not been determined.
O-GlcNAcylation Facilitates Monoubiquitination
Monoubiquitination is a dynamic and reversible PTM involved in nonproteolytic functions (62). Histones are well-known targets of monoubiquitination. H2B monoubiquitination at K120 has been shown to regulate transcription initiation and elongation (62). Several recent studies suggest that O-GlcNAc is also part of the histone code (63–67). Therefore, it is appealing to determine the interactions between O-GlcNAcylation and other histone markers. Fujiki et al. have demonstrated that O-GlcNAcylation of H2B at S112, which is sensitive to glucose availability, promotes K120 monoubiquitination by anchoring the BRE1A/1B complex, an E3 ligase (Fig. 1C) (64). Genome-wide analysis reveals that H2B S112 O-GlcNAcylation is associated with transcribed gene loci, many of which overlap with K120 monoubiquitination (64). It will be interesting to determine whether O-GlcNAcylation regulates the monoubiquitination of non-histone proteins.
O-GlcNAcylation Regulates the Ubiquitination Process
The preceding sections outline individual proteins that are covalently modified and regulated by O-GlcNAcylation and ubiquitination. There is also evidence that O-GlcNAcylation modulates global ubiquitination (68). Thermal stress induces a rapid increase in both O-GlcNAcylation and ubiquitination. Increasing O-GlcNAc levels via glucosamine or PUGNAc promotes ubiquitination, whereas decreasing O-GlcNAc levels via forskolin, glucose deprivation, or OGT knockdown reduces ubiquitination (68). However, increasing ubiquitination levels via the proteasome inhibitor has no obvious effect on global O-GlcNAcylation. These results suggest that O-GlcNAcylation affects ubiquitination, but not vice versa. The authors also found that the E1 enzyme Uba1 is O-GlcNAcylated, suggesting the possible regulation of protein ubiquitination by E1 O-GlcNAcylation (68).
A large number of O-GlcNAcylated proteins/peptides and O-GlcNAcylation sites have been discovered using proteomic approaches. We searched published O-GlcNAcylated protein datasets for those involved in the ubiquitination process including ubiquitin precursors, E1s, E2s, E3s, and DUBs, and we found that many of theses proteins are modified by O-GlcNAc (Table III). Although the functions of O-GlcNAcylation of these proteins have not been fully characterized, it is conceivable that O-GlcNAcylation regulates protein ubiquitination through multiple nodes of the UPS (Fig. 1D).
Table III. O-GlcNAcylated proteins involved in ubiquitination.
| Family | Symbol | Description | Species | Site | Reference |
|---|---|---|---|---|---|
| E1 | Uba1 | Ubiquitin activating enzyme 1 | Human, Drosophila | N/D | (68, 87, 88) |
| E3a | Cbl | Casitas B-lineage lymphoma proto-oncogene | Human | S601 | (89) |
| Cnot4 | CCR4-NOT transcription complex subunit 4 | Mouse | S316, T331, T573 | (90, 91) | |
| Fbxo2 | F-box protein 2 | Rat | N/D | (92) | |
| Fbxo41 | F-box protein 41 | Mouse | S386, T387 | (90) | |
| Hecd1 | HECT domain-containing protein 1 | Mouse | N/D | (91) | |
| Hectd1 | HECT domain containing 1 | Mouse | S1350 | (90) | |
| Herc1 | HECT domain and RCC1-like domain-containing protein 1 | Mouse | S3011 | (90) | |
| Hic1 | Hypermethylated in cancer 1 | Human | N/D | (93) | |
| Huwe1 | HECT, UBA, and WWE domain containing 1 | Rat | N/D | (92) | |
| Kcmf1 | Differentially expressed in branching tubulogenesis 91 | Mouse | S262 | (91) | |
| Nedd4 | Neural precursor cell expressed, developmentally down-regulated 4 | Mouse | S371, T375 | (90, 91, 94) | |
| Prpf19 | Pre-mRNA-processing factor 19 | Human | T169 | (95) | |
| Rc3h2 | Ring finger and CCCH-type zinc finger domains 2 | Mouse | T471, T472, S592, T841 or S844, S901 | (90, 91) | |
| Ring1 | RING finger protein 1 | Human | N/D | (96) | |
| Rnf2 | RING finger protein 2 | Human | N/D | (96) | |
| Rnf123 | Ring finger protein 123 | Mouse | S1078 | (90) | |
| Sf3b3 | Splicing factor 3b, subunit 3 | Human | N/D | (97) | |
| Sh3rf1 | SH3 domain-containing RING finger protein 1 | Mouse | T512, T91, T92, S526, T527 | (90, 91) | |
| Trim33 | Tripartite motif-containing 33 | Mouse | S650 | (90) | |
| Tulp4 | Tubby like protein 4 | Mouse | T943 | (90) | |
| Ubr4 | Ubiquitin protein ligase E3 component n-recognin 4 | Mouse | S2577 | (90) | |
| Unkl | RING finger protein unkempt-like | Mouse | S451, T459 | (90, 91) | |
| Zbtb20 | Zinc finger and BTB domain containing 20 | Mouse | S268, T465, T480 | (90) | |
| Zfp598 | Zinc finger protein 598 | Mouse | One site among S560, T563, and T564 | (90) | |
| DUBb | Bap1 | BRCA1 associated protein 1 | Human | N/D | (96) |
| Usp11 | Ubiquitin specific peptidase 11 | Mouse | Two sites between S617 and S619 | (90) | |
| Usp24 | Ubiquitin specific peptidase 24 | Mouse | S3 | (90) | |
| Usp31 | Ubiquitin specific peptidase 31 | Mouse | One site between T1163 and T1165 | (90) | |
| Usp5 | Ubiquitin specific peptidase 5 | Rat | N/D | (92) | |
| Usp8 | Ubiquitin specific peptidase 8 | Mouse | S218 and one site among S227/T231/S233 | (90) | |
| Vcpip1 | Valosin containing protein (p97)/p47 complex interacting protein 1 | Mouse | T1072, S1075 | (90, 91) |
O-GlcNAc Signaling Modulates Proteasome Activity
The 26S proteasome is responsible for the destruction of polyubiquitinated proteins. In addition to its roles in regulating protein ubiquitination, O-GlcNAc has long been proposed to directly modulate proteasome activity. Both the 20S catalytic particle and the 19S regulatory particle of the proteasome are known to be O-GlcNAcylated (69). Zhang et al. demonstrated that O-GlcNAcylation of Rpt2 ATPase in the 19S proteasome inhibits proteasome function, which may serve as a mechanism controlling cellular levels of amino acids in response to metabolic changes such as starvation and nutrient overload (70) (Fig. 1D). It should be noted that many of the studies listed in Table I were performed using indiscriminate approaches (i.e. OGT overexpression or OGA inhibition) that affect global O-GlcNAc levels. Thus, the stabilization of certain proteins by O-GlcNAc signaling could be, at least in part, attributed to proteasomal inhibition (Table I).
The short form of O-GlcNAcase (OGA-S), which lacks the histone acetyltransferase domain, accumulates on the surface of lipid droplets (71). Selective knockdown of OGA-S results in global proteasome inhibition and increased levels of perilipin-2 and -3. These findings suggest that proteasomal modulation links O-GlcNAc signaling to lipid droplet maturation (71).
O-GlcNAcylation and Ubiquitination in Physiology and Pathogenesis
During starvation, increased proteasomal degradation in the muscle supplies amino acids to the liver for de novo glucose synthesis (gluconeogenesis). In the starved muscle, reduced O-GlcNAc signaling promotes the ability of the proteasome to degrade polyubiquitinated proteins (72). Meanwhile, O-GlcNAc signaling in the liver promotes gluconeogenesis by deubiquitinating and stabilizing PGC-1α and CRTC2 (13, 37, 73). Thus, O-GlcNAcylation acts in concert with ubiquitination in multiple organs to control glucose metabolism.
Obesity and diabetes are associated with increased O-GlcNAcylation in multiple tissues (73). Glucotoxicity, which is caused by diabetic hyperglycemia, leads to hepatic steatosis, β cell failure, cardiomyopathy, and atherosclerosis. As shown in Table I, O-GlcNAcylation regulates the ubiquitination and/or stability of ChREBP, SirT1, Nkx2.5, and A20. Therefore, glucotoxcity may contribute to metabolic syndrome through aberrant O-GlcNAcylation and ubiquitination of those proteins in various tissues.
Many neurodegenerative diseases are associated with the formation of ubiquitin-conjugated protein aggregates in pathological inclusion bodies. For example, hyperphosphorylated Tau is the main component of neurofibrillary tangles, and amyloid-β is the major component of senile plaques in Alzheimer disease. α-synuclein aggregates form Lewy bodies in Parkinson disease, mutant Huntington proteins form inclusion bodies in Huntington disease, and skein-like inclusions exist in amyotrophic lateral sclerosis (74–77). The common cellular mechanism for these pathological conditions is the defective UPS. O-GlcNAc is also involved in many neurodegenerative diseases (10, 78, 79). O-GlcNAc signaling modulates phosphorylation of Tau and the processing of the amyloid-β precursor protein, and treatment of the OGA inhibitor hinders the progression of Alzheimer disease (78, 80). A recent study shows that O-GlcNAc cycling influences proteasome and autophagy pathways in C. elegans models of neurodegenerative diseases (81). Further studies should define the casual role of O-GlcNAcylation and ubiquitination in neurodegeneration, ideally using murine models and human patient samples.
The defective UPS causes the accumulation of misfolded or mutated proteins, which in turn contribute to tumor formation and progression (82). E3 ligases such as Cbl and DUBs such as BAP1 and A20 have been implicated in cancer development (82, 83). Many drugs that target the UPS machinery have shown promise in clinical trials of cancer therapy (82). Notably, the oncogene Cbl and the tumor suppressor BAP1 are O-GlcNAcylated (Table III). O-GlcNAc has also been implicated in cancer biology by targeting transcription factors such as p53, c-Myc, Sp1, and NF-κB (73, 84). Many proteins involved in cell proliferation, apoptosis, and adhesion, such as p53, Δ-lactoferrin, Snail1, β-catenin, FoxM1, and Sp1, are regulated by O-GlcNAcylation and ubiquitination (Table I). A better understanding of the regulation of the UPS by O-GlcNAcylation will provide crucial insight into cancer diagnosis and therapy.
Acknowledgments
We thank all members of the Yang laboratory for stimulating discussions.
Footnotes
* This work was supported by NIH R01 DK089098, American Diabetes Association, and Ellison Medical Foundation to X.Y.; a Brown-Coxe Postdoctoral Fellowship to H.B.R.; and NSFC 81120108005/81225003 to Y.N.
1 The abbreviations used are:
- ChREBP
- carbohydrate-response element-binding protein
- CK2
- casein kinase 2
- CRTC2
- cAMP-response element-binding-protein-regulated transcription coactivator
- DUB
- deubiquitinase
- ERβ
- estrogen receptor β
- HCF-1
- host cell factor C1
- O-GlcNAc
- O-linked β-N-acetylglucosamine
- OGT
- O-GlcNAc transferase
- PGC-1α
- peroxisome proliferator-activated receptor gamma co-activator 1-α
- PTM
- post-translational modification
- UPS
- ubiquitin-proteasome system.
REFERENCES
- 1. Goldstein G., Scheid M., Hammerling U., Schlesinger D. H., Niall H. D., Boyse E. A. (1975) Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells. Proc. Natl. Acad. Sci. U.S.A. 72, 11–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kimura Y., Tanaka K. (2010) Regulatory mechanisms involved in the control of ubiquitin homeostasis. J Biochem. 147, 793–798 [DOI] [PubMed] [Google Scholar]
- 3. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 4. Ikeda F., Dikic I. (2008) Atypical ubiquitin chains: new molecular signals. EMBO Rep. 9, 536–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hicke L. (2001) Protein regulation by monoubiquitin. Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 6. Hunter T. (2007) The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 7. Gao M., Karin M. (2005) Regulating the regulators: control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli. Mol. Cell 19, 581–593 [DOI] [PubMed] [Google Scholar]
- 8. Caron C., Boyault C., Khochbin S. (2005) Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. Bioessays 27, 408–415 [DOI] [PubMed] [Google Scholar]
- 9. Shukla A., Chaurasia P., Bhaumik S. R. (2009) Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell. Mol. Life Sci. 66, 1419–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hanover J. A., Krause M. W., Love D. C. (2012) Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 13, 312–321 [DOI] [PubMed] [Google Scholar]
- 12. Wells L., Vosseller K., Hart G. W. (2003) A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell. Mol. Life Sci. 60, 222–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ruan H. B., Han X., Li M. D., Singh J. P., Qian K., Azarhoush S., Zhao L., Bennett A. M., Samuel V. T., Wu J., Yates J. R., 3rd, Yang X. (2012) O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1alpha stability. Cell Metab. 16, 226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang X., Su K., Roos M. D., Chang Q., Paterson A. J., Kudlow J. E. (2001) O-linkage of N-acetylglucosamine to Sp1 activation domain inhibits its transcriptional capability. Proc. Natl. Acad. Sci. U.S.A. 98, 6611–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang X., Zhang F., Kudlow J. E. (2002) Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: coupling protein O-GlcNAcylation to transcriptional repression. Cell 110, 69–80 [DOI] [PubMed] [Google Scholar]
- 16. Yang X., Ongusaha P. P., Miles P. D., Havstad J. C., Zhang F., So W. V., Kudlow J. E., Michell R. H., Olefsky J. M., Field S. J., Evans R. M. (2008) Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451, 964–969 [DOI] [PubMed] [Google Scholar]
- 17. Torres C. R., Hart G. W. (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 [PubMed] [Google Scholar]
- 18. Sakabe K., Hart G. W. (2010) O-GlcNAc transferase regulates mitotic chromatin dynamics. J. Biol. Chem. 285, 34460–34468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Allison D. F., Wamsley J. J., Kumar M., Li D., Gray L. G., Hart G. W., Jones D. R., Mayo M. W. (2012) Modification of RelA by O-linked N-acetylglucosamine links glucose metabolism to NF-kappaB acetylation and transcription. Proc. Natl. Acad. Sci. U.S.A. 109, 16888–16893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fujiki R., Chikanishi T., Hashiba W., Ito H., Takada I., Roeder R. G., Kitagawa H., Kato S. (2009) GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature 459, 455–459 [DOI] [PubMed] [Google Scholar]
- 21. Deplus R., Delatte B., Schwinn M. K., Defrance M., Mendez J., Murphy N., Dawson M. A., Volkmar M., Putmans P., Calonne E., Shih A. H., Levine R. L., Bernard O., Mercher T., Solary E., Urh M., Daniels D. L., Fuks F. (2013) TET2 and TET3 regulate GlcNAcylation and H3K4 methylation through OGT and SET1/COMPASS. EMBO J. 32, 645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vousden K. H. (2000) p53: death star. Cell 103, 691–694 [DOI] [PubMed] [Google Scholar]
- 23. Kruse J. P., Gu W. (2008) SnapShot: p53 posttranslational modifications. Cell 133, 930–30.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., Cho J. W. (2006) Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat. Cell Biol. 8, 1074–1083 [DOI] [PubMed] [Google Scholar]
- 25. Mariller C., Hardiville S., Hoedt E., Huvent I., Pina-Canseco S., Pierce A. (2012) Delta-lactoferrin, an intracellular lactoferrin isoform that acts as a transcription factor. Biochem. Cell Biol. 90, 307–319 [DOI] [PubMed] [Google Scholar]
- 26. Mariller C., Hardiville S., Hoedt E., Benaissa M., Mazurier J., Pierce A. (2009) Proteomic approach to the identification of novel delta-lactoferrin target genes: characterization of DcpS, an mRNA scavenger decapping enzyme. Biochimie (Paris) 91, 109–122 [DOI] [PubMed] [Google Scholar]
- 27. Hardiville S., Hoedt E., Mariller C., Benaissa M., Pierce A. (2010) O-GlcNAcylation/phosphorylation cycling at Ser10 controls both transcriptional activity and stability of delta-lactoferrin. J. Biol. Chem. 285, 19205–19218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee J. M., Dedhar S., Kalluri R., Thompson E. W. (2006) The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J. Cell Biol. 172, 973–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xu Y., Lee S. H., Kim H. S., Kim N. H., Piao S., Park S. H., Jung Y. S., Yook J. I., Park B. J., Ha N. C. (2010) Role of CK1 in GSK3beta-mediated phosphorylation and degradation of snail. Oncogene 29, 3124–3133 [DOI] [PubMed] [Google Scholar]
- 30. Park S. Y., Kim H. S., Kim N. H., Ji S., Cha S. Y., Kang J. G., Ota I., Shimada K., Konishi N., Nam H. W., Hong S. W., Yang W. H., Roth J., Yook J. I., Cho J. W. (2010) Snail1 is stabilized by O-GlcNAc modification in hyperglycaemic condition. EMBO J. 29, 3787–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harris H. A. (2007) Estrogen receptor-beta: recent lessons from in vivo studies. Mol. Endocrinol. 21, 1–13 [DOI] [PubMed] [Google Scholar]
- 32. Cheng X., Hart G. W. (2001) Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor beta: post-translational regulation of turnover and transactivation activity. J. Biol. Chem. 276, 10570–10575 [DOI] [PubMed] [Google Scholar]
- 33. St-Denis N. A., Litchfield D. W. (2009) Protein kinase CK2 in health and disease: from birth to death: the role of protein kinase CK2 in the regulation of cell proliferation and survival. Cell. Mol. Life Sci. 66, 1817–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tarrant M. K., Rho H. S., Xie Z., Jiang Y. L., Gross C., Culhane J. C., Yan G., Qian J., Ichikawa Y., Matsuoka T., Zachara N., Etzkorn F. A., Hart G. W., Jeong J. S., Blackshaw S., Zhu H., Cole P. A. (2012) Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat. Chem. Biol. 8, 262–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Koo S. H., Flechner L., Qi L., Zhang X., Screaton R. A., Jeffries S., Hedrick S., Xu W., Boussouar F., Brindle P., Takemori H., Montminy M. (2005) The CREB coactivator TORC2 is a key regulator of fasting glucose metabolism. Nature 437, 1109–1111 [DOI] [PubMed] [Google Scholar]
- 36. Dentin R., Liu Y., Koo S. H., Hedrick S., Vargas T., Heredia J., Yates J., 3rd, Montminy M. (2007) Insulin modulates gluconeogenesis by inhibition of the coactivator TORC2. Nature 449, 366–369 [DOI] [PubMed] [Google Scholar]
- 37. Dentin R., Hedrick S., Xie J., Yates J., 3rd, Montminy M. (2008) Hepatic glucose sensing via the CREB coactivator CRTC2. Science 319, 1402–1405 [DOI] [PubMed] [Google Scholar]
- 38. Fernandez-Marcos P. J., Auwerx J. (2011) Regulation of PGC-1alpha, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 93, 884S–890S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Misaghi S., Ottosen S., Izrael-Tomasevic A., Arnott D., Lamkanfi M., Lee J., Liu J., O'Rourke K., Dixit V. M., Wilson A. C. (2009) Association of C-terminal ubiquitin hydrolase BRCA1-associated protein 1 with cell cycle regulator host cell factor 1. Mol. Cell. Biol. 29, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Machida Y. J., Machida Y., Vashisht A. A., Wohlschlegel J. A., Dutta A. (2009) The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J. Biol. Chem. 284, 34179–34188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bass J. (2012) Circadian topology of metabolism. Nature 491, 348–356 [DOI] [PubMed] [Google Scholar]
- 42. Ko C. H., Takahashi J. S. (2006) Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 15 Spec No 2, R271–R277 [DOI] [PubMed] [Google Scholar]
- 43. Bass J., Takahashi J. S. (2010) Circadian integration of metabolism and energetics. Science 330, 1349–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim E. Y., Jeong E. H., Park S., Jeong H. J., Edery I., Cho J. W. (2012) A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 26, 490–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ma Y. T., Luo H., Guan W. J., Zhang H., Chen C., Wang Z., Li J. D. (2013) O-GlcNAcylation of BMAL1 regulates circadian rhythms in NIH3T3 fibroblasts. Biochem. Biophys. Res. Commun. 431, 382–387 [DOI] [PubMed] [Google Scholar]
- 46. Li M. D., Ruan H. B., Hughes M. E., Lee J. S., Singh J. P., Jones S. P., Nitabach M. N., Yang X. (2013) O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 17, 303–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kaasik K., Kivimae S., Allen J. J., Chalkley R. J., Huang Y., Baer K., Kissel H., Burlingame A. L., Shokat K. M., Ptacek L. J., Fu Y. H. (2013) Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 17, 291–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Postic C., Dentin R., Denechaud P. D., Girard J. (2007) ChREBP, a transcriptional regulator of glucose and lipid metabolism. Annu. Rev. Nutr. 27, 179–192 [DOI] [PubMed] [Google Scholar]
- 49. Guinez C., Filhoulaud G., Rayah-Benhamed F., Marmier S., Dubuquoy C., Dentin R., Moldes M., Burnol A. F., Yang X., Lefebvre T., Girard J., Postic C. (2011) O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes 60, 1399–1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ido-Kitamura Y., Sasaki T., Kobayashi M., Kim H. J., Lee Y. S., Kikuchi O., Yokota-Hashimoto H., Iizuka K., Accili D., Kitamura T. (2012) Hepatic FoxO1 integrates glucose utilization and lipid synthesis through regulation of Chrebp O-glycosylation. PLoS One 7, e47231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim S., Coulombe P. A. (2007) Intermediate filament scaffolds fulfill mechanical, organizational, and signaling functions in the cytoplasm. Genes Dev. 21, 1581–1597 [DOI] [PubMed] [Google Scholar]
- 52. Omary M. B., Ku N. O., Tao G. Z., Toivola D. M., Liao J. (2006) “Heads and tails” of intermediate filament phosphorylation: multiple sites and functional insights. Trends Biochem. Sci. 31, 383–394 [DOI] [PubMed] [Google Scholar]
- 53. Srikanth B., Vaidya M. M., Kalraiya R. D. (2010) O-GlcNAcylation determines the solubility, filament organization, and stability of keratins 8 and 18. J. Biol. Chem. 285, 34062–34071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shrikhande G. V., Scali S. T., da Silva C. G., Damrauer S. M., Csizmadia E., Putheti P., Matthey M., Arjoon R., Patel R., Siracuse J. J., Maccariello E. R., Andersen N. D., Monahan T., Peterson C., Essayagh S., Studer P., Guedes R. P., Kocher O., Usheva A., Veves A., Kaczmarek E., Ferran C. (2010) O-glycosylation regulates ubiquitination and degradation of the anti-inflammatory protein A20 to accelerate atherosclerosis in diabetic ApoE-null mice. PLoS One 5, e14240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Olivier-Van Stichelen S., Guinez C., Mir A. M., Perez-Cervera Y., Liu C., Michalski J. C., Lefebvre T. (2012) The hexosamine biosynthetic pathway and O-GlcNAcylation drive the expression of beta-catenin and cell proliferation. Am. J. Physiol. Endocrinol. Metab. 302, E417–E424 [DOI] [PubMed] [Google Scholar]
- 56. Datta R., Choudhury P., Ghosh A., Datta B. (2003) A glycosylation site, 60SGTS63, of p67 is required for its ability to regulate the phosphorylation and activity of eukaryotic initiation factor 2alpha. Biochemistry 42, 5453–5460 [DOI] [PubMed] [Google Scholar]
- 57. Caldwell S. A., Jackson S. R., Shahriari K. S., Lynch T. P., Sethi G., Walker S., Vosseller K., Reginato M. J. (2010) Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene 29, 2831–2842 [DOI] [PubMed] [Google Scholar]
- 58. Hu P., Berkowitz P., Madden V. J., Rubenstein D. S. (2006) Stabilization of plakoglobin and enhanced keratinocyte cell-cell adhesion by intracellular O-glycosylation. J. Biol. Chem. 281, 12786–12791 [DOI] [PubMed] [Google Scholar]
- 59. Han I., Kudlow J. E. (1997) Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol. Cell. Biol. 17, 2550–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kim H. S., Woo J. S., Joo H. J., Moon W. K. (2012) Cardiac transcription factor Nkx2.5 is downregulated under excessive O-GlcNAcylation condition. PLoS One 7, e38053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lafontaine-Lacasse M., Dore G., Picard F. (2011) Hexosamines stimulate apoptosis by altering SIRT1 action and levels in rodent pancreatic beta-cells. J. Endocrinol. 208, 41–49 [DOI] [PubMed] [Google Scholar]
- 62. Weake V. M., Workman J. L. (2008) Histone ubiquitination: triggering gene activity. Mol. Cell 29, 653–663 [DOI] [PubMed] [Google Scholar]
- 63. Sakabe K., Wang Z., Hart G. W. (2010) Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. U.S.A. 107, 19915–19920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fujiki R., Hashiba W., Sekine H., Yokoyama A., Chikanishi T., Ito S., Imai Y., Kim J., He H. H., Igarashi K., Kanno J., Ohtake F., Kitagawa H., Roeder R. G., Brown M., Kato S. (2011) GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 480, 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang S., Roche K., Nasheuer H. P., Lowndes N. F. (2011) Modification of histones by sugar beta-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J. Biol. Chem. 286, 37483–37495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fong J. J., Nguyen B. L., Bridger R., Medrano E. E., Wells L., Pan S., Sifers R. N. (2012) beta-N-acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J. Biol. Chem. 287, 12195–12203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen Q., Chen Y., Bian C., Fujiki R., Yu X. (2013) TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guinez C., Mir A. M., Dehennaut V., Cacan R., Harduin-Lepers A., Michalski J. C., Lefebvre T. (2008) Protein ubiquitination is modulated by O-GlcNAc glycosylation. FASEB J. 22, 2901–2911 [DOI] [PubMed] [Google Scholar]
- 69. Sumegi M., Hunyadi-Gulyas E., Medzihradszky K. F., Udvardy A. (2003) 26S proteasome subunits are O-linked N-acetylglucosamine-modified in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 312, 1284–1289 [DOI] [PubMed] [Google Scholar]
- 70. Zhang F., Su K., Yang X., Bowe D. B., Paterson A. J., Kudlow J. E. (2003) O-GlcNAc modification is an endogenous inhibitor of the proteasome. Cell 115, 715–725 [DOI] [PubMed] [Google Scholar]
- 71. Keembiyehetty C. N., Krzeslak A., Love D. C., Hanover J. A. (2011) A lipid-droplet-targeted O-GlcNAcase isoform is a key regulator of the proteasome. J. Cell Sci. 124, 2851–2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Zhang F., Paterson A. J., Huang P., Wang K., Kudlow J. E. (2007) Metabolic control of proteasome function. Physiology (Bethesda) 22, 373–379 [DOI] [PubMed] [Google Scholar]
- 73. Ruan H. B., Singh J. P., Li M. D., Wu J., Yang X. (2013) Cracking the O-GlcNAc code in metabolism. Trends Endocrinol. Metab. 24, 301–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Deng H. X., Chen W., Hong S. T., Boycott K. M., Gorrie G. H., Siddique N., Yang Y., Fecto F., Shi Y., Zhai H., Jiang H., Hirano M., Rampersaud E., Jansen G. H., Donkervoort S., Bigio E. H., Brooks B. R., Ajroud K., Sufit R. L., Haines J. L., Mugnaini E., Pericak-Vance M. A., Siddique T. (2011) Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Huang Q., Figueiredo-Pereira M. E. (2010) Ubiquitin/proteasome pathway impairment in neurodegeneration: therapeutic implications. Apoptosis 15, 1292–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dennissen F. J., Kholod N., van Leeuwen F. W. (2012) The ubiquitin proteasome system in neurodegenerative diseases: culprit, accomplice or victim? Prog. Neurobiol. 96, 190–207 [DOI] [PubMed] [Google Scholar]
- 77. Gong B., Kielar C., Morton A. J. (2012) Temporal separation of aggregation and ubiquitination during early inclusion formation in transgenic mice carrying the Huntington's disease mutation. PLoS One 7, e41450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Gong C. X., Liu F., Iqbal K. (2012) O-GlcNAc cycling modulates neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 109, 17319–17320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Shan X., Vocadlo D. J., Krieger C. (2012) Reduced protein O-glycosylation in the nervous system of the mutant SOD1 transgenic mouse model of amyotrophic lateral sclerosis. Neurosci. Lett. 516, 296–301 [DOI] [PubMed] [Google Scholar]
- 80. Yuzwa S. A., Shan X., Macauley M. S., Clark T., Skorobogatko Y., Vosseller K., Vocadlo D. J. (2012) Increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation. Nat. Chem. Biol. 8, 393–399 [DOI] [PubMed] [Google Scholar]
- 81. Wang P., Lazarus B. D., Forsythe M. E., Love D. C., Krause M. W., Hanover J. A. (2012) O-GlcNAc cycling mutants modulate proteotoxicity in Caenorhabditis elegans models of human neurodegenerative diseases. Proc. Natl. Acad. Sci. U.S.A. 109, 17669–17674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Fulda S., Rajalingam K., Dikic I. (2012) Ubiquitylation in immune disorders and cancer: from molecular mechanisms to therapeutic implications. EMBO Mol. Med. 4, 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Dey A., Seshasayee D., Noubade R., French D. M., Liu J., Chaurushiya M. S., Kirkpatrick D. S., Pham V. C., Lill J. R., Bakalarski C. E., Wu J., Phu L., Katavolos P., LaFave L. M., Abdel-Wahab O., Modrusan Z., Seshagiri S., Dong K., Lin Z., Balazs M., Suriben R., Newton K., Hymowitz S., Garcia-Manero G., Martin F., Levine R. L., Dixit V. M. (2012) Loss of the tumor suppressor BAP1 causes myeloid transformation. Science 337, 1541–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Slawson C., Hart G. W. (2011) O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer 11, 678–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Li W., Bengtson M. H., Ulbrich A., Matsuda A., Reddy V. A., Orth A., Chanda S. K., Batalov S., Joazeiro C. A. (2008) Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One 3, e1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nijman S. M., Luna-Vargas M. P., Velds A., Brummelkamp T. R., Dirac A. M., Sixma T. K., Bernards R. (2005) A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 [DOI] [PubMed] [Google Scholar]
- 87. Drougat L., Olivier-Van Stichelen S., Mortuaire M., Foulquier F., Lacoste A. S., Michalski J. C., Lefebvre T., Vercoutter-Edouart A. S. (2012) Characterization of O-GlcNAc cycling and proteomic identification of differentially O-GlcNAcylated proteins during G1/S transition. Biochim. Biophys. Acta 1820, 1839–1848 [DOI] [PubMed] [Google Scholar]
- 88. Sprung R., Nandi A., Chen Y., Kim S. C., Barma D., Falck J. R., Zhao Y. (2005) Tagging-via-substrate strategy for probing O-GlcNAc modified proteins. J. Proteome Res. 4, 950–957 [DOI] [PubMed] [Google Scholar]
- 89. Hahne H., Sobotzki N., Nyberg T., Helm D., Borodkin V. S., van Aalten D. M., Agnew B., Kuster B. (2013) Proteome wide purification and identification of O-GlcNAc-modified proteins using click chemistry and mass spectrometry. J. Proteome Res. 12, 927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., Burlingame A. L. (2012) Global identification and characterization of both O-GlcNAcylation andphosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Alfaro J. F., Gong C. X., Monroe M. E., Aldrich J. T., Clauss T. R., Purvine S. O., Wang Z., Camp D. G., 2nd, Shabanowitz J., Stanley P., Hart G. W., Hunt D. F., Yang F., Smith R. D. (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U.S.A. 109, 7280–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Clark P. M., Dweck J. F., Mason D. E., Hart C. R., Buck S. B., Peters E. C., Agnew B. J., Hsieh-Wilson L. C. (2008) Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J. Am. Chem. Soc. 130, 11576–11577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Lefebvre T., Pinte S., Guerardel C., Deltour S., Martin-Soudant N., Slomianny M. C., Michalski J. C., Leprince D. (2004) The tumor suppressor HIC1 (hypermethylated in cancer 1) is O-GlcNAc glycosylated. Eur. J. Biochem. 271, 3843–3854 [DOI] [PubMed] [Google Scholar]
- 94. Zaro B. W., Yang Y. Y., Hang H. C., Pratt M. R. (2011) Chemical reporters for fluorescent detection and identification of O-GlcNAc-modified proteins reveal glycosylation of the ubiquitin ligase NEDD4–1. Proc. Natl. Acad. Sci. U.S.A. 108, 8146–8151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hahne H., Moghaddas Gholami A., Kuster B. (2012) Discovery of O-GlcNAc-modified proteins in published large-scale proteome data. Mol. Cell. Proteomics 11, 843–850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Teo C. F., Ingale S., Wolfert M. A., Elsayed G. A., Not L. G., Chatham J. C., Wells L., Boons G. J. (2010) Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat. Chem. Biol. 6, 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Nandi A., Sprung R., Barma D. K., Zhao Y., Kim S. C., Falck J. R., Zhao Y. (2006) Global identification of O-GlcNAc-modified proteins. Anal. Chem. 78, 452–458 [DOI] [PubMed] [Google Scholar]


