Abstract
The ubiquitin system is essential for the maintenance of proper protein homeostasis function across eukaryotic species. Although the general enzymatic architecture for adding and removing ubiquitin from substrates is well defined, methods for the comprehensive investigation of cellular ubiquitylation targets have just started to emerge. Recent advances in ubiquitin-modified peptide enrichment have greatly increased the number of identified endogenous ubiquitylation targets, as well as the number of sites of ubiquitin attachment within these substrates. Herein we evaluate current strategies using mass-spectrometry-based proteomics to characterize ubiquitin and ubiquitin-like modifications. Using existing data, we describe the characteristics of the ubiquitin-modified proteome and discuss strategies for the biological interpretation of existing and future ubiquitin-based proteomic studies.
Ubiquitin modification of cellular proteins is a highly conserved pathway catalyzed by a hierarchical system involving ubiquitin activating (E1), ubiquitin conjugating (E2), and ubiquitin ligase (E3) enzymes (1). Deubiquitylating enzymes (DUBs)1 oppose the effects of ubiquitylation by removing ubiquitin modifications on substrates. Ubiquitin can be covalently conjugated to substrates in several ways: a single ubiquitin can be conjugated to a single site (monoubiquitylation), to multiple sites (multiple monoubiquitylation), or as a polymeric chain (polyubiquitylation). Ubiquitin can form various isopeptide linkages with itself by utilizing one of seven internal lysine residues, as well as linear ubiquitin chains through head-to-tail attachment of ubiquitin, allowing for a diversity of chain topologies (1, 2). Since its initial discovery, there has been an increasingly sophisticated mechanistic understanding of the enzymology within the ubiquitin system. Yet there is still much about the system that remains elusive. How ubiquitylation of the diverse array of substrates is precisely controlled on both cellular and enzymatic levels remains a central question. In addition, relatively few E3s and DUBs have been matched with their substrates. It is also unclear how the ubiquitin system determines which specific lysine residues are targeted, and so far no consensus sequence that determines this specificity has been identified. Pinpointing the lysine residue(s) used for ubiquitin conjugation is essential for the molecular understanding of ubiquitylation, and until the recent development of antibody-based peptide enrichment strategies, our knowledge of specific lysine ubiquitylation sites was limited.
Protein-based Ubiquitin Proteomics
The need for the development of sensitive and reliable methods for monitoring spatial and temporal patterns of ubiquitylation in vivo has driven innovation in the field of mass spectrometry (MS)-based proteomics (3–5). MS is an ideal platform for studying protein modifications such as ubiquitylation because it allows for the simultaneous identification of endogenous ubiquitylation substrates and the precise lysine residue that is modified (6). Ubiquitylation of a substrate lysine ablates its recognition by trypsin and results in the cleavage of the attached ubiquitin after residue R74, generating signature peptides with a Gly-Gly tag (GG or diGLY) on modified lysine residues. Ubiquitin profiling was first pioneered with the expression of His- or His/biotin-tagged ubiquitin in cells allowing for the purification of target proteins by means of Ni-chelate chromatography under denaturing conditions (6–8) (Fig. 1A). Other epitope tags have been used to purify ubiquitylated proteins, including Myc, HA, and FLAG-tags, but the purification of these tagged proteins is more challenging using denaturing conditions (9, 10). It is important to note that exogenous expression of epitope-tagged ubiquitin might subvert endogenous ubiquitin-modification pathways, resulting in the modification of non-physiological substrates. For example, overexpression of the ubiquitin-like protein NEDD8 can result in spurious activation by the ubiquitin E1 enzyme to modify proteins with NEDD8 that are normally modified with ubiquitin (11). Moreover, expressing tagged ubiquitin in animal tissues or pathological specimens can be difficult or unfeasible.
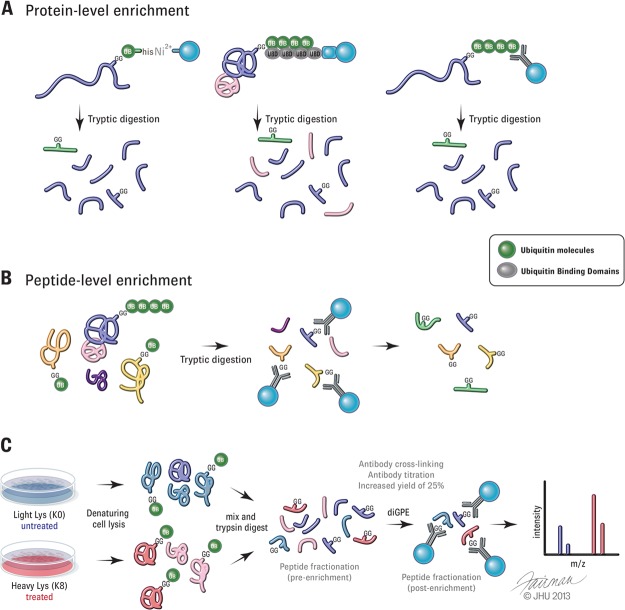
Fig. 1.
Strategies to isolate ubiquitylated substrates. A, protein-level enrichment of ubiquitylated proteins by purification of tagged ubiquitin (left), tandem ubiquitin binding domains (middle), or ubiquitin antibodies (right). B, schematic depicting peptide-level enrichment with diGLY remnant antibodies (diGPE). C, outline of SILAC-based proteomic approach for quantification of ubiquitylation sites using diGPE and subsequent analysis via MS. Text in the gray highlight technical improvements that have been shown to increase enrichment yield and specificity, including antibody usage (30) and fractionation of peptides either before or after immunoenrichment (26, 27, 29–32).
Methods that do not rely on ubiquitin overexpression include the use of tandem repeats of ubiquitin binding domains, which can be used as an affinity agent to isolate ubiquitylated substrates (12, 13) (Fig. 1A). Because these enrichments are not performed in denaturing conditions, analysis is complicated by co-purifying contaminants. Ubiquitin antibodies have also been used to isolate endogenous ubiquitylated substrates (14, 15) (Fig. 1A). However, antibody-based approaches have been complicated by differences in the affinity of antibodies toward different forms of ubiquitin. Recently, several linkage-specific antibodies have been developed (16–19). Whether or not these antibodies can efficiently profile linkage-specific substrates has not been demonstrated. Moreover, substrates may be modified by more than one type of polyubiquitin chain, complicating analysis (20).
As previous studies indicate that only a small portion of the total protein population is ubiquitylated at any given time on one or a few lysines, more than 98% of identified peptides from protein-level enrichments do not contain signature diGLY-modified peptides, making it difficult to determine the sites of modification (10). There is also uncertainty as to whether unmodified peptides are derived from the non-ubiquitylated portion of the protein or from co-precipitated non-ubiquitin-modified proteins.
diGLY-modified Peptide Enrichment
A number of studies have recently described methods utilizing a single-step immunoenrichment of diGLY-modified peptides. Also known as ubiquitin remnant profiling or diGLY-modified peptide enrichment (herein called diGPE), this method relies on antibodies that recognize the diGLY remnant (21) (Fig. 1B). Several recent studies have profiled the ubiquitin-modified proteome using two different commercially available antibodies (21–32). Antibody preference for certain amino acids adjacent to the diGLY-modified lysines has been reported, suggesting that the commonly used monoclonal antibodies might be biased to specific sequences (3, 5, 22, 31, 32). Although each antibody does enrich distinct subsets of diGLY sequences, overlapping sets of modified peptides were identified when used side by side, indicating that both antibodies recognize modified lysines within a range of sequence contexts (32). A mixture of diGLY remnant antibodies might prove to be an effective approach for profiling ubiquitylated substrates, and complementary use has been shown to increase the depth of ubiquitin site coverage (32).
In contrast to protein-level enrichment, through which up to 750 unique ubiquitylation sites have been identified in a single study (10), direct enrichment of diGLY-modified peptides from cellular lysates results in the identification of thousands of unique ubiquitylation sites (22, 24, 26, 27, 29–32). The increased depth and sensitivity achieved when using diGPE have allowed for the detection of known low-abundance ubiquitin-modified proteins, but like most MS-based approaches, this approach is biased toward more abundant proteins. Proteasome inhibitors have been utilized in several studies to improve the detection of potentially low-abundance labile substrates (22–25, 28–31). DUB inhibitors can also be used to augment protein ubiquitylation and increase substrate detection; however, proteasome impairment was found to be a more effective approach for increasing levels of ubiquitylation sites (29, 30). Nonetheless, these studies demonstrate that pharmacological DUB inhibition results in the widespread accumulation of substrates, similar to proteasome inhibition, which is contrary to what might be predicted upon DUB inhibition. This is because many studies have reported a decrease in substrate abundance upon RNAi-based knockdown of individual DUBs (33–37). This discrepancy may be explained by the fact that although knockdown results in protein depletion, an inhibitor inactivates the enzyme, possibly allowing for sustained substrate binding and possible sequestration. Also, typical knockdown experiments are done over multiple days, leading to possible compensatory mechanisms of ubiquitin-dependent protein turnover not seen with acute inhibition. Finally, broad-specificity DUB inhibitors inactivate proteasome-associated DUBs that are essential for substrate degradation and can lead to impaired rates of substrate proteolysis. These differences between knockdown and inhibitor-based studies raise questions about the cellular response to commonly used, broad-based DUB inhibitors.
A complicating issue with regard to the diGPE approach is that tryptic digestion of substrates modified by the ubiquitin-like proteins NEDD8 and ISG15 generates diGLY signatures indistinguishable from those of ubiquitin. Early experiments using multiple approaches found that no more than 6% of identified diGLY peptides resulted from neddylation, at least in the cell type utilized for those studies (22). Because the ubiquitin E1 enzyme UBA1 is capable of charging NEDD8 (38), one should take caution when interpreting data under conditions where the ubiquitin pool is depleted, as it is possible that NEDD8 may substitute for ubiquitin to modify proteins (22).
Another drawback observed when using trypsin-based ubiquitin proteomics is that proteolytic digestion results in a loss of structural information that distinguishes the different ubiquitin chain topologies. The use of antibodies recognizing specific ubiquitin linkages or tandem ubiquitin binding domains followed by diGPE might be able to enrich peptides modified with specific chain types. Also, determination of whether diGLY modifications of nonadjacent lysines on a single protein co-exist or represent modifications on separate members of a population is not feasible with diGPE.
One challenge associated with peptide-based enrichments is the possibility that some ubiquitylation sites might be analytically inaccessible when using trypsin-based proteomics. In such cases, utilizing alternative digestion strategies might increase the coverage of diGLY-modified sites. Protein-level enrichments in combination with diGPE might ensure the highest degree of sequence coverage when investigating ubiquitylation of an individual protein (25). Harper and colleagues used affinity-purification MS to validate PARKIN-dependent ubiquitylated proteins found using diGPE strategies (28). Indeed, several proteins found to associate with PARKIN were found to be ubiquitylated in a PARKIN-dependent manner.
Despite its shortcomings, diGPE has proven to be a powerful approach with the ability to map thousands of ubiquitin-modified lysines in a single experiment. Global profiling of site-specific ubiquitylation changes under different biological conditions is now possible when coupled with stable isotope labeling by amino acids in cell culture (SILAC) (22–25, 27–31) (Fig. 1C). The ability to quantitatively monitor protein ubiquitylation at the site level will pave the way for a more comprehensive understanding of how ubiquitin-mediated signaling orchestrates a variety of biological processes.
Technical Improvements to diGPE
Initially, large amounts of starting material of up to 35 mg and/or multiple experimental replicates were needed in order to achieve large numbers of diGLY-modified peptides in a single experiment (22, 31). However, recent technical advancements have improved the depth of ubiquitylation site coverage (Fig. 1C). Improvements in antibody usage, including chemical cross-linking of the diGLY antibody to beads prior to immunoprecipitation and the optimization of antibody-to-input lysate ratios, have been shown to increase enrichment yield and specificity (30). Crosslinking the antibody to beads reduced contaminating proteins, likely from the antibody itself, and increased the detection of diGLY-modified peptides by ∼25% (30). In addition, reducing complexity via the fractionation of peptides either before or after immunoaffinity enrichment has been found to increase the number of observed diGLY-modified peptides obtained. Work from the both the Peng and Carr groups reported an increase in diGLY peptide yields with pre-immunoaffinity fractionation of samples using strong cation exchange chromatography (SCX) (26, 29). Applying SCX fractionation to rat brain lysates prior to immunoprecipitation, Peng and colleagues reported an almost 2-fold improvement in unique diGLY peptides detected relative to direct enrichment (1769 unique peptides versus 915, respectively, with 20 mg of starting material). Carr and colleagues saw similar enhancements in the detection of diGLY species when they employed SCX fractionation on SILAC triple-encoded Jurkat cell samples. Starting with only 5 mg of protein per SILAC state, limited fractionation of samples into four SCX fractions prior to enrichment enabled the identification of up to 3300 unique diGLY-containing peptides in a single experiment. This is over a 3-fold increase in detected unique diGLY peptides over parallel experiments using only immunoaffinity enrichment (29). More recently, the Carr group used basic pH reverse-phase chromatography as a fractionation method prior to diGLY enrichment (30). As this method does not require sample desalting prior to analysis, sample handling is simplified, resulting in improved peptide recovery. When this fractionation method was used on triple-encoded SILAC samples, over 20,000 distinct diGLY sites were identified in a single replicate (30). However, the increased depth of ubiquitin site coverage seen cannot be completely attributed to the fractionation method, as other improvements were utilized, including antibody optimization and advanced MS and liquid chromatography conditions. Isoelectric focusing and SCX fractionation performed after diGPE have led to the identification of several thousand unique ubiquitylation sites in a single experiment (27, 31, 32). It is difficult to determine the degree of improvement with post-immunoprecipitation fractionation because none of the studies utilizing this method reported parallel experiments without fractionation. Additionally, because of variances in experimental setup, yields obtained from different studies cannot be directly compared, and one cannot generate conclusions on overall improvement with regard to sensitivity. Although both pre- and post-fractionation methods recover large numbers of ubiquitylation sites, this recovery comes at a cost of increased sample handling and instrument time needed for data acquisition. Fractionation post-diGPE has the advantage of saving reagent costs, as only one immunoprecipitation reaction is needed per condition.
Alterations and advancements in peptide and protein separation techniques prior to and after diGPE have led to deeper descriptions of the ubiquitin-modified proteome from increasingly smaller amounts of material. However, current techniques will need continued refinement to be able to detect and measure endogenous ubiquitylation events on a scale compatible with conditions in which the sample amount is limiting, such as human biopsy samples.
Ubiquitylation Promiscuity
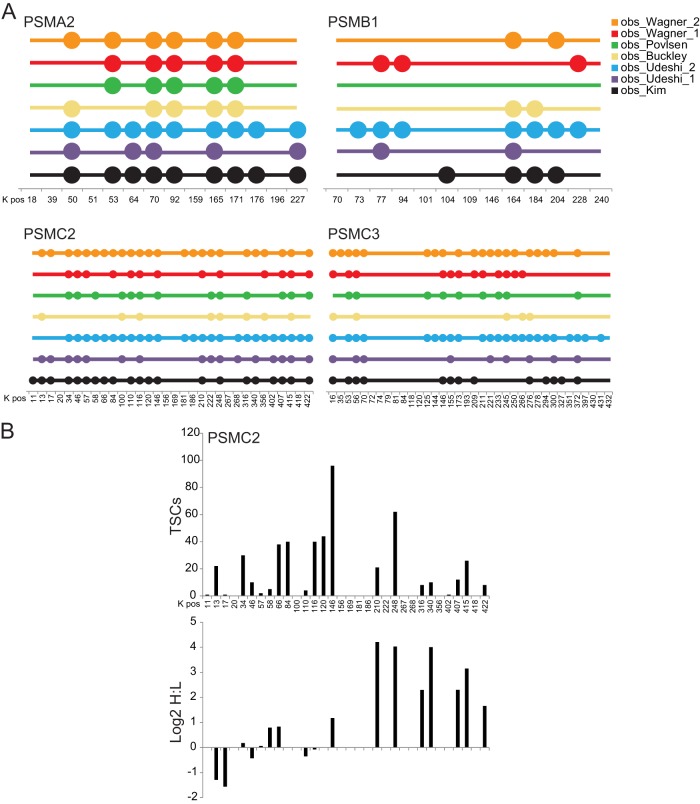
Technological advancements in both the sensitivity and the speed of mass spectrometers coupled with the development of diGPE strategies have led to a landslide of data documenting endogenous ubiquitylation events on a wide swath of the proteome. One of the large advantages realized upon the implementation of diGPE approaches is the ability to easily catalogue specific ubiquitin-modified lysines within a single protein or protein group. Inspection of the growing lists of ubiquitin-modified peptides from a collection of recent studies reveals a striking degree of promiscuity with regard to which lysine residues within proteins are utilized as ubiquitin-acceptor sites. It is common to observe more than five different modified lysines within a single protein, and some larger proteins contain dozens of modified lysines. It has been demonstrated that there is little correlation between overall protein abundance and the ability to detect ubiquitylated peptides, arguing that highly expressed proteins are not, by default, highly ubiquitylated (22). However, some highly expressed proteins, like the subunits of the proteasome, contain many modified lysines that have been observed in numerous studies (22, 23, 27, 29–32) (Fig. 2A). Within four separate proteasomal core subunits, 75% of all possible lysines have been observed to be ubiquitin-modified (Fig. 2A). It is likely that a portion of the remaining lysines reside in peptides that are difficult to detect via trypsin-based proteomic methods. Despite the high degree of site heterogeneity, there exists some specificity as to how the abundance of each modified site is altered in response to proteasome inhibition, suggesting a functional divergence of ubiquitin modifications within a single protein population (Fig. 2B). In the case of the 19S proteasomal subunit PSMC2, we observed 21 of the possible 32 lysine residues to be diGLY-modified. The spectral counts, which can be used to infer cellular abundance, vary from a single observation (K402) to those observed more than 50 times (K146, K248) across multiple studies (Figs. 2A, 2B). How each individual site is altered in response to proteasome inhibition cannot be predicted from the abundance of each modified lysine in untreated cells. This is exemplified by K13 and K17 in PSMC2, whose diGLY modification decreased in abundance after proteasome inhibition, whereas other similarly abundant modified peptides (K34, K116) were unaltered in response to proteasome inhibition (Fig. 2B). This indicates that modifications on distinct lysine residues within a single protein may result in different functional outcomes for that modified population.
Fig. 2.
Ubiquitin-modified site heterogeneity. A, the position of each lysine residue for the indicated proteins is indicated on the x-axis. Each colored circle signifies that the lysine indicated was observed to be ubiquitin-modified within the studies of Kim et al. (black) (22), Wagner et al. (red) (31), Povlsen et al. (green) (27), Udeshi et al. (purple) (29), Udeshi et al. (blue) (30), Wagner et al. (orange) (32), and Buckley et al. (yellow) (23). B, total spectral counts (TSCs; top) and the log2 SILAC ratio (heavy (H) = treated with 1 μm bortezimib; light (L) = untreated) for all lysine residues within PSMC2.The number indicates the position of the lysine residue, starting with the initiator methionine. All primary data used for this figure were extracted from the published diGLY-modification data set of Kim et al. (22).
Functional Redundancy of Ubiquitylated Lysines
The large number of proteins within the proteome that have now been demonstrated to be targets of ubiquitin modification suggests that ubiquitylation might be a common event on nearly every protein within the proteome, similar to what is now appreciated with protein phosphorylation. Where ubiquitylation and phosphorylation differ is that the sites of phosphorylation seem to be more conserved across species than ubiquitin-modified lysines. A careful examination of ubiquitin-modified lysine conservation by Krogan and colleagues revealed little evolutionary constraint of acceptor lysines, which supports previous observations (39, 40). The separation of modified lysines into those within structured and unstructured domains revealed some differences in the degree of conservation (40). Further separation of the data into functional categories (i.e. degradation versus signaling or protein–protein interaction) might reveal that sites used for non-degradative purposes are more conserved than others. There are several possible reasons that might underlie this lack of ubiquitin-modified lysine conservation. Functional redundancy of ubiquitin acceptor lysines might be one explanation. This is supported by many studies, both published and unpublished, in which the mutation of a single ubiquitin-modified lysine had little effect on either protein half-life or ubiquitylation (41, 42). This functional redundancy might be the result of multiple ligases directing the ubiquitylation of distinct lysines within a single protein. However, in some instances mutation of a single lysine has been demonstrated to block ubiquitylation (43, 44). Site-specificity may be imparted through structural mechanisms that limit the available lysines accessible to a bound ligase. One example of this might be cullin-RING ligase complexes that utilize phosphorylation-dependent degrons to target substrates for ubiquitylation (45). Another possible reason for the observed heterogeneity and lack of conservation of ubiquitylated lysines might be that the signal for degradation for some proteins might consist of multiple lysines that are simultaneously modified with a single ubiquitin moiety or short ubiquitin chains. In support of this, recent work has demonstrated that multiple monoubiquitylation events can serve as a degradation signal for both naturally occurring and model ubiquitin substrates (46–48). This idea is further substantiated by quantitative proteomic measurements of ubiquitin pools suggesting that overall chain lengths might be shorter than what is observed during in vitro ubiquitylation reactions (49, 50). It must be noted that these measurements represent cellular averages and that specific substrates may become modified with long chains during a single ubiquitin ligase-binding event. Indeed, differential substrate modification via processive or distributive mechanisms has been demonstrated to be a strategy utilized to achieve ordering of substrate ubiquitylation by the anaphase promoting complex (51). It remains possible that a large number of ubiquitin substrates utilize distributive signals across many lysine residues as a common degradation signal. An examination of how the abundance and utilization of individual modified lysine residues are altered upon the mutation of a single lysine residue will enable evaluation of possible distributive ubiquitylation mechanisms.
Ubiquitin-site Occupancy
The very fact that enrichment strategies are needed in order to observe and quantify endogenous ubiquitylation events argues that the ubiquitylated fraction of a given protein population is quite small. However, as much as 10% of the first demonstrated ubiquitin-modified substrate, histone H2A, has been observed to be monoubiquitylated (52, 53). Other monoubiquitylated substrates such as PCNA and histone H2B can be easily detected by immunoblotting with antibodies against the unmodified protein, suggesting that, at least for some proteins, the ubiquitin-modified fraction is relatively abundant (54). Further, some lysine residues appear to be more often utilized than others within a single protein, suggesting that individual site occupancy might differ depending on the functional outcome of particular ubiquitylation events (Fig. 2B). Determining ubiquitin-site occupancy may be further complicated by the fact that lysine residues are targets for a wide range of posttranslational modifications in vivo (5). Indeed, significant site-specific overlap was detected between lysine ubiquitylation and acetylation (10, 31). Conserved ubiquitylation sites might have lower occupancy because of acetylation-based protection of lysines from proteasome-dependent degradation (55, 56). Despite these observations, a careful examination and quantification of ubiquitin-site occupancy remains to be achieved for a vast majority of the ubiquitin-modified proteome.
Studies attempting to measure site occupancy for ubiquitin-modified lysines will be aided by previous studies in which phosphorylation site occupancies were determined using global proteomics techniques. One such study utilized a computational approach combining SILAC measurements of phosphopeptides, the corresponding unmodified peptide, and total protein to calculate site occupancies (57). Another approach integrated phosphatase treatment and quantitative SILAC-based proteomics to determine phosphorylation-site occupancy by comparing the nonphosphorylated form of a previously observed phosphopeptide with or without phosphatase treatment (58). Similar strategies to determine ubiquitin-site occupancy using nonspecific deubiquitylating enzymes to remove ubiquitin conjugates could be employed. Studies detailing ubiquitin-site occupancy might reveal that lysines with higher occupancy modifications are more conserved than other ubiquitylation events.
Specificity in Ubiquitylation-site Utilization
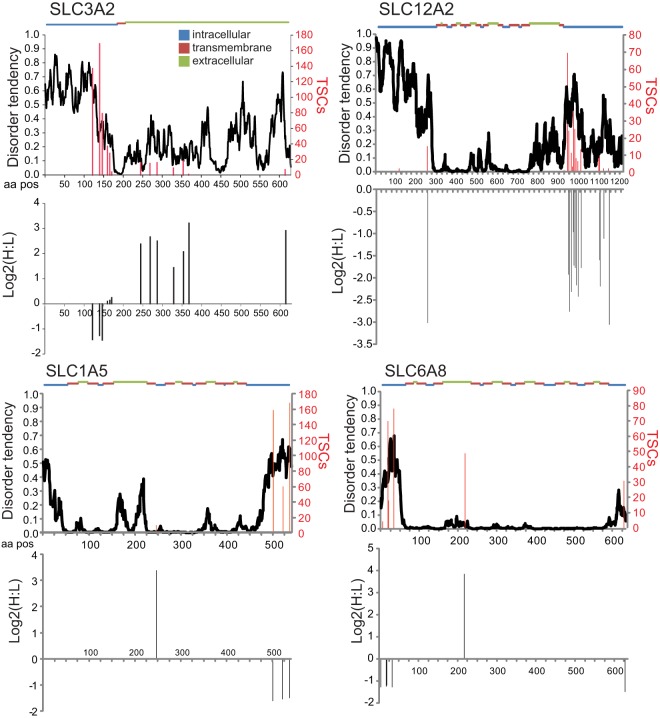
Layered within the heterogeneity of ubiquitin-site utilization is the observation that individual modified lysines within a single protein differ in both their overall abundance and how that abundance is altered upon proteasome inhibition. This is exemplified by examination of the observed sites of ubiquitylation within four different plasma-membrane-localized solute transporters (22) (Fig. 3). Using spectral counts as a proxy for abundance, it was found that the most abundant ubiquitylation sites for these proteins occur within regions that were predicted to be highly disordered. Modifications within predicted intracellular domains typically decreased in abundance upon inhibition of the proteasome, whereas the less often observed sites that occur within transmembrane or extracellular regions increased more than 4-fold upon proteasome inhibition. It has been previously demonstrated that some plasma-membrane proteins use monoubiquitylation as a mechanism to regulate their endocytosis and trafficking (59). This, coupled with the observation that most known monoubiquitylated proteins display a decrease in their ubiquitin modification levels in response to proteasome inhibition, makes it possible that the sites that decrease in abundance within the depicted proteins represent sites of monoubiquitylation (Fig. 3). However, because the diGPE method cannot discriminate between the various ubiquitin linkages formed at a particular site, it is possible that these sites are modified by polyubiquitylation in a manner that is reversed upon proteasome inhibition. Interestingly, the sites that increase in abundance after proteasome inhibition, especially those not predicted to sample the intracellular environment and thus should not be exposed to the cellular ubiquitylation machinery, might represent quality-control events. It is uncertain whether these ubiquitylation events occur at the plasma membrane and are characteristic of the recently described plasma membrane quality control pathway or are merely modified shortly after biosynthesis prior to delivery at the plasma membrane (60). Because of the possible dichotomous nature of ubiquitin modification within a single protein in response to proteasome inhibition, Western blotting of an averaged protein population would not be able to discriminate between ubiquitin-dependent mechanisms resulting in protein trafficking or degradation. Ubiquitin site-specific tools will need to be developed in order to properly evaluate the regulation of a particular ubiquitin-modified lysine. Investigations into phosphorylation-based modes of regulation were greatly aided by the development of phospho-specific antibodies that could be used to interrogate a specific modification within a variety of biological contexts. Although differences in both the chemical nature and the size of the ubiquitin modification represent a challenge for site-specific antibody development, a small number of ubiquitin-specific antibodies have already been developed that recognize the ubiquitylated populations of histones H2A and H2B (22, 61). It is also possible to develop quantitative proteomic methods to interrogate single or small groups of ubiquitylation events using multiplexed targeted approaches. However, the amount of sample required and the sensitivity of these types of assays usually are not favorable relative to assays utilizing site-specific antibodies. Careful biological analysis and the development of new analytical tools will be necessary in order to unravel the complexities of ubiquitylation that are emerging from the deluge of ubiquitin-modification proteomic data pouring in from global profiling studies.
Fig. 3.
Site specificity within proteins. Total spectral counts (TSCs; red lines) for specific ubiquitin-modified lysines are plotted against the linear peptide sequence for the indicated protein. The log2 SILAC ratios (heavy (H) = treated with 1 μm Bortezomib; light (L) = untreated) for the same ubiquitin-modified lysines are shown in the bottom half of the protein-specific plots. The disorder tendency (black line) as predicted by IUPred (71) and the predicted membrane topology are plotted along the protein sequence. All primary data used for this figure were extracted from the published diGLY-modification data set of Kim et al. (22).
Application of Ubiquitin Proteomics to Understand Biology
Although global proteomic profiling studies investigating and cataloging specific posttranslational modification events have had a large effect on a wide array of biological disciplines, the abundance of the data can be somewhat overwhelming to many biologists. Most are left wondering, “What does it all mean?” Initial investigations of the ubiquitin-modified proteome were somewhat surprising, as many known ubiquitylated substrates that have a short half-life were not observed, whereas a plethora of ubiquitylated peptides originating from highly abundant proteins such as ribosomal subunits, actin, and fatty acid synthase were readily observed and quantified in lysates generated from unperturbed cells (22, 23, 27, 29–32). The addition of proteasome inhibitors to cells can increase the number of identified ubiquitin-modified peptides up to 4-fold, aiding in the identification of low-abundance, short-lived proteins. However, care should be taken when interpreting data using proteasome inhibitors, as overt inhibition of the proteasome can have pleiotropic cellular consequences, including alterations in protein trafficking and translation.
The ubiquitin-modified proteome provides a snapshot of the unstable proteome, which is likely what most modified sites on the highly abundant proteins whose abundance increases in response to proteasome inhibition represent. In fact, most ubiquitin-modified peptides require continual protein translation to be observed, indicating that a large fraction of the unstable proteome arises shortly after or even concurrent with protein synthesis (22). Protein ubiquitylation is a highly dynamic process, and determining the origin and fate of a particular diGLY-modified protein likely requires intensive investigation far beyond a simple analysis of specific site dynamics in response to a biological stimulus. For example, a particular ubiquitin-modified protein might be abundantly detected because it is highly ubiquitylated (i.e. a large fraction of the total protein population is modified) or because it is a highly abundant protein in which a small portion is modified. Further, alterations within the modified population may arise from alterations in total protein levels or from differences in either E3 ligase or DUB activity in response to the chosen cellular perturbation. Although care must be taken when interpreting data from ubiquitin-profiling experiments, comparison of newly identified ubiquitin-modified proteins and how they are dynamically regulated to well-characterized ubiquitin substrates can assist in categorizing similarly behaved proteins for further study.
Application of the diGPE technology to investigate alterations in the ubiquitin-modified proteome upon specific cell stimuli or within a specific tissue type or cellular state will likely shed enormous light on how ubiquitin-dependent processes regulate various pathways. One recent study aimed at uncovering novel ubiquitin-dependent regulatory events in response to UV-based DNA damage exemplifies the use of the diGPE technology to investigate ubiquitin-dependent signaling under specific biological contexts (27). In this study, over 400 ubiquitin-modified sites were demonstrated to increase in abundance after UV exposure, including the well-characterized ubiquitylation on PCNA and FANCD2. Thorough follow-up studies characterized the function of ubiquitylation for a single substrate, PAF15. These studies were greatly aided by the fact that a substantial portion of PAF15 is modified by ubiquitin at steady state, allowing the use of antibodies against PAF15 to detect the ubiquitylated portion of PAF15 and to verify the loss of ubiquitylation upon UV irradiation. This study highlights the power of the diGPE approach to uncover new ubiquitin-dependent mechanisms that regulate specific biological processes.
Deciphering the biological implication of ubiquitin modification for many of the newly identified ubiquitylation substrates will require careful interrogation of the ubiquitylation dynamics for proteins of interest after perturbation of specific biological pathways. Further, combining quantitative diGLY profiling datasets with datasets utilizing global techniques to investigate protein turnover, such as the fluorescence-based global protein stability assay (62, 63) or proteomics-based heavy-label metabolic pulse-chase techniques (64), will assist in interpretation of the biological consequence of protein-specific ubiquitylation. It should be noted that one advantage of using diGLY proteomics to investigate protein turnover is that the diGLY-modified population of proteins is a distinct subpopulation that signifies the unstable (or at least modified) portion that might be overlooked when examining the turnover of an averaged protein population. Given such, the half-life of the ubiquitin-modified population might be quite distinct from the half-life of the total protein population (22). Because a large portion of the ubiquitin-modified proteome seemingly arises in close proximity to the translational machinery, fusion of ribosome-profiling techniques with diGLY profiling might be a particularly powerful approach for understanding the underlying principles governing co-translational ubiquitylation.
To gain a better understanding of the spatial and temporal control of ubiquitin networks, diGPE can be used in combination with techniques such as “spatial proteomics” (65) or ubiquitin binding domain–based fluorescence sensors (66). “Spatial proteomics” combines common fractionation techniques and SILAC to label the proteins in each cellular fraction with isotopes that can be distinguished via MS. This technique, when coupled to diGPE, would allow for characterization of dynamic changes in protein localization of ubiquitin-modified proteins in response to experimental perturbations. In addition, ubiquitin binding domain–based sensors have been shown to interfere with specific ubiquitin-mediated processes, such as the NFκB pathway (66), and when combined with diGPE one can measure alterations of the ubiquitin-modified proteome when linkage-specific pathways are perturbed.
Future Perspectives for Ubiquitin-based Proteomics
Diversification in ubiquitin signaling is accomplished at many levels, including the type and nature of the ubiquitin attachment to each substrate. As noted above, one large disadvantage of the peptide-based identification of ubiquitin substrates is that the required trypsin digestion of the cellular extract removes any information regarding the type of ubiquitin-chain linkage used on each substrate. Numerous studies have detailed the abundance of each chain linkage, some in exquisitely quantitative detail, but the number of substrates that utilize each chain linkage is completely unknown (20, 49, 67–70). With the exception of lysine-63 linked chains, all chain types can be utilized as in vivo proteasome degradation signals (1). Interestingly, in response to proteasome inhibition, ∼40% of quantified diGLY-modified peptides either decrease in abundance or are unchanged, arguing that a substantial fraction of the ubiquitin-modified proteome is modified by non-canonical, non-proteasomal targeting ubiquitin linkages (22). It is possible that this result can be explained by contributions from other ubiquitin-like proteins or a balance of DUB and ligase activity upon proteasome inhibition. However, it raises the possibility that monoubiquitylation and other non-degradative chains are more prevalent than previously anticipated. Combinations of linkage-specific antibodies or ubiquitin binding domains, the expression of ubiquitin-linkage mutants, and diGPE approaches will likely be necessary in order to decipher the ubiquitin-linkage code for each identified target.
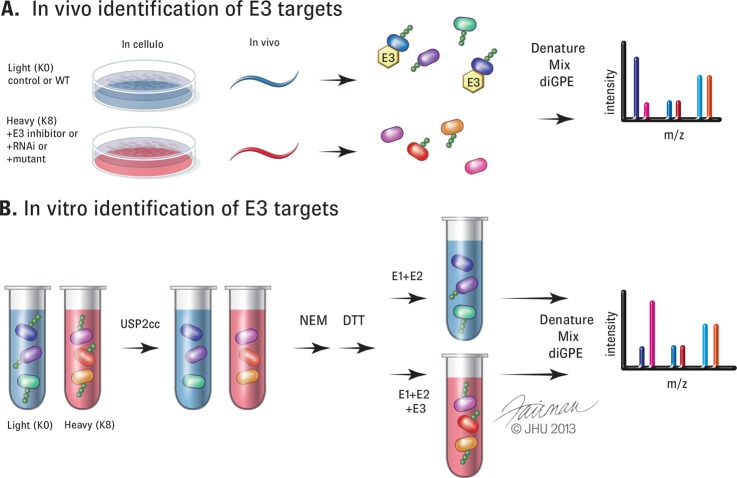
Now that detection of the ubiquitin-modified proteome has reached a certain level of depth and sophistication, it is possible to begin to decipher the substrate repertoire for individual ubiquitin-modifying enzymes (Fig. 4). The sheer diversity and heterogeneity of reported ubiquitylated substrates make identification of the relevant ubiquitin ligases a potentially difficult task. However, application of the diGPE strategy has been successfully utilized to identify putative substrates for either single ligases or a larger ligase family, the Cullin-RING ligases (22, 24, 25). For all studies to date, only a small percentage of candidate substrates were validated using orthogonal approaches. Further, in cases when substrate ubiquitylation does not result in overt changes in total protein abundance, validation at the site level, likely utilizing proteomic methods, might be required. Pharmacological compounds targeting individual DUBs or ligases with high specificity would greatly aid in substrate identification (Fig. 4A). Although there has been significant advancement in the development of small molecules that target ubiquitin-pathway components, it is likely that RNAi-based approaches would have to be implemented to reduce the levels of the DUB or ligase of interest prior to the application of diGPE strategies. A more robust approach might be to utilize either cell lines with inactivating deletions of the enzyme of interest or technologies aimed at the targeted somatic genomic deletion of a gene of interest in order to avoid complicating issues with off-target RNAi effects and inefficient knockdown (Fig. 4A). This type of approach was leveraged to identify PARKIN substrates upon mitochondrial membrane depolarization (28). In the study, the authors capitalized on the fact that HeLa cells contain little to no endogenous PARKIN to assist in the validation and prioritization of potential PARKIN substrates using quantitative diGPE approaches. Lastly, innovative in vitro approaches could be used to identify substrates for ligases, DUBs, and ubiquitin-binding-domain-containing proteins (Fig. 4B). To mitigate potential artifacts from in vitro additions of ubiquitin-modifying enzymes to cell extracts, careful validation of candidate substrates will be critical.
Fig. 4.
Utilization of diGPE to identify substrates of E3 ligases. A, schematic representation of in vivo strategies to identify substrates for E3 ligases. Inhibition of an E3 enzyme of interest (heavy (red)) by use of a pharmacological compound, RNA interference (RNAi), or genetic mutant in cells or an animal model (here represented by C. elegans) would decrease the abundance of diGLY peptides for the ligase substrate. B, schematic representation of in vitro strategies to identify E3 substrates. Treatment of non-denatured cellular lysates with USP2cc would strip ubiquitin modifications on all proteins. The addition of N-ethylmaleimide (NEM) to inhibit USP2cc activity would be followed by quenching with excess DTT. Recombinant E1 and E2 alone (light) or with active E3 (heavy) would be added to the respective lysates before denaturing, mixing, and diGPE. Increased diGLY peptides would be seen with the addition of active E3.
There remains much to be elucidated from the current datasets obtained using diGPE approaches. Continued refinement of current approaches and the development of more sensitive proteomic techniques will only improve the detection of ubiquitylated substrates, which will help us fully decipher the molecular mechanisms of ubiquitin-dependent biological processes.
Footnotes
1 The abbreviations used are:
- DUB
- deubiquitylating enzyme
- MS
- mass spectrometry
- SCX
- strong cation exchange chromatography
- SILAC
- stable isotope labeling by amino acids in cell culture
- diGPE
- diGLY-modified peptide enrichment
- TSCs
- total spectral counts
- NEM
- N-ethylmaleimide.
REFERENCES
- 1. Komander D., Rape M. (2012) The ubiquitin code. Annu. Rev. Biochem. 81, 203–229 [DOI] [PubMed] [Google Scholar]
- 2. Williamson A., Werner A., Rape M. (2013) The colossus of ubiquitylation: decrypting a cellular code. Mol. Cell. 49, 591–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bustos D., Bakalarski C. E., Yang Y., Peng J., Kirkpatrick D. S. (2012) Characterizing ubiquitination sites by peptide-based immunoaffinity enrichment. Mol. Cell. Proteomics 11, 1529–1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kirkpatrick D. S., Denison C., Gygi S. P. (2005) Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat. Cell. Biol. 7, 750–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sylvestersen K. B., Young C., Nielsen M. L. (2013) Advances in characterizing ubiquitylation sites by mass spectrometry. Curr. Opin. Chem. Biol. 17, 49–58 [DOI] [PubMed] [Google Scholar]
- 6. Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 7. Kirkpatrick D. S., Weldon S. F., Tsaprailis G., Liebler D. C., Gandolfi A. J. (2005) Proteomic identification of ubiquitinated proteins from human cells expressing His-tagged ubiquitin. Proteomics 5, 2104–2111 [DOI] [PubMed] [Google Scholar]
- 8. Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P. (2006) A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell. Proteomics 5, 737–748 [DOI] [PubMed] [Google Scholar]
- 9. Kim J. Y., Anderson E. D., Huynh W., Dey A., Ozato K. (2011) Proteomic survey of ubiquitin-linked nuclear proteins in interferon-stimulated macrophages. J. Interferon Cytokine Res. 31, 619–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Danielsen J. M., Sylvestersen K. B., Bekker-Jensen S., Szklarczyk D., Poulsen J. W., Horn H., Jensen L. J., Mailand N., Nielsen M. L. (2011) Mass spectrometric analysis of lysine ubiquitylation reveals promiscuity at site level. Mol. Cell. Proteomics 10, M110.003590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hjerpe R., Thomas Y., Kurz T. (2012) NEDD8 overexpression results in neddylation of ubiquitin substrates by the ubiquitin pathway. J. Mol. Biol. 421, 27–29 [DOI] [PubMed] [Google Scholar]
- 12. Hjerpe R., Aillet F., Lopitz-Otsoa F., Lang V., England P., Rodriguez M. S. (2009) Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 10, 1250–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shi Y., Chan D. W., Jung S. Y., Malovannaya A., Wang Y., Qin J. (2011) A data set of human endogenous protein ubiquitination sites. Mol. Cell. Proteomics 10, M110.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsumoto M., Hatakeyama S., Oyamada K., Oda Y., Nishimura T., Nakayama K. I. (2005) Large-scale analysis of the human ubiquitin-related proteome. Proteomics 5, 4145–4151 [DOI] [PubMed] [Google Scholar]
- 15. Vasilescu J., Smith J. C., Ethier M., Figeys D. (2005) Proteomic analysis of ubiquitinated proteins from human MCF-7 breast cancer cells by immunoaffinity purification and mass spectrometry. J. Proteome Res. 4, 2192–2200 [DOI] [PubMed] [Google Scholar]
- 16. Matsumoto M. L., Dong K. C., Yu C., Phu L., Gao X., Hannoush R. N., Hymowitz S. G., Kirkpatrick D. S., Dixit V. M., Kelley R. F. (2012) Engineering and structural characterization of a linear polyubiquitin-specific antibody. J. Mol. Biol. 418, 134–144 [DOI] [PubMed] [Google Scholar]
- 17. Matsumoto M. L., Wickliffe K. E., Dong K. C., Yu C., Bosanac I., Bustos D., Phu L., Kirkpatrick D. S., Hymowitz S. G., Rape M., Kelley R. F., Dixit V. M. (2010) K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol. Cell 39, 477–484 [DOI] [PubMed] [Google Scholar]
- 18. Newton K., Matsumoto M. L., Wertz I. E., Kirkpatrick D. S., Lill J. R., Tan J., Dugger D., Gordon N., Sidhu S. S., Fellouse F. A., Komuves L., French D. M., Ferrando R. E., Lam C., Compaan D., Yu C., Bosanac I., Hymowitz S. G., Kelley R. F., Dixit V. M. (2008) Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134, 668–678 [DOI] [PubMed] [Google Scholar]
- 19. Wang H., Matsuzawa A., Brown S. A., Zhou J., Guy C. S., Tseng P. H., Forbes K., Nicholson T. P., Sheppard P. W., Hacker H., Karin M., Vignali D. A. (2008) Analysis of nondegradative protein ubiquitylation with a monoclonal antibody specific for lysine-63-linked polyubiquitin. Proc. Natl. Acad. Sci. U.S.A. 105, 20197–20202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Phu L., Izrael-Tomasevic A., Matsumoto M. L., Bustos D., Dynek J. N., Fedorova A. V., Bakalarski C. E., Arnott D., Deshayes K., Dixit V. M., Kelley R. F., Vucic D., Kirkpatrick D. S. (2011) Improved quantitative mass spectrometry methods for characterizing complex ubiquitin signals. Mol. Cell. Proteomics 10, M110.003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu G., Paige J. S., Jaffrey S. R. (2010) Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., Sowa M. E., Rad R., Rush J., Comb M. J., Harper J. W., Gygi S. P. (2011) Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44, 325–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Buckley S. M., Aranda-Orgilles B., Strikoudis A., Apostolou E., Loizou E., Moran-Crusio K., Farnsworth C. L., Koller A. A., Dasgupta R., Silva J. C., Stadtfeld M., Hochedlinger K., Chen E. I., Aifantis I. (2012) Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell 11, 783–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Emanuele M. J., Elia A. E., Xu Q., Thoma C. R., Izhar L., Leng Y., Guo A., Chen Y. N., Rush J., Hsu P. W., Yen H. C., Elledge S. J. (2011) Global identification of modular cullin-RING ligase substrates. Cell 147, 459–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee K. A., Hammerle L. P., Andrews P. S., Stokes M. P., Mustelin T., Silva J. C., Black R. A., Doedens J. R. (2011) Ubiquitin ligase substrate identification through quantitative proteomics at both the protein and peptide levels. J. Biol. Chem. 286, 41530–41538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Na C. H., Jones D. R., Yang Y., Wang X., Xu Y., Peng J. (2012) Synaptic protein ubiquitination in rat brain revealed by antibody-based ubiquitome analysis. J. Proteome Res. 11, 4722–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Povlsen L. K., Beli P., Wagner S. A., Poulsen S. L., Sylvestersen K. B., Poulsen J. W., Nielsen M. L., Bekker-Jensen S., Mailand N., Choudhary C. (2012) Systems-wide analysis of ubiquitylation dynamics reveals a key role for PAF15 ubiquitylation in DNA-damage bypass. Nat. Cell. Biol. 14, 1089–1098 [DOI] [PubMed] [Google Scholar]
- 28. Sarraf S. A., Raman M., Guarani-Pereira V., Sowa M. E., Huttlin E. L., Gygi S. P., Harper J. W. (2013) Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Udeshi N. D., Mani D. R., Eisenhaure T., Mertins P., Jaffe J. D., Clauser K. R., Hacohen N., Carr S. A. (2012) Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol. Cell. Proteomics 11, 148–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Udeshi N. D., Svinkina T., Mertins P., Kuhn E., Mani D. R., Qiao J. W., Carr S. A. (2013) Refined preparation and use of anti-K-{epsilon}-GG antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol. Cell. Proteomics 12, 825–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteomics 10, M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wagner S. A., Beli P., Weinert B. T., Scholz C., Kelstrup C. D., Young C., Nielsen M. L., Olsen J. V., Brakebusch C., Choudhary C. (2012) Proteomic analyses reveal divergent ubiquitylation site patterns in murine tissues. Mol. Cell. Proteomics 11, 1578–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goncharov T., Niessen K., de Almagro M. C., Izrael-Tomasevic A., Fedorova A. V., Varfolomeev E., Arnott D., Deshayes K., Kirkpatrick D. S., Vucic D. (2013) OTUB1 modulates c-IAP1 stability to regulate signalling pathways. EMBO J. 32, 1103–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han K. J., Foster D. G., Zhang N. Y., Kanisha K., Dzieciatkowska M., Sclafani R. A., Hansen K. C., Peng J., Liu C. W. (2012) Ubiquitin-specific protease 9x deubiquitinates and stabilizes the spinal muscular atrophy protein-survival motor neuron. J. Biol. Chem. 287, 43741–43752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li J., D'Angiolella V., Seeley E. S., Kim S., Kobayashi T., Fu W., Campos E. I., Pagano M., Dynlacht B. D. (2013) USP33 regulates centrosome biogenesis via deubiquitination of the centriolar protein CP110. Nature 495, 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tong X., Buelow K., Guha A., Rausch R., Yin L. (2012) USP2a protein deubiquitinates and stabilizes the circadian protein CRY1 in response to inflammatory signals. J. Biol. Chem. 287, 25280–25291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang W. C., Shih H. M. (2012) The deubiquitinating enzyme USP37 regulates the oncogenic fusion protein PLZF/RARA stability. Oncogene ePub: doi: 10.1038/onc.2012.537 [DOI] [PubMed] [Google Scholar]
- 38. Whitby F. G., Xia G., Pickart C. M., Hill C. P. (1998) Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem. 273, 34983–34991 [DOI] [PubMed] [Google Scholar]
- 39. Beltrao P., Albanese V., Kenner L. R., Swaney D. L., Burlingame A., Villen J., Lim W. A., Fraser J. S., Frydman J., Krogan N. J. (2012) Systematic functional prioritization of protein posttranslational modifications. Cell 150, 413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hagai T., Toth-Petroczy A., Azia A., Levy Y. (2012) The origins and evolution of ubiquitination sites. Mol. Biosyst. 8, 1865–1877 [DOI] [PubMed] [Google Scholar]
- 41. Gao D., Wan L., Inuzuka H., Berg A. H., Tseng A., Zhai B., Shaik S., Bennett E., Tron A. E., Gasser J. A., Lau A., Gygi S. P., Harper J. W., DeCaprio J. A., Toker A., Wei W. (2010) Rictor forms a complex with Cullin-1 to promote SGK1 ubiquitination and destruction. Mol. Cell. 39, 797–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol. Cell. 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 43. Flick K., Ouni I., Wohlschlegel J. A., Capati C., McDonald W. H., Yates J. R., Kaiser P. (2004) Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat. Cell. Biol. 6, 634–641 [DOI] [PubMed] [Google Scholar]
- 44. Scherer D. C., Brockman J. A., Chen Z., Maniatis T., Ballard D. W. (1995) Signal-induced degradation of I kappa B alpha requires site-specific ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 92, 11259–11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Deshaies R. J. (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467 [DOI] [PubMed] [Google Scholar]
- 46. Dimova N. V., Hathaway N. A., Lee B. H., Kirkpatrick D. S., Berkowitz M. L., Gygi S. P., Finley D., King R. W. (2012) APC/C-mediated multiple monoubiquitylation provides an alternative degradation signal for cyclin B1. Nat. Cell. Biol. 14, 168–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kravtsova-Ivantsiv Y., Cohen S., Ciechanover A. (2009) Modification by single ubiquitin moieties rather than polyubiquitination is sufficient for proteasomal processing of the p105 NF-kappaB precursor. Mol. Cell 33, 496–504 [DOI] [PubMed] [Google Scholar]
- 48. Shabek N., Herman-Bachinsky Y., Buchsbaum S., Lewinson O., Haj-Yahya M., Hejjaoui M., Lashuel H. A., Sommer T., Brik A., Ciechanover A. (2012) The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol. Cell 48, 87–97 [DOI] [PubMed] [Google Scholar]
- 49. Ziv I., Matiuhin Y., Kirkpatrick D. S., Erpapazoglou Z., Leon S., Pantazopoulou M., Kim W., Gygi S. P., Haguenauer-Tsapis R., Reis N., Glickman M. H., Kleifeld O. (2011) A perturbed ubiquitin landscape distinguishes between ubiquitin in trafficking and in proteolysis. Mol. Cell. Proteomics 10, M111.009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kirkpatrick D. S., Hathaway N. A., Hanna J., Elsasser S., Rush J., Finley D., King R. W., Gygi S. P. (2006) Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell. Biol. 8, 700–710 [DOI] [PubMed] [Google Scholar]
- 51. Rape M., Reddy S. K., Kirschner M. W. (2006) The processivity of multiubiquitination by the APC determines the order of substrate degradation. Cell 124, 89–103 [DOI] [PubMed] [Google Scholar]
- 52. West M. H., Bonner W. M. (1980) Histone 2A, a heteromorphous family of eight protein species. Biochemistry 19, 3238–3245 [DOI] [PubMed] [Google Scholar]
- 53. Goldknopf I. L., Busch H. (1977) Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc. Natl. Acad. Sci. U.S.A. 74, 864–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 55. Wang F., Chan C. H., Chen K., Guan X., Lin H. K., Tong Q. (2011) Deacetylation of FOXO3 by SIRT1 or SIRT2 leads to Skp2-mediated FOXO3 ubiquitination and degradation. Oncogene 31, 1546–1557 [DOI] [PubMed] [Google Scholar]
- 56. Gronroos E., Hellman U., Heldin C. H., Ericsson J. (2002) Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell 10, 483–493 [DOI] [PubMed] [Google Scholar]
- 57. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal 3, ra3. [DOI] [PubMed] [Google Scholar]
- 58. Wu R., Haas W., Dephoure N., Huttlin E. L., Zhai B., Sowa M. E., Gygi S. P. (2011) A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat. Methods 8, 677–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Clague M. J., Urbe S. (2010) Ubiquitin: same molecule, different degradation pathways. Cell 143, 682–685 [DOI] [PubMed] [Google Scholar]
- 60. Okiyoneda T., Barriere H., Bagdany M., Rabeh W. M., Du K., Hohfeld J., Young J. C., Lukacs G. L. (2010) Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science 329, 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Minsky N., Shema E., Field Y., Schuster M., Segal E., Oren M. (2008) Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell. Biol. 10, 483–488 [DOI] [PubMed] [Google Scholar]
- 62. Yen H. C., Xu Q., Chou D. M., Zhao Z., Elledge S. J. (2008) Global protein stability profiling in mammalian cells. Science 322, 918–923 [DOI] [PubMed] [Google Scholar]
- 63. Benanti J. A., Cheung S. K., Brady M. C., Toczyski D. P. (2007) A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat. Cell Biol. 9, 1184–1191 [DOI] [PubMed] [Google Scholar]
- 64. Trotschel C., Albaum S. P., Wolff D., Schroder S., Goesmann A., Nattkemper T. W., Poetsch A. (2012) Protein turnover quantification in a multilabeling approach: from data calculation to evaluation. Mol. Cell. Proteomics 11, 512–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Boisvert F. M., Lam Y. W., Lamont D., Lamond A. I. (2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell. Proteomics 9, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van Wijk S. J., Fiskin E., Putyrski M., Pampaloni F., Hou J., Wild P., Kensche T., Grecco H. E., Bastiaens P., Dikic I. (2012) Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Mol. Cell 47, 797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bennett E. J., Shaler T. A., Woodman B., Ryu K. Y., Zaitseva T. S., Becker C. H., Bates G. P., Schulman H., Kopito R. R. (2007) Global changes to the ubiquitin system in Huntington's disease. Nature 448, 704–708 [DOI] [PubMed] [Google Scholar]
- 68. Dammer E. B., Na C. H., Xu P., Seyfried N. T., Duong D. M., Cheng D., Gearing M., Rees H., Lah J. J., Levey A. I., Rush J., Peng J. (2011) Polyubiquitin linkage profiles in three models of proteolytic stress suggest the etiology of Alzheimer disease. J. Biol. Chem. 286, 10457–10465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kaiser S. E., Riley B. E., Shaler T. A., Trevino R. S., Becker C. H., Schulman H., Kopito R. R. (2011) Protein standard absolute quantification (PSAQ) method for the measurement of cellular ubiquitin pools. Nat. Methods 8, 691–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Dosztanyi Z., Csizmok V., Tompa P., Simon I. (2005) IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21, 3433–3434 [DOI] [PubMed] [Google Scholar]