Abstract
Brassinosteroids (BRs) are essential phytohormones for plant growth and development. BRs are perceived by the cell surface receptor kinase BRI1, and downstream signal transduction through multiple components leads to activation of the transcription factors BZR1 and BZR2/BES1. BZR1 activity is highly controlled by BR through reversible phosphorylation, protein degradation, and nucleocytoplasmic shuttling. To further understand the molecular function of BZR1, we performed tandem affinity purification of the BZR1 complex and identified BZR1-associated proteins using mass spectrometry. These BZR1-associated proteins included several known BR signaling components, such as BIN2, BSK1, 14–3-3λ, and PP2A, as well as a large number of proteins with previously unknown functions in BR signal transduction, including the kinases MKK5 and MAPK4, histone deacetylase 19, cysteine proteinase inhibitor 6, a DEAD-box RNA helicase, cysteine endopeptidases RD21A and RD21B, calmodulin-binding transcription activator 5, ubiquitin protease 12, cyclophilin 59, and phospholipid-binding protein synaptotagmin A. Their interactions with BZR1 were confirmed by in vivo and in vitro assays. Furthermore, MKK5 was found to phosphorylate BZR1 in vitro. This study demonstrates an effective method for purifying proteins associated with low-abundance transcription factors, and identifies new BZR1-interacting proteins with potentially important roles in BR response.
Brassinosteroids (BRs)1 are growth-enhancing phytohormones that affect a wide range of cell physiological processes and plant developmental programs, including cell elongation and division, photomorphogenesis, stress responses, disease resistance, male gametophyte development, vascular bundle differentiation, and lateral organ development (1–3). Arabidopsis mutants with defects in BR biosynthesis or signal transduction display broad developmental phenotypes, including dwarfism, compact rosette leaves, photomorphogenesis in the dark, male sterility, late flowering, and delayed senescence. Many key components of the BR signaling pathway have been characterized by molecular genetic strategies, including the BR receptor kinase BR-insensitive 1 (BRI1) (4), BR coreceptor kinase BRI1-associated receptor kinase 1 (BAK1) (5), GSK/SHAGGY-like protein kinase BR-insensitive 2 (BIN2) (6), PP1 class phosphatase BRI1 suppressor 1 (BSU1) (7), and the transcription factor brassinazole-resistant 1 (BZR1) and its homolog BZR2 (also named BRI1-EMS-suppressor 1 [BES1]) (8, 9). Yeast two-hybrid screens using these genetically characterized components as bait identified BAK1 and BRI1 kinase inhibitor 1 (BKI1) as BRI1-interacting proteins (10, 11) and 14–3-3 as a BZR1-interacting protein (12, 13). Further, proteomic studies identified BR-signaling kinase 1 (BSK1) based on BR-induced phosphorylation in the plasma membrane fraction (14).
Biochemical studies have elucidated the function of each BR signaling component and outlined the complete signal transduction pathway, from BR perception at the cell surface to gene expression in the nucleus (15). In the absence of BRs, BRI1 is inactive, partly because of inhibition by BKI1, and BIN2 is active and phosphorylates BZR1 and BZR2. Phosphorylated BZR1 and BZR2 lose their DNA binding activity, and are retained in the cytoplasm by the phosphopeptide-binding protein 14–3-3 or degraded by the proteasome through unknown factors (16). Following BR binding, BRI1 is activated on its disassociation from BKI1 and association with the coreceptor BAK1. It then phosphorylates its substrate BSK1 and constitutive differential growth 1 (CDG1) (17). CDG1 in turn phosphorylates and activates BSU1, which dephosphorylates and inactivates BIN2 kinase. Consequently, the phosphorylation of BZR1 and BZR2/BES1 is attenuated, and, because of dephosphorylation by protein phosphatase 2A (PP2A) (18), unphosphorylated BZR1 and BZR2/BES1 accumulate in the nucleus and regulate a large number of BR-responsive genes (19, 20). A dominant mutation, present in bzr1-1D and bes1-D, increases binding to PP2A and causes BR-independent dephosphorylation of BZR1 and BZR2 and activation of the BR response (18).
BZR1 and BZR2 are key transcription factors that control most BR-responsive genes and a wide range of BR-regulated developmental processes (3, 19, 20); they also interact with additional factors to control gene expression. For example, BZR1 interacts with DELLA family proteins to mediate gibberellin-regulated gene expression and cell elongation (21–23), and it interacts with phytochrome-interacting factor 4 (PIF4) to mediate BR-regulated light responses (24). BZR2 also interacts with several factors involved in transcription and histone modification (25–28). These studies suggest that BZR1 and BZR2 are central factors that interact with a wide range of regulators and cofactors to mediate diverse developmental processes.
To gain a comprehensive understanding of the molecular functions of BZR1, we performed tandem affinity purification (TAP) followed by mass spectrometry analysis of BZR1-associated proteins. We identified not only several known components of the BR signaling pathway, but also a number of new BZR1-interacting proteins, which suggest new molecular functions and mechanisms of the BR pathway.
EXPERIMENTAL PROCEDURES
Construction of Epitope-Tagged BZR1 and bzr1-1D Expression Vectors
To produce a Gateway destination vector harboring tandem affinity tag, the Gateway cassette and upstream 35S promoter were amplified from pGWB20 using the forward primer PBIf (5′-GGAAACAGCTATGACCATGATTACGCC-3′) and reverse primer PBIr (5′-ggactagtGGGGAAATTCGAGCTGTAAGC-3′) and introduced into the binary vector pCAMBIA1390 between the HindIII and SpeI sites, creating seven tandem repeats of the Myc tag from pGWB20 in-frame with six histidines from pCAMBIA1390. The destination vector, named pG7MH1, harbored a 7Myc-6His tandem affinity tag (MH). To construct the 35S::BZR1-MH and 35S::bzr1-1D-MH expression vectors, the BZR1 and bzr1-1D (8) coding sequences without stop codons were amplified by two rounds of PCR, first using the primer pair P1f (5′-aaaaagcaggct ATGACTTCGGATGGAGCT-3′) and P1r (5′-agaaagctgggtaACCACGAGCCTTCCCATT-3′) and then using the primer pair attB1(f) (5′-ggggacaagtttgtacaaaaaagcaggct-3′) and attB2(r) (5′-ggggaccactttgtacaagaaagctgggt-3′). The products were cloned into pDONR201 (Invitrogen, Carlsbad, CA) by the BP reaction to generate BZR1 and bzr1-1D entry vectors. The BZR1 and bzr1-1D coding sequences were then cloned into pG7MH1 using LR recombination to produce the 35S::BZR1-MH and 35S::bzr1-1D-MH vector (Fig. 1A). The amino acid sequences of the fusion proteins are shown in supplemental Fig. S1A.
Fig. 1.

The phenotype of epitope-tagged bzr1-1D and BZR1 transgenic plants. A, Diagrammatic representation of the constructs used to transform wild-type Col plants. bzr1-1D or BZR1 was fused to 7Myc-6His (MH) under the control of the 2×35S promoter. Blue boxes indicate Gateway recombination sites. B and C, Three-week-old 35S::bzr1-1D-MH (B) and 35S::BZR1-MH (C) plants displayed a similar phenotype to that of bzr1-1D or pBZR1::bzr1-1D-CFP, and Col-0 or pBZR1::BZR1-CFP control plants respectively. D, Dark growth of the indicated seedlings on 0.5× MS medium containing 2 μm propiconazole (PPZ) for 6 days revealed that the 35S::BZR1-MH seedlings were sensitive to PPZ whereas the 35S::bzr1-1D-MH seedlings were insensitive to PPZ, similar to pBZR1::BZR1-CFP or Col-0, and pBZR1::bzr1-1D-CFP or bzr1-1D plants, respectively.
To construct the pBZR1::bzr1-1D-TAP-6His (bzr1-1D-TAPH) expression vector, a classical TAP cassette comprising one calmodulin (CaM)-binding peptide (CBP) and two repeats of the IgG-binding domain from pBS1539 (29) was introduced into pCAMBIA1390 between the BamHI and EcoRI sites. The TAP-His fusion was formed after removing the stop codon using EcoRI and SpeI. The resulting vector was further converted to the Gateway-compatible destination vector pGTAPH by inserting the Gateway cassette rfc.1 (Invitrogen) at the SalI site. A 2.5 kb fragment, including the 1.5 kb promoter sequence upstream of the start codon and BZR1 or bzr1-1D genomic DNA, was amplified and cloned into pENTR/SD/D-TOPO (Invitrogen) to produce the entry vector. The expression vector pBZR1::bzr1-1D-TAPH (supplemental Fig. S4A) was produced by LR recombination according to the manufacturer's protocol. The amino acid sequence of the tagged fusion protein is shown in supplemental Fig. S1B.
Transgenic Plants and Growth Conditions
The expression vectors were introduced into Arabidopsis thaliana ecotype Columbia (Col) through Agrobacterium-mediated transformation. Transgenic plants were screened on hygromycin plates and identified by Western blotting using anti-c-Myc monoclonal antibodies (M4439; Sigma, St. Louis, MO, USA) for the expression of BZR1/bzr1-1D-MH, and peroxidase-anti-peroxidase (PAP) soluble complex (P1291; Sigma) for the expression of bzr1-1D-TAPH. Homozygous T3 plants were grown on agar plates containing 0.5× Murashige and Skoog (MS) mineral salts and 3% sucrose (pH 5.7) in a light chamber at 22 °C for 16–18 days. Whole seedlings were harvested, ground in liquid nitrogen, and stored at −80 °C until purification. Seedlings expressing pBZR1::BZR1-CFP or pBZR1::bzr1-1D-CFP (cyan fluorescent fusion) were used as a control and grown side by side with the 35S::BZR1-MH or 35S::bzr1-1D-MH and pBZR1::bzr1-1D-TAP seedlings, respectively, and subjected to the same treatment throughout the experiment.
TAP
For His tag purification, 15 g of tissue powder was mixed with 30 ml of protein extraction buffer (50 mm Na2HPO4, 50 mm NaH2PO4, 150 mm NaCl, 1% Triton X-100, 15% glycerol, 5 mm imidazole, 25 mm NaF, 2 mm PMSF, and 10 μm MG132, pH 7.4), thoroughly suspended, and centrifuged at 20,000 × g for 10 min at 4 °C. About 40 ml of supernatant containing 148 mg of protein was filtered through Miracloth (Calbiochem, San Diego, CA) and incubated with 1 ml of settled Ni-NTA resin (Qiagen, Valencia, CA), which was pre-equilibrated with at least a 10-bed-volume of equilibration buffer (50 mm Na2HPO4, 50 mm NaH2PO4, 150 mm NaCl, 0.1% Triton X-100, 10% glycerol, and 5 mm imidazole, pH 7.4) on an orbital shaker at 60 rpm for 30 min at 4 °C. The resin was rinsed rapidly ten times with 1 ml of equilibration buffer each time, and the bound proteins were eluted by a 20-min incubation with 1 ml of elution buffer (50 mm Na2HPO4, 50 mm NaH2PO4, 150 mm NaCl, 0.1% Triton X-100, 10% glycerol, and 80 mm imidazole, pH 7.4) at 4 °C. The elution was repeated three times, and six eluates from two parallel purifications were pooled. Around 8.64 mg of the protein resulting from the His tag purification procedure was further incubated with 100 μl of settled anti-c-Myc agarose beads (A7470; Sigma), which had been pre-equilibrated with 10 ml of binding buffer (50 mm Na2HPO4, 50 mm NaH2PO4, 150 mm NaCl, 0.1% Triton X-100, and 10% glycerol, pH 7.4) at 4 °C for 30 min. After 20 washes with 1 ml of binding buffer each, the proteins bound by anti-c-Myc-agarose were eluted by heating in 200 μl of 1% SDS at 90 °C for 5 min. The supernatant was collected after a short spin and used as the final purified protein.
For bzr1-1D-TAPH purification ∼7.2 ml of eluate from the Ni-charged resin was incubated with 250 μl of pre-equilibrated and settled IgG Sepharose 6 Fast Flow (17–0969-01; GE Healthcare Bio-Sciences, Uppsala, Sweden) at 4 °C for 25 min. The bound proteins were washed thoroughly with 30 ml of binding buffer and then with 10 ml of cleavage buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 10% glycerol, 0.1% Triton-100, 1 mm DTT, and 0.5 mm EDTA). Around 3 ml of IgG-bound proteins were incubated with 200 U of AcTEV protease (12575–015; Invitrogen) at 16 °C for 1.5 h to release the target proteins. The supernatant was cleaned by incubation with Ni-NTA agarose to remove the His-tagged AcTEV protease. The resulting flow-through contained the final purified protein.
Western Blotting
At each step of purification, a 15-μl aliquot of protein sample was saved in liquid nitrogen and analyzed by Western blotting to monitor the enrichment and recovery of the target protein. The BZR1-MH and bzr1-1D-MH fusion proteins were probed with anti-c-Myc monoclonal antibodies, then with IRDye 800-conjugated anti-mouse IgG (610–631-002; Rockland Immunochemicals, Inc., Gilbertsville, PA), and developed using an Odyssey Infrared Imaging System (LI-COR, Lincoln, NE). The bzr1-1D-TAPH fusion protein was probed with PAP and developed with 0.3 mg/L 3,3′-diaminobenzidine supplemented with 0.03% H2O2.
Separation of the Purified Proteins by SDS-PAGE
Protein samples from five purifications were pooled in a centrifugal filter unit (Amicon Ultra-4; Millipore, Billerica, MA) and centrifuged at 4,000 × g to remove all SDS and small proteins (<5 kDa) and to concentrate the protein samples. Around 0.5 mg of protein from each purification was loaded side-by-side on a 5–20% gradient gel (18 × 16 × 1 mm). Based on staining pattern, the gel was cut into 21 and 27 unequal bands (narrower band for strong staining signals and wider bands for gel area of low signal) in the bzr1-1D-MH and BZR1-MH experiment respectively (Fig. 2, arrows). All the bands, except the IgG bands (b9, b20, B14, and B25), were analyzed by in-gel digestion followed by liquid chromatography-electrospray tandem mass spectrometry (LC-MS/MS).
Fig. 2.
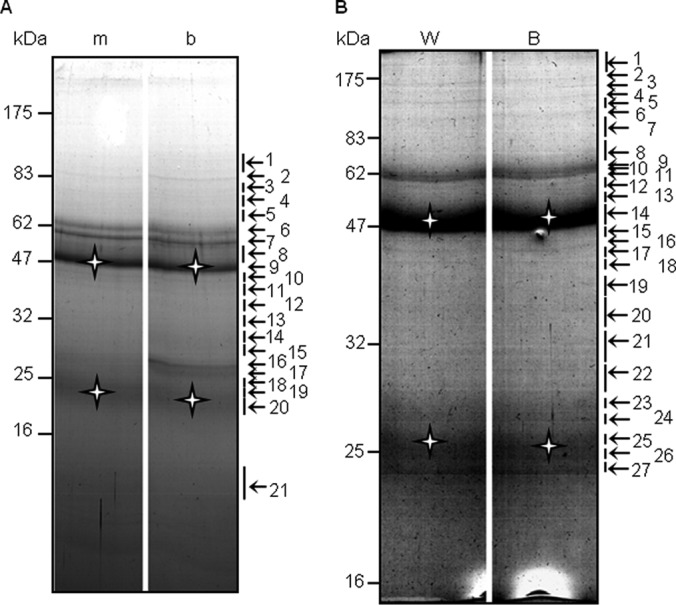
Separation of purified proteins by SDS-PAGE. bzr1-1D-MH (A) and BZR1-MH (B) copurified proteins were separated on a 5–20% gradient gel then stained with Coomassie blue. The numbers to the left indicate the sizes of the protein markers (kDa), whereas those to the right indicate the gel bands used for LC-MS/MS. m, bzr1-1D-CFP; b, bzr1-1D-MH; W, BZR1-CFP; B, BZR1-MH; star, heavy and light chains of IgG.
In-Gel Tryptic Digestion, Mass Spectrometric, and Database Searching
Protein in-gel digestion was performed as described previously with some modifications (see donatello.ucsf.edu/ingel.html). In brief, gel slices were cut up in cubic pieces 1 mm long, washed twice with 50% acetonitrile in 25 mm ammonium bicarbonate (NH4HCO3) and vacuum dried. The gel samples were then reduced with DTT (10 mm in 25 mm NH4HCO3, 56 °C for 1h), alkylated with iodoacetamide (55 mm in 25 mm NH4HCO3, room temperature for 1 h), and vacuum dried again. Samples were rehydrated in at least 32 μl of digestion buffer (10 ng/μl trypsin in 25 mm NH4HCO3) (actual volume was based on the original size of the gel slice), and covered with the minimum volume of NH4HCO3. After an overnight digestion at 37 °C, peptides were extracted twice with a solution containing 50% acetonitrile and 5% formic acid. The extracted digests were vacuum-dried and resuspended in 20 μl of 1% formic acid in water. The digests were analyzed by nano-flow High-Performance Liquid Chromatography (HPLC)-electrospray tandem mass spectrometry (LC-MS/MS) as described (30, 31). The separation was performed using a 75 μm×150 mm C18 PepMap column (LC Packings, Sunnyvale, CA) in an Ultimate HPLC system linked with a FAMOS autosampler (LC Packings, San Francisco, CA). Solvent A was 0.1% formic acid in water, solvent B was 0.1% formic acid in acetonitrile, the flow rate was ∼330 nl/min. Five microliter of the digests was injected at 5% B, and then the organic content of the mobile phase was increased linearly to 50% B over 30 min. The column effluent was directed to a microionspray ion source attached to either a hybrid linear ion trap-Fourier transform mass spectrometer (LTQ-FT, Thermo Scientific, San Jose, CA) or a hybrid linear ion trap-Orbitrap mass spectrometer (LTQ-Orbitrap XL, Thermo Scientific, San Jose, CA). Peptides were analyzed in positive ion mode and in information-dependent acquisition mode to automatically switch between MS and MS/MS acquisition. In the LTQ-Orbitrap, MS spectra were acquired in profile mode using the Orbitrap analyzer in the m/z range between 350 and 1500. For each MS spectrum, the 6 most intense multiple charged ions over a threshold of 5000 counts were selected to perform CID experiments. Product ions were analyzed on the linear ion trap in centroid mode. The CID collision energy was automatically set to 35%. In the LTQ-FT, MS spectra were acquired in profile mode using the ICR analyzer in the m/z range between 310 and 1600. For each MS spectrum, the five most intense multiple charged ions over a threshold of 200 counts were selected to perform CID experiments. Product ions were analyzed on the linear ion trap in profile mode. CID collision energy was automatically set to 35%. A dynamic exclusion window of 1 Da was applied that prevented the same m/z from being selected for 60 s after its acquisition.
In some cases, the LC/MS/MS analyses were performed on a QSTAR Pulsar mass spectrometer (Applied Biosystems/MDS Sciex, South San Francisco, CA). Peptides were analyzed in positive ion mode. MS spectra were acquired for 1 s in the m/z range between 310 and 1400. MS acquisitions were followed by 3 s CID experiments in information-dependent acquisition mode. For each MS spectrum, the most intense multiple charged peaks over a threshold of 30 counts were selected for generation of CID mass spectra. The CID collision energy was automatically set according to m/z ratio and charge state of the precursor ion. A dynamic exclusion window was applied that prevented the same m/z from being selected for 1 min after its acquisition.
LTQ-FT and LTQ-Orbitrap raw data were converted to ASCII peak lists by Mascot Distiller software v2.1.0.0 (Matrix Science, Boston, MA). Parameters for MS processing were set as follows: peak half width: 0.02; data point per Da: 100. For MS/MS data: peak half width: 0.02; data point per Da: 100. QSTAR pulsar data were analyzed with Analyst QS 1.1 software (Applied Biosystems/MDS Sciex, South San Francisco, CA) and peak lists were generated using the mascot.dll script (Mascot. Dll 1.6b18, Matrix Sciences). Precursor mass tolerance for grouping was set to 0.2. MS centroiding parameters were 50% peak height and 0.05 amu merge distance. MS/MS centroiding parameters were 50% peak height and 0.05 amu merge distance.
The peak lists were searched against the Arabidopsis subset of the NCBInr database as of July 10, 2009 (249,067 entries searched), using in-house Protein Prospector version 5.3.0 (a public version is available on line at http://prospector.ucsf.edu). The maximum number of missed enzyme cleavages per peptide was set at two. Variable modifications selected for searching include oxidation of methionine, N-terminal acetylation, N-terminal pyroglutamate, and modification of cysteine by carbamidomethylation or propionamide. In some cases phosphorylation was also taken into consideration for queries. Mass tolerance in searches was 100 ppm for precursor and 0.2 Da for product ions in QSTAR data, and 30 ppm for precursor and 0.8 Da for product ions for LTQFT or Orbitrap data. In all protein identifications, a minimal protein score of 22, a peptide score of 15, and a minimal discriminant score threshold of 0.0 were used for initial identification criteria. Maximum expectation values (number of different peptides with scores equivalent to or better than the result reported that are expected to occur in the database search by chance) for accepting individual spectra were set to 0.05. Control searches of some of the files against the whole database confirmed the absence of contamination from human keratins or other non-vegetal proteins, so searches were routinely performed on the aforementioned subset. When several accession numbers in the database matched the same set of peptides identified, the entries with the most descriptive name were reported. Individual isoforms of proteins were reported according to the detection of peptides unique to their sequences. If several isoforms shared the same set of peptides identified, they were all reported.
Those proteins identified only in the BZR1-MH samples and not in the BZR1-CFP samples were considered to be putative BZR1-interacting proteins. Proteins identified in both samples were considered to be nonspecific; this included proteins with the same ID, and proteins with different IDs but possessing a similar sequence.
Yeast Two-Hybrid Assay for Protein-Protein Interactions
The coding sequence of BZR1 or bzr1-1D and of the putative BZR1- and bzr1-1D-interacting proteins were cloned into pGADT7 or pGBKT7, respectively. The bait and prey vectors were cotransformed into yeast strain EHA109. Those transformants that grew on SD/-Leu-Trp-His (50 mm 3AT for BD-BZR1) plates were considered to exhibit direct interactions. In mating-based split-ubiquitin system tests, the coding sequence of synaptotagmin A (SYTA) was inserted into the bait vector Y-Cub, whereas BZR1/bzr1-1D, PP2A-A1, PP2A-A2, and PP2A-A3 were cloned into NubG-X, respectively, through in vivo recombination in yeast strains THY-AP5 and THY-AP4. Diploid cells resulting from the mating of AP5 and AP4 were selected on SC/-Trp-Leu-Ura medium (32). The interactions were tested by the replica plating of cells on four plates containing synthetic minimal medium plus 0, 75, 150, or 1000 μm Met (methionine). After 2–3 days, the cells were tested for β-galactosidase activity using an X-Gal overlay assay (32).
Subcellular Localization and Bimolecular Fluorescence Complementation (BiFC) Tests
To monitor subcellular localization, the coding sequence of UBP12 was introduced into pMDC83 to produce a GFP fusion protein, whereas Histone Deacetylase 19 (HDA19), CaM-binding Transcription Activator 5 (CAMTA5), the MAP kinase MAPK4, the MAP kinase kinase MKK5, the Cyclophilin 59 (CYP59), and Cysteine Proteinase Inhibitor 6 (CYSB) were cloned into pEarleyGate101 to produce a YFP fusion under the control of the 35S promoter through Gateway cloning. The constructs were introduced into tobacco (Nicotiana benthamiana) leaves through the injection of Agrobacteria strain GV3101. BiFC was assayed as described previously (13). The coding sequences of HDA19, CAMTA5, UBP12, CYSB, and Met were individually introduced into 35S::X-NYFP, and then cotransformed with 35S::BZR1-CCFP into tobacco leaves. MAPK4, MKK5, and CYP59 were introduced into 35S::X-CCFP then cotransformed with 35S::BZR1-NYFP. Fluorescence from the GFP and YFP fusion proteins and BiFC signals were observed under a confocal microscope (Meta510-LCM; Zeiss, Jena, Germany). For nuclear stain, 10 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI) in water was injected into N. benthamiana leaves 2 h before observation.
Pull-Down Assay, Protoplast Transfection, and Co-Immunoprecipitation (Co-IP) Assays
Pull-down assays, protoplast transfection, and Co-IP assays were performed as described previously (21).
Quantitative RT-PCR (qRT-PCR) Identification of BR-Regulated Gene Expression
Total RNA was isolated using Trizol reagent (Invitrogen). The transcription of BR-responsive genes was quantified by qRT-PCR with SYBR Premix EX Taq (Takara Bio Inc., Shiga, Japan) using specific primers (Table SII). PP2A A3 (At1g13320) was used as an internal control to normalize the data.
Generation of MKK5K99M
The kinase dead MKK5 was generated by replacing the lysine in ATP-binding loop with a methionine (33), using the Fast Mutagenesis System (FM111, Transgen Biotech).
RESULTS
Transgenic Plants Expressing 35S::bzr1-1D-MH in Arabidopsis Displayed a bzr1-1D Phenotype
Previous studies have shown that a single amino acid change, P234L, in bzr1-1D increased accumulation of BZR1 protein and a phenotype of BR activation (8). With the aim of identifying BZR1-interacting proteins in Arabidopsis, we first generated transgenic plants expressing the epitope-tagged fusion protein bzr1-1D-MH (Fig. 1A). As expected, the transgenic plants displayed a similar phenotype to that of bzr1-1D (8) and control plants expressing pBZR1::bzr1-1D-CFP, including dark green and downward-curving leaves in the light (Fig. 1B), and reduced sensitivity to the BR biosynthesis inhibitor propiconazole (PPZ) in the dark (i.e. longer hypocotyls than in wild type; Fig. 1D). Together, these findings indicate that the bzr1-1D-MH fusion protein functions in the same way as bzr1-1D does in planta, and that the MH at the C terminus did not interfere with the function of the protein. We therefore generated transgenic plants expressing 35S::BZR1-MH in wild-type Col plants (Fig. 1C); these plants had the same appearance as the untransformed wild type and transgenic plants expressing pBZR1::BZR1-CFP (Figs. 1C and 1D).
Purification of the BZR1 Protein Complex
Both BZR1-MH and bzr1-1D-MH were detected in the extracts prepared from corresponding transgenic plants but not in extracts prepared from control pBZR1::BZR1-CFP and pBZR1::bzr1-1D-CFP plants (supplemental Fig. S2). Altogether, about 295 mg of crude protein was applied to a 2 ml settled Ni column, and around 8.64 mg of protein was eluted and mixed with 100 μl of settled anti-c-Myc-agarose. Around 0.093 mg was eluted after washing. A significant amount of the BZR1-MH and bzr1-1D-MH proteins flowed through the beads (supplemental Figs. S2B and S2C), suggesting that the binding capacity of the columns was saturated at both steps.
When the purified proteins were separated by SDS-PAGE, most of the bands were the same between the BZR1-MH and BZR1-CFP, or between the bzr1-1D-MH and bzr1-1D-CFP samples (Fig. 2). Nevertheless, LC/MS-MS analysis identified 32 proteins specifically in the bzr1-1D-MH purification (Table I), and 27 proteins non-specifically in both the sample and control (supplemental Table SI). Following BZR1-MH purification (Fig. 2B), LC-MS/MS analysis identified 31 unique proteins that were not detected in the BZR1-CFP control (Table I), and 18 nonspecific proteins present in both, among them 13 proteins were also detected in the bzr1-1D-CFP control (supplemental Table SI). Those proteins that specifically copurified with BZR1-MH or bzr1-1D-MH included previously known BR signaling components such as BIN2 and its homologs, SHAGGY-related protein kinases (AtSK11, -12, -22, -23, and -31), 14–3-3-λ, BSK1, and several subunits of PP2A (Table I). The role of PP2A in dephosphorylating BZR1 and activating BR signaling has been reported (18). The identification of several known BZR1-interacting proteins demonstrates that the purification of BZR1/bzr1-1D-MH was successful.
Table I. BZR1/bzr1-1D co-purified proteins.a Y: yeast two hybrid assay, Bi: BiFC assay, PD: pull down assay, IP: Co-IP, ND: Non-determined.
| Gene locus | Protein name | BZR1 |
bzr1-1D |
Confirmation | ||||
|---|---|---|---|---|---|---|---|---|
| Unique peptides | Coverage (%) | Gel band | Unique peptides | Coverage (%) | Gel band | |||
| At1g75080 | BZR1 | 15 | 56.8 | B7-27 | 15 | 56.3 | b1-21 | Ref (18) |
| At1g25490 | PP2AA1 (RCN1,α)b | 1 | 2 | b7 | ||||
| At3g25800 | PP2AA2 (β) | 1 | 2.7 | B10 | 6 | 14 | b6 | |
| At1g13320 | PP2AA3 (γ) | 2 | 3.9 | b7 | ||||
| At3g09800 | PP2AB' βc | 1 | 2.2 | b8 | ||||
| At3g26020 | PP2AB' ηc | 2 | 2.5 | B12 | 2 | 4.5 | b8 | |
| At1g13460 | PP2AB' θc | 2 | 2.6 | B12 | 2 | 4.5 | b8 | |
| At4g18710 | AtSK21 (BIN2)d | 2 | 6.1 | B19/8 | Ref (16) | |||
| At1g03690 | AtSK22d | 2 | 5.7 | B19/8 | Ref (15) | |||
| At2g30980 | AtSK23d | 2 | 5.6 | B19/8 | ||||
| At3g05840 | AtSK12 | 4 | 10 | b11 | ||||
| At3g61160 | AtSK31b | 1 | 2.1 | B16 | ||||
| At5g26751 | AtSK11 | 2 | 4.2 | b11 | ||||
| At5g10450 | 14-3-3 λb | 1 | 6.5 | b16 | Ref (12) | |||
| At4g35230 | BSK1 | 2 | 5.5 | b3 | Ref (14) | |||
| At3g21220 | MKK5b | 1 | 4 | B19 | Y, Bi, PD, IP | |||
| At4g38130 | HDA19b | 1 | 3.4 | B9 | Y, Bi, PD, IP | |||
| At3g12490 | CYSB | 3 | 17.9 | B23-26 | 2 | 15.8 | b17/18 | Bi, PD, IP |
| At4g01370 | MAPK4b | 1 | 4.8 | B18 | Bi, PD, | |||
| At5g06600 | Ubiquitin-specific protease 12 (UBP12) | 12 | 15.1 | B3 | 15 | 14.3 | b1 | Bi |
| At1g53720 | Cyclophilin 59 (CYP59)b | 1e | 3.2 | B8 | 1e | 2.2 | b5 | Bi |
| At4g16150 | CaM-binding transcription activator 5 (CAMTA5) | 2 | 2.7 | B3 | 4 | 5.6 | b1/4 | Bi |
| At4g00660 | RNA Helicase like 8 (RH8)b | 1 | 2.8 | B11 | 1 | 2.8 | b9 | Y |
| At1g47128 | Cysteine protease (RD21A) | 5 | 13.9 | b12/13 | Y | |||
| At5g43060 | Cysteine protease (RD21B)b | 1 | 4.1 | b13 | Y | |||
| At2g20990 | Synaptotagmin A (SYTA) | 4 | 9.8 | b8 | Y(PP2A A) | |||
| At5g65010 | Asparagine synthetase (ASP) | 11 | 17.6 | b6 | No Y | |||
| At3g11910 | UBP13 | 8 | 8.1 | B2 | ND | |||
| At1g15750 | TOPLESS (TPL) | 3 | 3.2 | B3 | 5 | 6.1 | 1b | ND |
| At5g43470 | Disease resistance protein (RPP8) | 2 | 3 | B4 | ND | |||
| At5g20830 | Sucrose synthase 1 (ATSUS1) | 3 | 17.6 | B6 | ND | |||
| At4g24795 | Uncharacterized protein | 4 | 8.1 | B7 | ND | |||
| At1g04010 | Phospholipid Sterol Acyl Transferase 1 (PSAT1) | 2 | 4.3 | B7 | ND | |||
| At4g00740 | Qusimodo 3 (QUA3) | 2 | 4.7 | B8 | ND | |||
| At4g32250 | Putative protein kinase | 2 | 5.2 | B9 | ND | |||
| At5g19770 | Tubulin α 3/5 | 3 | 10.9 | B15 | ND | |||
| At2g28080 | UDP-glycosyltransferase-like protein | 2 | 6.2 | B15 | ND | |||
| At2g38670 | Phosphorylethanolamine cytidylyltransferase1 (PECT1) | 2 | 9.7 | B15 | ND | |||
| At1g70830 | MLP-Like protein 28 (MLP28) | 2 | 9.7 | B19 | ND | |||
| At5G26670 | Pectinacetylesterase family protein (F21E10) | 2 | 3.8 | B19 | ND | |||
| At3g14420 | Glycolate oxidase 1 (GOX1) | 2 | 7.5 | B19 | ND | |||
| At5g47500 | Pectin methylesterase 5 (PME5) | 2 | 6.6 | B23 | ND | |||
| At5g48620 | Probable disease resistance RPP8-like protein 4 (RPP8L4) | 4 | 5.2 | b2 | ND | |||
| At1g15130 | Programmed cell death 6-interacting protein | 2 | 2.7 | b2 | ND | |||
| At4g16990 | Disease resistance RPP5 like protein | 2 | 3.3 | b3 | ND | |||
| At5g60790 | ABC transporter F family member 1 (ATGCN1) | 3 | 5.9 | b5 | ND | |||
| At5g44340 | Tublin-β-4 | 2 | 6.3 | b9 | ND | |||
| At1g43190 | Polypyrimidine tract-binding protein 3 (PTB3) | 2 | 7.6 | b9 | ND | |||
| At3g18780 | Actin 2 | 6 | 25.5 | b11 | ND | |||
| At1g49240 | Actin-8 | 6 | 25.5 | b11 | ND | |||
| At3g08580 | ADP/ATP carrier1 (AAC1) | 3 | 8.1 | b15-18 | ND | |||
| At1g13430 | Sulfotransferase 4C (ATST4C) | 2 | 7.4 | b13 | ND | |||
| At3g21211 | Hypothetical protein/RNA binding | 3 | 7.1 | b13 | ND | |||
a The proteins with ≥ 2 unique peptides or 1 peptide but confirmed were included.
b The mass spectrometry data were showed in Supplemental Fig. S3.
c ,d Share the same peptide.
e Peptide is different.
The copurified proteins also included new putative BZR1-interacting proteins such as MKK5, MAPK4, HDA19, CYP59, CAMTA5, the RNA-binding DEAD-box protein (RH8), two ubiquitin-specific proteases (UBP12 and UBP13), two cysteine endopeptidases (RD21A and RD21B), CYSB, SYTA, and TOPLESS (TPL). These proteins have not been previously shown to function in the BR pathway. CYSB, UBP12, CYP59, CAMTA5, RH8, and TPL were copurified with both bzr1-1D and BZR1, as were BIN2 homologues and PP2A members. Those detected only in the BZR1 or bzr1-1D purification (Table I) may have different binding abilities with BZR1 and bzr1-1D or were because of incomplete detection of the components of the complex. The spectra of specific proteins identified by only one peptide are shown in supplemental Fig. S3. The nonspecific proteins pulled down in both the experimental samples and controls were mostly abundant metabolic enzymes and structural proteins (supplemental Table SI).
The Classical TAP Tag is Less Effective Than the Myc-His Tag in Arabidopsis
We also performed purification using transgenic plants expressing pBZR1::bzr1-1D-TAPH, which displayed similar phenotype to the control plants expressing pBZR1::bzr1-1D-CFP, with kinks at the base of the shoot branches (supplemental Figs. S4A and S4B). Western blotting of the bzr1-1D-TAPH purification revealed that, (1) the target protein was enriched by the Ni-charged resin but was partly cleaved (supplemental Fig. S4C, arrows); (2) The application of the protease AcTEV led to bzr1-1D-TAPH degradation such that the fusion protein was undetectable in gel blots by biotin-CaM (supplemental Fig. S4D, lane R-G); 3) Many bands in the elution of Ni-column were strongly recognized by biotin-CaM (supplemental Fig. S4D, triangles) but weakly by PAP (supplemental Fig. S4D, arrows), suggesting that large amounts of protein were enriched through association with the CBP tag; 4) The purified bzr1-1D-TAPH complex displayed more protein bands in SDS-PAGE than the purified bzr1-1D-MH complex (compare supplemental Fig. S4E with Fig. 2A). LC-MS/MS of one of the obviously enriched bands in the bzr1-1D-TAPH sample (supplemental Fig. S4E, arrow 2) identified a heat shock protein 70 (Hsc70) (data not shown), which was reported by other groups in classic TAP-tag purification in tobacco and rice (34, 35), and also appeared as a nonspecific protein in our MH tag purification (Table SI). Hsc70 was likely pulled down through its association with the TAP tag or with other proteins but not with bzr1-1D, because bzr1-1D was not detectable in elute. Overall, purification of bzr1-1D-TAPH was unsuccessful.
BZR1 Interacts With and is Phosphorylated by Components of the MAPK Complex
To confirm our affinity purification-mass spectrometric data, we tested BZR1 interactions with selected proteins using combinations of several assays, including yeast two-hybrid, in vitro pull-down, bi-molecular florescence complementation (BiFC), and co-immunoprecipitation (Co-IP) assays.
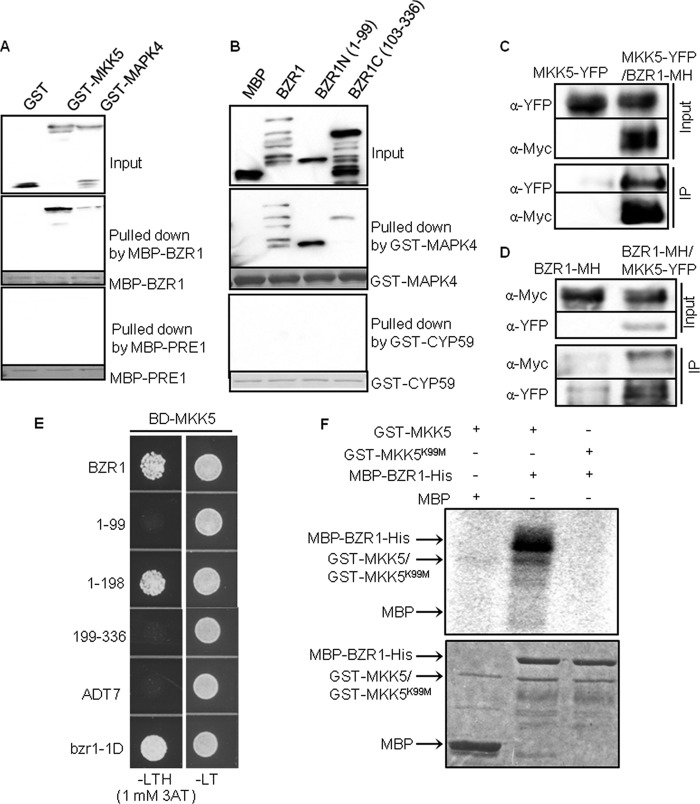
The interactions of BZR1 with MKK5 and MAPK4, which are components of the MAPK module (36), were analyzed further. First, in vitro pull-down assays showed that the MBP-BZR1 protein strongly interacts with MKK5, but only weakly with MAPK4; whereas PRE1 (37), a HLH protein used as negative control, showed no interaction with neither MKK5 nor MAPK4 (Fig. 3A). To further test the specificity of MAPK4 interaction, we tested the interactions with the two halves of BZR1, and the results showed that the N-terminal domain (aa 1–99, DNA-binding) interacts more strongly with MAPK4 than the C-terminal domain (aa 103–336), indicating specific MAPK4 interaction with a particular domain of BZR1. In contrast, the GST-CYP59 protein showed no interaction with BZR1 in this assay (Fig. 3B). Both MKK5 and MAPK4 interacted with BZR1 in BiFC assays (supplemental Fig. 6A). However, interaction between MAPK4 and BZR1 was not detected in yeast or Co-IP assays (data not shown), suggesting that the interaction is weak or modulated in eukaryotic cells. On the other hand, Co-IP assays using protoplasts showed that MKK5-YFP was immunoprecipitated by BZR1-MH (Fig. 3C), and that BZR1-MH was immunoprecipitated by MKK5-YFP (Fig. 3D). A yeast two-hybrid assay showed that MKK5 interacted with the N-terminal half of BZR1 (aa 1–198) but not the C-terminal half (aa 199–336) of BZR1 (Fig. 3E). In vitro kinase assay showed that MKK5 possesses autophosphorylation activity and can phosphorylate BZR1 (Fig. 3F, lanes 1 and 2), whereas the kinase dead form of GST-MKK5K99M did not (Fig. 3F, lanes 3). These results provide evidence for BZR1 regulation by a MAPK pathway.
Fig. 3.
BZR1 interacts with and is phosphorylated by components of the MAPK complex. A, MBP-BZR1, but not MBP-PRE1 (PRE1, a HLH protein used as a negative control) pulled down GST-MKK5 and GST-MAPK4. The blots were probed with anti-GST antibodies. B, GST-MAPK4, but not GST-CYP59, pulled down MBP-BZR1, MBP-BZR1N, and (weakly) MBP-BZR1C. The blots were probed with anti-MBP antibodies. Ponceau S staining shows the amount of bait protein on the beads. C and D, Co-immunoprecipitation (IP) of BZR1 with MKK5. Arabidopsis protoplasts transfected with 35S::MKK5-YFP and 35S::BZR1-MH were immunoprecipitated using anti-c-Myc antibody (C) or anti-YFP antibodies (D), and the immunoblot was probed with anti-YFP or anti-c-Myc antibodies. E, A yeast two-hybrid assay showed that MKK5 interacted with BZR1, amino acids 1–198 of BZR1, and bzr1-1D. Yeast grown on leucine-, tryptophan-, and histidine-deficient medium (-LTH) supplemented with 1 mm 3AT revealed the interaction. Empty ADT7 was used as a negative control. F, In vitro kinase assay of MBP-BZR1 phosphorylation by GST-MKK5 but not by kinase dead GST-MKK5K99M. Upper panel, autoradiograph; bottom panel, Coomassie blue staining of the protein gel.
BZR1 Interacts With a Histone Deacetylase, HDA19
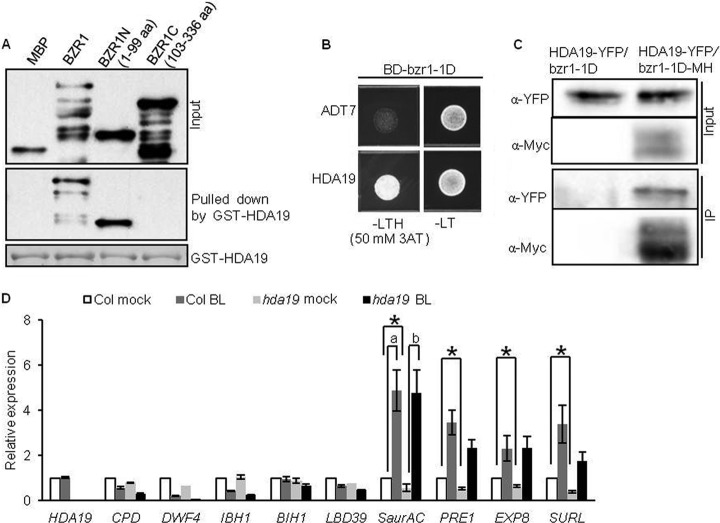
The BZR1 interaction with HDA19, a histone deacetylase also named AtHD1 (38), was confirmed by yeast two-hybrid and BiFC assays (supplemental Fig. S5A and supplemental Fig. S6A), and by in vitro pull-down assay (Fig. 4A). Furthermore, HDA19 interacted with BZR1N (aa 1–99), but not BZR1C (aa 103–336) in vitro (Fig. 4A). Consistent with interaction of BZR1N, bzr1-1D containing P234L substitution in the BZR1C region (8), interacted with HDA19 in yeast (Fig. 4B). Co-IP assays were therefore carried out using bzr1-1D-MH and HDA19-YFP transgenic Arabidopsis plants, because bzr1-1D is more stable than BZR1 (8). The result demonstrated the interaction between HDA19 and bzr1-1D in vivo (Fig. 4C). To test the effect of HDA19 on BZR1 function, we analyzed the expression levels of several BZR1 target genes in response to BR in the null allele mutant hda19 (19, 21, 39, 40). Compared with wild-type Col, the hda19 mutant showed significantly reduced expression (ratio <0.67 and p < 0.05) of SaurAC, PRE1, EXP8 and SURL before BR treatment. Expression levels of CPD, DWF4 and LBD39 were also slightly reduced in hda19 (p < 0.05). SaurAC also showed higher fold change in response to BR in the hda19 mutant compared with wild-type (p < 0.05) (Fig. 4D). These results suggest that the interaction with HDA19 is required for normal function of BZR1 in regulating target gene expression.
Fig. 4.
HDA19 interacts with BZR1 in vitro and in vivo. A, Pull-down assay showing that GST-HDA19 interacts with MBP-BZR1 and MBP-BZR1N, but not MBP-BZR1C. The blot was probed with anti-MBP antibodies. Ponceau S staining shows the amount of bait protein on the beads. B, HDA19 interacts with bzr1-1D in yeast. Yeast grown on -LTH medium supplemented with 50 mm 3AT revealed the interaction. C, Interaction of HDA19 with bzr1-1D in planta as shown by a co-immunoprecipitation (IP) assay. Extracts of transgenic Arabidopsis seedlings expressing 35S::HDA19-YFP in a bzr1-1D or 35S::bzr1-1D-MH background were immunoprecipitated with anti-c-Myc antibodies, and the immunoblot was probed using anti-YFP antibodies. D, Relative expression of BZR1 target genes in the HDA19 null allele mutant hda19 and wild type. Seven-day-old seedlings grown on 2 μm BRZ were treated with mock solution or 100 nm BL for 1 h. *: significant difference (<0.67 ratio, and p < 0.05) between wild-type and hda19 plants before BR treatment. (a) and (b) represent a significant difference (>1.5 ratio, and p < 0.05) in fold change between wild-type and hda19 plants.
BZR1 Interacts With a Cysteine Protease Inhibitor
CYSB is a cysteine protease inhibitor (41, 42). Its interaction with BZR1 was further confirmed through an in vitro pull-down assay (Fig. 5A), co-immunoprecipitation in transgenic Arabidopsis plants (Fig. 5B), and BiFC assays (supplemental Fig. S6A). Although both phosphorylated and unphosphorylated BZR1 were present in the input extracts, only dephosphorylated BZR1 was detected after immunoprecipitation, indicating that CYSB-YFP preferentially interacts with unphosphorylated BZR1 (Fig. 5B). BR treatment induced dephosphorylation of BZR1 (16), and increased the interaction of CYSB with unphosphorylated BZR1 (Fig. 5B, +BL). Consistent with the co-immunoprecipitation assay, BiFC assay showed that CYSB interacted with BZR1 in the nucleus, where unphosphorylated BZR1 is localized, whereas a CYSB-YFP fusion protein was localized mainly to the cytoplasm (supplemental Figs. S6A and S6B). It seems that interaction with BZR1 can retains a fraction of CYSB into the nucleus.
Fig. 5.
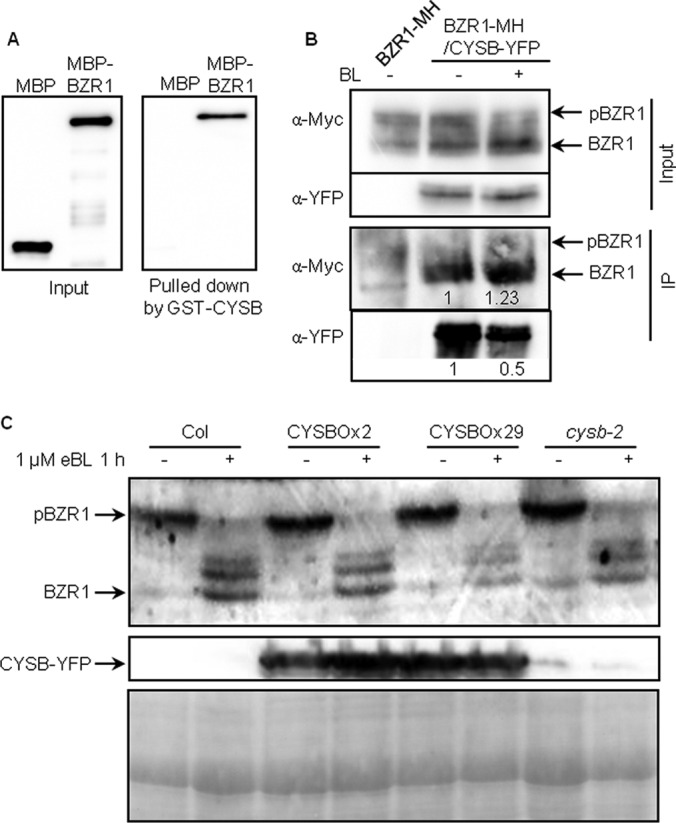
CYSB interacts with BZR1 in vivo and in vitro. A, Pull-down assay results showing the interaction of GST-CYSB with MBP-BZR1, but not with MBP alone. The immunoblot was probed with anti-MBP antibodies. B, Co-immunoprecipitation (IP) assay results showing that CYSB interacts with BZR1 in vivo. Protein extracts from Arabidopsis seedlings co-expressing 35S:: :bzr1-1D-MH and 35S::CYSB-YFP treated with 100 nm BL or mock for 1 h, or expressing 35S:: :bzr1-1D-MH alone, were immunoprecipitated with anti-YFP antibodies, and the immunoblot was probed using anti-c-Myc antibodies. Arrows indicate phosphorylated and dephosphorylated BZR1. Numbers below indicate the relative intensity of BZR1-MH and CYSB-YFP bands. C, Detection of the BZR1 level in Col, CYSB overexpression and cysb-2 plants exposed to 1 μm eBL or mock for 1 h using anti-BZR1 antibodies. Ponceau S (PS) staining indicates loading control.
As a protease inhibitor, CYSB is likely to affect BZR1 proteolysis. We therefore analyzed BZR1 using immunoblots in the CYSB loss-of-function and over-expression plants before or after 1 h BR treatment. The results showed that the accumulation level and phosphorylation status of BZR1 were not significantly different in the cysb-2 null allele and CYSB-overexpressing plants compared with wild type (Fig. 5C, and supplemental Fig. S7). CYSB is a member of gene family, and it is possible that the lack of phenotype and effect on BZR1 in the cysb-2 single mutant is because of genetic redundancy. The function of CYSB-BZR1 interaction remains to be elucidated in future studies.
Other Candidate Proteins Interact With BZR1
In addition to the above interacting proteins confirmed by both in vivo and in vitro assay, RH8, SYTA, and RD21A/B were also verified to interact with BZR1 by selected methods. The putative RNA helicases, RH8 and RH12, but not RH38, interacted with BZR1 and bzr1-1D in yeast (supplemental Fig. S8A). Furthermore, the N-terminal half of BZR1 (aa 1–198) interacted with RH8 (supplemental Fig. S8B). In addition, CAMTA5, UBP12, and CYP59, but not Met, interacted with BZR1 in BiFC assays (supplemental Fig. S6A). Most of these putative interacting proteins were localized to the nucleus (supplemental Fig. S6B), and the interactions with BZR1 were also detected by BiFC in the nucleus (supplemental Fig. S6A). SYTA is a phospholipid binding protein, which did not interact with BZR1 or bzr1-1D in yeast two-hybrid assay. However, SYTA was identified as an interacting protein of PP2A subunit A in a split ubiquitin yeast two-hybrid assay (supplemental Fig. S9). Thus, it is likely that SYTA associates with BZR1 and bzr1-1D through direct binding with PP2A. Together the confirmed and potential BZR1-interacting proteins are summarized in Table I. These proteins identified in this study, together with BZR1-interacting proteins characterized previously, and their binding domains of BZR1 are shown in a diagram of BZR1 complex in Fig. 6.
Fig. 6.
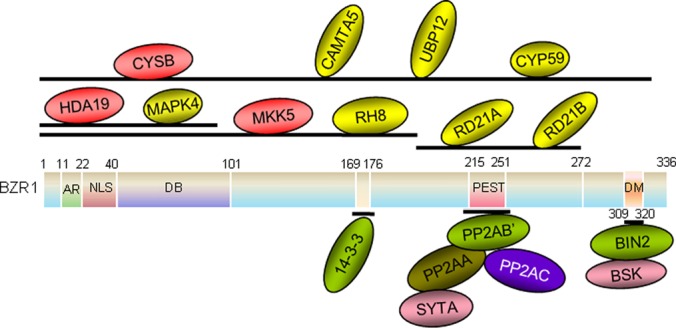
Diagrammatic representation of the BZR1-co-purified proteins identified in this study and their binding positions. 14–3-3, PP2A, and BIN2 (in green) are known BZR1-binding proteins. CYSB, HDA19, and MKK5 (in red) were shown by in vitro and in vivo experiments to be new BZR1-binding proteins. CAMTA5, UBP12, CYP59, MAPK4, RH8, RD21A, and RD21B (in yellow) were shown by in vitro experiments to be BZR1-interacting proteins. BSK1 and SYTA (in pink) interact indirectly with BZR1. The BZR1-binding regions for the interacting proteins are given above the bar.
DISCUSSION
Functional Proteomics is an Effective Approach for Identifying Signaling Components
Proteomic studies have made important contributions to the study of the BR signaling pathway. Genetic screening for BR-insensitive or -hypersensitive mutants has identified several key components of the BR signal transduction pathway (4–9, 17). However, some components were not identified by genetic approaches despite extensive effort, largely because of genetic redundancy. In Arabidopsis, each BR signaling component is encoded by members of a small gene family, and loss-of-function mutations in a single gene usually do not cause an obvious phenotype, with the exception of BRI1 (4). Several components were identified by dominant alleles that increased the level or activity of the protein, including BAK1, BIN2, BSU1, BZR1, BES1, and CDG1 (5–9, 17). A key component, BSK1, which fills the gap between the BRI1 receptor kinase and cytoplasmic components, was identified by quantitative proteomic studies of BR-induced changes in the plasma membrane proteome (14, 31). The proteomic identification of BZR1-associated proteins described here complements genetic analysis and provides important insight into BR signal transduction mechanisms.
TAP is believed to improve data specificity compared with single-step purification, and it has been widely used in yeast and other systems (43, 44). However, according to our results (supplemental Fig. S4) and previous reports (45), the original TAP tag, which contains the CBP and IgG-binding domain of protein A, is not very effective in Arabidopsis, likely because of high abundance of the endogenous CaMs and CBPs (46, 47). Nonspecific interaction with the TAP tag has been reported, e.g. HSP70 has been shown to bind with CaM in vitro (48). We have thus developed a new tandem affinity tag using 7×Myc and 6×His tags. The robust and inexpensive 6His tag allows for a large amount of starting material, which is required for the detection of low-abundance proteins, to be concentrated during the first step of purification. The higher affinity and specificity of 7Myc tag allows for tight binding of the low abundant transcription factor in second step purification. A similar tag has been reported recently as effective for use in plants (45). Despite the two-step purification procedure, large numbers of nonspecific proteins were detected in both the sample and negative control. In fact, 45.8% (27 of 59) of the proteins detected in the bzr1-1D-MH sample and 36.7% (18 of 49) of the proteins detected in the BZR1-MH sample were also identified in the bzr1-1D-CFP and BZR1-CFP control respectively, demonstrating that such a negative control is essential for distinguishing noise in affinity purification experiments.
Among the BZR1-associated proteins identified in our BZR1-MH experiment were several proteins that have been previously reported to have important functions in regulating BZR1 activity. These include BIN2 and several other GSK3-like kinases, which phosphorylate BZR1 (16); 14–3-3λ protein, which bind to BZR1 following its phosphorylation at S173 (12); and several subunits of PP2A, which dephosphorylates and activates BZR1. We have previously demonstrated that PP2A binds to the PEST domain of BZR1, and the bzr1-1D mutation (P234L) enhances BZR1 binding to PP2A (18). Two classes of known BZR1-interacting proteins, DELLA-domain proteins (21–23) and PIFs (24), were not detected in this study, likely because these proteins accumulate to high levels only under gibberellin-deficient conditions and in the dark, respectively (49, 50). The tissues used in this study were grown under light and not deficient in gibberellin. Apparently, selecting a tissue that accumulates the protein of interest is important for identification by proteomics. Therefore, the isolation of known BZR1 regulators not only demonstrates that the affinity purification method described here is effective, it also suggests that the additional proteins identified in this study are real BZR1-associated proteins that play a role in BR responses.
We found that five of the newly identified proteins interacted directly with BZR1 in yeast two-hybrid assays (Table I), and some of these interactions were further confirmed by in vitro assays. Interestingly, these proteins were not identified in previous yeast two-hybrid screens for BZR1- or BZR2/BES1-interacting proteins (12, 25). These two methods are apparently complementary. For example, a yeast two-hybrid screen using BZR1 as bait identified a large number of clones of 14–3-3 proteins (12, 13), but not any clones of PP2A subunit B', which was identified by affinity purification of the BZR1 complex and subsequently shown to interact with BZR1 in a yeast two-hybrid assay as well as other in vitro and in vivo assays (18). Furthermore, some proteins that associate with BZR1 indirectly through other BZR1-binding proteins or in a modification-dependent manner were identified by affinity purification but not by yeast two-hybrid assays. For example, SYTA showed no direct interaction with BZR1, but it showed an interaction with PP2A in yeast (supplemental Fig. S9B); Therefore, those BZR1-associated proteins that were identified by affinity purification but did not show an interaction in yeast might interact indirectly through other BZR1-interacting proteins, and thus still have a function in BR-regulated processes. The copurification of BSK1 with BZR1 suggests that BR-signaling components exist in a multi-protein complex, possibly involving BIN2 interaction with BRI1, BSU1 (15), and BSK1 (51).
Possible Functions of BZR1-associated Proteins
As a key transcription factor downstream of the BR pathway, BZR1 regulates the expression of a large number of BR-responsive genes (19). Reversible phosphorylation of BZR1 is a key determinant of its stability, subcellular localization, and transcriptional activity (12, 16, 18). In addition to 25 putative BIN2 phosphorylation sites, BZR1 has 11 putative phosphorylation sites for other kinases predicted using Omiga 2.0 software. We found that MKK5 and MAPK4 copurified with BZR1, and we confirmed the interactions using several in vitro and in vivo assays. Furthermore, we showed that MKK5 phosphorylates BZR1 in vitro (Fig. 3E). Interestingly, a recent study showed that BIN2 directly phosphorylates and inactivates both the MAPK kinase kinase YODA and its substrates MKK5 and MKK4 (the closest homolog of MKK5) (53, 54). The effects of MKK5 phosphorylation on BZR1 molecular function and possible cross-talk with other signaling pathways that regulate the MAPK module remain to be clarified.
The EAR motif (LxLxL), which is present in many plant transcription factors, has been shown to mediate interactions with the TOPLESS (TPL) repressor complex and to indirectly recruit HDA19; thus, it mediates transcriptional repression (55). The BZR1 C terminus has one putative EAR motif (LELTL, aa 325–329) (supplemental Fig. S1), and it is particularly interesting that HDA19 and TPL were copurified with BZR1. We confirmed the interaction between BZR1 and HDA19 in a yeast two-hybrid, BiFC, Co-IP, and in vitro pull-down assay (Fig. 4). EAR motifs have been reported to recruit HDA19 through an interaction with SAP18 (56). Our in vitro pull-down assay showed that HDA19 binds to the N-terminal domain of BZR1 but not the EAR-containing C-terminal domain. In addition, we observed slightly reduced expression of some of the BZR1 target genes that we tested in the hda19 mutant before BR treatment (Fig. 4D). Although such negative effects of hda19 on BZR1 target genes are in contrast to the function in transcriptional repression reported previously for HDA19 (55), similar negative effects of hda19 have been observed for several genes involved in pathogen and ethylene responses (39) The function of HDA19 in the regulation of gene expression by BZR1 requires further investigation.
CAMTA5, which has been reported to positively regulate CBF2 expression (57) and a V-PPase type proton pump (58), was identified in the BZR1 complex. CBF2 is a BR-repressed gene and a direct target of both BZR1 and BZR2/BES1 (19, 20). CAMTA5 may act as a cofactor of BZR1 to regulate target gene expression.
Two closely related DEAD-box RNA helicase-like proteins, RH8 and RH12, interact with BZR1 and bzr1-1D, and the interactions were confirmed in yeast two-hybrid assay, implying that BZR1 might be involved in pre-mRNA alternative splicing, processing, or metabolism. RH8 has been reported to confer pathogen resistance to plants by binding with the viral protein VPg (59).
Phosphorylated BZR1 is supposed to be targeted to the 26S proteasome for degradation (16), but the machinery has not been identified. In this work, we did not find any E3 ligase that mediates BZR1 ubiquitination, but we found two deubiquitinating enzymes (UBP12 and UBP13) and we confirmed that UBP12 interacts with BZR1 in the nucleus of tobacco cells (supplemental Fig. 6A). In addition to 26S proteasome-mediated degradation, BZR1 may also be subject to protease cleavage (supplemental Figs. S2B and S2C, red arrow). We found the cysteine endopeptidases RD21A and RD21B in the BZR1 and bzr1-1D complex (Table I), and we confirmed their interaction with BZR1 in yeast (supplemental Fig. S5A). On the other hand, CYSB was also found to interact with BZR1 in vitro and to preferentially interact with dephosphorylated BZR1 in vivo (Figs. 5A and 5B). We presume that CYSB counteracts cysteine peptidases and protects BZR1 from cleavage in vivo. However, the mutation or overexpression of CYSB had no obvious effect on BZR1 accumulation (Fig. 5C). Whether CYSB acts downstream of BZR1 remains to be tested. It was reported that CYSB overexpression increases plant tolerance to abiotic stress (41, 42). Whether the increase in tolerance is related to BZR1 and whether the CYSB-BZR1 interaction is functional under conditions of stress requires further evaluation.
In summary, using TAP combined with sensitive mass spectrometric analysis, we identified a large number of BZR1-associated proteins. Some of these proteins have been characterized in detail to explain key mechanisms of BZR1 regulation. The interactions of BZR1 with many newly identified proteins, confirmed in this study by in vivo and in vitro assays, will offer novel insight into the molecular functions of BZR1 (Fig. 6). Future functional studies of these BZR1-interacting proteins will enhance our understanding of the BZR1 functional mechanism and the BR signaling network.
Acknowledgments
We thank Dr. Zhongchi Liu at the University of Maryland for hda19 seeds (Salk_139445). T-DNA knock-out mutant cysb-2 was provided by Salk institute and Arabidopsis Stock Center (ABRC).
Footnotes
* This work was supported by National Basic Research Program of China (2013CB126903) and National Science Foundation of China (30740073, 31170264), by the Ph.D. Training Program of Education Ministry of China (20050094006), by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through Grant DE-FG02–08ER15973, and by NIH (R01GM066258); Mass spectrometry analysis was provided by the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A.L. Burlingame, Director) supported by the Biomedical Research Technology Program of the NIH National Center for Research Resources, NIH NCRR P41RR001614, 1S10RR012961, and 1S10RR019934.
 This article contains supplemental Figs. S1 to S9 and Table SI to SII.
This article contains supplemental Figs. S1 to S9 and Table SI to SII.
Author Contributions: Y.S. and Z.-Y.W. conceived the project and designed the experiments.Y.S. and C.W. performed BZR1 purification, Z.D. carried out in-gel digestion, and J.A.O-P. and A.L.B. did the mass spectrometry analysis. J.-X.S. contributed data in Figs 3A-E, 4, 5 and Supplemental Figs. S5B, S6, S7 and Table SII. Q.-X.C. made contributions to Figs. S6A and S8B. M.-Y.B. and C.-Q.W. contributed to Fig 3F. Y.Y. and Y.-L.Z. contributed to Supplemental Fig. S8A, M.Y. contributed to Supplemental Fig. S9, C.-C.M. contributed to Supplemental Fig. S5A.
1 The abbreviations used are:
- BR
- brassinosteroid
- PPZ
- propiconazole
- BZR1
- brassinazole-resistant 1
- LC-MS/MS
- liquid chromatography-electrospray tandem mass spectrometry
- PAP
- peroxidase-anti-peroxidase
- BIN2
- BR-insensitive 2
- BSK1
- BR-signaling kinase 1
- BiFC
- bimolecular fluorescence complementation
- TAP
- tandem affinity purification.
REFERENCES
- 1. Vert G., Nemhauser J. L., Geldner N., Hong F., Chory J. (2005) Molecular mechanisms of steroid hormone signaling in plants. Annu. Rev. Cell Dev. Biol. 21, 177–201 [DOI] [PubMed] [Google Scholar]
- 2. Wang Z. Y., Bai M. Y., Oh E., Zhu J. Y. (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 46, 701–724 [DOI] [PubMed] [Google Scholar]
- 3. Gendron J. M., Liu J. S., Fan M., Bai M. Y., Wenkel S., Springer P. S., Barton M. K., Wang Z. Y. (2012) Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 21152–21157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li J., Chory J. (1997) A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 90, 929–938 [DOI] [PubMed] [Google Scholar]
- 5. Li J., Wen J., Lease K. A., Doke J. T., Tax F. E., Walker J. C. (2002) BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 110, 213–222 [DOI] [PubMed] [Google Scholar]
- 6. Li J., Nam K. H. (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 295, 1299–1301 [DOI] [PubMed] [Google Scholar]
- 7. Mora-Garcia S., Vert G., Yin Y., Caño-Delgado A., Cheong H., Chory J. (2004) Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 18, 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang Z. Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujioka S., Yoshida S., Asami T., Chory J. (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2, 505–513 [DOI] [PubMed] [Google Scholar]
- 9. Yin Y., Wang Z. Y., Mora-Garcia S., Li J., Yoshida S., Asami T., Chory J. (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109, 181–191 [DOI] [PubMed] [Google Scholar]
- 10. Nam K. H., Li J. (2002) BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 110, 203–212 [DOI] [PubMed] [Google Scholar]
- 11. Wang X., Chory J. (2006) Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 313, 1118–1122 [DOI] [PubMed] [Google Scholar]
- 12. Gampala S. S., Kim T. W., He J. X., Tang W., Deng Z., Bai M. Y., Guan S., Lalonde S., Sun Y., Gendron J. M., Chen H., Shibagaki N., Ferl R. J., Ehrhardt D., Chong K., Burlingame A. L., Wang Z. Y. (2007) An essential role for 14–3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell. 13, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bai M. Y., Zhang L. Y., Gampala S. S., Zhu S. W., Song W. Y., Chong K., Wang Z. Y. (2007) Functions of OsBZR1 and 14–3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. U.S.A. 104, 13839–13844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang W., Kim T. W., Oses-Prieto J. A., Sun Y., Deng Z., Zhu S., Wang R., Burlingame A. L., Wang Z. Y. (2008) BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 321, 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim T. W., Guan S., Sun Y., Deng Z., Tang W., Shang J. X., Sun Y., Burlingame A. L., Wang Z. Y. (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell. Biol. 11, 1254–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. He J. X., Gendron J. M., Yang Y., Li J., Wang Z. Y. (2002) The GSK3-like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 99, 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim T. W., Guan S., Burlingame A. L., Wang Z. Y. (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell. 43, 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang W., Yuan M., Wang R., Yang Y., Wang C., Oses-Prieto J. A., Kim T. W., Zhou H. W., Deng Z., Gampala S. S., Gendron J. M., Jonassen E. M., Lillo C., DeLong A., Burlingame A. L., Sun Y., Wang Z. Y. (2011) PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell. Biol. 13, 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun Y., Fan X. Y., Cao D. M., Tang W., He K., Zhu J. Y., He J. X., Bai M. Y., Zhu S., Oh E., Patil S., Kim T. W., Ji H., Wong W. H., Rhee S. Y., Wang Z. Y. (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 19, 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu X., Li L., Zola J., Aluru M., Ye H., Foudree A., Guo H., Anderson S., Aluru S., Liu P., Rodermel S., Yin Y. (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J. 65, 634–646 [DOI] [PubMed] [Google Scholar]
- 21. Bai M. Y., Shang J. X., Oh E., Fan M., Bai Y., Zentella R., Sun T. P., Wang Z. Y. (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell. Biol. 14, 810–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li Q. F., Wang C., Jiang L., Li S., Sun S. S., He J. X. (2012) An interaction between BZR1 and DELLAs mediates direct signaling crosstalk between brassinosteroids and gibberellins in Arabidopsis. Sci. Signal. 5, ra72. [DOI] [PubMed] [Google Scholar]
- 23. Gallego-Bartolome J., Minguet E. G., Grau-Enguix F., Abbas M., Locascio A., Thomas S. G., Alabadi D., Blázquez M. A. (2012) Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 13446–13451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oh E., Zhu J. Y., Wang Z. Y. (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat. Cell. Biol. 14, 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. (2005) A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259 [DOI] [PubMed] [Google Scholar]
- 26. Yu X., Li L., Guo M., Chory J., Yin Y. (2008) Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 7618–7623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ye H., Li L., Guo H., Yin Y. (2012) MYBL2 is a substrate of GSK3-like kinase BIN2 and acts as a corepressor of BES1 in brassinosteroid signaling pathway in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 20142–20147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li L., Yu X., Thompson A., Guo M., Yoshida S., Asami T., Chory J., Yin Y. (2009) Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 58, 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 30. Deng Z., Zhang X., Tang W., Oses-Prieto J. A., Suzuki N., Gendron J. M., Chen H., Guan S., Chalkley R. J., Peterman T. K., Burlingame A. L., Wang Z. Y. (2007) A proteomics study of brassinosteroid response in Arabidopsis. Mol. Cell. Proteomics 6, 2058–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tang W., Deng Z., Oses-Prieto J. A., Suzuki N., Zhu S., Zhang X., Burlingame A. L., Wang Z. Y. (2008) Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol. Cell. Proteomics 7, 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Obrdlik P., El-Bakkoury M., Hamacher T., Cappellaro C., Vilarino C., Fleischer C., Ellerbrok H., Kamuzinzi R., Ledent V., Blaudez D., Sanders D., Revuelta J. L., Boles E., André B., Frommer W. B. (2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc. Natl. Acad. Sci. U.S.A. 101, 12242–12247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gomez-Gomez L., Boller T., Ausubel F. M., Sheen J. (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415, 977–983 [DOI] [PubMed] [Google Scholar]
- 34. Rohila J. S., Chen M., Cerny R., Fromm M. E. (2004) Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 38, 172–181 [DOI] [PubMed] [Google Scholar]
- 35. Rohila J. S., Chen M., Chen S., Chen J., Cerny R., Dardick C., Canlas P., Xu X., Gribskov M., Kanrar S., Zhu J. K., Ronald P., Fromm M. E. (2006) Protein-protein interactions of tandem affinity purification-tagged protein kinases in rice. Plant J. 46, 1–13 [DOI] [PubMed] [Google Scholar]
- 36. I. K., et al. (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Science 7, 8. [DOI] [PubMed] [Google Scholar]
- 37. Zhang L. Y., Bai M. Y., Wu J., Zhu J. Y., Wang H., Zhang Z., Wang W., Sun Y., Zhao J., Sun X., Yang H., Xu Y., Kim S. H., Fujioka S., Lin W. H., Chong K., Lu T., Wang Z. Y. (2009) Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21, 3767–3780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fong P. M., Tian L., Chen Z. J. (2006) Arabidopsis thaliana histone deacetylase 1 (AtHD1) is localized in euchromatic regions and demonstrates histone deacetylase activity in vitro. Cell. Res. 16, 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou C., Zhang L., Duan J., Miki B., Wu K. (2005) HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17, 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hollender C., Liu Z. (2008) Histone deacetylase genes in Arabidopsis development. J Integr Plant Biol. 50, 875–885 [DOI] [PubMed] [Google Scholar]
- 41. Zhang X., Liu S., Takano T. (2008) Two cysteine proteinase inhibitors from Arabidopsis thaliana, AtCYSa and AtCYSb, increasing the salt, drought, oxidation and cold tolerance. Plant Mol Biol. 68, 131–143 [DOI] [PubMed] [Google Scholar]
- 42. Hwang J. E., Hong J. K., Je J. H., Lee K. O., Kim D. Y., Lee S. Y., Lim C. O. (2009) Regulation of seed germination and seedling growth by an Arabidopsis phytocystatin isoform, AtCYS6. Plant Cell. Rep. 28, 1623–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 44. Knuesel M., Wan Y., Xiao Z., Holinger E., Lowe N., Wang W., Liu X. (2003) Identification of novel protein-protein interactions using a versatile mammalian tandem affinity purification expression system. Mol. Cell. Proteomics 2, 1225–1233 [DOI] [PubMed] [Google Scholar]
- 45. Rubio V., Shen Y., Saijo Y., Liu Y., Gusmaroli G., Dinesh-Kumar S. P., Deng X. W. (2005) An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 41, 767–778 [DOI] [PubMed] [Google Scholar]
- 46. McCormack E., Tsai Y. C., Braam J. (2005) Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389 [DOI] [PubMed] [Google Scholar]
- 47. Reddy V. S., Ali G. S., Reddy A. S. (2002) Genes encoding calmodulin-binding proteins in the Arabidopsis genome. J. Biol. Chem. 277, 9840–9852 [DOI] [PubMed] [Google Scholar]
- 48. Sun X. T., Li B., Zhou G. M., Tang W. Q., Bai J., Sun D. Y., Zhou R. G. (2000) Binding of the maize cytosolic Hsp70 to calmodulin, and identification of calmodulin-binding site in Hsp70. Plant Cell. Physiol. 41, 804–810 [DOI] [PubMed] [Google Scholar]
- 49. Sun T. P. (2010) Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol. 154, 567–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Castillon A., Shen H., Huq E. (2007) Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12, 514–521 [DOI] [PubMed] [Google Scholar]
- 51. Sreeramulu S., Mostizky Y., Sunitha S., Shani E., Nahum H., Salomon D., Hayun L. B., Gruetter C., Rauh D., Ori N., Sessa G. (2013) BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 74, 905–919 [DOI] [PubMed] [Google Scholar]
- 52. Deleted in proof.
- 53. Kim T. W., Michniewicz M., Bergmann D. C., Wang Z. Y. (2012) Brassinosteroid regulates stomatal development by GSK3-mediated inhibition of a MAPK pathway. Nature 482, 419–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Khan M., Rozhon W., Bigeard J., Pflieger D., Husar S., Pitzschke A., Teige M., Jonak C., Hirt H., Poppenberger B. (2013) Brassinosteroid-regulated GSK3/Shaggy-like kinases phosphorylate mitogen-activated protein (MAP) kinase kinases, which control stomata development in Arabidopsis thaliana. J. Biol. Chem. 288, 7519–7527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kagale S., Rozwadowski K. (2011) EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Song C. P., Galbraith D. W. (2006) AtSAP18, an orthologue of human SAP18, is involved in the regulation of salt stress and mediates transcriptional repression in Arabidopsis. Plant Mol. Biol. 60, 241–257 [DOI] [PubMed] [Google Scholar]
- 57. Doherty C. J., Van Buskirk H. A., Myers S. J., Thomashow M. F. (2009) Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mitsuda N., Isono T., Sato M. H. (2003) Arabidopsis CAMTA family proteins enhance V-PPase expression in pollen. Plant Cell. Physiol. 44, 975–981 [DOI] [PubMed] [Google Scholar]
- 59. Li D., Liu H., Zhang H., Wang X., Song F. (2008) OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J. Exp. Bot. 59, 2133–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]