Abstract
Programmed cell death is an event displayed by many different organisms along the evolutionary scale. In plants, programmed cell death is necessary for development and the hypersensitive response to stress or pathogenic infection. A common feature in programmed cell death across organisms is the translocation of cytochrome c from mitochondria to the cytosol. To better understand the role of cytochrome c in the onset of programmed cell death in plants, a proteomic approach was developed based on affinity chromatography and using Arabidopsis thaliana cytochrome c as bait. Using this approach, ten putative new cytochrome c partners were identified. Of these putative partners and as indicated by bimolecular fluorescence complementation, nine of them bind the heme protein in plant protoplasts and human cells as a heterologous system. The in vitro interaction between cytochrome c and such soluble cytochrome c-targets was further corroborated using surface plasmon resonance. Taken together, the results obtained in the study indicate that Arabidopsis thaliana cytochrome c interacts with several distinct proteins involved in protein folding, translational regulation, cell death, oxidative stress, DNA damage, energetic metabolism, and mRNA metabolism. Interestingly, some of these novel Arabidopsis thaliana cytochrome c-targets are closely related to those for Homo sapiens cytochrome c (Martínez-Fábregas et al., unpublished). These results indicate that the evolutionarily well-conserved cytosolic cytochrome c, appearing in organisms from plants to mammals, interacts with a wide range of targets on programmed cell death. The data have been deposited to the ProteomeXchange with identifier PXD000280.
Programmed cell death (PCD)1 is a fundamental event for the development of multicellular organisms and the homeostasis of their tissues. It is an evolutionarily conserved mechanism present in organisms ranging from yeast to mammals (1–3).
In mammals, cytochrome c (Cc) and dATP bind to apoptosis protease-activating factor-1 (Apaf-1) in the cytoplasm, a process leading to the formation of the Apaf-1/caspase-9 complex known as apoptosome. This apoptosome subsequently activates caspases-3 and -7 (4, 5). In other organisms, such as Caenorhabditis elegans or Drosophila melanogaster, however, Cc is not essential for the assembly and activation of the apoptosome (6) despite the presence of proteins homologous to Apaf-1—cell death abnormality-4 (CED-4) in C. elegans and Drosophila Apaf-1-related killer (Dark) in D. melanogaster—which have been found to be essential for caspase cascade activation. Furthermore, other organisms such as Arabidopsis thaliana lack Apaf-1 (7). In fact, only highly distant caspase homologues (metacaspases) (8, 9), serine proteases (saspases) (10), phytaspases (11) and VEIDases (12–14) with caspase-like activity have been detected in plants; however, their targets remain veiled and whether they are activated by Cc remains unclear.
Intriguingly, the release of Cc from mitochondria into the cytoplasm during the onset of PCD is an evolutionarily conserved event found in organisms ranging from yeast (15) and plants (16) to flies (17), and mammals (18). However, understanding of the roles of this phenomenon in different species can be said to be uneven at best. In fact, the release of Cc from mitochondria has thus far been considered a random event in all organisms, save mammals. Thus, the participation of Cc in the onset and progression of PCD needs to be further elucidated.
Even in the case of mammals, the role(s) of Cc in the cytoplasm during PCD remain(s) controversial. Recently, new putative functions of Cc, going beyond the already-established apoptosome assembly process, have been proposed in the nucleus (19, 20) and the endoplasmic reticulum (21–23). Neither these newly proposed functions nor other arising functions, such as oxidative stress (24), are as yet fully understood. This current state of affairs demands deeper exploration of the additional roles played by Cc in nonmammalian species.
In this study, putative novel Cc-partners involved in plant PCD were identified. For this identification, a proteomic approach was employed based on affinity chromatography and using Cc as bait. The Cc-interacting proteins were identified using nano-liquid chromatography tandem mass spectrometry (NanoLC-MS/MS). These Cc-partners were then further confirmed in vivo through bimolecular fluorescence complementation (BiFC) in A. thaliana protoplasts and human HEK293T cells, as a heterologous system. Finally, the Cc-GLY2, Cc-NRP1 and Cc-TCL interactions were corroborated in vitro using surface plasmon resonance (SPR).
These results indicate that Cc is able to interact with targets in the plant cell cytoplasm during PCD. Moreover, they provide new ways of understanding why Cc release is an evolutionarily well-conserved event, and allow us to propose Cc as a signaling messenger, which somehow controls different essential events during PCD.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Plasmid pCytA (25), containing the coding region for A. thaliana Cc, was used to obtain the Cc mutant A111C, in which the C-terminal alanine was replaced by a cysteine, through mutagenic PCR. The oligonucleotides designed to build the A111C mutant were 5′-gaaggcacctgttgatgaattc-3′ and 3′-cttccgtggacaactacttaag-5′. The A111C mutant was expressed and further purified using ionic exchange chromatography, a process previously described for wild-type Cc by Rodríguez-Roldán et al. (25).
A. thaliana Cell Cultures and PCD Induction
A. thaliana MM2d cell suspension cultures (Bayer CropScience) were grown in 1 × Murashige and Skoog (MS) medium (Duchefa Biochemie) supplemented with 30 g/L sucrose (Sigma-Aldrich), 0.5 mg/L NAA (Sigma-Aldrich), 0.05 mg/L kinetin (Sigma-Aldrich), 200 mg/L cefotaxime (Duchefa Biochemie) and 200 mg/L penicillin (Duchefa Biochemie) at 100 rpm and 25 °C.
PCD was then induced according to the procedure described by De Pinto et al. (26). Explained briefly, a stationary phase culture was diluted 5:100 (v/v). Following 3 days of growth under normal conditions, 35 mm H2O2 was added to 100 ml cell suspension cultures.
Cell Viability and Morphology
Cell viability was measured using the trypan blue dye exclusion test as described by De Pinto et al. (27) and cells were counted with a hemocytometer. The MM2d cell viability rate was calculated dividing the number of viable cells by the total number of cells. Following the collection of MM2d cells through centrifugation at 1000 × g for 10 min, cell morphology was analyzed and visualized using an Olympus BX60 fluorescence microscope.
Protein and Chlorophyll Determination
Protein content was determined using the Bradford assay (28), whereas chlorophyll determination carried out according to MacKinney's protocol (29).
Cell Extract Preparation for Purification by Affinity Chromatography
Cell extracts from 0.5 L of culture containing either untreated or 35 mm H2O2-treated cells were prepared for affinity chromatography purification. In both cases, cells were harvested following centrifugation at 1000 × g for 5 min, washed twice in PBS, pelleted again and resuspended to be further lysed by sonication in buffer I (50 mm Tris-HCl (pH 7.5), 50 mm NaCl, 0.25% Triton X-100) supplemented with 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, 10 μg·ml−1 leupeptin and 10 μg/ml of soybean trypsin inhibitor. Cellular debris was then removed through centrifugation at 20,000 × g for 30 min at 4 °C. Protein aliquots were stored at −80 °C.
Purification by Affinity Chromatography
As previously described in Azzi et al. (30), affinity chromatography was carried out in a column prepared by the covalent linkage of the Cc mutant A111C to the Thiol-Sepharose 4B (TS-4B) matrix (Pharmacia). As a control, a TS-4B matrix devoid of Cc (Blank TS-4B) was also prepared.
MM2d cell extracts, both those untreated and treated with 35 mm H2O2, were loaded into the columns, both with and without Cc. The columns were washed with 30 ml of buffer I and 30 ml of buffer II (50 mm Tris-HCl (pH 7.5), 75 mm NaCl) to remove nonspecifically bound proteins. Proteins interacting with greater strength were then eluted with 30 ml of buffer III (50 mm Tris-HCl (pH 7.5), 300 mm NaCl), collected, lyophilized, and stored at −80 °C before being analyzed using NanoLC-MS/MS.
Four sets of samples were thus obtained: (1) untreated cell extracts loaded into Blank TS-4B column, (2) untreated cell extracts loaded into the Cc TS-4B column, (3) cell extracts treated with H2O2 and purified with the Blank TS-4B column, and (4) cell extracts treated with H2O2 and purified using the Cc TS-4B column.
NanoLC-MS/MS
Before the performance of NanoLC-MS/MS analysis, the purified protein samples above were digested with trypsin. Thus, samples were treated with 8 m urea and 10 mm dithiothreitol (DTT). Following 1 h of incubation at 37 °C, iodoacetamide was added until a final concentration of 55 mm was reached and incubated for 1 h in the dark at room temperature. The samples were diluted with ammonium bicarbonate 4× until obtaining a final concentration of 2 m urea. Finally, 25 mg of recombinant trypsin was added and the mixture was incubated overnight at 37 °C.
The resulting peptides were analyzed using NanoLC-MS/MS on a linear trap quadrupole (LTQ; Thermo Electron), a linear ion trap mass spectrometer. The peptides were separated in a BioBasic C-18 PicoFrit column (75 μm (internal hamester) by 10 cm; New Objective) at a flow rate of 200 nL/min. Water and acetonitrile, both containing 0.1% formic acid, were used as solvents A and B, respectively. The peptides were trapped and desalted in the trap column for 5 min. The gradient was started and maintained for 5 min at 5% B, then ramped to 50% B over 120 min, ramped to 70% over 10 min and finally maintained at 95% B for another 10 min. The mass spectrometer was operated in data-dependent mode to automatically switch between full MS and MS/MS acquisition. Parameters for ion scanning were the following: full-scan MS (400–1800 m/z) plus top seven peaks Zoom/MS/MS (isolation width 2 m/z), normalized collision energy 35%.
Peak lists from all MS/MS spectra were extracted from the Xcalibur RAW files using a freely available program DTAsupercharge v1.19 (http://msquant.sourceforge.net).
Bioinformatics
For protein identification, the UniProt Arabidopsis protein database 100323 (90895 sequences, 33249465 residues) was searched using a local license for MASCOT 2.1. Database search parameters used were the following: trypsin as enzyme; peptide tolerance, 300 ppm; fragment ion tolerance, 0.6 Da; missed cleavage sites,1, and fixed modification, carbamidomethyl cysteine and variable modifications, methionine oxidation. In all protein identification, probability scores were greater than the score established by MASCOT (30) as significant, with a p value less than 0.05.
Design of Vectors for BiFC Assays
The cDNA coding available for nine out of the 10 Cc potential targets previously identified by the proteomic approach were purchased (ABRC Stocks, The Ohio State University, Columbus, OH, USA). The cDNA of Cc and its novel protein partners were fused with the C-end fragment of the yellow fluorescent protein (cYFP) of the pSPYCE vector and with the N-end part of the YFP (nYFP) of the pSPYNE vector, respectively (31). As a negative control, protoplasts were transfected with the chromatin-remodeling complex element SWI3B, which is unable to interact with Cc. The oligonucleotides indicated in the supplemental Data (supplemental Fig. S1A) were used to amplify the cDNAs while introducing proper restriction sites by PCR. In supplemental Fig. S1B, a scheme is shown of the vector constructs used for the BiFC assays.
Similarly, supplemental Fig. S2A represents the oligonucleotides required for cloning cDNAs into YFP vectors. BiFC experiments in human HEK293T cells were assayed after cloning Cc cDNA into the cYFP vector and the cDNA of its targets into the nYFP vector (supplemental Fig. S2B) (32). As discussed by Hu et al. (33), pBiFC-bJunYN155 and pBiFC-bFosYC155 were used as positive controls, whereas pBiFC-bJunYN155 and pBiFC-bFosΔZipYC155 were employed as negative controls.
BiFC Assays in A. thaliana Protoplasts: Cell Cultures, Protoplast Preparation, Cellular Transfection and Fluorescence Microscopy
Protoplasts were generated from 1-week-old A. thaliana MM2d cell cultures grown in MS medium. A 50 ml aliquot of cells were collected using centrifugation at 1500 rpm for 5 min and then resuspended in 50 ml of MS-Glucose/Mannitol (0.34 m), cellulose 1% and macerozyme 0.2%. Cells were incubated in this buffer for 3 h at 50 rpm in the dark to facilitate the digestion of the cell wall. Resulting protoplasts were collected following two, 5-min rounds of centrifugation at 800 rpm with a wash with 25 ml of MS-Glucose/Mannitol (0.34 m) carried out between centrifugations. The final pellet was resuspended in MS-Sucrose (0.28 m) and centrifuged at 800 rpm for 5 min. The A. thaliana protoplasts were recovered from supernatant.
Following Sheen's protocol (34), protoplasts were transiently transfected with the pSPYCE/pSPYNE BiFC vectors and incubated overnight; on PCD induction with 35 mm H2O2, the resulting fluorescence was monitored.
BiFC Assays in Human HEK293T: Cell Cultures, Cellular Transfection, and Fluorescence Microscopy
HEK293T cells were grown in Dulbecco's modified Eagle's medium (DMEM; PAA, Etobicoke, ON) supplemented with 2 mm l-glutamine (Invitrogen, Carlsbad, CA), 100 U/ml streptomycin (Invitrogen), 100 μg/ml penicillin (Invitrogen) and 10% heat-inactivated fetal bovine serum (PAA) at 37 °C in a humidified atmosphere of 5% CO2/95% air. HEK293T cells were grown to 80% confluence in 24-well plates with 500 μl of DMEM, containing 20 mm coverslips. Cells were transfected with the YFP BiFC vectors using the Lipofectamine 2000 Transfection Reagent (Invitrogen) following the manufacturer's instructions. To favor the protein expression of both constructs, the transfected cells were then incubated for 24 h at 37 °C. Apoptosis was further induced with 10 μm CPT (camptothecin) for 6 h and in vivo binding was assessed through YFP reconstitution visualized with fluorescence microscopy. Nuclei were then stained with 4′,6-diamidino-2-phenylindole (DAPI).
Western Blot Analysis
The HEK293T cells were harvested 48 h after transfection through centrifugation at 1500 rpm for 5 min. Total cell extracts were obtained through repeated freeze-thaw cycles. SDS-PAGE was performed using 12% polyacrylamide gels. Proteins were transferred onto nitrocellulose membranes (BioRad, Hercules, CA) using a semidry transfer system and immunoblotted with a rabbit anti-EGFP polyclonal antibody (1:1,000; Biovision Research Products). A horseradish-peroxidase (HRP)-conjugated goat anti-rabbit IgG (1:12,000; Sigma-Aldrich) was then used for detection. The immunoreactive bands were developed using ECL Plus Western blotting Detection System (Amersham Biosciences).
Cloning, Expression and Purification of A. thaliana Cc and Its Protein Partners
Wild-type A. thaliana Cc was cloned in the pBTR vector under lac promoter and expressed in E. coli BL-21. For this, 25 ml of precultures were grown overnight at 37 °C in LB medium. 2.5 ml of preculture was used to inoculate 2.5 L of the same medium in a 5 L Erlenmeyer flask. The culture was shaken at 30 °C for 24 h, after which further protein purification was carried out as indicated in Rodríguez-Roldán et al. (25).
Proteins interacting with A. thaliana Cc—GAPDC1, GLY2, NRP1, and TCL—were cloned in the pET-28a vector under the T7 promoter. cDNAs coding for Cc targets were purchased from ABRC. These constructs were used to express the Cc-targets in the E. coli BL-21 (DE3) RIL strain. 250 ml precultures in LB medium supplemented with 50 μg/ml kanamycin were grown overnight and used to inoculate 2.5 L of LB medium in 5 L flasks. Following the induction of cultures (1 mm IPTG) and growth at 30 °C for 24 h, cells were harvested at 6000 rpm for 10 min and resuspended in 40 ml lysis buffer (20 mm Tris-HCl buffer (pH 8), 0.8 m NaCl, 10 mm imidazole, 0.01% phenylmethylsulphonyl fluoride (PMSF), 0.2 mg/ml lysozyme, 5 mm DTT and 0.02 mg/ml DNase), sonicated for 4 min and then centrifuged at 20,000 rpm for 20 min. Proteins were further purified by means of an Ni-column (GE Healthcare).
SPR Measurements
The formation of complexes between A. thaliana Cc and its protein partners—GAPDC1, GLY2, NRP1, and TCL—was assayed with SPR using a BiaCore 3000 and CM4 Chips. An automated desorption procedure was performed before each experiment to ensure the cleanliness of the BiaCore tubing, channels, and sample injection port. The initial electrostatic attraction of A. thaliana Cc to the CM4 Sensor Chip surface was assessed by taking into account its isoelectric point and was optimized to pH 5.8. The plant Cc was then covalently attached to the matrix using standard amine-coupling chemistry, as previously described (35). A reference flow cell was used as a control in which the chip surface was treated as described above, but without the injection of plant Cc.
The binding measurements were performed at 25 °C using HBS-EP buffer containing 10 mm HEPES, 150 mm NaCl, 3 mm EDTA, and 0.005% surfactant P20, adjusted to pH 7.4. Interactions between plant Cc and its protein partners were analyzed by flowing several GAPDC1, GLY2, NRP1 and TCL proteins at different concentrations (from 0.1 to 10 μm) over the Cc-modified surface at a flow rate of 10 μl/min. Each concentration was injected at least three times. In each sensogram, the signals from the reference flow cell surface were subtracted.
RESULTS
PCD Induction in A. thaliana Cells
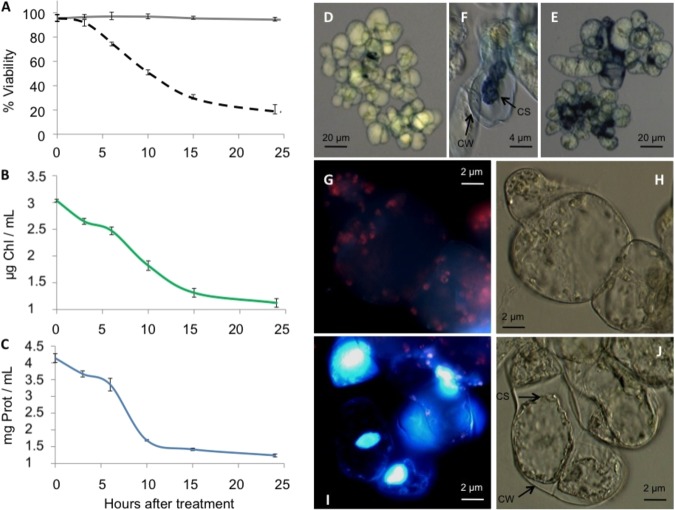
Oxidative stress has emerged as an important signal in the activation of plant PCD. Thus, following Desikan et al. (36), PCD was induced in A. thaliana MM2d cells growing in MS medium with 35 mm H2O2. Following a 6 h period during which Cc is known to be released into the cytosol (37), the cells were collected and morphological hallmarks occurring after PCD induction in plants (38) were analyzed. Fig. 1A shows the cellular viability (75%) after 6 h H2O2 treatment, a calculation obtained from the performance of the trypan blue dye exclusion test (Fig. 1D). Correspondingly, chlorophyll (Fig. 1B) and protein (Fig. 1C) contents decreased after 6 h.
Fig. 1.
PCD hallmarks in A. thaliana MM2d cell cultures treated with 35 mm H2O2. A, Cell viability measured using trypan blue dye exclusion assay. Untreated and H2O2-treated cells are represented by solid and dashed lines, respectively. Data is the result of three independent experiments, each including 500 cells. B, Effect of H2O2 treatment on chlorophyll concentration. One milliliter of H2O2-treated cell cultures was collected at the indicated times and the chlorophyll amount was calculated according to MacKinney's protocol (29). C, Effect of H2O2 treatment on protein concentration. Similar to (B), but the protein amount was determined using the Bradford assay (28). D, Bright-field microscope image showing cells treated with H2O2 for 6 h. Cells were stained with trypan blue dye. E, Similar to (D), but after 24 h of H2O2 treatment. F, Bright-field microscope image for 35 mm H2O2 treated cells, stained with trypan blue dye indicating a cell wall (CW) and cell shrinkage (CS). G–J, Changes in the cellular morphology of H2O2-treated cells analyzed by DAPI nuclear staining, chlorophyll fluorescence and bright-field images. Cells were observed under fluorescence (Left Panels G and I) and bright-field (Right panels H and J) microscopy. Upper (G and H) and lower (I and J) panels correspond to 0 and 24 h of H2O2 treatment, respectively. Apoptotic nuclei are stained in blue dye and, having undergone shrinkage, lack red fluorescence.
At 24 h after the H2O2 treatment, the amount of chlorophyll and protein decreased substantially, suggesting the activation of proteolysis (Figs. 1B and 1C) and coinciding with an important increase of cell death to 80% (Figs. 1A and 1E). The morphological features of PCD were visualized as indicated by Houot et al. (39) with cytoplasmic and nuclear shrinkage clearly appreciable (Figs. 1E and 1F). The degradation of chlorophyll and condensation of nuclei concur with the cytoplasmic shrinkage in the H2O2–treated cells (Figs. 1I and 1J) and not in the untreated cells (Figs. 1G and 1H).
Exploring Novel Cc Protein Partners during PCD Using a Proteomic Approach Based on Affinity Chromatography and NanoLC-MS/MS
As explained earlier, understanding the role of Cc during plant PCD is necessary to grasp the evolution of heme protein-dependent PCD pathways. Hence, novel Cc protein partners during PCD were identified using a proteomic approach relying on affinity chromatography and mass spectrometry (supplemental Fig. S3). In affinity chromatography, the thiol-Sepharose matrix (TS-4B) was used, which covalently binds proteins displaying solvent-exposed cysteines. Therefore, to attach Cc to the TS-4B matrix, the C-terminal alanine residue of Cc was replaced by a cysteine through mutagenic PCR. The resulting A111C Cc mutant, expressed and purified as described in Rodríguez-Roldán et al. (25), was able to bind covalently the TS-4B matrix through a disulfide bridge. The other two cysteine residues in Cc were unreactive because they were already binding the heme group (40). A TS-4B column devoid of A111C Cc (Blank TS-4B) was used as a control.
A. thaliana MM2d cell extracts, both untreated and H2O2-treated, were loaded into both types of TS-4B columns. Thus, proteins interacting with Cc were eluted by increasing ionic strength. Given the differing experimental conditions, four different protein samples were produced: (a) proteins from untreated cells purified using the control column; (b) proteins from H2O2-treated cells purified with the control column; (c) proteins from untreated cells purified using a column with Cc covalently bound to the matrix; (d) proteins from H2O2-treated cells purified using a column with Cc covalently bound to the matrix. All samples were separately lyophilized, with the mixture of proteins present in each being analyzed by NanoLC-MS/MS and the putative new Cc partners identified in this work summarized in Table I.
Table I. Cc protein partners identified with NanoLC-MS/MS. Up to 10 novel Cc partners identified. MW: molecular weight, pI: isoelectric point. #Cell location of the Cc-interacting proteins is that reported in the literature.
| Protein name | Uniprot ID | Score | Cell location# | MW (kDa)/pI |
|---|---|---|---|---|
| Apoptosis inhibitory 5 | O22957 | 47 | ND | 61.9/8.05 |
| Hydroxyacylglutathione hydrolase (GLY2) | Q0WQY6 | 62 | Cytoplasm | 29.2/5.93 |
| Glyceraldehyde 3-phosphate dehydrogenase (GAPDC1) | Q0WVE7 | 100 | Cytoplasm | 37.0/6.62 |
| Translation initiation factor eIF2γ | Q8LAP5 | 45 | Cytoplasm | 51.4/8.96 |
| Nucleosome assembly protein 1-related protein 1 (NRP1) | Q9CA59 | 404 | Nucleus, cytoplasm | 29.5/4.23 |
| Small nuclear ribonucleoprotein Sm/D1 | Q9SY09 | 96 | Nucleus, cytoplasm | 12.7/11.23 |
| Transcriptional coactivator-like (TCL) | Q8L773 | 186 | Nucleus, cytoplasm | 25.7/9.83 |
| Cysteine proteinase RD21 | P43297 | 34 | ER Bodies | 52.1/5.26 |
| Luminal-binding protein 1 (BiP1) | Q9LKR3 | 47 | Endoplasmic reticulum | 73.8/5.08 |
| Luminal-binding protein 2 (BiP2) | Q39043 | 47 | Endoplasmic reticulum | 73.8/5.11 |
Up to 10 novel Cc partners were identified (supplemental Data 1). Table I shows a list of these Cc-targets, indicating their molecular weight, isoelectric point, as well as their cellular localization as described in the literature.
Verification of Cc Interactions In vivo
The BiFC approach is a widely used technique permitting the analysis of protein–protein interactions in their biological environment, as well as the localization of the protein complexes in living cells (33). For this purpose, A. thaliana Cc cDNA as well as the cDNAs available for nine out of 10 Cc potential targets—excluding Apoptosis Inhibitory 5—were fused to the C-end and N-end fragments of YFP, respectively.
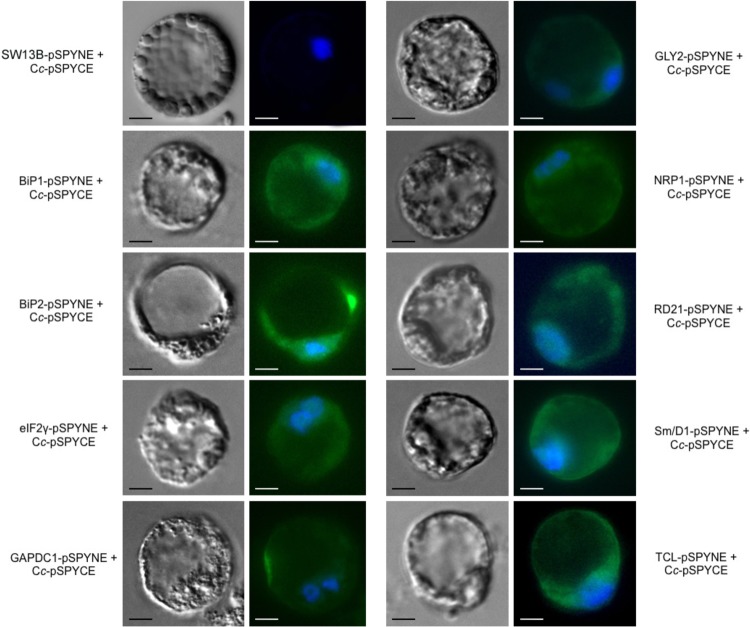
The resulting vectors were then codelivered into A. thaliana protoplasts, following the protocol described by Sheen (34). Fluorescence images showing the YFP reconstitution on Cc-target binding were captured 6 h following PCD induction (Fig. 2). In each case, representative bright-field (left) and fluorescence microscopy (right) images are shown. The YFP reconstitution is represented as green fluorescence and the nuclei appear in blue as a result of DAPI staining. Protoplasts transfected with chromatin-remodeling complex element SWI3B, a protein unable to interact with Cc, were used as a negative control. The observable YFP complementation for all constructs tested clearly indicates that the interactions previously identified in vitro through affinity chromatography also occur in vivo at the onset of PCD. In contrast, no YFP fluorescence signal was detected in cotransfected plant protoplasts before the H2O2-mediated PCD induction because the Cc-cYFP fusion protein is targeted to the mitochondria (see supplemental Fig. S4, for the target BIP2). To test the ability of the purported targets to interact with Cc in an independent heterologous system, BiFC assays were also performed on HEK293T cells. Plant cDNA was cloned into BiFC mammalian vectors nYFP and cYFP (32). HEK293T cells were efficiently cotransfected with both constructs and PCD was induced with 10 μm of CPT for 6 h, as reported in Johnson et al. (41).
Fig. 2.
BiFC assays in A. thaliana protoplasts. A. thaliana protoplasts were transfected with pSPYCE/pSPYNE vectors, as described in Sheen (34), to corroborate the in vivo interaction of Cc and its potential targets in BiFC. Images were captured 24 h following transient transfection and after 6 h of treatment with 35 mm H2O2. Reconstruction of eYFP leads to the obtainment of green fluorescence signal emission, indicative of interaction between Cc and its partners. Protoplasts transfected with chromatin-remodeling complex element SWI3B, a protein unable to interact with Cc, were used as a negative control. The nucleus was stained in blue using DAPI dye. Scale bar is 10 μm.
It had been previously reported that YFP fragments may complement each other with low efficiency, yet still yield fluorescent complexes, even in the absence of a specific interaction (42). Thus, to ensure that the interactions performed by Cc were not the result of spontaneous YFP complementation, several precise controls were designed. In particular, the most appropriate controls for BiFC assays are based on the expression of two fusion proteins that, being expressed in the same cellular compartment, are unable to interact. Therefore, pBiFC-bFosYC155 and pBiFC-bJunYN155 were used as a positive control and pBiFC-bFosΔZIPYC155 and pBiFC-bJunYN155 were used as a negative control (43).
Fluorescence results are shown in supplemental Fig. S5. Overall, the interaction between Cc and the 9 targets was also corroborated in human cells. Some of these interactions, like those involving eIF2γ, BiP1, BiP2, GAPDC1, and RD21, occur in the cytoplasm, whereas others like those involving GLY2 and Sm/D1 whose YFP fluorescence overlaps with DAPI staining, are characterized by nucleo-cytoplasmic localization. Interestingly, NRP1-Cc and TCL-Cc complexes take place inside the nucleus.
Notably, apo-Cc needs to be translocated from the cytosol to mitochondria to assemble its heme group and form a holoenzyme. Under homeostatic conditions, the punctuate pattern of fluorescence for the distribution of Cc, following the transfection of HEK293T cells with both the Cc-cYFP vector and empty nYFP vector, indicates mitochondrial localization of Cc (supplemental Fig. S6) (44). In contrast, CPT-treated cells show a diffuse fluorescence pattern, consistent with the release of Cc from mitochondria into the cytosol (supplemental Fig. S6). Transient expression of the Cc-targets fused to nYFP in the BiFC assays was confirmed by immunoblotting with a rabbit anti-EGFP polyclonal antibody, as shown in supplemental Fig. S5. All constructs assayed together with Cc yielded a band of the expected molecular mass for each target.
In vitro Validation of Cc Adducts: SPR Measurements
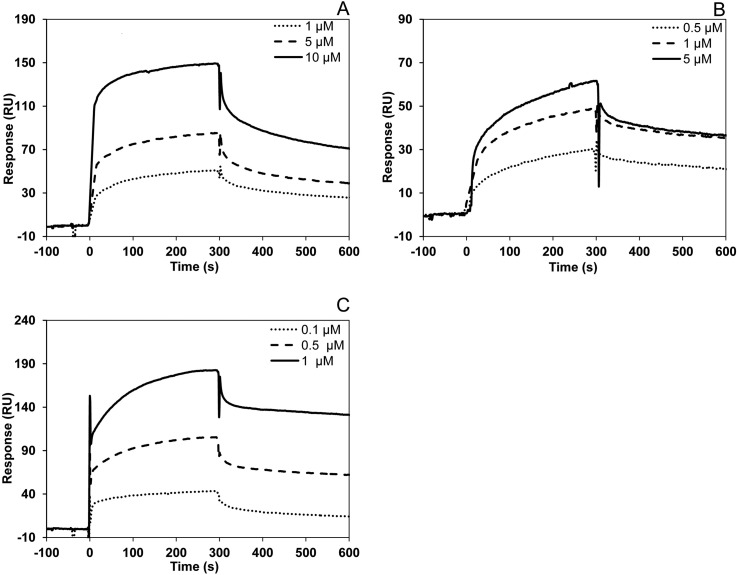
The interactions between plant Cc and the novel partners presented here were further analyzed through SPR in vitro. Of the nine Cc-targets, GAPDC1, GLY2, NRP1 and TCL were overexpressed as soluble recombinant proteins. Fig. 3 shows the SPR-sensograms resulting from increased flowing concentrations of GLY2, NRP1 or TCL on a Cc-immobilized chip. The background response was subtracted from the sample sensogram to obtain the actual binding response. The Cc-GAPDC1 interaction could not be detected by this technique. Values for dissociation constants (KD), association and dissociation rate constants (kon and koff, respectively) are shown in supplemental Data 2.
Fig. 3.
SPR Measurements. A, Sensograms recorded for the binding of plant Cc with GLY2. Three replicate injections were performed for each protein concentration. In each sensogram, the signals from the control surface were subtracted. B, Similar to (A), but for the plant Cc-NRP1 complex. C, Similar to (A), but for the plant Cc-TCL interaction.
DISCUSSION
Mammalian Cc acts as an electron shuttle in the respiratory chain. In addition, it is a constituent piece of the apoptosome platform during PCD. This moonlighting character of Cc has been underscored in recent studies revealing additional functions of the heme protein (19–24, 45).
As mentioned previously, during PCD, Cc is released from mitochondria into the cytosol in a wide variety of organisms including yeasts (15), plants (16), flies (17), and mammals (18). Nevertheless, a function for this cytoplasmic pool of Cc has, thus far, been described only in mammals (4, 5). This evolutionarily conserved release of Cc, as well as the additional and less-understood functions of mammalian Cc during PCD suggest the existence of a conserved signaling network hovering around Cc. Such would also seem to explain why Cc is a highly conserved protein (46) and, furthermore, why the mitochondria-to-cytosol translocation of the heme protein is a common, evolutionarily conserved event.
Based on a proteomic approach combined with BiFC, 9 Cc interacting proteins have been identified in A. thaliana. These novel Cc-targets are divided into seven main categories, according to their cellular functions (Fig. 4):
Fig. 4.
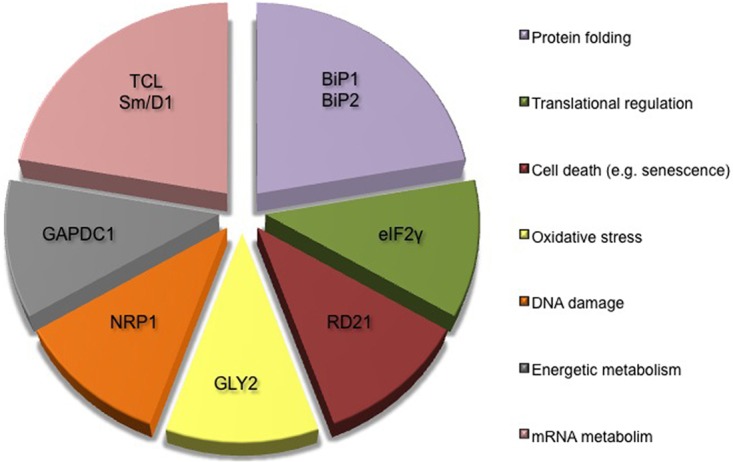
Principal functions ascribed to novel Cc protein partners. Diagram showing the principal functions of novel plant Cc protein partners identified in vitro with proteomics and corroborated in vivo with BiFC. All targets have been grouped into seven functional categories.
1. Protein Folding
Luminal binding proteins BiP1 and BiP2 have been related to endoplasmic reticulum (ER) stress, drought tolerance and leaf senescence (47). Moreover, the overexpression of BiP proteins in tobacco protoplasts increases cell tolerance to ER stress (48). BiP1 and BiP2 are also known to be close homologs of human HSPA5, which has been related to caspase inhibition (49) and the regulation of survival pathways and cell proliferation (50–52). Furthermore, translocation from the ER to cytoplasm has been previously described under ER stress conditions (51). Notably, HSPA5 is also targeted by Cc in human cells (Martínez-Fábregas et al., unpublished).
2. Translational Regulation
Eukaryotic initiation factor 2 (eIF2) is a heterotrimeric complex formed by eIF2α, eIF2β, and eIF2γ (53). In mammals, the inhibition of protein synthesis enhances the induction of PCD through different stimuli (54). Under apoptotic conditions, PKR phosphorylates eIF2α, leading to eIF2 dissociation and thereby hindering translation (55), an essential event for autophagy initiation (56). Notably, it was found that eIF2α (Martínez-Fábregas et al., unpublished) and eIF2γ bind Cc in human and A. thaliana cells, respectively, indicating eIF2 to be a common target of the heme protein in eukaryotes.
3. Cell Death
In animal cells, the activation of cysteine proteases is an important step for PCD (57). These enzymes have also been detected in plant cells undergoing PCD (58–60).
RD21 (Responsive to Dehydration 21) is a cysteine protease synthesized as a 57 kDa inactive precursor which matures into a 33 kDa active form (61). RD21 contains a redox-sensitive catalytic site, GxCGSCW, with two cysteine residues capable of forming a disulfide bond (62). It has been recently proposed that protein disulfide-isomerase-5 (PDI5) sequesters plant cysteine proteases in the protein storage vacuoles of endothelial cells, thereby blocking their protease activity until the onset of PCD. According to yeast two-hybrid assays, PDI5 interacts with RD21 and inhibits recombinant RD21 activity in vitro (63). Hence, PDI5 seems to be involved in regulating the timing of PCD (64).
4. Oxidative Stress
The glyoxalase system consists of two enzymes, GLY1 and GLY2, involved in methylglyoxal (MG) detoxification, though novel glyoxalases have been recently described (65). MG is produced in all living organisms and its levels in plants are enhanced on exposure to different abiotic stresses (66). Excessive MG formation leads to ROS production, causing oxidative stress (67). Increased levels of GLY2 have been detected in mammalian tumor cells and GLY2 inhibitors have been used to slow the growth of tumor cells in vitro (68, 69). Moreover, MG has been demonstrated to induce apoptosis in different types of mammalian cells (70–73).
5. DNA Damage
Nucleosome assembly protein 1 (NAP-1)-related proteins (NRP) are well conserved in all kingdoms. In A. thaliana, the nrp1–1 nrp2–1 double loss-of-function mutant is highly sensitive to genotoxic stress and shows increased levels of DNA damage, being essential for cell proliferation (74).
NRP1 is also a homolog of human SET (74), which has been related to DNA repair after single-strand breaks during oxidative stress (75). Nevertheless, this function has not yet been attributed to plant NRP1. Interestingly, SET has been identified as a human Cc target (Martínez-Fábregas et al., unpublished).
6. Energetic Metabolism
GAPDC1 is homologous to mammal GAPDH, a soluble multitasking protein involved in glycolysis, apoptosis induction, cell signaling, tRNA export and DNA repair, among other functions (76). Recently, new anti-PCD functions for plant GAPDH have begun to emerge (e.g. suppression of reactive oxygen species) (77). Notably, Cc does not target the same protein in human cells, but rather another, ALDOA (Martinez-Fábregas et al., unpublished), affecting the same metabolite, GAL-3P.
7. mRNA Metabolism
The spliceosome, a macromolecular machine containing several uridine-rich small nuclear ribonucleoproteins (U snRNPs) and many non-RNP splicing factors, is a “major player” in splicing (78). Although there are different U snRNP complexes, all share core components such as Sm proteins (e.g. Sm/D1) (79). Alternative splicing has been recently linked to apoptosis in mammals by different groups (80, 81). The mutation, deletion or knockdown of core spliceosomal proteins can result in altered splicing patterns in yeast cells (82–85), fly cells (86), and mammalian cells (87–91).
The TREX (transcription/export) complex has a conserved role in coupling transcription to mRNA export in yeast and metazoan. It consists of two export factors, Yra1/ALY/REF and Sub2/UAP56, along with the THO transcription elongation complex (92, 93). The A. thaliana genome contains at least one gene (At5g59950, TCL) homologous to Yra1/ALY/REF and two genes (At5g11170 and At5g11200) homologous to Sub2/UAP56 (94). The existence of the THO complex in plants, as well, underscores the importance of THO-related proteins in the context of plant development.
It is worth mentioning that most of these new Cc-interacting proteins, or their homologs in humans, play an anti-PCD role in plant cells, except in the cases of RD21 and API5. Finally, SPR measurements permitted us to corroborate the in vitro interaction of three targets—GLY2, NRP1, and TCL—with Cc.
In summary, the new data produced regarding the poorly understood process of nonmammalian PCD suggests that Cc targets analogous functions in different organisms. Furthermore, the data points to the conservation of the role of Cc during PCD even in organisms devoid of an apoptosome. This supports the theory proposed by D.R. Green et al. that the PCD signaling role of Cc emerged early in the evolutionary timeline (95). Thus, although the Cc-targets likely vary according to the organism, the results recorded here indicate that cytoplasmic Cc targets processes essential for cell life and may ensure the correct progress of PCD.
Supplementary Material
Acknowledgments
We thank Dr. A. Orea (Seville, Spain) for her work with fluorescence microscopy and P. Alcántara (Seville, Spain) for her technical support. N-YFP and C-YFP vectors were kindly provided by Dr. P. Ciruela (Barcelona, Spain), positive and negative controls for BiFC assays in HEK293T cells were kindly offered by Dr. T.K. Kerppola (Ann Arbor, Michigan, USA), pSPYCE and pSPYNE vectors as well as negative controls were contributed by Dr. P.L. Rodríguez (Valencia, Spain). The proteomic analyses were carried out at the “Proteomics Facility UCM-FPCM', a member of the Spanish ProteoRed Network.”
Footnotes
* This study was funded by the Spanish Ministry of Economy and Competitiveness (BFU2009-07190 and BFU2012-31670) and the Regional Government of Andalusia (BIO198). Additional funding for J. Martínez-Fábregas was provided by a Spanish Ministry of Science and Innovation FPI grant (BES-2007-16156). Support for the recording of SPR measurements at Saarland University (Saarbrücken, Germany) was provided to K. González-Arzola by an IUBMB Wood-Whelam Fellowship. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (96) with the dataset identifier PXD000280 and DOI 10.6019/PXD000280.
 This article contains supplemental Figs. S1 to S6, Data Files S1 and S2, and Text.
This article contains supplemental Figs. S1 to S6, Data Files S1 and S2, and Text.
1 The abbreviations used are:
- PCD
- programmed cell death
- Cc
- Cytochrome c
- Apaf-1
- apoptosis protease-activating factor-1
- CED-4
- cell death abnormality-4
- Dark
- Drosophila Apaf-1-related killer
- BiFC
- Bimolecular fluorescence complementation
- CPT
- Camptothecin
- SPR
- surface plasmon resonance
- NAA
- naphthalene acetic acid
- MS
- Murashige and Skoog medium
- TS-4B
- Thiol-Sepharose 4B
- YFP
- yellow fluorescent protein
- DAPI
- 4′,6-diamidino-2-phenylindole
- DMEM
- Dulbecco's modified Eagle's medium.
REFERENCES
- 1. Vander Heiden M. G., Chandel N. S., Williamson E. K., Schumacker P. T., Thompson C. B. (1997) Bcl-XL regulates the membrane potential and volume homeostasis of mitochondria. Cell 91, 627–637 [DOI] [PubMed] [Google Scholar]
- 2. Matsuyama S., Nouraini S., Reed J. C. (1999) Yeast as a tool for apoptosis research. Curr. Opin. Microbiol. 2, 618–623 [DOI] [PubMed] [Google Scholar]
- 3. Sundström J. F., Vaculova A., Smertenko A. P., Savenkov E. I., Golovko A., Minina E., Tiwari B. S., Rodriguez-Nieto S., Zamyatnin A. A., Välineva T., Saarikettu J., Frilander M. J., Suarez M. F., Zavialov A., Stahl U., Hussey P. J., Silvennoinen O., Sundberg E., Zhivotovsky B., Bozhkov P. V. (2009) Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat. Cell Biol. 11, 1347–1354 [DOI] [PubMed] [Google Scholar]
- 4. Li P., Nijhawan D., Budihardjo I., Srinivasula S. M., Ahmad M., Alnemri E. S., Wang X. (1997) Cytochrome c and dATP-dependent formation of Apaf-1/Caspase-9 complex initiates an apoptotic protease cascade. Cell 91, 479–489 [DOI] [PubMed] [Google Scholar]
- 5. Yu X., Acehan D., Ménétret J. F., Booth C. R., Ludtke S. J., Riedl S. J., Shi Y., Wang X., Akey C. W. (2005) A structure of the human apoptosome at 12.8 Å resolution provides insights into this cell death platform. Structure 13, 1725–1735 [DOI] [PubMed] [Google Scholar]
- 6. Adams J. M., Cory S. (2002) Apoptosomes: engines for caspase activation. Curr. Opin. Cell Biol. 14, 715–720 [DOI] [PubMed] [Google Scholar]
- 7. van Nocker S., Ludwig P. (2003) The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim S. M., Bae C., Oh S. K., Choi D. (2013) A pepper (Capsicum annuum L.) metacaspase 9 (Camc9) plays a role in pathogen-induced cell death in plants. Mol. Plant Pathol. 14, 557–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Watanabe N., Lam E. (2011) Arabidopsis metacaspase 2d is a positive mediator of cell death induced during biotic and abiotic stresses. Plant J. 66, 969–982 [DOI] [PubMed] [Google Scholar]
- 10. Coffeen W. C., Wolpert T. J. (2004) Purification and characterization of serine proteases that exhibit caspase-like activity and are associated with programmed cell death in Avena sativa. Plant Cell 16, 857–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chichkova N. V., Shaw J., Galiullina R. A, Drury G. E., Tuzhikov A. I., Kim S. H., Kalkum M., Hong T. B., Gorshkova E. N., Torrance L., Vartapetian A. B., Taliansky M. (2010) Phytaspase, a relocalisable cell death promoting plant protease with caspase specificity. EMBO J. 29, 1149–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bosch M., Franklin-Tong V. E. (2007) Temporal and spatial activation of caspase-like enzymes induced by self-incompatibility in Papaver pollen. Proc. Natl. Acad. Sci. U.S.A. 104, 18327–18332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borén M., Höglund A. S., Bozhkov P, Jansson C. (2006) Developmental regulation of a VEIDase caspase-like proteolytic activity in barley caryopsis. J. Exp. Bot. 57, 3747–3753 [DOI] [PubMed] [Google Scholar]
- 14. Bozhkov P. V., Filonova L. H., Suarez M. F., Helmersson A., Smertenko A. P., Zhivotovsky B., von Arnold S. (2004) VEIDase is a principal caspase-like activity involved in plant programmed cell death and essential for embryonic pattern formation. Cell Death Differ. 11, 175–182 [DOI] [PubMed] [Google Scholar]
- 15. Giannattasio S., Atlante A., Antonacci L., Guaragnella N., Lattanzio P., Passarella S., Marra E. (2008) Cytochrome c is released from coupled mitochondria of yeast en route to acetic acid-induced programmed cell death and can work as an electron donor and a ROS scavenger. FEBS Lett. 582, 1519–1525 [DOI] [PubMed] [Google Scholar]
- 16. Balk J., Leaver C. J., McCabe P. F. (1999) Translocation of cytochrome c from the mitochondria to the cytosol occurs during heat-induced programmed cell death in cucumber plants. FEBS Lett. 463, 151–154 [DOI] [PubMed] [Google Scholar]
- 17. Arama E., Bader M., Srivastava M., Bergmann A., Steller H. (2006) The two Drosophila cytochrome c proteins can function in both respiration and caspase activation. EMBO J. 25, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Robertson J. D., Enoksson M., Suomela M., Zhivotovsky B., Orrenius S. (2002) Caspase-2 acts upstream of mitochondria to promote cytochrome c release during etoposide-induced apoptosis. J. Biol. Chem. 277, 29803–29809 [DOI] [PubMed] [Google Scholar]
- 19. Ruíz-Vela A., González de Buitrago G., Martínez-A C. (2002) Nuclear Apaf-1 and cytochrome c redistribution following stress-induced apoptosis. FEBS Lett. 517, 133–138 [DOI] [PubMed] [Google Scholar]
- 20. Nur-E-Kamal A., Gross S. R., Pan Z., Balklava Z., Ma J., Liu L. F. (2004) Nuclear translocation of cytochrome c during apoptosis. J. Biol. Chem. 279, 24911–24914 [DOI] [PubMed] [Google Scholar]
- 21. Boehning D., Patterson R. L., Sedaghat L., Glebova N. O., Kurosaki T., Snyder S. H. (2003) Cytochrome c binds to inositol (1,4,5) trisphosphate receptors, amplifying calcium-dependent apoptosis. Nat. Cell Biol. 5, 1051–1061 [DOI] [PubMed] [Google Scholar]
- 22. Boehning D., van Rossum D. B., Patterson R. L., Snyder S. H. (2005) A peptide inhibitor of cytochrome c/inositol 1,4,5-trisphosphate receptor binding blocks intrinsic and extrinsic cell death pathways. Proc. Natl. Acad. Sci. U.S.A. 102, 1466–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Szado T., Vanderheyden V., Parys J. B., De Smedt H., Rietdorf K., Kotelevets L., Chastre E., Khan F., Landegren U., Söderberg O., Bootman M. D., Roderick H. L. (2008) Phosphorylation of inositol 1,4,5-trisphosphate receptors by protein kinase B/Akt inhibits Ca2+ release and apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 2427–2432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hüttemann M., Pecina P., Rainbolt M., Sanderson T. H., Kagan V. E., Samavati L., Doan J. W., Lee I. (2011) The multiple functions of cytochrome c and their regulation in life and death decisions of the mammalian cell: From respiration to apoptosis. Mitochondrion 11, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rodríguez-Roldán V., García-Heredia J. M., Navarro J. A., Hervás M., De la Cerda B., Molina-Heredia F. P., De la Rosa M. A. (2006) A comparative analysis of the reactivity of plant, horse, and human respiratory cytochrome c towards cytochrome c oxidase. Biochem. Biophys. Res. Commun. 346, 1108–1113 [DOI] [PubMed] [Google Scholar]
- 26. De Pinto M. C., Paradiso A., Leonetti P., De Gara L. (2006) Hydrogen peroxide, nitric oxide and cytosolic ascorbate peroxidase at the crossroad between defence and cell death. Plant J. 48, 784–795 [DOI] [PubMed] [Google Scholar]
- 27. De Pinto M. C., Francis D., De Gara L. (1999) The redox state of the ascorbate-dehydroascorbate pair as a specific sensor of cell division in tobacco BY-2 cells. Protoplasma 209, 90–97 [DOI] [PubMed] [Google Scholar]
- 28. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 29. MacKinney G. (1941) Absorption of light by chlorophyll solutions. J. Biol. Chem. 140, 315–322 [Google Scholar]
- 30. Azzi A., Bill K., Broger C. (1982) Affinity chromatography purification of cytochrome c binding enzymes. Proc. Natl. Acad. Sci. U.S.A. 79, 2447–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004) Visualization of protein interactions in living plants cells using bimolecular fluorescence complementation. Plant J. 40, 428–438 [DOI] [PubMed] [Google Scholar]
- 32. Gandia J., Galino J., Amaral O. B., Soriano A., Lluís C., Franco R., Ciruela F. (2008) Detection of higher-order G protein-coupled receptor oligomers by a combined BRET-BiFC technique. FEBS Lett. 582, 2979–2984 [DOI] [PubMed] [Google Scholar]
- 33. Hu C. D., Grinberg A. V., Kerppola T. K. (2006) Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC) analysis. Curr. Protoc. Cell Biol. (Chapter 21, Unit 21.3) [DOI] [PubMed] [Google Scholar]
- 34. Sheen J. (2001) Signal transduction in Maize and Arabidopsis Mesophyll protoplasts. Plant Physiol. 127, 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- 35. Janocha S., Bichet A., Zöllner A., Bernhardt R. (2011) Substitution of lysine with glutamic acid at position 193 in bovine CYP11A1 significantly affects protein oligomerization and solubility but not enzymatic activity. Biochim. Biophys. Acta 1814, 126–131 [DOI] [PubMed] [Google Scholar]
- 36. Desikan R., Reynolds A., Hancock J. T., Neill S. J. (1998) Harpin and hydrogen peroxide both initiate programmed cell death but have differential effects on defence gene expression in Arabidopsis suspension cultures. Biochem. J. 330, 115–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. García-Heredia J. M., Hervás M., De la Rosa M. A., Navarro J. A. (2008) Acetylsalicylic acid induces programmed cell death in Arabidopsis cell cultures. Planta 228, 89–97 [DOI] [PubMed] [Google Scholar]
- 38. Hörtensteiner S. (2006) Chlorophyll degradation during senescence. Ann. Rev. Plant Biol. 57, 55–77 [DOI] [PubMed] [Google Scholar]
- 39. Houot V., Etienne P., Petitot A. S., Barbier S., Blein J. P., Suty L. (2001) Hydrogen peroxide induces programmed cell death features in cultured tobacco BY-2 cells, in a dose-dependent manner. J. Exp. Bot. 52, 1721–1730 [PubMed] [Google Scholar]
- 40. Allen J. W., Tomlinson E. J., Hong L., Ferguson S. J. (2002) The Escherichia coli cytochrome c maturation (Ccm) system does not detectably attach heme to single cysteine variants of an apocytochrome c. J. Biol. Chem. 277, 33559–33563 [DOI] [PubMed] [Google Scholar]
- 41. Johnson N., Ng T. T., Parkin J. M. (1997) Camptothecin causes cell cycle perturbations within T-Lymphoblastoid cells followed by dose dependent induction of apoptosis. Leuk. Res. 21, 961–972 [DOI] [PubMed] [Google Scholar]
- 42. Kerppola T. K. (2006) Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1, 1278–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hu C. D., Chinenov Y., Kerppola T. K. (2002) Visualization of interactions among bZip and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol. Cell 9, 789–798 [DOI] [PubMed] [Google Scholar]
- 44. Goldstein J. C., Muñoz-Pinedo C., Ricci J. E., Adams S. R., Kelekar A., Schuler M., Tsien R. Y., Green D. R. (2005) Cytochrome c is released in a single step during apoptosis. Cell Death Differ. 12, 453–462 [DOI] [PubMed] [Google Scholar]
- 45. Godoy L. C., Muñoz-Pinedo C., Castro L., Cardaci S., Schonhoff C. M., King M., Tórtora V., Marín M., Miao Q., Jiang J. F., Kapralov A., Jemmerson R., Silkstone G. G., Patel J. N., Evans J. E, Wilson M. T., Green D. R., Kagan V. E., Radi R., Mannick J. B. (2009) Disruption of the M80-Fe ligation stimulates the translocation of cytochrome c to the cytoplasm and nucleus in nonapoptotic cells. Proc. Natl. Acad. Sci. U.S.A. 106, 2653–2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baba M. L., Darga L. L., Goodman M., Czelusniak J. (1981) Evolution of cytochrome c investigated by the maximum parsimony method. J. Mol. Evol. 17, 197–213 [DOI] [PubMed] [Google Scholar]
- 47. Valente M. A., Faria J., Soares-Ramos J., Reis P. A., Pinheiro G. L., Piovesan N. D., Morais A. P., Menezes C. C., Cano M. A., Fietto L. G., Loureiro M. E., Aragao F. J., Fontes E. P. (2009) The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J. Exp. Bot. 60, 533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leborgne-Castel N., Jelitto-Van Dooren E. P., Crofts A. J., Denecke J. (1999) Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell 11, 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reddy R. K., Mao C., Baumeister P., Austin R. C., Kaufman R. J., Lee A. S. (2003) Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: Role of ATP binding site in suppression of caspase 7 activation. J. Biol. Chem. 278, 20915–20924 [DOI] [PubMed] [Google Scholar]
- 50. Jin S., Zhuo Y., Guo W., Field J. (2005) p21-activated kinase 1 (Pak1)-dependent phosphorylation of Raf-1 regulates its mitochondrial localization, phosphorylation of BAD, and Bcl-2 association. J. Biol. Chem. 280, 24698–24705 [DOI] [PubMed] [Google Scholar]
- 51. Shkoda A., Ruiz P. A., Daniel H., Kim S. C., Rogler G., Sartor R. B., Haller D. (2007) Interleukin-10 blocked endoplasmic reticulum stress in intestinal epithelial cells: impact on chronic inflammation. Gastroenterology 132, 190–207 [DOI] [PubMed] [Google Scholar]
- 52. Shu C. W., Sun F. C., Cho J. H., Lin C. C., Liu P. F., Chen P. Y., Chang M. D., Fu H. W., Lai Y. K. (2008) GRP78 and Raf-1 cooperatively confer resistance to endoplasmic reticulum stress-induced apoptosis. J. Cell. Physiol. 215, 627–635 [DOI] [PubMed] [Google Scholar]
- 53. Sugarani R. N., Kamindla R., Ehtesham N. Z., Ramaiah K. V. (2005) Interaction of recombinant human eIF2 subunits with eIF2B and eIF2α kinases. Biochem. Biophys. Res. Commun. 338, 1766–1772 [DOI] [PubMed] [Google Scholar]
- 54. Polunovsky V. A., Wendt C. H., Ingbar D. H., Peterson M. S., Bitterman P. B. (1994) Induction of endothelial cell apoptosis by TNF-α: modulation by inhibitors of protein synthesis. Exp. Cell Res. 214, 584–594 [DOI] [PubMed] [Google Scholar]
- 55. Saelens X., Kalai M., Vandenabeele P. (2001) Translation inhibition in apoptosis: Caspase-dependent PKR activation and eIF2-α phosphorylation. J. Biol. Chem. 276, 41620–41628 [DOI] [PubMed] [Google Scholar]
- 56. Kouroku Y., Fujita E., Tanida I., Ueno T., Isoai A., Kumagai H., Ogawa S., Kaufman R. J., Kominami E., Momoi T. (2007) ER stress (PERK/eIF2-α phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 14, 230–239 [DOI] [PubMed] [Google Scholar]
- 57. Jacobson M. D., Weil M., Raff M. C. (1997) Programmed cell death in animal development. Cell 88, 347–354 [DOI] [PubMed] [Google Scholar]
- 58. Minami A., Fukuda H. (1995) Transient and specific expression of a cysteine endopeptidases associated with autolysis during differentiation of Zinnia mesophyll cells into tracheary elements. Plant Cell Physiol. 26, 1599–1606 [PubMed] [Google Scholar]
- 59. Solomon M., Belenghi B., Delledone M., Menachem E., Levine A. (1999) The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11, 431–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Watanabe N., Lam E. (2005) Two Arabidopsis metacaspases AtMCP1b and AtMCP2b are Arginine/Lysine-specific cysteine proteases and activate apoptosis-like cell death in yeast. J. Biol. Chem. 280, 14691–14699 [DOI] [PubMed] [Google Scholar]
- 61. Yamada K., Matsushima R., Nishimura M., Hara-Nishimura I. (2001) A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 127, 1626–1634 [PMC free article] [PubMed] [Google Scholar]
- 62. Balmer Y., Koller A., del Val G., Manieri W., Schürmann P., Buchanan B. B. (2003) Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proc. Natl. Acad. Sci. U.S.A. 100, 370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Farquharson K. L. (2008) A protein disulfide isomerase plays a role in programmed cell death. Plant Cell 20, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ondzighi A., Christopher D. A., Cho E. J., Chang S. C., Staehelin L. A. (2008) Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell 20, 2205–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kwon K., Choi D, Hyun J. K., Jung H. S., Baek K., Park C. (2013) Novel glyoxalases from Arabidopsis thaliana. FEBS J. 280, 3328–3339 [DOI] [PubMed] [Google Scholar]
- 66. Mustafiz A., Sahoo K. K., Singla-Pareek S. L., Sopory S. K. (2010) Metabolic engineering of glyoxalase pathway for enhancing stress tolerance in plants. Methods Mol. Biol. 639, 95–118 [DOI] [PubMed] [Google Scholar]
- 67. Desai K. M., Chang T., Wang H., Banigesh A., Dhar A., Liu J., Untereiner A., Wu L. (2010) Oxidative stress and aging: is methylglyoxal the hidden enemy? Can. J. Physiol. Pharmacol. 88, 273–284 [DOI] [PubMed] [Google Scholar]
- 68. Chyan M. K., Elia A. C., Principato G. B., Giovannini E., Rosi G., Norton S. J. (1994) S-fluorenylmethoxycarbonyl glutathione and diesters: inhibition of mammalian glyoxalase II. Enz. Prot. 48, 164–173 [DOI] [PubMed] [Google Scholar]
- 69. Rulli A., Carli L., Romani R., Baroni T., Giovannini E., Rosi G., Talesa V. (2001) Expression of glyoxalase I and II in normal and breast cancer tissues. Breast Cancer Res. Treat. 66, 67–72 [DOI] [PubMed] [Google Scholar]
- 70. Kang Y., Edwards L. G., Thornalley P. J. (1996) Effect of methylglyoxal on human leukemia 60 cell growth: modification of DNA G1 growth arrest and induction of apoptosis. Leuk. Res. 20, 397–405 [DOI] [PubMed] [Google Scholar]
- 71. Du J., Suzuki H., Nagase F., Akhand A. A., Yokoyama T., Miyata T., Kurokawa K., Nakashima I. (2000) Methylglyoxal induces apoptosis in Jurkat leukemia T cells by activating c-Jun N-terminal kinase. J. Cell. Biochem. 77, 333–344 [DOI] [PubMed] [Google Scholar]
- 72. Du J., Suzuki H., Nagase F., Akhand A. A., Ma X. Y., Yokoyama T., Miyata T., Nakashima I. (2001) Superoxide-mediated early oxidation and activation of ASK1 are important for initiating methylglyoxal-induced apoptosis process. Free Radic. Biol. Med. 31, 469–478 [DOI] [PubMed] [Google Scholar]
- 73. Fukunaga M., Miyata S., Liu B. F., Miyazaki H., Hirota Y., Higo S., Hamada Y., Ueyama S., Kasuga M. (2004) Methylglyoxal induces apoptosis through activation of p38 MAPK in rat Schawnn cells. Biochem. Biophys. Res. Commun. 320, 689–695 [DOI] [PubMed] [Google Scholar]
- 74. Zhu Y., Dong A., Meyer D., Pichon O., Renou J. P., Cao K., Shen W. H. (2006) Arabidopsis NRP1 and NRP2 encode histone chaperones and are required for maintaining postembryonic root growth. Plant Cell 18, 2879–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chowdhury D., Beresford P. J., Zhu P., Zhang D., Sung J. S., Demple B., Perrino F. W., Lieberman J. (2006) The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol. Cell. 23, 133–142 [DOI] [PubMed] [Google Scholar]
- 76. Sirover M. A. (2005) New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J. Cell. Biochem. 95, 45–52 [DOI] [PubMed] [Google Scholar]
- 77. Baek D., Jin Y., Jeong J. C., Lee H. J., Moon H., Lee J., Shin D., Kang C. H., Kim D. H., Nam J., Lee S. Y., Yun D. J. (2008) Suppression of reactive oxygen species by glyceraldehyde-3-phosphate dehydrogenase. Phytochemistry 69, 333–338 [DOI] [PubMed] [Google Scholar]
- 78. Will C. L., Lührmann R. (2001) Spliceosomal UsnRNP biogenesis, structure and function. Curr. Opin. Cell Biol. 13, 290–301 [DOI] [PubMed] [Google Scholar]
- 79. Klein-Gunnewiek J. M., van de Putte L. B., van Venrooij W. J. (1997) The U1 snRNP complex: an autoantigen in connective tissue diseases. An update. Clin. Exp. Rheumatol, 15, 549–560 [PubMed] [Google Scholar]
- 80. Schwerk C., Schulze-Osthoff K. (2005) Regulation of apoptosis by alternative pre-mRNA splicing. Mol. Cell 19, 1–13 [DOI] [PubMed] [Google Scholar]
- 81. Moore M. J., Wang Q., Kennedy C. J., Silver P. A. (2010) An alternative splicing network links cell-cycle control to apoptosis. Cell 142, 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Clark T. A., Sugnet C. W., Ares M., Jr. (2002) Genomewide analysis of mRNA processing in yeast splicing-specific microarrays. Science 296, 907–910 [DOI] [PubMed] [Google Scholar]
- 83. Pleiss J. A., Withworth G. B., Bergkessel M., Guthrie C. (2007) Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 5, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kawashima T., Pellegrini M., Chanfreau G. F. (2009) Nonsense-mediated mRNA decay mutes the splicing defects of spliceosome component mutations. RNA 15, 2236–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Campion Y., Neel H., Gostan T., Soret J., Bordonné R. (2010) Specific splicing defects in S. pombe carrying a degron allele of the survival of motor neuron gene. EMBO J. 29, 1817–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Park J. W., Parisky K., Celotto A. M., Reenan R. A., Graveley B. R. (2004) Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 101, 15974–15979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Massiello A., Roesser J. R., Chalfant C. E. (2006) SAP155 binds to ceramide-responsive RNA cis-element 1 and regulates the alternative 5′ splice site selection of Bcl-x pre-mRNA. FASEB J. 20, 1680–1682 [DOI] [PubMed] [Google Scholar]
- 88. Pacheco T. R., Moita L. F., Gomes A. Q., Hacohen N., Carmo-Fonseca M. (2006) RNA interference knockdown of hU2AF35 impairs cell cycle progression and modulates alternative splicing of Cdc25 transcripts. Mol. Biol. Cell 17, 4187–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Hastings M. L., Allemand E., Duelli D. M., Myers M. P., Krainer A. R. (2007) Controls of pre-mRNA splicing by the general splicing factors PUF60 and U2AF65. PLoS One 2, e538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. (2008) SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133, 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Baumer D., Lee S., Nicholson G., Davies J. L., Parkinson N. J., Murray L. M., Gillingwater T. H., Ansorge O., Davies K. E., Talbot K. (2009) Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet. 5, e1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Sträβer K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondón A. G., Aguilera A., Struhl K., Reed R., Hurt E. (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417, 304–308 [DOI] [PubMed] [Google Scholar]
- 93. Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R. (2005) Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 19, 1512–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Furumizu C., Tsukaya H., Komeda Y. (2010) Characterization of EMU, the Arabidopsis homolog of the yeast THO complex member HPR1. RNA 16, 1809–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Oberst A., Bender C., Green D. R. (2008) Living with death: the evolution of the mitochondrial pathway of apoptosis in animals. Cell Death Differ. 15, 1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Vizcaíno J. A., Côté R. G., Csordas A., Dianes J. A., Fabregat A., Foster J. M., Griss J., Alpi E., Birim M., Contell J., O'Kelly G., Schoenegger A., Ovelleiro D., Pérez-Riverol Y., Reisinger F., Ríos D., Wang R., Hermjakob H. (2013) The Proteomics Identifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 41(D1), D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.