Abstract
Verticillium wilt causes massive annual losses of cotton yield, but the mechanism of cotton resistance to Verticillium dahliae is complex and poorly understood. In this study, a comparative proteomic analysis was performed in resistant cotton (Gossypium barbadense cv7124) on infection with V. dahliae. A total of 188 differentially expressed proteins were identified by mass spectrometry (MALDI-TOF/TOF) analysis and could be classified into 17 biological processes based on Gene Ontology annotation. Most of these proteins were implicated in stimulus response, cellular processes and metabolic processes. Based on the proteomic analysis, several genes involved in secondary metabolism, reactive oxygen burst and phytohormone signaling pathways were identified for further physiological and molecular analysis. The roles of the corresponding genes were further characterized by employing virus-induced gene silencing (VIGS). Based on the results, we suggest that the production of gossypol is sufficient to affect the cotton resistance to V. dahliae. Silencing of GbCAD1, a key enzyme involving in gossypol biosynthesis, compromised cotton resistance to V. dahliae. Reactive oxygen species and salicylic acid signaling may be also implicated as regulators in cotton responsive to V. dahliae according to the analysis of GbSSI2, an important regulator in the crosstalk between salicylic acid and jasmonic acid signal pathways. Moreover, brassinosteroids and jasmonic acid signaling may play essential roles in the cotton disease resistance to V. dahliae. The brassinosteroids signaling was activated in cotton on inoculation with V. dahliae and the disease resistance of cotton was enhanced after exogenous application of brassinolide. Meanwhile, jasmonic acid signaling was also activated in cotton after inoculation with V. dahliae and brassinolide application. These data provide highlights in the molecular basis of cotton resistance to V. dahliae.
Cotton (Gossypium spp.) is one of the most important economic crops globally. However, the yield of cotton is restricted by many unfavorable environmental conditions including biotic and abiotic stresses. Among these stresses, Verticillium wilt, a soil-borne vascular disease caused by Verticillium dahliae, is a devastating disease of cotton worldwide (1), reducing the quality and yield of the fiber; up to 30% yield reductions can occur during a severe outbreak of the disease (2). Few germplasms have been found with resistance to V. dahliae in upland cotton (Gossypium hirsutum), which contributes ∼95% of the total cotton yield (3), and the complex genetics of resistance to V. dahliae in upland cotton cultivars has prevented the efficient breeding of disease-resistant cotton (4–6). Moreover, the fungus can survive in soil for many years even in the absence of hosts (2). These issues make it difficult to develop an effective and practical management plan for the control of V. dahliae in cotton production.
Plants have evolved a complete, multilayered immune system that includes constitutive and inducible defenses to counteract colonization by pathogens (7). Several endogenous signal molecules, such as salicylic acid (SA),1 ethylene (ET), and jasmonic acid (JA), are synthesized and activate distinct defense pathways involved in complex defense signaling networks (8). Among these molecules, JA usually acts with ethylene to induce resistance against necrotrophic pathogens, whereas SA-mediated defense responses are effective against hemi-biotrophs and biotrophs and are critical for systemic acquired resistance (9). In addition, defense signaling pathways mediated by SA and JA frequently act antagonistically to mediate defense against specific types of pathogens (10–12). For example, SA accumulation and SA-derived signaling are induced by virulent Pseudomonas syringae infection, which enhances susceptibility to Alternaria brassicicola by inhibiting JA-mediated defense responses in Arabidopsis (11). Nevertheless, the phytotoxin coronatine, a structural analog of JA produced by Pseudomonas syringae, can suppress SA-derived responses in the host (10). Other endogenous signal molecules, such reactive oxygen species (ROS), auxin and brassinosteroids (BRs), can also influence defense signaling and resistance (13–17). Plants have evolved regulatory defense mechanisms to adapt efficiently to changes in their complex environment during pathogen invasion. Crosstalk among signal molecules provides plants with a powerful capacity to finely regulate the immune response. The molecular mechanisms involved in plant immunity are complex.
During the past two decades, extensive studies have enriched our understanding of the molecular mechanism of cotton resistance to V. dahliae. Production of phytoalexins, including terpenoids, and phenylpropanoid substances, is induced quickly in cotton on infection by V. dahliae (18, 19). Gossypol is one of the most important sesquiterpene phytoalexin, which exists specifically in cotton and plays a crucial role in the defense against the invasion of pathogens and insects (20). Though many phytoalexin-related genes have been shown to be important in mediating cotton defense, the molecular mechanism is unknown (21, 22). As sequencing technology develops, a number of genes related to disease resistance (e.g. aerobic metabolism enzymes, pathogen-related proteins, ethylene biosynthesis and response genes, etc.) have been identified from resistant cotton cultivars (Gossypium barbadense cv7124 or Pima 90) through suppression subtractive hybridization (23, 24). Using RNA-Seq-dependent transcriptional analysis, a subset of genes participating in lignin metabolism was demonstrated to be very important in the resistance of cotton to V. dahliae (19). Additionally, defense- and stress-related proteins, such as pathogenesis-related proteins and proteins likely to be involved in the oxidative burst, sugars, ethylene signaling, and isoprenoid synthesis, have recently been suggested to be involved in cotton response to V. dahliae (25, 26). Most of the candidate genes involving in disease resistance are isolated from transcriptomic analysis, whereas only few genes have been functionally characterized (27, 28). Ve1 is the only R gene isolated using map-based cloning from tomato (Solanum lycopersicum) and has been shown to provide race-specific resistance to race 1 strains of V. dahliae and V. albo-atrum in tomato and Arabidopsis (17, 29). Although several genes homologous to Ve1 have been cloned from cotton (30, 31), it is unclear whether Ve1-mediated resistance signaling also exists in cotton.
In this study, we performed a comparative proteomic analysis on Mock and V. dahliae inoculated roots of G. barbadense cv7124, which shows high resistance to V. dahliae, at different time points. A number of differentially expressed proteins were identified. Furthermore, three classes of genes, involved in gossypol metabolism, BR signaling, and JA signaling were characterized using virus-induced gene silencing (VIGS). Our data suggest that gossypol, BRs and JA act as important factors in contributing resistance of cotton to V. dahliae. For the first time, proteomics was combined with VIGS to discover and validate defense-related genes in cotton. Our research provides new insights into the molecular basis of cotton defense against V. dahliae.
EXPERIMENTAL PROCEDURES
Plant Growth and Pathogen Inoculation
Seeds of G. barbadense cv7124 (resistant) and G. hirsutum cvYZ-1 (susceptible) were grown in a controlled environment chamber under a 14 h light/10 h dark cycle at 28 °C for 2 weeks.
The defoliating isolate V991 of V. dahliae was grown on a potato-dextrose agar medium for 4 d; the fungus was then incubated in Czapek's medium (NaNO3, 0.3% w/v; MgSO4, 0.1% w/v; KH2PO4, 0.1% w/v; FeSO4, 0.0002% w/v; KCl, 0.1% w/v; sucrose, 3% w/v; pH 6.0) at 25 °C for 5 d. The concentration of spores was adjusted to ∼106 conidia per ml with deionized water for inoculation. The cotton seedlings were removed from the soil and dip-infected with the liquid containing V. dahliae spores. The seedlings were incubated at 25 °C under a 14 h/10 h light/dark photoperiod, and the roots were harvested at 1, 6, 12, 24, 48, and 72 h after inoculation. Seedlings treated with sterile distilled water in the same manner were used as a Mock treatment. Roots were stored at −80 °C until protein extraction was performed.
Protein Extraction and 2D Electrophoresis
For 2D-PAGE, proteins from cotton roots were prepared according to Yao and Pan (32, 33) with minor modifications. Frozen root tissues were ground in liquid nitrogen to a fine powder and incubated in extraction buffer (−20 °C precooled acetone, 12% w/v trichloroacetic acid, and 0.07% w/v dithiothreitol (DTT)). After washing twice with cold acetone containing 0.07% (w/v) DTT, the vacuum-dried powder was suspended in extraction buffer [30% sucrose, 50 mm Tris-HCl (pH 8.0), 2% SDS, 2 mm PMSF, 0.07% DTT, and an equal volume of Tris-saturated phenol (pH 8.0)]. The phenol phase was then precipitated with 5 volumes of 0.1 m ammonium acetate in methanol at −20 °C. The collected protein pellets were washed twice with 80% methanol and 80% acetone. After air-drying, the pellets were dissolved in lysis buffer (8 m urea, 2 m thiourea, 2% CHAPS, 1% DTT, and 2% v/v immobilized pH gradient (IPG) buffer, pH 4–7). Protein concentration was determined using a 2-D quant kit (Bio-Rad, Hercules, CA).
The first dimensional gel separation was performed according to the manufacturer's protocol with modifications (Bio-Rad). Total proteins (1.0 mg) were diluted in 450 μl rehydration buffer (8 m urea, 2 m thiourea, 2% CHAPS, 1% DTT, and 2% v/v IPG buffer, pH 4–7) and loaded on IPG strips (24 cm, pH 4–7 nonlinear) (Bio-Rad). The IPG strips were rehydrated for 12 h at room temperature and focused at gradient steps of 50 V for 1 h, 500 V for 1 h, 1000 V for 1 h, and 10,000 V for 4 h, with a final step of 10,000 V toward a total of 90 kVh. Before second dimension analysis, IPG strips were incubated in equilibration buffer (50 mm Tris-HCl, pH 8.8; 6 m urea; 30% glycerol; and 2% SDS) containing 1% DTT and then in equilibration buffer containing 2% (w/v) iodoacetamide. For the second dimension, IPG strips were fixed on 12.5% acrylamide gels. Then, 2D-PAGE was performed using the Ettan DALT six electrophoresis unit (GE Healthcare) at 5 W per gel for 1 h and then 15 W per gel for 6 h until the bromphenol blue dye front reached the bottom of the gels.
Gels were stained with Coomassie brilliant blue solution (0.2% Coomassie brilliant blue G250, 20% methanol, 10% phosphoric acid, and 10% ammonium sulfate). The 2D gels were scanned using a GS-800 Calibrated Densitometer (Bio-Rad). Protein spots were detected using PDQuest software (Bio-Rad). After volumetric quantification and matching, differences in protein content between Mock and inoculated samples were analyzed using a Student t test and calculated as the fold ratio. Data was from three biological replicates; a threshold of p ≤ 0.05 and fold change of ≥2 or ≤0.5 was used to identify significantly differentially expressed protein spots.
MALDI-TOF/TOF Analysis
Differentially expressed proteins were excised from the gels and destained with 25 mm NH4HCO3 in 50% acetonitrile (ACN) until the Coomassie brilliant blue disappeared. After being washed twice with 100% ACN for 10 min, proteins were digested in-gel using trypsin (Promega, Madison, WI) overnight at 37 °C. The gel pieces were extracted once with extraction buffer (67% ACN and 5% TFA). After being dried completely, the samples were resuspended in 0.1% TFA and then mixed in a 1:1 ratio with a matrix consisting of a saturated solution of CHCA in 50% ACN containing 0.1% TFA. Samples were then spotted onto a freshly cleaned target plate. After air-drying, the crystallized spots were analyzed using an ABI 4800 MALDI-TOF/TOF Plus mass spectrometer (Applied Biosystems, Foster City, CA). Both the MS and MS/MS data were integrated and performed using GPS Explorer V3.6 software (Applied Biosystems). Proteins were successfully identified with a 95% or higher confidence interval using the MASCOT V2.3 search engine (Matrix Science, London, UK) and searching in the Gossypium EST database (release data 20120128; 2476590 sequences; 555009942 residues). The other search parameters were the enzyme trypsin; one missed cleavage site; partial modifications of cysteine carbamido methylation and methionine oxidization; no fixed modifications; peptide tolerance of 100 ppm; and fragment mass tolerance of 0.3 Da.
RNA Isolation, Reverse Transcription-PCR (RT-PCR) and Quantitative Real-time PCR (qPCR)
Total RNA was isolated from cotton as previously described (34). First strand cDNA was synthesized from 5 μg of total RNA using the Superscript first-strand synthesis system (Invitrogen, Foster City, CA). For RT-PCR, an aliquot of the reverse transcription product was used as the template. The ubiquitin7 gene (DQ116441) of cotton was used as an internal control. Primers were designed as shown in supplemental Table S1. qPCR was performed according to the guidelines of the Minimum Information for Publication of Quantitative Real Time PCR Experiments (35). Diluted cDNA was used for qPCR with SYBR green using ABI 7500 Real Time PCR system (Applied Biosystems). The expression analysis data are presented as the means ± S.D. from three biologically independent experiments. All primers for qPCR were designed using Primer Express 5.0 software (Applied Biosystems) and are shown in supplemental Table S2.
VIGS in Cotton Followed by Pathogen Inoculation
The TRV vectors and Agrobacterium tumefaciens for VIGS were prepared according to Fradin et al. (17). Inserts to generate TRV:GbCAD1, TRV:Gb14-3-3c, TRV:Gb14-3-3d, TRV:GbSSI2 and positive control TRV:GbCLA1 (cloroplastos alterados 1) were amplified from the cDNA of G. barbadense cv7124. Primer pairs to generate TRV vectors are shown in supplemental Table S3. PCR fragments were digested with BamHI and KpnI and then ligated into the TRV:00 plasmid (36). The constructs were transformed into A. tumefaciens GV3101 by electroporation. TRV vectors were agro-infiltrated as described (31) into the cotyledons of 10-day-old seedlings of G. barbadense cv7124 or G. hirsutum cvYZ-1. The seedlings were then grown at 25 °C with a 16 h/8 h light/dark photoperiod cycle in a controlled environment chamber. As shown in supplemental Fig. S1, the leaf bleaching phenotype was observed 2 weeks after infiltration in TRV:GbCLA1 plants. Therefore, inoculation with V. dahliae isolate V991 was also performed 2 weeks after infiltration. The rate of diseased plants and disease index were scored with at least 16 plants per treatment and repeated at least for three times. Plant disease index (DI) is calculated as the following formula: INSERT EQUATION HERE, n denotes disease level, cotton seedlings were divided into five levels based on their disease severity after V. dahliae inoculation (level 0, 1, 2, 3, 4) according to Xu et al. (37), DI reflects the disease infection status of a population, not an individual plant; higher DI means more serious infection.
Treatments with Methyl Jasmonate (MeJA) and Brassinolide (BL)
For treatments with MeJA and BL, the concentrations were 200 μm and 5 μg/pot, respectively. Cotton seedlings were cultured in a pot (four or five plants per pot) in a greenhouse and three-leaf stage plants were treated with MeJA or BL by soil drench application.
Measurement of Total SA and JA Levels
The extraction and measurement of the endogenous SA and JA of cotton seedlings were performed as described (38). Three replicates of each frozen sample (∼100 mg for each replicate) were ground to a fine powder in liquid nitrogen and mixed with 750 μl cold extraction buffer (methanol/water/acetic acid, 80:19:1, v/v/v). After shaking for 16 h at 4 °C in the dark, the supernatants were collected and then filtered using a syringe-facilitated 13-mm diameter nylon filter with a pore size of 0.22 μm (Nylon 66; Jin Teng Experiment Equipment Co., Ltd, Tian jin, China). Filtrates were dried using nitrogen gas at room temperature and then dissolved in 200 μl methanol. An aliquot of dissolved sample was further diluted 100 times using methanol for the quantification of SA because cotton contains high levels of SA. Supernatants were analyzed on an HPLC-MS/MS (1200L LC-MS system, Varian).
DAB Staining and H2O2 Measurement
For H2O2 detection, leaves were incubated in 1 mg/ml pH 3.8 DAB-HCl (Sigma-Aldrich) in the dark for 8 h. The leaves were then cleared by boiling in alcoholic lactophenol (95% ethanol/lactophenol, 2:1 v/v) for 20 min. The reddish color of the leaves as evidence of H2O2 was visualized by light microscopy. Hydrogen peroxide content was measured using an H2O2 Quantitative Assay Kit (Sangon Biotech, China).
Trypan Blue Staining
Trypan blue staining was performed as described by Choi and Hwang (39). Leaves were stained by boiling in lactophenol-trypan blue (10 ml lactic acid, 10 ml glycerol, 10 g phenol, and 10 mg trypan blue dissolved in 10 ml distilled water) followed by destaining with chloral hydrate (2.5 g/ml).
Gossypol Visualization and Measurement
Gossypol was visualized by photography within 5 min after dropping a saturated solution of antimony trichloride (SbCl3) in 40% perchloric acid (HClO4) on fresh free-hand sections (∼100 mm thick) from the same sites of Mock and infected cotton roots. Images were captured using an Olympus light microscope equipped with an Olympus U-PMTVC adapter and a Leica DC300F camera, and processed using Leica IM50 Image Manager V1.20 software.
Sesquiterpene aldehydes (gossypol equivalents) were extracted and quantitated using the phloroglucinol/HCl method (22).
Statistical Analysis
For all generated data, at least three biological replicates were performed. The data are presented as the mean ± S.D. Statistical significance was determined using two-tailed unpaired Student's t-tests, and p values <0.05 were considered statistically significant.
RESULTS
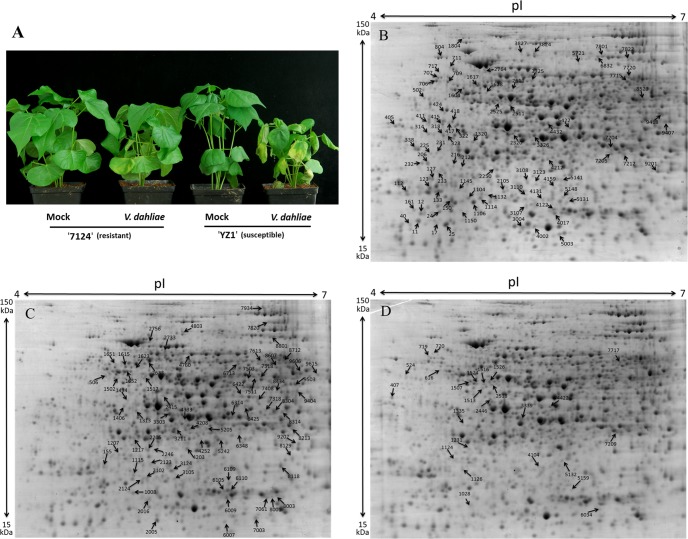
Two Cotton Cultivars, G. barbadense cv7124 (resistant) and G. hirsutum cvYZ-1 (susceptible) were inoculated with a highly aggressive defoliating fungal pathogen V. dahliae strain V991 (Fig. 1A). Approximately 2 weeks later, susceptible plants exhibited visual symptoms of cotyledon wilting, leaf chlorosis, and severe stunting, whereas resistant plants only showed slight stunting compared with the control. Therefore, the potential presence of a special mechanism and related defense response to V. dahliae in 7124 were investigated using comparative proteomics.
Fig. 1.
Disease symptoms on two kinds of cotton cultivars and representative 2-DE maps of total differential expression proteins isolated from the resistant cotton cultivar. A, Infection of cotton seedlings with V. dahliae. After seeding for 2 weeks, the seedlings of G. barbadense cv7124 (resistant) and G. hirsutum cv.YZ-1 (susceptible) were dip-infected with the liquid containing V. dahliae spores and treated with sterile distilled water as a Mock treatment. At least 20 plants were used for V. dahliae inoculation per experiment and the experiment was repeated for three times at the same condition. The Mock and infected roots of 7124 harvested at 1, 6, 12, 24, 48, and 72 hpi were used for protein and RNA extraction. Photos were taken 14 d post inoculation. B, Protein spots of down-regulation, C, up-regulation and (D) other regulatory patterns are indicated in the representative 2-DE maps. The differently expressed proteins were labeled in the images.
Proteomics Analysis and Identification of Cotton Defense-related Proteins
Proteins were isolated from the roots of 7124 during V991 inoculation and screened using 2-DE gels. Approximately 1500 protein spots were detected on each 2-DE gel of Mock and infected cotton roots at 1, 6, 12, 24, 48, and 72 hpi. Differences in protein content between Mock and inoculated samples were analyzed using a Student t test and calculated as the fold ratio. Data was from three biological replicates; a threshold of p ≤ 0.05 and fold change of ≥2 or ≤0.5 was used to identify significantly differentially expressed protein spots. A total of 274 proteins with significant expression changes after V. dahliae inoculation compared with Mock were successfully identified using MALDI-TOF/TOF (Figs. 1B–1D). These differently expressed proteins were mostly observed at 1, 24, and 48 hpi (22.63%, 27.01%, and 24.09%, respectively); 130 proteins were up-regulated whereas 144 proteins were down-regulated (Table I).
Table I. Number of differentially expressed proteins at different times after inoculation.
| Time after inoculation (h) |
Total | ||||||
|---|---|---|---|---|---|---|---|
| 1 h | 6 h | 12 h | 24 h | 48 h | 72 h | ||
| Up | 26 | 24 | 12 | 42 | 22 | 4 | 130 |
| % up | 20.00% | 18.46% | 9.23% | 32.31% | 16.92% | 3.08% | |
| Down | 36 | 5 | 9 | 32 | 44 | 18 | 144 |
| % down | 25.00% | 3.47% | 6.25% | 22.22% | 30.56% | 12.50% | |
| Total | 62 | 29 | 21 | 74 | 66 | 22 | 274 |
| % total | 22.63% | 10.58% | 7.66% | 27.01% | 24.09% | 8.03% | |
Among these 274 proteins, several were detected at multiple time points. Thus, 188 unique proteins with significant expression changes during V. dahliae infection were obtained after removing redundant proteins. Detailed information about these proteins is shown in supplemental Table S4. All these proteins were divided into three groups according to their expression patterns (up-regulated, down-regulated, and other patterns of regulation; Figs. 1B–1D); 47% of the 188 proteins were down-regulated during infection (supplemental Fig. S2A). To globally understand the potential function of these 188 proteins, GO functional classification analysis was performed to calculate the functional category distribution based on level 2 biological processes (supplemental Fig. S2B). As a result, all identified proteins could be classified into 17 biological processes, including metabolism, cellular process, response to stimulus, biogenesis, immune system, etc. In addition to the response to stimulus-related proteins, more proteins involved in metabolic and cellular processes were found to be down-regulated, which may indicate that the normal development of cotton was retarded by the infection of the fungus.
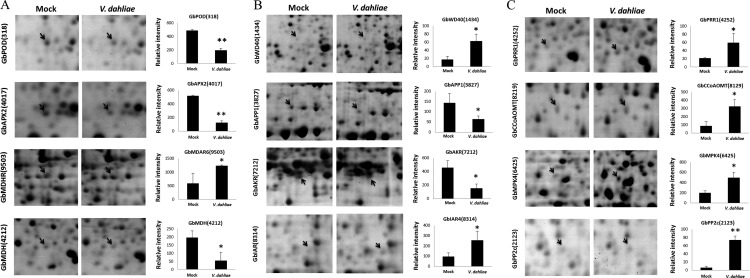
A set of disease-related genes involved in the complex biological processes of cotton defense against V. dahliae was identified, including oxidative burst-, auxin response-, secondary metabolism- and pathogenesis-related genes. The expression level of several proteins participating in the regulation of redox homeostasis, including peroxidase (GbPOD, ssp318), ascorbate peroxidase (GbAPX2, ssp4017), monodehydroascorbate reductase (GbMDAR6, ssp9503), and malate dehydrogenase (GbMDH, ssp4212), were found to be clearly influenced by the inoculation with V. dahliae (Fig. 2A). Meanwhile, a class of genes potentially involved in disease resistance was a group of the auxin signaling-related genes, such as the WD40 repeat-like superfamily protein (GbWD40, ssp1434), amino peptidase P1 (GbAPP1, ssp3827), auxin-induced protein pcnt115 (GbAKR, ssp7212), and pyruvate dehydrogenase E1a-like subunit (GbIAR4, ssp8314). These proteins are involved in the accumulation, polar transport or homeostasis of auxin (40–42), and their expression levels changed dramatically in cotton after inoculation with V. dahliae (Fig. 2B). Furthermore, secondary metabolism-related proteins, such as related to lignin metabolism, were found to be activated during the inoculation. Two important proteins participating in lignin metabolism, phenylcoumaran benzylic ether reductase-like protein (GbPRR1, ssp4252) and caffeoyl-CoAO-methyltransferase (GbCCoAOMT, ssp8129) (19), were increased significantly on infection with the fungus (Fig. 2C). Several other proteins involved in signal transduction, such as mitogen-activated protein kinase (GbMAPK4, ssp6425) and protein phosphatase 2c (GbPP2C, ssp2123), were also clearly induced during the interaction between cotton and V. dahliae (Fig. 2C).
Fig. 2.
Representative protein spots shown on 2-DE maps and quantification of the signal intensities. A, Oxidative burst-related proteins. GbPOD, peroxidase; GbAPX2, ascorbate peroxidase; GbMDAR6, monodehydro ascorbate reductase; GbMDH, malate dehydrogenase. B, Auxin signaling-related proteins. GbWD40, WD40 repeat-like superfamily protein; GbAPP1, amino peptidase P1; GbAKR, auxin-induced protein pcnt115; GbIAR4, pyruvate dehydrogenase E1a-like subunit. C, Other disease-related proteins. GbPRR1, phenylcoumaran benzylic ether reductase-like protein; GbCCoAOMT, caffeoyl-CoAO-methyltransferase; GbMAPK4, mitogen-activated protein kinase; GbPP2C, protein phosphatase 2c. Proteins with differential expression were shown in 2-DE maps and the signal intensities obtained from three independent 2-DE gels. Error bars represent the standard deviation of three biological replicates; asterisks indicate statistically significant differences, as determined by the Student t test (*p < 0.05; **p < 0.01).
Gossypol is Involved in the Resistance of Cotton to V. dahliae
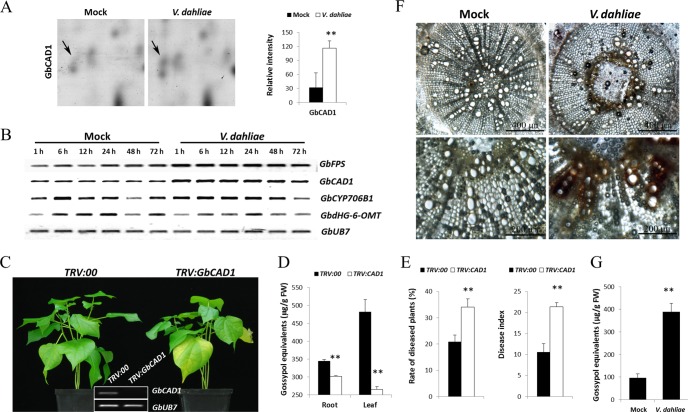
Among these identified proteins, the protein GbCAD1 (ssp2124), related to the biosynthesis of the terpenoid phytoalexin gossypol (18), was clearly up-regulated in the roots after inoculation with V. dahliae (Fig. 3A). To investigate and verify the participation of gossypol in response to V. dahliae, the expression of genes involved in gossypol biosynthesis was determined by RT-PCR. As shown in Fig. 3B, the expression of GbCAD1 was enhanced after inoculation with V. dahliae compared with mock-treated samples, which was consistent with the expression pattern at the protein level. Furthermore, two other key genes (farnesyl diphosphate synthase, GbFPS; cytochrome P450 mono-oxygenase, GbCYP706B1) were also up-regulated, whereas GbdHG-6-OMT, a negative regulator of gossypol biosynthesis, was down-regulated, indicating that inoculation with V. dahliae could activate the biosynthesis of the terpenoid phytoalexin gossypol in cotton.
Fig. 3.
Gossypol is involved in the resistance of cotton to V. dahliae. A, Shown are representative protein spot (GbCAD1) on inoculation with V. dahliae and quantification of the signal intensities obtained from three independent 2-DE maps. Error bars represent the standard deviation of three biological replicates; asterisks indicate statistically significant differences, as determined by the Student t test (**p < 0.01). B, RT-PCR analysis of gossypol biosynthesis-related genes at the transcriptional level in G. barbadense cv7124 on inoculation with V. dahliae. PCR was performed by 28 cycles of amplification for GbUB7 and 29 cycles for other genes. GbFPS, farnesyl diphosphate synthase; GbCYP706B1, cytochrome P450 mono-oxygenase. C, Disease symptoms induced on empty vector control (TRV:00) or GbCAD1-silenced (TRV:GbCAD1) cotton plants after inoculation with V. dahliae strain V991. Ten-day-old G. barbadense cv7124 seedlings were hand-infiltrated with Agrobacterium carrying individual genes in the VIGS vector. Two weeks after infiltration, the seedlings were dip-inoculated with V. dahliae. Photos were taken at 12 d after inoculation. D, Accumulation of sesquiterpene aldehydes (gossypol equivalents) of control (TRV:00) and GbCAD1-silenced (TRV:GbCAD1) cotton plants. The roots and leaves of control and GbCAD1-silenced cotton plants were collected for sesquiterpene aldehydes extraction and quantitation 12 d after VIGS-infiltration. Error bars represent the standard deviation of three replicates (n = 12); asterisks indicate statistically significant differences, as determined by the Student t test (**p < 0.01). E, Rate of diseased plants and disease index measurement of control (TRV:00) and GbCAD1-silenced (TRV:GbCAD1) cotton plants after inoculation with conidial suspension of V991. The rate of diseased plants and disease index were measured at 4 d after the plants beginning to present disease symptoms. Error bars represent the standard deviation of three biological replicates (n ≥ 16); asterisks indicate statistically significant differences, as determined by the Student t test (**p < 0.01). F, Detection of gossypol accumulation in cotton roots inoculated with V991. Radial sections of uninfected cotton roots display less orange-staining gossypol than infected cotton roots. Scale bars: 400 μm and 200 μm, respectively. G, Accumulation of sesquiterpene aldehydes (gossypol equivalents) of G. barbadense cv7124 inoculated with V991. After inoculation with V991 for 5 d, the Mock and infected roots of 7124 were harvested for sesquiterpene aldehydes extraction and quantitation. Error bars represent the standard deviation of three replicates (n = 12); asterisks indicate statistically significant differences, as determined by the Student t test (**p < 0.01).
VIGS was employed to better understand the function of GbCAD1 in the cotton defense response. Two weeks after Agrobacterium infiltration in 7124, the transcripts of GbCAD1 were significantly reduced in TRV:GbCAD1 plants compared with TRV:00 plants, as shown by RT-PCR analysis (Fig. 3C), indicating that GbCAD1 was effectively silenced in cotton. On inoculation with V991, more wilting and etiolated leaves were observed in TRV:GbCAD1 plants compared with the control (Fig. 3C). To investigate whether the gossypol production was altered after GbCAD1 was silenced, the phloroglucinol/HCl method was used to measure the content of gossypol equivalents (22). Gossypol equivalents in both roots and leaves of TRV:GbCAD1 plants was significantly lower compared with the control 12 d after VIGS infiltration (Fig. 3D). Statistical analysis of data for the rate of diseased plants and the disease index showed that deficiency of GbCAD1 compromises resistance to V. dahliae (Fig. 3E).
Because GbCAD1 has been shown to be an important enzyme in gossypol biosynthesis, the changes in gossypol production in cotton after pathogen inoculation were evaluated. Plants were harvested 5 d after inoculation and stained with SbCl3-HClO4, and the results show the presence of gossypol in vascular bundles more frequently in inoculated plants compared with uninfected plants, and the production of gossypol equivalents in infected seedlings was enhanced compared with uninfected plants (Figs. 3F, 3G). These results indicate that gossypol production was significantly increased on V. dahliae infection, and inhibiting the biosynthesis of gossypol impairs cotton resistance to V. dahliae.
BR Signaling Positively Regulates the Resistance of Cotton to V. dahliae
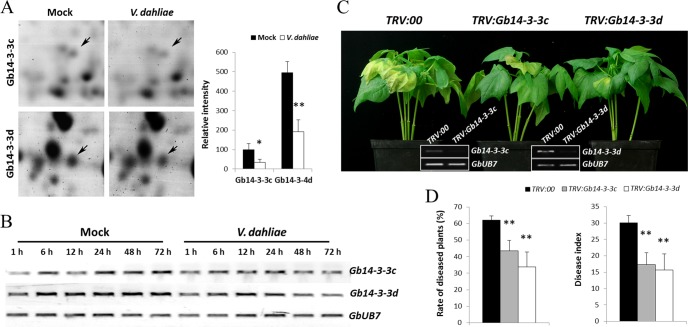
Four Gb14-3-3 proteins, the homologous genes of which in Arabidopsis were reported to interact with the BIN2-phosphorylated targets in the BZR1 protein (43), were all down-regulated significantly in the protein expression profile (Fig. 4A). The transcripts of four Gb14-3-3 protein-encoding genes were confirmed using RT-PCR analysis. The transcriptional levels of Gb14-3-3c (ssp127) and Gb14-3-3d (ssp4002) were suppressed in cotton after inoculation with V. dahliae, which is consistent with the protein analysis (Fig. 4B). Although the transcripts of Gb14-3-3a (ssp12) and Gb14-3-3b (ssp123) were not significantly changed, there were no detectable effect on disease resistance of cotton when silencing these two genes with VIGS (data not shown). Therefore, Gb14-3-3c (ssp127) and Gb14-3-3d (ssp4002) were chosen for further study.
Fig. 4.
Silencing of Gb14-3-3 enhances cotton resistance to V. dahliae. A. Shown are representative protein spots (Gb14-3-3c, Gb14-3-3d) on inoculation with V. dahliae and quantification of the signal intensities obtained from three independent 2-DE maps. Error bars represent the standard deviation of three biological replicates; asterisks indicate statistically significant differences, as determined by the Student t test (*p < 0.05; **p < 0.01). B, RT-PCR analysis of the expression pattern of Gb14-3-3c and Gb14-3-3d at the transcriptional level in G. barbadense cv.7124 on inoculation with V. dahliae. PCR was performed by 28 cycles of amplification for GbUB7 and 30 cycles for other genes. C, Disease symptoms induced on TRV:00, TRV:Gb14-3-3c or TRV:Gb14-3-3d cotton after inoculation with V. dahliae strain V991. Ten-day-old G. hirsutum cvYZ-1 seedlings were hand-infiltrated with Agrobacterium carrying individual genes in the VIGS vector. Two weeks after infiltration, the seedlings were dip-inoculated with V. dahliae. Photos were taken at 9 d after inoculation. D, Rate of diseased plants and disease index measurement of TRV:00, TRV:Gb14-3-3c and TRV:Gb14-3-3d cotton plants after inoculation with conidial suspension of V991 by root dipping method. The rate of diseased plants and disease index were measured at 4 d after the plants beginning to present disease symptoms. Error bars represent the standard deviation of three biological replicates (n ≥ 16); asterisks indicate statistically significant differences, as determined by the Student t test (**p < 0.01).
Seedlings of susceptible cultivar YZ-1 infiltrated with Agrobacterium carrying TRV: Gb14-3-3c and TRV:Gb14-3-3d were prepared to investigate the roles of these two Gb14-3-3 proteins in cotton. The results of the RT-PCR analysis showed that the transcripts of Gb14-3-3c and Gb14-3-3d were successfully suppressed in seedlings 2 weeks after VIGS infiltration (Fig. 4C). The rate of diseased plants and the disease index were calculated, and the results show that silencing of Gb14-3-3c and Gb14-3-3d improves resistance to the pathogen based on the reduced wilting and etiolated leaves observed in TRV:Gb14-3-3c and TRV:Gb14-3-3d seedlings (Fig. 4D). This suggests that the 14-3-3 proteins function as negative regulators of resistance to V. dahliae in cotton.
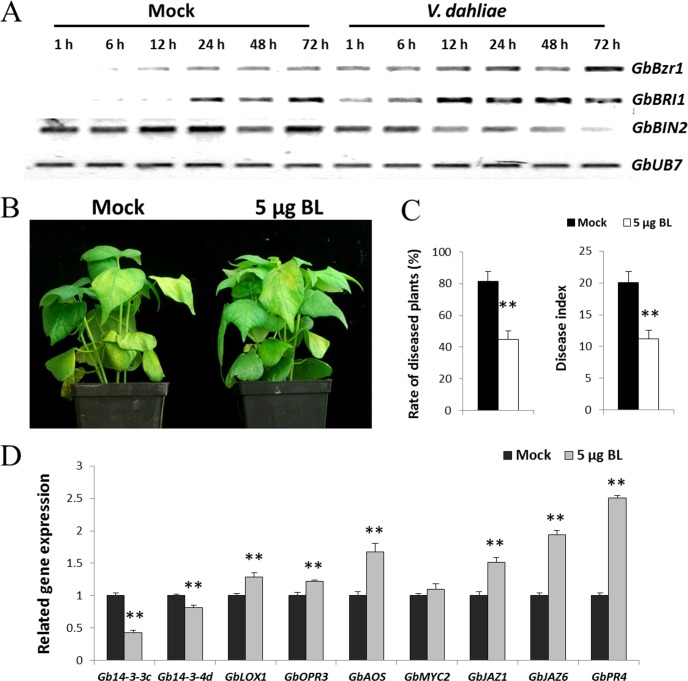
Based on previous studies, the 14-3-3 genes described above are involved in the negative regulation of the BR signaling pathway in plants (43–45). It is possible that BR signaling is involved in regulating the resistance of cotton to V. dahliae. To verify this hypothesis, the expression patterns of three genes involved in the BR signaling pathway were analyzed in roots after inoculation with V991 (Fig. 5A). Brassinosteroid insensitive 1 (BRI1), a BR receptor gene, and Brassinazole resistant 1 (BZR1), a positive BR response factor, were up-regulated, whereas Brassinosteroid insensitive 2 (BIN2), a negative response receptor of BR, was down-regulated in roots after inoculation with V. dahliae.
Fig. 5.
BL enhances disease resistance caused by V. dahliae in cotton. A, RT-PCR analysis of BR signaling pathway-related genes at the transcriptional level in G. barbadense cv7124 on inoculation with V. dahliae. PCR was performed by 28 cycles of amplification for GbUB7 and 29 cycles for other genes. BZR1, Brassinazole resistant 1; BRI1, Brassinosteroid insensitive 1; BIN2, Brassinosteroid insensitive 2. B, Effect of 5 μg/pot BL application on G. hirsutum cvYZ-1 seedlings inoculated with V. dahliae. Each experiment contains 16 plants and the experiment repeats for three times. Photograph of representative disease symptoms taken 9 d after inoculation. C, Rate of diseased plants and disease index of water- and BL-treated cotton plants. Plants were treated with BL by soil drenching 24 h before challenge inoculation with V. dahliae. The rate of diseased plants and disease index were measured at 4 d after the plants beginning to present disease symptoms. Error bars represent the standard deviation of three biological replicates (n ≥ 16); asterisks indicate statistically significant differences, as determined by the Student t test (**p < 0.01). D, qPCR analysis of the Gb14-3-3c and Gb14-3-3d transcripts in control and BL treatments. JA signaling-related genes were detected at the transcriptional levels by qPCR. Error bars represent the standard deviation for three independent experiments, and three technical replicates were analyzed; asterisks indicate statistically significant differences, as determined by the Student t test (*p < 0.05; **p < 0.01).
To characterize the role of BR in the cotton plant response to V. dahliae, exogenous BL (brassinolide, a biologically active BR; 5 μg/pot) was applied to seedlings of YZ-1 24 h before inoculation with V991. Seedlings treated with exogenous BL exhibited significantly more resistance to V. dahliae than mock-treated plants (Figs. 5B, 5C). Meanwhile, the expression of Gb14-3-3c and Gb14-3-3d was significantly inhibited after the seedlings were treated with BL (Fig. 5D). This demonstrates a role for BR as a positive regulator of cotton resistance to V. dahliae.
SA and JA have been reported to play important roles in plant immunity, so we also analyzed expression of SA and JA signaling pathway-related genes in cotton roots after treatment of BL. Surprisingly, most of genes involved in JA signaling were up-regulated in cotton plants after treatment of BL (Fig. 5D), but there were no obvious changes in the transcripts of SA signaling pathway-related genes (data not shown).
GbSSI2 Influences the Cotton Resistance by Altering SA- and JA-mediated Defense Signaling
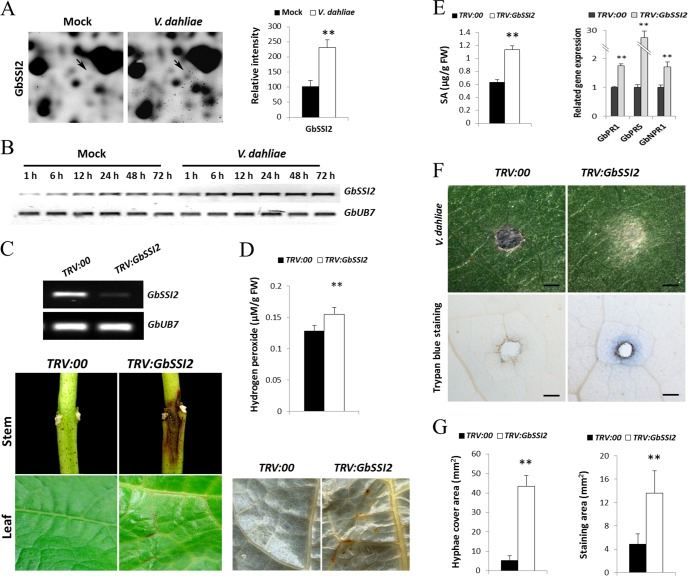
A novel protein identified as stearoylacyl-carrier protein desaturase (ssp4333, gi 48813830), which shares 78% amino acid sequence identity with SSI2 (AT2G43710) in Arabidopsis, was significantly induced by V. dahliae and named GbSSI2 (Fig. 6A). The transcript of this gene also accumulated within 1 h after inoculation with V. dahliae, as shown by RT-PCR analysis (Fig. 6B). To further investigate the function of GbSSI2 in cotton, VIGS was performed to silence the expression of GbSSI2 in the resistant cultivar 7124. The RT-PCR analysis showed that GbSSI2 transcripts were significantly reduced in cotton after infiltration with Agrobacterium containing TRV:GbSSI2 vector (Fig. 5C). Furthermore, spontaneous lesions in stems and yellow leaf veins were found in GbSSI2-silenced plants (Fig. 6C). These spontaneous lesions expanded during plant growth and severely blocked cotton development, which is similar to the phenotype of ssi2 in Arabidopsis. Spontaneous lesions in the Arabidopsis mutant ssi2 are caused by intense hypersensitive cell death because of the accumulation of ROS (46). Biochemical quantitative determination of the H2O2 content of GbSSI2-silenced cotton revealed levels significantly higher than those in control plants (Fig. 6D). Supporting this, accumulation of H2O2 as visualized by DAB stainingconfirmed the higher levels of H2O2 in the silenced plants (Fig. 6D).
Fig. 6.
Activation of ROS and SA enhances the susceptibility of GbSSI2-silenced cotton to V. dahliae. A, Shown are representative protein spot (GbSSI2) on inoculation with V. dahliae and quantification of the signal intensities obtained from three independent 2-DE maps. Error bars represent the standard deviation of three biological replicates; asterisks indicate statistically significant differences, as determined by the Student t test (*p < 0.05; **p < 0.01). B, RT-PCR analysis of the expression pattern of GbSSI2 at the transcriptional level in G. barbadense cv7124 on inoculation with V. dahliae. PCR was performed by 28 cycles of amplification for GbUB7 and 30 cycles for GbSSI2. C, Spontaneous lesion formation on the stems and leaves of TRV:GbSSI2 plants. The cotyledons of 10-day-old seedlings of G. barbadense cv7124 were hand-infiltrated with Agrobacterium carrying either TRV:GbSSI2 or VIGS-vector control (TRV:00). Spontaneous lesion was present on the stems and leaves of GbSSI2-silenced plants 12 d after infiltration. RT-PCR analysis indicated that the transcripts of GbSSI2 were reduced 12 d after infiltration. D, DAB staining and measurement of H2O2 accumulation in TRV:00 and TRV:GbSSI2 leaves. Error bars represent the standard deviation of three replicates (n = 12); asterisks indicate statistically significant differences, as determined by the Student t test (**p < 0.01). E, Detection of SA signaling pathway-related genes and alteration of SA levels in TRV:00 and TRV:GbSSI2 plants. Error bars of qRT analysis represent the standard deviation for three independent experiments, and three technical replicates were analyzed; Error bars of SA levels represent the standard deviation of three biological replicates (n = 10); asterisks indicate statistically significant differences, as determined by the Student t test (**p < 0.01). F, Disease symptoms induced on the leaves of TRV:00 and TRV:GbSSI2 cotton plants 3 d after inoculation with spore suspension of V. dahliae (106 conidia per ml). Mycelia growth on leaves inoculated with V991 (Fig. 6F, upper panel). Trypan blue staining of V991-infected leaves (Fig. 6F, lower panel). Scale bars: 1 mm. G, Hyphae cover area and trypan blue staining area of V991-infected leaves. Error bars represent the standard deviation of three replicates (n = 12); asterisks indicate statistically significant differences, as determined by the Student t test (**p < 0.01).
Hypersensitive cell death is generally considered a major result of plant disease resistance mediated by SA signaling (47). The endogenous SA content was measured and found to be significantly increased in GhSSI2-silenced plants compared with control plants (Fig. 6E). Meanwhile, expression of GbNPR1, GbPR1, and GbPR5, three important genes involved in the SA signaling pathway, was analyzed using qPCR, and results show that the transcripts of all three genes were up-regulated significantly after silencing of GbSSI2 in cotton (Fig. 6E).
Leaves from GbSSI2-silenced cotton were used for fungal inoculation with V991. Mycelia were observed 3 d after dropping spore suspension (106 conidia per ml) onto the leaf surface. Silencing of GbSSI2 greatly increased the mycelia growth of V. dahliae and accelerated cell death at the infected location, as indicated by trypan blue staining (Fig. 6F). After inoculation with V. dahliae, statistical analysis showed that hyphal cover area and trypan blue staining area of TRV:SSI2 plant leaves were significantly larger than those of TRV:00 plant leaves (Fig. 6G). These results show an essential role of GbSSI2 participating in cotton resistance to V. dahliae and demonstrated that the accumulation of H2O2 and the activation of the SA signaling pathway led to enhanced infection of cotton by V. dahliae.
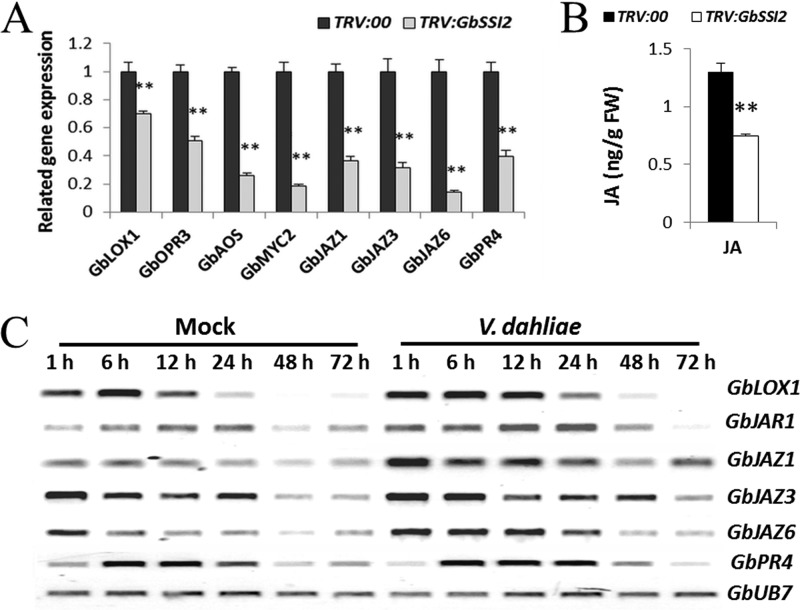
Defense signaling pathways mediated by SA and JA frequently act antagonistically to mediate defense against specific types of pathogens (10–12). To explore whether activation of the SA pathway in TRV:GbSSI2 plants influences JA signal transduction, the expression of genes participating in the biosynthesis of JA (GbLOX1, GbAOS, and GbOPR3) and its signal transduction (GbJAZ1, GbJAZ6, GbMYC2, and GbPR4) was analyzed. qPCR results show that the transcripts of genes involved in both JA biosynthesis and JA signal transduction were inhibited in TRV:GbSSI2 plants (Fig. 7A). Moreover, the content of JA was significantly decreased in TRV:GbSSI2 plants compared with the control (Fig. 7B), which was in accordance with the expression patterns of related genes (Fig. 7A). All the mentions above indicate that GbSSI2 may influence the cotton resistance by altering SA- and JA-mediated defense signaling.
Fig. 7.
The JA signaling pathway is suppressed in GbSSI2-silenced cotton and activated in cotton after inoculation with V. dahliae. A, Detection of JA signaling pathway-related genes in TRV:00 and TRV:GbSSI2 plants by qPCR. Error bars represent the standard deviation for three independent experiments, and three technical replicates were analyzed; asterisks indicate statistically significant differences, as determined by the Student t test (**p < 0.01). B, Detection of JA levels in TRV:00 and TRV:GbSSI2 plants. Error bars represent the standard deviation of three biological replicates (n = 10); asterisks indicate statistically significant differences, as determined by the Student t test (**p < 0.01). C, RT-PCR analysis of JA signaling pathway-related genes in G. barbadense cv7124 on inoculation with V. dahliae. PCR was performed by 28 cycles of amplification for GbUB7 and 29 cycles for other genes.
DISCUSSION
Verticillium wilt is a serious disease that significantly affects the yield and quality of cotton. Understanding the molecular mechanism of cotton-V. dahliae interactions is a key aim to support for disease resistance breeding in cotton. As the development of sequencing technologies progresses, diverse differentially expressed genes following cotton inoculation with V. dahliae have been identified at the transcriptional level (19, 24), whereas the functional roles of few candidate genes have been confirmed. Despite the relative simplicity and high throughput, the transcriptional data is complex, and so difficult to draw meaningful conclusions, and does not readily provide new functional insights. For example, it has been shown that the transcriptional level does not necessarily correlate with the protein level (48). Here, a complete analysis of protein expression using 2-DE and MALDI-TOF/TOF, combined with expression analysis and functional studies using VIGS and infection phenotyping, offers a useful approach to reveal the molecular basis of cotton defense against V. dahliae.
Using the resistant cultivar 7124 from G. barbadense, we identified 188 differentially expressed proteins through a proteomics approach from roots inoculated with V. dahliae. Among these proteins, those that were down-regulated by the fungus were the majority of differentially expressed proteins, which may imply that metabolism is repressed to in turn repress the plant immune network. This idea is consistent with our previous results obtained using transcriptional analysis by RNA-seq (19). According to the results of GO functional classification, the three largest biological processes, including “cellular process,” “metabolism,” and “stimulus response,” which together represent the principal fundamental cell functional processes, were involved in the response of cotton to V. dahliae. This suggests that the host undergoes major physiological changes in response to the plant-pathogen interaction.
Secondary metabolites become especially abundant in plant cells when plants are threatened by diverse pathogens in nature (19, 24, 25). Generally, the production of secondary metabolites, such as phytoalexins and lignin, can inhibit the growth of pathogens directly or provide a physical barrier against the invading plant pathogens. Our previous studies have demonstrated the importance of lignin involvement in the phenylpropanoid pathway; lignin was found to contribute to cotton disease resistance (19). In this study, GbCCoAOMT, a protein involved in lignin biosynthesis, was also identified and was significantly up-regulated at the protein level on infection of V. dahliae.
Gossypol, the major type of sesquiterpenoid, is a cotton-specific secondary metabolite that has been shown to be associated with disease resistance (18, 20, 21, 49–51). The involvement of gossypol metabolites in the interaction between cotton and V. dahliae was further confirmed in the current study. GbCAD1, an important enzyme involved in gossypol biosynthesis (18, 51), was identified through our proteomics analysis. Gossypol content was dramatically increased by fungal infection, and silencing of GbCAD1 by VIGS enhanced cotton susceptibility to the pathogen. Moreover, it was speculated that down-regulation of dHG-6-OMT could maintain a relatively high terpenoid content and significantly increase the resistance of G. hirsutum to fungi and pests (51). In our study, GbdHG-6-OMT (ssp1320), a gene negatively regulating resistance, was suppressed in cotton roots during V. dahliae inoculation. Other genes involved in gossypol biosynthesis, including GbFPS and GbCYP706B1, were up-regulated on infection of V. dahliae, as shown by RT-PCR analysis and so leading to enhanced resistance. These results suggest that the accumulation of gossypol is an effective strategy for the response of cotton to V. dahliae.
Recent advances in plant immunity research have provided new insights into the underlying defense signaling network by diverse small-molecule hormones, such as auxin, BR, JA, and SA (8, 16). These signaling pathways cross-communicate in an antagonistic or synergistic manner, providing the plant with a powerful capacity to finely regulate its immune response (16). However, evidence is accumulating that pathogens can manipulate hormone-regulated signaling pathways to evade host immune responses (16). We isolated a number of proteins involved in the phytohormone signaling network with diverse expression patterns during V. dahliae infection, which implies that complicated cross-talk of phytohormone signaling pathways is involved in the incompatible interaction between cotton and V. dahliae.
Over the past decade, extensive research efforts have demonstrated that BRs can act as regulatory factors in biotic stress responses in plants (52–56). BZR1 and BZR2 are two key transcription factors that mediate BR signals by regulating downstream gene expression. In addition, 14-3-3 proteins have been demonstrated to be negative regulators of BR signaling by regulating the subcellular localization and activity of both BZR1 and BZR2 (44). Four Gb14-3-3 proteins involved in BR signaling were identified in our proteomic analysis to have decreased in abundance in cotton after inoculation with V. dahliae. Our RT-PCR data also showed that the genes positively regulating BR signaling were induced in cotton after inoculation with V. dahliae. Silencing of Gb14-3-3c and Gb14-3-3d in cotton through VIGS enhanced the resistance of cotton to V. dahliae. Meanwhile, exogenous BL treatment has a similar effect on cotton disease resistance to V. dahliae. Moreover, SERK3/BAK1 is a coreceptor that physically associates with BRI1 for BR-dependent signaling and has been shown to be required for Ve1-mediated resistance in tomato and Arabidopsis (17, 29). The above results demonstrate that BRs and the BR signaling pathway positively regulate the resistance of cotton during V. dahliae infection. This study therefore provides new information about the function of BR in cotton disease resistance.
In plants, ROS play a dual role as both toxic byproduct of normal cell metabolism and regulatory molecules in stress perception and signal transduction (57). The expression levels of several proteins that participate in the regulation of redox homeostasis, GbPOD, GbAPX2, GbMDAR6, and GbMDH, were found to be influenced, and most were down-regulated, in cotton on infection with V. dahliae. At the same time, SSI2, a stearoyl-ACP-desaturase, was induced during the interaction between cotton and V. dahliae. Silencing of GbSSI2 in cotton led to the accumulation of H2O2 and impaired the resistance of cotton to V. dahliae. These results imply that ROS in cotton may be negatively correlated with disease resistance to V. dahliae. However, ROS with other signaling molecules are complex in plant cells (14, 47), and the exact role of ROS involved in cotton defense against V. dahliae invasion remains to be discovered.
Two phytohormones SA and JA are known to play major roles in regulating plant defense response against various pathogens. SA participates in the activation of the defense response against biotrophic and hemi-biotrophic pathogens, while JA/ET mediates plant defenses against necrotrophic pathogens (16, 58). Previous studies have also shown that SA is a potent suppressor of JA-mediated defenses against necrotrophs (8, 16). A balance between glycerol-3-phosphate (G3P) and oleic acid levels mediated by SSI2 is critical for the regulation of SA- and JA-mediated defense signaling in the plant (59). Silencing of GbSSI2 in cotton through VIGS simultaneously up-regulates SA synthesis and SA-mediated responses and inhibits JA-inducible defenses, resulting in increased susceptibility to V. dahliae, which is similar to the results from a mutation or silencing of SSI2 in Arabidopsis or rice to pathogens (60–62). When cotton plants were invaded by V. dahliae, protein levels of GbSSI2 increased significantly, as shown by proteomic analysis. Therefore, we speculated that the JA signaling pathway may be activated in this process. Our speculation was verified by the up-regulation of transcripts of JA signaling pathway-related genes in the roots of 7124 inoculation by V. dahliae (Fig. 7C).
GaWRKY1, which participates in the regulation of gossypol biosynthesis in cotton, may also be induced by JA (22). Arabidopsis MYC2, a positive regulator in the JA signaling pathway, is involved in the induction of sesquiterpene synthase genes (63). Our results also confirm this, as GbFPS, GbCYP706B1, GbCAD1 and WRKY1 were significantly induced in Gossypium barbadense cv7124 treated with MeJA (supplemental Fig. S3). All these result indicate that gossypol, the sesquiterpene specifically synthesized in cotton, is induced and regulated by JA. Recently, BAK1 was shown to regulate the accumulation of jasmonic acid, whereas BAK1-silenced plants showed attenuated JA and JA-isoleucine bursts (52, 64), and JA application to the BAK1-silenced plants restored the induction of defensive trypsin proteinase inhibitors to higher levels. Furthermore, Ve1-mediated resistance could be compromised in the bak1–4, coi1–16 and jar1–1 mutants in Arabidopsis (29). We also monitored the expression pattern of JA synthesis- and signaling pathway-related genes in cotton roots treated with BL. All these genes are induced by BL, indicating JA signaling pathway is active in cotton plants treated with BL (Fig. 5D). These data demonstrate that cross-talk between BR and JA signaling may exist in cotton and may positively contribute to the disease resistance of cotton to V. dahliae. However further research is required to define the precise underlying molecular mechanisms.
In summary, comparative proteomics was employed in our study to identify disease response proteins and to understand the mechanism of cotton resistance to the fungal pathogen V. dahliae. Combined with VIGS, we revealed that gossypol, BRs and JA act as important factors in the resistance of cotton to V. dahliae. These data provide important information to highlight the molecular processes of disease resistance in cotton and facilitate further studies in cotton breeding for disease resistance.
Supplementary Material
Acknowledgments
We thank Dr. Bart P. H. J. Thomma (Laboratory of Phytopathology, Wageningen University, The Netherlands) for providing the VIGS vectors, Dongqin Li (National Key Laboratory of Crop Genetic Improvement, China) for technical advice in measuring hormone content and Liebo Shu for technical assistance in MS analysis (Shanghai Boyuan Biotechnology, China).
Footnotes
* This work was supported by the National High-tech program (2013AA102601–4) and the National Natural Science Foundation of China (31271772).
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S4.
1 The abbreviations used are:
- SA
- salicylic acid
- 2-DE
- two-dimensional gel electrophoresis
- qPCR
- quantitative real time PCR
- At
- Arabidopsis thaliana
- Gh
- Gossypium hirsutum
- Gb
- Gossypium barbadense
- hpi
- hours past inoculation
- ssp
- sample spot protein
- NCBI
- National Center for Biotechnology information
- VIGS
- virus-induced gene silencing
- ROS
- reactive oxygen species
- JA
- jasmonic acid
- BRs
- brassinosteroids
- BL
- brassinolide
- DI
- disease index.
REFERENCES
- 1. Sal'kova E. G., Guseva N. N. (1965) The role of pectolytic enzymes of the verticillium dahliae fungus in the development of cotton wilt. Dokl. Akad. Nauk SSSR 163, 515–522 [PubMed] [Google Scholar]
- 2. Cai Y. F., He X. H., Mo J. C., Sun Q., Yang J. P., Liu J. G. (2009) Molecular research and genetic engineering of resistance to Verticillium wilt in cotton: A review. Afr. J. Biotechnol. 8, 7363–7372 [Google Scholar]
- 3. Zhang J., Sanogo S., Flynn R., Baral J., Bajaj S., Hughs S. E., Percy R. (2012) Germplasm evaluation and transfer of Verticillium wilt resistance from Pima (Gossypium barbadense) to Upland cotton (G. hirsutum). Euphytica 187, 147–160 [Google Scholar]
- 4. Aguado A., Santos B. D. L., Blanco C., Romero F. (2008) Study of gene effects for cotton yield and Verticillium wilt tolerance in cotton plant (Gossypium hirsutum L.). Field Crops Res. 107, 78–86 [Google Scholar]
- 5. Jiang F., Zhao J., Zhou L., Guo W., Zhang T. (2009) Molecular mapping of Verticillium wilt resistance QTL clustered on chromosomes D7 and D9 in upland cotton. Sci. China C Life Sci. 52, 872–884 [DOI] [PubMed] [Google Scholar]
- 6. Zhang J. F., Lu Y., Adragna H., Hughs E. (2005) Genetic improvement of New Mexico Acala cotton germplasm and their genetic diversity. Crop Sci. 45, 2363 [Google Scholar]
- 7. Jones J. D., Dangl J. L. (2006) The plant immune system. Nature 444, 323–329 [DOI] [PubMed] [Google Scholar]
- 8. Bari R., Jones J. D. (2009) Role of plant hormones in plant defence responses. Plant Mol. Biol. 69, 473–488 [DOI] [PubMed] [Google Scholar]
- 9. Dempsey D. A., Klessig D. F. (2012) SOS - too many signals for systemic acquired resistance? Trends Plant Sci. 17, 538–545 [DOI] [PubMed] [Google Scholar]
- 10. Brooks D. M., Bender C. L., Kunkel B. N. (2005) The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Mol. Plant Pathol. 6, 629–639 [DOI] [PubMed] [Google Scholar]
- 11. Spoel S. H., Johnson J. S., Dong X. (2007) Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc. Natl. Acad. Sci. U.S.A. 104, 18842–18847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spoel S. H., Koornneef A., Claessens S. M., Korzelius J. P., Van Pelt J. A., Mueller M. J., Buchala A. J., Métraux J. P., Brown R., Kazan K., Van Loon L. C., Dong X., Pieterse C. M. (2003) NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grant M. R., Jones J. D. (2009) Hormone (dis)harmony moulds plant health and disease. Science 324, 750–752 [DOI] [PubMed] [Google Scholar]
- 14. Li A., Zhang R., Pan L., Tang L., Zhao G., Zhu M., Chu J., Sun X., Wei B., Zhang X., Jia J., Mao L. (2011) Transcriptome analysis of H2O2-treated wheat seedlings reveals a H2O2-responsive fatty acid desaturase gene participating in powdery mildew resistance. PLoS ONE 6, e28810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lamb C., Dixon R. A. (1997) The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275 [DOI] [PubMed] [Google Scholar]
- 16. Pieterse C. M., Leon-Reyes A., Van der Ent S., Van Wees S. C. (2009) Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316 [DOI] [PubMed] [Google Scholar]
- 17. Fradin E. F., Zhang Z., Juarez Ayala J. C., Castroverde C. D., Nazar R. N., Robb J., Liu C. M., Thomma B. P. (2009) Genetic dissection of Verticillium wilt resistance mediated by tomato Ve1. Plant Physiol. 150, 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu C. J., Heinstein P., Chen X. Y. (1999) Expression pattern of genes encoding farnesyl diphosphate synthase and sesquiterpene cyclase in cotton suspension-cultured cells treated with fungal elicitors. Mol. Plant. Microbe Interact. 12, 1095–1104 [DOI] [PubMed] [Google Scholar]
- 19. Xu L., Zhu L., Tu L., Liu L., Yuan D., Jin L., Long L., Zhang X. (2011) Lignin metabolism has a central role in the resistance of cotton to the wilt fungus Verticillium dahliae as revealed by RNA-Seq-dependent transcriptional analysis and histochemistry. J. Exp. Bot. 62, 5607–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Luo P., Wang Y. H., Wang G. D., Essenberg M., Chen X. Y. (2001) Molecular cloning and functional identification of (+)-delta-cadinene-8-hydroxylase, a cytochrome P450 mono-oxygenase (CYP706B1) of cotton sesquiterpene biosynthesis. Plant J. 28, 95–104 [DOI] [PubMed] [Google Scholar]
- 21. Townsend B. J., Poole A., Blake C. J., Llewellyn D. J. (2005) Antisense suppression of a (+)-delta-cadinene synthase gene in cotton prevents the induction of this defense response gene during bacterial blight infection but not its constitutive expression. Plant Physiol. 138, 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu Y. H., Wang J. W., Wang S., Wang J. Y., Chen X. Y. (2004) Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-delta-cadinene synthase-A. Plant Physiol. 135, 507–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zuo K., Wang J., Wu W., Chai Y., Sun X., Tang K. (2005) Identification and characterization of differentially expressed ESTs of Gossypium barbadense infected by Verticillium dahliae with suppression subtractive hybridization. Mol. Biol. 39, 191–199 [PubMed] [Google Scholar]
- 24. Xu L., Zhu L. F., Tu L. L., Guo X. P., Long L., Sun L. Q., Gao W., Zhang X. L. (2011) Differential gene expression in cotton defence response to Verticillium dahliae by SSH. J. Phytopathol. 159, 606–615 [Google Scholar]
- 25. Wang F. X., Ma Y. P., Yang C. L., Zhao P. M., Yao Y., Jian G. L., Luo Y. M., Xia G. X. (2011) Proteomic analysis of the sea-island cotton roots infected by wilt pathogen Verticillium dahliae. Proteomics 11, 4296–4309 [DOI] [PubMed] [Google Scholar]
- 26. Zhao F. A., Fang W., Xie D., Zhao Y., Tang Z., Li W., Nie L., Lv S. (2012) Proteomic identification of differentially expressed proteins in Gossypium thurberi inoculated with cotton Verticillium dahliae. Plant Sci. 185–186, 176–184 [DOI] [PubMed] [Google Scholar]
- 27. Munis M. F., Tu L., Deng F., Tan J., Xu L., Xu S., Long L., Zhang X. (2010) A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem. Biophys. Res. Commun. 393, 38–44 [DOI] [PubMed] [Google Scholar]
- 28. Shi J., An H. L., Zhang L., Gao Z., Guo X. Q. (2010) GhMPK7, a novel multiple stress-responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol. Biol. 74, 1–17 [DOI] [PubMed] [Google Scholar]
- 29. Fradin E. F., Abd-El-Haliem A., Masini L., van den Berg G. C., Joosten M. H., Thomma B. P. (2011) Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang B., Yang Y., Chen T., Yu W., Liu T., Li H., Fan X., Ren Y., Shen D., Liu L., Dou D., Chang Y. (2012) Island cotton Gbve1 gene encoding a receptor-like protein confers resistance to both defoliating and non-defoliating isolates of Verticillium dahliae. PLoS ONE 7, e51091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao X., Wheeler T., Li Z., Kenerley C. M., He P., Shan L. (2011) Silencing GhNDR1 and GhMKK2 compromises cotton resistance to Verticillium wilt. Plant J. 66, 293–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yao Y., Yang Y. W., Liu J. Y. (2006) An efficient protein preparation for proteomic analysis of developing cotton fibers by 2-DE. Electrophoresis 27, 4559–4569 [DOI] [PubMed] [Google Scholar]
- 33. Pang C. Y., Wang H., Pang Y., Xu C., Jiao Y., Qin Y. M., Western T. L., Yu S. X., Zhu Y. X. (2010) Comparative proteomics indicates that biosynthesis of pectic precursors is important for cotton fiber and Arabidopsis root hair elongation. Mol. Cell Proteomics 9, 2019–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu L., Tu L., Zeng F., Liu D., Zhang X. (2005) An improved simple protocol for isolation of high quality RNA from Gossypium spp. suitable for cDNA library construction. Acta Agronomic Sinica 31, 1657–1659 [Google Scholar]
- 35. Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M. W., Shipley G. L., Vandesompele J., Wittwer C. T. (2009) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 [DOI] [PubMed] [Google Scholar]
- 36. Liu Y., Schiff M., Marathe R., Dinesh-Kumar S. P. (2002) Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429 [DOI] [PubMed] [Google Scholar]
- 37. Xu F., Yang L., Zhang J., Guo X., Zhang X., Li G. (2012) Prevalence of the defoliating pathotype of Verticillium dahliae on cotton in central china and virulence on selected cotton cultivars. J. Phytopathol. 160, 369–376 [Google Scholar]
- 38. Bowling S. A., Guo A., Cao H., Gordon A. S., Klessig D. F., Dong X. (1994) A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell 6, 1845–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Choi du S., Hwang B. K. (2011) Proteomics and functional analyses of pepper abscisic acid-responsive 1 (ABR1), which is involved in cell death and defense signaling. Plant Cell 23, 823–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parry G., Ward S., Cernac A., Dharmasiri S., Estelle M. (2006) The Arabidopsis suppressor of auxin resistance proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18, 1590–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sieburth L. E., Muday G. K., King E. J., Benton G., Kim S., Metcalf K. E., Meyers L., Seamen E., Van Norman J. M. (2006) SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 18, 1396–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quint M., Barkawi L. S., Fan K. T., Cohen J. D., Gray W. M. (2009) Arabidopsis IAR4 modulates auxin response by regulating auxin homeostasis. Plant Physiol. 150, 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Vries S. C. (2007) 14-3-3 proteins in plant brassinosteroid signaling. Dev. Cell 13, 162–164 [DOI] [PubMed] [Google Scholar]
- 44. Gampala S. S., Kim T. W., He J. X., Tang W., Deng Z., Bai M. Y., Guan S., Lalonde S., Sun Y., Gendron J. M., Chen H., Shibagaki N., Ferl R. J., Ehrhardt D., Chong K., Burlingame A. L., Wang Z. Y. (2007) An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 13, 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang H., Yang C., Zhang C., Wang N., Lu D., Wang J., Zhang S., Wang Z. X., Ma H., Wang X. (2011) Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 21, 825–834 [DOI] [PubMed] [Google Scholar]
- 46. Kachroo P., Kachroo A., Lapchyk L., Hildebrand D., Klessig D. F. (2003) Restoration of defective cross talk in ssi2 mutants: role of salicylic acid, jasmonic acid, and fatty acids in SSI2-mediated signaling. Mol. Plant. Microbe Interact. 16, 1022–1029 [DOI] [PubMed] [Google Scholar]
- 47. Govrin E. M., Levine A. (2000) The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10, 751–757 [DOI] [PubMed] [Google Scholar]
- 48. Gygi S. P., Rochon Y., Franza B. R., Aebersold R. (1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu J., Benedict C. R., Stipanovic R. D., Bell A. A. (1999) Purification and characterization of S-adenosyl-L-methionine: desoxyhemigossypol-6-O-methyltransferase from cotton plants. An enzyme capable of methylating the defense terpenoids of cotton. Plant Physiol. 121, 1017–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tan X. P., Liang W. Q., Liu C. J., Luo P., Heinstein P., Chen X. Y. (2000) Expression pattern of (+)-delta-cadinene synthase genes and biosynthesis of sesquiterpene aldehydes in plants of Gossypium arboreum L. Planta 210, 644–651 [DOI] [PubMed] [Google Scholar]
- 51. Liu J., Benedict C. R., Stipanovic R. D., Magill C. W., Bell A. A. (2002) Cloning and expression of desoxyhemigossypol-6-O-methyltransferase from cotton (Gossypium barbadense). J. Agric. Food Chem. 50, 3165–3172 [DOI] [PubMed] [Google Scholar]
- 52. Choudhary S. P., Yu J. Q., Yamaguchi-Shinozaki K., Shinozaki K., Tran L. S. (2012) Benefits of brassinosteroid crosstalk. Trends Plant Sci. 17, 594–605 [DOI] [PubMed] [Google Scholar]
- 53. Krishna P. (2003) Brassinosteroid-Mediated Stress Responses. J. Plant Growth Regul. 22, 289–297 [DOI] [PubMed] [Google Scholar]
- 54. Nakashita H., Yasuda M., Nitta T., Asami T., Fujioka S., Arai Y., Sekimata K., Takatsuto S., Yamaguchi I., Yoshida S. (2003) Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 33, 887–898 [DOI] [PubMed] [Google Scholar]
- 55. Vriet C., Russinova E., Reuzeau C. (2012) Boosting crop yields with plant steroids. Plant Cell 24, 842–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wang H., Nagegowda D. A., Rawat R., Bouvier-Navé P., Guo D., Bach T. J., Chye M. L. (2012) Overexpression of Brassica juncea wild-type and mutant HMG-CoA synthase 1 in Arabidopsis up-regulates genes in sterol biosynthesis and enhances sterol production and stress tolerance. Plant Biotechnol. J. 10, 31–42 [DOI] [PubMed] [Google Scholar]
- 57. Gadjev I., Vanderauwera S., Gechev T. S., Laloi C., Minkov I. N., Shulaev V., Apel K., Inzé D., Mittler R., Van Breusegem F. (2006) Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141, 436–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bernoux M., Ellis J. G., Dodds P. N. (2011) New insights in plant immunity signaling activation. Curr. Opin. Plant Biol. 14, 512–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kachroo A., Venugopal S. C., Lapchyk L., Falcone D., Hildebrand D., Kachroo P. (2004) Oleic acid levels regulated by glycerolipid metabolism modulate defense gene expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 5152–5157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gao Q. M., Venugopal S., Navarre D., Kachroo A. (2011) Low oleic acid-derived repression of jasmonic acid-inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiol. 155, 464–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kachroo A., Lapchyk L., Fukushige H., Hildebrand D., Klessig D., Kachroo P. (2003) Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell 15, 2952–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jiang C. J., Shimono M., Maeda S., Inoue H., Mori M., Hasegawa M., Sugano S., Takatsuji H. (2009) Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol. Plant. Microbe Interact. 22, 820–829 [DOI] [PubMed] [Google Scholar]
- 63. Hong G. J., Xue X. Y., Mao Y. B., Wang L. J., Chen X. Y. (2012) Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell 24, 2635–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang D. H., Hettenhausen C., Baldwin I. T., Wu J. (2011) BAK1 regulates the accumulation of jasmonic acid and the levels of trypsin proteinase inhibitors in Nicotiana attenuata's responses to herbivory. J. Exp. Bot. 62, 641–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.