Abstract
Epithelial-mesenchymal transition (EMT) is a key contributor in tumor progression and metastasis. EMT produces cellular heterogeneity within head and neck squamous cell carcinomas (HNSCC) by creating a phenotypically distinct mesenchymal subpopulation that is resistant to conventional therapies. In this study, we systematically characterized differences in the secretomes of E-cadherin high epithelial-like and E-cadherin low mesenchymal-like subpopulations using unbiased and targeted proteomics. A total 1765 proteins showed significant changes with 177 elevated in the epithelial subpopulation and 173 elevated in the mesenchymal cells. Key nodes in affected networks included NFκB, Akt, and ERK, and most implicated cellular components involved various aspects of the extracellular matrix. In particular, large changes were observed in multiple collagens with most affected collagens at much higher abundance levels in the mesenchymal subpopulation. These cells also exhibited a secretome profile resembling that of cancer-associated fibroblastic cells (CAF). S100A4, a commonly used marker for cancer-associated fibroblastic cells, was elevated more than 20-fold in the mesenchymal cells and this increase was further verified at the transcriptome level. S100A4 is a known mediator of EMT, leading to metastasis and EMT has been proposed as a potential source of cancer-associated fibroblastic cells in solid tumors. S100A4 knockdown by small interfering RNA led to decreased expression, secretion and activity of matrix metalloproteinase 2, as verified by quantitative PCR, multiple reaction monitoring and zymography analyses, and reduced invasion in collagen-embedded spheroids. Further confirmation in three-dimensional organotypic reconstructs showed less invasion and advanced differentiation in the S100A4 RNA interference samples. Orthotopic metastasis model, developed to validate the findings in vivo, demonstrated a decrease in spontaneous metastasis and augmented differentiation in the primary tumor in siS100A4 xenografts. These results demonstrate the value of secretome profiling to evaluate phenotypic conversion and identify potential novel therapeutic targets such as S100A4.
Head and neck squamous cell carcinoma (HNSCC)1 is the sixth most common cancer worldwide with high morbidity and mortality. Treatment options are limited; surgery, radiation and conventional chemotherapy are generally associated with considerable impairment to quality of life (1). Currently cetuximab, the anti-epidermal growth factor receptor (EGFR) monoclonal antibody, is the only FDA-approved molecularly targeted drug for HNSCC. Several clinical trials are underway that target EGFR or other tyrosine kinases, but many have failed to progress beyond Phase II (2). One reason behind the failure of new treatment options for HNSCC is resistance to therapy, which tumor heterogeneity is thought to provide.
We have previously shown that an E-cadherin low mesenchymal subpopulation (MSP) within a predominantly epithelial carcinoma confers resistance to both conventional chemotherapy and cetuximab treatment (3, 4). Epithelial and mesenchymal subpopulations within these tumors exhibited phenotypic plasticity, indicative of epithelial-mesenchymal transition (EMT) (3). EMT is a known contributor of tumor heterogeneity in carcinomas, enabling an invasion-metastasis cascade (5). Scheel et al. (6) reported a spontaneously arising MSP in immortalized human mammary epithelial cells (HMLE). Activation of several signaling pathways, cooperating with each other in the EMT program, was necessary to empirically induce MSP cells from HMLEs.
Within primary tumors these signals are obtained by autocrine and paracrine manner. The tumor microenvironment, providing the paracrine signals, includes angiogenic vascular cells, infiltrating immune and inflammatory cells, and cancer-associated fibroblastic cells (CAFs) (7). Despite the recent investigations into the origins of tumor-associated stromal cells, the precise origin of CAFs remains unclear; EMT has been proposed as a potential source in conjunction with local fibroblasts (8). CAFs secrete a variety of factors that promote both tumor growth and progression, such as growth factors, cytokines, extracellular matrix (ECM) components and matrix metalloproteinases (MMP) (9). They are highly heterogeneous, thus no single marker can distinguish them. Commonly used markers are alpha-smooth muscle actin (α-SMA), platelet-derived growth factor receptors alpha and beta (PDGFRα/β), integrin alpha-5 (ITGA5), fibroblast activation protein (FAP, also known as seprase) and S100A4 (also known as fibroblast-specific protein 1, FSP1) (10). S100A4 is a member of the S100 calcium-binding protein superfamily. It is a known modulator of ECM and cytoskeletal integrity, thus promoting metastasis (11). S100A4 has also been shown to regulate the expression of MMPs (12–14). In addition to the intracellular form, S100A4 is also secreted from cells through an unknown mechanism (15).
Since EMT is proposed as a possible origin for cancer-associated fibroblastic cells, we wanted to investigate whether the MSP of heterogeneous HNSCCs resembles CAFs. As the first step we characterized the phenotypic properties of the epithelial and mesenchymal subpopulations in vitro and in vivo. Liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) was used to analyze the secretomes of the epithelial and mesenchymal subpopulations in an unbiased manner. Following this approach we further validated the expression of several CAF markers at the transcriptome level and identified S100A4 as an important mediator of the phenotypic conversion. Knockdown of S100A4 by small interfering RNA (siRNA), verified by targeted proteomics using multiple reaction monitoring (MRM), led not only to reduced metastasis burden, but also to advanced differentiation of the primary tumor, demonstrating a novel finding with potential implications for therapeutic strategy.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
OCTT2 cell line, derived from a human HNSCC primary surgical specimen, and FEF3 primary fibroblast strain, isolated from human fetal esophagus and stably transduced with green fluorescent protein, have been described previously and were cultured as detailed earlier (3, 4, 16). For preparation of conditioned medium (CM), cells were cultured overnight in serum-free conditions after which supernatant was collected, centrifuged, and filtered through low protein binding filters (EMD Millipore Corporation, Billerica, MA) to remove cell debris. Genetic purity of cell lines was confirmed based on microsatellite DNA markers using an Identify Mapping Kit (Coriell Institute for Medical Research) and cells were routinely tested for mycoplasma (University of Pennsylvania School of Medicine, Cell Culture Services).
Antibodies and Reagents
The following antibodies were used: mouse monoclonal anti-S100A4 (Thermo Fisher Scientific, Waltham, MA), goat polyclonal anti-S100A4 (Everest Biotech, Ramona, CA), mouse monoclonal anti-MMP2 (Cell Signaling Technology, Danvers, MA), rabbit polyclonal anti-GAPDH (Sigma-Aldrich, St. Louis, MO), rabbit polyclonal anti-vimentin (AnaSpec, Fremont, CA) rabbit polyclonal anti-involucrin (Thermo Fisher Scientific), IRDye 680LT goat anti-mouse, IRDye 800CW goat anti-rabbit, IRDye 800CW donkey anti-goat (all LI-COR Biosciences, Lincoln, NE), allophycocyanin anti-human E-cadherin (BioLegend, San Diego, CA), allophycocyanin mouse IgG control (BD Biosciences, Bedford, MA), goat IgG control (Merck & Co, Whitehouse, NJ), donkey anti-goat (Jackson Immunoresearch, West Grove, PA). Alexa Fluor 568 goat anti-rabbit, Alexa Fluor 568 phalloidin and Hoechst 33342 were from Invitrogen (Grand Island, NY).
Proliferation Assay, Fluorescence-activated Cell Sorting (FACS) and Flow Cytometry
CellTiter cell proliferation assays (Promega, Madison, WI) were performed by seeding 10,000 cells/well of the 72h RNAi-treated OCTT2 cells onto 96-well plates. After 72 h, MTS reagent was added and incubated for 2 h at +37 °C in 5% CO2 according to manufacturer's protocol. Optical density in each well was measured at 490 nm using EL800 (BioTek, Winooski, VT) plate reader.
FACS was performed as previously described (3). Briefly, E-cadherin high and E-cadherin low cell populations were sorted based on allophycocyanin-E-cadherin stain and propidium iodide was used to exclude dead cells. FACSAria (BD Biosciences) sorter was used for FACS. Flow cytometry data were acquired by BD LSR II and analyzed using FlowJo, V 10.1 (Tree Star, Ashland, OR).
Immunoblotting, Immunofluorescence, Immunohistochemistry and Imaging
Immunoblotting was performed as previously described (17). Immunoreactive proteins were visualized with appropriate primary and secondary antibodies using Odyssey Imaging System (LI-COR Biosciences). Band quantification was performed using Odyssey Image Studio software (LI-COR Biosciences). Expression levels were normalized to GAPDH.
Immunofluorescence staining protocol has been described previously (18). Cells were fixed with Prefer fixative (Anatech, Battle Creek, MI), organotypic reconstructs were fixed with formalin. Images were obtained using Nikon TE2000 inverted and E600 upright microscopes and processed with ACT-1 software (Nikon, Melville, NY) and ImagePro-Plus v6.2 (Media Cybernetics, Rockville, MD). To quantify the invasion in collagen-embedded spheroids, the area of the spheroid core (red line) was subtracted from the total area (green line) to obtain the area of invasion.
For immunohistochemistry, formalin fixed paraffin embedded tissue sections were deparaffinized with xylene substitute and rehydrated through 100%, 100%, 95%, 70%, and 50% ethanol. After three washes in PBS, the slides were incubated in antigen retrieval buffer (2.5% trypsin, 10% immunobuffer, and 10 mm CaCl2) for 20 min at 37 °C. After rinsing with dH2O, the sections were blocking with 3% BSA for 30 min at RT. Primary and isotype control antibodies were added on sections and incubated overnight at 4 °C. After PBS washes, anti-goat IgG was added on the sections and incubated 45 min at RT, followed by washing in PBS. ABC reagent (Vector Laboratories, Burlingame, CA) was added and incubated 30 min and the visualization signal was developed with AEC (Vector Laboratories). The slides were counterstained with hematoxylin.
In Vitro Three-dimensional Assays
Multicellular spheroids were obtained as previously described (17). Three-day old spheroids were embedded into bovine type I collagen (Organogenesis, Canton, MA) and images were taken at 0, 24, 48, and 72 h time points. Organotypic reconstructs were obtained using modified protocol from Li et al. (19). Briefly, a single cell suspension of FEF3 fibroblasts (0.45 × 106 cells in 1.5 ml) were embedded into bovine type I collagen/Matrigel (BD) matrix. Gels were allowed to contract for 48 h, after which the HNSCC cells (0.8 × 106/1 ml) were seeded on top of the matrix. Reconstructs were air-lifted after 48h culture and grown in air-liquid interface for an additional 2 weeks. Three-dimensional lattices were fixed with 10% formalin for 2 h and embedded into paraffin.
Comparative Secretome Analysis
For comparative secretome analysis of epithelial and mesenchymal subpopulations, filtered conditioned media from three biological replicates (normalized to cell counts) were combined to achieve sufficient total protein for an in-depth protein profile. The pool from each cell type was concentrated to 20 μl using an Amicon Ultra 10K filter (Millipore) before separation for 3 cm on an SDS gel. Each gel lane was sliced into 30 fractions that were then digested with trypsin. LC-MS/MS analyses were performed using a nanoLC system (Eksigent Technologies, Dublin, CA) coupled to a LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific). Peptides were separated using a 75 μm i.d. × 25 cm PicoFrit (New Objective Inc., Woburn, MA) column packed with 3 μm MAGIC C18-AQ resin, and eluted using a gradient formed by solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile) as follows: 3–28% B over 40 min, 28–50% B over 25.5 min, 50–80% B over 5 min, and constant 80% B over 5 min. A 30-min blank gradient was run between sample injections to minimize carryover. Eluted peptides were analyzed by the mass spectrometer set to repetitively scan m/z from 350 to 2000. The full MS scan was collected at 60,000 resolution in the Orbitrap in profile mode followed by data-dependent MS/MS scans in the linear trap on the six most abundant ions exceeding a minimum threshold of 1000. Monoisotopic precursor selection was enabled and charge-state screening was enabled to reject singly charged ions. Ions subjected to MS/MS were excluded from repeated analysis for 45 s.
MS/MS data were searched using MaxQuant (version 1.3.0.5) against the human UniRef 100 protein database (March 2013, Protein Information Resource, Georgetown University, Washington, DC) combined with a list of common contaminants (trypsin, keratins, etc.), a Bovine database (May 2012), and a reverse database. The combined database has a total of 234,906 entries. MS/MS spectra were searched using trypsin specificity without the Proline restriction with up to three missed cleavages, a 10 ppm precursor mass tolerance, 0.5 amu fragment ion mass tolerance, static modification of Cys (carboxamidomethylation +57.02146 Da), and up to three variable modifications for Met oxidation and protein N-terminal acetylation. Minimum peptide length was set at seven residues. Match between runs and label-free quantification were enabled with a time window of 4 min. Consensus protein lists were generated with False Discovery Rates (FDR) of <1% at both peptide and protein levels using posterior error probability (PEP), a measure of confidence of identification that was computed and used to rank identified peptides as described (20). Proteins were also required to be identified by at least two razor peptides (peptides common between multiple protein groups were re-assigned solely to the group with the highest number of peptides) in at least one sample (epithelial or mesenchymal). Protein fold changes were calculated based on the label-free quantification after normalizing values so that the median protein fold change was 1.0.
Multiple Reaction Monitoring
For MRM, conditioned media from three independent RNAi experiments was pooled for each sample to achieve sufficient total protein and concentrated to 20 μl as described above. BCA (Bio-Rad, Hercules, CA) assay was performed to quantitate the total protein amount after which 20 μg of each sample was loaded onto a preparative gel. Based on the secretome analysis the gel slices containing S100A4 (P26447) and MMP2 (P08253) were determined and the corresponding slices were subjected to label-free targeted MS2 analysis using a Q Exactive mass spectrometer (Thermo Fisher Scientific). Data were acquired with 17,500 resolution, AGC target value of 2E5, maximum ion time of 120 ms, and isolation window of 2 m/z. The following peptides (transitions) were targeted and quantitated: P26447 LMSNLDSNR (525.2558/805.3799), ALDVMVSTFHK (416.5532/619.3198), ELPSFLGK (445.7527/324.6894) and TDEAAFQK (455.2192/693.3566), and for P08253 WCGTTEDYDR (651.7564/956.3955), AFQVWSDVTPLR (709.8749/973.5101), WEHGDGYPFDGK (469.8703/563.2823), EYNSCTDTGR (601.7407/796.3253), YESCTSAGR (515.7165/738.3199), IDAVYEAPQEEK (696.3380/630.3093) and TYIFAGDK (457.7345/650.3508). For methionine-containing peptides, the oxidized form was also targeted. Label-free quantitation based on peak area was performed as previously described (21) using Skyline 1.3 software (MacCoss Lab, University of Washington School of Medicine). The reproducibility, accuracy, and sensitivity of our label-free quantitation approach have been previously demonstrated (22–24). Protein levels were reported as percent expression relative to siN level set at 100%. Standard deviations were determined from the targeted peptide levels for each protein.
Gene Expression Profiling and Real-time Quantitative PCR
HumanHT-12v3 Beadchips (Illumina, San Diego, CA) gene expression analysis of E-cadherin high epithelial and E-cadherin low mesenchymal HNSCC cell subpopulations has been published previously (3). In brief, E-cad hi and E-cad lo subpopulations, from three independent experiments, were sorted and biotinylated cRNA was generated. Bioconductor Lumi package was used for raw data variance-stabilizing transformation and quantile normalization. Bioconductor Limma package was used to identify differential expression with Bayes-adjusted variance analysis from data where probes below background level were excluded (p < 0.01). False discovery rate adjusted p values of <0.05 was used for gene identifications. The raw data is available under Gene Expression Omnibus #18722.
Isolation of RNA and cDNA synthesis were performed as described previously (17). Real-time quantitative PCR was done using Applied Biosystems' 7500 Fast Real-Time PCR System with Power SYBR Green PCR Master Mix (Invitrogen). The following primers, purchased from Integrated DNA Technologies (Coralville, IA), were used: S100A4 forward 5′-AACTAAAGGAGCTGCTGACCC and reverse 5′-TGTTGCTGTCCAAGTTGCTC; ITGA5 forward 5′-CTCAGTGGAGTTTTACCGGC and reverse 5′-AGGTAGACAGCACCACCCTG; FAP forward 5′-GCTGTGCTTGCCTTATTGGT and reverse 5′-GTGTGAGTGCTCTCATTGTAT; MMP2 forward 5′- ATGCCGCCTTTAACTGGAG and reverse 5′- GGAAGCCAGGATCCATTTT and GAPDH forward 5′-ATGGGGAAGGTGAAGGTCG and reverse 5′-TAA AAG CAG CCC TGG TGA CC. Relative expression of target gene mRNA referenced to GAPDH was calculated with the REST-MSC software using the Pair-Wise Fixed Reallocation Randomization Test (25, 26).
Luciferase Assay, RNA Interference and Zymography
S100A4 luciferase reporter pGl4.10 S100A4 (-632/+1010) was kindly provided by Dr. Kathleen O'Connor (University of Kentucky). Cells were grown in six-well plates and cotransfected with 1 μg of the S100A4 reporter along with 0.1 μg of pRL-TK Renilla control reporter (Promega) for 24 h. Cells were then induced by control or 24 h FEF3 conditioned medium for 48 h. Cells were collected, and luciferase activity was measured by the Dual-Luciferase Reporter Assay System (Promega) according to manufacturers' instructions. The data are presented as a relative value to internal control.
For RNAi, OCTT2 cells were plated onto 6-well plates (2 × 105/well) and small interfering RNA against S100A4 and nontargeting negative control oligos (OriGene Technologies, Rockville, MD) were transiently transfected into the cells using Lipofectamine 2000 (Invitrogen) at a final concentration of 12.5 nm. After siRNA treatment for 72 h, cells were subjected to different experiments.
Zymography was performed to detect MMP activity using commercially available Novex Zymogram gels (Invitrogen) as per manufacturer's instructions. In brief, 72 h conditioned medium of siRNA transfected cells were collected and protein concentration determined using Bio-Rad Protein Assay kit. Conditioned media were adjusted for equivalent concentrations (∼10 μg/ml) and mixed with Novex Tris-glycine SDS native sample buffer (Invitrogen) and loaded into precast 10% Novex zymogram gelatin gels (Invitrogen). The gel electrophoresis was set to a constant voltage of 125 V for ∼100 mins, followed by a quick rinse in distilled water to remove SDS in the buffer. Then gels were incubated in a Novex zymogram renaturing buffer (Invitrogen) for 1 h at 37 °C to reactivate MMPs followed by an incubation on a rotary shaker in Novex zymogram developing buffer (Invitrogen) for 4 h at 37 °C to allow denatured MMPs to digest the gelatin substrate. To visualize proteolytic digestion, the gels were rinsed and stained by incubation with Safe Stain (Invitrogen) for 2 h and then destained with a solution of acetic acid, methanol, and water (10:50:40). Proteolytic activity was visualized as areas of clear bands against a dark blue background. The identity of the proteases was determined by molecular weight of the band sizes.
Animals and In Vivo Experiments
Nonobese diabetic/severe combined immunodeficient/interleukin-2 receptor γ-chain-deficient mice (NSG) were used in all experiments. The animals, originally obtained from the Jackson Laboratory (Bar Harbor, ME), were bred and used at the Wistar Institute animal facility under protocols approved by the Institutional Animal Care and Use Committee. Xenografts were generated by subcutaneous injection of 1 × 105 cells in 100 μl reduced growth factor Matrigel near the right back limb (n = 6 per group). Tumor volumes were measured as [length x width2]. Orthotopic xenografts (n = 4 per group, total of 12 mice for siS100A4) were generated by subcutaneous injection of 2 × 105 cells in 50 μl growth factor reduced Matrigel into the floor of the tongue. This model metastasizes spontaneously into the lungs of the NSG mice within 3 weeks. Animals were sacrificed after 3 weeks and primary tumors as well as lungs were collected. Prior to fixation with 10% formalin, weight of the tumors were measured. Samples were then embedded into paraffin. The pathologist assessing the H&E slides was blinded to the groups, to prevent biased result interpretation.
Statistical Analyses
All in vitro experiments were done in duplicates and repeated three times. Two-tailed unpaired t tests were used in all statistical tests using GraphPad Prism software (version 5; GraphPad Software Inc, La Jolla, CA). The mean and S.D. of experiments are shown and p values of less than 0.05 were considered to be statistically significant. Network analysis was carried out using the Ingenuity Pathways Analysis software (IPA, Ingenuity Systems, Redwood City, CA). Data sets containing gene identifiers and corresponding intensity values were uploaded into the application and each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. A cutoff of 4.0 was set to identify genes whose expression was significantly differentially regulated and network limit was set to 35 molecules. Networks generated by the IPA network generation algorithm were ranked by score; the score reflects the statistical significance of association between the genes and the networks by the Fisher's exact test. A score of 3 indicates a 1/1000 chance that the focus genes are in a network because of random chance. DAVID functional annotation tool (27) was used to analyze Gene Ontology (GO) terms for biological processes, molecular functions and cellular components. We chose to use a Poisson distribution to calculate the statistical probability of the individual metastasis burden based on the observed numbers, as described by Kendal (28).
RESULTS
Functional Characterization of Epithelial and Mesenchymal HNSCC Subpopulations
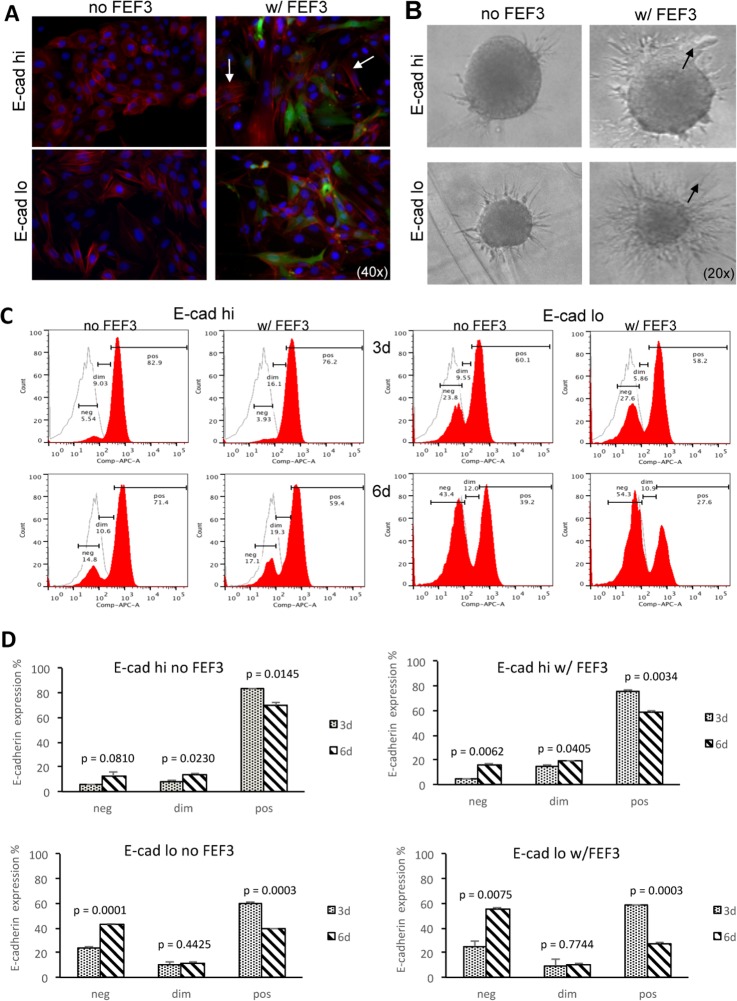
Considering the concept that carcinomas initially depend on local fibroblasts for activation of EMT, and later lose this dependence (29), we investigated the role of fibroblasts in affecting the epithelial and mesenchymal subpopulations of the heterogeneous OCTT2 cell line. To define these changes, GFP-tagged fibroblasts were cocultured with the OCTT2 cells for 24 h and analyzed for re-arrangement of cytoskeleton, observed by phalloidin staining. Typical epithelial cell cobblestone morphology was seen in the FACS-sorted E-cadherin high cells without the FEF3 fibroblasts, whereas in the cocultures, stress fiber formation, a sign of reorganization of the actin cytoskeleton, was observed in the OCTT2 cells (Fig. 1A, top panels). MSP cells exhibited elongated bi-polar spindle-like shape and the coculture had no effect on the morphology of these cells (Fig. 1A, lower panels). Pre-formed E-cadherin high and E-cadherin low multicellular spheroids were embedded into collagen in the presence of control or FEF3 conditioned media to study invasion (Fig 1B). Increased invasion was observed in both subpopulations when cultured with the fibroblast-CM. Mode of invasion was collective in the epithelial subset, whereas the MSP cells invaded in a single-cell fashion, paralleling the known invasion modes of these cell types (30). Number of collective events increased greatly with fibroblast-CM in the epithelial subpopulation, whereas no collective invasion was observed in the MSP with or without fibroblast-CM (supplemental Fig. S1).
Fig. 1.
Fibroblast-derived factors promote EMT and invasion. A, Stress fiber formation was induced in FACS-segregated E-cad hi OCTT2 cells c0cultured with FEF3 fibroblasts for 24 h. Representative image of three independent experiments (blue Hoechst, red phalloidin, green GFP).) B, Fibroblast-CM induced phenotypic migration modes of collective invasion in epithelial subpopulation and single-cell invasion in mesenchymal subset of E-cadherin expression-based segregated cells (arrows). Representative 48 h images of three independent experiments. C, Expression of E-cadherin was down-modulated both in E-cad high and low subpopulations cocultured with FEF3 fibroblasts for 3 and 6 days, as shown by representative flow cytometry histograms. D, Statistical comparison of 3 and 6 day cultures, with and without fibroblasts (n = 3).
To quantify the EMT response, the respective OCTT2 subpopulations were cultured with or without FEF3 cells for 3 and 6 days and the levels of E-cadherin were measured by flow cytometry. As shown in Fig. 1C, the percentage of strongly E-cadherin positive cells (pos) decreased from 76.2% to 59.4% and the percentage of E-cadherin negative fraction increased from 3.93% to 17.1% in the epithelial subset cocultured with the fibroblasts (E-cad hi, right panels). In the MSP/FEF3 cocultures, the percentage of strongly E-cadherin positive cells (pos) decreased from 58.2% to 27.6% with concomitant increase in the E-cadherin negative fraction (neg) from 27.6% to 54.3% (E-cad lo, right panels). Statistical comparison of day 3 and day 6 with and without the fibroblasts (Fig. 1D) revealed a significant decrease in the percentage of the E-cadherin positive fraction even without the fibroblasts, indicating an ongoing EMT process in both E-cad hi and E-cad lo subpopulations. However, the fibroblasts augmented the down-modulation of E-cadherin in both subpopulations even further.
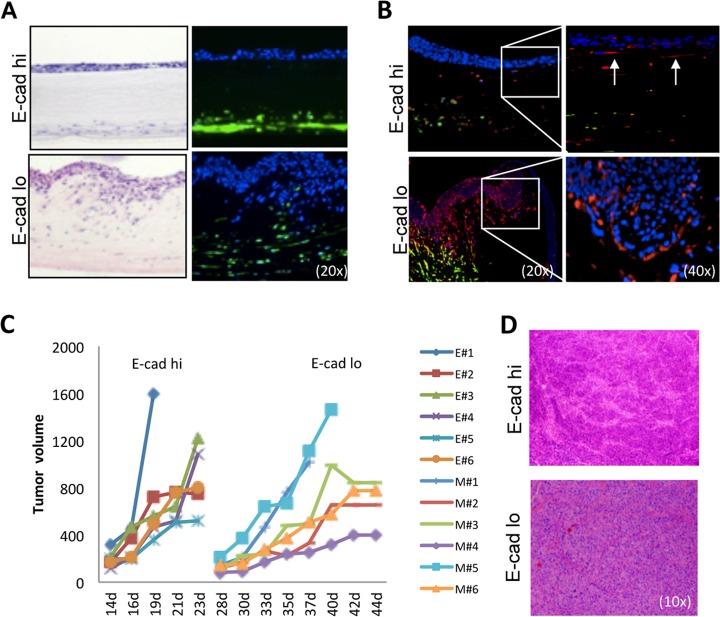
Next we used organotypic reconstructs containing FEF3 cells in collagen/Matrigel matrix to mimic the three-dimensional tumor microenvironment. The epithelial subset was not as invasive as the MSP (Fig. 2A); however immunofluorescence staining revealed that cells which had lost polarization were positive for vimentin (Fig. 2B, top panels, arrows). The MSP exhibited bi-directional migration with the FEF3 cells and vimentin was expressed at the invasive fronts (Fig. 2B, lower panels). Based on these results, we asked whether the sorted HNSCC subsets exhibited differences in tumor growth in vivo. Subcutaneous injection of 1 × 105 E-cadherin-sorted cells showed that the epithelial subset started to form tumors within 2 weeks of injection, whereas the MSP took almost a month to grow (Fig. 2C). However, as shown in the representative H&E images (Fig. 2D), the resulting tumors were nearly identical, correlating with our previous in vitro observation of dynamic reversibility of the epithelial and mesenchymal phenotypes at clonal level (3).
Fig. 2.
Role of the three-dimensional tumor microenvironment on epithelial and mesenchymal subpopulation growth and invasion properties. A, In organotypic reconstructs, H&E staining (left panels) showed a clear separation of the epithelial cells from the fibroblasts, whereas the MSP was highly invasive. Direct GFP-fluorescence of FEF3 cells (right panels) revealed bi-directional migration of MSP cells and fibroblasts (blue Hoechst, green GFP). Representative image of three independent experiments. B, Immunofluorescence staining of organotypic reconstructs indicated that epithelial cells, which have lost polarization, stain positive for vimentin (top panels, 40× magnification of the 20× image shown in right side). In the MSP (bottom panels) vimentin-positive red cells were observed mainly in the invasive front. The FEF3 fibroblasts are seen as yellow, copositive for GFP and vimentin. Representative image of three independent experiments. C, Growth curves of the subcutaneous xenografts of 1 × 105 cells sorted based on E-cadherin expression demonstrated a clear difference in the tumor growth kinetics. At day 14, the epithelial tumors started to form, whereas tumors containing MSP cells start forming at day 28 (n = 6/group, E = epithelial subset, M = mesenchymal subset). D, H&E images of respective tumors.
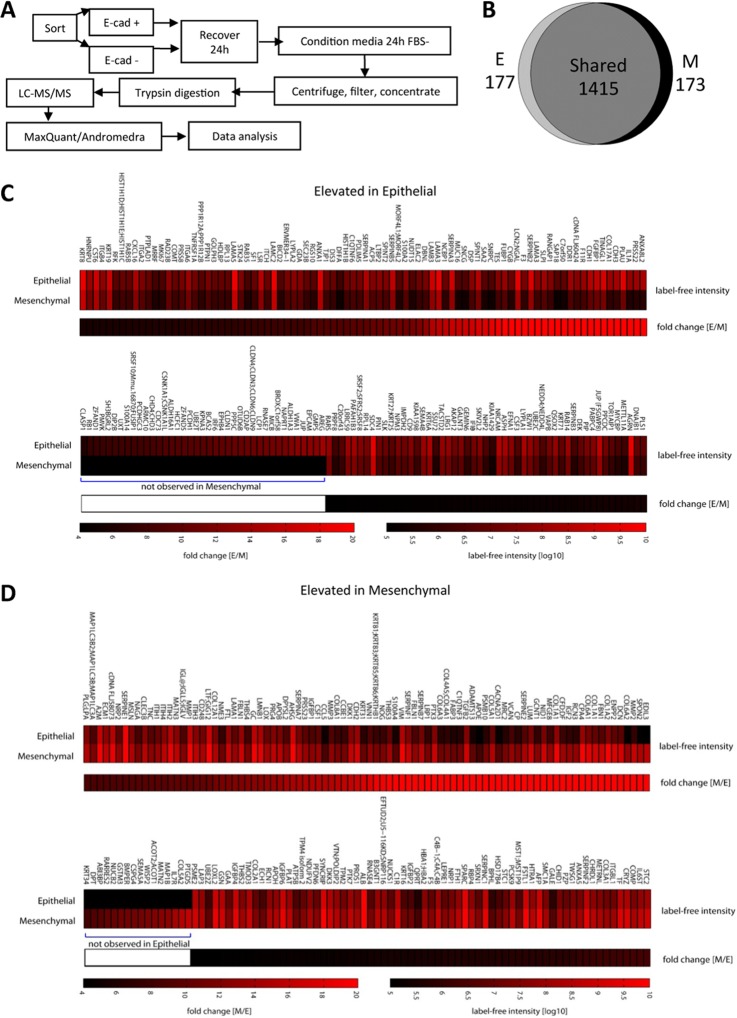
Comparison of the Epithelial and Mesenchymal Secretomes of HNSCC Cells
Since CAFs are known to secrete a variety of factors promoting tumorigenesis (9), we conducted a comparative LC-MS/MS proteomic analyses of the epithelial and MSP secretomes using the workflow shown in Fig 3A. For each subpopulation, conditioned media from three batches of cells were pooled because of the low amount of total protein shed by the cells, the need to restrict incubation in serum-free media to 24 h, and the limited number of cells that could be sorted while maintaining good cell viability. Because it was not practical to perform replicate proteome analyses, a stringent fourfold cutoff and exclusion of single-peptide hits were used. Based on using a similar workflow on other cell lines where sample amounts were not limiting, biological replicates typically exhibited 3.0- to 3.5-fold differences at three standard deviations. On this basis, changes greater than fourfold were considered significant. A total of 1765 proteins were identified and 177 showed elevated levels in the epithelial subpopulation secretome and 173 were elevated in the MSP secretome, as summarized in an area-proportional Venn diagram (Fig. 3B). The complete list of identified proteins with quantities and fold changes are shown in supplemental Table S1. Heat maps of the significantly elevated proteins in the epithelial cells are shown in Fig. 3C and proteins elevated in the MSP cells are shown in Fig. 3D. Proteins known to be important in CAF biology that are elevated in the MSP secretome are shown in Table I.
Fig. 3.
Comparative secretome analysis of the epithelial and mesenchymal subpopulations. A, Schematic workflow of the LC-MS/MS secretome analysis of the cell subpopulations. B, Area-proportional Venn diagram depicting total protein distribution; 1415 proteins were not significantly different between subpopulations, 177 were elevated in the epithelial subpopulation and 173 were elevated in the MSP using a fourfold cutoff for significant differences. C, Heat maps of the significantly elevated proteins in the epithelial subset. D, Heat maps of the proteins significantly elevated in the mesenchymal cells.
Table I. CAF-like factors secreted by the subsets. Fold change calculated from intensities.
| ID | Symbol | Protein name | Peptide count | Sequence coverage (%) | Mass (kDa) | Intensity epithelial | Intensity mesenchymal | Fold change [M/E] |
|---|---|---|---|---|---|---|---|---|
| P26447 | S100A4 | S100 calcium binding protein A4 | 2 | 19.8 | 11.728 | 2.7E+05 | 5.6E+06 | 20.6 |
| O95965 | ITGBL1 | Integrin, beta-like 1 | 3 | 8.3 | 53.921 | 1.6E+07 | 1.3E+08 | 8.1 |
| B2R5J8 | CCL5 | Chemokine (C-C motif) ligand 5 | 2 | 22 | 10.018 | 2.0E+05 | 3.1E+06 | 15.3 |
| P09603 | CSF1 | Colony-stimulating factor 1 (macrophage) | 5 | 13.7 | 60.179 | 6.6E+06 | 1.0E+08 | 15.2 |
| P08253 | MMP2 | Matrix metallopeptidase 2 | 14 | 31.7 | 73.881 | 1.1E+06 | 8.0E+08 | 727.0 |
| P08254 | MMP3 | Matrix metallopeptidase 3 | 2 | 4.4 | 53.977 | 4.3E+06 | 6.7E+07 | 15.5 |
| P08833 | IGFBP1 | Insulin-like growth factor binding protein 1 | 5 | 20.8 | 27.903 | 4.2E+06 | 6.3E+07 | 15.1 |
| G3QMZ4 | IGF2 | Insulin-like growth factor 2 | 4 | 22.9 | 26.343 | 1.1E+06 | 9.9E+07 | 87.7 |
| B2R7T2 | TGFB2 | Transforming growth factor, beta 2 | 13 | 37 | 47.777 | 4.4E+07 | 1.4E+09 | 30.9 |
| B2R6I6 | STC1 | Stanniocalcin 1 | 6 | 26.7 | 27.591 | 9.3E+06 | 5.9E+07 | 6.3 |
| O76061 | STC2 | Stanniocalcin 2 | 5 | 22.2 | 33.248 | 1.3E+08 | 1.1E+09 | 8.5 |
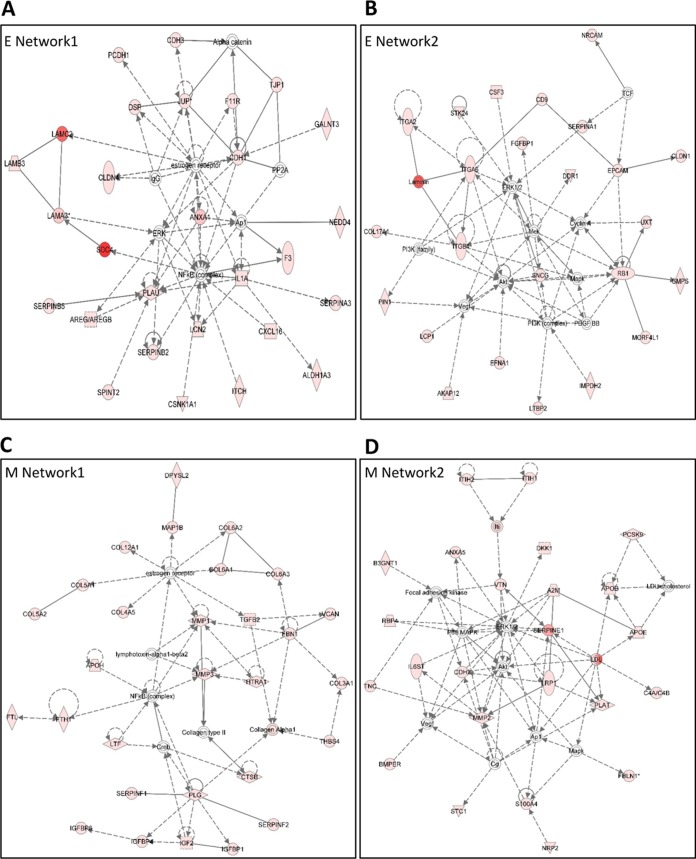
IPA network analyses were performed for the unique proteins found in the epithelial and mesenchymal secretomes; the two top-ranking networks for both secretomes are shown in Fig. 4. These networks were very highly significant, as the scores were greater than 20 in all of them. In epithelial Network 1, the score was 47 and number of focus molecules 28, and in Network 2, 37 and 24, respectively. In the mesenchymal Network 1, the score was 49 and number of focus molecules 29, and in Network 2, 37, and 24, respectively. In both secretomes, key signaling nodes of NFκB, ERK and Akt were indicated, reflecting the identical origin of these subpopulations. However, majority of the focus molecules were different between the epithelial and mesenchymal networks, indicating a diversion acquired via EMT.
Fig. 4.
IPA-generated network analyses of proteins found in epithelial and mesenchymal secretomes. A, epithelial network 1, B, epithelial network 2, C, mesenchymal network 1, and D, mesenchymal network 2. The analysis settings were 35 focus molecules per network, score above 20.
Increased deposit of collagens is a hallmark of tumor stroma (31) and supporting this, increased secretion of multiple collagens was observed for the MSP (Table II) and was also reflected in the mesenchymal Network 1 (Fig. 4C). Furthermore, most of the significant enriched cellular component GO terms involved collagen and the extracellular matrix as defined by the DAVID functional annotation tool (Table III).
Table II. Collagen levels in the secretomes.
| ID | Symbol | Protein name | Peptide count | Sequence coverage (%) | M.W. [kDa] | PEPa | Intensity E | Intensity M | Fold change [M/E] |
|---|---|---|---|---|---|---|---|---|---|
| P02452 | COL1A1 | Collagen alpha-1(I) chain | 32 | 32.8 | 138.94 | 0 | 7.4E+07 | 6.3E+09 | 86.0 |
| P08123 | COL1A2 | Collagen alpha-2(I) chain | 48 | 58.5 | 129.31 | 0 | 3.9E+07 | 8.0E+09 | 202.9 |
| G3QNV9 | COL2A1 | Collagen alpha-1(II) chain; Collagen alpha-1(II) chain; Chondrocalcin | 3 | 2.5 | 142.26 | 5.87E-22 | 8.5E+06 | 3.7E+07 | 4.4 |
| P02461 | COL3A1 | Collagen alpha-1(III) chain | 4 | 3.3 | 138.56 | 1.78E-60 | 1.5E+07 | 1.2E+08 | 8.0 |
| P29400–2 | COL4A5; COL4A1 | Collagen alpha-5(IV) chain; Collagen alpha-1(IV) chain; Arresten | 4 | 4.2 | 161.63 | 5.34E-08 | 1.0E+06 | 2.9E+07 | 28.4 |
| B2ZZ86 | COL5A1 | Collagen alpha-1(V) chain | 12 | 9 | 183.59 | 1.16E-75 | 9.0E+06 | 3.4E+08 | 38.2 |
| P05997 | COL5A2 | Collagen alpha-2(V) chain | 15 | 15.3 | 144.91 | 1.97E-105 | 0 | 3.6E+08 | N/A |
| P12109 | COL6A1 | Collagen alpha-1(VI) chain | 31 | 44.3 | 108.53 | 0 | 1.9E+07 | 2.8E+09 | 147.2 |
| P12110–2 | COL6A2 | Collagen alpha-2(VI) chain | 11 | 15.1 | 97.418 | 2.31E-73 | 1.3E+05 | 8.6E+07 | 655.0 |
| P12111 | COL6A3 | Collagen alpha-3(VI) chain | 5 | 1.8 | 343.67 | 4.32E-11 | 6.6E+05 | 1.8E+07 | 27.2 |
| A1KY36 | COL8A1 | Collagen alpha-1(VIII) chain; Vastatin | 2 | 3.4 | 73.452 | 8.32E-07 | 1.6E+06 | 2.5E+07 | 15.8 |
| Q99715 | COL12A1 | Collagen alpha-1(XII) chain | 52 | 23 | 333.14 | 4.28E-191 | 2.7E+07 | 3.0E+08 | 11.3 |
| D3DRA2 | COL17A1 | Collagen alpha-1(XVII) chain; 120 kDa linear IgA disease antigen; 97 kDa linear IgA disease antigen | 10 | 10 | 150.45 | 4.98E-95 | 1.1E+08 | 8.2E+05 | −132.5 |
a Posterior error probability (see Methods).
Table III. MSP GO terms analyzed by DAVID.
| Count | % | P-Value | |
|---|---|---|---|
| Cellular components term | |||
| Extracellular region | 77 | 62.60163 | 1.51E-38 |
| Extracellular region part | 54 | 43.90244 | 8.25E-33 |
| Extracellular space | 39 | 31.70732 | 5.61E-23 |
| Proteinaceous extracellular matrix | 26 | 21.13821 | 1.31E-18 |
| Extracellular matrix | 26 | 21.13821 | 7.88E-18 |
| Extracellular matrix part | 14 | 11.38211 | 4.71E-12 |
| Collagen | 9 | 7.317073 | 1.76E-10 |
| Platelet alpha granule lumen | 7 | 5.691057 | 6.18E-07 |
| Cytoplasmic membrane-bounded vesicle lumen | 7 | 5.691057 | 9.53E-07 |
| Vesicle lumen | 7 | 5.691057 | 1.25E-06 |
| Biological processes term | |||
| Response to external stimulus | 32 | 26.01626 | 1.73E-12 |
| Response to wounding | 25 | 20.3252 | 2.36E-12 |
| System development | 47 | 38.21138 | 3.63E-10 |
| Extracellular structure organization | 14 | 11.38211 | 4.53E-10 |
| Collagen fibril organization | 8 | 6.504065 | 2.08E-09 |
| Wound healing | 14 | 11.38211 | 3.19E-09 |
| Cell adhesion | 24 | 19.5122 | 4.07E-09 |
| Biological adhesion | 24 | 19.5122 | 4.18E-09 |
| Anatomical structure development | 47 | 38.21138 | 5.45E-09 |
| Organ development | 38 | 30.89431 | 5.86E-09 |
| Molecular function term | |||
| Calcium ion binding | 32 | 26.01626 | 1.93E-12 |
| Growth factor binding | 13 | 10.56911 | 3.01E-11 |
| Endopeptidase inhibitor activity | 13 | 10.56911 | 1.34E-09 |
| Serine-type endopeptidase inhibitor activity | 11 | 8.943089 | 2.37E-09 |
| Peptidase inhibitor activity | 13 | 10.56911 | 2.49E-09 |
| Insulin-like growth factor binding | 7 | 5.691057 | 3.09E-08 |
| Pattern binding | 12 | 9.756098 | 3.16E-08 |
| Polysaccharide binding | 12 | 9.756098 | 3.16E-08 |
| Carbohydrate binding | 16 | 13.00813 | 1.03E-07 |
| Enzyme inhibitor activity | 14 | 11.38211 | 1.81E-07 |
Transcriptomic Verification of Known CAF Markers
Previously conducted gene expression analysis of the epithelial and mesenchymal subsets of patient-derived tumor cell lines had indicated that the E-cadherin low MSP had gone through a cadherin switch from E- to N-cadherin and gained de novo expression of fibronectin and vimentin (3), indicative that these cells have gone through complete EMT (32). Using the secretome results, we validated the expression of CAF markers in the MSP. Gene expression analysis showed significant up-regulation of S100A4, ITGA5, FAP, and PDGFRβ in the MSP compared with the epithelial subpopulation (Fig. 5A and supplemental Fig. S2). Notably, α-SMA, the most commonly used CAF marker, was not elevated in the MSP secretome, supporting data that using α-SMA as a sole marker will identify myofibroblasts, but not all CAFs (10). The expression of the top three genes, S100A4, ITGA5, and FAP, was further verified by quantitative real-time PCR in the OCTT2 cell line (Fig. 5B): S100A4 expression was 8-fold higher in the MSP compared with the epithelial subpopulation at the gene expression level, compared with more than 20-fold higher at the protein level in the secretome. To elucidate the role of fibroblast conditioned media in up-regulating S100A4, a luciferase reporter assay was used. Significant increase in the signal was seen when unsorted OCTT2 cells were cultured in fibroblast-CM, indicating activation of S100A4 in these cells by fibroblast-derived factors (Fig. 5C).
Fig. 5.
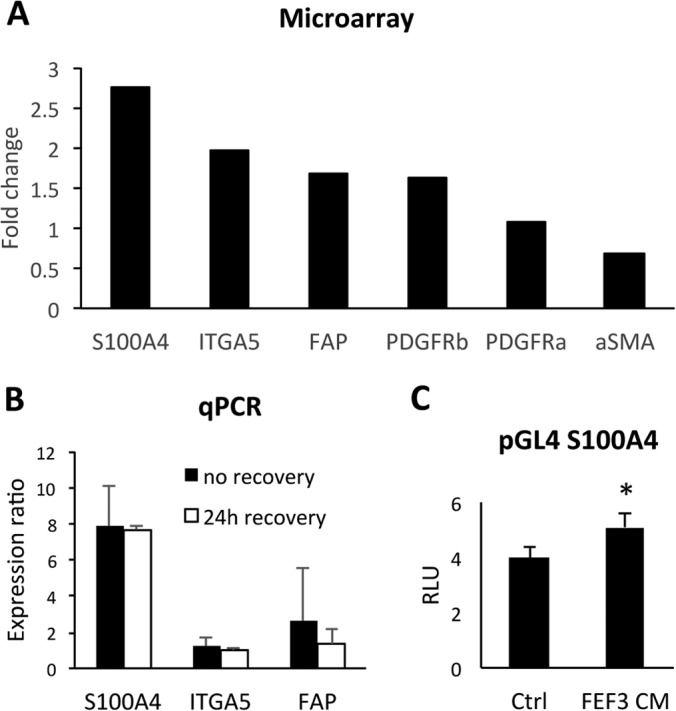
Verification of selected CAF markers on transcriptome level. A, Expression of genes up-regulated in E-cad lo MSP displayed as fold-change relative to E-cad high epithelial cells using Illumina HumanHT-12v3 Beadchips microarray in OCTT2 cells. S100A4 (2.77-fold, p = 1.32 × 109), ITGA5 (1.98-fold, p = 4.99 × 1010), FAP (1.69-fold, p = 4.22 × 107), and PDGFRβ (1.63-fold, p = 8.46 × 108). B, Validation of S100A4, ITGA5 and FAP mRNA expression by real-time qPCR. Epithelial and mesenchymal subsets from OCTT2 cell line were FASC-segregated and RNA was isolated either directly after cell sort (solid bars) or after 24 h recovery period (open bars). GAPDH was used for normalization, and relative gene expression in MSP was compared with the epithelial subpopulation using REST-MSC software. (S100A4 = 7.89- and 7.77-fold; ITGA5 = 1.2- and 0.99-fold; FAP = 2.59- and 1.35-fold). C, OCTT2 cells transfected with S100A4 promoter construct for 24h, after which culture media was changed to either control or FEF3-CM. Luciferase activity was measured after 48h and normalized to Renilla expression. n = 3, *p < 0.05.
Evaluation of S100A4 for HNSCC Targeted Therapy
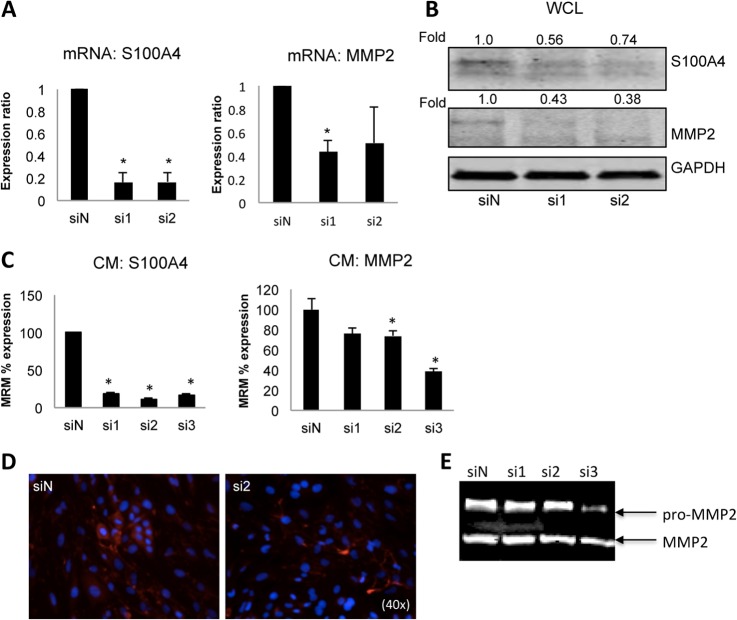
Based on the aforementioned results, we choose S100A4 as a target for siRNA-based therapy. Two independent siS100A4 oligos repressed S100A4 expression significantly at the mRNA level (Fig. 6A, left graph). Quantification of S100A4 immunoblot indicated a 44% reduction in the total S100A4 protein level (Fig. 6B). As S100A4 is known to affect matrix metalloproteinases, and MMPs were found in the MSP secretome, we analyzed the effect of S100A4 RNAi to the expression of MMP2 and MMP3. Paralleling the S100A4 levels, MMP2 expression at both mRNA and protein level was decreased in the siS100A4 samples compared with the negative nontargeting control siRNA (Fig. 6A, right graph and Fig. 6B). MMP3 was not affected (data not shown).
Fig. 6.
S100A4 knockdown by siRNA leads to MMP2 down-regulation. Unsorted OCTT2 cells were transfected either with nontargeting siRNA (siN) or siS100A4 oligos si1 and si2, respectively. Knockdown of S100A4 was quantitated by qPCR (A, left graph), Western blot of whole cell lysates (WCL) (B), MRM analysis (C) and immunofluorescence (D) 72 h after transfection. Down-regulation of S100A4 led to concomitant decrease in MMP2 expression, as measured by qPCR (A, right graph) Western blot (B), and MRM (C). For the MRM analysis a third siS100A4 oligo was included (si3). MRM confirmed that the amount of soluble S100A4 was significantly decreased in the siS100A4 samples compared with the siN oligo, and MMP2 was significantly decreased with si2 and si3. Representative zymographic analysis shows decreased proteolytic activity in CM collected from si3-transfected cells. n = 3, *p < 0.05.
Next we wanted to know whether the S100A4 RNAi affected the secretion of these two proteins. For these experiments a third independent siS100A4 oligo was added (si3), whose effectiveness was validated before the multiple reaction monitoring (MRM) experiment (data not shown). Fig. 6C shows MRM data for the CM of three pooled RNAi experiments. The amount of soluble S100A4 was decreased significantly in the siS100A4 samples compared with the siN oligo (Fig. 6C, right panel), consistent with the qPCR results. Confirmatory immunofluorescence showed that in particular the diffuse staining, indicative of soluble S100A4, was lost in the RNAi samples (Fig. 6D, right panel). In addition, MRM detecting MMP2 showed decreased levels by si2 and si3 S100A4 RNAi (Fig. 6C, right panel). To confirm the effect of S100A4 knockdown on MMP2 activity, a zymography assay was performed. On the zymogram gelatin gel (Fig 6E), suppressed proteolytic activity was seen in sample that was obtained from si3 S100A4 siRNA-transfected cells. Supporting these findings, in the mesenchymal Network 2 an indirect interaction between S100A4 and MMP2 was indicated (Fig. 4D). Collectively these results suggest a mechanistic role for S100A4 regulating MMP2 protein level and activity.
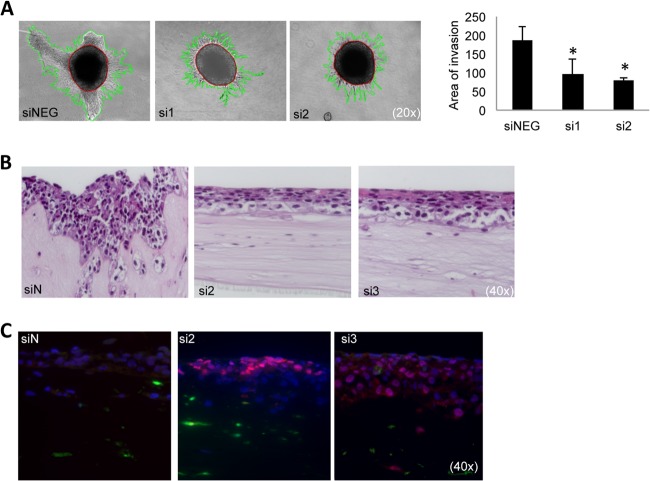
Depletion of S100A4 Reduces Invasion and Induces Advanced Differentiation In Vitro
Next we assessed the biological function of S100A4 knockdown. Invasion of collagen-embedded spheroids composed of unsorted OCTT2 cells was significantly reduced by the S100A4 targeting oligos (Fig. 7A). In organotypic reconstructs (Fig. 7B) a clear reduction in invasion was observed in the RNAi samples. Interestingly, the cells also looked more differentiated in the H&E staining. Therefore, we did immunofluorescence staining of the differentiation marker involucrin (Fig. 7C), which showed an increased expression of this protein in the siS100A4 organotypic reconstructs.
Fig. 7.
Functional validation of S100A4 knockdown in vitro. A, OCTT2 cells were transfected with siS100A4 for 48 h, after which spheroids were formed for 72 h and embedded into type I collagen. Invasion was significantly repressed at 72 h time point compared with control, as quantified in the graph (right panel). Representative 20× images of three independent experiments. ImagePro was used to quantitate the area of invasion (mm2). B, OCTT2 cells transfected with siS100A4 (48 h) were over-laid on top of FEF3-GFP 3-D organotypic matrix. Reconstructs were grown for 2 weeks. 40× images of representative H&E slides. C, Reconstructs stained with involucrin showed increased differentiation in the siS100A4 samples. Representative image of three independent experiments. n = 3, *p < 0.05.
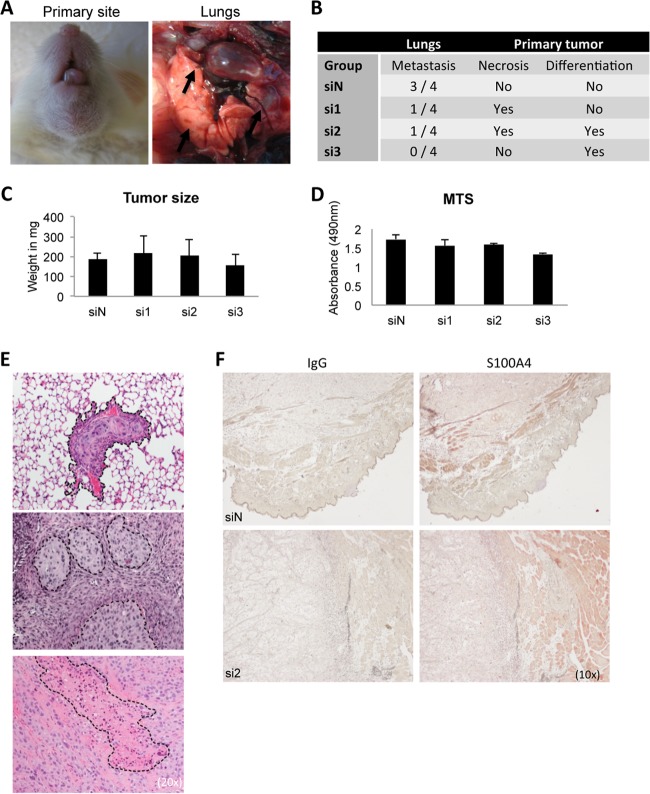
In Vivo, Knockdown of S100A4 Reduces Metastasis Burden and Causes Differentiation of Primary Tumor
Since S100A4 is a well-known mediator of metastasis, we tested the effect of S100A4 depletion to metastasis burden. We developed a model for orthotopic tumor growth that spontaneously metastasizes into the lungs of NSG mice within 3 weeks of tumor initiation (Fig. 8A). RNAi-treated OCTT2 cells (2 × 105 cells, n = 4 per group, total 12 mice for siS100A4) were injected subcutaneously into the base of tongue and the mice were sacrificed after 3 weeks. Results of a blinded analysis of H&E slides are shown in Fig. 8B. Decreased metastasis burden in the lungs of the siS100A4 mice with all three independent oligos was observed. To calculate the probability of seeing only one metastasis in the siN group, Poisson distribution was used which yielded a statistical possibility of 14.9%. Corroborating the in vitro reconstruct findings, increased levels of necrosis and differentiation were observed in the S100A4-RNAi containing xenografts. Primary tumor volumes were measured, and correlating with the MTS assay, no significant changes in the tumor weight were observed between groups (Figs. 8C and 8D). The representative H&E images are shown in Fig. 8E. Immunohistochemistry of S100A4 in the primary tumors (Fig. 8F) showed a slightly decreased staining in the si2-treated cancer cells compared with siN-treated cancer cells, indicating a sustained suppression of S100A4 expression by transient RNAi at 3 weeks after transfection. As expected, the mouse cells, in particular adipocytes, stain strongly positive for S100A4 in both xenografts.
Fig. 8.
In vivo characterization of S100A4 knockdown using an orthotopic spontaneous mouse model. OCTT2 cells were transfected with siS100A4 or siN for 48 h, after which 2 × 105 cells were injected into the base of the tongue of NSG mice. Tumors were grown for 3 weeks, mice were sacrificed, and primary tumors and lungs were collected. A, Representative images of the primary tumor and metastatic lungs. B, Results of the blinded histopathological analysis of H&E slides. C, Weight of the primary tumors, n = 4 per group. No significant differences in the growth of the primary tumor were observed between the groups. D, In vitro cell proliferation assay of OCTT2 cells transfected with siS100A4 showed no significant differences in proliferation rate between siRNA oligos, correlating with in vivo tumor volumes. n = 3. E, Representative images of lung metastasis, necrosis and advanced squamous differentitation. 20× images of the H&E slides. F, Representative images of S100A4 immunohistochemistry of the primary tumors, 10×.
DISCUSSION
The critical importance of the tumor microenvironment in driving carcinoma progression has emerged in recent years. Various cell types contribute to tumorigenesis and confer resistance to both chemo- and targeted therapy (7). In our model, conditioned medium from fibroblasts induced rapid remodeling of the actin cytoskeleton and down-modulation of E-cadherin, indicating that secreted factors in the fibroblast-CM promote EMT in heterogeneous HNSCC cells. We observed phenotypical modes of invasion when the epithelial and mesenchymal subpopulations were segregated and cultured in three-dimensional models containing fibroblasts or fibroblast-derived factors. Interestingly, when a low number of cells were injected subcutaneously, a 2-week latency period was observed in the MSP tumors when compared with the tumors of the epithelial subset. Yet, the resulting xenografts were indistinguishable from each other when assessed by H&E staining. In both cases the epithelial and mesenchymal subsets were visible, as in the original patient tumor (3). This suggests that the MSP tumors went through mesenchymal-epithelial transition (MET) to grow the primary tumor mass (33). A similar phenomenon is observed in melanoma, where the cells undergo transcriptional switching between proliferative and metastatic states, regulated by the tumor microenvironment (34, 35).
Comparative secretome approach was taken to dissect the differences on protein level of the epithelial and mesenchymal subpopulations. As the subpopulations were obtained from the same tumor, majority of the identified proteins and key signaling nodes overlapped between the secretomes. However, significant differences were also observed and interestingly we identified a CAF-like signature in the MSP. Cancer-associated fibroblastic cells are major contributors in carcinoma progression, but their origin and function has not been fully elucidated (36). Here we provide new evidence that in heterogeneous HNSCC, the mesenchymal cells arising through EMT have taken the role of CAFs. The MSP expressed several classical CAF markers on an mRNA level and secreted a variety of factors that have been characterized as stromal-derived, such as growth factors, chemokines and matrix metalloproteinases.
In addition to the paracrine-mediated modulation, CAFs mechanically modulate the tumor microenvironment. Collagen remodeling is a key to this force-mediated invasion (37) and to support this we found increased expression of several collagens in the secretome of the CAF-like MSP cells. Fibrillar collagens type I, II, III, and V were increased in the MSP. Type I collagen is a heterotrimer composed of two alpha-1(I) and one alpha-2(I) chains (38); however, in the MSP secretome the fold change was greater in the alpha-2(I) than in alpha-1(I) levels (203 versus 86-fold increase). Collagen type V is composed of alpha-1(V), alpha-2(V) and alpha-3(V) (38), but we detected only alpha-1(V) and alpha-2(V) in the MSP secretome. Microfibrillar collagen type VI, FACIT collagen type XII, short chain collagen type VIII and basement membrane collagen type IV were also increased in the MSP. All three chains of collagen type VI were found to be increased in the MSP secretome, suggesting a functional state for this particular collagen, in addition to type II and III, which are homotrimers. Interestingly, transmembrane type collagen XVII was decreased 132-fold in the MSP. Due to its transmembrane nature, this collagen functions as a cell-matrix adhesion molecule (39) and therefore its down-regulation in MSP cells is well-fitted with their more motile nature.
EMT has been linked to an important aspect of treatment resistance, cancer stem cells (CSCs), which are thought to originate from transdifferentiated epithelial cells that have gone through EMT (40). S100A4 is a well-characterized mediator of EMT and; here we show that its expression and secretion are maintained in the MSP. S100A4 also has been linked to CSCs (41), and its expression in HNSCC correlates with self-renewal and stemness properties (42). A high-throughput screening approach identified the anthelminthic drug, niclosamide, as an inhibitor of S100A4-mediated metastasis in colon cancer (43). Since niclosamide is not a specific S100A4 inhibitor, we chose RNAi specifically targeting S100A4 as a treatment modality in this study. S100A4 RNAi led mechanistically to a decrease in MMP2 expression and activity. Recent reports have indicated that S100A4 directly regulates MMP2 expression in pancreatic cancer (44) and esophageal squamous cell carcinoma (45), thus corroborating the findings presented here. However, whether in our case the regulation is direct or indirect, needs to be studied further. Importantly, the secretion of both S100A4 and MMP2 was decreased in the siS100A4 samples, as analyzed by MRM, a novel finding that could possibly be used as a biomarker approach for classification and treatment response (46).
Previous studies using RNAi against S100A4 have indicated that it reduces invasion and metastasis without retarding cell proliferation or primary tumor growth (47, 48) and our results correlate with these findings. However, we made a novel observation that S100A4 RNAi leads to a more differentiated phenotype, both in vitro and in vivo. In organotypic reconstructs, expression of the differentiation marker involucrin was induced in the siS100A4-treated cells. In OCTT2 xenografts, we observed increased tumor necrosis and well-defined islands of advanced differentiation in S100A4 RNAi-treated mice. Based on our observations, it is thus plausible that depletion of the S100A4-positive cells decreases the pool of CSCs and leads to differentiation. This could have potential for differentiation therapy (49) leading to increased chemosensitivity, as shown by Azzi et al. (50).
Several RNAi-based therapies, both stand-alone and in adjuvant settings, are currently in clinical trials (51). The use of synthetic nanoparticles containing human transferrin (TF) protein as targeting ligand engaging TF receptors on the surface of cancer cells and loaded with siRNA oligos against M subunit of ribonucleotide reductase has been shown to work in a first in-human phase I clinical trial (52). Expression of the TF receptor is up-regulated in many cancer types and the receptor is readily internalized from the cell surface upon ligand binding, making it an effective delivery system for a variety of cancer therapeutics (53). As S100A4 is expressed in many of cell types, such as fibroblasts, immune cells and adipocytes, and not just in cancer cells (54), a cancer-specific targeting would be required for S100A4 therapy. Thus to overcome this, one option would be to use nanoparticles containing siS100A4 and TF as the cancer-specific target.
In conclusion, our results show that in head and neck squamous cell carcinoma EMT drives heterogeneity by creating distinct epithelial and mesenchymal subpopulations and that the MSP acts in a CAF-like manner. The proteomic analyses used here demonstrate that the factors secreted by the tumor microenvironment should be taken into consideration for diagnostic and therapeutic approaches. Both intra- and extracellular forms of CAF-derived S100A4 are important mediators in tumorigenesis, and targeting it via RNAi has the potential of a viable new treatment modality worth exploring in further studies.
Supplementary Material
Acknowledgments
We thank the following Wistar Core Facilities: Proteomics, Flow Cytometry, Histotechnology, and Animal Facility. James Hayden and Fred Keeney of the Microscopy core are thanked for assistance with all aspects of imaging. Dr. Kathleen O'Connor of University of Kentucky is thanked for the S100A4 luciferase reporter construct. Dr. Ashani Weeraratna is thanked for critical review of this manuscript.
Footnotes
* This work was supported by the National Institutes of Health/NCI grants P01-CA098101 (KR, DB, AKS, AKR, MH), CA010815 and U01-CA14305603, National Institutes of Health/NIDDK Center for Molecular Studies in Digestive and Liver Diseases grant P30-DK050306, American Cancer Society grant RP-10-033-01-CCE, and the University of Pennsylvania, Faculty of Medicine Molecular Pathology and Imaging, Molecular Biology and Cell Culture Core Facilities.
 This article contains supplemental Figs. S1 and S2 and Table S1.
This article contains supplemental Figs. S1 and S2 and Table S1.
Conflict of interest: The authors declare no conflict of interest.
1 The abbreviations used are:
- HNSCC
- head and neck squamous cell carcinoma
- CAF
- cancer-associated fibroblastic cell
- CM
- conditioned media
- CSCs
- cancer stem cells
- ECM
- extracellular matrix
- EGFR
- epidermal growth factor receptor
- EMT
- epithelial-mesenchymal transition
- FAP
- fibroblast activation protein
- GFP
- green fluorescent protein
- GO
- gene ontology
- HMLE
- human mammary epithelial cells
- IPA
- Ingenuity Pathway Analysis
- ITGA5
- integrin alpha 5
- MET
- mesenchymal-epithelial transition
- MMP
- matrix metalloproteinase
- MSP
- mesenchymal subpopulation
- PDGFR
- platelet-derived growth factor receptor
- RNAi
- RNA interference
- siRNA
- small interfering RNA
- SMA
- smooth muscle actin
- TF
- transferrin.
REFERENCES
- 1. Masuda M., Toh S., Wakasaki T., Suzui M., Joe A. K. (2013) Somatic evolution of head and neck cancer – biological robustness and latent vulnerability. Mol. Oncol. 1, 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Worsham M. J., Ali H., Dragovic J., Schweitzer V. P. (2012) Molecular characterization of head and neck cancer: How close to personalized targeted therapy? Mol. Diagn. Ther. 16, 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Basu D., Nguyen T. T., Montone K. T., Zhang G., Wang L. P., Diehl J. A., Rustgi A. K., Lee J. T., Weinstein G. S., Herlyn M. (2010) Evidence for mesenchymal-like sub-populations within squamous cell carcinomas possessing chemoresistance and phenotypic plasticity. Oncogene 29, 4170–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Basu D., Montone K. T., Wang L., Gimotty P. A., Hammond R., Diehl J. A., Rustgi A. K., Lee J. T., Rasanen K., Weinstein G. S., Herlyn M. (2011) Detecting and targeting mesenchymal-like subpopulations within squamous cell carcinomas. Cell Cycle 10, 2008–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kalluri R., Weinberg R. A. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Invest. 119, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scheel C., Eaton E. N., Li S. H., Chaffer C. L., Reinhardt F., Kah K. J., Bell G., Guo W., Rubin J., Richardson A. L., Weinberg R. A. (2011) Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell 145, 926–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanahan D., Coussens L. M. (2012) Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell. 21, 309–322 [DOI] [PubMed] [Google Scholar]
- 8. Marsh T., Pietras K., McAllister S. S. (2013) Fibroblasts as architects of cancer pathogenesis. Biochim. Biophys. Acta 1832, 1070–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rasanen K., Vaheri A. (2010) Activation of fibroblasts in cancer stroma. Exp. Cell Res. 316, 2713–2722 [DOI] [PubMed] [Google Scholar]
- 10. Sugimoto H., Mundel T. M., Kieran M. W., Kalluri R. (2006) Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer. Biol. Ther. 5, 1640–1646 [DOI] [PubMed] [Google Scholar]
- 11. Sherbet G. V. (2009) Metastasis promoter S100A4 is a potentially valuable molecular target for cancer therapy. Cancer Lett. 280, 15–30 [DOI] [PubMed] [Google Scholar]
- 12. Saleem M., Kweon M. H., Johnson J. J., Adhami V. M., Elcheva I., Khan N., Bin Hafeez B., Bhat K. M., Sarfaraz S., Reagan-Shaw S., Spiegelman V. S., Setaluri V., Mukhtar H. (2006) S100A4 accelerates tumorigenesis and invasion of human prostate cancer through the transcriptional regulation of matrix metalloproteinase 9. Proc. Natl. Acad. Sci. U.S.A. 103, 14825–14830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olsen C. J., Moreira J., Lukanidin E. M., Ambartsumian N. S. (2010) Human mammary fibroblasts stimulate invasion of breast cancer cells in a three-dimensional culture and increase stroma development in mouse xenografts. BMC Cancer 10, 444–2407-10–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Andersen K., Mori H., Fata J., Bascom J., Oyjord T., Maelandsmo G. M., Bissell M. (2011) The metastasis-promoting protein S100A4 regulates mammary branching morphogenesis. Dev. Biol. 352, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Donato R., Cannon B. R., Sorci G., Riuzzi F., Hsu K., Weber D. J., Geczy C. L. (2013) Functions of S100 proteins. Curr. Mol. Med. 13, 24–57 [PMC free article] [PubMed] [Google Scholar]
- 16. Noma K., Smalley K. S., Lioni M., Naomoto Y., Tanaka N., El-Deiry W., King A. J., Nakagawa H., Herlyn M. (2008) The essential role of fibroblasts in esophageal squamous cell carcinoma-induced angiogenesis. Gastroenterology 134, 1981–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Räsänen K., Virtanen I., Salmenperä P., Grenman R., Vaheri A. (2009) Differences in the nemosis response of normal and cancer-associated fibroblasts from patients with oral squamous cell carcinoma. PLoS One 4, e6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rasanen K., Vaheri A. (2010) TGF-beta1 causes epithelial-mesenchymal transition in HaCaT derivatives, but induces expression of COX-2 and migration only in benign, not in malignant keratinocytes. J. Dermatol. Sci. 58, 97–104 [DOI] [PubMed] [Google Scholar]
- 19. Li L., Fukunaga-Kalabis M., Herlyn M. (2011) The three-dimensional human skin reconstruct model: A tool to study normal skin and melanoma progression. J. Vis. Exp. 54, pii: 2937 doi, 10.3791/2937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 21. Wang H., Tang H. Y., Tan G. C., Speicher D. W. (2011) Data analysis strategy for maximizing high-confidence protein identifications in complex proteomes such as human tumor secretomes and human serum. J. Proteome Res. 10, 4993–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tang H. Y., Beer L. A., Barnhart K. T., Speicher D. W. (2011) Rapid verification of candidate serological biomarkers using gel-based, label-free multiple reaction monitoring. J. Proteome Res. 10, 4005–4017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Beer L. A., Tang H. Y., Sriswasdi S., Barnhart K. T., Speicher D. W. (2011) Systematic discovery of ectopic pregnancy serum biomarkers using 3-D protein profiling coupled with label-free quantitation. J. Proteome Res. 10, 1126–1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tang H. Y., Beer L. A., Chang-Wong T., Hammond R., Gimotty P., Coukos G., Speicher D. W. (2012) A xenograft mouse model coupled with in-depth plasma proteome analysis facilitates identification of novel serum biomarkers for human ovarian cancer. J. Proteome Res. 11, 678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pfaffl M. W. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pfaffl M. W., Horgan G. W., Dempfle L. (2002) Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 28. Kendal W. S. (2007) Empirically-based estimates for the burden of subclinical metastases. Int. J. Radiat. Biol. 83, 383–393 [DOI] [PubMed] [Google Scholar]
- 29. Tsellou E., Kiaris H. (2008) Fibroblast independency in tumors: Implications in cancer therapy. Future Oncol. 4, 427–432 [DOI] [PubMed] [Google Scholar]
- 30. Friedl P., Wolf K. (2010) Plasticity of cell migration: A multiscale tuning model. J. Cell Biol. 188, 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dvorak H. F. (1986) Tumors: Wounds that do not heal. similarities between tumor stroma generation and wound healing. N. Engl. J. Med. 315, 1650–1659 [DOI] [PubMed] [Google Scholar]
- 32. Grunert S., Jechlinger M., Beug H. (2003) Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat. Rev. Mol. Cell Biol. 4, 657–665 [DOI] [PubMed] [Google Scholar]
- 33. Polyak K., Weinberg R. A. (2009); 2009) Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Rev. Cancer 9, 265–273 [DOI] [PubMed] [Google Scholar]
- 34. Hoek K. S., Eichhoff O. M., Schlegel N. C., Dobbeling U., Kobert N., Schaerer L., Hemmi S., Dummer R. (2008) In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 68, 650–656 [DOI] [PubMed] [Google Scholar]
- 35. Widmer D. S., Cheng P. F., Eichhoff O. M., Belloni B. C., Zipser M. C., Schlegel N. C., Javelaud D., Mauviel A., Dummer R., Hoek K. S. (2012) Systematic classification of melanoma cells by phenotype-specific gene expression mapping. Pigment Cell Melanoma Res. 25, 343–353 [DOI] [PubMed] [Google Scholar]
- 36. Hanahan D., Weinberg R. A. (2011) Hallmarks of cancer: The next generation. Cell 144, 646–674 [DOI] [PubMed] [Google Scholar]
- 37. Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J. F., Harrington K., Sahai E. (2007) Fibroblast-led collective invasion of carcinoma cells with differing roles for RhoGTPases in leading and following cells. Nat. Cell Biol. 9, 1392–1400 [DOI] [PubMed] [Google Scholar]
- 38. Gelse K., Poschl E., Aigner T. (2003) Collagens—structure, function, and biosynthesis. Adv. Drug Deliv. Rev. 55, 1531–1546 [DOI] [PubMed] [Google Scholar]
- 39. Powell A. M., Sakuma-Oyama Y., Oyama N., Black M. M. (2005) Collagen XVII/BP180: a collagenous transmembrane protein and component of the dermoepidermal anchoring complex. Clin. Exp. Dermatol. 30, 682–687 [DOI] [PubMed] [Google Scholar]
- 40. Singh A., Settleman J. (2010) EMT, cancer stem cells and drug resistance: An emerging axis of evil in the war on cancer. Oncogene 29, 4741–4751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen C., Zimmermann M., Tinhofer I., Kaufmann A. M., Albers A. E. (2013) Epithelial-to-mesenchymal transition and cancer stem(-like) cells in head and neck squamous cell carcinoma. Cancer Lett. 338, 47–56 [DOI] [PubMed] [Google Scholar]
- 42. Lo J. F., Yu C. C., Chiou S. H., Huang C. Y., Jan C. I., Lin S. C., Liu C. J., Hu W. Y., Yu Y. H. (2011) The epithelial-mesenchymal transition mediator S100A4 maintains cancer-initiating cells in head and neck cancers. Cancer Res. 71, 1912–1923 [DOI] [PubMed] [Google Scholar]
- 43. Sack U., Walther W., Scudiero D., Selby M., Kobelt D., Lemm M., Fichtner I., Schlag P. M., Shoemaker R. H., Stein U. (2011) Novel effect of antihelminthic niclosamide on S100A4-mediated metastatic progression in colon cancer. J. Natl. Cancer Inst. 103, 1018–1036 [DOI] [PubMed] [Google Scholar]
- 44. Li N., Song M. M., Chen X. H., Liu L. H., Li F. S. (2012) S100A4 siRNA inhibits human pancreatic cancer cell invasion in vitro. Biomed. Environ. Sci. 25, 465–470 [DOI] [PubMed] [Google Scholar]
- 45. Zhang H. Y., Zheng X. Z., Wang X. H., Xuan X. Y., Wang F., Li S. S. (2012) S100A4 mediated cell invasion and metastasis of esophageal squamous cell carcinoma via the regulation of MMP-2 and E-cadherin activity. Mol. Biol. Rep. 39, 199–208 [DOI] [PubMed] [Google Scholar]
- 46. Cohen A., Wang E., Chisholm K. A., Kostyleva R., O'Connor-McCourt M., Pinto D. M. (2013) A mass spectrometry-based plasma protein panel targeting the tumor microenvironment in patients with breast cancer. J. Proteomics 81, 135–147 [DOI] [PubMed] [Google Scholar]
- 47. Chen D., Zheng X. F., Yang Z. Y., Liu D. X., Zhang G. Y., Jiao X. L., Zhao H. (2012) S100A4 silencing blocks invasive ability of esophageal squamous cell carcinoma cells. World J. Gastroenterol. 18, 915–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sekine H., Chen N., Sato K., Saiki Y., Yoshino Y., Umetsu Y., Jin G., Nagase H., Gu Z., Fukushige S., Sunamura M., Horii A. (2012) S100A4, frequently overexpressed in various human cancers, accelerates cell motility in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 429, 214–219 [DOI] [PubMed] [Google Scholar]
- 49. Cruz F. D., Matushansky I. (2012) Solid tumor differentiation therapy. Oncotarget 3, 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Azzi S., Bruno S., Giron-Michel J., Clay D., Devocelle A., Croce M., Ferrini S., Chouaib S., Vazquez A., Charpentier B., Camussi G., Azzarone B., Eid P. (2011) Differentiation therapy: Targeting human renal cancer stem cells with interleukin 15. J. Natl. Cancer Inst. 103, 1884–1898 [DOI] [PubMed] [Google Scholar]
- 51. Davidson B. L., McCray P. B., Jr. (2011) Current prospects for RNA interference-based therapies. Nat. Rev. Genet. 12, 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Davis M. E., Zuckerman J. E., Choi C. H. J., Seligson D., Tolcher A., Alabi C. A., Yen Y., Ribas A. (2010) Evidence of RNAi in humans from systemically administered siRNA via targeted nanoparticles. Nat. Lett. 464, 1067–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Daniels T. R., Bernabeu E., Rodriguez J. A., Patel S., Kozman M., Chiappetta D. A., Holler E., Ljubimova J. Y., Helguera G., Penichet M. L. (2012) Tranferrin receptors and the targeted delivery of therapeutic agenst against cancer. Biochim. Biophys. Acta 1820, 291–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gross S. R., Sin C. G., Barraclough R., Rudland P. S. (2013) Joining S100 proteins and migration: for better or worse, in sickness and in health. Cell Mol. Life Sci. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.