Abstract
B lymphocytes play a pivotal role in multiple sclerosis pathology, possibly via both antibody-dependent and -independent pathways. Intrathecal immunoglobulin G in multiple sclerosis is produced by clonally expanded B-cell populations. Recent studies indicate that the complementarity determining regions of immunoglobulins specific for certain antigens are frequently shared between different individuals. In this study, our main objective was to identify specific proteomic profiles of mutated complementarity determining regions of immunoglobulin G present in multiple sclerosis patients but absent in healthy controls. To achieve this objective, we purified immunoglobulin G from the cerebrospinal fluid of 29 multiple sclerosis patients and 30 healthy controls and separated the corresponding heavy and light chains via SDS-PAGE. Subsequently, bands were excised, trypsinized, and measured with high-resolution mass spectrometry. We sequenced 841 heavy and 771 light chain variable region peptides. We observed 24 heavy and 26 light chain complementarity determining regions that were solely present in a number of multiple sclerosis patients. Using stringent criteria for the identification of common peptides, we found five complementarity determining regions shared in three or more patients and not in controls. Interestingly, one complementarity determining region with a single mutation was found in six patients. Additionally, one other patient carrying a similar complementarity determining region with another mutation was observed. In addition, we found a skew in the κ-to-λ ratio and in the usage of certain variable heavy regions that was previously observed at the transcriptome level. At the protein level, cerebrospinal fluid immunoglobulin G shares common characteristics in the antigen binding region among different multiple sclerosis patients. The indication of a shared fingerprint may indicate common antigens for B-cell activation.
Autoimmune mechanisms play a central role in the pathogenesis of multiple sclerosis (MScl).1 Recent trials indicate that B-lymphocyte depletion therapy can substantially reduce disease activity in relapsing-remitting MScl patients (1). Clinical amelioration after depletion seems to precede a reduction in autoantibody levels, possibly because this treatment rapidly affects the antigen-presenting cell functions of B cells (2). This finding has boosted interest in studies on the pathogenic role of autoreactive B cells. Despite the success in inhibiting antibody-independent functions of B cells, arguments remain for an additional chronic pathogenic role for autoantibodies within the central nervous system (CNS). This includes (a) the presence of antibodies in cerebrospinal fluid (CSF) and brain tissue (3), (b) depositions of antibody within areas of demyelination along with local complement activation (3), and (c) myelin oligodendrocyte glycoprotein specific antibodies in some subpopulations of MScl patients (4, 5). Additionally, KIR 4.1 was recently identified as a target of autoantibody response in a subgroup of persons with MScl (6).
It has been shown that the distribution of genes used to generate antibodies in B cells from CSF and lesions of MScl patients is skewed relative to naturally expected distributions. Several groups described clonal B-cell populations within the CNS, sometimes even skewed to certain families of variable heavy (VH) regions (7, 8). No common motifs have yet been found to be shared between different MScl patients. This would be in line with the classical immunological insight that suggests that it is extremely rare to find common sequences in the immunoglobulin G (Ig) variable regions among different individuals. However, this view has recently been challenged (9–11). Both after vaccination and in paraneoplastic syndromes such as anti-Hu, strikingly identical shared complementarity determining region (CDR) motifs were observed among patients (12). Of note is also a surprising study in which it was observed that malignant chronic lymphocytic leukemia B cells in different patients all recognized a single fungal antigen and showed shared use of CDR3 sequences among different individuals (13).
A novel approach for studying Ig gene usage in the biofluids of MScl patients is the use of proteomic sequencing. Obermeier and colleagues described overlap between Ig B-cell (CSF) transcriptomes and proteomes in four individual MScl cases, without interindividual overlapping sequences (14). However, this elegant proof-of-principle study was limited to four MScl patients, and there was no comparison between patients and controls. The possibility of sequencing CSF Ig at the protein level (10, 14, 15) may bring along some advantages. The genetic approaches used so far share the benefit that complete sequences can be identified at the single-cell level, but Ig derived from such clones does not necessarily represent the actual Ig repertoire found in CSF. Furthermore, whereas genomic studies are restricted to CSF cells, humoral CSF studies also include Ig proteins from other anatomical brain areas, such as parenchyma, meninges, and Virchow–Robin spaces. Finding common characteristics of the antigen binding sites of Ig among patients might provide leads regarding the question of whether common antigenic stimuli are responsible for the recruitment of intrathecal B cells in MScl. We previously described a new approach using advanced nano-scale liquid chromatography coupled online to a high-resolution mass spectrometer (LC-MS) (10, 16, 17), a reliable and powerful method for the sensitive detection of CDR peptides (18). Moreover, it can also be used to compare CDR peptide profiles among a relatively large number of patients and controls.
Our main question was whether we could detect specific proteomic profiles of CDR present in MScl patients but absent in controls. Here, we report a number of common CDR sequences in Ig of a group of MScl patients that were not observed in healthy controls. In addition, we show disturbed κ/λ chain ratios in CSF Ig of MScl patients and VH family usage in patients relative to controls.
MATERIALS AND METHODS
Clinical Samples: MScl Patients and Non-neurological Controls
CSF samples were collected from untreated MScl patients, which were selected by an experienced neurologist (R.Q.H.) and were followed prospectively by the Rotterdam Multiple Sclerosis Center ErasMS at the Department of Neurology at the Erasmus Medical Center (Rotterdam, The Netherlands). The procedure for CSF sample collection was as described previously (19). All MScl patients had defined relapsing-remitting MScl or a clinically isolated syndrome according to the 2005 McDonald criteria for MScl (20). The control individuals were free of any neurological disorders; therefore, these individuals are hereinafter referred to as healthy controls. They underwent minor, non-neurologically indicated surgeries, and CSF was taken prior to the administration of sedatives as part of the anesthesia procedure. Immediately after collection, the CSF samples were centrifuged (10 min at 3000 rpm) so that cells and cellular elements could be discarded, and the supernatant was aliquoted and stored at −80 °C until further use for this study. Blood-contaminated CSF samples were excluded based on the presence of erythrocytes detected via microscopic examination immediately after sampling. One aliquot of a sample was used for routine CSF diagnostics in the Clinical Chemistry Department of Erasmus Medical Center. This diagnostic procedure for the MScl patients included the quantification of total protein and albumin, assessment of the number of oligoclonal bands, and Ig index. Samples were also taken from another aliquot that had been studied previously to determine intraindividual variations in CSF protein abundances (37). This study was approved by the Institutional Ethical Committee of the Erasmus Medical Center, and written informed consent was obtained from all participants.
Ig Quantification Assay
An ELISA assay using 96-well plates (Immuno 96 MicroWellTM solid plate, Thermo Fisher, Bremen, Germany) was used to determine Ig concentrations in CSF samples. In this assay, AffiniPure F(ab′)2 fragment goat anti-human IgG (H + L) (Jackson ImmunoResearch Laboratories, Suffolk, UK) at a concentration of 1.3 mg/ml was used to coat the wells as a capture antibody. As a detection antibody, horseradish peroxidase conjugated polyclonal secondary antibody antihuman Fc goat (anti-human IgGHRP, Sigma-Aldrich, Saint Louis, MO) was used. Samples were incubated with antibodies for 5 min and gently shaken (400 rpm) at 4 °C on a thermo-cycler shaker (Eppendorf AG, Hamburg, Germany). 3,3′,5,5′-tetramethylbenzidine (Sigma-Aldrich, Saint Louis, MO) (100 μl per well) was used as a substrate for horseradish peroxidase and developed a soluble blue reaction product. The reaction was stopped with 100 μl 1 m hydrochloric acid. The Ig concentration was determined photometrically based on absorbance at 450 nm and was quantified using an eight-point calibration curve ranging from 1.0 × 10−5 to 0.5 μg/μl.
Ig Purification
Ig was purified from CSF samples using a Melon Gel IgG Spin Purification Kit (Pierce, Rockford, IL) according to the manufacturer's protocol for serum, with slight modifications. All CSF samples were diluted at a ratio of 1:3 with purification buffer. We used 100 μl CSF from patients and 200 μl from controls based on the Ig concentration assay (two times the CSF volume was used for controls in order to normalize for Ig concentration). Subsequently, the diluted CSF was added to a spin column containing Melon Gel resin. After 15 min of incubation, the spin column was centrifuged at 5000g, and the flow-through containing the purified Ig was collected. Ig concentrations were determined in the flow-through fractions after purification (with the above-described Ig ELISA). Equal amounts of Ig across all patients and controls were then taken and subsequently lyophilized (Sublimator 400, Zirbus Technology, Tiel, The Netherlands) for six hours. The lyophilized Ig fractions were then stored at −20 °C for one day before undergoing separation by SDS-PAGE.
For SDS-PAGE separation, loading buffer was added to each lyophilized sample and heated at 90 °C for 10 min. Purified Ig antibodies were resolved into heavy (IgH) and light (IgL) chains by means of reducing one-dimensional SDS-PAGE using Bio-Rad Mini-Protean electrophoresis system gels (10% polyacrylamide gels of 0.75-mm thickness). The gels were stained with Novex® Colloidal Blue Staining (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Overnight destaining was performed for visualization of IgH and IgL chain protein bands, and subsequently gels were scanned.
In-gel Trypsin Digestion
We excised gel bands manually in a laminar flow cabinet as a preventive measure to minimize environmental keratin and other contaminating protein-like materials. Protein bands were cut into plugs and transferred into Eppendorf tubes. We performed reduction by dithiothreitol and alkylation with iodoacetamide. Subsequently, in-gel digestion was performed overnight at 37 °C, and further peptide extraction procedures were performed (18). After peptide extraction, samples were dried for three hours in a vacuum centrifuge (SPD 1010, Thermo Savant, Holbrook, NY) and then stored at −80 °C until LC-MS measurements.
Chromatography Separation and Mass Spectrometric Measurement
Before LC-MS measurements, the dried peptide samples were dissolved in 40 μl of an aqueous solution of 0.1% TFA and sonified. The samples were measured with a nano-LC system (Ultimate 3000, Thermo Fisher Scientific, Amsterdam, The Netherlands) coupled online to a hybrid linear ion trap/Orbitrap mass spectrometer (LTQ-Orbitrap-XL, Thermo Fisher Scientific, Bremen, Germany). Samples were loaded onto a trap column (PepMap C18, 300-μm inner diameter by 5-mm length, 5-μm particle size, 100-Å pore size; Thermo Fisher Scientific) and washed and desalted for 10 min using 0.1% TFA (in water) as the loading solvent. Then the trap column was switched online with the analytical column (PepMap C18, 75 μm inner diameter by 250 mm, 3-μm particle size, 100-Å pore size; Thermo Fisher Scientific) and peptides were eluted with the following binary gradient: 100% solvent A, then from 0% to 25% solvent B in 60 min and from 25% to 50% solvent B in 30 min. Solvent A consisted of 2% acetonitrile and 0.1% formic acid in HPLC-grade water, and solvent B consisted of 80% acetonitrile and 0.08% formic acid in HPLC-grade water. All LC solvents were purchased from Biosolve (Valkenswaard, The Netherlands). The column flow rate was set at 300 nl/min, and eluting peptides were measured by a UV detector (at a wavelength of 214 nm in a 3-nl nanoflow cell (Thermo Fisher Scientific)) and consecutively introduced into the mass spectrometer. For electrospray ionization, metal-coated nano-electrospray ionization emitters (New Objective, Woburn, MA) were used and a spray voltage of 1.5 kV was applied. For MS detection, a data-dependent acquisition method was used: a high-resolution survey scan from 400–1800 Th was detected in the Orbitrap (target of automatic gain control = 106, resolution = 30,000 at 400 m/z, lock mass set to 445.120025 Th (protonated [Si(CH3)2O]6 (21))). On the basis of this full scan, the five most intensive ions were consecutively isolated (automatic gain control target set to 104 ions), fragmented via collision-activated dissociation (applying 35% normalized collision energy), and detected in the ion trap. Precursor masses within a tolerance range of ±5 ppm that were selected once for MS/MS were excluded for MS/MS fragmentation for 3 min or until the precursor intensity fell below a signal-to-noise ratio of 1.5 for more than five scans. Samples were prepared and measured in a randomized order. An internal quality-control sample was measured once in every five measurements. Before each run, a blank run was performed to monitor the background of the system.
Data Analysis and Peptide Identification
Peptide Identification by Mass and Fragmentation
Acquired LC-MS profiles for the separate purified IgH chain and IgL chain datasets were analyzed separately using the Progenesis LC-MS software package (version 2.6, Nonlinear Dynamics Ltd., Newcastle-upon-Tyne, UK). Individual runs were aligned with each other to compensate for variations in retention times (samples that could not be aligned by at least 200 vectors using the automated alignment option were excluded from further analysis as recommended by the manufacturer). Before peak selection, integration of the area of the peaks was performed. The resulting peaks could then be associated with the amino acid sequence information if corresponding fragmentation spectra were available.
From raw data files, MS/MS spectra were extracted and converted into MGF files using extract msn (part of Xcalibur version 2.0.7, Thermo Fisher Scientific). Sequencing of the fragmentation spectra was conducted via a Mascot MS/MS database search (version 2.3.01, Matrix Science Inc., London, UK) against the human subset of the NCBInr non-redundant sequence database (August 15, 2010, Homo sapiens taxonomy; 232,854 sequences). The following settings were used for the database search: a maximum of two missed cleavages, tryptic cleavage, oxidation as a variable modification of methionine (+15.995 u), and carbamidomethylation as a fixed modification of cysteine (+57.021 u). The peptide mass tolerance was set at 10 ppm, and the fragment mass tolerance at 0.5 Da. For peptide identification, a minimum ion score of 25 was required. The resulting peptide identifications were filtered using Scaffold (version 3.2.0, Proteome Software Inc., Portland, OR). Peptide false discovery rates were calculated by Scaffold as (false positives)/(false positives + true positives). On average, the false discovery rate determined was always lower than 0.1%. Filter criteria for the generated identification result table were a peptide probability and a protein probability greater than 95%. At this stage, all non-Ig proteins (albumin, transferrin, keratin, etc.) were filtered out on the basis of their protein names so we could focus solely on the Ig proteins. Subsequently, the identified peptides and proteins were imported into the Progenesis software package and linked to their corresponding peptide peaks. The Progenesis analysis matrix contained mass, charge, intensity, abundance, and MS/MS fragmentation spectra of the detected peptides.
The abundance listed for all peaks can be defined as the background or signal. To remove background from real peak signals, we used the following procedure: First, around 30 randomly chosen peaks in all samples with a low-intensity region were reviewed regarding their isotopic pattern. Peaks with more than two isotopes were classified as valid peaks, and peaks with no isotope or just one or two isotopes were classified as background. We performed manual checks on all peptides of interest to confirm the background level and the detection of the exact location and overlap of peaks among samples in the abovementioned Progenesis software package. The specific aim of this work was to find CDR peptide fragments that were present in MScl patients but not in the control group. Therefore, we filtered the datasets for candidates that were found in at least three MScl patients but were absent in control patients.
Manual Confirmation of Peptide Identifications
For further confirmation of the identification of peptides as described above, we assessed the MS/MS fragmentation spectra. If a peptide is common among patients, one should expect a similar spectral pattern (in terms of mass fragmentation and retention time window) to be present in different samples. Additionally, we evaluated the isotopic patterns in the mass spectrometry spectra in terms of the number and appearance of isotopes. Using the method described above, those marker peptides that did not pass our filter were not qualified as markers.
CDR Identification (Assigning Location in Ig Structure)
All identified amino acid sequences were aligned to a variable (V), diverse (D), joining (J), or constant (C) region human (Homo sapiens) Ig germ line sequence derived from the International ImMunoGeneTics Information System (IMGT) database (Montpellier, France) (22). As described previously (16, 17, 23), we used the BLASTp search algorithm (NCBI BLAST version 2.2.22) to align the identified peptide sequences to the corresponding Ig.
All peptide alignments with bit scores greater than 12.5 were selected for further analysis. Peptides aligned to the variable domain germ line were further submitted to the IMGT/domain gap alignment tool, which positions the peptide to the germ line sequence in the IMGT unique numbering residue system. The alignment with the most homologous germ line allele was provided by the IMGT tool and included in the data if the identity score was at least 70%. With this approach we were able to locate a peptide in a CDR or framework region. A CDR peptide was defined as a peptide containing a minimum of three amino acids in the CDR part, irrespective of framework length. An overview of the methodology is presented in the form of a flow chart in Fig. 1. The analysis matrix generated for IgH and IgL chain comparison by the Progenesis software included with the peptide alignment summary (derived from a BLAST– IMGT search) is presented in supplemental Files S1 and S2.
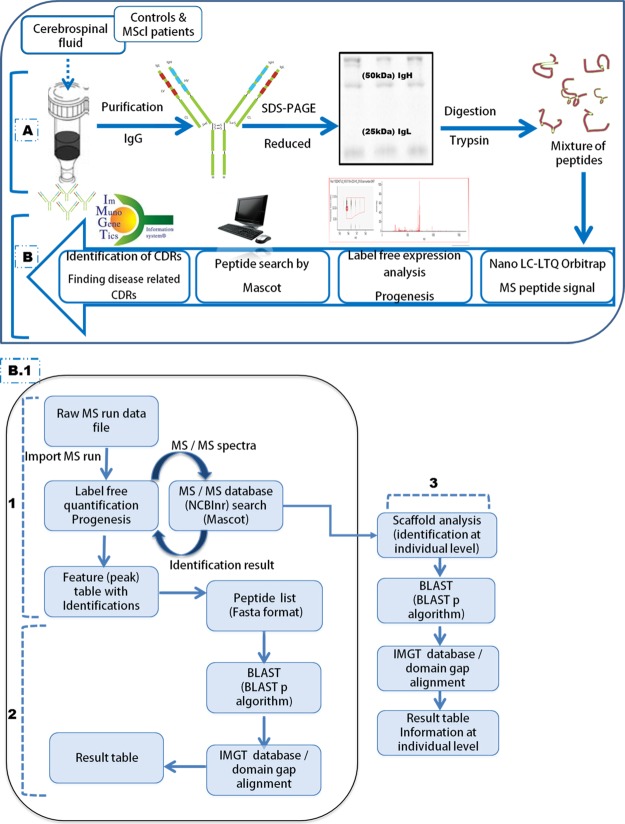
Fig. 1.
Schematic illustration of the proteomics-based methodology used to assign CDR presence exclusively in the CSF Ig of MScl patients. A, Ig was purified from the CSF of MScl patients (100 μl) and non-neurological controls (200 μl) based on the Ig concentration assay. Purification was performed using a Melon Gel IgG Spin Purification Kit. Purified Ig was separated into IgH and IgL chains via reducing one-dimensional SDS-PAGE gels. B, after in-gel trypsin digestion of excised IgH and IgL bands, the mixture of peptides was measured via nano-LC–LTQ-Orbitrap MS. Mass spectra were analyzed by Progenesis software, and the peptide search was performed by Mascot. Identified peptides were used for CDR identification using the IMGT database and BLASTp search algorithm to find MScl-specific CDRs. (1) Nano-LC–LTQ-Orbitrap MS-generated LC-MS profiles (raw MS run data) for the IgH and IgL datasets. These were analyzed separately using the Progenesis LC-MS software package (label-free quantification). Sequencing of the MS/MS spectra was executed via a Mascot MS/MS database search against the human subset of the NCBInr sequence database. Afterward, the identified peptides were imported into Progenesis and linked to their corresponding peaks. The peak abundance (UV area under the curve) information was used for analysis of CDR presence or absence. (2) Identified peptides were extracted and converted into FASTA format. They were aligned to V, D, J, and C elements of Ig (Homo sapiens) germline sequences derived from the IMGT database. We used the BLASTp search algorithm to align the identified peptide sequences to the corresponding Ig fragments. Next, they were submitted to the IMGT/domain gap alignment tool. This analysis provided alignment details of CSF Ig peptides relative to the Ig germline that included the gene name, homology match score, mutation/mismatch, and start and end positions. (3) Peptide identification details were uploaded in Scaffold. The Scaffold file contained a peptide identification view report based on the spectral counts (at MS/MS level). Peptide counts were obtained at the individual level for each sample and were exported to a spreadsheet containing detailed information about the protein and peptide hits. On the basis of the resulting combined peptide set, alignment summaries relative to the germline were assigned at the individual level (using a BLASTp algorithm and the IMGT database). This information was used for VH and VK family distribution analysis. BLAST, basic local alignment search tool; BLASTp, basic local alignment search tool for protein; IMGT, ImMunoGeneTics information system; Feature (or Peak), object of defined mass with an identified charge state, retention time, and isotopes characterized by the analysis software.
Statistical Analysis
We found a set of CDR peptides that were present in at least three MScl samples but not in any control samples (true result). To determine the probability that this finding was not due to chance alone, we performed permutation tests. We permutated (randomized) the sample group assignment (patient or control group) and reran the determination of exclusively present CDR peptides 5000 times (false hits). The relative frequency of the occurrence of exclusive CDR peptides in a randomized sample set was calculated by dividing the number of false hits exceeding the true result by the numbers of randomization trials. For these computations we used the statistics package R (version 2.15.2). Other statistical analyses of data and graphical presentations were performed using GraphPad Prism (GraphPad Software version 5.00) or by an Excel 2010 function.
RESULTS
In total, 59 samples were included for analysis of the IgH comparison set (n = 29 MScl versus n = 30 controls). The IgL comparison set included 55 samples (n = 29 MScl versus n = 26 controls). Four IgH samples (two MScl and two controls) and six IgL samples (one MScl and five controls) were excluded after label-free analysis because of weak alignment and UV information.
Patient Characteristics
All patient characteristics are shown in Table I. Significant differences in gender and age (Mann–Whitney test, p < 0.0001) were observed between MScl patients and controls, but no other parameters showed any differences between the groups (Mann–Whitney test, p > 0.05). MScl patients have slightly increased Ig concentrations in CSF. Therefore, we normalized the CSF Ig concentration. After normalization, the UV-quantified area (peptide abundance) obtained during LC-MS measurements did not show any significant difference (Mann–Whitney test, p = 0.80 IgH and p = 0.18 IgL) in the concentration of digested peptides between groups (supplemental Fig. S1).
Table I. Clinical characteristics of MScl patients and non-neurological controls. Clinical characteristics are presented as medians (25th and 75th percentiles). The differences were identified via Mann–Whitney test.
| IgH |
IgL |
|||||
|---|---|---|---|---|---|---|
| Control (n = 30) | MScl (n = 29) | p value | Control (n = 26) | MScl (n = 29) | p value | |
| Age (years) at sampling | 52 (39–68) | 31 (26–41) | < 0.0001 | 53 (41–67) | 33 (27–42) | < 0.0001 |
| Gender | 13 ♂ 17 ♀ | 2 ♂ 27 ♀ | < 0.0001 | 13 ♂ 13 ♀ | 2 ♂ 27 ♀ | < 0.0001 |
| Total protein (g/l) | 0.39 (0.33–0.50) | 0.37 (0.26–0.46) | 0.07 | 0.39 (0.33–0.51) | 0.36 (0.26–0.45) | 0.05 |
| Albumin (g/l)a | 0.19 (0.17–0.35) | 0.19 (0.14–0.24) | 0.61 | 0.18 (0.10–0.36) | 0.19 (0.15–0.24) | 0.87 |
| Abnormal CSF Ig indexb (OBs)c | N/A | 21 (23) | N/A | N/A | 21 (23) | N/A |
| Time from CIS to CSF samplingd (months) | N/A | 5 (2–38) | N/A | N/A | 5 (2–40) | N/A |
Statistical significance considered at p < 0.05.
IgH, heavy chain; IgL, light chain; N/A, not applicable; OB, oligoclonal band; ♀, female; ♂, male.
a Albumin concentration (g/l) is reported for n = 11 IgH and n = 8 IgL.
b Ig index, (CSF/serum Ig):(CSF/serum albumin), was denoted as increased if greater than 0.67.
c OBs: presence of CSF-specific OBs.
d Time from first attack to CSF sampling; first attack defined as the first neurological episode caused by inflammation or demyelination.
VH and VK Family Distribution in MScl Patients and Controls
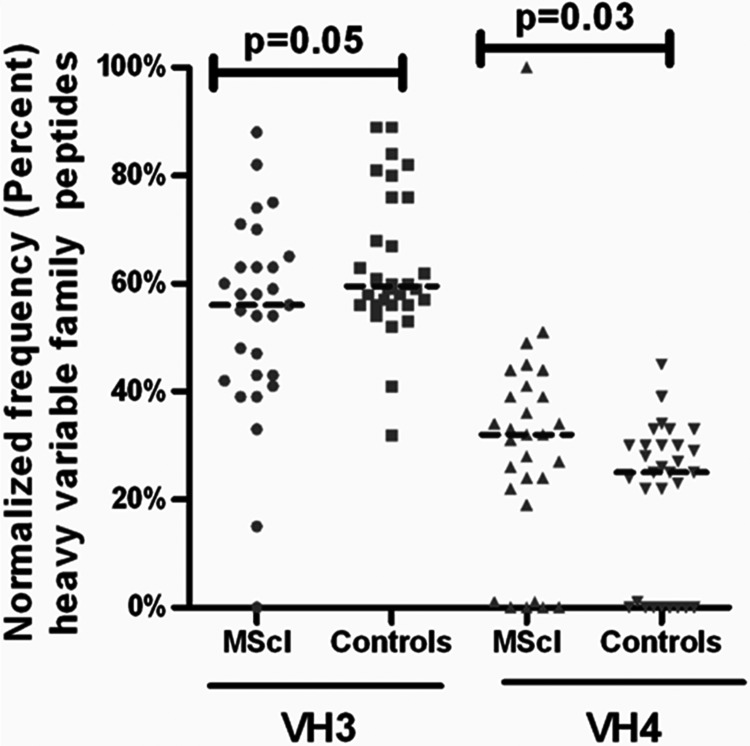
The peptide spectral count (based on MS/MS identification) information (Scaffold-based) acquired at the individual level (Fig. 1) was used to analyze the Ig VH family distribution between MScl patients and controls. An alignment match score of 70% was used as a cutoff value, and a mean score of 90% ± 8.5% was observed. The mean sequence length was 10 ± 2.1 amino acids (mean ± S.D.). Peptide counts were normalized at the individual patient level to the total number of peptides found for the given IgH chain family. The usage of peptides assigned to each family was compared between groups. Log-transformed data were not normally distributed (according to a D'Agostino and Pearson omnibus normality test); therefore, a non-parametric statistical test was used to determine statistical significance using raw data. In comparison to controls, VH 4 was found to be slightly but statistically significant (p = 0.03), and a trend toward increased VH 3 in MScl was observed (p = 0.05) (Fig. 2). No significant differences were found for the other six VH families (p > 0.05) (supplemental Fig. S2A). The difference in family usage of the VK chains between MScl patients and controls was also determined in a similar way. We found a peptide alignment match mean score of 86% ± 11%. The mean amino acid sequence length was 10 ± 2.6 (mean ± S.D.). Analysis did not show a significant difference between groups (supplemental Fig. S2B).
Fig. 2.
The family usage of VH3 and VH4 peptides in MScl in comparison to controls. The family usage of the IgH repertoire expressed in the CSF of MScl patients and controls. Horizontal lines represent median values. Peptide counts were normalized at the individual patient level to the total number of peptides in given family. Frequencies (percentage) of peptides are plotted along the y-axis. The usage of peptides assigned to each family was compared between the two groups. A slight overrepresentation of the VH4 family (Mann–Whitney test, p = 0.03) was observed in the MScl patient group (n = 29) relative to the control group (n = 30).
Disturbed κ/λ Ratio in MScl Patients
The κ/λ ratio was analyzed in controls (n = 26) and MScl patients (n = 29). The ratio of κ and λ light chains was determined from the abundance data for representative peptides from the LC-MS dataset (Progenesis). The peptides used were C-region peptides (n = 10) having an alignment match score of 100% ± 0% (mean ± S.D.) for κ (n = 6) and 97% ± 3% (n = 4) for λ peptides. The κ/λ mean ratio in controls was 2.52 ± 1.71, and in MScl it was 4.06 ± 2.76. A two-tailed t test was applied because on log scale transformed data a normal distribution was found by means of normality testing. The ratio showed significant elevation in the MScl group (p = 0.03) (Fig. 3).
Fig. 3.
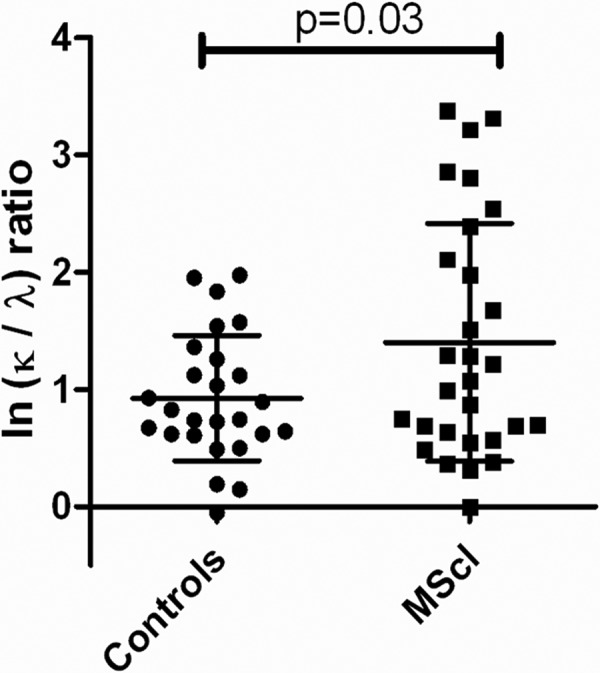
The κ-to-λ ratio in CSF Ig of MScl patients (n = 29) and controls (n = 26). The κ-to-λ ratio in CSF Ig of MScl patients and controls is expressed on a logarithmic (ln) scale and shown as the mean with S.D. Ratios showed elevation in the MScl group (unpaired t test, p = 0.03).
Identical CDR Peptide Identification in Several MScl Patients
As a result of nano-LC-LTQ Orbitrap MS measurements of tryptically digested Ig bands, we found 54,057 MS peaks in the IgH chain and 56,664 in the IgL chain comparison set from Progenesis. The database (NCBInr) analysis by Mascot identified 1086 peptides for IgH samples. Similarly, (via NCBInr) we identified 920 peptides for IgL samples. Next, the BLASTp algorithm aligned our peptide (experimental) sequences to a database (IMGT) of germline sequences that were present in naive B cells (as described in “Materials and Methods”). The best-matching germline allele was selected from the database. Comparison of the peptide with the best-matching germline allele also revealed which amino acids were most likely mutated during rearrangement and affinity maturation. In this way, from the 1086 IgH peptides, 809 sequences were assigned to the V region, 32 sequences to the J region, and 99 sequences to the C region. Similarly (by IMGT), from the 920 IgL peptides, 722 peptides were assigned to the V region, 49 sequences to the J region, and 99 to the C region. Next, within the IgH variable domain, we were able to assign 41 peptides to CDR1, 128 to CDR2, and 171 to CDR3. Within the IgL variable domain, we were able to assign 78 peptides to CDR1, 233 to CDR2, and 51 to CDR3. A summary of the identification is presented as a flowchart in supplemental Fig. S3. Many of the IgH and IgL peptides in our dataset were unassigned. A possible reason for this might be that rearranged CDR3 sequences do not have enough similarity to the germline sequence to allow for alignment.
We were interested in peptides that were observed only in the MScl group. Based on the presence or absence of these peptides in the MScl group, we found 24 IgH and 26 IgL peptides in the MScl group that could be identified only in the MScl group and not in controls. In contrast, as expected, peptides derived from the Ig C region were equally distributed in both patients and controls (data not shown). We searched for those CDR-related peptides in the group of peptides that were common among multiple MScl patients. Next, we zoomed into the CDR regions to assess for peptides that were shared in at least three MScl patients and absent in the controls. We found nine IgH peptides and six IgL peptides. At a lower threshold (i.e. shared in at least two MScl) we observed 14 IgH peptides and 13 IgL peptides (data not shown). The elucidated results here emphasized the fact that CDR peptides were from MScl patients and were not from three or more controls. As a confirmation step, a second round of assessment was performed by means of two-dimensional view analysis via the Progenesis software package. To check whether the fragmentation spectra of a peptide were similar in all patients in whom that peptide had been identified, we critically scrutinized and assessed the fragmentation spectra of peptides. We found five peptides that were proven to be shared by three or more MScl patients. The exact characteristics are shown in Table II.
Table II. Exclusive presence of common CDR motifs in CSF Ig of MScl patients. Table shows CDR peptide precursor mass, charge, confidence score for the sequence based on the MS/MS spectra, amino acid sequence, location details in Ig structure, expression in controls and MScl patients, gene information, and match score with Ig germline.
| m/z | z | Confidence | CDRsa (peptide by MS/MS) | CDRb | Mutation | Expression |
Germ lined | Gene namee (allele name) | Homology (%) | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MSclc | Control | ||||||||||
| 745.838 | 2 | 64 | QDGSETYYVDSVKa′ | CDR2 | Yes | 6/29 | 0/30 | QDGSEKYYVDSVK | IGHV 3–7 (IGHV 3–7*02) | 92.3 | (14, 33, 34) |
| 759.833 | 2 | 65 | QDGSEEYYVDSVKa′ | CDR2 | Yes | 3/29 | 0/30 | QDGSEKYYVDSVK | IGHV 3–7 (IGHV 3–7*02) | 92.3 | (14, 33, 34) |
| 583.605 | 3 | 43 | IDWDDDKYYSTSLKa″ | CDR2 | No | 4/29 | 0/30 | IDWDDDKYYSTSLK | IGHV 2–70 (IGHV 2–70*01) | 100 | (34, 35) |
| 490.786 | 2 | 47 | LLIHGASNR | CDR2 | Yes | 3/29 | 0/26 | LLIYGASSR | IGKV 3–20 (IGKV 3–20*02) | 77.8 | - |
| 656.830 | 2 | 52 | YNSAPLTFGGGTK | CDR3 | No | 3/29 | 0/26 | YNSAPLTFGGGTK | IGKV 1–27 and IGKJ4 (IGKV 1–27*01 and IGKJ4*01) | 100 | - |
CDR, complementarity determining region (CDR1, CDR2, CDR3); MS/MS, tandem mass spectrometry; z, charge. A CDR peptide was defined as a peptide containing a minimum of three amino acids in the CDR part irrespective of framework length.
a Amino acid sequence of IgH and IgL CDR obtained based on the MS/MS spectra. CDR is underlined; the mutation (or mismatch) in amino acid from the germline is shown in bold. a′ and a″ denote sequences resembling sequences with a different mutation, present exclusively in one patient, and are shown in supplementary Table S2.
b The position of the sequence in the Ig structure is indicated.
c The number of MScl patients in which these shared CDR motifs were found (number of patients sharing motif/total number of patients).
d Sequence translated from the Ig germline and derived from the IMGT database.
e The most homologous IgH and IgL germline gene.
To determine the probability that the exclusive presence of CDR in MScl was not due to chance alone, we permutated the entire dataset repeatedly 5000 times for CDR marker category. Analysis showed that CDR's exclusive presence in MScl was not due to chance in IgH (p = 0.0005) (see supplemental Fig. S4: (A) CDR 2 original occurrence histogram and (B) box plot after randomization) or in IgL (p = 0.0007) (see supplemental Fig. S4: (C) CDR 2 and 3 original occurrence and (D) after randomization as a box plot).
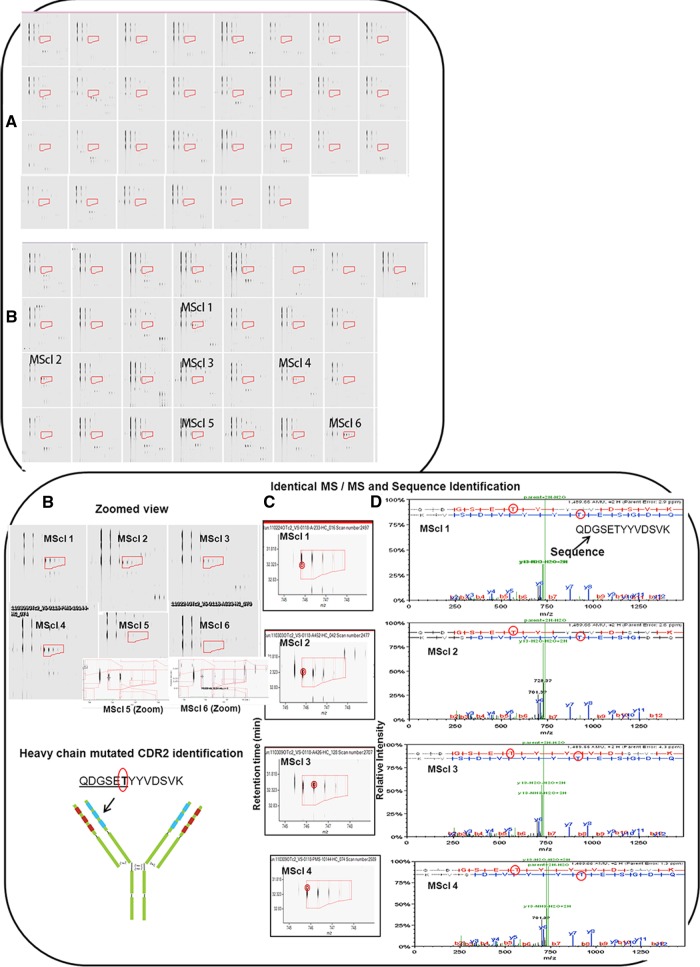
Five peptides were found to be specific to three or more patients in the MScl group. Interestingly, one of the mutated CDR2s commonly used was seen in seven different MScl patients (supplemental Table S1A). IMGT alignment analysis showed that this CDR peptide had different mutations (T, E, and N) or an insertion (F) at the same spot. The QDGSETYYVDSVK (amino acid in bold; mismatch/mutation from Ig germline) peptide was quantified in 6 MScl patients and not in 30 controls (Fig. 4). Furthermore, four out of the six MScl patients were identified via MS/MS (but not controls), providing additional support for proper sequence identification (Fig. 4). Sequence alignment with the human germline sequences (derived from the IMGT database) showed homology to IGHV 3–7, and threonine (T) was found mutated/mismatched from the lysine (K) of the germ line. A similar peptide, QDGSEEYYVDSVK, was identified in three MScl patients (supplemental Fig. S5). Alignment showed the mutation of a glutamic acid (E) at the same spot. In addition to the peptide described above, two CDRs resembling mutations, QDGSETFYVDSVK (the bold italic letter represents the insertion) and QDGSENYYVDSVK, were also observed solely in one MScl patient (supplemental Table S2 and supplemental Fig. S6). Next, the IDWDDDKYYSTSLK peptide was quantified exclusively in four MScl patients (supplemental Fig. S7A). For two of these MScl patients, fragmentation spectra were obtained that showed identical peptide MS/MS results, supporting the robustness of the peptide identification. Alignment analysis showed homology to IGHV 2–70 *01 (CDR2) for these peptides; another identical CDR, IDWDDDKYYTTSLK, with a mutation, was observed solely in one MScl patient (supplemental Table S2 and supplemental Fig. S7B). Supplemental Table S1B shows common uses of the same peptide in five MScl patients. Next, the YNSAPLTFGGGTK peptide was identified exclusively in three patients (supplemental Fig. S8). A homology search showed alignment to the CDR3 of gene IGKV 1–27 *01 and IGKJ 4 *01. Finally, the LLIHGASNR peptide was identified solely in three MScl patients (supplemental Fig. S9). Alignment showed homology to IGKV 3–20 *02 CDR2, and histidine (H) and asparagine (N) were found mutated from the tyrosine (Y) and serine (S) relative to the germ line.
Fig. 4.
QDGSETYYVDSVK mutated CDR2 identification in six MScl patients and not in 30 controls. Peptides were analyzed by Progenesis LC–MS, and comparison between MScl and control groups was performed. After the chromatograms had been aligned in the Progenesis software, two-dimensional profiles were compared with find-expression profiles between the two groups. A, precursor mass of the peptide not found in any of the 30 controls. B, precursor mass of the peptide identified in six out of 29 MScl patients. The region covered in A and B by red boxes/isotope boundaries indicates the area of interest and shows the precursor mass isotopic pattern. The peptide abundance was calculated as the sum of the peak areas within these isotope boundaries. C, example of identical MS/MS fragmentation spectra in different MScl samples. Red circles (left) in the figure indicate the location of the retention time and the mass (m/z) where the MS/MS scan was triggered. Almost identical MS/MS fragmentation spectra were observed in different patients. D, MS/MS spectra containing the m/z values and abundances of peptide ion products. A longer series of contiguous y and b ions shows a higher probability of correct identification. The sequence proposes a mutated CDR2 peptide. Red circles show a mutated (T) amino acid from the germline that was found during alignment using the IMGT database.
DISCUSSION
The main observation of this study was the shared use of identical mutated sequences in CDRs of purified Ig from the CSF of different MScl patients. In contrast, in a set of 30 healthy controls, no overlap in the tryptic peptide sequences of CDRs existed. To the best of our knowledge, this was the first study to use a proteomics approach to analyze and compare the sequences of purified Ig from the CSF of a significant, large set of MScl patients and controls. Most studies until now have investigated clonality at the transcriptome level (8, 24–26). In these previous studies, no common sequences between distinct individuals had been reported. These studies used FACS-based cell sorting methods or CSF cell isolation and ended up with a limited number of CD19+ B cells and/or CD138+ plasma cells (8, 25). Further, they used cloning at the nucleic-acid level by means of PCR-based technologies. Apart from the larger number of patients investigated, this study differs in that we targeted the CSF Ig at the protein level and did not investigate CSF B cells.
Although all MScl CSF samples tested here had signs of elevated intrathecal Ig production, we cannot claim that the public sequences shared here between distinct individuals are responsible for the oligoclonal bands seen in immunoelectric focusing used in routine hospital chemistry. It should be realized that the sophisticated MS technique applied here makes it possible to detect peptide sequences at concentrations far below the threshold for routine immunoelectric focusing (18).
The current paradigm in immunology is that the antigen specificity of B cells is determined via random mechanisms, and therefore one would expect different sequences of the antigen binding CDR in different individuals. This has recently been challenged by a number of observations. First, Scheid et al. cloned 576 new human immunodeficiency virus (HIV) antibodies from four unrelated individuals and found that despite extensive hypermutation, these antibodies shared consensus sequences in both framework and CDR V regions of IgH chains (9). In another study on Sjögren syndrome, secreted human Ro52 antibody from unrelated patients was found to share public V region sequences (27). Finally, our group observed 28 common Ig-derived sequences in paraneoplastic anti-Hu-syndrome that were specific for autoantigen and were found exclusively in samples from single autoantibody-defined clinical neurological entities (12).
Our study in MScl patients appears to follow the same paradigm-challenging pattern. A considerable set of 24 IgH and 26 IgL CDR peptides were exclusively present in MScl patients. We found nine peptides in the IgH set and six peptides shared among at least three MScl patients. These numbers were somewhat higher with a lower threshold for sharing Ig in at least two MScl patients. At the lower threshold (shared in two MScl patients), we observed 14 CDR peptides in IgH and 13 in IgL exclusively in MScl patients. This indicates that under (auto)antigenic pressure there may be common selection mechanisms for the production of intrathecal Ig production by B lymphocytes. The suggestion of shared sequences within different MScl individuals is reminiscent of what has been reported on recruited T lymphocytes in this disease (28). Although the reason remains unclear, it would not be farfetched to speculate that some CDR sequences might better survive the clonal selection process than others, perhaps because of a stronger binding of the three-dimensional structure of the CDR to the antigen. Better insight into common selection mechanisms for (auto)antibodies in MScl and the identification of interindividually shared specific CDR sequences might even deliver markers for subgroup identification.
Apart from focusing on the CDR regions, we also investigated the possible use of common VH and lgL chain families. VH repertoires can be divided into seven families based on sequence similarity (22). Previous studies at the genomic level indicated a skewed use of VH4 in MScl (8, 29). Although it is hard to draw firm conclusions here, it was striking to see the overrepresentation of peptide sequences (p = 0.03) of the VH4 family in MScl patients relative to controls. In addition, we observed a trend toward increased VH3 family usage in MScl patients (p = 0.05). No significant difference in the use of the VK chain was observed between the two groups.
An additional observation here was the increased κ/λ ratios in the MScl group. This observation is in agreement with previous studies (30) using conventional assays for IgL detection.
Compared with the other studies, a limitation of our study is that we do not know the antigen specificity of the V regions identified here. In fact, we do not show complete Ig sequences, because trypsin digestion is needed for this approach. In light of the shared sequences observed in known antigen-specific antibodies in HIV infection, anti-Hu paraneoplastic disease, and Ro52 autoimmunity, it would be of future relevance to investigate the possible use of public V-region sequences in purified specific antibodies against MScl candidate antigens such as anti-MOG, antineurofascin, and anti-KIR 4.1 (6, 31, 32).
In the available previous studies, no attempts were shown to match the identified sequences with those available in public databases such as BLAST (NCBI). We here performed such a cross-check and were surprised to notice that similar sequences were found in other studies investigating B cells from the CSF of MScl patients.
Although the technique used makes it impossible to show the sequence of complete Ig proteins, it is striking that these peptides have also been identified in other MScl studies. For example, our study showed a common CDR usage in seven different patients. This QDGSEKYYVDSVK peptide, which is part of CDR2, belongs to the IGHV 3–7 germline. We found different amino acid mutations (T, E, or N) and an insertion (F) at the same spot (K) as in the germline. QDGSETYYVDSVK and QDGSETFYVDSVK sequences were published in the database from a study assessing intrathecal CSF B-cell sequences in two MScl patients using an RT-PCR approach (33). The peptide QDGSETFYVDSVK was also shown by a different study that sequenced CSF oligoclonal bands in four MScl patients using a proteomics approach (14). Furthermore, gene IGHV 3–7 associated with the same has also been shown in previous studies performed on CSF B cells of MScl patients (14, 33, 34). Next, gene IGHV 2–70, related to IDWDDDKYYSTSLK and IDWDDDKYYTTSLK, has also been described in different studies performed on CSF B cells of MScl patients (34, 35). The peptide IDWDDDKYYSTSLK has no mutation; although it is plausible that this peptide may be found in the larger, healthy population, we presume that this peptide is enriched in the MScl population. Certain V genes may be preferentially selected in the antibody response against certain antigens (repertoire bias) (7). Thus, such peptides, especially as a part of a larger panel of peptides, help in the identification of patient populations even if they are not unique to that population. Next, peptide YNSAPLTFGGGTK does not contain amino acids introduced by somatic hypermutation, but it was generated by V (D) J recombination. Nevertheless, it is unique because of its particular deletion of two nucleotides of the V region in conjunction with one nucleotide of the J region and the lack of any additional N nucleotides. In all, three of the CDRs observed in three or more MScl patients have been linked to MScl in the past.
In studies involving MScl patients, the phenomenon of repertoire bias (7, 8, 29, 36) has been described, whereby specific genes from the germline repertoire are favored in the panel of antibodies that is produced during the immune process. These pressures drive antibodies in convergent directions. Indeed, one might expect many CDR3 sequences, as they are important for the specificity of an antibody. However, two aspects may favor CDR1/2 instead. First, CDR3 is generally highly mutated on the border of V, D, and J germline alleles. As such, it is difficult to identify based on homology to the germline sequences, and it may remain unidentified. Second, it might be that the highly mutated CDR3 is in fact mostly unique to an individual, and that motifs shared between individuals are instead found in the moderately mutated CDR1/2 regions.
To conclude, this proteomic study shows for the first time CDR peptides shared among individual MScl patients but not controls. There was striking overlap with a few CDR peptides identified in other studies that assessed B-cell clonality in the CSF of MScl patients at the nucleic-acid level. Whether such common B-cell responses are indeed driven by autoantigens remains to be determined. It will be of interest to study common V-region use in known autoreactive Igs that appear to play a role in MScl (6).
Supplementary Material
Footnotes
* This work was supported by a program grant provided by the EC 7th Framework Program of Marie Curie Initial Training Network–The United Europeans for the Development of Pharmacogenomics in Multiple Sclerosis (UEPHA-MS grant PITN-GA-2008212877), a grant from Netherlands Multiple Sclerosis Research Foundation to the Rotterdam MS center, ErasMS, the Netherlands Organization for scientific research (ZON-MW, RQH), Hersenstichting Nederland, and Zenith Grant No. 93511034 from the Netherlands Organisation for Scientific Research (NWO).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- BLAST
- basic local alignment search tool
- BLASTp
- basic local alignment search tool for protein
- C
- constant region
- CDR
- complementarity determining region
- CSF
- cerebrospinal fluid
- D
- diversity element of immunoglobulin sequence
- Ig
- immunoglobulin G
- IgH
- Ig heavy chain
- IgL
- Ig light chain
- IMGT
- International ImMunoGeneTics Information System
- J
- joining element of immunoglobulin sequence
- LC–MS
- liquid chromatography–mass spectrometry
- MScl
- multiple sclerosis
- MS/MS
- tandem mass spectrometry
- V
- variable element of immunoglobulin sequence
- VH
- variable region of immunoglobulin heavy chain
- VK
- variable region of immunoglobulin light chain κ.
REFERENCES
- 1. Hauser S. L., Waubant E., Arnold D. L., Vollmer T., Antel J., Fox R. J., Bar-Or A., Panzara M., Sarkar N., Agarwal S., Langer-Gould A., Smith C. H. (2008) B-cell depletion with rituximab in relapsing-remitting multiple sclerosis. N. Engl. J. Med. 358, 676–688 [DOI] [PubMed] [Google Scholar]
- 2. Barr T. A., Shen P., Brown S., Lampropoulou V., Roch T., Lawrie S., Fan B., O'Connor R. A., Anderton S. M., Bar-Or A., Fillatreau S., Gray D. (2012) B cell depletion therapy ameliorates autoimmune disease through ablation of IL-6-producing B cells. J. Exp. Med. 209, 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. von Budingen H. C., Bar-Or A., Zamvil S. S. (2011) B cells in multiple sclerosis: connecting the dots. Curr. Opin. Immunol. 23, 713–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. O'Connor K. C., Appel H., Bregoli L., Call M. E., Catz I., Chan J. A., Moore N. H., Warren K. G., Wong S. J., Hafler D. A., Wucherpfennig K. W. (2005) Antibodies from inflamed central nervous system tissue recognize myelin oligodendrocyte glycoprotein. J. Immunol. 175, 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kuhle J., Pohl C., Mehling M., Edan G., Freedman M. S., Hartung H. P., Polman C. H., Miller D. H., Montalban X., Barkhof F., Bauer L., Dahms S., Lindberg R., Kappos L., Sandbrink R. (2007) Lack of association between antimyelin antibodies and progression to multiple sclerosis. N. Engl. J. Med. 356, 371–378 [DOI] [PubMed] [Google Scholar]
- 6. Srivastava R., Aslam M., Kalluri S. R., Schirmer L., Buck D., Tackenberg B., Rothhammer V., Chan A., Gold R., Berthele A., Bennett J. L., Korn T., Hemmer B. (2012) Potassium channel KIR4.1 as an immune target in multiple sclerosis. N. Engl. J. Med. 367, 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bennett J. L., Haubold K., Ritchie A. M., Edwards S. J., Burgoon M., Shearer A. J., Gilden D. H., Owens G. P. (2008) CSF IgG heavy-chain bias in patients at the time of a clinically isolated syndrome. J. Neuroimmunol. 199, 126–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Owens G. P., Winges K. M., Ritchie A. M., Edwards S., Burgoon M. P., Lehnhoff L., Nielsen K., Corboy J., Gilden D. H., Bennett J. L. (2007) VH4 gene segments dominate the intrathecal humoral immune response in multiple sclerosis. J. Immunol. 179, 6343–6351 [DOI] [PubMed] [Google Scholar]
- 9. Scheid J. F., Mouquet H., Ueberheide B., Diskin R., Klein F., Oliveira T. Y., Pietzsch J., Fenyo D., Abadir A., Velinzon K., Hurley A., Myung S., Boulad F., Poignard P., Burton D. R., Pereyra F., Ho D. D., Walker B. D., Seaman M. S., Bjorkman P. J., Chait B. T., Nussenzweig M. C. (2011) Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science 333, 1633–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. VanDuijn M. M., Dekker L. J., Zeneyedpour L., Smitt P. A., Luider T. M. (2010) Immune responses are characterized by specific shared immunoglobulin peptides that can be detected by proteomic techniques. J. Biol. Chem. 285, 29247–29253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weinstein J. A., Jiang N., White R. A., 3rd, Fisher D. S., Quake S. R. (2009) High-throughput sequencing of the zebrafish antibody repertoire. Science 324, 807–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Maat P., VanDuijn M., Brouwer E., Dekker L., Zeneyedpour L., Luider T., Smitt P. S. (2012) Mass spectrometric detection of antigen-specific immunoglobulin peptides in paraneoplastic patient sera. J. Autoimmun. 38, 354–360 [DOI] [PubMed] [Google Scholar]
- 13. Hoogeboom R., van Kessel K. P., Hochstenbach F., Wormhoudt T. A., Reinten R. J., Wagner K., Kater A. P., Guikema J. E., Bende R. J., van Noesel C. J. (2013) A mutated B cell chronic lymphocytic leukemia subset that recognizes and responds to fungi. J. Exp. Med. 210, 59–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Obermeier B., Mentele R., Malotka J., Kellermann J., Kumpfel T., Wekerle H., Lottspeich F., Hohlfeld R., Dornmair K. (2008) Matching of oligoclonal immunoglobulin transcriptomes and proteomes of cerebrospinal fluid in multiple sclerosis. Nat. Med. 14, 688–693 [DOI] [PubMed] [Google Scholar]
- 15. Cheung W. C., Beausoleil S. A., Zhang X., Sato S., Schieferl S. M., Wieler J. S., Beaudet J. G., Ramenani R. K., Popova L., Comb M. J., Rush J., Polakiewicz R. D. (2012) A proteomics approach for the identification and cloning of monoclonal antibodies from serum. Nat. Biotechnol. 30, 447–452 [DOI] [PubMed] [Google Scholar]
- 16. de Costa D., Broodman I., Vanduijn M. M., Stingl C., Dekker L. J., Burgers P. C., Hoogsteden H. C., Sillevis Smitt P. A., van Klaveren R. J., Luider T. M. (2010) Sequencing and quantifying IgG fragments and antigen-binding regions by mass spectrometry. J. Proteome Res. 9, 2937–2945 [DOI] [PubMed] [Google Scholar]
- 17. Broodman I., de Costa D., Stingl C., Dekker L. J., VanDuijn M. M., Lindemans J., van Klaveren R. J., Luider T. M. (2012) Mass spectrometry analyses of kappa and lambda fractions result in increased number of complementarity-determining region identifications. Proteomics 12, 183–191 [DOI] [PubMed] [Google Scholar]
- 18. Dekker L. J., Zeneyedpour L., Brouwer E., van Duijn M. M., Sillevis Smitt P. A., Luider T. M. (2011) An antibody-based biomarker discovery method by mass spectrometry sequencing of complementarity determining regions. Anal. Bioanal. Chem. 399, 1081–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Teunissen C. E., Petzold A., Bennett J. L., Berven F. S., Brundin L., Comabella M., Franciotta D., Frederiksen J. L., Fleming J. O., Furlan R., Hintzen R. Q., Hughes S. G., Johnson M. H., Krasulova E., Kuhle J., Magnone M. C., Rajda C., Rejdak K., Schmidt H. K., van Pesch V., Waubant E., Wolf C., Giovannoni G., Hemmer B., Tumani H., Deisenhammer F. (2009) A consensus protocol for the standardization of cerebrospinal fluid collection and biobanking. Neurology 73, 1914–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Polman C. H., Reingold S. C., Edan G., Filippi M., Hartung H. P., Kappos L., Lublin F. D., Metz L. M., McFarland H. F., O'Connor P. W., Sandberg-Wollheim M., Thompson A. J., Weinshenker B. G., Wolinsky J. S. (2005) Diagnostic criteria for multiple sclerosis: 2005 revisions to the “McDonald Criteria.” Ann. Neurol. 58, 840–846 [DOI] [PubMed] [Google Scholar]
- 21. Olsen J. V., de Godoy L. M., Li G., Macek B., Mortensen P., Pesch R., Makarov A., Lange O., Horning S., Mann M. (2005) Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol. Cell. Proteomics 4, 2010–2021 [DOI] [PubMed] [Google Scholar]
- 22. Cook G. P., Tomlinson I. M. (1995) The human immunoglobulin VH repertoire. Immunol. Today 16, 237–242 [DOI] [PubMed] [Google Scholar]
- 23. Ehrenmann F., Kaas Q., Lefranc M. P. (2010) IMGT/3Dstructure-DB and IMGT/DomainGapAlign: a database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res. 38, D301–D307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Owens G. P., Ritchie A. M., Burgoon M. P., Williamson R. A., Corboy J. R., Gilden D. H. (2003) Single-cell repertoire analysis demonstrates that clonal expansion is a prominent feature of the B cell response in multiple sclerosis cerebrospinal fluid. J. Immunol. 171, 2725–2733 [DOI] [PubMed] [Google Scholar]
- 25. Colombo M., Dono M., Gazzola P., Roncella S., Valetto A., Chiorazzi N., Mancardi G. L., Ferrarini M. (2000) Accumulation of clonally related B lymphocytes in the cerebrospinal fluid of multiple sclerosis patients. J. Immunol. 164, 2782–2789 [DOI] [PubMed] [Google Scholar]
- 26. Qin Y., Duquette P., Zhang Y., Talbot P., Poole R., Antel J. (1998) Clonal expansion and somatic hypermutation of V(H) genes of B cells from cerebrospinal fluid in multiple sclerosis. J. Clin. Invest. 102, 1045–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arentz G., Thurgood L. A., Lindop R., Chataway T. K., Gordon T. P. (2012) Secreted human Ro52 autoantibody proteomes express a restricted set of public clonotypes. J. Autoimmun. 39, 466–470 [DOI] [PubMed] [Google Scholar]
- 28. Lee S. J., Wucherpfennig K. W., Brod S. A., Benjamin D., Weiner H. L., Hafler D. A. (1991) Common T-cell receptor V beta usage in oligoclonal T lymphocytes derived from cerebrospinal fluid and blood of patients with multiple sclerosis. Ann. Neurol. 29, 33–40 [DOI] [PubMed] [Google Scholar]
- 29. Owens G. P., Kraus H., Burgoon M. P., Smith-Jensen T., Devlin M. E., Gilden D. H. (1998) Restricted use of VH4 germline segments in an acute multiple sclerosis brain. Ann. Neurol. 43, 236–243 [DOI] [PubMed] [Google Scholar]
- 30. Link H., Zettervall O. (1970) Multiple sclerosis: disturbed kappa:lambda chain ratio of immunoglobulin G in cerebrospinal fluid. Clin. Exp. Immunol. 6, 435–438 [PMC free article] [PubMed] [Google Scholar]
- 31. Mathey E. K., Derfuss T., Storch M. K., Williams K. R., Hales K., Woolley D. R., Al-Hayani A., Davies S. N., Rasband M. N., Olsson T., Moldenhauer A., Velhin S., Hohlfeld R., Meinl E., Linington C. (2007) Neurofascin as a novel target for autoantibody-mediated axonal injury. J. Exp. Med. 204, 2363–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mayer M. C., Hohlfeld R., Meinl E. (2012) Viability of autoantibody-targets: how to tackle pathogenetic heterogeneity as an obstacle for treatment of multiple sclerosis. J. Neurol. Sci. 319, 2–7 [DOI] [PubMed] [Google Scholar]
- 33. Colombo M., Dono M., Gazzola P., Chiorazzi N., Mancardi G., Ferrarini M. (2003) Maintenance of B lymphocyte-related clones in the cerebrospinal fluid of multiple sclerosis patients. Eur. J. Immunol. 33, 3433–3438 [DOI] [PubMed] [Google Scholar]
- 34. von Budingen H. C., Harrer M. D., Kuenzle S., Meier M., Goebels N. (2008) Clonally expanded plasma cells in the cerebrospinal fluid of MS patients produce myelin-specific antibodies. Eur. J. Immunol. 38, 2014–2023 [DOI] [PubMed] [Google Scholar]
- 35. Owens G. P., Bennett J. L., Lassmann H., O'Connor K. C., Ritchie A. M., Shearer A., Lam C., Yu X., Birlea M., DuPree C., Williamson R. A., Hafler D. A., Burgoon M. P., Gilden D. (2009) Antibodies produced by clonally expanded plasma cells in multiple sclerosis cerebrospinal fluid. Ann. Neurol. 65, 639–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Baranzini S. E., Jeong M. C., Butunoi C., Murray R. S., Bernard C. C., Oksenberg J. R. (1999) B cell repertoire diversity and clonal expansion in multiple sclerosis brain lesions. J. Immunol. 163, 5133–5144 [PubMed] [Google Scholar]
- 37. Stoop M. P., Coulier L., Rosenling T., Shi S., Smolinska A. M., Buydens L., Ampt K., Stingl C., Dane A., Muilwijk B., Luitwieler R. L., Sillevis Smitt P. A., Hintzen R. Q., Bischoff R., Wijmenga S. S., Hankemeier T., van Gool A. J., Luider T. M. (2010) Quantitative proteomics and metabolomics analysis of normal human cerebrospinal fluid samples. Mol. Cell. Proteomics 9, 2063–2075 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.