Abstract
Objectives
Generalizable, high-throughput phenotyping methods based on supervised machine learning (ML) algorithms could significantly accelerate the use of electronic health records data for clinical and translational research. However, they often require large numbers of annotated samples, which are costly and time-consuming to review. We investigated the use of active learning (AL) in ML-based phenotyping algorithms.
Methods
We integrated an uncertainty sampling AL approach with support vector machines-based phenotyping algorithms and evaluated its performance using three annotated disease cohorts including rheumatoid arthritis (RA), colorectal cancer (CRC), and venous thromboembolism (VTE). We investigated performance using two types of feature sets: unrefined features, which contained at least all clinical concepts extracted from notes and billing codes; and a smaller set of refined features selected by domain experts. The performance of the AL was compared with a passive learning (PL) approach based on random sampling.
Results
Our evaluation showed that AL outperformed PL on three phenotyping tasks. When unrefined features were used in the RA and CRC tasks, AL reduced the number of annotated samples required to achieve an area under the curve (AUC) score of 0.95 by 68% and 23%, respectively. AL also achieved a reduction of 68% for VTE with an optimal AUC of 0.70 using refined features. As expected, refined features improved the performance of phenotyping classifiers and required fewer annotated samples.
Conclusions
This study demonstrated that AL can be useful in ML-based phenotyping methods. Moreover, AL and feature engineering based on domain knowledge could be combined to develop efficient and generalizable phenotyping methods.
Keywords: Active Learning, Phenotyping Algorithm, Machine Learning, Natural Language Processing, Electronic Health Records
Introduction
In the past decade, the increasing adoption of electronic health records (EHR) in the healthcare industry has made longitudinal practice-based clinical data available for clinical, genomic, and translational studies.1–4 One of the main challenges of EHR-based research is accurately and efficiently to extract phenotypic information (eg, records of disease status and treatments of patients) from heterogeneous clinical data. Manual chart review by domain experts can accurately identify disease cohorts, but it is time-consuming and costly. Specific disease phenotyping algorithms consider information from multiple sources including billing codes, clinical documents, laboratory data, and medication exposures. They have been developed and performed well, typically using combinations of Boolean logic with multiple modes of phenotype information.1 Generalizable high-throughput phenotyping methods based on supervised machine learning (ML) algorithms have recently gained greater attention.5 6 One limitation of ML-based phenotyping algorithms is that they often require large numbers of annotated training sets, which are costly and time-consuming to create. The goal of this study was to address this challenge by applying active learning (AL) approaches to ML-based phenotyping, which attempts to build better ML models using fewer annotated samples by intelligently selecting samples for annotation.
In this study, we used an uncertainty sampling approach with support vector machines (SVM) to generate phenotyping algorithms for three diseases: rheumatoid arthritis (RA), colorectal cancer (CRC), and venous thromboembolism (VTE). Each of these is a relatively common healthcare problem associated with significant morbidity and mortality, and has been the subject of genetic association studies, including some using EHR-linked biobanks.7 8 We have previously published phenotyping algorithms for two of these diseases, RA9–11 and CRC,12 using combinations of natural language processing (NLP), deterministic, and ML algorithms. We assessed the effect of AL on two different types of feature sets: unrefined features that contain at least all clinical concepts from notes and billing codes; and refined features that were selected by domain experts. Our evaluation showed that AL could reduce the number of annotated samples by up to 68% for a goal area under the curve (AUC) of 0.95 when using unrefined features. However, the improvement of AL on refined features was more limited.
Background
EHR contain detailed longitudinal information about patients’ disease diagnosis, prognosis, treatment, and response and have become an appealing data source for clinical and translational research. For example, institutions such as those in the electronic medical records and genomics (eMERGE) network5 have linked EHR data with DNA biobanks to facilitate genomic research.4 Importantly, studies have shown that once an EHR dataset has been genotyped, it can also be reused to analyze other phenotypes.13 As genotyping cost gets lower, phenotyping, the process by which one identifies cases and controls for a given disease or trait of interest (ie, the phenotype), becomes the main obstacle for EHR-based genomic research. Manual chart review to extract phenotypic information for clinical studies is not only costly and time-consuming, but also infeasible when a study involves a large number of subjects.
Automated phenotyping methods for EHR data have been extensively studied. For disease cohort identification, early studies have focused on billing codes such as International Classification of Disease (ICD) codes, but researchers often found that billing codes alone were not sufficiently granular or accurate enough.14 15 Current disease phenotyping methods often consider information from multiple sources, including billing codes, clinical text (via NLP or simple text searches), laboratory data, and medication exposures to identify cases and controls accurately in selective populations.1 Clinical documents that contain much more details about patients’ conditions have been recognized as a valuable source for phenotyping, in addition to coded data. Therefore, NLP methods16 that can extract clinical concepts from providers’ notes have been used in various phenotyping algorithms.1 For example, cTAKES17 has been used to discover peripheral arterial disease cases from radiology notes,18 and combinations of KnowledgeMap19 with section identification using SecTag20 have been used to extract additional information from clinical narratives to help identify RA patients21 and those with normal cardiac conduction.9
The eMERGE network is developing phenotyping algorithms for approximately 40 diseases and traits, including 12 currently publicly available algorithms for diseases such as peripheral arterial disease, normal cardiac conduction, cataracts, and type 2 diabetes (available at PheKB.org). They are rule-based algorithms and rely heavily on domain experts to define specific criteria for each disease. For example, Kho et al22 developed rules that combine diagnoses codes, medications, and A1C laboratory test to define type 2 diabetes patients. These algorithms achieved high accuracy and good portability across three institutions and replicated known genetic associations well. However, this approach requires significant interaction between domain experts and informaticians to create algorithms for each disease, which limits its scalability to different phenotypes. Therefore, high-throughput phenotyping algorithms that are generalizable to different diseases are of great interest to both the clinical and informatics community.23
Recent studies showed that supervised ML algorithms could potentially offer a generalizable approach for phenotyping. Wei et al6 developed an SVM classifier to identify a type 2 diabetes cohort using all concepts extracted from clinical notes, achieving F-scores over 0.95. Carroll et al9 applied SVM to identify RA cases and showed better performance than a previously published deterministic algorithm. More interestingly, their study showed that when the sample size was large enough, the SVM classifier trained on unrefined features (eg, all concepts from notes and ICD 9 codes) achieved similar performance as the classifier trained on refined features that were manually selected by domain experts. In another study, Carroll et al11 also demonstrated that the ML-based phenotyping method has good portability to identify RA from three EHR systems. These findings suggest that it is possible to develop generalizable phenotyping approaches with minimum domain knowledge-based features by using ML methods.
However, to achieve high performance, ML-based phenotyping models often require large numbers of annotated samples, which are costly to develop. AL24 actively selects the samples judged to be the ‘most informative’ for annotation (instead of random selection) when training a ML-based phenotyping algorithm. In the biomedical domain, AL has been applied to many classification tasks, including biomedical text classification,25 26 information extraction,27 imaging classification,28 gene expression analysis,29 etc. However, its utility in phenotyping has not been explored. Our hypothesis is that AL can reduce annotation effort while maintaining or improving the quality of ML-based phenotyping models. The pool-based AL framework25 with uncertainty sampling algorithm30 has been shown to be practical and efficient in domains with a large sample size but high annotation costs.
In this study, we assessed the use of AL in identifying three phenotypes: RA, CRC, and VTE, all of which are important human diseases with active ongoing clinical and genomic studies. RA is the most common inflammatory arthritis, affecting 0.5–1% of the world's population.31 CRC is the fourth most common cancer and the second leading cause of cancer death in the USA.32 VTE, also a major cause of mortality including sudden death, has an incidence of 7.1–11.7 persons per 10 000 person-years for community residents.33 34
Methods
Datasets
In this study, we used existing annotated datasets for RA, CRC, and VTE. We have developed ML-based phenotyping models for RA,9–11 CRC,12 and VTE (a manuscript is in preparation). Table 1 shows the distributions of three datasets with respect to sample size, case/control sample ratio, demographics, and feature dimension. For more details about the construction of these datasets, please refer to our previous publications.9 12
Table 1.
Sample size, case/control distribution, and demographics of RA, CRC, and VTE datasets
| Phenotypes | RA | CRC | VTE |
|---|---|---|---|
| Sample size | 376 | 300 | 703 |
| Case samples (%) | 185 (49) | 121 (40) | 502 (71) |
| Number of women (%) | |||
| Cases | 141 (76) | 54 (43) | 269 (54) |
| Controls | 148 (77) | 92 (51) | 104 (52) |
| Mean age (SD) | |||
| Cases | 52.9 (13.1) | 69.8 (12.3) | 63.6 (15.4) |
| Controls | 56.2 (16.5) | 63.1 (18.1) | 60.9 (16.1) |
CRC, colorectal cancer; RA, rheumatoid arthritis; VTE, venous thromboembolism.
ML-based phenotyping method
We used SVM35 36 to build supervised phenotyping models for all three phenotyping tasks. The parameters of SVM classifiers, such as kernels and regularization, were pre-selected based on previous research.9–11 We used the predicted distance of a sample to the SVM hyperplane in the AL querying algorithm for sample selection (described in the following section).
We investigated two different types of feature sets for each phenotype: unrefined features that included at least all billing codes (ICD-9, Current Procedural Terminology, etc.) and NLP-derived unified medical language system (UMLS) concept unique identifiers from clinical notes; and refined features, which included billing codes and UMLS concepts highly relevant to the specific phenotypes, as selected by domain experts. The NLP tools included KnowledgeMap concept identifier37 with SecTag,20 MedLEE,38–40 and MedEx.41 Table 2 summarizes the details of unrefined and refined features for RA, CRC, and VTE.
Table 2.
Unrefined and refined feature sets for RA, CRC, and VTE
| Task | Feature set | Description | No of features | Details |
|---|---|---|---|---|
| RA | Unrefined | All ICD-9 codes; all UMLS CUI, which are neither negated nor associated with other people; all medication names | 13 043 | Carroll et al9 |
| Refined | ICD-9 codes 714.*, 696.0 and 710.0; selected CUI of RA and related diseases; medications commonly used in treating RA | 23 | ||
| CRC | Unrefined | All ICD-9 and CPT billing codes and all UMLS CUI, which are neither negated nor associated with other people | 18 059 | Xu et al12 |
| Refined | CRC related ICD-9 and CPT codes and selected UMLS CUI related to CRC | 12 | ||
| VTE | Unrefined | All ICD-9 codes; all UMLS CUI, which are neither negated nor associated with other people; and all medication names | 13 862 | Online supplementary tables S1 and S2 |
| Refined | ICD-9 codes 453.*, 415.*, and V12.51; selected VTE-related CUI; medications including ‘lovenox’, ‘heparin’, ‘warfarin’, and ‘coumadin’ | 56 |
CPT, Current Procedural Terminology; CRC, colorectal cancer; CUI, concept unique identifiers; ICD, International Classification of Disease; RA, rheumatoid arthritis; UMLS, unified medical language system; VTE, venous thromboembolism.
AL for ML-based phenotyping
When building ML classifiers for phenotyping, the conventional method is to select a set of samples randomly for annotation, which is also called passive learning (PL). On the contrary, AL uses a querying algorithm actively to select the most informative samples for annotation in an iterative and interactive fashion. In this study, we simulated a pool-based AL strategy25 in ML-based phenotyping tasks using existing datasets. We assumed that all samples of a phenotype (eg, 376 samples for RA) were unlabeled and they formed the pool for AL. We then took the following steps:
Initialize—we randomly selected two samples from the pool (one control and one case) and used them to build the initial ML model.
Predict—we used the trained ML model to predict remaining samples in the pool.
Query—we used the uncertainty sampling algorithm30 to find most informative samples, which are samples that have the most uncertain predicted labels by the ML model. For binary SVM classifiers, uncertainty sampling-based algorithm queries the samples that are closest to the hyperplane, which separates cases and controls. We queried one sample in each iteration.
Annotate and re-train the ML model—we annotated the next most informative sample from step 3 (by assigning labels from gold standard) and re-built the ML model by combining the new training sample with previous ones.
Repeat and stop—we repeated steps 2–4 until the stop criterion was met. In this study, the learning process stopped when the unlabeled pool was empty.
For PL, we ran the same procedure using random sampling as its querying algorithm.
Evaluation
The AL experiments were conducted using fivefold stratified cross-validation, which controls the percentage of positive cases in each fold to be similar to the entire dataset. In each iteration, samples in four folds were used as the pool for AL and the remaining fold was used as the test set for independent evaluation of the phenotyping model. For each phenotype, method, and feature set, we generated learning curves by plotting the AUC score of a classifier on the test set as a function of the number of annotated training samples. The area under the learning curve (ALC) score was used to compare different learning curves.
As a baseline companion, a PL curve was generated by using random sampling and fivefold cross-validation. To improve the expectation estimate of the PL curves, this process was repeated 10 times and averaged. The final PL curve, serving as the baseline result, would compare with other learning curves generated by AL approaches. To evaluate whether the global performance of AL is significantly better than PL, the Wilcoxon rank sum test was performed to test the null hypothesis that the medians of AUC scores in the learning curves by AL and PL are equal.
Results
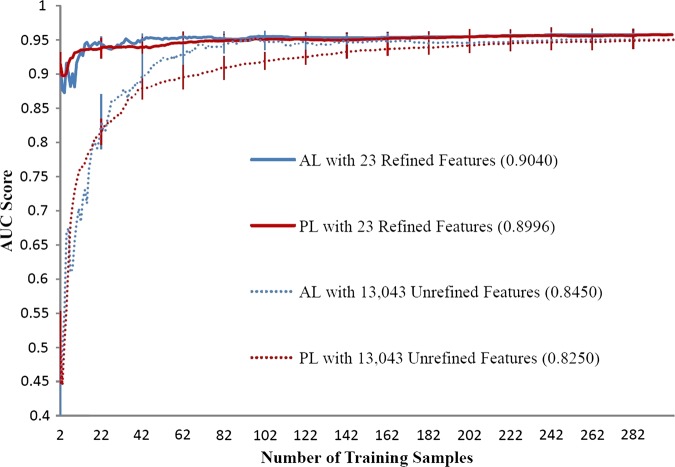
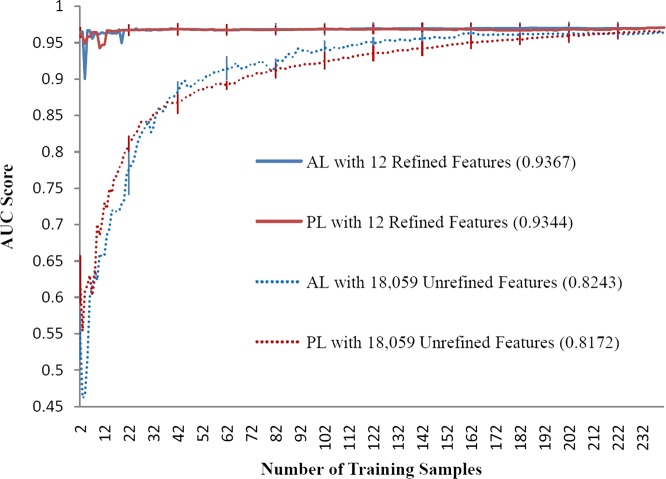
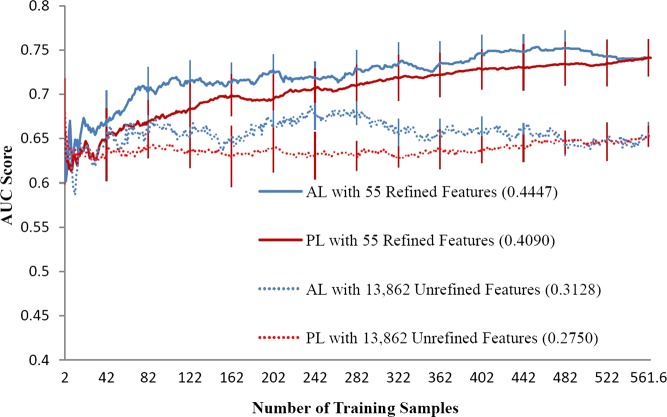
Figures 1–3 illustrate the AL and PL learning curves using refined or unrefined feature sets for RA, CRC, and VTE. The ALC score of each learning curve is shown in the legend of each figure. In all scenarios, ALC scores of AL learning curves were better than ALC scores of corresponding PL learning curves, indicating that AL is helpful in ML-based phenotyping methods. For all phenotypes, the learning curve using the refined feature set always performed better than the learning curve using the unrefined feature set, indicating that domain knowledge also improves phenotyping algorithm performance. AL improved ALC more when unrefined features were used than when refined features were used. For example, the improvement on the ALC score was 0.0044 (AL vs PL) for the RA refined feature set, compared to 0.0200 for the RA unrefined feature set.
Figure 1.
Learning curves of area under the curve (AUC) score for rheumatoid arthritis phenotyping by support vector machines. AL, active learning; PL, passive learning.
Figure 2.
Learning curves of area under the curve (AUC) score for colorectal cancer phenotyping by support vector machines. AL, active learning; PL, passive learning.
Figure 3.
Learning curves of area under the curve (AUC) score for venous thromboembolism phenotyping by support vector machines. AL, active learning; PL, passive learning.
Table 3 shows the p values for three phenotypes using refined and unrefined feature sets. AL significantly outperformed PL in all circumstances. Furthermore, the ML-based phenotyping method showed different optimal performance for the three different diseases. When all training samples were used, the SVM classifier achieved an optimal AUC of greater than 0.95 for RA and CRC. For VTE, the best performing model only achieved an AUC of 0.75 for VTE, suggesting that VTE is a more challenging phenotype.
Table 3.
p Values of the Wilcoxon rank sum test for the comparison of learning curves between AL and PL
| Phenotypes | Refined set | Unrefined set |
|---|---|---|
| RA | 3.2×10−11 | 1.6×10−18 |
| CRC | 1.2×10−22 | 4.4×10−8 |
| VTE | 9.8×10−29 | 1.2×10−129 |
AL, active learning; CRC, colorectal cancer; PL, passive learning; RA, rheumatoid arthritis; VTE, venous thromboembolism.
To illustrate further how AL helps improve ML models and reduces annotation cost, we compared the number of required samples for AL or PL in order to reach a specific AUC score (table 4). For RA, AL using the unrefined set achieved 0.95 AUC by training on 96 annotated samples, while PL needed 298 training samples (a 68% reduction). For the refined feature set, AL needed 42 samples versus 97 for PL (a 57% reduction). Similar results were observed for CRC and VTE. The maximum annotation reduction was also observed in VTE when refined features were used: to achieve an AUC of 0.70, AL needed 69 samples while PL required 213 samples (a 68% reduction). CRC demonstrated a smaller improvement in performance, with a 23% reduction in samples needed to review for an AUC of 0.95. Supplementary table S (available online only) shows the AUC scores with SE of AL versus PL for both refined and unrefined feature sets when different fixed numbers of training samples were used. For VTE, AL always showed better performance than PL at any sample size. For RA and CRC, the performance of AL and PL were close when the refined feature set was used. However, when the unrefined feature set was used, the performance of AL was worse than PL at small sample sizes (eg, 20–40 samples), but the performance of AL was better than PL with more samples (eg, 60–100 samples). These patterns could also be observed from the learning curves.
Table 4.
Performance comparisons of AL versus PL in annotation cost when AUC is fixed in RA, CRC, and VTE phenotyping tasks using both unrefined and refined feature sets
| AUC | ||||||||
|---|---|---|---|---|---|---|---|---|
| Phenotype+feature set | 0.60 | 0.65 | 0.70 | 0.75 | 0.80 | 0.85 | 0.90 | 0.95 |
| RA | ||||||||
| Unrefined set | ||||||||
| AL | 7 | 9 | 13 | 16 | 21 | 27 | 44 | 96 |
| PL | 5 | 6 | 7 | 10 | 18 | 32 | 70 | 298 |
| Refined set | ||||||||
| AL | – | – | – | – | – | – | 8 | 42 |
| PL | – | – | – | – | – | – | 5 | 95 |
| CRC | ||||||||
| Unrefined set | ||||||||
| AL | 6 | 10 | 14 | 21 | 25 | 33 | 53 | 124 |
| PL | 4 | 9 | 11 | 16 | 21 | 32 | 68 | 161 |
| Refined et | ||||||||
| AL | – | – | – | – | – | – | – | 5 |
| PL | – | – | – | – | – | – | – | 13 |
| VTE | ||||||||
| Unrefined set | ||||||||
| AL | 2 | 559 | – | – | – | – | – | – |
| PL | 2 | 559 | – | – | – | – | – | – |
| Refined set | ||||||||
| AL | 2 | 17 | 69 | 492 | – | – | – | – |
| PL | 2 | 42 | 213 | * | – | – | – | – |
*For PL on VTE with refined set, SVM did not achieve 0.75 AUC from the entire learning curve; the best AUC was 0.74 when all annotated samples were used.
AL, active learning; AUC, area under the curve; CRC, colorectal cancer; PL, passive learning; RA, rheumatoid arthritis; SVM, support vector machine; VTE, venous thromboembolism.
Discussion
In this study, we evaluated the impact of AL for ML-based phenotyping methods using physician-reviewed datasets for three different disease cohorts. AL effectively reduced the needed annotation set size and produced better classification models. This finding suggests that AL could be useful for developing high-throughput phenotyping methods using ML. To the best of our knowledge, this is the first study to evaluate AL for EHR phenotyping tasks. In addition, our study also revealed that some phenotypes are more challenging using simple or expert-derived features and demonstrated the value of expert-refined features for ML-based phenotyping algorithms.
As shown in figures 1–3, the best performance for VTE (AUC 0.75) was much worse than that of RA and CRC (AUC above 0.95), indicating that the identification of VTE was harder than RA and CRC. VTE may be more difficult to identify than the other phenotypes for several reasons. First, VTE is an acute disease such that the features based on the count of codes are not as efficient as the ones for chronic diseases, such as RA and CRC. Moreover, as VTE is a life-threatening disease requiring urgent treatment, a provider may include a bill for VTE and start appropriate medications before radiographic verification of the diagnosis. In addition, VTE codes and concepts are frequently mentioned in clinical documents as a ‘rule out’ differential diagnosis, or for prophylaxis (often found without the words ‘prophylaxis’). Therefore, false positives in ICD-9 codes, NLP-derived concepts, and even medications may occur for VTE. More advanced NLP techniques and patient modeling may allow the generation of more reliable features in ML-based methods. An interesting observation was that domain expert-selected features seemed to play a more critical role in difficult phenotypes than in easy phenotypes. In the example of VTE, the best AUC using refined features was much higher than the best AUC using unrefined features, even when all samples were used, but for RA and CRC, the best AUC for refined and unrefined features were similar when all samples were used. This finding suggests that the complexity of phenotypes needs to be considered when developing ML-based algorithms for specific diseases. More phenotypes need to be investigated to draw generalizable conclusions.
Domain expert-refined features dramatically improved the performance of ML-based phenotyping models. For RA and CRC, SVM classifiers using refined features could reach high AUC (over 0.90) with only one or two dozens of annotated samples. Therefore, efficient methods to select relevant features for specific phenotypes are important and need further investigation. However, selecting relevant features for specific phenotypes may sometimes not always be a trivial task, even for domain experts. Many samples may have to be reviewed before relevant features can be summarized. One possible solution is to combine automatic feature selection methods, such as HITON,42 with expert review to speed up the identification of relevant features.
AL reduced the number of annotated samples required to achieve optimal performance for unrefined features. Therefore, instead of feature engineering, another alternative solution for building efficient phenotyping models is to improve AL for unrefined features. We noticed that AL did not perform well with small numbers of training samples. In the early stages of RA and CRC learning curves using unrefined features, the AL learning curves were below PL ones and their amplitude of oscillation was high. A possible explanation is that the SVM models were not able to perform well with small sample sizes and high feature dimensionality. Moreover, uncertainty sampling relies on the quality of the model. In the early stage, the selected samples were not necessarily good because of a poor SVM model. In difficult phenotypes such as VTE, AL may probably not replace the role of defining relevant features. To achieve optimal performance for difficult phenotypes, we will have to combine feature engineering with AL approaches. It is also possible that a new class of features or modeling of features (eg, taking into account the temporality or persistence of a concept, for instance) will engender superior performance on more challenging phenotypes such as VTE.
This study has limitations. Uncertainty-based AL is very sensitive to the classifier and its parameters. In our study, the parameters of SVM, such as kernels and regularization, were pre-selected based on the previous research. Given a new task without previous knowledge about optimal parameters, a strategy that considers automatic parameter selection in different stages of AL may be beneficial. For example, to avoid over-fitting in the early stage when a few samples were used in training, we may use a simple model, such as linear kernel, with a high cost to penalize over-fitting. In addition, this study assumed that each sample requires the same effort to annotate. In reality, different samples take different amounts of time to annotate (eg, based on record size or clinical factors). Cost-sensitive AL, which also considers annotation time, is an interesting additional topic to consider for ML-based phenotyping tasks. In the future, we also plan to investigate other querying algorithms such as query-by-committee,43 expected model change,44 expected error reduction,45 and density-based sampling,27 as well as to develop a real-world AL-enabled system for phenotyping tasks.
Conclusion
In this study, we applied AL to ML-based phenotyping methods for RA, CRC, and VTE, and showed that AL could reduce annotation cost while maintaining high classification performance. In addition, we demonstrated that expert-refined features were also important and should be integrated with active leaning for developing generalizable and high performance phenotyping methods.
Footnotes
Contributors: YC, HX and JCD designed methods and experiments; RJC provided the dataset for the RA phenotyping task; AS generated the dataset for the CRC phenotyping task; ERMH and RJC guided the dataset generation for the VTE phenotyping task; AEE and JCD were case reviewers for RA; ERMH and JCD were reviewers for VTE; YC conducted active learning experiments for all tasks; YC, HX and JCD interpreted the results and drafted the main manuscript. All authors have approved the manuscript.
Funding: This study is supported in part by grants from the National Library of Medicine (R01-LM010685), the National Cancer Institute (R01-CA141307), the National Institute of General Medical Sciences (R01-GM102282), and patient-centered cognitive support under the Strategic Health IT Advanced Research Projects Program from the Office of the National Coordinator for Health Information Technology (10510592). The synthetic derivative, from which the annotated records were derived, is supported by National Center for Research Resources UL1 RR024975, which is now at the National Center for Advancing Translational Sciences, 2 UL1 TR000445.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Denny JC. Chapter 13: mining electronic health records in the genomics era. PLoS Comput Biol 2012;8:e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prokosch HU, Ganslandt T. Perspectives for medical informatics. Reusing the electronic medical record for clinical research. Methods Inf Med 2009;48:38–44 [PubMed] [Google Scholar]
- 3.Tannen RL, Weiner MG, Xie DW. Use of primary care electronic medical record database in drug efficacy research on cardiovascular outcomes: comparison of database and randomised controlled trial findings. BMJ 2009;338:b81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarty CA, Chisholm RL, Chute CG, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.eMERGE Network. https://www.mc.vanderbilt.edu/victr/dcc/projects/acc/index.php/Main_Page.
- 6.Wei WQ, Tao C, Jiang G, et al. A high throughput semantic concept frequency based approach for patient identification: a case study using type 2 diabetes mellitus clinical notes. AMIA Annu Symp Proc 2010;2010:857–61 [PMC free article] [PubMed] [Google Scholar]
- 7.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA 2009;106:9362–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurreeman F, Liao K, Chibnik L, et al. Genetic basis of autoantibody positive and negative rheumatoid arthritis risk in a multi-ethnic cohort derived from electronic health records. Am J Hum Genet 2011;88:57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll RJ, Eyler AE, Denny JC. Naive Electronic health record phenotype identification for rheumatoid arthritis. AMIA Annu Symp Proc 2011;2011:189–96 [PMC free article] [PubMed] [Google Scholar]
- 10.Ritchie MD, Denny JC, Crawford DC, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet 2010;86:560–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc 2012;19:e162–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H, Fu Z, Shah A, et al. Extracting and integrating data from entire electronic health records for detecting colorectal cancer cases. AMIA Annu Symp Proc 2011;2011:1564–72 [PMC free article] [PubMed] [Google Scholar]
- 13.Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet 2011;89:529–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li L, Chase HS, Patel CO, et al. Comparing ICD9-encoded diagnoses and NLP-processed discharge summaries for clinical trials pre-screening: a case study. AMIA Annu Symp Proc 2008:404–8 [PMC free article] [PubMed] [Google Scholar]
- 15.Elkin PL, Ruggieri AP, Brown SH, et al. A randomized controlled trial of the accuracy of clinical record retrieval using SNOMED-RT as compared with ICD9-CM. AMIA Annu Symp Proc 2001:159–63 [PMC free article] [PubMed] [Google Scholar]
- 16.Meystre SM, Savova GK, Kipper-Schuler KC, et al. Extracting information from textual documents in the electronic health record: a review of recent research. Yearb Med Inform 2008:128–44 [PubMed] [Google Scholar]
- 17.Savova GK, Masanz JJ, Ogren PV, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc 2010;17:507–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savova GK, Fan J, Ye Z, et al. Discovering peripheral arterial disease cases from radiology notes using natural language processing. AMIA Annu Symp Proc 2010;2010:722–6 [PMC free article] [PubMed] [Google Scholar]
- 19.Denny JC, Smithers JD, Miller RA, et al. “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc 2003;10:351–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denny JC, Spickard A, III, Johnson KB, et al. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc 2009;16:806–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao KP, Cai TX, Gainer V, et al. Electronic medical records for discovery research in rheumatoid arthritis. Arthritis Care Res 2010;62:1120–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc 2012;19:212–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc 2013;20:117–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Settles B. Active learning literature survey. University of Wisconsin-Madison; 2009, Computer Sciences Technical Report 1648 [Google Scholar]
- 25.Lewis DD, Gale WA. A sequential algorithm for training text classifiers. Proceedings of the ACM SIGIR Conference on Research and Development in Information Retrieval; 1994:3–12 [Google Scholar]
- 26.Tong S, Koller D. Support vector machine active learning with applications to text classification. J Machine Learn Res 2002;2:45–66 [Google Scholar]
- 27.Settles B, Craven M. An analysis of active learning strategies for sequence labeling tasks. Proceedings of the Conference on Empirical Methods in Natural Language Processing (EMNLP); 2008:1069–78 [Google Scholar]
- 28.Tong S, Chang E. Support vector machine active learning for image retrieval. Proceedings of the ACM International Conference on Multimedia; 2001:107–18 [Google Scholar]
- 29.Liu Y. Active learning with support vector machine applied to gene expression data for cancer classification. J Chem Inform Comput Sci 2004;44:1936–41 [DOI] [PubMed] [Google Scholar]
- 30.Lewis D, Catlett J. Heterogeneous uncertainty sampling for supervised learning. Proceedings of the Eleventh International Conference on Machine Learning; 1994 [Google Scholar]
- 31.Firestein GS. Etiology and pathogenesis of rheumatoid arthritis. Kelley's Textbook of Rheumatology. 7th edn Philadelphia, PA: WB Saunders, 2005 [Google Scholar]
- 32.Ries LAG, Eisner MP, Kosary CL, et al. SEER Cancer Statistics Review, 2004
- 33.Snow V, Qaseem A, Barry P, et al. Management of venous thromboembolism: a clinical practice guideline from the American College of Physicians and the American Academy of Family Physicians. Ann Fam Med 2007;5:74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spencer FA, Emery C, Joffe SW, et al. Incidence rates, clinical profile, and outcomes of patients with venous thromboembolism. The Worcester VTE study. J Thromb Thrombolysis 2009;28:401–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang CC, Lin CJ. LIBSVM: a library for support vector machines. ACM Transact Int Syst Technol 2011;2:1–27. [Google Scholar]
- 36.Fan RE, Chang KW, Hsieh CJ, et al. LIBLINEAR: a library for large linear classification. J Machine Learn Res 2008;9:1871–4 [Google Scholar]
- 37.Denny JC, Smithers JD, Miller RA, et al. “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc 2003;10:351–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hripcsak G, Friedman C, Alderson PO, et al. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med 1995;122:681–8 [DOI] [PubMed] [Google Scholar]
- 39.Friedman C, Alderson PO, Austin JH, et al. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc 1994;1:161–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hripcsak G, Austin JHM, Alderson PO, et al. Use of natural language processing to translate clinical information from a database of 889,921 chest radiographic reports. Radiology 2002;224:157–63 [DOI] [PubMed] [Google Scholar]
- 41.Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010;17:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aliferis CF, Statnikov A, Tsamardinos I, et al. Local causal and markov blanket induction for causal discovery and feature selection for classification part I: algorithms and empirical evaluation. J Mach Learn Res 2010;11:171–234 [Google Scholar]
- 43.Seung HS, Opper M, Sompolinsky H. Query by committee. Proceedings of the ACM Workshop on Computational Learning Theorym; 1992 [Google Scholar]
- 44.Settles B, Craven M, Ray S. Multiple-instance active learning, in advances in Neural Information Processing Systems (NIPS). MIT Press, 2008:1289–96 [Google Scholar]
- 45.Roy N, McCallum A. Toward optimal active learning through sampling estimation of error reduction. Proceedings of the International Conference on Machine Learning (ICML); Morgan Kaufmann, 2001 [Google Scholar]