Abstract
Objective
Mental illness is the leading cause of disability in the USA, but boundaries between different mental illnesses are notoriously difficult to define. Electronic medical records (EMRs) have recently emerged as a powerful new source of information for defining the phenotypic signatures of specific diseases. We investigated how EMR-based text mining and statistical analysis could elucidate the phenotypic boundaries of three important neuropsychiatric illnesses—autism, bipolar disorder, and schizophrenia.
Methods
We analyzed the medical records of over 7000 patients at two facilities using an automated text-processing pipeline to annotate the clinical notes with Unified Medical Language System codes and then searching for enriched codes, and associations among codes, that were representative of the three disorders. We used dimensionality-reduction techniques on individual patient records to understand individual-level phenotypic variation within each disorder, as well as the degree of overlap among disorders.
Results
We demonstrate that automated EMR mining can be used to extract relevant drugs and phenotypes associated with neuropsychiatric disorders and characteristic patterns of associations among them. Patient-level analyses suggest a clear separation between autism and the other disorders, while revealing significant overlap between schizophrenia and bipolar disorder. They also enable localization of individual patients within the phenotypic ‘landscape’ of each disorder.
Conclusions
Because EMRs reflect the realities of patient care rather than idealized conceptualizations of disease states, we argue that automated EMR mining can help define the boundaries between different mental illnesses, facilitate cohort building for clinical and genomic studies, and reveal how clear expert-defined disease boundaries are in practice.
Keywords: Electronic Medical Records, Autism, Schizophrenia, Bipolar Disorder, Data Mining, Network Analysis
Background
Fluid boundaries of mental illnesses
Over the course of the 20th century, scientists have struggled to refine the mental health classification system. Even as diagnostic manuals swell with new disorders and additional definitions of existing conditions, the underlying complexity of the mental illness landscape has continued to handicap effective diagnosis and treatment. Mental disorders are often characterized as distinct illnesses despite high rates of comorbidity, which indicate more fluid boundaries.1 Some have argued that even the extensively updated DSM-V (fifth edition of the Diagnostic and Statistical Manual of Mental Disorders) does little to address the need of an objective approach to disease classification.2 3 Diagnostic criteria continue to evolve; new categories are added, and some traditional categories have recently been dropped.4
Because electronic medical records (EMRs) reflect the realities of patient care rather than idealized conceptualizations of diseases, they could potentially serve as a new source of information for this classification task. By revealing the phenotypic signatures of different disorders as they are observed and recorded by clinicians, EMRs could allow a more data-driven approach to the classification of mental illnesses.5–7
Among mental illnesses, three related disorders—autism, bipolar disorder, and schizophrenia—have a particularly high impact on affected individuals and their families, and present a heavy economic burden for the healthcare system.8–11 These disorders are complex, with a significant degree of shared clinical presentation, and may share common genetic origins.12 For example, recent work exploring the genetic link between schizophrenia and autism has suggested that certain rare structural genetic variants, including copy number variants, play a role in both disorders.13 14 Bipolar disorder and schizophrenia have also been shown to overlap because of their shared presentation of certain ‘neurological soft signs’,15 as well as cognitive and memory impairments.16 17 Certain copy number variants are also common to the two disorders.18 Finally, case studies have suggested a genetic link between bipolar disorder and autism.19 20
Our approach
In an effort to elucidate the shared and distinct phenotypic features of autism, bipolar disorder, and schizophrenia as they are discussed in clinical records, we mined the records of over 7000 psychiatric patients from Stanford Hospital and the Palo Alto Medical Foundation (PAMF). After annotating these records with concepts from the Unified Medical Language System (UMLS) and other ontologies, we searched for concepts that were enriched in each disorder relative to the baseline population at each facility. We also identified highly associated concept pairs in each disorder, and we applied dimensionality-reduction techniques to the individual patient records to determine variations in the clinical presentations for these disorders. We present our findings with a focus on the specific areas of overlap among the different disorders. We demonstrate that EMRs present a rich, objective and expert-sourced repository of information for studies seeking to delineate the clinical, phenotypic boundaries between important neuropsychiatric disorders.
Methods
Data sources
Our data consisted of EMRs from Stanford University's STRIDE (Stanford Translational Research Integrated Database Environment) database and PAMF. Stanford Hospital is a 613-bed teaching hospital and tertiary care facility; it admits over 24 000 patients per year and receives over 558 000 outpatient visits. In contrast, PAMF consists of community-based clinics throughout the San Francisco Bay area, covering roughly 700 000 patients across Alameda, San Mateo, Santa Clara, and Santa Cruz counties. Because Stanford Hospital is a referral facility, its psychiatric patient population might not be representative of the general population of patients with autism, bipolar disorder, and schizophrenia. We therefore considered data from both facilities in all parts of our analysis.
All of the data in this study were deidentified using the method described in US Patent Application 13 420 402. We used this technology to process clinical notes that contained protected health information, keep only the medically relevant concepts, and transform the data into a deidentified patient–feature matrix.21 22 The Stanford Institutional Review Board determined that the analysis of patient data deidentified in this way was not human subjects research, and the PAMF Institutional Review Board approved the study under an expedited review protocol for deidentified data. The technology used to deidentify the data is freely available to academics, and can be licensed non-exclusively to commercial entities.
Patient cohorts
Our initial patient cohorts consisted of patients who had received International Classification of Diseases (ICD-9) diagnosis codes of autism, bipolar disorder, or schizophrenia at any visit. These patients and their notes constitute the ‘Before pruning’ column in table 2. We then narrowed the cohorts, keeping patients who had received two separate diagnoses for the same disorder within 1 year and had more than 1 year between their earliest and latest notes. While there is no standard way to identify patients with a specific mental diagnosis based on EMR data alone, the best-performing algorithm for extracting depressed patients from EMRs requires two separate diagnoses and at least 1 year of follow-up.23 24
Table 2.
Patients and records (notes) represented in the STRIDE and PAMF corpora, before and after pruning
| Before pruning | After pruning | |||
|---|---|---|---|---|
| Patients | Notes | Patients | Notes | |
| STRIDE, autism | 2037 | 30 718 | 533 | 6598 |
| PAMF, autism | 1474 | 108 769 | 610 | 15 941 |
| STRIDE, bipolar | 5901 | 139 455 | 2946 | 55 700 |
| PAMF, bipolar | 5299 | 677 980 | 2296 | 129 980 |
| STRIDE, schizophrenia | 2198 | 45 292 | 270 | 1886 |
| PAMF, schizophrenia | 1018 | 136 758 | 449 | 27 937 |
| STRIDE, total | 10 136 | 215 465 | 3749 | 64 184 |
| PAMF, total | 7791 | 923 507 | 3355 | 173 858 |
PAMF, Palo Alto Medical Foundation; STRIDE, Stanford Translational Research Integrated Database Environment.
After identifying candidate patients, we extracted only those notes that were associated with patient visits. For STRIDE, the final set of notes included clinic notes, consultation notes, progress/discharge/transfer summaries, letters (such as referral letters) written by physicians, and patient histories/physicals. Pathology results and radiology transcripts were excluded. Detailed initial psychological evaluations were also excluded in the interest of patient privacy. For PAMF, the final set of notes included letters, history/physical notes, procedure notes, and problem/visit notes. Patient instructions and transcripts of phone calls were excluded. The pruned set of patients and notes constitutes the ‘After pruning’ column in table 2.
Annotation pipeline
The final set of notes was processed with the annotation pipeline described previously.25–27 We used an optimized version of the NCBO (National Center for Biomedical Ontology) Annotator28 to annotate the patient notes with concepts from 22 clinically relevant ontologies. The pipeline involved:
Generating a corpus of ∼5.6 million strings from 22 relevant ontologies and all trigger terms from NegEx29 and ConText.30
Pruning by term frequency and syntactic type information (eg, predominant noun phrases) from Medline to create a clean lexicon,31–33 which was used for dictionary-based concept recognition to produce annotations.
Using NegEx and ConText rules to filter out negated terms and terms found in the family history.
Normalizing terms into concepts based on interontology mappings and semantic grouping by drug, disease, device, or procedure.
Normalizing drugs into ingredients using RxNorm,34 so that (for example) ‘fluoxetine’ and ‘Prozac’ were both normalized to ‘fluoxetine’. Brand name mentions were normalized to their generic names, and combination drugs to their constituent substances.
Concept enrichment calculations
We first identified those concepts that were (a) significantly enriched among patients with a particular mental disorder relative to the general population and (b) not indicative of the patient population at a particular facility, but rather, indicative of the disorder itself. Because our six patient cohorts (table 2) were of different sizes, statistical power calculations were critical: we wanted to ensure that the minimum effect size required for concept enrichment was the same across all three disorders, as well as across both facilities. For each disorder, we carried out the following:
Randomly selected 1000 notes from patients with that disorder. This process involved first selecting a patient at random, then selecting one of that patient's notes (again at random). Thus patients with longer histories were not over-represented.
Randomly selected 1000 notes from patients without a mental disorder diagnosis at the same facility in the same fashion.
For each UMLS concept found in a note from a patient with the disorder, performed Fisher's exact test to determine if the term was significantly enriched among patients with the disorder (p<0.05).
Repeated steps 1–3 twenty times for each facility.
Using the statistical software package G*Power,35 we determined that a cohort size of 1000 ‘cases’ and 1000 ‘controls’ would yield a power of 0.80 for detecting effect sizes of ∼6% difference in proportions between the cases and controls. We therefore considered a term ‘enriched’ for a particular mental disorder if Fisher's test found it so with p<0.05 in at least 80% of our trials from both facilities. It is important to note that this sampling procedure did not guarantee that each note in the sample came from a unique patient, even when the number of patients was much greater than 1000; it simply ensured that sampling was uniform across patients, regardless of length of history.
Finding associated concepts
We defined the clinical presentation of a disease along two dimensions: the concepts enriched for that disease, and the significant associations among those concepts. Power and effect size calculations were less straightforward for the association analysis because of the widely varying numbers of notes in which each concept occurred. We wanted to ensure that we did not see an increased number of significant concept–concept associations for a given disorder, such as bipolar disorder, simply because the larger number of notes for that disorder conferred greater power (the ability to detect associations with smaller effect sizes).
If we consider two concepts, C1 and C2, the ‘effect size’ was the difference in proportions of occurrence of C2 between notes where (a) C1 occurred and (b) C1 did not occur. If C1 and C2 were highly associated, we would expect C2 to occur in a high proportion of notes that contained C1 and a lower proportion of notes that did not contain C1. Initial power calculations revealed that, with an overall sample size of 1866 (the size of our smallest set of notes, for schizophrenia/STRIDE), we could observe an effect size of 8% with a power of 0.8 at a significance level of 0.05, even if the distribution of notes containing C1 versus notes without C1 were skewed as much as 4:1.
We used Fisher's exact test to find enriched associations between all pairs of concepts for each disorder where at least one of the concepts was among our set of 123 ‘enriched’ drugs or phenotypes for autism, bipolar disorder, and schizophrenia (figure 1). We required that the effect size be at least 8% for an association to be significant. This meant that, even if a given association was significant with p<0.05, it would not be accepted if the post hoc estimated effect size was not large enough to assure us that we would see the same association in the schizophrenia/STRIDE group with 80% power. Doing so reduced the chance that we would call an association significant for one disorder but not for a second disorder simply because the second disorder had a smaller sample size.
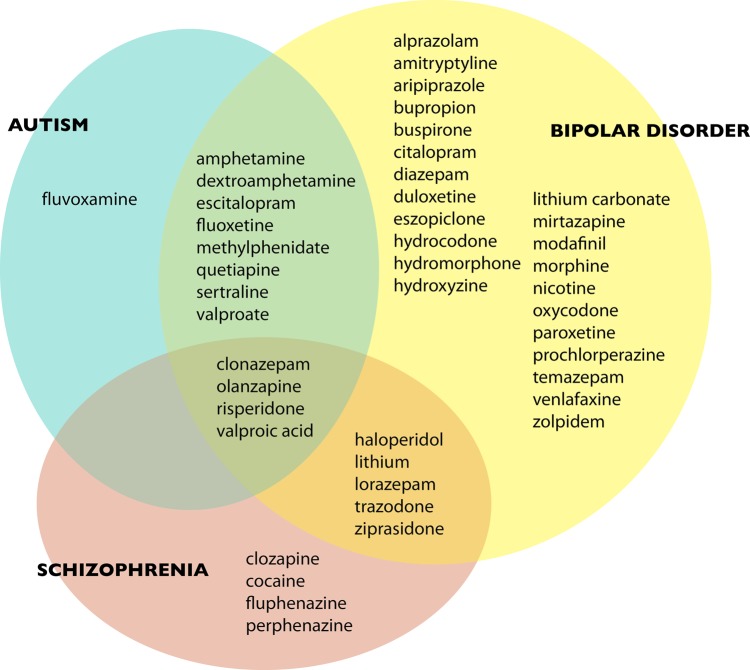
Figure 1.
Significantly enriched psychoactive drug terms for each disease, and their overlap.
Finally, for each pair of concepts, we actually measured two separate associations: the chance of observing C2, depending on whether C1 was present in the note, and the chance of observing C1, depending on whether C2 was present. The two effect sizes may be different, and the two directional associations were treated as separate in our analysis and viewed in our subsequent network visualization as directed edges between C1 and C2.
Network visualization and analysis were performed using Cytoscape 2.8.36 A ‘neighbor’ of a node in a network is any node that is connected to it via an edge, ignoring edge direction. The ‘mean degree’ is the average number of neighbors of a node in the network. The ‘clustering coefficient’ is defined as the number of edges among neighbors of a specific node, divided by the total number of edges that could possibly exist among those neighbors, averaged over all nodes in the network. It is a measure of transitivity: how likely it is that ‘a friend of my friend is also my friend’.
Practically speaking, these network metrics provide different, though related, perspectives on similar phenomena. A high mean degree indicates the presence of a large number of significant associations among concepts for a particular disorder; the concepts are not distributed independently throughout patient notes, but tend to occur preferentially with certain other concepts: the higher the mean degree, the greater the number of significant associations. The clustering coefficient is related to the presence of community structure in the network.37 Networks with high clustering coefficients are often distinguished by the presence of ‘hubs’: concepts with a disproportionately large number of connections.
Patient-level analysis
To visualize the overlap among patients with the three disorders, we consolidated each patient's record into a feature vector of length 123: one element for each of the 45 enriched drugs and 78 enriched phenotypes shown in figure 1 and figure 2. The value at each location was the number of mentions of the term in each patient's record, divided by the total number of notes in the patient's record. We normalized by the number of notes in the patient's record to ensure that two patients with similar distributions of concepts looked similar, even if one had a greater number of total notes (perhaps because of a longer follow-up time). We then performed principal components analysis38 on the patient–concept matrix and graphed the patients’ locations along the first three principal components.
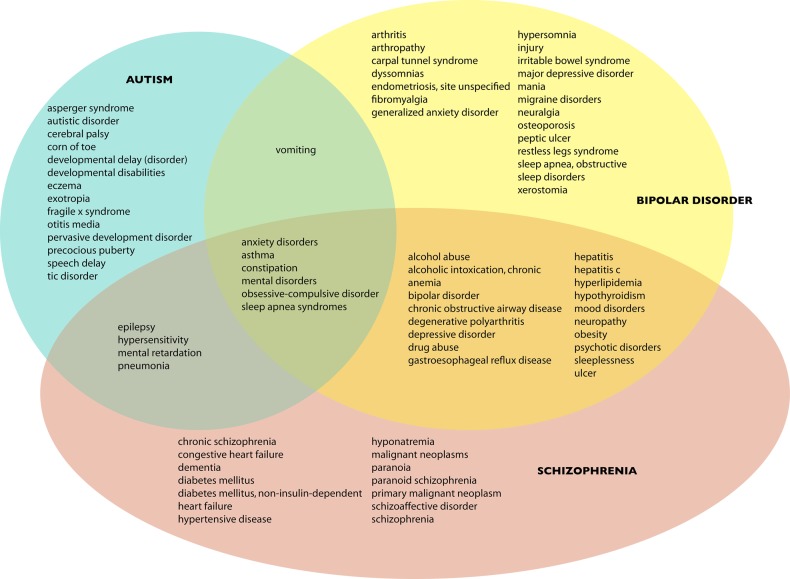
Figure 2.
Significantly enriched phenotypes for each disease, and their overlap.
Results
Patient demographics and dataset characteristics
Age and gender information for the population in our study is shown in table 1. Autism shows a bias toward younger ages, while there is substantial overlap of age ranges for schizophrenia and bipolar disorder. The gender ratios for the three disorders differ, with autism enriched for males and bipolar for females, while schizophrenia is more evenly distributed.
Table 1.
Demographic information about the study population in the two centers
| Sex ratio (male:female) | Age (median; range) | |
|---|---|---|
| STRIDE, autism | 80:20 | 15; 4–63 |
| PAMF, autism | 82:18 | 10; 2–53 |
| STRIDE, bipolar | 33:67 | 51; 14–102 |
| PAMF, bipolar | 34:66 | 49; 10–103 |
| STRIDE, schizophrenia | 51:49 | 56; 19–98 |
| PAMF, schizophrenia | 44:56 | 53; 11–96 |
PAMF, Palo Alto Medical Foundation; STRIDE, Stanford Translational Research Integrated Database Environment.
Table 2 shows the numbers of patients and notes in each dataset, before and after preprocessing. Schizophrenia was the least common disorder, and the schizophrenia/STRIDE cohort our smallest dataset, containing 270 patients and 1886 notes. In contrast, bipolar disorder was the largest cohort; the bipolar/PAMF dataset contained 2296 patients and 129 980 notes.
Enriched concepts
We grouped the significantly enriched terms for each disease into drugs and phenotypes (symptoms or clinical findings). The enriched set of drugs for each disorder is shown in figure 1, and the enriched phenotypes are shown in figure 2.
While the overall pattern of drug use in figure 1 is not surprising, it serves as a useful validation for the phenotype enrichment and concept association calculations. Note that because of our note sampling procedure, the number of drugs found enriched for bipolar disorder is not an artifact of its larger sample size: there are in fact 40 different drugs significantly associated with bipolar disorder, compared with 13 for schizophrenia and 13 for autism. Bipolar patients overlap with schizophrenic patients mainly because of their use of antipsychotics, such as haloperidol (Haldol), and they overlap with autistic patients mainly because of their use of antidepressants, such as sertraline (Zoloft), and stimulants, such as methylphenidate (Ritalin). There are no drugs associated with both schizophrenia and autism that are not also associated with bipolar disorder. If we interpreted these disorders solely in terms of their associated drugs, we might conclude that bipolar disorder is the most variable in its presentation, and that autism and schizophrenia each overlap with distinct aspects of bipolar disorder.
Figure 2 shows the overlap among the phenotypes associated with each disorder. There is substantial phenotypic overlap between bipolar disorder and schizophrenia: these patients are more likely to abuse alcohol and suffer from chronic health problems such as obesity, chronic obstructive pulmonary disease, neuropathy, and hepatitis. (For example, approximately 10–15% of patients with bipolar disorder are also chronically infected with hepatitis C.39) These associations may reflect these patients’ more advanced age as well as various social and environmental factors associated with mental illness; elevated rates of nicotine dependence, for example, are well known in both schizophrenia and bipolar disorder.40 41 Epilepsy and mental retardation are both common to autism and schizophrenia but not bipolar disorder.
From a diagnostic standpoint, the most interesting findings are those phenotypic concepts uniquely enriched in each disorder. If we assume that some of these specific phenotypes lead physicians to prefer one diagnosis over another, we may consider them ‘signature’ concepts for each disorder. For example, enriched phenotypes unique to autism include developmental delays (eg, in speech) or certain repetitive behaviors such as tics. (Although the concept ‘corn of toe’ seems incongruous here, the terms ‘corn’ and ‘callous’ both map to it and these conditions could be the result of repetitive behaviors common in autistic children.) Enriched phenotypes in bipolar disorder include symptoms associated with depression and anxiety, such as migraines, irritable bowel syndrome, sleep disorders, and ulcers, as well as the core diagnostic criterion of bipolar disorder itself: mania. Finally, schizophrenic patients exhibit both paranoia and schizoaffective traits, but also suffer from chronic problems such as heart failure, cancers, dementia, diabetes, and hypertension. These probably reflect the advanced age of schizophrenic patients as well as the physical toll of living with schizophrenia.
Patient-level analysis
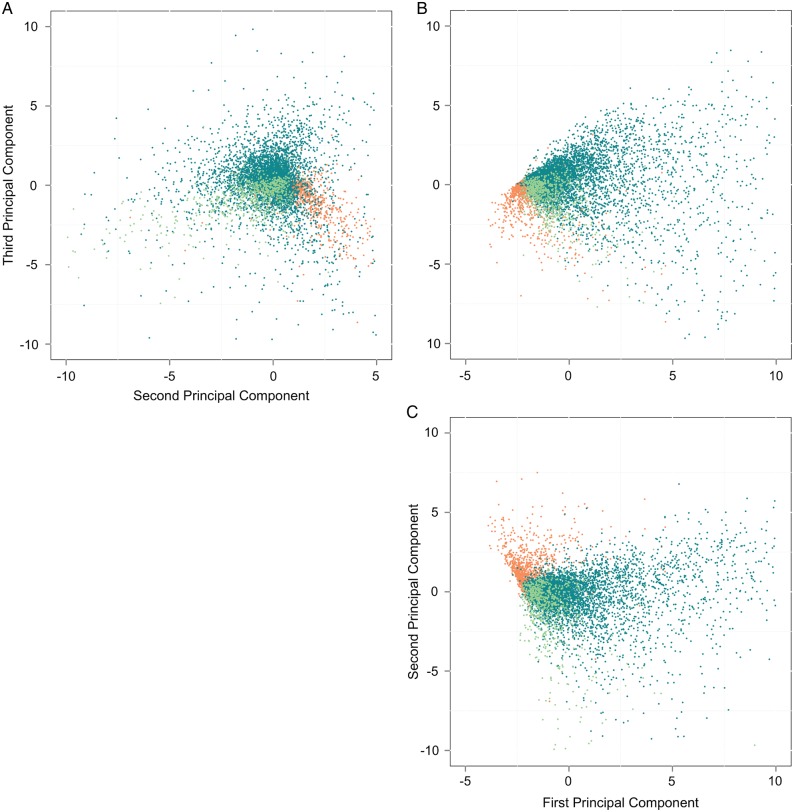
Figure 3 shows the result of the patient-level principal components analysis. Each dot represents a single patient. We see that there is significant overlap between schizophrenic and bipolar patients; in fact, the schizophrenic patient cluster is nearly encompassed by the bipolar cluster in all subplots. In contrast, autistic patients form a distinct cluster.
Figure 3.
Patient-level plots of the first three principal components (PC1 vs PC2 in (C), PC2 vs PC3 in (A), and PC1 vs PC3 in (B)). Autistic patients are shown in orange, schizophrenic patients in light green, and bipolar patients in dark turquoise.
The first principal component (PC1) is a vector in the space of covariates (drugs, phenotypes) from the original dataset; it captures the direction along which most of the variability in the dataset lies.38 The second principal component (PC2) captures the direction of next highest variability (orthogonal to the first); the others follow in turn. It is therefore interesting to look at the first few principal components from our data to see which drug and phenotype terms contribute the most to each.
The major contributors to PC1 are, in order, ‘depressive disorder’, ‘anxiety disorders’, ‘mental disorders’, ‘drug abuse’, ‘sleeplessness’, ‘mood disorders’, and ‘bipolar disorder’. These contributing terms all have a positive sign, which means that the more each term occurs the more it pulls the associated patient along the positive PC1 axis. PC1 includes terms indicating the presence or absence of broad sets of conditions and could perhaps represent the variability in a patient's primary presentation. Bipolar patients extend beyond the rest along the positive PC1 axis; their records appear to discuss a wider variety of different potential diagnoses. (Notably, the term ‘bipolar disorder’ itself also plays a prominent role in this component.)
PC2 includes the following major contributors in the positive direction: ‘autistic disorder’, ‘developmental delay’, ‘pervasive development disorder’, ‘developmental disabilities’, ‘valproate’, and ‘valproic acid’, followed closely by ‘dextroamphetamine’ and ‘methylphenidate’. All of these terms are highly associated with autism. Conversely, the major negative contributors to PC2 are ‘heart failure’, ‘congestive heart failure’, ‘chronic obstructive airway disease’, ‘hypertensive disease’, ‘schizophrenia’, ‘anemia’, and ‘degenerative polyarthritis’. PC2 might be considered the ‘autism/schizophrenia axis’; patients are situated further along the positive axis if they exhibit autistic traits and further along the negative axis if they exhibit schizophrenic traits or chronic health disorders commonly associated with schizophrenia. PC2 explains roughly half as much variance in the original data as PC1; a patient can be autistic-like or schizophrenic-like, but not both. (Of course, much of this variability can be explained by the relative ages of the two groups of patients.)
The third principal component (PC3) contains the following major positive contributing terms: ‘major depressive disorder’, ‘depressive disorder’, ‘sleep apnea syndromes’, ‘duloxetine’, ‘bupropion’, ‘sleep apnea, obstructive’, and ‘anxiety disorders’. Major negative contributing terms include ‘valproic acid’, ‘valproate’, ‘bipolar disorder’, ‘haloperidol’, ‘mania’, ‘risperidone’, ‘schizoaffective disorder’, and ‘psychotic disorders’. We might conclude that PC3 represents a tradeoff between depressive/anxious symptoms and psychotic/manic symptoms. A patient sits further along the positive PC3 axis if he or she is depressed, has trouble sleeping, is anxious, or is taking duloxetine or bupropion (both antidepressants). A patient moves toward the negative axis if he or she is taking valproate/valproic acid (a mood stabilizer and anticonvulsant), an antipsychotic such as haloperidol or risperidone, or has been diagnosed with mania or psychotic symptoms. There is considerable overlap between autistic and schizophrenic patients along PC3, while bipolar patients exhibit high variance in this aspect of the clinical presentation.
After considering the terms that contribute the most to each principal component, therefore, the picture in figure 3 becomes clearer. First, we see that diversity of clinical presentation is greater for bipolar patients than autistic or schizophrenic patients: bipolar patients show a wide spread across the PC1 axis, and extend off in the positive direction. (Interestingly, inter-rater reliability studies of diagnoses from the initial DSM-V field trials showed a κ statistic of 0.52 for bipolar I disorder in children compared with 0.69 for autism spectrum disorder in children,3 indicating higher inter-rater agreement for autism vs bipolar diagnoses.) Second, we see that patients can also be organized along an ‘autism/schizophrenia axis’, PC2, and along a ‘depressive/psychotic scale’, PC3. Autism and schizophrenia exhibit considerable separation along PC2 but not PC3. Bipolar overlaps with schizophrenia, but not autism, along PC2, and extends much further than either schizophrenia or autism into ‘depressive territory’ along PC3.
Enriched associations
If we consider the significant associations for each disorder as a network of directed edges (arcs) between concepts, the autism network encompasses 204 unique concepts and 363 arcs. The bipolar network includes 1160 unique concepts and 4306 arcs. The schizophrenia network includes 219 unique concepts and 485 arcs. There were 25 concept–concept associations that occurred in all three disorders, two that occurred for autism and schizophrenia but not bipolar, 136 that occurred for bipolar disorder and schizophrenia but not autism, and 112 that occurred for autism and bipolar disorder but not schizophrenia.
The full network files are available as online supplementary material for this paper, and can be viewed using Cytoscape or similar software.
We visualized each disease in terms of its characteristic phenotypes and the associations among them ignoring drugs, as well as phenotypes that are merely side effects of drugs (figure 4). We removed drug names from the networks, as well as all phenotype concepts that were connected to the networks only through their interactions with drugs. Table 3 contains a summary of each network's properties.
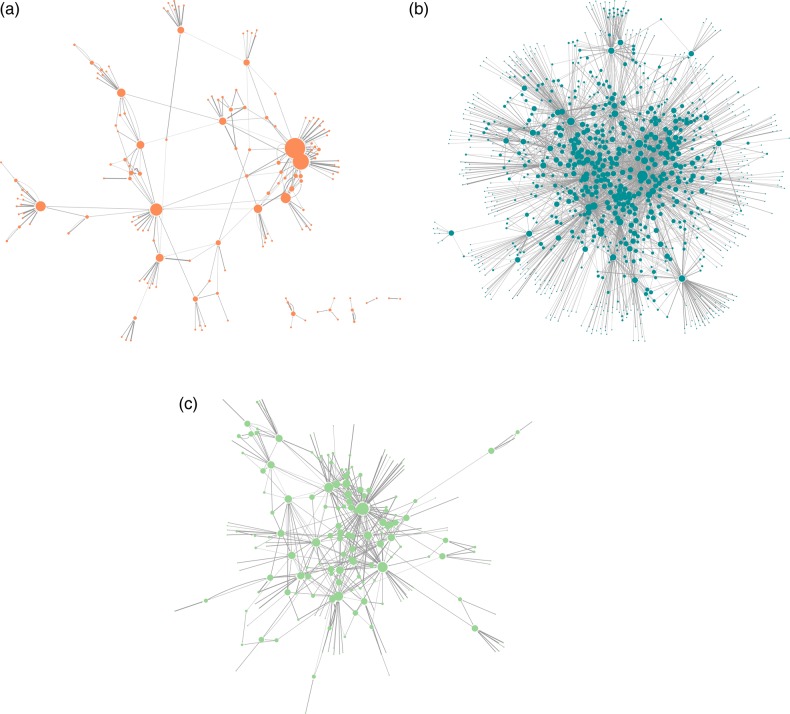
Figure 4.
Network representations of phenotype–phenotype associations for (A) autism, (B) bipolar disorder, and (C) schizophrenia. The nodes represent phenotypic concepts, and the node sizes are proportional to the number of connections for each node (disregarding directionality). The edge widths are proportional to the strength of the interaction between the two concepts they connect.
Table 3.
A summary of some basic network parameters for the phenotype networks shown in figure 4
| Parameter | Autism value | Bipolar value | Schizophrenia value |
|---|---|---|---|
| Clustering coefficient | 0.154 | 0.306 | 0.249 |
| Mean degree (ignoring edge direction) | 2.699 | 5.177 | 4.074 |
| Hubs (nodes with highest degrees) |
|
|
|
For the purposes of this analysis, the networks are treated as undirected.
The differences among the three networks are apparent even by inspection. The network for bipolar disorder is the largest and has the highest clustering coefficient and the highest mean degree. It appears to form one connected ‘community’ of edges without any fragmentation. On the opposite end of the spectrum is autism, a much smaller network with the lowest clustering coefficient and lowest mean degree. The autism network consists of six separate fragments, and there exist nodes that, if removed, would fragment it further. From a topological standpoint, the autism network consists of phenotypic ‘communities’ that are loosely bound to each other, reflecting the heterogeneity of autism and the fact that it may actually be a group of disorders.42 Schizophrenia's network lies between these two extremes.
Discussion
Phenotypic signatures in cohort building
The annotation and statistical analysis methods described here serve two main purposes. First, recognition of ontological concepts in unstructured EMR text followed by statistical enrichment analysis identifies a set of UMLS concepts, and associations among concepts, that are indicative of one or more disorders of interest. This process defines phenotypic ‘signatures’ for disorders, enabling researchers interested in cohort building for clinical or genomic studies to identify patients that may meet the phenotypic criteria for a particular disease. Importantly, phenotypic signatures can be ‘built’ using EMR text from one facility and then applied to text from a different facility to identify phenotypically similar patients, even if specific diagnosis codes are applied differently at the two facilities. Such signatures could also play a crucial role in the development of a new ‘taxonomy’ of diseases, as suggested by the National Academy of Sciences.43
Second, once the phenotypic signatures are identified, individual patients can be situated relative to the principal conceptual ‘axes’ within the overall landscape of a disorder. Considering an individual as a point in a high-dimensional feature space—where each feature is one of the ‘signature’ concepts for a disorder—analyses such as ours enable researchers to pick out ‘matched’ patients from different disorder groups, and to identify patients who are more or less representative of different disorders; both of which are useful steps in cohort building. Although we have focused on mental disorders, the methods described here can be applied to other disorder(s) of interest.
Related work
Several groups have explored ways to define phenotypic signatures for diseases from EMRs,44 although most have used structured EMR data (diagnosis codes, etc) rather than unstructured text. For example, Hanauer et al45 examined temporal associations among ICD-9 codes, and Kohane et al46 showed that patients with autism have significantly increased rates of other comorbidities. Denny et al47 were the first to conceptualize the PheWAS, or Phenome-Wide Association Study, where EMR data are used to identify specific phenotypes of interest, and then genetic data from those patients are examined to identify associated genetic variations. Data-driven phenotyping methods similar to ours are therefore essential to nuanced PheWAS studies for neuropsychiatric disorders. Finally, recent work on ‘EMR phenotyping’ has resulted in a web knowledge base that collects algorithms designed to extract patients with specific phenotypes from EMRs (http://phekb.org). Our approach is complementary to this effort; the annotation pipeline and enrichment statistics framework can be applied to develop phenotypic signatures for multiple diseases of interest.
Study limitations
As with any observational study, especially one where much about the underlying data collection process is unknown, ours suffers from several important limitations. Foremost among these is the difficulty we faced in validating our findings, since there is no such thing as a training set of all known UMLS concepts associated with schizophrenia, for example, or all known associations among different UMLS concepts for these different disorders. The most we could hope for was that a psychiatrist would deem our findings reasonable. We did present this work to several psychiatrists and other researchers interested in mental illness, but it would be impossible to ask an expert to produce a list of all relevant concepts and associations that exist so we could estimate our recall. Therefore, our study is necessarily somewhat qualitative and descriptive in nature.
In addition, it is possible that clinicians might base their descriptions of patients on what is in the DSM, in an attempt to provide support for a particular diagnosis. If this is the case, we might miss rarer associations with concepts that are not found in the DSM for a particular disorder. We may also have biased our results somewhat because of a lack of knowledge (as a result of privacy concerns) about the specialties of the particular physicians who wrote the clinical notes; an internist might record a longer and more detailed history for a patient who has seen a psychiatrist, for example, than for one who has not.
Future directions
Beyond cohort building and the identification of phenotypic signatures for specific diseases, our approaches could be extended to other problems related to EMR-based phenotyping. For example, the use of EMR-based text mining in combination with network-based analyses will likely have broad utility for uncovering new associations between clinical entities48 and potentially in analyzing patient outcomes.49 In addition, it is known that psychiatry patients have a higher mortality and morbidity from ‘usual’ causes, such as heart failure, diabetes, etc.50 Knowing the strength of associations among comorbidities, as we have calculated in our association networks, would assist in devising care management protocols similar to those in Petri et al.51
Conclusion
By annotating, and then statistically exploring, the text of EMRs associated with mental illness, we have examined the phenotypic signatures of autism, bipolar disorder, and schizophrenia from three perspectives: the enriched concepts for each disorder, the networks of associations among those concepts, and the clustering of patients based on the concept mentions in their records. We have also examined the combinations of concepts that account for the most variance in patient records, uncovering a ‘diagnostic variability’ axis, an ‘autistic/schizophrenic’ axis, and a ‘depressive/psychotic’ axis. Our experiences analyzing these data underscore the importance of statistical power calculations to reduce the chances of introducing artifacts when comparing disease populations of very different sizes.
We have demonstrated that EMR mining can extract relevant drugs and phenotypes associated with three important psychiatric disorders, and can reveal reasonable phenotypic signatures. Examination of the first three principal components appears to isolate autism as a separate disorder, while revealing significant overlap between schizophrenia and bipolar disorder. We conclude that such automated EMR mining can elucidate the phenotypic boundaries between different mental illnesses, reveal how clear the defined diagnostic boundaries are in practice, assist in data-driven cohort building, and assist in the classification of psychiatric disorders in an objective manner.
Acknowledgments
We acknowledge Cliff Olson and Tanya Podchiyska for assistance with data extraction. We would also like to thank the members of the Conte Collaboration for their helpful comments on this work.
Footnotes
Contributors: SL performed data preprocessing, database querying, and statistical analysis of the data and wrote the first draft of the manuscript. BP provided technical guidance for the development of the methods, performed the statistical power calculations, and edited the manuscript to produce the final version. RBA developed the initial idea for the project, chose the three disorders, and provided technical feedback as well as a medical perspective. PJL and SVI obtained the original data and performed the annotation of clinical documents with medical concepts. NHS conceived and devised the clinical note annotation pipeline, provided technical guidance on data normalization using ontologies, and edited the manuscript.
Funding: NHS, PJL and SVI acknowledge support from NIH grant U54-HG004028 for the National Center for Biomedical Ontology. SL, BP and RBA acknowledge support from NIH grant P50 MH094267 for the Conte Center for computational systems genomics of neuropsychiatric phenotypes. BP also acknowledges support from a research grant by the Oracle Health Sciences Institute.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Hyman SE. The diagnosis of mental disorders: the problem of reification. Annu Rev Clin Psych 2010;6:155–79 [DOI] [PubMed] [Google Scholar]
- 2.Sachdev PS. Is DSM-5 defensible? Aust N Z J Psychiatry 2013;47:10–11 [DOI] [PubMed] [Google Scholar]
- 3.Freedman R, Lewis DA, Michels R, et al. The initial field trials of DSM-5: new blooms and old thorns. Am J Psychiatry 2013;170:1–5 [DOI] [PubMed] [Google Scholar]
- 4.Braff DL, Ryan J, Rissling AJ, et al. Lack of use in the literature from the last 20 years supports dropping traditional schizophrenia subtypes from DSM-5 and ICD-11. Schizophr Bull 2013;39:751–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spaulding W, Deogun J. A pathway to personalization of integrated treatment: informatics and decision science in psychiatric rehabilitation. Schizophr Bull 2011;37(Suppl 2):S129–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frayne SM, Miller DR, Sharkansky EJ, et al. Using administrative data to identify mental illness: what approach is best? Am J Med Qual 2010;25:42–50 [DOI] [PubMed] [Google Scholar]
- 7.Newton KM, Peissig PL, Kho AN, et al. Validation of electronic medical record-based phenotyping algorithms: results and lessons learned from the eMERGE network. J Am Med Inform Assoc 2013;20:e147–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Druss BG, Zhao L, Von Esenwein S, et al. Understanding excess mortality in persons with mental illness: 17-year follow up of a nationally representative US survey. Med Care 2011;49:599–604 [DOI] [PubMed] [Google Scholar]
- 9.Newschaffer CJ, Curran LK. Autism: an emerging public health problem. Public Health Reports 2003;118:393–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ketter TA. Diagnostic features, prevalence, and impact of bipolar disorder. J Clin Psychiatry 2010;71:e14. [DOI] [PubMed] [Google Scholar]
- 11.Wu EQ, Birnbaum HG, Shi L, et al. The economic burden of schizophrenia in the United States in 2002. J Clin Psych 2005;66:1122–9 [DOI] [PubMed] [Google Scholar]
- 12.Carroll LS, Owen MJ. Genetic overlap between autism, schizophrenia and bipolar disorder. Genome Med 2009;1:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilman SR, Chang J, Xu B, et al. Diverse types of genetic variation converge on functional gene networks involved in schizophrenia. Nature Neurosci 2012;15:1723–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doherty JL, O'Donovan MC, Owen MJ. Recent genomic advances in schizophrenia. Clin Genet 2012;81:103–9 [DOI] [PubMed] [Google Scholar]
- 15.Zhao Q, Ma YT, Lui SS, et al. Neurological soft signs discriminate schizophrenia from major depression but not bipolar disorder. Prog Neuropsychopharmacol Biol Psychiatry 2012;43C:72–8 [DOI] [PubMed] [Google Scholar]
- 16.Chan RC, Lui SS, Wang Y, et al. Patients with bipolar disorders share similar but attenuated prospective memory impairments with patients with schizophrenia. Psychol Med 2012;26:1–11 [DOI] [PubMed] [Google Scholar]
- 17.Meesters PD, Schouws S, Stek M, et al. Cognitive impairment in late life schizophrenia and bipolar I disorder. Int J Geriatr Psychiatry 2013;28:82–90 [DOI] [PubMed] [Google Scholar]
- 18.Craddock N, O'Donovan MC, Owen MJ. Psychosis genetics: modeling the relationship between schizophrenia, bipolar disorder, and mixed (or “schizoaffective”) psychoses. Schizophr Bull 2009;35:482–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerbeshian J, Burd L, Randall T, et al. Autism, profound mental retardation and atypical bipolar disorder in a 33-year-old female with a deletion of 15q12. J Ment Defic Res 1990;34:205–10 [DOI] [PubMed] [Google Scholar]
- 20.Leyfer OT, Folstein SE, Bacalman S, et al. Comorbid psychiatric disorders in children with autism: interview development and rates of disorders. J Autism Dev Disord 2006;36:849–61 [DOI] [PubMed] [Google Scholar]
- 21.Leeper NJ, Bauer-Mehren A, Iyer SV, et al. Practice-based evidence: profiling the safety of cilostazol by text-mining of clinical notes. PLoS One 2013;8:e63499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LePendu P, Iyer SV, Bauer-Mehren A, et al. Pharmacovigilance using clinical notes. Clin Pharmacol Ther 2013;93:547–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Townsend L, Walkup JT, Crystal S, et al. Mini-sentinel systematic evaluation of health outcome of interest definitions for studies using administrative data. http://www.mini-sentinel.org/work_products/HealthOutcomes/MS_HOI_DepressionReport.pdf.
- 24.Platt R, Carnahan RM, Brown JS, et al. The U.S. Food and Drug Administration's Mini-Sentinel program: status and direction. Pharmacoepidemiol Drug Saf 2012;21(Suppl 1):1–8 [DOI] [PubMed] [Google Scholar]
- 25.Lependu P, Iyer SV, Fairon C, et al. Annotation analysis for testing drug safety signals using unstructured clinical notes. J Biomed Semantics 2012;3(Suppl 1):S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Lependu P, Iyer S, et al. Using temporal patterns in medical records to discern adverse drug events from indications. AMIA Summits Transl Sci Proc 2012;2012:47–56 [PMC free article] [PubMed] [Google Scholar]
- 27.Lependu P, Iyer SV, Bauer-Mehren A, et al. Pharmacovigilance using clinical notes. Clin Pharm & Therapeut 2013;93:547–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jonquet C, Shah NH, Musen MA. The open biomedical annotator. Summit on Translat Bioinform 2009;2009:56–60 [PMC free article] [PubMed] [Google Scholar]
- 29.Chapman WW, Bridewell W, Hanbury P, et al. A simple algorithm for identifying negated findings and diseases in discharge summaries. J Biomed Inform 2001;34:301–10 [DOI] [PubMed] [Google Scholar]
- 30.Harkema H, Dowling JN, Thornblade T, et al. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. J Biomed Inform 2009;42:839–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu ST, Liu H, Li D, et al. Unified Medical Language System term occurrences in clincial notes: a large-scale corpus analysis. J Am Med Inform Assoc 2012;19:e149–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu R, Musen MA, Shah NH. A comprehensive analysis of five million UMLS Metathesaurus terms using eighteen million MEDLINE citations. AMIA Annu Symp Proc 2010;2010:907–11 [PMC free article] [PubMed] [Google Scholar]
- 33.Parai GK, Jonquet C, Xu R, et al. The Lexicon Builder web service: building custom lexicons from two hundred biomedical ontologies. AMIA Annu Symp Proc 2010;2010:587–91 [PMC free article] [PubMed] [Google Scholar]
- 34.RxNorm. http://www.nlm.nih.gov/research/umls/rxnorm/
- 35.Faul F, Erdfelder E, Lang AG, et al. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 2007;39:175–91 [DOI] [PubMed] [Google Scholar]
- 36.Cline MS, Smoot M, Cerami E, et al. Integration of biological networks and gene expression data using Cytoscape. Nature Protocols 2007;2:2366–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Orman K, Labatut V, Cherifi H. An empirical study of the relation between community structure and transitivity. Springer Berlin Heidelberg: Complex Networks, 2013:99–110 [Google Scholar]
- 38.Ch. 8: Principal Components In: Johnson RA, Wichern DW. eds. Applied Multivariate Statistical Analysis. Upper Saddle River, NJ: Prentice Hall, 2002:430–80 [Google Scholar]
- 39.Rifai MA. Hepatitis C treatment of patients with bipolar disorder: a case series. Prim Care Companion J Clin Psychiatry 2006;8:361–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Leon J, Diaz FJ, Rogers T, et al. Initiation of daily smoking and nicotine dependence in schizophrenia and mood disorders. Schizophrenia Research 2002;56:47–54 [DOI] [PubMed] [Google Scholar]
- 41.Dalack GW, Healy DJ, Meador-Woodruff JH. Nicotine dependence in schizophrenia: clinical phenomena and laboratory findings. Am J Psychiatry 1998;155:1490–501 [DOI] [PubMed] [Google Scholar]
- 42.Carter M, Scherer S. Autism spectrum disorder in the genetics clinic: a review. Clin Genet 2013;83:399–407 [DOI] [PubMed] [Google Scholar]
- 43.National Academy of Sciences Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease, 2011 [PubMed]
- 44.Pillai RR, Divekar R, Brasier A, et al. Strategies for molecular classification of asthma using bipartite network analysis of cytokine expression. Curr Allergy Asthma Rep 2012;12:388–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hanauer DA, Ramakrishnan N. Modeling temporal relationships in large clinical associations. J Am Med Inform Assoc 2013;20:332–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kohane IS, McMurry A, Weber G, et al. The co-morbidity burden of children and young adults with autism spectrum disorders. PLoS One 2012;7:e33224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics 2010;26:1205–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanauer DA, Rhodes DR, Chinnaiyan AM. Exploring clinical associations using “-omics” based enrichment analyses. PLoS One 2009;4:e5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bauer-Mehren A, LePendu P, Iyer SV, et al. Network analysis of unstructured EHR data for clinical research. San Francisco, CA: AMIA Summit on Clinical Research Informatics, 2013 [PMC free article] [PubMed] [Google Scholar]
- 50.Miller MD, Paschall I, Svendsen MD. Mortality and medical comorbidity among patients with serious mental illness. Psychiatric Services 2006;57:1482–7 [DOI] [PubMed] [Google Scholar]
- 51.Petri H, Maldonato D, Robinson NJ. Data-driven identification of co-morbidities associated with rheumatoid arthritis in a large US health plan claims database. BMC Musculoskelet Disord 2010;11:247. [DOI] [PMC free article] [PubMed] [Google Scholar]