Abstract
Background and aim
Celiac disease (CD) is a lifelong immune-mediated disease with excess mortality. Early diagnosis is important to minimize disease symptoms, complications, and consumption of healthcare resources. Most patients remain undiagnosed. We developed two electronic medical record (EMR)-based algorithms to identify patients at high risk of CD and in need of CD screening.
Methods
(I) Using natural language processing (NLP), we searched EMRs for 16 free text (and related) terms in 216 CD patients and 280 controls. (II) EMRs were also searched for ICD9 (International Classification of Disease) codes suggesting an increased risk of CD in 202 patients with CD and 524 controls. For each approach, we determined the optimal number of hits to be assigned as CD cases. To assess performance of these algorithms, sensitivity and specificity were calculated.
Results
Using two hits as the cut-off, the NLP algorithm identified 72.9% of all celiac patients (sensitivity), and ruled out CD in 89.9% of the controls (specificity). In a representative US population of individuals without a prior celiac diagnosis (assuming that 0.6% had undiagnosed CD), this NLP algorithm could identify a group of individuals where 4.2% would have CD (positive predictive value). ICD9 code search using three hits as the cut-off had a sensitivity of 17.1% and a specificity of 88.5% (positive predictive value was 0.9%).
Discussion and conclusions
This study shows that computerized EMR-based algorithms can help identify patients at high risk of CD. NLP-based techniques demonstrate higher sensitivity and positive predictive values than algorithms based on ICD9 code searches.
Keywords: algorithm; artificial intelligence; celiac; decision support system, clinical; inflammation
Background and significance
Celiac disease (CD) is triggered by exposure to gluten in genetically predisposed individuals. On gluten exposure, these patients develop inflammation and villous atrophy in the proximal small intestine.1 The disease occurs in about 1% of the Western population,2 3 which equals almost two million Americans. Most individuals with CD go undiagnosed, with increased risk of developing fractures4 5 and suffering from adverse pregnancy outcome,6 despite the readily available and simple serological detection test. Once the disease is identified, typical treatment consists of a gluten-free diet.
Classical CD includes patients with malabsorptive symptoms and signs, but this presentation may not be the dominant one nowadays,7 and many patients have other symptoms (non-classical), or no symptoms at all (asymptomatic).1 For instance, iron-deficiency anemia occurs at CD diagnosis in some 30–50% of patients.8 9 Certain risk groups have also been established, including individuals with type 1 diabetes10 and thyroid disease.11 Although it has been argued that simple rules are better than complicated algorithms for the detection of CD,12 computerized models can allow for complexity. In particular, if such models are implemented within an electronic medical record (EMR) environment, we believe that EMR-based algorithms could serve as an active part of healthcare, and remind physicians of the need to test patients for CD when clinical and laboratory data suggest that the patient is at high risk of CD.
A number of studies have used algorithms to identify gastrointestinal disorders,13–16 but we are only aware of one study examining whether artificial intelligence techniques can facilitate the diagnosis of CD.17
The aim of this study was to test two algorithms of symptoms, signs and diagnostic codes to construct a model that will identify individuals at high risk of CD and who would benefit from CD screening.
Materials and methods
This study was carried out by a study group consisting of one pediatric gastroenterologist (JFL), one gastroenterologist (JAM), two statisticians (ER, PSK), two bioinformaticians (JP, CGC), and two IT technicians (SM, MD). All data in this study were obtained from the Mayo Clinic's EMR systems.
Outcome measure: CD
Through a local database at the Mayo clinic (Rochester, Minnesota, USA) maintained by JAM, we identified 237 patients with an incident physician-assigned diagnosis of CD (International Classification of Disease, ICD9 code 579.0) and villous atrophy (Marsh histopathology stage 3) on small intestinal biopsy between 1995 and 2012. Data on histopathology were obtained from the pathology department of the Mayo Clinic. Of these patients, 233 had a record in Mayo's EMR data and constituted the basis of the present study. We queried the EMR data via ICD9 codes as well as processing of provider clinical notes via text mining. The celiac diagnosis in participating patients was manually adjudicated by a trained research assistant in CD. With each patient, an anchor date was provided that indicated the encounter date when the CD was diagnosed. We used an arbitrary time span of 1 week before and after the anchor date when pulling EMRs for each patient.
Controls
For each individual with CD, we randomly selected five controls that were matched for age, sex, birth year and statehood at time of diagnosis (and corresponding year in controls) (n=1168). Controls were also obtained using EMR data from Mayo's data warehouse (called MCLSS; Mayo Clinic Life Sciences System) using the Data Discovery and Query Builder (DDQB) tool. The ratio between controls and cases (5:1) was chosen arbitrarily. Because of the matching procedure, the reasons for attending the Mayo Clinic varied and controls may have had either mild or severe disease not related to CD. None of the controls had a diagnosis of CD before the end of follow-up. A similar date range was used to pull EMRs for each control subject.
Detecting CD via natural language processing (NLP)
We constructed an EMR-based algorithm that comprised NLP of clinical notes as well as searches for ICD codes signaling conditions associated with CD. As with most NLP pipelines and classifiers that annotate patient data using named entity recognition, a key aspect that eventually affects the performance of the classifier is to define a dictionary comprising named entities of interest. Search terms were included according to the clinical experience of two of the coauthors (JFL and JAM). These terms were matched exactly in the text.
In order to maximize sensitivity, we searched notes by gastroenterologists, but also notes from other specialties, including general internal medicine, within Mayo's clinical practice. The NLP search terms did not include any terms for family history of disease, since this is not systematically recorded in the EMR. None of the data used for the algorithm were collected directly from patients for the purpose of this study. In addition, all definitive terms are removed from the dictionary, except for special cases that are mentioned in the following dialog.
By limiting the dictionary to those words and phrases that represent symptomatic descriptive terms —for example, ‘diarrhea’, ‘iron deficiency,’ and ‘weight loss’—but excluding terms that were more definitive of the disease itself, a balance needed to be achieved to return the most effective recall/precision ratio.
Aside from the term-extraction component, the pipeline was also able to determine the context of the phrase as it relates to the patient historically, family history and negation. Since genetics have a significant factor in the probability of incurring the disease, family history contexts are allowed access to the definitive descriptions of the disease. For example, the phrase ‘Mother had CD’ will be of interest, but not ‘the patient has symptoms of sprue’.
The trade-off of using the more symptomatic terms is the prevalence of these phrases for other comorbidities. Therefore, the system was able to keep ‘false positives’ at a minimum while maximizing the rate of ‘true positives’ by applying a voting procedure. Using this technique, a mention from the phrases of interest would be counted as a vote once, and only once, if it were discovered as a non-negated phrase anywhere in the document.
Using ‘voting’, all symptomatic terms are given equal weight for determining relevance to the disorder in order to keep the procedure simple. Within the same document, the presence of unique mentions from the list of the following were considered: ‘weight loss (or loss of weight), underweight, diabetes type 1 (or type 1 diabetic), diarrhea (or frequent stools or watery stools), iron deficiency (or anemia, low ferritin, microcytic or iron supplement), osteoporosis (or Fosamax), depression, anorexia, hyperthyreosis (or hyperthyroidism, Hashimoto or autoimmune thyroid), Down syndrome, malabsorption, short stature, growth failure (or failure to thrive), small-bowel, irritable bowel syndrome (or ibs), abdominal’.
Thus no consideration is given to the relative location of either the term or relationship to another term in a given time span. After several iterations, it was determined that two votes was the optimum value for finding prospective cases without introducing those patients with non-related illnesses.
Selected NLP terms are listed in table 1.
Table 1.
Characteristics of study participants
| Characteristic | NLP search |
ICD9 search |
||
|---|---|---|---|---|
| Celiac disease N=216 |
Controls N=280 |
Celiac disease N=202 |
Controls N=524 |
|
| Age at CD diagnosis, mean±SD | 42.2±23.4 (4/26/1969 average DOB) | 70.1±13.7 | 40.5±23.0 | 41.1±24.9 |
| Male sex | 74 (34.2%) | 94 (33.6%) | 68 (33.7%) | 156 (29.8%) |
| Selected NLP search terms* | ||||
| Weight loss | 16 (7.4%) | 9 (3.2%) | – | – |
| Iron deficiency | 48 (22.2%) | 1 (0.3%) | – | – |
| Anemia | 44 (20.4%) | 6 (2.1%) | – | – |
| Underweight | 0 (0%) | 0 (0%) | – | – |
| Osteoporosis | 17 (7.9%) | 12 (4.3%) | – | – |
| Selected ICD9 codes† | ||||
| Type 1 diabetes | – | – | 18 (8.9%) | 34 (6.5%) |
| Chronic diarrhea | – | – | 113 (55.9%) | 316 (60.3%) |
| Weight loss | – | – | 10 (5.0%) | 9 (1.7%) |
| Autoimmune thyroid disease | – | – | 16 (7.9%) | 22 (4.2%) |
*We also used the following terms in our NLP search: anorexia, hyperthyreosis/hypothyreosis, hyperthyroidism, Hashimoto, Down syndrome, malabsorption, abnormal weight loss, short stature, growth failure, failure to thrive, poor growth, frequent stools, watery stools, diabetes type 1/type 1 diabetic, ibs, small-bowel, irritable bowel syndrome, abdominal, autoimmune thyroid, diarrhea, low ferritin, microcytic, iron supplement, depression, Fosamax.
†We also used the following ICD9 codes in our search: fatigue, growth failure, Addison's disease, microscopic colitis, Down syndrome, osteoporosis, IgA deficiency, anemia, bloody diarrhea.
CD, celiac disease; DOB, date of birth; ICD, International Classification of Disease; NLP, natural language processing.
Detecting CD through ICD9 code search
This algorithm was based on a system of assigning points for each of the following diagnoses: chronic diarrhea, weight loss, growth failure (age <18 years), fatigue, type 1 diabetes, Addison's disease, autoimmune thyroid disease, microscopic colitis, Down syndrome, osteoporosis (age <50 years), IgA deficiency, and iron deficiency. Negative points were assigned for the presence of bloody diarrhea or if patients were African-American. Each criterion was counted as a single point, with the points adding together. Patients with at least three points were classified as positive. Selected ICD9 codes are listed in table 1.
Statistical analysis
For the NLP search algorithm, CD cases and controls were randomly divided into two datasets (two-thirds of training set for developing the algorithm and one-third of testing set for validating the results). Using the training set, the optimal number of hits was chosen by maximizing the sum of sensitivity and specificity. Performance of the algorithm using the optimal number of hits was assessed in the testing dataset, by calculating sensitivity, specificity, and F-measure. A similar approach was used for the ICD9 code search. We used SAS (V.9.2) for the analyses.
Results
Patient characteristics
In a group of 233 patients with confirmed CD, 216 had an adequate date range for an NLP search (ie, they had a record containing free text at time of first diagnosis), while 202 patients were eligible for an ICD9 code search (ie, they had at least one ICD9 diagnosis within the specified time span). Similarly, of 1168 matched controls, 280 and 524 subjects were included for NLP and ICD9 search, since they had a record of free text or at least one ICD9 diagnosis, respectively, in the stipulated time frame. Most of the randomly selected controls from the Mayo Clinic EMR database system did not hence fulfill our time span criteria.
Age and sex distribution of CD patients and control subjects are given in table 1. Our study included both children and adults. For subjects used in the NLP search, the mean age was 42.2 years for CD cases and 70.1 years for controls. Subjects used in the ICD9 search had a similar age distribution between CD cases and controls. Both search groups had fewer male subjects (34.2% vs 33.6% for NLP; 33.7% vs 29.8%).
Determination of optimum number of hits
In order to determine the optimum number of hits for finding prospective cases of CD, we compared sensitivity and specificity of one to four hits for the NLP search and the ICD9 search (table 2). On the basis of these iterations, we chose two hits as the cut-off for the NLP algorithm (figure 1) and three hits for the ICD9 algorithm (figure 2).
Table 2.
Sensitivity and specificity with different number of hits in training set
| Number of hits | NLP | ICD-9 | ||
|---|---|---|---|---|
| Sensitivity (%) | Specificity (%) | Sensitivity (%) | Specificity (%) | |
| 1 | 90.4 | 69.1 | 98.5 | 10.6 |
| 2 | 78.1 | 94.5 | 53.8 | 50.0 |
| 3 | 64.4 | 97.2 | 23.5 | 88.0 |
| 4 | 50.7 | 97.8 | 10.7 | 97.4 |
ICD, International Classification of Disease; NLP, natural language processing.
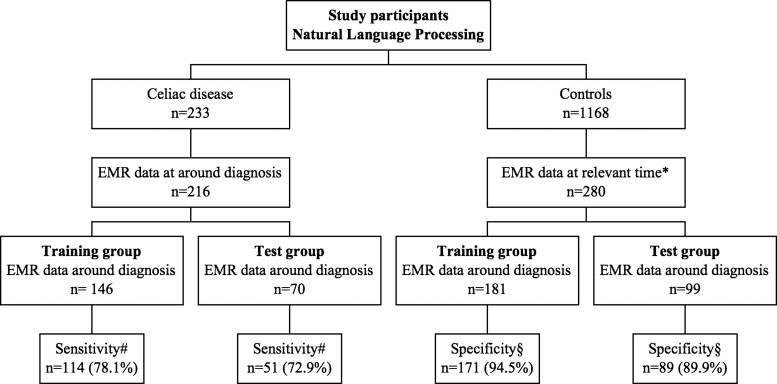
Figure 1.
Natural language processing (NLP): flowchart of study participants. EMR, electronic medical record. *At the same time as the matched individuals with celiac disease were diagnosed. #Number and percentage of individuals detected by the NLP algorithm using two hits. §Number and percentage of individuals not detected by the NLP algorithm using two hits.
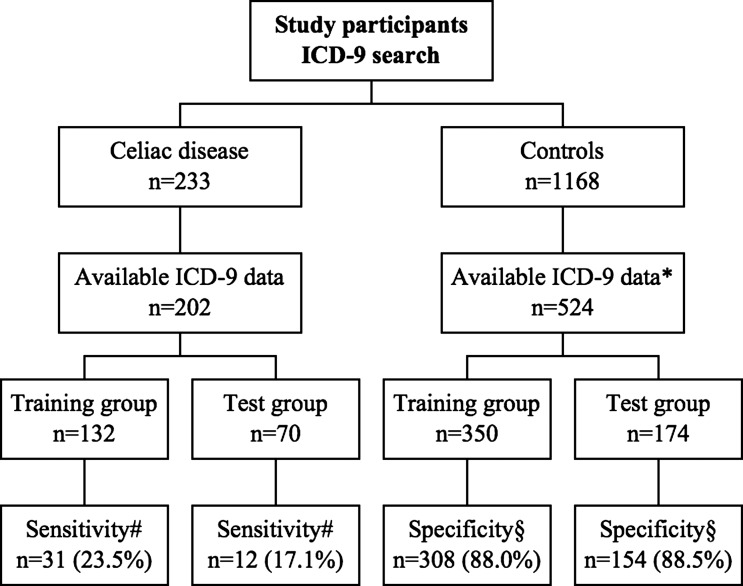
Figure 2.
ICD9 code search: flowchart of study participants. ICD, International Classification of Disease. *At the same time as the matched individuals with celiac disease were diagnosed. #Number and percentage of individuals detected by the ICD algorithm using three hits. §Number and percentage of individuals not detected by the ICD algorithm using three hits.
Natural language processing
The training group comprised 146 CD cases and 181 control subjects (figure 1). Table 2 shows the sensitivity and specificity of the algorithm based on different numbers of hits, and two hits as the optimal number (figure 1). With two hits as the cut-off, 114/146 patients were classified as positive (sensitivity=78.1%) and 171/181 control subjects were classified as negative (specificity=94.5%). In the validation set, two hits provided a sensitivity of 72.9% and specificity of 89.9%. The F-measure for accuracy of the NLP algorithm using two hits was 84.5% in the training set and 78.0% in the validation set. In a representative US population of individuals without a prior celiac diagnosis (assuming that 0.6% had undiagnosed CD3), this NLP algorithm could identify a group of individuals where 4.2% would have CD (positive predictive value based on the testing dataset).
Detecting CD through ICD9 code search
Using the training set of 132 CD cases and 350 control subjects, the ICD9 search algorithm was optimized with three hits, which provided a sensitivity of 88.1% and specificity of 23.5% (table 2 and figure 2). In the validation set, sensitivity and specificity of the algorithm using three hits were 17.1% and 88.5%, respectively, and the positive predictive value was 0.9%. The F-measure for accuracy of the ICD9 code search using three hits was 30.0% in the training set and 23.3% in the validation set.
Discussion
CD occurs in 1% of the Caucasian population, and patient and doctor's delay is often substantial.18 Part of that delay is due to unawareness of CD. The main contribution of this paper is that it presents data on both sensitivity and specificity for two algorithms, one using NLP and the other ICD9 code searches. We a priori divided the true positive cases into two groups: a training dataset used for testing both free text searches and ICD9 searches, and a validation test dataset where our final models were applied.
Clinical decision support systems (CDSSs)19 can improve clinical practice, and can be used for several purposes such as recommendations for testing, but also to predict the prognosis of healthy and non-healthy individuals.20 Predictors of future disease do not necessarily have to cause the disease that is being predicted; they may just reflect underlying factors.20 Among well-known predictors are the Framingham Cardiovascular Risk Score21 and the neonatal score introduced by Dr Virginia Apgar, and now referred to as the Apgar score.22
Using two hits as the cut-off, our computerized NLP algorithm identified 72.9% of all patients with CD and had a specificity of 89.9%. The accuracy of the algorithm was 78.0% based on the F-measure. The British NIHR (National Institute of Health Research) recently reviewed 24 studies on computerized decision support systems for ordering laboratory tests.23 The general conclusion of the review was that CDSSs have a role in modern healthcare. While CDSSs may lead to increased testing for disease, they can also direct physicians to the correct test, and thereby lead to a decrease in economic costs.23 Our final NLP model included 21 text terms, and several of these variables were also included in the final CD model by Tenorio et al (diarrhea and weight loss).17 That several of the included symptoms and signs also occurred with a high prevalence in the controls (table 1) confirms the notion that CD is difficult to diagnose and features different phenotypes.1
The current algorithm should not be seen as an attempt to escape serology testing and ultimately small intestinal biopsy, but rather as a means of identifying patients at high risk of CD, where CD serology is feasible. The diagnosis of CD cannot be made exclusively on the basis of phenotype.
Applying the NLP algorithm to a population of 10 000 individuals without a prior diagnosis of CD, and where undiagnosed CD occurs in 0.6%3 (n=60 true CD patients), we would detect 44 of the CD patients but also 1004 controls (the latter being false positive). Hence the rate of CD among individuals defined as ‘positive’ by our NLP algorithm was 4.2% (44/(44+1004)). This rate is similar to the CD prevalence among first-degree relatives,24 25 a group currently recommended for celiac screening by both NICE (National Institute for Health and Clinical Excellence, UK)26 and WGO-OMGE (World Gastroenterology Organization).27 Hence our NLP could be used as a tool to identify a high-risk group where screening with tissue transglutaminase is appropriate.
In contrast, our ICD9 code algorithm yielded lower sensitivity (17.1% for ICD9 vs 72.9% for NLP), specificity (88.5% for ICD9 vs 89.9% for NLP) and overall accuracy based on F-measure (23.3% for ICD9 vs 78.0% for NLP). While subjects used in these two algorithms did not completely overlap, results using the same subjects (173 CD cases and 153 controls) were not significantly different from those reported above for each algorithm. Thus, the ICD9 code algorithm currently cannot be recommended for large-scale use to identify individuals at risk of CD.
This paper has some strengths and weaknesses. A strength of our paper is that we used an out-sample to eliminate the risk of overfitting our model to our case population. We divided our CD patients into two groups and tested them separately. Testing an algorithm in two independent datasets increases the generalizability. Of note, both sensitivity and specificity decreased slightly in the validation test group.
Our computerized approach means that we were not limited to a small number of variables since no patient or physician will have to enter any data into the model. Such data will be retained from existing clinical registers. As opposed to that of Tenorio et al,17 our algorithm is not intended for individual physicians. Our study differs from that of Tenorio et al17 on other accounts as well. While we agree that CDSSs can play a role in diagnosing CD, Tenorio et al17 achieved their highest precision using a Bayesian classifier implemented into a web-based decision system, while we focused on a retrospective EMR-driven algorithm that can be implemented into large-scale healthcare.
We cannot rule out the possibility that our patients with CD differ from the general CD population with regard to symptoms and signs, since the Mayo clinic is a tertiary center. This may also explain why we could only identify 237 incident cases with CD where the small intestinal biopsy had been obtained at the Mayo Clinic. We did not include CD cases where the biopsy had taken place outside Mayo since the appearance of the biopsy specimen could not be verified in those cases.
Finally, there were age differences between celiac cases (42 years) and controls (70 years) in our NLP model. An algorithm based on free text search (NLP) can only work when the patient chart contains text. More older than younger people had been in touch with healthcare during a specified time span, and this led to a higher average age in the control group and is likely to decrease both specificity and the positive predictive value. However, it should be noted that our celiac algorithms are only intended to identify a group of individuals at high risk of CD who would then undergo testing for CD.
In conclusion, our NLP algorithm had a reasonable sensitivity and specificity, while ICD9 codes could not be used to identify patients at risk of having CD. Computerized support that enables the automated review of electronic health records, if coupled to testing, is likely to increase the diagnostic rate of CD. Ideally, our NLP algorithm should be tested in other populations and settings.
Acknowledgments
Research assistant Carol van Dyke for help adjudicating the celiac diagnosis in patients.
Footnotes
Contributors: JFL contributed to the study design, interpretation of data, and wrote the first draft of the article. JP contributed to the study design, revising the article and study supervision. SM contributed to the study design, the data analysis and revising the article. MD contributed to the study design and revision of the article. PSK contributed to the study design, the data analysis and revising the article. CGC contributed to the study design and revision of the article. ER contributed to the study design including statistical advice, data analysis and revising the article. JAM contributed to the study design, the interpretation of data, revising the article and study supervision.
Funding: JFL was supported by grants from the Swedish Society of Medicine, the Swedish research council (522-2A09-195), the Swedish Celiac Society, and the Fulbright Commission.
Competing interests: JAM: Grant support,Alba Therapeutics (>US$50 000); Advisory board, Alvine Pharmaceuticals (<US$10 000), Nexpep (<US$10 000); Consultant (none above 10 000 USD), Ironwood, Flamentera, Actogenix, Ferring Research Institute, Bayer Healthcare Pharmaceuticals, Vysera Biomedical, 2G Pharma, ImmunosanT, and Shire US. The National Institutes of Health—DK057892.
Ethics approval: Mayo Clinic institutional review board.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Ludvigsson JF, Leffler DA, Bai JC, et al. The Oslo definitions for coeliac disease and related terms. Gut 2013;62:43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Walker MM, Murray JA, Ronkainen J, et al. Detection of celiac disease and lymphocytic enteropathy by parallel serology and histopathology in a population-based study. Gastroenterology 2010;139:112–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rubio-Tapia A, Ludvigsson JF, Brantner TL, et al. The prevalence of celiac disease in the United States. Am J Gastroenterol 2012;107:1538–44; quiz 37, 45 [DOI] [PubMed] [Google Scholar]
- 4.Stenson WF, Newberry R, Lorenz R, et al. Increased prevalence of celiac disease and need for routine screening among patients with osteoporosis. Arch Intern Med 2005;165:393–9 [DOI] [PubMed] [Google Scholar]
- 5.Ludvigsson JF, Michaelsson K, Ekbom A, et al. Coeliac disease and the risk of fractures—a general population-based cohort study. Aliment Pharmacol Ther 2007;25:273–85 [DOI] [PubMed] [Google Scholar]
- 6.Ludvigsson JF, Montgomery SM, Ekbom A. Celiac disease and risk of adverse fetal outcome: a population-based cohort study. Gastroenterology 2005;129:454–63 [DOI] [PubMed] [Google Scholar]
- 7.Rampertab SD, Pooran N, Brar P, et al. Trends in the presentation of celiac disease. Am J Med 2006;119:355.e9–14 [DOI] [PubMed] [Google Scholar]
- 8.Bottaro G, Cataldo F, Rotolo N, et al. The clinical pattern of subclinical/silent celiac disease: an analysis on 1026 consecutive cases. Am J Gastroenterol 1999;94:691–6 [DOI] [PubMed] [Google Scholar]
- 9.Ludvigsson JF, Brandt L, Montgomery SM, et al. Validation study of villous atrophy and small intestinal inflammation in Swedish biopsy registers. BMC Gastroenterol 2009;9:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bao F, Yu L, Babu S, et al. One third of HLA DQ2 homozygous patients with type 1 diabetes express celiac disease-associated transglutaminase autoantibodies. J Autoimmun 1999;13:143–8 [DOI] [PubMed] [Google Scholar]
- 11.Elfstrom P, Montgomery SM, Kampe O, et al. Risk of thyroid disease in individuals with celiac disease. J Clin Endocrinol Metab 2008;93:3915–21 [DOI] [PubMed] [Google Scholar]
- 12.Catassi C, Fasano A. Celiac disease diagnosis: simple rules are better than complicated algorithms. Am J Med 2010;123:691–3 [DOI] [PubMed] [Google Scholar]
- 13.Horowitz N, Moshkowitz M, Halpern Z, et al. Applying data mining techniques in the development of a diagnostics questionnaire for GERD. Dig Dis Sci 2007;52:1871–8 [DOI] [PubMed] [Google Scholar]
- 14.Sakai S, Kobayashi K, Nakamura J, et al. Accuracy in the diagnostic prediction of acute appendicitis based on the Bayesian network model. Methods Inf Med 2007;46:723–6 [PubMed] [Google Scholar]
- 15.Lahner E, Intraligi M, Buscema M, et al. Artificial neural networks in the recognition of the presence of thyroid disease in patients with atrophic body gastritis. World J Gastroenterol 2008;14:563–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pace F, Riegler G, de Leone A, et al. Is it possible to clinically differentiate erosive from nonerosive reflux disease patients? A study using an artificial neural networks-assisted algorithm. Eur J Gastroenterol Hepatol 2010;22:1163–8 [DOI] [PubMed] [Google Scholar]
- 17.Tenorio JM, Hummel AD, Cohrs FM, et al. Artificial intelligence techniques applied to the development of a decision-support system for diagnosing celiac disease. Int J Med Inform 2011;80:793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green PHR, Stavropoulos SN, Panagi SG, et al. Characteristics of adult celiac disease in the USA: results of a national survey. Am J Gastroenterol 2001;96:126–31 [DOI] [PubMed] [Google Scholar]
- 19.Kawamoto K, Houlihan CA, Balas EA, et al. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ 2005;330:765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moons KG, Royston P, Vergouwe Y, et al. Prognosis and prognostic research: what, why, and how? BMJ 2009;338:b375. [DOI] [PubMed] [Google Scholar]
- 21.Kannel WB, McGee D, Gordon T. A general cardiovascular risk profile: the Framingham Study. Am J Cardiol 1976;38:46–51 [DOI] [PubMed] [Google Scholar]
- 22.Apgar V. A proposal for a New Method of Evaluation of the Newborn Infant. Curr Res Anesth Analg 1953;32:260–7 [PubMed] [Google Scholar]
- 23.Main C, Moxham T, Wyatt JC, et al. Computerised decision support systems in order communication for diagnostic, screening or monitoring test ordering: systematic reviews of the effects and cost-effectiveness of systems—Health Technology Assessment. Southampton: NETSCC, 2010 [DOI] [PubMed] [Google Scholar]
- 24.Rubio-Tapia A, Van Dyke CT, Lahr BD, et al. Predictors of family risk for celiac disease: a population-based study. Clin Gastroenterol Hepatol 2008;6:983–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fasano A, Berti I, Gerarduzzi T, et al. Prevalence of celiac disease in at-risk and not-at-risk groups in the United States: a large multicenter study. Arch Intern Med 2003;163:286–92 [DOI] [PubMed] [Google Scholar]
- 26.NICE Coeliac disease. Recognition and assessment of celiac disease. London: NICE clinical guideline No 86, 2009 [Google Scholar]
- 27.Bai J, Fried M, Corazza GR, et al. Celiac disease, World Gastroenterology Organisation Global Guidelines. Milwaukee, WI, USA: WGO-OMGE, 2012 [Google Scholar]