Abstract
Objective
We define and validate an architecture for systems that identify patient cohorts for clinical trials from multiple heterogeneous data sources. This architecture has an explicit query model capable of supporting temporal reasoning and expressing eligibility criteria independently of the representation of the data used to evaluate them.
Method
The architecture has the key feature that queries defined according to the query model are both pre and post-processed and this is used to address both structural and semantic heterogeneity. The process of extracting the relevant clinical facts is separated from the process of reasoning about them. A specific instance of the query model is then defined and implemented.
Results
We show that the specific instance of the query model has wide applicability. We then describe how it is used to access three diverse data warehouses to determine patient counts.
Discussion
Although the proposed architecture requires greater effort to implement the query model than would be the case for using just SQL and accessing a data-based management system directly, this effort is justified because it supports both temporal reasoning and heterogeneous data sources. The query model only needs to be implemented once no matter how many data sources are accessed. Each additional source requires only the implementation of a lightweight adaptor.
Conclusions
The architecture has been used to implement a specific query model that can express complex eligibility criteria and access three diverse data warehouses thus demonstrating the feasibility of this approach in dealing with temporal reasoning and data heterogeneity.
Keywords: Cohort Selection, Eligibility Criteria, Electronic Health Records, Modelling, Architecture
Background and significance
A key challenge facing researchers in health informatics is developing techniques to identify patient cohorts for clinical trials and other studies from electronic health records (EHR) systems that were not designed with that purpose in mind. The task of repurposing is made more difficult by the heterogeneity of patient data. It is always possible to access an individual source on an ad hoc basis by fashioning specific queries for a particular study. However, this approach can be very expensive to conduct across several data sources and certainly does not scale to meet the increasing demands of clinical research. There is a greater need for an approach that allows users to formulate their queries based on a generic model that provides users with a common view of data hailing from multiple diverse sources, in particular:
Researchers need not concern themselves as to how the data are represented or stored in each source.
It is possible to query and combine data from multiple sources in a consistent way, so that a single query may be sent to multiple sources and the results aggregated.
A researcher-friendly interface may be built on top of the generic model so that non-technical researchers can operate such a system.
To achieve this, the criteria used to query the data or select cohorts must be framed in an unambiguous way and evaluated automatically against structured data that will be available from at least some of the sources available. For example, a criterion to exclude patients with ‘abnormal liver function tests’ (LFT) would not meet this condition unless first, the specific LFT are identified with predefined reference ranges; second, at least some of the sources contain the results of these tests in a structured (as opposed to free-text) form; and third LFT can be semantically identified and unambiguously distinguished.
Taweel et al1 identify six types of heterogeneity in patient data: system, syntactic, structural, semantic, chronology and security. For the problem specified above, primarily two of these concern us in this paper: semantic and structural. By semantic heterogeneity, we mean differences in how particular attributes of the patient and associated events are expressed. We shall assume that any data source has a vocabulary that is used to define the semantic representation such as the coding system for clinical concepts and units of measurement for physical quantities. By structure, we mean the way in which attributes relating to a patient or event are linked to one another; for example, in schematic representation. There are limitless potential implementations of both vocabulary and structure. In as far as standards do exist for the representation of patient data2–4 they are not widely followed and many legacy systems predate these standards.
In addition to the problem of data heterogeneity, there is the issue of how to provide a common view of data to identify the cohort of interest to the researcher. For clinical trials and many other studies, this is defined by a set of eligibility criteria (EC), usually framed using natural language. Many attempts have been made to formalize EC to reduce ambiguity and afford formal reasoning.5–7 Clearly, for EC to be evaluated automatically, some formal model will be required. According to Ross et al,8 a survey of 1000 (natural language) EC, showed that 47% included some temporal condition, 14% contained some arithmetic condition and 53% some Boolean connector. So, in addition to dealing with a formal representation of clinical concepts (eg, by the use of coding systems), to be widely applicable, any such model should also address the following:
Boolean connectors such as AND, OR and NOT;
comparison of values to reference ranges; for example, hemoglobin A1c result greater than 7%;
semantics of temporal concepts/aspects.
However, computing EC across different data sources is also dependent on the capability of their underlying querying technologies or languages through which their data can be interrogated. For a number of demanding EC these will also determine and limit the complexity of the queries that can be answered by a data source.
Although systems for selecting or counting eligible patients have been constructed such as informatics for integrating biology and the bedside (i2b2)/SHRINE,9–11 the electronic patient care research network (ePCRN),12 13 FARSITE,14 and VISAGE,15 such set-ups presume the use of a specific model and/or homogeneous data sources. ePCRN uses a local grid-based software service, named Gateway, in which clinical data made available by the institution are placed. The Gateway structure is based on the continuity of care record (CCR) standard3 as the common model. ePCRN uses an extraction–transform–load (ETL) process to transfer the local raw data to Gateway, with which other ePCRN user tools communicate. ePCRN uses a graphical interface tool to formulate queries and transform them into CCR-compliant XPath queries. Similarly, i2b2 adopts a local data warehouse approach, with a specific data model at its core. It uses a service-based layer with a programmable interface to communicate with its tools. SHRINE11 is used as a graphical user interface (GUI) tool to formulate and run queries for i2b2, which are eventually rendered locally in SQL. FARSITE similarly relies on its own data model and a GUI tool. These tools, ePCRN, SHRINE and FARSITE, formulate reasonably complex queries but are limited in their temporal reasoning capabilities and ability to access heterogeneous data sources.
Various solutions to the problem of expressing temporal semantics have been proposed. TSQL216 allows the user to query a database with known schema using an extension to SQL. However, this system is not implemented on the database platforms used in hospitals. Chronus17 provides a temporal query language intended for clinical use. This is built on SQL but again assumes that the database schema is known. Deshpande et al18 proposed a system for evaluating EC with temporal semantics again based on SQL but use a predefined database schema. Thus, these systems do not address semantic and structural heterogeneity. Therefore, ensuring such a model is capable of using data from heterogeneous sources while implementing temporal semantics is a far from trivial problem and one that we address. Both its expressivity and reasoning capability should be agnostic and independent of that of the data sources. Indeed, the separation of the reasoning about clinical data and the extraction of that data have already been employed in the RetroGuide system19 20 for the construction of workflows rather than cohort identification, so that data from diverse EHR systems can be extracted while presenting an identical interface to the user.
This paper proposes a model-based approach combined with configurable template-based techniques for the full cycle of EC capture, transmission, translation into computable queries to interrogate and interoperate with diverse data sources. The approach ensures semantic consistency throughout the process by faithfully translating the researcher's EC requirements into corresponding executable queries on respective data sources. The approach enables an adaptable architecture for the interrogation of heterogeneous data sources by separating the functions of computational reasoning from those of data extraction reducing the complexity of interoperability into a set of adaptable templates. This approach is currently being adopted in the EHR for clinical research (EHR4CR), a European Union innovative medicines initiative project that aims to support clinical research within the secondary care domain.
Objective
We define and validate an architecture for systems that identifies patient cohorts for clinical trials from multiple heterogeneous data sources. This architecture has the following properties:
Users can represent a set of EC as a query to interrogate diverse data sources by means of a common representation of that query—thus we require an explicit query model.
Such a query model can support temporal reasoning as well as be powerful enough to express predicates based on the required range of clinical attributes of patients.
It should be capable of accessing multiple data sources with diverse structures and vocabularies.
The effort required to adapt the system to new data sources is reduced by separating the source-specific part from the remainder of the system that is source agnostic.
In the next section, we present the architecture and its components and describe the generic approach. We then validate this architectural approach by implementing a specific but widely applicable query model and show that it can connect to multiple heterogeneous data sources. The proposed model is evaluated with 10 trials from the EHR4CR project. It is then shown that this model can access using three different heterogeneous data sources including the EHR4CR data warehouse, an i2b2 warehouse10 and the general practice research database of the UK's medicines and healthcare products regulatory agency.
Materials and methods
The key aim of this approach is to enable clinical researchers to identify eligible patient cohorts from data sources without much knowledge about their semantic or structural representations. At the data source level, an information model defines both the attributes of patients or events associated with them and the structure that relates them together, in terms of entities and relationships. To access the data, the query model specifies and limits what questions may be asked of the information available. We argue that the query model and information model are actually always present; it is just that they are often implicit and bound together. For a simple architecture, the information model can be the database schema; the query model is implied by the query language; for example, SQL. However, to achieve our aim, we explicitly separate the information models for each data source from the common query model used by the researcher. The query model is therefore independent of the specific information models that may be used in the data sources, which may conform to one of the many published models2–4 21 22 or be peculiar to the specific data source. We do require, however, that clinical concepts are represented unambiguously by the query vocabulary and the only current way to do this is using a coding system.
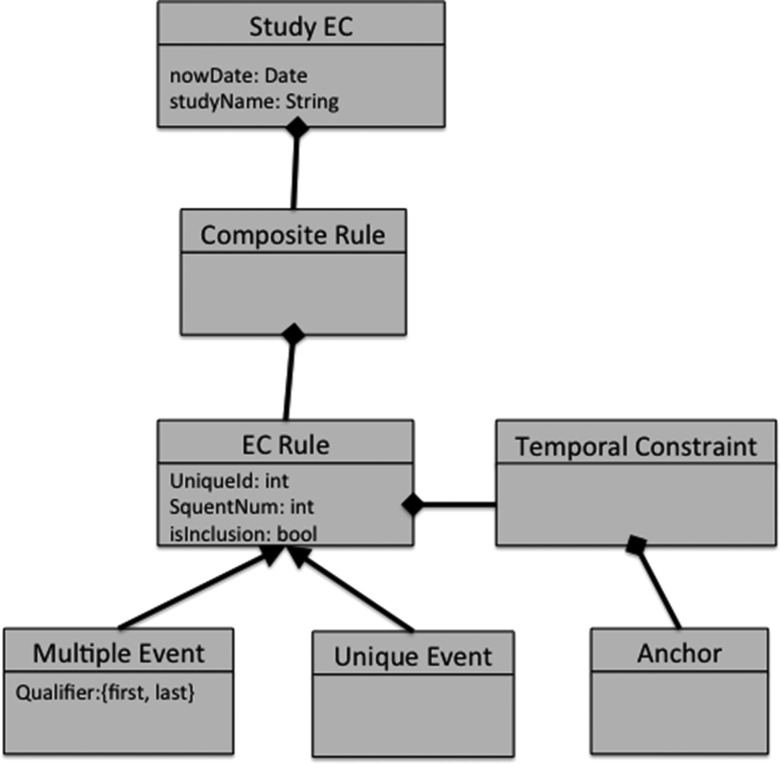
Given their diversity, it would be impractical for clinical researchers to comprehend the distinct information models of each individual data source. What clinical researchers actually need is to express the characteristics of eligible patients, which is related to the query model not the information model. Therefore, our approach abstracts the query model into a form that clinical researchers can understand and use to express their eligibility needs, yet can be applied to the data source information models without losing its structural or semantic consistency. We refer to this query model as an abstract query model (AQM) and an implementation, the query model implementation (QMI) that can compute results. The AQM (and thus QMI) is agnostic to source information models of individual data sources. To meet clinical research needs, it should express the above-mentioned characteristics including semantics of clinical and temporal concepts with possible logical and Boolean constructs. Figure 1 below illustrates the example of AQM, which is later used to evaluate our approach.
Figure 1.
Part of the electronic health records for clinical research (EHR4CR) eligibility criteria (EC) model used as an example of an abstract query model.
Before proceeding further, we need to make two assumptions about the diverse data sources, without which querying target data sources consistently using the proposed approach may not be possible.
Each source contains at least some patient data specified in the information model in a structured (not free text) representation.
Mappings between the relevant vocabularies, contained in a dictionary, are computable.
In relation to this second point, once mappings between coding systems exist in electronic form they may be automated. However, the process of preparing vocabulary mappings is an arduous one and has only been accomplished in specific cases.
High-level architecture
Fundamentally, the query model lies at the heart of the architecture we propose. The general architecture itself is consistent with any number of query models but henceforth we shall assume that a particular AQM has been defined and chosen.
To apply an actual query defined by the AQM to a particular data source, while staying agnostic to any underlying information model, it is essential to separate the functions of processing the data. Computationally, executing queries to identify eligible patients can be seen as two distinct functions: extracting the relevant data items about the patients and then reasoning over them. The key feature of the proposed architecture is that these two functions are separated because the reasoning can be conducted independently of the structure and semantic representation of the patient data whereas extracting the data cannot. This gives rise to a four-step process implemented at the data source end:
Use the query expressed by the AQM to determine the necessary data items needed to compute the query and then generate source-independent representations of these data as in the form of mini-query templates.
Use the mini-query templates to create a number of corresponding mini-queries in the source query language; for example, SQL (pre-processing).
Run these mini-queries on the specific patient database and obtain the raw results.
Use the QMI and raw results to reason over the data automatically and generate results that meet the AQM query, that is, identify eligible patients (post-processing).
Figure 2 shows a high-level view of the architecture, in which queries defined by the AQM can be transmitted to several selected data sources. The researcher accesses the system only by the workbench that allows him to compose an instance of an AQM query and view the results. At the data source end, the link between the data source and QMI are connected by an adaptor that is specific to each data source. The adaptor is a lightweight component that can be constructed for the specific data source for the sole purpose of extracting data. By ensuring that most of the reasoning functionality is placed in the QMI, the adaptor can become a configurable component, reducing the cost of connecting to new data sources. Given the paper is focused on the latter part, the rest of the paper will focus on the data key parts of the architecture, illustrated in figure 3.
Figure 2.
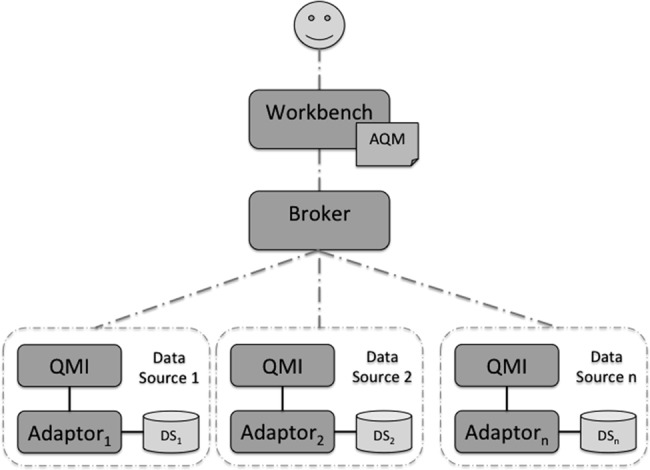
High-level architecture for multiple data sources. AQM, abstract query model; QMI, query model implementation.
Figure 3.
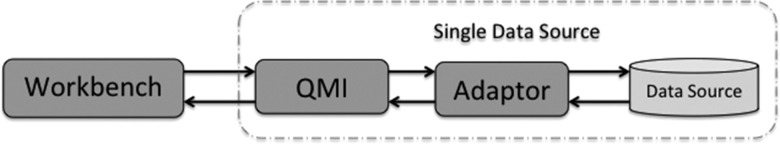
High-level architecture for a single data source. QMI, query model implementation.
Detailed view
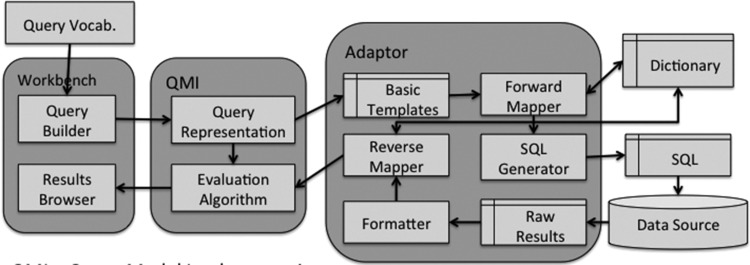
The GUI workbench will consist of both a query builder that is used to compose the query using a query vocabulary, which may be distinct from the vocabulary used by the data source. A results browser allows the results to be displayed. A detailed view of the architecture is shown in figure 4.
Figure 4.
Detailed architecture. QMI, query model implementation.
The query model has two components:
A query representation more powerful than general query languages such as SQL so that it can perform more advanced temporal reasoning.
An evaluation algorithm to produce end results (ie, cohort identification) for the user to browse.
Both would be developed in an application programming language, such as Java, rather than using database technology. The query representation is used to hold the query composed by the user and generate a set of elementary mini-queries that only extract the required data to perform the reasoning. They perform no reasoning of themselves. The mini-queries will contain clinical concepts expressed in the query vocabulary; for example, a diagnosis or laboratory test. This has to be translated into the data source vocabulary using the forward mapper before they can be executed on the database. As vocabulary mappings between systems are often not one to one, at the forward mapping stage a single code in the query vocabulary may be mapped to many codes in the data source vocabulary. The translated mini-queries are then mapped to the data source query language; for example, SQL, according to the database schema. These steps comprise the pre-processing.
The raw results from the data source are post-processed in three stages. First, as the format of the raw results may vary between different data sources, a formatter is required to create a canonical representation. Second, the results will be expressed in the source-specific vocabulary and so will need to be translated into the query vocabulary so that they can be evaluated against the original AMQ query. This is done by the reverse mapper. Third, the evaluation algorithm uses both the QMI that holds the user's query and processed data to perform reasoning to produce the results; for example, eligible patient identification, which can be returned to the user GUI workbench. We note that Das and Musen17 also use post-processing to deal with temporal reasoning in a similar context but do not deal with differences in structural and semantic representation.
The pre/post-processing paradigm requires defining a small set (typically fewer than a dozen) of mini-query templates related to the high-level clinical concepts that would be defined in the AQM such as gender, diagnosis or a numerical reading. Thus defining the required set of templates requires identifying each high-level concept in the AQM and ensuring that they return the appropriate data items needed to evaluate each query. The number and nature of the templates is determined by the data requirements of the query model; a different query model would require a different set. The adaptor shown in figure 4, containing the source-specific components, is lightweight because the query templates are conceptually simple and mapping between vocabularies is straightforward once the mappings have been defined. The QMI is more heavyweight because reasoning using the evaluation algorithm must now be implemented explicitly rather than relying on the data-based management system (DBMS) to perform this role. The difficulty of implementing the evaluation algorithm is a function of the complexity of the chosen query model. The great advantage is that it separates computational reasoning from the data extraction function making it information model and data source agnostic and thus addressing heterogeneity using a lightweight adaptable architecture. An example that illustrates the computational steps can be found in the supplementary appendix (available online only).
Evaluation
We first demonstrate that the specific instance of the query model, the eligibility criteria model (ECM), is sufficiently powerful to be used on actual clinical trials and thus shows a practical application of the architecture proposed. We then describe how the source-independent part of the platform, the GUI and query model was connected with three data warehouses with their respective adaptors.
Results
The EHR4CR platform
The proposed approach has been implemented as part of the EHR4CR project to construct a platform for determining patient counts and then identifying specific patients for clinical trials at secondary care sites. At the time of writing only the patient count functionality was fully implemented. A query builder is used to compose a query representing a set of EC and this is then send to a number of remote hospital sites to calculate patient counts from their respective data warehouses. The communication technology used is beyond the scope of this paper but is described in Chen et al.23 Results of the counts from each site are displayed both for each hospital site and aggregated over all sites. The hospitals used heterogeneous warehouse implementations as detailed in the next section.
The query model AQM has been realized as the ECM. The ECM expresses the query as a set of rules, each of which defines a phenotype by one of the clinical events, drawn from CCR3 and shown in table 1. Each rule has a patient attribute defined by a clinical concept and expressed as a code in the query vocabulary. This is embedded within a predicate evaluated as true or false for each patient. The predicate may detect the existence of some attribute such as a diagnosis or compare a value (numerical or categorical) to a reference range. Each rule must specify whether the first or last reading is used and optionally has a temporal constraint requiring that it is before or after some temporal anchor; for example, the event from a previous rule. The rules are then combined using the Boolean logical operators to form a set of EC for each trial.
Table 1.
Clinical events used in the ECM
| Name| | CCR equivalent | Template used | Predicate type |
|---|---|---|---|
| Born | DateOfBirth | General | Existential |
| Codedstatus | FunctionalStatus | Coded observation | Categorical |
| Deceased | n/a | Dead | Existential |
| Diagnosis | Problem | Diagnosis | Existential |
| Gender | Gender | General | Categorical |
| Medication | Medication | Medication | Existential |
| Numericstatus | FunctionalStatus | Numeric observation | Numeric |
| Procedure | Procedure | Procedure | Existential |
| Result | Result | Numeric observation | Numeric |
| Vitalsign | VitalSign | Numeric observation | Numeric value |
CCR, continuity of care record; ECM, eligibility criteria model.
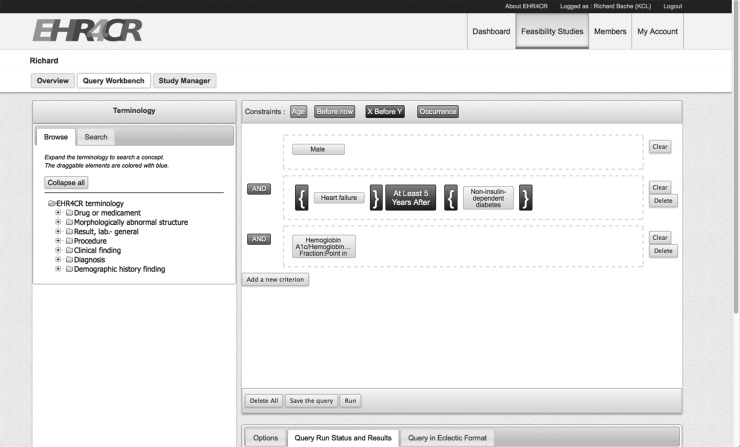
A drag-and-drop query builder has been constructed23 by the University of Rennes, which is usable by non-technical users to compose queries and this GUI is shown in figure 5. The user composes rules by first selecting concepts identified in the query vocabulary and when appropriate the range of permitted values. He may also add a temporal constraint. Once a set of rules has been composed graphically, a clinician-readable notation, eligibility criteria language for European clinical trial investigation and construction (ECLECTIC), is generated. Figure 6 shows the ECLECTIC for the example given in figure 5.
Figure 5.
Screenshot of drag-and-drop query builder (monochrome rendering). EHR4CR, electronic health records for clinical research.
Figure 6.

Example of European clinical trial investigation and construction notation.
The ECM generates mini-queries based on seven templates, as shown in table 2; these are mapped to SQL for the respective data warehouse. The set of seven templates was shown to be sufficient as each of the events given in table 1 will map onto exactly one template.
Table 2.
Elementary query templates used by EHR4CR platform
| Template name | Parameters | Data returned | Entity selected |
|---|---|---|---|
| General | – | Patient id., date of birth, gender | Patients |
| Dead | – | Patient id., date of death | Dead patients |
| Diagnosis | List of diagnosis codes | Patient id., event date | Diagnoses |
| Procedure | List of procedure codes | Patient id., event date | Procedures |
| Medication | List of medication codes | Patient id., event date | Administrations of medications |
| Numeric observation | List of observation codes | Patient id., event date, value, measurement unit | Vital signs, lab tests, numeric observations and measurements |
| Coded observation | List of observation codes | Patient id., event date, value | Observations and measurements with a categorical value |
EHR4CR, electronic health records for clinical research.
Evaluation of the ECM
Ten previously published clinical trials comprising 82 EC were used to evaluate the expressive power of the ECM before its implementation as part of the EHR4CR platform. By attempting to encode each set of EC using the text-based representation of the ECM, ECLECTIC,23 it would demonstrate sufficiency for its intended purpose. The original 10 studies, expressed in natural language, were cleaned by a clinician, first to remove or amend subjective criteria, including concepts for which no clinical code could exist, and second to resolve ambiguity. The task of transforming the 82 criteria into 110 ECM rules was performed by a non-clinician, seeking clinical advice as needed. Writing the EC in a formal notation without any tools support needed to be performed by a computer scientist with an understanding of formal grammars because the GUI had not at that stage been built.. Some criteria addressed more than one attribute and needed more than one rule, hence the increase from 82 to 110. The only obstacle encountered when expressing the EC in the ECM was that there were clinical concepts for which no code could be found using the UMLS concept unique identifier codes. Expressive sufficiency was demonstrated in any case when the clinical concept was codeable. Only four criteria could not be coded at all and four could only be partly coded because of a lack of codes. Details of the studies are listed in table 3.
Table 3.
Summary of 10 clinical trials used to evaluate ECM
| Company name | Study id | Condition | Inclusion rules | Exclusion rules | Total |
|---|---|---|---|---|---|
| Sanofi | EFC11785 | Treatment of metastatic castration resistant prostate cancer | 5 | 3 | 8 |
| Roche | NC25113 | Patients with inadequately controlled diabetes mellitus type 2 and cardiovascular disease | 8 | 11 | 19 |
| Novartis | CSPP100A2368 | Patients with acute decompensated heart failure | 10 | 8 | 18 |
| Novartis | CENA713B2315 | Mild to moderately severe dementia associated with Parkinson's disease | 9 | 16 | 25 |
| Merck | 27919 | Idiopathic Parkinson's disease with motor fluctuations | 6 | 6 | 12 |
| Janssen | COU-AA-301 | Castration-resistant prostate cancer previously treated with docetaxel-based chemotherapy | 6 | 0 | 6 |
| GSK | OMB112517 | Relapsed CLL responding to induction therapy | 4 | 7 | 11 |
| Bayer | 11899 | Symptomatic deep-vein thrombosis or pulmonary embolism | 4 | 1 | 5 |
| GSK | BIO111482 | Patients with melanoma, after surgical removal of their tumor | 3 | 0 | 3 |
| AstraZeneca | D4320C00015 | Non-metastatic hormone-resistant prostate cancer | 2 | 1 | 3 |
| Total | 57 | 53 | 110 | ||
CCL, chronic lymphocytic leukemia; ECM, eligibility criteria model.
Accessing the data warehouses
The native data warehouse was developed by the EHR4CR project and is based on the HL7 RIM.2 There was no need to map clinical codes to the query vocabulary as this had been done during the ETL process, except for units of measurement, which were converted by the adaptor. Three hospital sites in the EHR4CR project already had i2b2 warehouses10 with their corresponding ETL processes in place and so this was an opportunity to test the architecture with another source. The project's information model was designed from the outset to be compatible with i2b2, so creating new SQL mappings for the templates was straightforward. The i2b2 warehouses used their own vocabularies rather than the query vocabulary and so semantic conversion at query time was required. These warehouses have been extensively tested by many non-technical users from Pharma to determine patient counts using actual clinical trial protocols.
An anonymized warehouse of general practice research database data with 5000 patients was used to demonstrate its use in a primary care setting. Vocabulary mappings also need to be set up, and although the system used standard READ and Multilex codes, for which standard conversion exists, some system-specific codes also need to be mapped by hand. Although not as comprehensively tested as the two previous examples, EC have been used as queries to obtain patient counts.
Discussion
The creation of an AQM and the pre/post-processing paradigm offers two advantages:
Queries can be more complex than would be afforded by the sources specific query languages and, in particular, can represent temporal semantics.
Many data sources can be accessed with a low-cost adaptor being constructed for each new data source.
It is worth noting here that the components with the adaptor are to some extent themselves re-usable. By designing the SQL queries carefully, all three adaptors actually used the same formatter. However, we cannot guarantee that this would always be the case.
Using the pre/post-processing paradigm requires defining an explicit query model and implementing an evaluation algorithm in an imperative programming language to handle potentially very large datasets. Modern hardware makes cost in processing time and memory easily affordable. The approach does require additional development effort over the alternative of using SQL to perform both data extraction and reasoning. However, the QMI needs only be implemented once no matter how many data sources are accessed. If the queries used for cohort identification are not directly expressible in SQL because they require complex temporal semantics or if we wish to access sources with diverse schemata, this effort is necessary.
We cannot guarantee that mapping to any data source is always possible. Separating the extraction of patient data from subsequent reasoning offers great flexibility and we have found that the template paradigm is more widely applicable, specifically to general practice data, than was originally intended because the ECM was devised with hospital data warehouses in mind. However, as Rector et al24 point out, it is difficult to separate entirely the semantic and structural issues and we note that the mappings from the templates to the general practice warehouse did require some simple semantic processing because codes were used to specify the structure of data in certain tables within the database. This processing was embedded in the SQL.
Thus far we have assumed that the warehouse is implemented by a relational DBMS system (queriable with SQL) and populated by an ETL process from EHR systems. If warehouses were accessible by some other means; for example, an API, it would, in principle, be possible to create an adaptor to access this. Such a scenario is entirely consistent with the principles of the architecture.
Although the approach handles only computable and coded criteria and concepts, non-computable criteria or un-coded concepts that often require human judgment are not ignored. By their very nature they require human input to perform filtering and this could be applied to a shortlist of patients generated automatically.
Conclusions
The architecture proposed here has been demonstrated to work for a query model instance, the ECM. This model has been shown to have a wide range of applicability for codifying EC as a query on structured data, while hiding the structural and semantic differences from the end user. The pre/post-processing paradigm allows a lightweight adaptor to be readily constructed for each new data source, while re-using the same QMI. There is development effort required to construct the query model and in particular the evaluation algorithm, because we are now only relying on the DBMS to extract data and not perform any reasoning, but when temporal reasoning is required or there are heterogeneous data sources or, as shown here, both, the architecture proposed here provides a viable solution.
Acknowledgments
The authors would like to thank the many members of the EHR4CR project who helped with the development of the EHR4CR platform. They acknowledge the work of Mark McGilchrist in devising the original SQL mapping, and also the work of David Ouagne, Eric Sadou and Eric Zapatal in constructing and testing the i2b2 adaptor and of Isabelle Stevant and Pascal van Hille in constructing the Query Builder. The authors further acknowledge the work of Marc Cuggia for disambiguating the EC used to evaluate the ECM.
Footnotes
Contributors: RB was involved in the design of the architecture and ECM and implemented both the specific QMI and two of the three adaptors. SM and AT contributed to the design of the architecture and ECM. All authors were involved in drafting and approving the paper.
Funding: This work was part funded by the IMI-funded collaborative project EHR4CR (electronic healthcare records for clinical research), grant agreement no. 115189.
Competing interests: None.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Taweel A, Delaney B, Speedie S. Towards achieving semantic interoperability in e-Health services. In: Watfa M. ed E-Healthcare systems and wireless communications: current and future challenges. IGI, 2012:388–401 ISBN: 978-1-61350-123-8 [Google Scholar]
- 2.Benson T. Principles of Health Interoperability HL7 and SNOMED: Chapter 7. Springer, 2009 [Google Scholar]
- 3.Standard Specification for Continuity of Care Record (CCR), ASTM E2369-05e2 West Conshohocken, PA, USA, 2010 [Google Scholar]
- 4.Kush RD.2012. Current status and future scope of CDISC standards. CDISC Journal. Oct.
- 5.Sordo M, Boxwala A, Ogunyemi O, et al. Description and status update on GELLO: a proposed standardized object-oriented expression language for clinical decision support. Stud Health Technol Inform 2004;107:164–8 [PubMed] [Google Scholar]
- 6.Weng C, Tu SW, Sim I, et al. Formal representation of eligibility criteria: a literature review. J Biomed Inform 2010;43:451–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Austin T, Kalra A, Tapuria A, et al. Implementation of a query interface for a generic record server. Int J Med Inform 2008;77:754–64 [DOI] [PubMed] [Google Scholar]
- 8.Ross J, Tu S, Carini S, et al. Analysis of eligibility criteria complexity in clinical trials. AMIA Summits on Translational Science Proceedings 2010:46–50 [PMC free article] [PubMed] [Google Scholar]
- 9.SHRINE http://catalyst.harvard.edu/services/shrine/ (accessed 25 Mar 2013).
- 10.i2b2—Informatics for Integrating Biology and the Bedside, National Centre for Biomedical Computing. https://www.i2b2.org (accessed 1 Mar 2013).
- 11.Weber GM, Murphy SN, McMurrey AJ, et al. The shared health research information network (shrine): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc 2009;16:24–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ePCRN The electronic patient care research network. http://www.epcrn.bham.ac.uk/ (accessed 25 Mar 2013).
- 13.Delaney BC, Peterson KA, Speedie S, et al. Envisioning a learning health care system: the Electronic Primary Care Research Network, a case study. Ann Fam Med 2012;10:54–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thew S, Leeming G, Ainsworth J, et al. FARSITE: evaluation of an automated trial feasibility assessment and recruitment tool. Trials 2011;12(Suppl. 1):A113 [Google Scholar]
- 15.Zhang G, Siegler T, Saxman P, et al. VISAGE: a query interface for clinical research. AMIA Summits on Translational Science Proceedings 2010:76–80 [PMC free article] [PubMed] [Google Scholar]
- 16.Snodgrass RT. ed The TSQL2 temporal query language. Kluwer Academic Publishers, 1995 [Google Scholar]
- 17.Das AK, Musen M. A temporal query system for protocol-directed decision support. Methods Inf Med 1994;33:358–70 [PubMed] [Google Scholar]
- 18.Deshpande AM, Brandt C, Nadkarni PM. Temporal query of attribute-value patient data: utilizing the constraints of clinical studies. Int J Med Inform 2003;70:59–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huser V, Narus SP, Rocha RA. Evaluation of a flowchart-based EHR query system: a case study of RetroGuide. J Biomed Inform 2010;43:41–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huser V, Rasmussen LV, Oberg R, et al. Implementation of workflow engine technology to deliver basic clinical decision support functionality. BMC Med Res Methodol 2011;11:43 http://www.biomedcentral.com/1471-2288/11/43 (accessed 30 May 2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Quality Forum, Quality Data Model December 2012. http://www.qualityforum.org/WorkArea/linkit.aspx?LinkIdentifier=id&ItemID=72517 (accessed 29 May 2013).
- 22.Standards and Interoperability Framework Health eDecision—Clinical Decision Support Guidance Service (Use Case 2). 2013. http://wiki.siframework.org/file/view/SIFramework_HeD_UC2_CDSGuidanceService_v1.0.docx/420514032/SIFramework_HeD_UC2_CDSGuidanceService_v1.0.docx (accessed 29 May 2013).
- 23.Chen Y, Bache R, Miles S, et al. A SOA-based Platform for Automating Clinical Trial Feasibility Study, to appear e-Health conference. Prague, July 2013 [Google Scholar]
- 24.Rector AL, Qamar R, Marley T. Binding ontologies and coding systems to electronic health records and messages. Appl Ontol 2009;4:51–69 [Google Scholar]