With the completion of the Human Genome Project1 as well as recent advances in genomic science and comparative biological studies, a new era of individualized medicine is evolving where novel biomedical discoveries are leading to more effective prevention, treatment, and diagnosis of disease. Although altered phenotypes are one of the most reliable manifestations of altered gene functions, research in extracting, representing, and analyzing phenotype–genotype relationships is still evolving. This has led to the emergence of a trans-discipline field, called ‘Phenomics,’2 that aims to capitalize on high-throughput computation and informatics technologies for the systematic study of phenotypes and how they might influence personal genomics.3 Many comparative phenomics studies in the recent past4 5 have demonstrated the power of positively correlating phenotypes with several measures of gene functions. However, despite the advances, research in phenomics is presented with various challenges, including (i) developing approaches for high-throughput extraction and representation of phenotypes, (ii) building techniques for storing, integrating, and querying phenotype data, and (iii) advancing phenotypic-driven analysis to derive phenotype–genotype associations. A significant barrier in the discovery of new genetic variants is the requirement to obtain the large sample sizes needed for an effective study (since variants may be rare within a population) leading to time-consuming and onerous sample collection efforts.
Electronic health records (EHRs) can accelerate clinical research and genomic medicine, but are hindered by the limited number of validated processes and tools to enable accurate and rapid phenotype extraction.6 EHRs are increasing in ubiquity, functionality, and comprehensiveness across the USA, in part due to Meaningful Use standards7 implemented as part of the Health Information Technology for Economic and Clinical Health (HITECH) Act. One recent advance has been the coupling of DNA biorepositories to EHR data,8–13 combined with advances in informatics techniques, such as natural language processing (NLP), to enable genomic discoveries. The Electronic Medical Records and Genomics (eMERGE14 15) consortium—a network of nine academic medical centers—has demonstrated the effectiveness of EHR-derived phenotyping algorithms for cohort identification to conduct genome- and phenome-wide association studies.16–20 Algorithms to define phenotypes in eMERGE have typically followed an iterative path, and highlight the importance of intermittent chart review to validate phenotype accuracy, typically at more than one site (when a multisite implementation is planned16). Once finalized, eMERGE phenotype algorithms (as well as non-eMERGE algorithms) can be viewed at http://phekb.org.
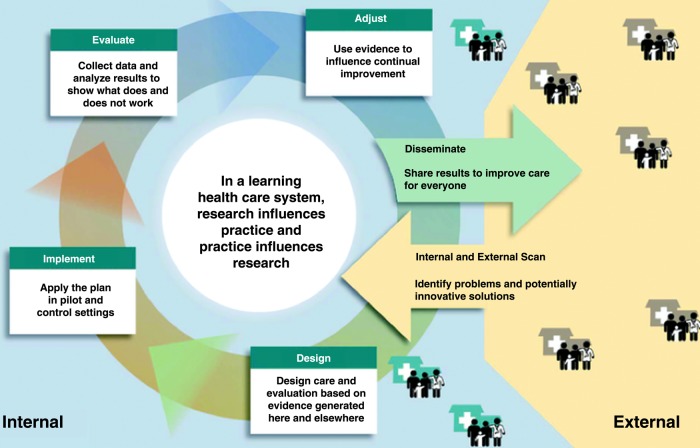
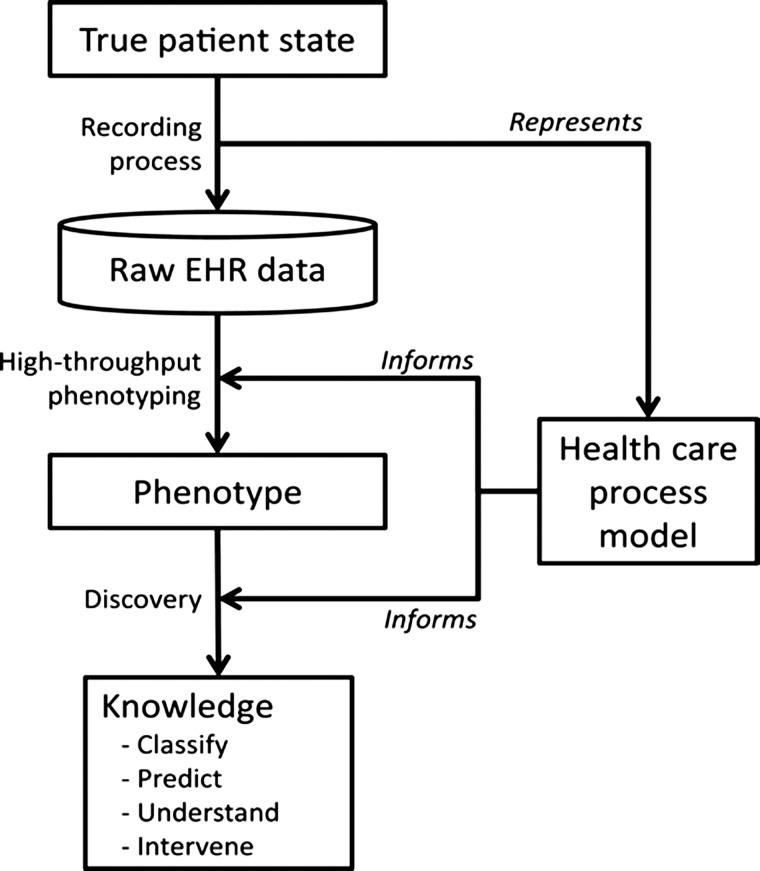
While eMERGE presents an exciting and encouraging demonstration of secondary use of EHR data, evaluating the strengths and limitations for EHRs has important implications for clinical and translational research, including clinical trials, observational cohorts, outcomes research, and comparative effectiveness research. These issues are further amplified within the realm of a learning healthcare system21 that emphasizes the ability to have real-time access to knowledge, digital capture of care experience, engaged and empowered patients, alignment of incentives to value, and a leadership-instilled culture of learning (figure 1). Hripcsak and Albers6 highlight several such challenges in leveraging EHRs for research, including data that is often incomplete, inaccurate, highly complex, and biased, and propose a combination of top-down knowledge engineering and bottom-up learning and analysis to address these issues (figure 2). Specifically, understanding the inherent complexity involved in a healthcare process model is critically important towards achieving a scalable and high-throughput phenotyping process.
Figure 1.
Different phases in a learning healthcare system (figure adapted with permission from Greene et al21).
Figure 2.
Electronic health record (EHR)-driven phenotyping and knowledge discovery (figure adapted with permission from Hripcsak and Albers6).
Several have acknowledged that there is a recognized tension between facilitating data entry by allowing narrative text in EHRs, and utilizing these data for research.22 A key component for identifying patient cohorts in the EHR is to define inclusion and exclusion criteria that algorithmically select sets of patients based on stored clinical data. This process allows the definition of phenotypes over structured data (eg, demographics, diagnoses, medications, and laboratory measurements)23 as well as unstructured clinical text (eg, radiology reports,22 24 encounter notes, and discharge summaries25–30). In general, this process can be quite complex, involving heuristics encoded as rules or machine learning algorithms.31 32 Several NLP techniques33 have been developed specifically for clinical text, and include solutions to address concept extraction,25–29 32–39 coreference resolution,31 37 40–51 word sense disambiguation,52 53 and temporal relations,54–62 to name just a few. The creation of annotated corpora63 64 to help develop and test these algorithms has also been the focus of the biomedical and clinical NLP community.
A notable challenge for general-purpose NLP systems applied to phenotyping is the requirement for high precision across many records. Thus, another important effort in phenotyping is purpose-developed NLP solutions designed to extract specific features over entire records with high accuracy. In addition, researchers have generated focused NLP and machine learning systems for common tasks such as general medication extraction65 66 and smoking determination.67
Several efforts have demonstrated the portability of phenotype algorithms across sites and EHR platforms,17 19 68 although implementation still requires significant manual effort. Efforts such as popHealth69 and SHRINE70 are attempting to provide common platforms to enable federated querying, although currently using simpler methodologies than invoked through networks such as eMERGE. The National Quality Forum's Quality Data Model may also represent one such standard.71 As in any other process, use of EHR phenotypes across institutions and across applications will not be complete without standardization. While some standards that are specific for research applications are continually being refined,72 existing standards that were designed for clinical applications also have high relevance in EHR phenotyping.37 73
Summary of papers in the special issue
The focus of this special issue of the journal is to provide a forum for presenting methodologies, tools, and algorithms to enable high-throughput phenotype extraction from EHR data. The journal has traditionally published articles related to EHR use for clinical decision support associated with patient safety and screening,74–84 as well as therapy management,78 80 85–92 but the extension of EHR use for research is relatively recent, reflecting the growth of clinical research informatics.6 33 93 94
Judging by the response, interest in EHR-based phenotyping is strong: of over 60 papers submitted for consideration in this issue, 20 articles95–109 were accepted for publication. These articles were evaluated through a rigorous process involving three guest associate editors (JP, JD, AK) in addition to the editor and one additional associate editor who dealt with any submissions a guest associate editor could not handle due to conflicts of interest.
Phenotyping algorithm development, implementation, and ecology emerged as major themes within the accepted manuscripts, along with applications in clinical trials and clinical decision support. The first article by Overby et al103 presents a collaborative approach for developing an EHR-driven phenotyping algorithm for drug-induced liver injury. This work, done within the eMERGE consortium, highlights the challenges in algorithm development and portability across multiple EHR systems, and emphasizes the need for robust validation methods complementing local, institution-specific algorithm implementation processes. The article by Tian et al107 presents a similar approach for identifying patients with chronic pain at a multi-site community health center. Here the authors demonstrate that by combining multiple different classes of data—diagnosis, medications, and pain scores—one can achieve much higher performance in terms of sensitivity and specificity. Finally, the article by Ludvigsson et al101 also argues that a combination of structured data with information extracted from unstructured text via NLP—in this case, for detecting patients with celiac disease—achieves a much higher performance.
With respect to applying more advanced machine learning, text mining, and statistical approaches for phenotype extraction, Chen et al96 demonstrated that active learning was useful in the identification of patient cohorts for rheumatoid arthritis, colorectal cancer, and venous thromboembolism. Not only did active learning methods outperform passive learning techniques, but the authors concluded that machine learning and feature engineering principles can be combined to develop efficient and generalizable phenotyping algorithms on a larger scale. The article by Klann et al99 explored Bayesian structured learning methods for population phenotyping to prioritize and tailor pediatric preventative care services. This work demonstrates how population phenotyping models can be built automatically, using prior data, without any human intervention for prioritizing pediatric screening questions and reminders in a patient-tailored manner. Similarly, the article by Deleger et al illustrates the application of conditional random fields to risk stratify abdominal pain patients based on pediatric appendicitis scores. The information driven approach demonstrated performance comparable to physician chart reviews, and represents a promising new approach for future computerized decision support applications. On the application of statistical based approaches, Lyalina et al102 applied automated text processing pipelines to annotate clinical notes with Unified Medical Language System concepts, and used dimensionality reduction to study patient-level phenotypic variations for neuropsychiatric disorders. The authors argue that such methods enable large-scale cohort building for clinical and genomic studies. Similarly, Gundlapalli et al demonstrated the high-throughput annotation process for extracting psychosocial concepts from approximately 1 billion documents from the medical facilities of the Department of Veterans Affairs. This work further illustrates the need to leverage high-yield documents and clinical notes, as opposed to the entire unstructured text corpus within an EHR system. Finally, in a similar effort, the article by Davis et al demonstrates the applicability of using NLP methods and scalable annotations to identify patients with multiple sclerosis and the key clinical traits for the disease course.
A key aspect for portability of phenotyping algorithms is standardization. The article by Richesson et al105 highlights this need by comparing seven different algorithms to identify diabetes mellitus. These algorithms yielded different cohorts when applied to the same population within the Duke University Health System. Similar results were observed in the article by Fan et al wherein an algorithm for identifying patients with peripheral arterial disease performed differently depending on the medical specialties involved. Not only do such differences lead to different interpretations of results and data, but multiple phenotype definitions can potentially affect their application within a healthcare organization. The article by Pathak et al attempts to address this challenge by leveraging standardized information modeling and Meaningful Use standards for representation of phenotyping algorithms. This work has led to the development of a publicly accessible library for standardized phenotype definitions. In addition to the standardized definitions, there is also the need for uniform infrastructure to enable cohort identification. The article by Fernandez-Breis et al97 proposes the use of standardized EHR models, archetypes, and ontologies for colorectal cancer screening. The authors argue that emerging semantic web technologies can facilitate the much needed interoperability among EHR data and systems. Finally, the article by Bache et al109 proposes an architecture for identifying patient cohorts by explicit query modeling and support for temporal reasoning. The authors illustrate that such an approach, while initially burdensome to establish (eg, compared to direct SQL-based queries), is more scalable and adaptable to heterogeneous data sources.
Understanding the inherent complexities associated with the healthcare process is vital for achieving scalable and high-throughput phenotyping. Along these lines, the article by Hripcsak and Albers98 studies correlation between EHR variables and healthcare process events. The authors illustrate that variable groups represent not only clinical and physiological properties, but also characteristics related to the way the information is gathered and recorded during the healthcare process. Similarly, the article by Boland et al95 introduces the concept of a ‘verotype’ (the Latin word ‘verus’ means ‘true’ or ‘actual’) to represent the ‘true’ population of similar patients for treatment purposes through the integration of genotype, phenotype, and disease subtype (eg, specific glucose value pattern in patients with diabetes) information. Both these works have implications for how phenotype extraction methods are used for real-world applications. The article by Richesson et al104 highlights these challenges within the context of clinical trials for the NIH Health Care Systems Collaboratory initiative. Of equal importance is the ability not only to implement and execute phenotyping algorithms, but also to understand the associated healthcare process events to achieve meaningful phenotype extraction. This issue is specifically highlighted in the article by London et al100 whereby the authors use a research data mart—powered by i2b2—for identifying patients eligible for clinical trials. In a similar effort, Warner et al108 describe a temporal phenome analysis to create a visual analysis of phenomic associations and healthcare process events. This work presents a new methodology for visual analytics and testable hypothesis generation from EHR data, which reveals patterns of context-specific complications with clinical implications.
Finally, the article by Rosenbloom et al106 discusses the ethical and legal implications of opt-out biobanks, such as Vanderbilt's DNA biobank, BioVU.8 Opt-out models have the advantage of rapid and inexpensive data collection; however, the characteristics of patients who opt-out versus those included in the biobank is not currently known, nor are the reasons why patients opt out. The importance of such research is highlighted by a recent Advance Notice of Proposed Rule-Making announced by the Department of Health and Human Services.110
Next steps and the future
The articles presented in this issue provide a glimpse into the opportunities that lie ahead. The rapid proliferation of EHRs, and the ability to connect and integrate data across multiple EHR systems through the robust application of data standards, will create ever richer clinical data for high-throughput phenotyping algorithms. These same algorithms can and are being adapted for direct clinical applications, such as automation of clinical quality measures111 and of clinical documentation.23 Across large populations, EHR data have potential for novel discovery of associations between disease and genetic, environmental, or process measures. Increasingly, electronic data will be available from what have been considered non-clinical sources, such as patient behavior/activity (eg, Fitbit) or social networks, and these can be combined with EHR-derived data to create more comprehensive ecological views of patients. These opportunities will naturally uncover issues and challenges around integration, analysis, interpretation, and sharing of ‘big data.’ It is hoped that this issue of the journal will serve as a useful reference and guide over the next few years. The technologies presented here will mature and evolve towards scalable and high-throughput integrative phenotyping that is needed to facilitate research, patient care, and healthcare management.
Footnotes
Funding: This work was supported in part by National Institutes of Health grant R01-GM105688, eMERGE Network grants (U01-HG006379, U01-HG006388, U01-HG006378), R01-LM010685, and a Pharmacogenomics Research Network grant (U19-GM061388).
Competing interests: None.
Provenance and peer review: Commissioned; internally peer reviewed.
References
- 1.Watson J. The human genome project: past, present, and future. Science 1990;248:44–9 [DOI] [PubMed] [Google Scholar]
- 2.Freimer N, Sabatti C. The human phenome project. Nat Genet 2003;34:15–21 [DOI] [PubMed] [Google Scholar]
- 3.Yngvadottir B, MacArthur D, Jin H, et al. The promise and reality of personal genomics. Genome Biol 2009;10:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Butte A, Kohane I. Creation and implications of a phenome-genome network. Nat Biotechnol 2006;24:55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Himes BE, Klanderman B, Kohane IS, et al. Assessing the reproducibility of asthma genome-wide association studies in a general clinical population. J Allergy Clin Immunol 2011;127:1067–9 [DOI] [PubMed] [Google Scholar]
- 6.Hripcsak G, Albers DJ. Next-generation phenotyping of electronic health records. J Am Med Inform Assoc 2013;20:117–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. N Engl J Med 2010;363:501–4 [DOI] [PubMed] [Google Scholar]
- 8.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther 2008;84:362–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarty CA, Peissig P, Caldwell MD, et al. The Marshfield Clinic Personalized Medicine Research Project: 2008 scientific update and lessons learned in the first 6 years. Personalized Med 2008;5:529–42 [DOI] [PubMed] [Google Scholar]
- 10.Scott CT, Caulfield T, Borgelt E, et al. Personal medicine[mdash]the new banking crisis. Nat Biotech 2012;30:141–7 [DOI] [PubMed] [Google Scholar]
- 11.Bielinski SJ, Chai HS, Pathak J, et al. Mayo Genome Consortia: a genotype-phenotype resource for genome-wide association studies with an application to the analysis of circulating bilirubin levels. Mayo Clinic Proc 2011;86:606–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wolf W, Doyle M, Aufox S, et al. DNA banking study in an ethnically diverse urban university hospital. Am J Hum Genet 2003;73:423 [Google Scholar]
- 13.Kohane IS. Using electronic health records to drive discovery in disease genomics. Nat Rev Genet 2011;12:417–28 [DOI] [PubMed] [Google Scholar]
- 14.McCarty C, Chisholm R, Chute C, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics 2011;4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottesman O, Kuivaniemi H, Tromp G, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med 2013;15:761–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denny JC. Chapter 13: mining electronic health records in the genomics era. PLoS Comput Biol 2012;8:e1002823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc 2012;19:212–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ritchie MD, Denny JC, Zuvich RL, et al. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation 2013;127:1377–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Denny JC, Crawford DC, Ritchie MD, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. Am J Hum Genet 2011;89:529–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shameer K, Denny J, Ding K, et al. A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum Genet 2013. [Epub ahead of print 12 Sep 2013] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greene SM, Reid RJ, Larson EB. Implementing the learning health system: from concept to action. Ann Inter Med 2012;157:207–10 [DOI] [PubMed] [Google Scholar]
- 22.Garvin JH, DuVall SL, South BR, et al. Automated extraction of ejection fraction for quality measurement using regular expressions in Unstructured Information Management Architecture (UIMA) for heart failure. J Am Med Inform Assoc 2012;19:859–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright A, Pang J, Feblowitz JC, et al. A method and knowledge base for automated inference of patient problems from structured data in an electronic medical record. J Am Med Inform Assoc 2011;18:859–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Percha B, Nassif H, Lipson J, et al. Automatic classification of mammography reports by BI-RADS breast tissue composition class. J Am Med Inform Assoc 2012;19:913–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang M, Chen Y, Liu M, et al. A study of machine-learning-based approaches to extract clinical entities and their assertions from discharge summaries. J Am Med Inform Assoc 2011;18:601–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minard AL, Ligozat AL, Ben Abacha A, et al. Hybrid methods for improving information access in clinical documents: concept, assertion, and relation identification. J Am Med Inform Assoc 2011;18:588–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patrick JD, Nguyen DH, Wang Y, et al. A knowledge discovery and reuse pipeline for information extraction in clinical notes. J Am Med Inform Assoc 2011;18:574–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rink B, Harabagiu S, Roberts K. Automatic extraction of relations between medical concepts in clinical texts. J Am Med Inform Assoc 2011;18:594–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Torii M, Wagholikar K, Liu H. Using machine learning for concept extraction on clinical documents from multiple data sources. J Am Med Inform Assoc 2011;18:580–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng K, Mei Q, Hanauer DA. Collaborative search in electronic health records. J Am Med Inform Assoc 2011;18:282–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Hong K, Tsujii J, et al. Feature engineering combined with machine learning and rule-based methods for structured information extraction from narrative clinical discharge summaries. J Am Med Inform Assoc 2012;19:824–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D'Avolio LW, Nguyen TM, Goryachev S, et al. Automated concept-level information extraction to reduce the need for custom software and rules development. J Am Med Inform Assoc 2011;18:607–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kahn MG, Weng C. Clinical research informatics: a conceptual perspective. J Am Med Inform Assoc 2012;19(e1):e36–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clark C, Aberdeen J, Coarr M, et al. MITRE system for clinical assertion status classification. J Am Med Inform Assoc 2011;18:563–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts K, Harabagiu SM. A flexible framework for deriving assertions from electronic medical records. J Am Med Inform Assoc 2011;18:568–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu H, Jiang M, Oetjens M, et al. Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. J Am Med Inform Assoc 2011;18:387–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ware H, Mullett CJ, Jagannathan V, et al. Machine learning-based coreference resolution of concepts in clinical documents. J Am Med Inform Assoc 2012;19:883–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strauss JA, Chao CR, Kwan ML, et al. Identifying primary and recurrent cancers using a SAS-based natural language processing algorithm. J Am Med Inform Assoc 2013;20:349–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y, Wang Y, Liu T, et al. Joint segmentation and named entity recognition using dual decomposition in Chinese discharge summaries. J Am Med Inform Assoc 2013. Published Online First: 9 Aug 2013. doi: 10.1136/amiajnl-2013-001806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bodnari A, Szolovits P, Uzuner O. MCORES: a system for noun phrase coreference resolution for clinical records. J Am Med Inform Assoc 2012;19:906–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dai HJ, Chen CY, Wu CY, et al. Coreference resolution of medical concepts in discharge summaries by exploiting contextual information. J Am Med Inform Assoc 2012;19:888–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jonnalagadda SR, Li D, Sohn S, et al. Coreference analysis in clinical notes: a multi-pass sieve with alternate anaphora resolution modules. J Am Med Inform Assoc 2012;19:867–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramesh BP, Prasad R, Miller T, et al. Automatic discourse connective detection in biomedical text. J Am Med Inform Assoc 2012;19:800–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rink B, Roberts K, Harabagiu SM. A supervised framework for resolving coreference in clinical records. J Am Med Inform Assoc 2012;19: 875–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uzuner O, Bodnari A, Shen S, et al. Evaluating the state of the art in coreference resolution for electronic medical records. J Am Med Inform Assoc 2012;19:786–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu Y, Liu J, Wu J, et al. A classification approach to coreference in discharge summaries: 2011 i2b2 challenge. J Am Med Inform Assoc 2012;19:897–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zheng J, Chapman WW, Miller TA, et al. A system for coreference resolution for the clinical narrative. J Am Med Inform Assoc 2012;19:660–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu ST, Liu H, Li D, et al. Unified Medical Language System term occurrences in clinical notes: a large-scale corpus analysis. J Am Med Inform Assoc 2012;19(e1):e149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu Y, Tsujii J, Chang EI. Named entity recognition of follow-up and time information in 20,000 radiology reports. J Am Med Inform Assoc 2012;19:792–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen P, Hinote D, Chen G. A rule based solution to co-reference resolution in clinical text. J Am Med Inform Assoc 2013;20:891–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li Q, Zhai H, Deleger L, et al. A sequence labeling approach to link medications and their attributes in clinical notes and clinical trial announcements for information extraction. J Am Med Inform Assoc 2013;20:915–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stevenson M, Agirre E, Soroa A. Exploiting domain information for word sense disambiguation of medical documents. J Am Med Inform Assoc 2012;19:235–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garla VN, Brandt C. Knowledge-based biomedical word sense disambiguation: an evaluation and application to clinical document classification. J Am Med Inform Assoc 2013;20:882–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanauer DA, Ramakrishnan N. Modeling temporal relationships in large scale clinical associations. J Am Med Inform Assoc 2013;20:332–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Y, Wang Y, Liu T, et al. An end-to-end system to identify temporal relation in discharge summaries: 2012 i2b2 challenge. J Am Med Inform Assoc 2013;20:849–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cherry C, Zhu X, Martin J, et al. A la Recherche du Temps Perdu: extracting temporal relations from medical text in the 2012 i2b2 NLP challenge. J Am Med Inform Assoc 2013;20:843–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sohn S, Wagholikar KB, Li D, et al. Comprehensive temporal information detection from clinical text: medical events, time, and TLINK identification. J Am Med Inform Assoc 2013;20:836–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun W, Rumshisky A, Uzuner O. Evaluating temporal relations in clinical text: 2012 i2b2 Challenge. J Am Med Inform Assoc 2013;20:806–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tang B, Wu Y, Jiang M, et al. A hybrid system for temporal information extraction from clinical text. J Am Med Inform Assoc 2013;20:828–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kovacevic A, Dehghan A, Filannino M, et al. Combining rules and machine learning for extraction of temporal expressions and events from clinical narratives. J Am Med Inform Assoc 2013;20:859–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun W, Rumshisky A, Uzuner O. Temporal reasoning over clinical text: the state of the art. J Am Med Inform Assoc 2013;20:814–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roberts K, Rink B, Harabagiu SM. A flexible framework for recognizing events, temporal expressions, and temporal relations in clinical text. J Am Med Inform Assoc 2013;20:867–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Savova GK, Chapman WW, Zheng J, et al. Anaphoric relations in the clinical narrative: corpus creation. J Am Med Inform Assoc 2011;18:459–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Albright D, Lanfranchi A, Fredriksen A, et al. Towards comprehensive syntactic and semantic annotations of the clinical narrative. J Am Med Inform Assoc 2013;20:922–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu H, Stenner SP, Doan S, et al. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc 2010;17:19–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uzuner Ö, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc 2010;17:514–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Savova GK, Ogren PV, Duffy PH, et al. Mayo Clinic NLP system for patient smoking status identification. J Am Med Inform Assoc 2008;15:25–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J Am Med Inform Assoc 2012;19(e1):e162–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.popHealth. http://projectpophealth.org/ (accessed 13 Mar 2012)
- 70.Weber GM, Murphy SN, McMurry AJ, et al. The Shared Health Research Information Network (SHRINE): a prototype federated query tool for clinical data repositories. J Am Med Inform Assoc 2009;16:624–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Conway M, Berg R, Carrell D, et al. Analyzing the heterogeneity and complexity of electronic health record oriented phenotyping algorithms. American Medical Informatics Association (AMIA) Annual Symposium, 2011 [PMC free article] [PubMed] [Google Scholar]
- 72.Overhage JM, Ryan PB, Reich CG, et al. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc 2012;19:54–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tao C, Jiang G, Oniki TA, et al. A semantic-web oriented representation of the clinical element model for secondary use of electronic health records data. J Am Med Inform Assoc 2013;20:554–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carroll AE, Biondich PG, Anand V, et al. Targeted screening for pediatric conditions with the CHICA system. J Am Med Inform Assoc 2011;18:485–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carroll NM, Ellis JL, Luckett CF, et al. Improving the validity of determining medication adherence from electronic health record medications orders. J Am Med Inform Assoc 2011;18:717–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoeksema LJ, Bazzy-Asaad A, Lomotan EA, et al. Accuracy of a computerized clinical decision-support system for asthma assessment and management. J Am Med Inform Assoc 2011;18:243–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Landis Lewis Z, Mello-Thoms C, Gadabu OJ, et al. The feasibility of automating audit and feedback for ART guideline adherence in Malawi. J Am Med Inform Assoc 2011;18:868–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sohn S, Kocher JP, Chute CG, et al. Drug side effect extraction from clinical narratives of psychiatry and psychology patients. J Am Med Inform Assoc 2011;18(Suppl 1):i144–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zlabek JA, Wickus JW, Mathiason MA. Early cost and safety benefits of an inpatient electronic health record. J Am Med Inform Assoc 2011; 18:169–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Niland JC, Stiller T, Neat J, et al. Improving patient safety via automated laboratory-based adverse event grading. J Am Med Inform Assoc 2012;19:111–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feldman MJ, Hoffer EP, Barnett GO, et al. Presence of key findings in the medical record prior to a documented high-risk diagnosis. J Am Med Inform Assoc 2012;19:591–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mathias JS, Gossett D, Baker DW. Use of electronic health record data to evaluate overuse of cervical cancer screening. J Am Med Inform Assoc 2012;19(e1):e96–e101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Parsons A, McCullough C, Wang J, et al. Validity of electronic health record-derived quality measurement for performance monitoring. J Am Med Inform Assoc 2012;19:604–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Middleton B, Bloomrosen M, Dente MA, et al. Enhancing patient safety and quality of care by improving the usability of electronic health record systems: recommendations from AMIA. J Am Med Inform Assoc 2013;20(e1):e2–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Austrian JS, Adelman JS, Reissman SH, et al. The impact of the heparin-induced thrombocytopenia (HIT) computerized alert on provider behaviors and patient outcomes. J Am Med Inform Assoc 2011;18:783–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haynes K, Linkin DR, Fishman NO, et al. Effectiveness of an information technology intervention to improve prophylactic antibacterial use in the postoperative period. J Am Med Inform Assoc 2011;18:164–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Strom BL, Schinnar R, Jones J, et al. Detecting pregnancy use of non-hormonal category X medications in electronic medical records. J Am Med Inform Assoc 2011;18(Suppl 1): i81–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Were MC, Shen C, Tierney WM, et al. Evaluation of computer-generated reminders to improve CD4 laboratory monitoring in sub-Saharan Africa: a prospective comparative study. J Am Med Inform Assoc 2011;18:150–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Connelly DP, Park YT, Du J, et al. The impact of electronic health records on care of heart failure patients in the emergency room. J Am Med Inform Assoc 2012;19:334–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dowding DW, Turley M, Garrido T. The impact of an electronic health record on nurse sensitive patient outcomes: an interrupted time series analysis. J Am Med Inform Assoc 2012; 19:615–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Griffey RT, Lo HG, Burdick E, et al. Guided medication dosing for elderly emergency patients using real-time, computerized decision support. J Am Med Inform Assoc 2012;19: 86–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Herwehe J, Wilbright W, Abrams A, et al. Implementation of an innovative, integrated electronic medical record (EMR) and public health information exchange for HIV/AIDS. J Am Med Inform Assoc 2012;19:448–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weng C, Appelbaum P, Hripcsak G, et al. Using EHRs to integrate research with patient care: promises and challenges. J Am Med Inform Assoc 2012;19:684–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hurdle JF, Haroldsen SC, Hammer A, et al. Identifying clinical/translational research cohorts: ascertainment via querying an integrated multi-source database. J Am Med Inform Assoc 2013;20:164–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Boland MR, Hripcsak G, Shen Y, et al. Defining a comprehensive verotype using electronic health records for personalized medicine. J Am Med Inform Assoc 2013;20:e232–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Y, Carroll RJ, Hinz ER, et al. Applying active learning to high-throughput phenotyping algorithms for electronic health records data. J Am Med Inform Assoc 2013;20:e253–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fernandez-Breis JT, Maldonado JA, Marcos M, et al. Leveraging electronic healthcare record standards and semantic web technologies for the identification of patient cohorts. J Am Med Inform Assoc 2013;20:e288–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hripcsak G, Albers DJ. Correlating electronic health record concepts with healthcare process events. J Am Med Inform Assoc 2013;20:e311–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Klann JG, Anand V, Downs SM. Patient-tailored prioritization for a pediatric care decision support system through machine learning. J Am Med Inform Assoc 2013;20:e267–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.London JW, Balestrucci L, Chatterjee D, et al. Design-phase prediction of potential cancer clinical trial accrual success using a research data mart. J Am Med Inform Assoc 2013;20:e260–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ludvigsson JF, Pathak J, Murphy S, et al. Use of computerized algorithm to identify individuals in need of testing for celiac disease. J Am Med Inform Assoc 2013;20:e306–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lyalina S, Percha B, Lependu P, et al. Identifying phenotypic signatures of neuropsychiatric disorders from electronic medical records. J Am Med Inform Assoc 2013;20:e297–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Overby CL, Pathak J, Gottesman O, et al. A collaborative approach to developing an electronic health record phenotyping algorithm for drug-induced liver injury. J Am Med Inform Assoc 2013;20:e243–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Richesson RL, Hammond WE, Nahm M, et al. Electronic health records based phenotyping in next-generation clinical trials: a perspective from the NIH Health Care Systems Collaboratory. J Am Med Inform Assoc 2013;20:e226–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Richesson RL, Rusincovitch SA, Wixted D, et al. A comparison of phenotype definitions for diabetes mellitus. J Am Med Inform Assoc 2013;20:e319–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rosenbloom ST, Madison JL, Brothers KB, et al. Ethical and practical challenges to studying patients who opt out of large-scale biorepository research. J Am Med Inform Assoc 2013;20:e221–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tian TY, Zlateva I, Anderson DR. Using electronic health records data to identify patients with chronic pain in a primary care setting. J Am Med Inform Assoc 2013;20:e275–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Warner JL, Zollanvari A, Ding Q, et al. Temporal phenome analysis of a large electronic health record cohort enables identification of hospital-acquired complications. J Am Med Inform Assoc 2013;20:e281–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bache R, Miles S, Taweel A. An adaptable architecture for patient cohort identification from diverse data sources. J Am Med Inform Assoc 2013;20:e327–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Human subjects research protections: enhancing protections for research subjects and reducing burden, delay, and ambiguity for investigators. Federal Register 2011;76 \http://www.gpo.gov/fdsys/pkg/FR-2011-07-26/pdf/2011-18792.pdf [Google Scholar]
- 111.Garrido T, Kumar S, Lekas J, et al. e-Measures: insight into the challenges and opportunities of automating publicly reported quality measures. J Am Med Inform Assoc 2013. Published Online First: 5 Aug 2013. doi: 10.1136/amiajnl-2013-001789 [DOI] [PMC free article] [PubMed] [Google Scholar]