Abstract
Regeneration of cardiac tissue has the potential to transform cardiovascular medicine. Recent advances in stem cell biology and direct reprogramming, or transdifferentiation, have produced powerful new tools to advance this goal. In this Review we examine key developments in the generation of new cardiomyocytes in vitro as well as the exciting progress that has been made toward in vivo reprogramming of cardiac tissue. We also address controversies and hurdles that challenge the field.
A myocardial infarction, or heart attack, results in the death of approximately 1 billion cardiomyocytes in the left ventricle within a matter of hours1. Damage to cardiac function can be progressive and often leads to congestive heart failure, which is the leading cause of death in the industrialized world2. For this reason, the replacement of lost cardiomyocytes is a primary target of regenerative medicine research. Potential sources of replacement cells include autologous cardiac stem cells, as used in the Cardiac Stem Cell Infusion in Patients with Ischemic Cardiomyopathy (SCIPIO)3,4 and Cardiosphere-derived Autologous Stem Cells to Reverse Ventricular Dysfunction (CADUCEUS)5 clinical trials, as well as myocytes derived from pluripotent stem cells (PSCs). The advent of direct reprogramming approaches (also known as transdifferentiation or direct conversion) to change one terminally differentiated cell type into another without first producing a pluripotent intermediate is a potential new approach for replacing lost cardiomyocytes.
Cardiomyocytes generated by direct reprogramming are termed induced cardiomyocytes, or iCMs, in keeping with the terminology established for induced pluripotent stem cells (iPSCs) generated by reprogramming6 (iCMs have also been referred to as induced cardiac-like myocytes, or iCLMs). iCMs have substantial promise as patient-specific tools for drug dosing and toxicity testing, for transplantation to replace cells lost during myocardial infarction, for the generation of contractile myocardial patches and to model cardiac development and disease in vitro. When compared to iPSC-derived cardiomyocytes, iCMs have potential advantages that include a reduced chance of teratoma formation and minimization of alternative cell fates, as well as the potential to more specifically target the type of cardiomyocyte that is produced (for example, atrial, ventricular or nodal), although this remains theoretical. Unlike pluripotent cell–based strategies, direct reprogramming also opens up an exciting new avenue of heart regeneration research that does not involve cell transplantation—the direct in vivo conversion of cardiac fibroblasts and scar tissue to contractile cardiac muscle. In this Review we briefly summarize the state of the art in the production of cardiac cells from PSCs before turning our attention to direct reprogramming (Fig. 1). We examine the history of transdifferentiation to cardiac cells, highlighting the many recent advances that have been made with both in vitro and in vivo cardiac reprogramming.
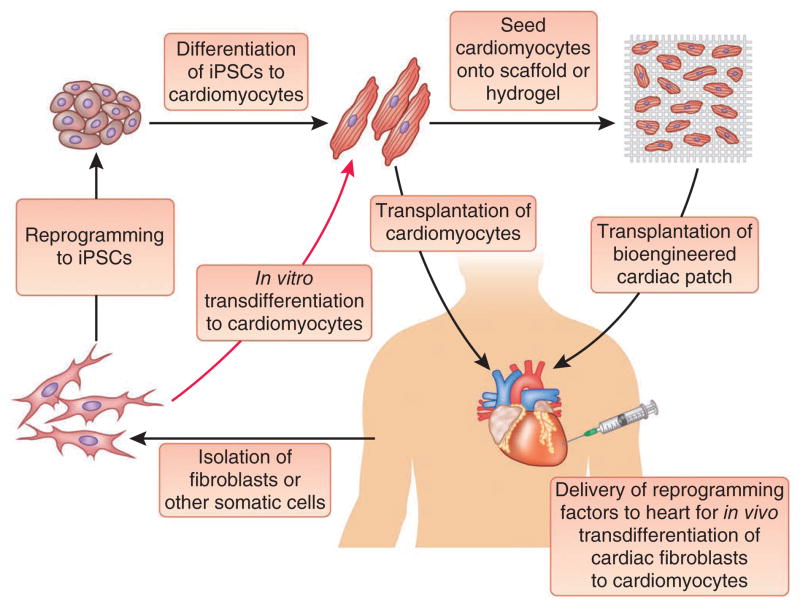
Figure 1.
Therapeutic approaches to regenerate cardiac tissue. A schematic representation of the various approaches under investigation to produce new cardiac muscle is shown.
Generation of cardiomyocytes from pluripotent stem cells
Cardiomyocytes were first generated from embryonic stem cells (ESCs) by spontaneous differentiation, as reported by Doetschman et al.7 in 1985. Since that time, protocols for the directed differentiation of pluripotent stem cells, whether ESCs or iPSCs, have become increasingly effective as more and more lessons from developmental biology have been applied (Fig. 2). In one of the more widely used protocols for PSC differentiation, Kattman et al.8 described how to drive cells toward mesoderm specification by adding activin A, bone morphogenetic protein 4 (BMP4) and basic fibroblast growth factor (FGF2) to mimic the signaling environment of the primitive streak in a post-gastrulation embryo. Cardiac progenitor cells are isolated by sorting for the dual expression of the surface markers kinase insert domain receptor (KDR, also known as fetal liver kinase 1, FLK1) and platelet-derived growth factor receptor α (PDGFR-α). Cardiac differentiation is subsequently induced by inhibiting Wnt and transforming growth factor β (TGF-β) signaling, yielding cardiomyocytes at a high efficiency (>80% of total cells). Several additional groups9–14 have reported success in generating cardiomyocytes from human PSCs (recently reviewed by Burridge et al.15). Although different modes of cell culture have been used (that is, either embryoid body formation or monolayer growth), the theme of developmental mimicry remains the same.
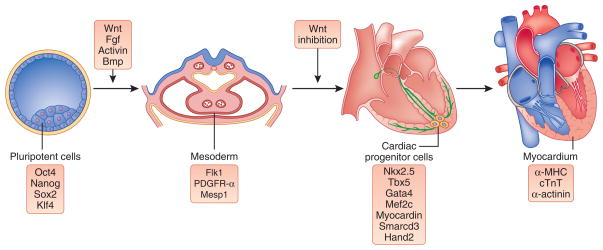
Figure 2.
Development of cardiac muscle. The progression of lineage restriction and specification of cardiac muscle is depicted moving from the pluripotent inner cell mass (from which ESCs are derived) on the left to mature myocardial tissue on the right, with some of the key signaling pathways (top) and gene expression characteristics (bottom) shown. The factors that are listed represent examples that are discussed in the text and are not intended to be inclusive.
Just as important as improving the efficiency of PSC differentiation is the need for protocols to isolate relatively pure populations of newly generated cardiomyocytes for subsequent use. Substantial progress has been reported to this end, including human ESCs that are engineered to express GFP after differentiation to cardiac cells11 and the identification of vascular cell adhesion molecule 1 (VCAM1)13 and signal-regulatory protein α (SIRPA)16, which are markers on the surface of cardiomyocytes that permit their antibody-mediated purification. More recently, Tohyama et al.17 took advantage of the fact that cardiomyocytes, unlike most other cell types, can survive in a glucose- poor, lactate-rich environment. This allowed for the nongenetic enrichment of PSC-derived cardiomyocytes with up to 99% purity.
Cardiomyocytes generated from PSCs are an attractive source of cells to replace damaged myocardium. Shiba et al.18 recently demonstrated that human ESC-derived myocytes can integrate into infarcted guinea pig hearts to improve cardiac function and suppress arrhythmias. However, cells that are directly transplanted to the myocardium often have poor survival19. It is therefore probable that cardiac tissue engineering will play a vital part in transplantation efforts (recently reviewed by Vunjak-Novakovic et al.20 and Segers and Lee19). Myocytes implanted as cell sheets21 or as patches of tissue grown on a scaffold22 or delivered in hydrogels or other biomaterials23 have substantially greater survival and effectiveness than those injected as single cells. Despite the promise of PSC-derived cardiomyocytes for heart repair, the use of a pluripotent cell source may always carry some inherent risk of teratoma formation. Although it is remarkable that iPSCs can be generated from easily obtained cell sources such as dermal fibroblasts24, peripheral blood25–27 and even urine28, direct transdifferentiation may obviate the need for a pluripotent step altogether. The remainder of this Review will focus on the rapidly moving field of direct reprogramming to cardiomyocytes.
Transdifferentiation to cardiac cells in embryos
The first example of transcription factor–mediated transdifferentiation to cardiac cells was reported in 1999 by Reiter et al.29, who demonstrated that overexpression of GATA-binding protein 5 (gata5) in zebrafish embryos was sufficient to generate ectopic regions of beating cardiomyocytes. A few years later, Latinkić et al.30 showed that both gata5 and gata4 could direct cardiomyogenesis in Xenopus embryonic ectoderm explants. David et al.31 subsequently delivered a plasmid expressing a different transcription factor, mesoderm posterior 1 (mesp1), to one of two blastomeres in two-cell Xenopus embryos. The tadpoles that developed had regions of beating cardiomyocytes throughout their bodies. MESP1 has been labeled a master regulator of cardiac development, as it is able to efficiently direct the differentiation of embryonic stem cells to the cardiac lineage32,33. Takeuchi and Bruneau showed that although Mesp1 does not have the same capacity to mediate transdifferentiation to cardiac tissue in mouse as it does in Xenopus, a set of three factors could achieve the same effect—the combination of Gata4, Tbx5 and Baf60c (also known as Smarcd3) directed the conversion of mouse mesoderm tissue into functional cardiomyocytes in cultured embryos34. The requirement of Baf60c, a component of the BAF chromatin remodeling complex, underscored the point that making the genomic sequence accessible to the appropriate transcription factors can be just as important in directing cell fates as choosing the transcription factors themselves. The authors used chromatin immunoprecipitation to demonstrate that only in the presence of Baf60c could Gata4 bind to cardiomyocyte-specific promoters in transfected mesodermal cells.
Transdifferentiation of terminally differentiated cells
Although these studies provide an intriguing glimpse at the potency of transdifferentiation, a clear distinction can be made between reprogramming that is effected while embryonic development is ongoing as opposed to the conversions of cells that are already terminally differentiated in the adult. The latter has been reported for an increasingly long list of cell types, including PSCs6, skeletal muscle cells35, smooth muscle cells36, pancreatic beta cells37, blood cells38, neurons39–41, neural progenitor cells42, hepatocytes43,44, sensory hair cells45,46 and Sertoli cells47. In cell fusion experiments, Blau et al.48 demonstrated that fusions (heterokaryons) between nonmuscle cells and skeletal muscle adopted the fate of the skeletal muscle cell, indicating that the skeletal muscle fate was dominant (later shown to be due to the action of MyoD35). Evans et al.49 showed that a dominant muscle fate was not the case for heterokaryons of fibroblasts and cardiomyocytes, perhaps portending that the transdifferentiation of fibroblasts to cardiomyocytes would be a challenging endeavor. The generation of cardiomyocytes from terminally differentiated cells was first reported by Ieda et al.50, who showed that three transcription factors—Gata4, myocyte enhancer factor 2C (Mef2c) and Tbx5 (this combination was called GMT)—were sufficient to directly convert mouse fibroblasts to cardiomyocytes. The authors used a strategy to identify an effective factor combination that was first reported by Takahashi et al.6 to generate iPSCs and has since been used in nearly all reprogramming efforts: start with a list of genes known to be important for the development, maintenance or both of the desired cell type, express these genes in the target cells and systematically subtract or add factors until the optimal combination is reached. For their factor screen, Ieda et al.50 targeted neonatal cardiac fibroblasts isolated from mice with an α-myosin heavy chain (α-MHC)-GFP reporter. Using flow cytometry to quantify α-MHC–GFP+ cells, they were able to whittle a list of 14 transcription factors down to 3. The efficiency of reprogramming was reported as a combination of outcome measures; the fraction of α-MHC–GFP+ cells reached a maximum of about 20% after 10 days of GMT treatment. Of the GFP+ cells, less than a third were initially positive for cardiac troponin T (cTnT), but this fraction increased to 45% after 4 weeks. The authors further reported that approximately 30% of the α-MHC–GFP+ cells (around 6% of the total population) showed some degree of the spontaneous Ca2+ oscillations that are characteristic of functional cardiomyocytes. Beating iCMs were reportedly detected at a low frequency.
The effectiveness of the GMT combination was challenged by the report of Chen et al.51, who found those factors to be insufficient to generate cardiomyocytes from either cardiac fibroblasts or tail-tip fibroblasts. Using the same lentiviral expression vectors that were published by Ieda et al.50, Chen et al.51 were able to achieve expression of the GMT factor cocktail but could not detect activation of cardiomyocyte-specific promoters such as those of α-MHC and Nkx2.5. Substantial upregulation of cTnT was observed, but functional cardiomyocytes were not detected. The cause of the discrepancy between the reports of Ieda et al.50 and Chen et al.51 is not clear but may perhaps be found in the seemingly subtle differences between their experimental procedures. Chen et al.51 did not describe the culture medium used in their reprogramming experiments, and neither group reported the titers of viruses used to deliver GMT, the substrates on which cells were plated (that is, gelatin, poly-lysine or others) or the densities at which cells were plated. It is possible, therefore, that there were considerable variations in the experimental protocols. This could have resulted in, for example, different levels of expression of individual reprogramming factors or differences in the relative ratios of factor expression. To this point, Srivastava and Ieda52 reported that the lentiviral vectors used by Chen et al.51 could not express GMT at the high levels achieved using retroviral vectors. Expression of GMT by lentiviruses was reported to be just 1–10% of that driven by retroviruses53. Furthermore, although Ieda et al.50 reported successful conversion of adult cardiac fibroblasts and tail-tip fibroblasts to cardiomyocytes, the majority of their experiments involved reprogramming neonatal cardiac fibroblasts. In contrast, the cardiac fibroblasts used by Chen et al.51 were from older mice (3–6 weeks of age). This difference in the developmental stage of the starting population of cells might have a substantial impact on the efficiency of reprogramming; in cases of reprogramming to other cell types, such as iPSCs, cells isolated from embryos and neonates are more amenable to direct reprogramming than cells isolated from adult tissues54.
Since the original publication of Ieda et al.50, in vitro transdifferentiation of fibroblasts to cardiomyocytes (or, to use a more conservative term, cardiomyocyte-like cells) has been reported by several additional groups (listed in Table 1). Song et al.55 used an α-MHC–GFP reporter system to screen for transcription factor combinations that activated the reporter, as Ieda et al.50 had. They went on to show that addition of the transcription factor heart and neural crest derivatives expressed transcript 2 (Hand2) to the GMT combination led to the generation of cardiomyocytes from adult tail-tip fibroblasts and cardiac fibroblasts. A report by Protze et al.56 took a different approach. They evaluated transcription factor combinations expressed in mouse embryonic fibroblasts by measuring the expression of a panel of cardiomyocyte genes to assess conversion. Although they did not report the generation of functional cardiomyocytes, they reported that the three-factor combination of myocardin, Mef2c and Tbx5 led to greater expression of cardiomyocyte genes than did GMT. Jayawardena et al.57 demonstrated that transdifferentiation of neonatal and adult cardiac fibroblasts to cardiomyocytes could be achieved without the use of any transcription factors. Instead, the authors used a combination of microRNAs (miR-1, miR-133, miR-208 and miR-499) and a chemical JAK inhibitor to mediate reprogramming. They further demonstrated that miR-1 alone was sufficient to induce the cardiac phenotype. The fact that reprogramming to cardiomyocytes can be achieved by a variety of different factor combinations is similar to what has been observed in reprogramming to other cell types: iPSCs6,58,59, dopaminergic neurons40,60,61 and hepatocytes43,44 are among the targets that have been reached with multiple sets of reprogramming factors.
Table 1.
Published reports of transdifferentiation to cardiac cells
| Reference | In vitro or in vivo | Reprogramming factors | Organism | Starting cell population | Comments |
|---|---|---|---|---|---|
| 29 | In vivo | gata5 | Zebrafish | Embryos at 25 h after fertilization | |
| 30 | In vitro | gata4 or gata5 | Frog | Embryonic ectodermal explants | |
| 31 | In vivo | mesp1 | Frog | Two-cell embryos | |
| 34 | In vivo | Gata4, Tbx5 and Baf60c | Mouse | Embryonic mesoderm | |
| 50 | In vitro | Gata4, Mef2c and Tbx5 (GMT) | Mouse | Neonatal and adult cardiac fibroblasts; tail-tip fibroblasts | The first report of transdifferentiation of somatic cells to cardiomyocytes |
| 77 | In vitro | Oct4, Sox2, Klf4 and c-Myc | Mouse | Embryonic fibroblasts | |
| 67 | In vitro | Whole transcriptome | Mouse | Embryonic fibroblasts and astrocytes | |
| 51 | In vitro | GMT | Mouse | Adult cardiac and tail-tip fibroblasts | Demonstrated inability of GMT to drive efficient reprogramming |
| 55 | In vitro and in vivo | GMT and Hand2 | Mouse | Adult cardiac and tail-tip fibroblasts | |
| 57 | In vitro and in vivo | miR-1, miR-133, miR-208 and miR-499; JAK inhibitor I | Mouse | Neonatal and adult cardiac fibroblasts | One of the first three reports of in vivo reprogramming to iCMs |
| 64 | In vivo | GMT | Mouse | Adult cardiac fibroblasts | One of the first three reports of in vivo reprogramming to iCMs |
| 56 | In vitro | Myocardin, Mef2c and Tbx5 | Mouse | Embryonic fibroblasts and neonatal cardiac fibroblasts | One of the first three reports of in vivo reprogramming to iCMs |
| 65 | In vivo | GMT | Mouse | Adult cardiac fibroblasts | |
| 73 | In vitro | MESP1 and ETS2 | Human | Neonatal foreskin fibroblasts | The first report of human fibroblasts reprogrammed to cardiac cells |
| 66 | In vivo | GMT and Vegf | Rat | Adult cardiac fibroblasts | |
| 74 | In vitro | GATA4, HAND2, myocardin, TBX5, miR-1 and miR-133 | Human | Neonatal foreskin, adult cardiac and adult dermal fibroblasts | |
| 78 | In vitro | GMT, Mesp1, myocardin, Smarcd3 (Baf60c) and Srf | Mouse | Embryonic fibroblasts | |
| 69 | In vitro | GMT, Hand2 and Nkx2.5 | Mouse | Embryonic fibroblasts and adult cardiac fibroblasts | Used a functional measure (calcium oscillation) to screen factor combinations |
When comparing cardiomyocyte generation through in vitro reprogramming to the differentiation of pluripotent stem cells, it is evident that the stem cell approach is considerably further advanced. Whether one examines the efficiency of direct conversion compared to PSC differentiation, the sheer numbers of cardiomyocytes generated, the characterization of myocyte subtypes (atrial, ventricular or nodal)62, efforts to drive maturation63 or the performance of transplanted cells in animal models of heart disease18, stem cell differentiation currently has a sizeable advantage. Continued advances in direct reprogramming will close this gap, and the approach will probably be more cost effective and scalable in humans. Even now, however, transdifferentiation offers a striking advantage over stem cell therapies: the potential to replace lost cardiomyocytes without cell transplantation. By delivering reprogramming factors directly to the damaged heart to induce regeneration in situ, many of the pitfalls of cell-based therapies are avoided, including expansion and delivery of a sufficient number of cells, efficient engraftment, survival of transplanted cells, potential tumorigenicity of residual stem cells and possible immune rejection (not to mention lingering ethical controversies) when using ESC derivatives. We now turn our attention to recent advances in direct in vivo reprogramming.
In vivo conversion of cardiac fibroblasts to cardiomyocytes
Three of the reports listed above55,57,64 demonstrated that transdifferentiation to cardiomyocytes could be achieved in vivo as well as in vitro. To demonstrate that cardiomyocytes were indeed derived from cardiac fibroblasts, each group used a Cre recombinase driven by one or more fibroblast-specific promoters to permanently label cardiac fibroblasts and any cells to which they gave rise. All three reports used the fibroblast-specific protein 1 (Fsp1) promoter; Qian et al.64 also used the promoter of periostin, another fibroblast-enriched gene, whereas Song et al.55 used the transcription factor 21 (Tcf21, also called epicardin) promoter, which is specifically expressed in nonmyocyte heart cells. This particular labeling approach had the advantage of using an inducible Cre recombinase system that limited the chance of ectopic promoter activation in cardiomyocytes, which is a substantial concern in this type of lineage fate–mapping experiment. A more recent publication from Inagawa et al.65 supported the results of Qian et al.64 by demonstrating GMT-mediated reprogramming in infarcted mouse hearts. Mathison et al.66 showed that the proangiogenic factor vascular endothelial growth factor (Vegf) can enhance the efficiency of GMT reprogramming in rat hearts. Together these reports provide proof of principle that endogenous cardiac fibroblasts can be induced to transdifferentiate into cardiomyocytes in vivo. Both Qian et al.64 and Song et al.55 reported modest yet statistically significant functional recovery of infarcted hearts after the reprogramming treatment, including an increased ejection fraction and reduced infarct size. Notably, however, all of the groups delivered reprogramming factors at the same time that the infarct was being generated, a scenario that does not mimic the clinical setting.
It remains unclear precisely which set of reprogramming factors is the most effective at transdifferentiation to cardiomyocytes, whether in vitro or in vivo, largely because the different combinations reported have not been compared side by side in the same system. Advances in single-cell analysis, which were recently applied to cardiac reprogramming by Kim et al.67, will probably be invaluable in efforts to compare and optimize various reprogramming approaches and to determine the optimal transcript levels and relative ratios for efficient transdifferentiation.
Defining cardiomyocyte identity and efficient conversion
In all direct reprogramming strategies, whether they are used to generate cardiomyocytes, neurons or other cell types, it is important to delineate the key characteristics that define the desired cell type. These may include gene expression signatures, cellular organization of structural proteins, morphology, epigenetic marks and functional attributes such as resting membrane potential, the capacity to have action potentials and the ability to secrete the appropriate factors or matrix. In reports of transdifferentiation to iCMs, a wide range of characteristics have been used as criteria to evaluate successful cell conversion (Box 1). It is apparent that not all criteria are equal; the expression of a single gene or fluorescent reporter is less stringent an outcome measure than the demonstration of contracting iCMs that have spontaneous or induced action potentials. There is currently no universally accepted set of standards for defining an iCM, but this issue is not entirely a new one—a great deal can be learned from the efforts to characterize cardiomyocytes generated by differentiation of PSCs (reviewed by Mummery et al.68).
BOX 1. Criteria to evaluate transdifferentiation to iCMs.
Although there is no clear consensus within the field as to what characteristics must be present to consider a cell an iCM, some combination of the attributes listed below must be demonstrated. Functional attributes (firing action potentials or oscillating calcium) are desirable, as they require the coordination of complex sets of molecules. A quantitative measurement of reprogramming efficiency should be reported, whether by flow cytometric analysis for cardiomyocyte markers or a fluorescent reporter driven by a cardiomyocyte-specific gene promoter, manual counting of immunostained cells showing sarcomere formation or quantifiable functional measures such as the percentage of cells that show calcium transients. The number of converted cells as a percentage of the starting cell number should be reported.
Lower stringency
RT-PCR to demonstrate upregulation of cardiomyocyte-specific genes and downregulation of genes specific to the starting cell type
Activation of a reporter transgene, such as GFP, driven by a cardiomyocyte-specific gene promoter
Cardiomyocyte-specific protein expression: immunostaining for proteins such as cTnT, α-MHC and α-actinin should reveal proper structural organization of sarcomeres
Higher stringency
Global transcriptome analysis using microarray or RNA-seq to compare iCMs to authentic subtypes of immature and mature cardiomyocytes
Contraction of iCMs: a bona fide myocyte should show contraction (and release), either spontaneously if it is a pacemaker cell or an immature cardiomyocyte or as a result of electrical or chemical stimulation if it is a mature cardiomyocyte
Electrophysiological characterization: cardiac action potentials should be detected using patch clamp analysis for intracellular recordings or microelectrode arrays to make extracellular measurements on a large number of cells at once. If the iCMs are sufficiently mature, electrophysiological analysis can also be used to determine the subtype of a given cardiomyocyte (that is atrial, ventricular or nodal)
Calcium transients: the oscillation of intracellular Ca2+ concentrations is the functional link between membrane excitation and cell contraction79. Ca2+ flux can be directly visualized using Ca2+-sensitive dyes such as fura-2 and rhodamine-3 or genetically encoded calcium indicators such as GCaMP80,81
Presence of cardiomyocyte-specific epigenetic marks: Ieda et al.50 reported patterns of methylation at cardiomyocyte-specific gene promoters in iCMs that were consistent with patterns seen in authentic cardiomyocytes
Ability of iCMs to generate force76, preferably using three dimensional bioengineered constructs, with quantitative assessment of static and dynamic tension
Genetic or lineage tracking evidence that iCMs are derived from fibroblasts or another starting cell type
Ability of in vitro–generated iCMs to form gap junctions and electrically couple with host cardiomyocytes
Functional improvement (for engrafted iCMs or in vivo reprogramming in infarcted hearts)—for example, improved ejection fraction, reduced infarct size and/or increased cardiac mass
As they have a higher level of stringency, functional outcome measures probably provide the most accurate assessment of a given reprogramming method (Fig. 3). Our group recently reported the evaluation of several transcription factor combinations using the induction of calcium oscillation—the phenomenon that couples cell membrane excitation to contraction—as a functional measure of success69. Using a transgenic calcium reporter driven by a cardiomyocyte- specific gene promoter, we were able to quantify the generation of functional iCMs and found that the addition of Nkx2.5 to the combination of GMT and Hand2 led to more efficient transdifferentiation of fibroblasts. It is interesting to note that the laboratories of both Srivastava and Olson have reported that Nkx2.5 reduces the number of cells that express GFP using the α-MHC–GFP reporter50,55. It may be the case that the addition of Nkx2.5 reduces the number of partially reprogrammed iCMs while increasing the number of functional iCMs. In future mid- to high-throughput screens, we predict that functional outcome measures will provide superior results to those obtained with GFP or luciferase reporters when additional transcription factors, microRNAs and small molecules, for example, are sought to enhance transdifferentiation to iCMs.
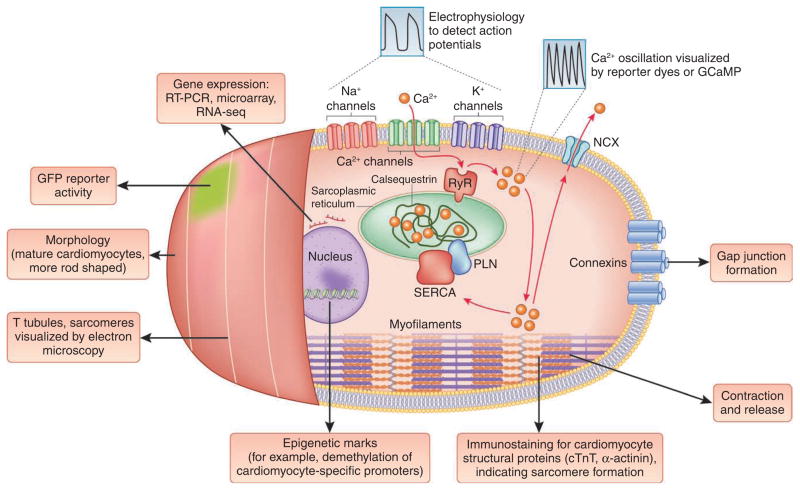
Figure 3.
Defining cardiomyocyte identity. A wide range of phenotypic features can be assayed to determine whether a reprogrammed cell is an iCM. These range from expression of a panel of genes or the activation of a reporter transgene (such as α-MHC–GFP) to more complex functional characteristics, such as the ability to fire action potentials and show calcium oscillation. As functional attributes require the orchestration of a complex collection of parts, they are probably more reliable indicators of successful transdifferentiation to iCMs. Other testable characteristics include the presence of cardiomyocyte-specific epigenetic marks, the ability of iCMs to generate force76, the ability of in vitro–generated iCMs to form gap junctions and electrically couple with host cardiomyocytes after transplantation and genetic or lineage tracing to show that iCMs are derived from fibroblasts or another starting cell type. T tubule, transverse tubule; RyR, ryanodine receptor; SERCA, sarco/endoplasmic reticulum Ca2+-ATPase; NCX, sodium-calcium exchanger; PLN, phospholamban; GCaMP, a genetically encoded calcium indicator consisting of a fusion of GFP and calmodulin (CaM).
Perhaps the most striking phenotypic feature that a cardiomyocyte can show is spontaneous contraction. Indeed, the identification of beating foci has been a standard measure of success for cardiomyocyte generation from various stem cell populations and by direct reprogramming. However, it is important to consider that spontaneous contraction, or automaticity, is a not a trait of all mature, adult atrial or ventricular cardiomyocytes. Primary myocytes isolated from embryonic hearts beat spontaneously in vitro70, whereas those isolated from adult hearts do not beat in normal conditions (that is, without stimulation)68. This is due, at least in part, to the increased density of inward rectifier currents as myocytes mature71, which raises the energetic threshold that must be reached for contraction-inducing action potentials to occur72. Thus, the spontaneously beating iCMs identified in several reports of direct reprogramming are probably immature, as is often the case when cardiomyocytes are generated by ESC differentiation. It remains to be seen whether fully mature cardiomyocytes can be obtained by transdifferentiation.
Defining what it means to be a cardiomyocyte is of particular relevance when evaluating the relative efficiencies of different reprogramming approaches. For simplicity, a single outcome measure is often chosen to report the efficiency of transdifferentiation. Not surprisingly, the choice of a less stringent criterion generally leads to a higher reported efficiency. Further complicating matters is the fact that in many reprogramming experiments, the nonreprogrammed cells continue to proliferate. Ieda et al.50 report that the percentage of α-MHC–GFP+ cells follows a bell-shaped curve when plotted over time—there is an initial increase in GFP+ cells after a few days, reaching a maximum at about 10 days, after which the percentage gradually drops. This decrease is attributed to the increased proliferation rate of GFP− cells (presumably cardiac fibroblasts that did not reprogram) compared to GFP+ cells. By contrast, the percent efficiency of reprogramming to iPSCs is generally reported by a different metric: the number of colonies generated relative to the number of cells in the starting population. If an iPSC experiment begins with 100 cells and produces 3 iPSC clones after 10 days, the efficiency is reported as 3%. But if a direct conversion experiment begins with the same 100 cells and produces 3 cardiomyocytes, the efficiency may be variably calculated. As a result of proliferation, those 3 iCMs might be present among 1,000 total cells, leading to an efficiency of 0.3%. Depending on which cells are proliferating and when, the effectiveness of transdifferentiation may be over- or under-reported. It is important to consider these factors when attempting to compare and appraise different reprogramming protocols, and we suggest that one measure of reprogramming efficiency should be reported as the number of reprogrammed cells divided by the number of cells initially treated with the reprogramming factors.
Furthermore, we propose the adoption of a standard set of characteristics that can be used to define an iCM. To be designated an iCM, a reprogrammed cell should show: (i) gene expression patterns (determined by RT-PCR, microarray or transcriptome sequencing (RNA-seq)) that are more closely related to those of cardiomyocytes than any other cell type, as well as downregulation of genes specific to the starting cell type; (ii) proper structural organization of sarcomeres shown by immunostaining for cardiomyocyte-specific proteins such as cTnT, α-MHC and α-actinin; and (iii) at least one functional attribute of cardiomyocytes—this may be the demonstration of action potentials using patch clamping or microelectrode arrays, calcium oscillation visualized by reporter dyes or genetically encoded calcium indicators or contraction and release (that is, beating, either spontaneous or induced). As a hypothetical example, consider a report in which human dermal fibroblasts are transduced with a reporter lentivirus that drives GFP expression under the control of the myosin light chain 2V (MLC2v, encoded by MYL2) promoter. After 4 weeks of treatment with reprogramming factors, 30% of cells are GFP+. Immunostaining shows that essentially all of the GFP+ cells coexpress α-MHC and α-actinin, with sarcomeres clearly visible. When GFP+ cells are plated on microelectrode arrays, one in three have action potential firing in response to field stimulation. By the standards we propose, 10% of the total cell population should then be classified as iCMs.
Challenges ahead
As noted above, the use of transdifferentiation to generate cardiomyocytes currently lags behind stem cell approaches. This gap is even more pronounced when evaluating protocols using human cells. However, substantial progress has been made recently: Islas et al.73 reported that MESP1 and ETS2 can convert neonatal human foreskin fibroblasts into cardiac progenitor cells, whereas Nam et al.74 used a combination of transcription factors (GATA4, HAND2, myocardin and Tbx5) and microRNAs (miR-1 and miR-133) to push adult human cardiac and dermal fibroblasts toward a cardiac fate. Notably, as of the writing of this Review, no set of reprogramming factors has been shown to work robustly on both mouse and human cells. This was not the case in reprogramming to iPSCs, in which the same set of factors is potent in multiple species, including humans. This difference may be important as attempts are made to translate the results of in vivo transdifferentiation strategies to therapeutic applications. If conversion of cardiac fibroblasts to cardiomyocytes requires a markedly different protocol in human compared to mouse cells, then additional animal models will need to be considered to bridge mouse in vivo reprogramming results to the clinic. Perhaps larger animals, such as pigs or sheep, or animals more closely related to humans, such as monkeys, will be informative. To have confidence that a particular procedure for in vivo reprogramming can be translated to humans, it will be necessary to first demonstrate that the procedure works well in vitro in both human cells and cells isolated from the large animal model. Even if a reprogramming strategy is found to work well in both mouse and human cells, large animal models will still be needed to test the safety and effectiveness of this therapeutic approach in hearts that more closely resemble human hearts in size and physiology.
If iCMs generated in vitro are to be used for personalized medicine—that is, to test whether an individual patient’s cardiomyocytes are responsive and not harmed in drug dosing and toxicology studies—fully mature, adult myocytes will be required. This challenge is shared by cells derived from PSCs, and considerable progress has already been made toward the in vitro maturation of cardiomyocytes63. Another challenge shared by both in vitro–generated iCMs and PSC-derived cells is that if they are to be transplanted into damaged hearts, tissue engineering will probably play a key part. It will be important to determine whether the reprogramming process can occur within the context of three-dimensional scaffolds and bioreactors. If so, it may be advantageous to first seed an easily cultured and expanded cell population such as fibroblasts before the initiation of transdifferentiation. As noted above, the field of cardiac bioengineering is moving rapidly, and iCMs offer a valuable new source of cells to seed onto scaffolds for the production of myocardial patches75.
With regard to in vivo reprogramming, several of the challenges mirror those seen in the field of gene therapy—reprogramming factors must be delivered in a highly targeted fashion, preferably using vectors that do not integrate into the host’s genome. It may also be important to use an expression system that can be regulated (and turned off), as the continuous expression of reprogramming factors may be detrimental or have unwanted effects in cells that are unintentionally transduced. The issue of cardiomyocyte maturation, discussed above, may also have a major role in determining the success of in vivo reprogramming strategies. The possibility that immature myocytes might be disruptive or arrhythmogenic is an important consideration that can only be addressed through continued experimentation. Although there is clearly much work to be done, the rapid pace of the cardiac reprogramming field provides cause for considerable optimism that new strategies for heart repair may be on the horizon.
Acknowledgments
This work was supported by the American Heart Association Jon Holden DeHaan Cardiac Myogenesis Research Center, US National Institutes of Health grant NIH U01 HL100405 and the Spain Fund for Regenerative Medicine.
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Murry CE, Reinecke H, Pabon LM. Regeneration gaps: observations on stem cells and cardiac repair. J Am Coll Cardiol. 2006;47:1777–1785. doi: 10.1016/j.jacc.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Go AS, et al. Heart disease and stroke statistics—2013 update: a report from the American Heart Association. Circulation. 2013;127:e6–e245. doi: 10.1161/CIR.0b013e31828124ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chugh AR, et al. Administration of cardiac stem cells in patients with ischemic cardiomyopathy: the SCIPIO trial: surgical aspects and interim analysis of myocardial function and viability by magnetic resonance. Circulation. 2012;126:S54–S64. doi: 10.1161/CIRCULATIONAHA.112.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolli R, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378:1847–1857. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Makkar RR, et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379:895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 8.Kattman SJ, et al. Stage-specific optimization of activin/nodal and BMP signaling promotes cardiac differentiation of mouse and human pluripotent stem cell lines. Cell Stem Cell. 2011;8:228–240. doi: 10.1016/j.stem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Willems E, et al. Small-molecule inhibitors of the Wnt pathway potently promote cardiomyocytes from human embryonic stem cell–derived mesoderm. Circ Res. 2011;109:360–364. doi: 10.1161/CIRCRESAHA.111.249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burridge PW, et al. A universal system for highly efficient cardiac differentiation of human induced pluripotent stem cells that eliminates interline variability. PLoS ONE. 2011;6:e18293. doi: 10.1371/journal.pone.0018293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott DA, et al. NKX2-5(eGFP/w) hESCs for isolation of human cardiac progenitors and cardiomyocytes. Nat Methods. 2011;8:1037–1040. doi: 10.1038/nmeth.1740. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Q, et al. Direct differentiation of atrial and ventricular myocytes from human embryonic stem cells by alternating retinoid signals. Cell Res. 2011;21:579–587. doi: 10.1038/cr.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uosaki H, et al. Efficient and scalable purification of cardiomyocytes from human embryonic and induced pluripotent stem cells by VCAM1 surface expression. PLoS ONE. 2011;6:e23657. doi: 10.1371/journal.pone.0023657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hudson J, Titmarsh D, Hidalgo A, Wolvetang E, Cooper-White J. Primitive cardiac cells from human embryonic stem cells. Stem Cells Dev. 2012;21:1513–1523. doi: 10.1089/scd.2011.0254. [DOI] [PubMed] [Google Scholar]
- 15.Burridge PW, Keller G, Gold JD, Wu JC. Production of de novo cardiomyocytes: human pluripotent stem cell differentiation and direct reprogramming. Cell Stem Cell. 2012;10:16–28. doi: 10.1016/j.stem.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubois NC, et al. SIRPA is a specific cell-surface marker for isolating cardiomyocytes derived from human pluripotent stem cells. Nat Biotechnol. 2011;29:1011–1018. doi: 10.1038/nbt.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tohyama S, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell–derived cardiomyocytes. Cell Stem Cell. 2013;12:127–137. doi: 10.1016/j.stem.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Shiba Y, et al. Human ES-cell–derived cardiomyocytes electrically couple and suppress arrhythmias in injured hearts. Nature. 2012;489:322–325. doi: 10.1038/nature11317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segers VF, Lee RT. Biomaterials to enhance stem cell function in the heart. Circ Res. 2011;109:910–922. doi: 10.1161/CIRCRESAHA.111.249052. [DOI] [PubMed] [Google Scholar]
- 20.Vunjak-Novakovic G, Lui KO, Tandon N, Chien KR. Bioengineering heart muscle: a paradigm for regenerative medicine. Annu Rev Biomed Eng. 2011;13:245–267. doi: 10.1146/annurev-bioeng-071910-124701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda S, Shimizu T, Yamato M, Okano T. Cell sheet engineering for heart tissue repair. Adv Drug Deliv Rev. 2008;60:277–285. doi: 10.1016/j.addr.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 22.Li RK, et al. Survival and function of bioengineered cardiac grafts. Circulation. 1999;100:II63–II69. doi: 10.1161/01.cir.100.suppl_2.ii-63. [DOI] [PubMed] [Google Scholar]
- 23.Kofidis T, et al. Injectable bioartificial myocardial tissue for large-scale intramural cell transfer and functional recovery of injured heart muscle. J Thorac Cardiovasc Surg. 2004;128:571–578. doi: 10.1016/j.jtcvs.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 24.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 25.Staerk J, et al. Reprogramming of human peripheral blood cells to induced pluripotent stem cells. Cell Stem Cell. 2010;7:20–24. doi: 10.1016/j.stem.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loh YH, et al. Reprogramming of T cells from human peripheral blood. Cell Stem Cell. 2010;7:15–19. doi: 10.1016/j.stem.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seki T, et al. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Zhou T, et al. Generation of induced pluripotent stem cells from urine. J Am Soc Nephrol. 2011;22:1221–1228. doi: 10.1681/ASN.2011010106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reiter JF, et al. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Latinkić BV, Kotecha S, Mohun TJ. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development. 2003;130:3865–3876. doi: 10.1242/dev.00599. [DOI] [PubMed] [Google Scholar]
- 31.David R, et al. MesP1 drives vertebrate cardiovascular differentiation through Dkk-1–mediated blockade of Wnt-signalling. Nat Cell Biol. 2008;10:338–345. doi: 10.1038/ncb1696. [DOI] [PubMed] [Google Scholar]
- 32.Bondue A, et al. Mesp1 acts as a master regulator of multipotent cardiovascular progenitor specification. Cell Stem Cell. 2008;3:69–84. doi: 10.1016/j.stem.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 33.Lindsley RC, et al. Mesp1 coordinately regulates cardiovascular fate restriction and epithelial-mesenchymal transition in differentiating ESCs. Cell Stem Cell. 2008;3:55–68. doi: 10.1016/j.stem.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci USA. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng R, et al. PU. 1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci USA. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vierbuchen T, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Addis RC, et al. Efficient conversion of astrocytes to functional midbrain dopaminergic neurons using a single polycistronic vector. PLoS ONE. 2011;6:e28719. doi: 10.1371/journal.pone.0028719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Caiazzo M, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 42.Kim J, et al. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci USA. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang P, et al. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 44.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 45.Kelly MC, Chang Q, Pan A, Lin X, Chen P. Atoh1 directs the formation of sensory mosaics and induces cell proliferation in the postnatal mammalian cochlea in vivo. J Neurosci. 2012;32:6699–6710. doi: 10.1523/JNEUROSCI.5420-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu Z, et al. Age-dependent in vivo conversion of mouse cochlear pillar and Deiters’ cells to immature hair cells by Atoh1 ectopic expression. J Neurosci. 2012;32:6600–6610. doi: 10.1523/JNEUROSCI.0818-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buganim Y, et al. Direct reprogramming of fibroblasts into embryonic sertoli-like cells by defined factors. Cell Stem Cell. 2012;11:373–386. doi: 10.1016/j.stem.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blau HM, et al. Plasticity of the differentiated state. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 49.Evans SM, Tai LJ, Tan VP, Newton CB, Chien KR. Heterokaryons of cardiac myocytes and fibroblasts reveal the lack of dominance of the cardiac muscle phenotype. Mol Cell Biol. 1994;14:4269–4279. doi: 10.1128/mcb.14.6.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ieda M, et al. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen JX, et al. Inefficient reprogramming of fibroblasts into cardiomyocytes using gata4, mef2c, and tbx5. Circ Res. 2012;111:50–55. doi: 10.1161/CIRCRESAHA.112.270264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srivastava D, Ieda M. Critical factors for cardiac reprogramming. Circ Res. 2012;111:5–8. doi: 10.1161/CIRCRESAHA.112.271452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Inagawa K, Ieda M. Direct reprogramming of mouse fibroblasts into cardiac myocytes. J Cardiovasc Transl Res. 2013;6:37–45. doi: 10.1007/s12265-012-9412-5. [DOI] [PubMed] [Google Scholar]
- 54.Wang B, et al. Reprogramming efficiency and quality of induced pluripotent stem cells (iPSCs) generated from muscle-derived fibroblasts of mdx mice at different ages. PLoS Curr. 2011;3:RRN1274. doi: 10.1371/currents.RRN1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Song K, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Protze S, et al. A new approach to transcription factor screening for reprogramming of fibroblasts to cardiomyocyte-like cells. J Mol Cell Cardiol. 2012;53:323–332. doi: 10.1016/j.yjmcc.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 57.Jayawardena TM, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anokye-Danso F, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 60.Kim J, et al. Functional integration of dopaminergic neurons directly converted from mouse fibroblasts. Cell Stem Cell. 2011;9:413–419. doi: 10.1016/j.stem.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pfisterer U, et al. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci USA. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res. 2003;93:32–39. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 63.Otsuji TG, et al. Progressive maturation in contracting cardiomyocytes derived from human embryonic stem cells: qualitative effects on electrophysiological responses to drugs. Stem Cell Res. 2010;4:201–213. doi: 10.1016/j.scr.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 64.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Inagawa K, et al. Induction of cardiomyocyte-like cells in infarct hearts by gene transfer of gata4, mef2c, and tbx5. Circ Res. 2012;111:1147–1156. doi: 10.1161/CIRCRESAHA.112.271148. [DOI] [PubMed] [Google Scholar]
- 66.Mathison M, et al. In vivo cardiac cellular reprogramming efficacy is enhanced by angiogenic preconditioning of the infarcted myocardium with vascular endothelial growth factor. J Am Heart Assoc. 2012;1:e005652. doi: 10.1161/JAHA.112.005652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim TK, et al. Transcriptome transfer provides a model for understanding the phenotype of cardiomyocytes. Proc Natl Acad Sci USA. 2011;108:11918–11923. doi: 10.1073/pnas.1101223108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mummery CL, et al. Differentiation of human embryonic stem cells and induced pluripotent stem cells to cardiomyocytes: a methods overview. Circ Res. 2012;111:344–358. doi: 10.1161/CIRCRESAHA.110.227512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Addis RC, et al. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J Mol Cell Cardiol. 2013;60:97–106. doi: 10.1016/j.yjmcc.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.DeHaan RL, Gottlieb SH. The electrical activity of embryonic chick heart cells isolated in tissue culture singly or in interconnected cell sheets. J Gen Physiol. 1968;52:643–665. doi: 10.1085/jgp.52.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davies MP, et al. Developmental changes in ionic channel activity in the embryonic murine heart. Circ Res. 1996;78:15–25. doi: 10.1161/01.res.78.1.15. [DOI] [PubMed] [Google Scholar]
- 72.Satin J, et al. Mechanism of spontaneous excitability in human embryonic stem cell derived cardiomyocytes. J Physiol (Lond) 2004;559:479–496. doi: 10.1113/jphysiol.2004.068213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Islas JF, et al. Transcription factors ETS2 and MESP1 transdifferentiate human dermal fibroblasts into cardiac progenitors. Proc Natl Acad Sci USA. 2012;109:13016–13021. doi: 10.1073/pnas.1120299109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nam YJ, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci USA. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chiu LL, Iyer RK, Reis LA, Nunes SS, Radisic M. Cardiac tissue engineering: current state and perspectives. Front Biosci. 2012;17:1533–1550. doi: 10.2741/4002. [DOI] [PubMed] [Google Scholar]
- 76.Boudou T, et al. A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues. Tissue Eng Part A. 2012;18:910–919. doi: 10.1089/ten.tea.2011.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Efe JA, et al. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 78.Christoforou N, et al. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS One. 2013;8:e63577. doi: 10.1371/journal.pone.0063577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bers DM. Cardiac excitation-contraction coupling. Nature. 2002;415:198–205. doi: 10.1038/415198a. [DOI] [PubMed] [Google Scholar]
- 80.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tallini YN, et al. Imaging cellular signals in the heart in vivo: cardiac expression of the high-signal Ca2+ indicator GCaMP2. Proc Natl Acad Sci USA. 2006;103:4753–4758. doi: 10.1073/pnas.0509378103. [DOI] [PMC free article] [PubMed] [Google Scholar]