Abstract
The bacterial 5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase (MTAN) enzyme is a multifunctional enzyme that catalyzes the hydrolysis of the N-ribosidic bond of at least four different adenosine-based metabolites: S-adenosylhomocysteine (SAH), 5’methylthioadenosine (MTA), 5’-deoxyadenosine (5’-DOA) and 6-amino-6-deoxyfutalosine. These activities place the enzyme at the hub of seven fundamental bacterial metabolic pathways: S-adenosylmethionine (SAM) utilization, polyamine biosynthesis, the purine salvage pathway, the methionine salvage pathway, the SAM radical pathways, autoinducer-2 biosynthesis, and menaquinone biosynthesis. The last pathway makes MTAN essential for H. pylori viability. Although structures of various bacterial and plant MTANs have been described, the interactions between the homocysteine moiety of SAH and the 5’-alkylthiol binding site of MTAN have never been resolved. We have solved crystal structures of an inactive mutant form of Helicobacter pylori MTAN bound to MTA and SAH to 1.63 and 1.20 Å, respectively. The active form of MTAN was also crystallized in the presence of SAH allowing structure determination of a ternary enzyme-product complex resolved at 1.50 Å. These structures identify interactions between the homocysteine moiety and the 5’-alkylthiol binding site of the enzyme. This information can be leveraged for the development of species-specific MTAN inhibitors that prevent the growth of H. pylori.
Keywords: Enzyme-substrate interactions, X-ray crystallography, Helicobacter pylori, Enzyme structure, Kinetics, Nucleoside nucleotide metabolism
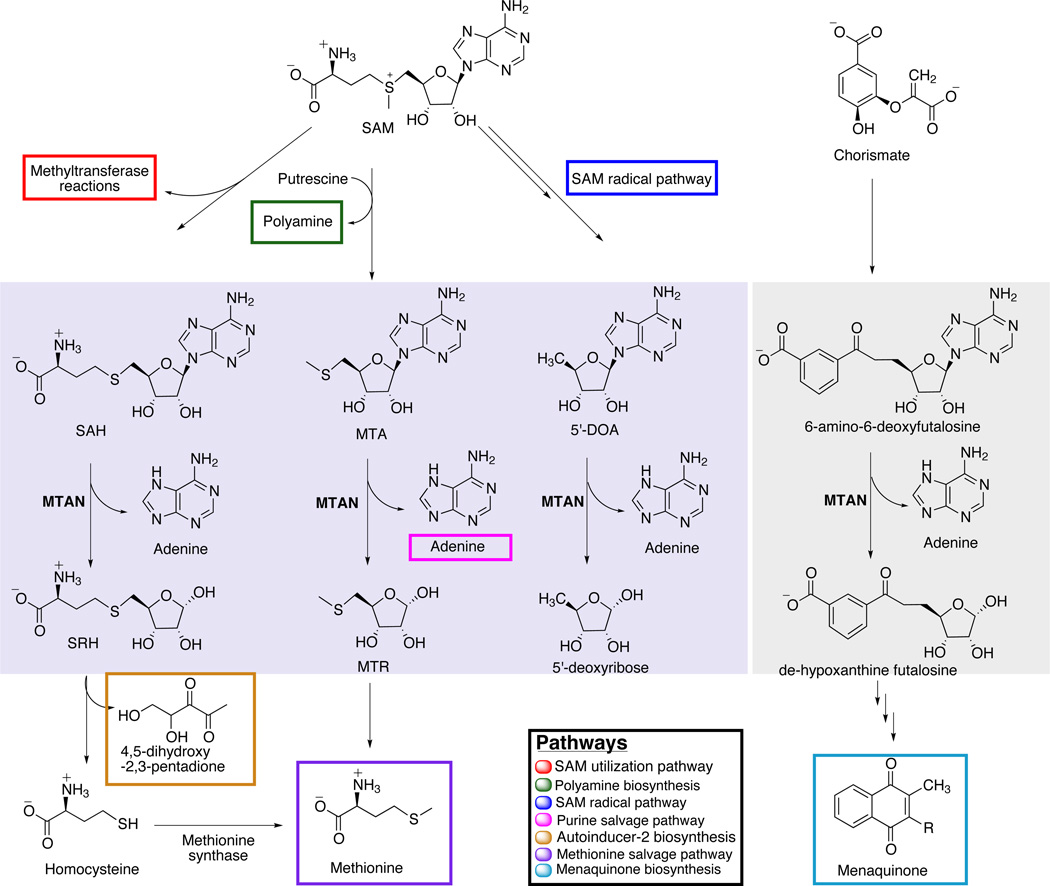
5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase plays a key role in multiple pathways in bacterial cells. It catalyzes the hydrolysis of the N-ribosidic bond of four different adenosine based metabolites releasing adenine and the ribose-containing products (Figure 1). It has been known that it utilizes SAH, MTA and 5’-DOA from the various SAM utilization pathways.1–3 SAH is a product of every SAM-dependent methyl transfer reaction. These biochemical transformations encompass methylation of the 3 common biological polymers: proteins, nucleic acids, and complex carbohydrates as well as various lipids.4 Methylation of these polymers affects fundamental properties in all living organisms such as gene expression, cell signaling and cellular metabolism. In all of these cases, SAM-dependent methylation is susceptible to product inhibition if SAH accumulates to high levels.5 Continuous breakdown of MTA is also required for the proper functioning of spermidine and spermine synthases that are otherwise inhibited by the accumulation of MTA.6 MTAN activity is also linked to important downstream metabolic pathways such as adenine and methionine salvage and autoinducer I (AI1) and II (AI2) production, which are important quorum-sensing signaling molecules.7 AI2 regulates the expression of genes that are required for interspecies communication, biofilm formation, and bacterial virulence and are therefore important facilitators of bacterial pathogenesis.8, 9 AI2 concentration in the cell depends on the transformation of S-ribosylhomocysteine (SRH) into homocysteine and 4,5-dihydroxy-2,3-pentanedione, a precursor for AI2.10 AI1 compounds are N-acylhomoserine lactones (AHLs) that allow intraspecies communication in primarily Gram-negative bacteria. AHL synthase catalyzes the transformation of SAM into AHL and MTA, so the activity of AHL synthase is sensitive to the accumulation of MTA.11 Choi-Rhee and Cronan have reported that MTAN is required for the catalytic hydrolysis of 5’-DOA and that accumulation of 5’-DOA inhibits SAM radical enzymes such as Biotin synthase.1 Recently, Li et al. determined that the Campylobacter jejuni and H. pylori MTAN (HpMTAN) enzymes play an essential role in an alternative menaquinone biosynthetic pathway.12 They reported that, unlike E. coli and T. thermophillus, C. jejuni and H. pylori biosynthesize menaquinone though an intermediate step that requires MTAN to catalyze the hydrolysis of 6-amino-6-deoxyfutalosine. This role, combined with previous data, suggest that MTAN represents a target for compounds capable of affecting bacterial metabolism and bacterial communication.7, 13, 14 Indeed, very recently, Wang et al. have shown that powerful inhibitors of MTAN inhibit the growth of H. pylori.15
FIGURE 1.
MTAN is a multifunctional enzyme. MTAN has been shown to use three different adenosine-based substrates in diverse bacterial systems: SAH, MTA and 5’-DOA (light violet solid box). In Campylobacter, it also hydrolyzes the N-ribosidic bond of 6-amino-6-deoxyfutalosine (grey solid box). Colored boxes encompass the compounds relevant to the pathways directly affected by MTAN activity. Only one adenine is boxed, but all four MTAN catalyzed reactions allow adenine salvage.
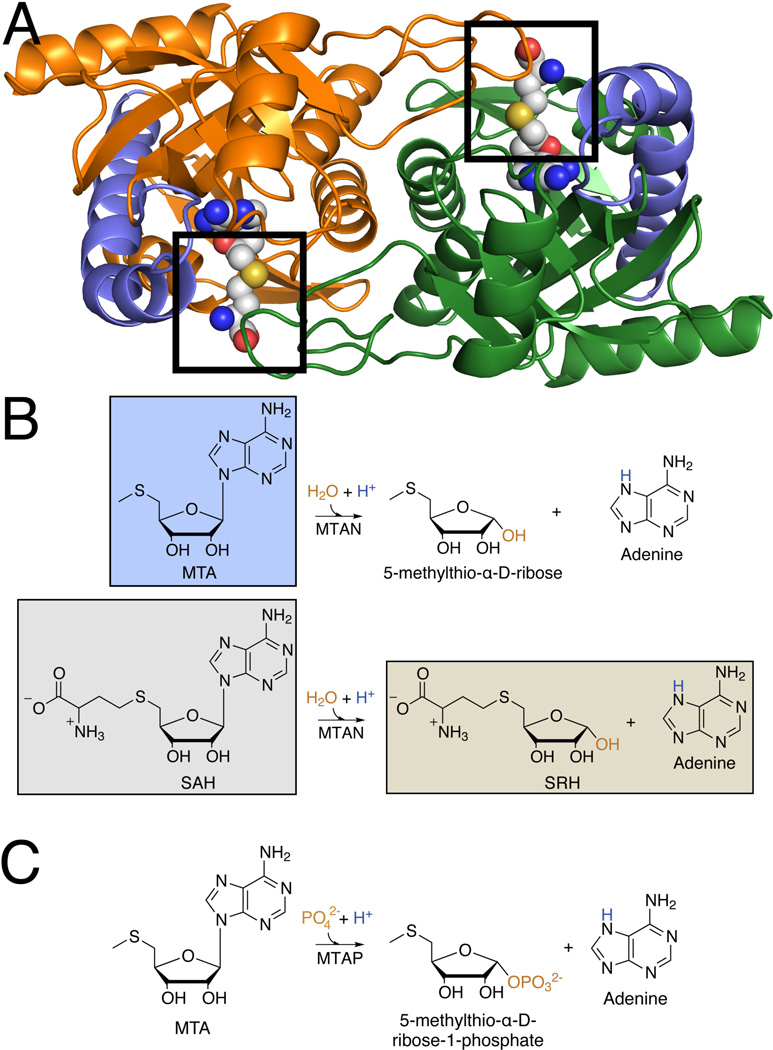
Previously determined bacterial and plant MTAN structures show that MTAN is an obligatory homodimer and the two molecules form a set of two shared active sites where residues of one molecule form the adenyl and ribosyl binding pockets and residues of the other molecule form the majority of the 5’alkylthio binding pocket (Fig. 2A).16–19 The catalytic mechanism has been thoroughly characterized as well as the interactions required to initiate that reaction (Fig. 2B and 2C).20–23 Binding of the substrate to the open form of the enzyme stimulates a conformational change characterized predominately by the kinking of helix α6. This closes the active site and positions residue D198 (HpMTAN numbering) to interact with N7 and the N6 exocyclic amine of the adenine moiety. It has been suggested that D198 acts as a general acid by donating a proton to N7 of the substrate resulting in the stretching of the N-ribosyl bond and subsequent formation of an oxocarbenium intermediate that undergoes nucleophilic attack by a water molecule.16 Release of the adenine and SRH products appears facile following hydrolysis and relaxation of helix α6.
FIGURE 2.
Shared active site in the HpMTAN homodimer and the enzyme mechanism. A. The closed form of the HpMTAN-D198N active site is shown with SAH (spheres colored by CPK) bound. The cartoons (orange and green) represent the HpMTAN homodimer. The position of helix α6 is indicated for each monomer in light blue. The boxed regions indicate the location of the 5’-alkylthio binding subsite that interacts with the homocysteine moiety of SAH. B. Reactions proposed for the two most commonly studied MTAN substrates are shown. The ligands for each of the determined structures are highlighted in boxes. The substrates are shown in black, the nucleophile in orange, and the MTAN-derived proton is in blue. C. Proposed reaction for human MTAP enzyme. The coloring of the components is the same as in panel B.
Although much is known about the structure of MTAN and its interactions with adenosine-containing substrates, specifically inhibiting the bacterial MTANs without also inhibiting homologous human enzymes is a challenge. The human genome encodes both a purine nucleoside phosphorylase and the 5’-MTA phosphorylase (MTAP) that are inhibited by the current generation of MTAN inhibitors.24, 25 The MTAN inhibitors with the tightest binding are either transition state or nucleoside analogues that interact with residues forming the ribosyl and adenyl binding pockets within the active site.22, 26 These regions are largely conserved between the bacterial MTANs and MTAPs.16 As a result, some of these compounds possess lower Ki values for human MTAP than for MTAN.27 Comparison of the MTAN and MTAP active site structures shows that the latter possesses a truncated 5’-alkylthio binding pocket that permits use of MTA as a substrate but not SAH.28 Since the larger 5’-alkylthio binding subsite of MTAN is an important feature that differentiates it from human MTAP, characterization of the specific interactions between the 5’-alkylthio binding site of MTAN and the homocysteine moiety of SAH may offer insight toward the design of MTAN inhibitors with improved selectivity for the bacterial enzymes as a basis for new drugs to treat H. pylori infections.
EXPERIMENTAL PROCEDURES
Mutagenesis
Site directed mutagenesis was performed to make the D198N mutant using a pET-32 based plasmid containing the wild-type pfs gene as a template. The template plasmid encodes a polyhistidine-tagged thioredoxin-HpMTAN fusion protein containing a prescission protease cut site immediately N-terminal to the first residue of wild-type HpMTAN (wt-HpMTAN). Proteolysis with prescission protease produces a polyhistidine tagged thioredoxin and untagged HpMTAN. Sequencing of the plasmid resulting from the mutagenesis experiment confirmed the presence of the D918N mutation.
MTAN Expression and Purification
Wild-type and HpMTAN mutants were expressed and purified as previously described.18 The plasmid containing the mutated gene was used to transform BL21 (DE3) Rosetta cells (EMD Biosciences). Cultures of LB media containing 0.1 mM chloramphenicol and 0.3 mM ampicillin were incubated at 37 °C. Cells were induced after reaching an Abs600nm = 0.6–0.8 by the addition of 0.1 mM IPTG and incubated for 18–20 hours at 16 °C. Cells were harvested by centrifugation and resuspended in buffer A (20 mM HEPES pH 7.5, 0.5 M NaCl, 5 mM β-mercaptoethanol and 25 mM imidazole). The resuspended cells were lysed by sonication and then centrifuged at 15,000g. The supernatant was applied to a 5 mL HisTrap column equilibrated with buffer A (GE Healthcare). Elution of recombinant proteins was performed using a linear gradient of imidazole from 25 to 250 mM over 20 column volumes. The fractions containing the purified proteins were treated with Prescission Protease and dialyzed overnight against buffer A. This protein sample was again applied to a HisTrap column (GE Healthcare) to selectively bind the cleaved histidine tag and the protease. The fractions containing HpMTAN were then subjected to size exclusion chromatography on a Hi-Load Superdex 200 column as a polishing step (GE Healthcare). All protein samples were analyzed for purity using SDS-PAGE. The absorbance at 280 nm was used to determine protein concentration using an extinction coefficient of 3105 M−1 cm−1.
Crystallization
For the crystallization studies, purified protein samples were dialyzed against the crystallization buffer (20 mM HEPES pH 7.5, 0.2 mM TCEP, and 1 mM EDTA) and concentrated to 16 mg/ml by ultrafiltration (Millipore). Crystals of the MTAN-D198N mutant complexed with either MTA or SAH were grown by the hanging-drop vapor diffusion method. Crystallization drops containing 1 µl of well solution, 1 µl of MTAN-D198N (16 mg/ml) and 0.5 µl of SAH/MTA (10 mM) were equilibrated with 100 µl of well solution. The well solution for producing MTAN-D198N/MTA complex crystals contained 0.2 M magnesium chloride, 0.1 M HEPES pH 7.5 and 25 % (w/v) PEG 3350. The well solution for producing MTAN-D198N/SAH complex crystals contained 0.05 M magnesium chloride hexahydrate, 0.1 M HEPES pH 7.5 and 30 % (v/v) PEG-MME 550. Crystals of the wt-HpMTAN/SRH/adenine complex were produced in the same conditions as the MTAN-D198N/SAH complex crystals. X-ray diffraction experiments were performed at the LS-CAT ID-D beamline at the Advanced Photon Source, Argonne National Labs. Data were integrated and scaled using HKL2000.29 Although these crystals were isomorphous with previously determined HpMTAN structures, molecular replacement was necessary and performed with a monomer of HpMTAN (pdb accession 3NM6) using the program EPMR30 Structure refinement proceeded using Phenix and Coot.31–33
Kinetic characterization of wt-MTAN
All assays were performed in triplicate in 100 mM HEPES and 50 mM KCl buffer at pH 7.2. For determining kinetic parameters, stocks of 1 mM SAH and 1 mM MTA were prepared in the same buffer. The substrates were serially diluted to produce a final concentration range of 5 µM to 125 µM in 100 µl of assay solution. The reduction of absorbance at a wavelength of 274 nm was monitored for 15 minutes at 37 ºC using a Biotek (Winooski, VT) Synergy H4 plate reader. The extinction coefficient of 1.6 mM−1 cm−1 was used for assays with both SAH and MTA. Non-linear regression analysis of kinetic data was performed using Prism 5.
RESULTS AND DISCUSSION
The homocysteine moiety of SAH forms specific interactions with the HpMTAN active site
Although much is known about the interactions between MTAN and the adenosine moiety of the variety of functional substrates, the mechanism by which MTAN interacts with components of the 5’alkylthio moiety of those substrates is not. The strategy used to characterize these interactions was to create an inactive mutant capable of binding substrate but lacking enzymatic activity. Based on the previously reported E. coli MTAN (EcMTAN) mutant/MTA complex, an HpMTAN-D198N mutant was created.23 The crystal structure of the HpMTAN-D198N/SAH complex was solved and refined to 1.2 Å resolution (Table 1) allowing for clear assessment of the interactions between the SAH substrate and the 5’-alkythio binding subsite.
Table 1.
X-ray diffraction and refinement statistics. The structures of the enzyme bound compound are shown for clarity. Parentheses indicate units. Brackets indicate additional information regarding that value. A.S.U., asymmetric unit; r.m.s., root mean square. Rsym = Σ|Ii − <Ii>| / Σ|Ii|, where I is the intensity of a given reflection and <I> is the average intensity for that reflection. Rwork and Rfree = Σ||Fobs| − |Fcalc||/Σ|Fobs|, where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rfree is calculated from a set of reflections chosen randomly. These data are equal to 5% of the unique reflections and were not used for model refinement or calculation of the Rwork value.
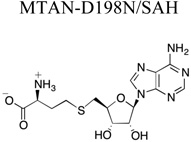 |
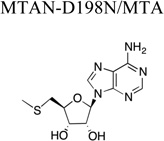 |
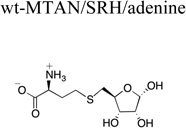 |
|
|---|---|---|---|
| Resolution range (Å) [highest shell] | 50.00-1.20 [1.22-1.20] | 50.00-1.63 [1.67-1.63] | 50.00-1.50 [1.53-1.50] |
| Space group | P3221 | P3221 | P3221 |
| a, b (Å) | 80.7 | 80.7 | 81.2 |
| c (Å) | 67.2 | 67.2 | 67.4 |
| Total reflections [unique reflections] | 822470 [75266] | 348061 [32308] | 237786 [40655] |
| Completeness (%) [highest shell] | 95.0 [92.6] | 99.7 [99.2] | 98.5 [97.5] |
| Redundancy [highest shell] | 10.9 [10.7] | 10.8 [10.1] | 5.8 [5.7] |
| Average I/σ (I) [highest shell] | 16.0 [4.1] | 11.7 [3.3] | 13.9 [3.6] |
| Rsym (%) [highest shell] | 5.0 [30.0] | 6.5 [38.1] | 6.2 [44.9] |
| Atoms/A.S.U | 2,218 | 2,127 | 2,010 |
| Rwork (%) | 15.4 | 13.5 | 18.2 |
| Rfree (%) | 17.5 | 17.8 | 20.5 |
| Average B-factor protein (Å2) | 16.6 | 20.9 | 22.2 |
| Average B-factor ligand (Å2) | 14.1 | 18.9 | 21.3 |
| Average B-factor solvent (Å2) | 27.0 | 31.3 | 29.5 |
| Ligand occupancy (%) | 100 | 100 | 100 |
| r.m.s. bonds (Å) | 0.005 | 0.005 | 0.006 |
| r.m.s. angles (°) | 1.09 | 0.95 | 0.99 |
| Ramachandran favored (%) | 96.9 | 96.9 | 96.9 |
| Ramachandran disallowed (%) | 0.0 | 0.0 | 0.0 |
| Coordinate Error (Å) | 0.14 | 0.17 | 0.34 |
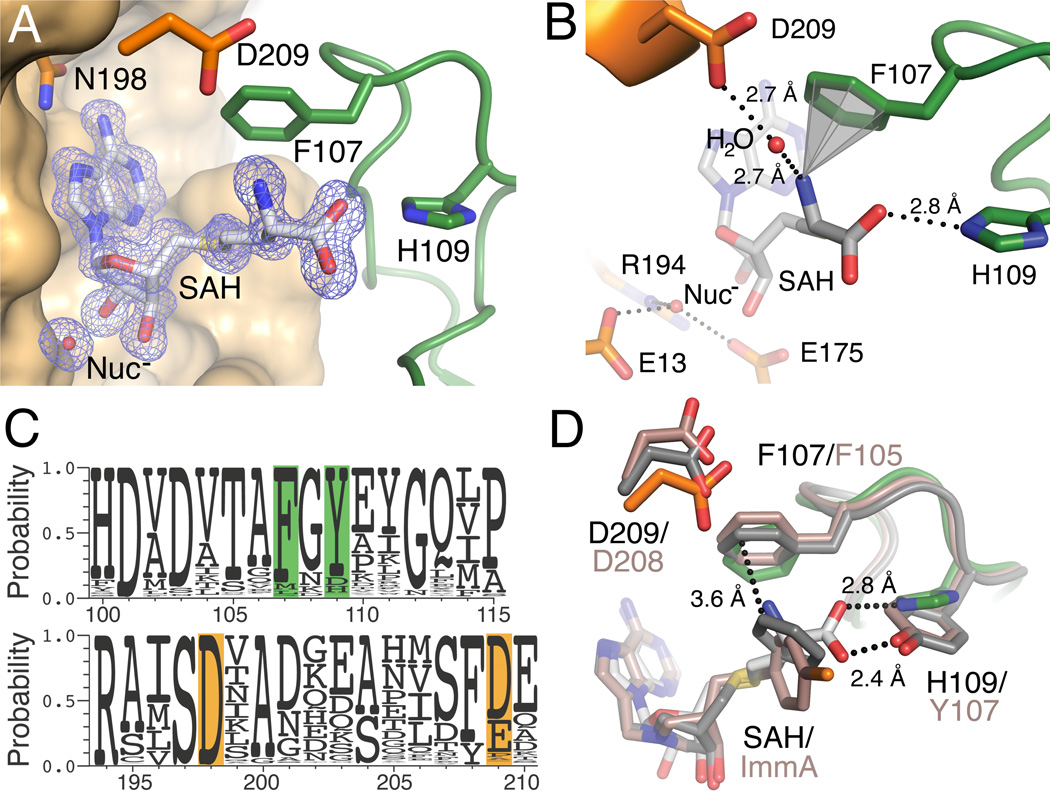
The difference maps within the region enveloped by the HpMTAN-D198N active site exhibited electron density that allowed unambiguous fitting of each non-hydrogen atom of SAH, including the previously unobserved α-amino and carboxyl groups of the homocysteine moiety (Fig. 3A). The α-amino group of the homocysteine moiety forms two types of interactions with the 5’-alkythio binding subsite. The first is a cation-π interaction with the aromatic side chain of F107, which is positioned at distance of 3.3 Å between the α-amino nitrogen of SAH and the six carbons of the phenylalanine aromatic ring (Fig. 3B). This type of interaction has been observed in other proteins that specifically bind ligands or substrates that contain ammonium moieties and disruption of the cation-π interaction is known to affect the binding affinity within those systems.34 Excellent examples of this are represented by the structures of the γ-aminobutyric-acid (GABA) receptor and acetylcholine binding protein (AChBP) bound to their respective ligands or ligand analogs.34–36 In the AChBP/carbamylcholine structure, the aromatic residues found within the AChBP ligand binding pocket form multiple cation-π interactions with the quaternary ammonium of carbamylcholine.35 Recently, this type of interaction was observed in an insect GABA receptor and incorporation of artificial fluorophenylalanine amino acids within the ligand binding site of the acetylcholine receptor eliminates a cation-π interaction and produces a consistent increase in the measured EC50 values when compared to the wild-type acetylcholine receptor.34
FIGURE 3.
SAH active site interactions. A. Shown is the HpMTAN-D198N active site. The electron density of an Fo-Fc omit map is shown contoured at 3σ, where both SAH and the nucleophilic water (Nuc−) were omitted during map calculation. The orange molecular surface represents most of HpMTAN-D198N molecule 1 with the exception of residues N198 and D209 (shown with orange carbons). N198 is the residue mutated to inactivate the enzyme. The green ribbon and carbon atoms correspond to molecule 2 of the homodimer with residues F107 and H109 forming direct interactions with the substrate. Heteroatoms are colored by CPK. B. Colors are as indicated for panel a. Dashed lines indicate bonding interactions between. The gray cone signifies the cation-π interaction. The nucleophilic water and the conserved residues that coordinate it are positioned on the lower left and labeled. C. A sequence alignment of 1000 MTAN sequences is shown as a logo alignment. Residues are numbered according to that of HpMTAN. The orange indicates important residues originating from molecule 1 and green highlights those originating from molecule 2. D. Superposition of the wt-EcMTAN/Immucillin and the HpMTAN-D198N/SAH complexes highlights the interactions within the 5’-alkylthio binding subsite. The atoms of the HpMTAN-D198N/SAH complex are as shown previously. The dark gray atoms indicate those from the wt-EcMTAN/BnT-DADMeImmA complex (3DF9), while the rose colored atoms indicates those from the wt-EcMTAN/(4-Chlorophenyl)thio-DADMe-Immucillin A complex (3O4V). The rose labels indicate residues from wt-EcMTAN/inhibitor complexes while black labels indicate residues from the HpMTAN-D198N/SAH complex. The potential hydrogen bond between the α-carboxyl of SAH and Y107 of EcMTAN is shown as a dashed bond with a length of 2.4 Å. The interaction between BnT-DADMe-Immucillin and F105 of EcMTAN is indicated with a 3.6 Å dashed bond.
The second interaction between HpMTAN-D198N and the α-amino moiety is through a water-mediated hydrogen bond to D209. This residue is located near the N-terminus of helix α6, which is disordered in ligand-free MTAN structures but takes a helical form as a consequence of substrate binding.18, 23 One consequence of this structural organization is to position D198 near the adenyl moiety to initiate proton transfer to the substrate, while the other consequence is the formation of the through-water hydrogen bond. The distance from the water molecule to the side chain of D209 and to the α-amino moiety of SAH is equidistant with each hydrogen bond measuring 2.7 Å (Fig. 3B). Additionally, the 4.4 Å distance between the side chain of D209 and the α-amino moiety of SAH suggests a possible ionic interaction. The α-carboxyl moiety of SAH in the HpMTAN-D198N/SAH complex also forms a specific interaction with the enzyme active site. Specifically, the Nε atom of H109 forms a 2.8 Å bonding interaction with SAH.
To assess the potential importance of these interactions on substrate recognition, we performed a sequence alignment of the 1000 MTAN protein sequences in the UniProt knowledgebase most closely related to that of H. pylori (Fig. 3C). This sequence comparison showed that more than 85% of these MTANs possess a phenylalanine at the position corresponding to F107 in HpMTAN suggesting that the cation-π interaction is an important feature that supports specific enzyme/substrate interactions. The sequence alignment also suggests that the charge of the residue at position 209 is important, as it is either an aspartate or glutamate in roughly 90% of the 1000 analyzed MTAN sequences. If any hydrogen bond acceptor or donor were sufficient at this position one would expect to observe asparagine and glutamine with roughly equivalent probability. H109 is conserved in bacteria of the order Campylobacterales, which represents approximately 10% of the 1000 MTAN sequences inspected in this study. In contrast, 80% of the sequences possess a tyrosine residue at the analogous position, which is common to all of the previously determined bacterial MTAN structures. Superposition of the HpMTAN with the EcMTAN (Fig. 3D) suggests that a tyrosine side chain could maintain a hydrogen bond with the α-carboxyl of SAH. This highlights the importance of MTAN possessing a hydrogen bond donor or acceptor at this position. In addition, the lack of a resolved SAH/MTAN complex prior to this study suggests that a hydrogen bond between the tyrosine of EcMTAN and the SAH α-carboxylate is insufficient to structurally resolve this interaction, whereas the interaction between the α-carboxyl moiety of SAH and H109 of HpMTAN-D198N is of sufficient strength to fix the conformation of SAH. To ensure that these interactions are observed in the wild-type enzyme and to identify any structural changes in the 5’-alkylthio binding region following substrate hydrolysis, we solved the crystal structure of wt-HpMTAN complexed with adenine and SRH to 1.54 Å resolution.
Active site interactions with the homocysteine moiety are unchanged during catalysis
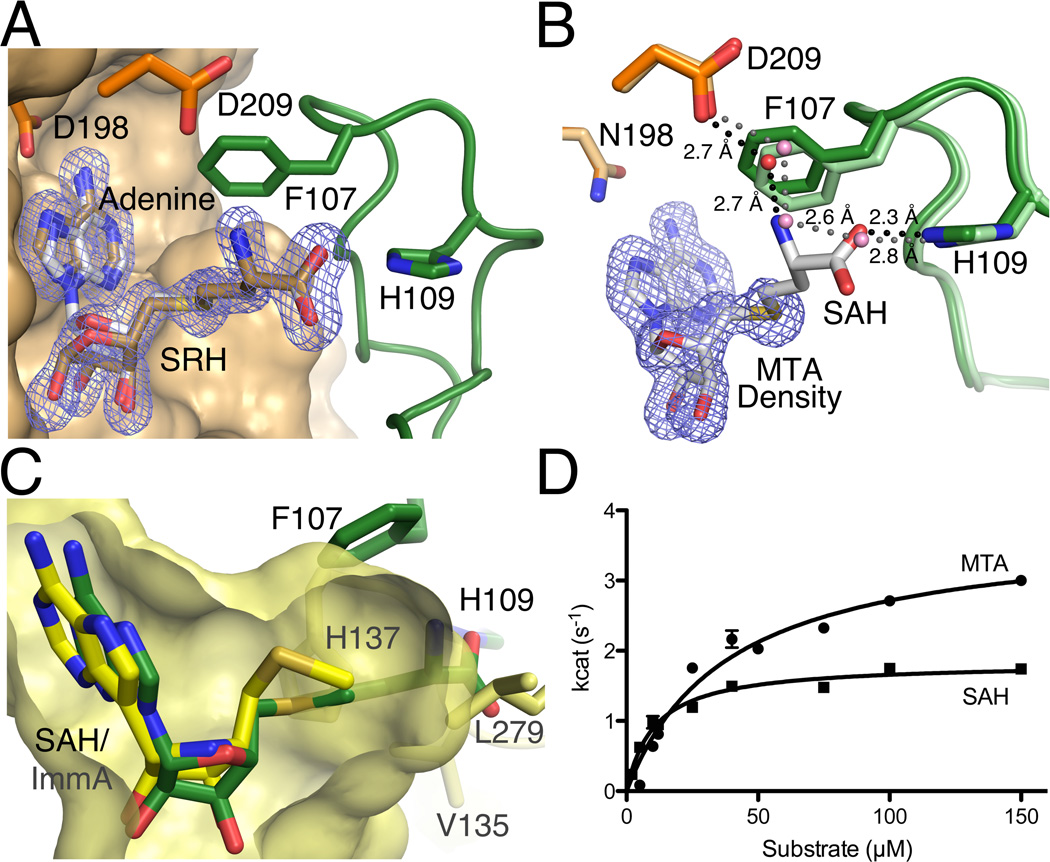
Previous attempts to form a wt-MTAN/product complex resulted in a complex with adenine and tris in the active site.18 The crystallization experiments described here produced wt-MTAN crystals in the closed form with both adenine and the α-anomer of SRH bound in the active site as evidenced by two discrete regions of density in an Fo-Fc difference map (Fig. 4A). The presence of the α-anomer is consistent with the proposed catalytic mechanism where breaking of the N-ribosidic bond produces an oxocarbenium intermediate that undergoes a nucleophilic attack on the α face of the ribosyl group. This nucleophilic attack is from an ordered water molecule coordinated by conserved residues E13, E175, and R194, which has been observed in all MTAN crystal structures to date (Fig. 3B).16, 18 In the wt-HpMTAN/adenine/SRH structure, it is clear that conversion from the β-ribosyl substrate to the α-ribosyl product also alters the ribose conformation from C4’-endo to C2’-endo. This stems from maintaining the location of the nucleophilic water, which is now O1 of the SRH product, through continued coordination by residues E13, E175, and R194. This conformational change in the ribosyl moiety contrasts with the fixed conformation of the homocysteine moiety and the interactions it forms with the 5’-alkylthio binding pocket. This suggests that interactions defined by the 5’-alkylthio binding subsite allow for less conformational flexibility than those in the ribose binding subsite and that the interactions between the homocysteine moiety and the 5’-alkythio binding subsite likely remain unchanged throughout the enzymatic reaction.
FIGURE 4.
Comparison of SAH and MTA as substrates. A. Shown is the superposition of the HpMTAN-D198N/SAH and wt-HpMTAN/SRH/adenine complex structures. The Fo-Fc difference density from the wt-HpMTAN/SRH/adenine diffraction data is shown contoured at 3σ. Both SRH and adenine were omitted during map calculation. Coloring of the protein and SAH is the same as previous figures. The bronze carbon atoms represent the SRH and adenine products. B. Shown is the superposition of the HpMTAN-D198N/SAH and MTA complex structures. The protein atoms from the HpMTAN-D198N/SAH structure is colored as in previous figures and the carbon atoms of the enzyme of the MTA complex structure are slightly lighter in color. Since SAH and MTA atom positions superimpose perfectly, the atoms of MTA are not shown but the location of MTA is indicated by an Fo-Fc omit map contoured at 3σ where MTA was omitted from the map calculation. The hydrogen bonded water network in the MTA bound structure is indicated by the dashed gray lines and that of the SAH complex structure is indicated by the dashed black lines. C. HpMTAN and human MTAP 5’-alkylthio binding subsites differ in structure. A superposition of the HpMTAN-D198N/SAH complex active site with the human MTAP active site showing bound methylthio-Immucillin-A (1K27). Components of the HpMTAN-D198N/SAH complex are colored as shown previously. The corresponding 5’-alkylthio binding subsite of human MTAP is shown in yellow. The yellow carbon atoms and surface indicate the MTAP/methylthio-Immucillin-A (ImmA) complex and the active site cavity, respectively. H137, V135 and L279 form the boundary of the 5’-alkylthio binding subsite in MTAP. H109, F107 and SAH from the HpMTAN-D198N/SAH complex are also shown. The homocysteine moiety of SAH protrudes more than 4Å beyond the confines of the human MTAP active site. D. Comparison of steady-state kinetics using either MTA or SAH shows a slightly higher affinity for SAH but lower enzyme turnover when compared to the use of MTA. Error bars are the result of reactions performed in triplicate.
The HpMTAN 5’-alkylthio binding subsite structure is substrate independent
The first X-ray crystal structures of MTAN highlighted the conformational change in helix α6 resulting from the induced fit between MTAN and its substrates. However, inspection of the 5’-alkylthio binding subsites from those structures and subsequent structures suggests that this region is insensitive to the substrate-bound state of the enzyme. To determine if the variability of the 5’-alkylthio moieties of the known MTAN substrates affects differences in the structure of the 5’-alkylthio binding subsite, we determined the crystal structure of an HpMTAN-D198N/MTA complex to 1.6 Å resolution and compared this structure to that of the HpMTAN/SAH complex structure.
When superimposing the MTA and SAH complex structures (rmsd for Cα atoms = 0.067 Å), the MTA substrate superimposes perfectly with the corresponding atoms of SAH. Additionally, the 5’-alkylthio binding pockets of both structures maintain the same conformation (Fig. 4B). In particular, the loop containing residues F107 and H109 maintains the identical conformation in both structures. Another similarity between the SAH and MTA complexes is the solvent structure and the hydrogen bonded network within the 5’-alkylthio binding pocket. Specifically, the hydrogen bonded network formed by D209, a water molecule, and the α-amino group of SAH is observed in the MTA structure as a D209-water-water network. These solvent interactions and the lack of structural changes in the 5’-alkylthio binding pocket suggest that MTAN follows a lock-and-key model for binding the homocysteine moiety that contrasts with the dynamics observed in the residues that form the adenyl and ribosyl binding sites.37 Based on the findings described here, it is possible to draw parallels between the interactions observed in the HpMTAN-D198N/SAH complex and those interactions likely formed in a complex of the HpMTAN with the recently identified MTAN substrate, 6-amino-6-deoxyfutalosine. Primarily, the benzoate moiety of 6-amino-6-deoxyfutalosine would be positioned analogously to the α-amino group of SAH within the 5’-alkylthio binding subsite and interact with F107 through a π-π stacking interaction. Since 6-amino-6-deoxyfutalosine, similarly to SAH, terminates with a carboxylate moiety it likely forms an interaction with H109 similar to that observed in the HpMTAN-D198N/SAH and wt-HpMTAN/SRH/adenine complexes.
To date, two wt-MTAN complex structures have been solved with a ligand containing an aromatic functional group bound within the 5’-alkylthio binding pocket but none possessing a carboxylate moiety. The EcMTAN complexed with two different methyl-immucillin inhibitors: BnT-DADMeImmA (pdb accession code 3DF9) and (4-Chlorophenyl)thio-DADMe-ImmA (pdb accession code 3O4V). These structures show that the phenyl moieties bind in the same location but possess slightly different orientations, which results from differences in conformation of the pyrrolidine moieties of these transition-state analogs. In the 3DF9 structure, the phenyl moiety forms a herringbone interaction with the residue analogous to F107 and overlaps with the position of the α-amino moiety of SAH when bound to HpMTAN. However, in 3O4V, the chlorophenyl moiety does not form any π-π stacking interactions between the inhibitor and the 5’-alkylthio binding pocket suggesting that other interactions are predominating and forcing an alternative inhibitor conformation or the presence of the chlorine atom coupled to the phenyl moiety is sterically hindering the π-π stacking interaction.
The HpMTAN/ligand interactions observed within the 5’-alkylthio binding subsite differ markedly from those observed in the analogous region of the human MTAP/methylthio-Immucillin-A complex structure (accession code 1K27).38 The structural differences between EcMTAN and human MTAP have been previously detailed.28 This comparison clearly exhibited that MTAP has a truncated, hydrophobic 5’-alkylthio binding pocket that can accommodate a methylthio moiety but lacked sufficient volume to accommodate longer or bulkier 5’ substituents. In contrast, bacterial MTANs possess a longer 5’-alkylthio binding pocket that extends to the protein surface, which would allow for the binding of compounds with a more hydrophilic and significantly larger 5’ substituent. This is also true for HpMTAN, but the structures described here shine new light on the importance of these differences.
While the 5’-alkylthio binding pockets of HpMTAN and human MTAP each possess similar residues in the 5’-alkylthio binding pockets, the relative positions of these residues are significantly different (Figure 4C). H137, V135 and L279 of human MTAP form the end of the 5’-alkylbinding pocket and form primarily van der Waals interactions with the methylthio moiety of methylthio Immucillin-A. From the superposition, it is clear that there is not sufficient volume in the human MTAP active site to accommodate the larger 5’-homocysteine moiety, which extends roughly 4.5 Å beyond the boundary of the MTAP active site surface. Additional evidence for possible steric hindrance in this region comes from the relative positions of the sulfur atoms in the two ligands. The sulfur of SAH abuts the MTAP active site envelope whereas the sulfur of the Immucillin is positioned near the center of the 5’alkylthio binding subsite due to a roughly 110 ° difference in the 4’-5’ dihedral. This suggests that design of an inhibitor with a fixed structure at the 5’ carbon of ribose that approximates the conformation of SAH in the HpMTAN/SAH complex structure should be quite selective for the HpMTAN.
5’-alkythiol binding pocket-homocysteine interactions affect HpMTAN kinetics
The data presented here suggest a role for the HpMTAN 5’-alkylthio binding subsite in specifically coordinating the homocysteine moiety of SAH and that the structure of the 5’-alkylthio binding region is independent of substrate identity. Because these interactions play an important role in substrate recognition and the structural evidence suggests that the homocysteine moiety contributes only a minor conformational entropy penalty to binding, it is expected that evidence of the impact on substrate binding could be observed using steady-state kinetics. In particular, the three additional interactions formed between HpMTAN and the SAH substrate should produce a lower KM for the HpMTAN/SAH reaction when compared with the HpMTAN/MTA reaction.
To perform these experiments, the hydrolysis of either MTA or SAH by wt-HpMTAN was monitored by measuring the absorbance decrease at 274 nm. Fitting the initial velocity data allowed calculation of the kinetic parameters (Fig. 4D). The KM values for SAH and MTA were 10 ± 1 µM and 39 ± 5 µM, respectively (Table 2). The kcat values for SAH and MTA were 1.8 ± 0.1 sec−1 and 3.8 ± 0.2 sec−1, respectively. These values are in accordance with those determined for the wild-type Streptococcus pnueumoniae MTAN, where Singh et al. showed that wild-type S. pnueumoniae MTAN has a 1.8-fold lower KM when using SAH versus using MTA.38 However, results for both HpMTAN and S. pnueumoniae MTAN contrast with those observed for EcMTAN.39 Lee et al. showed that wt-EcMTAN exhibits a 1.6-fold lower KM when using MTA than when using SAH as the substrate, but kcat values are similar for each substrate (2.6 ± 0.1 and 3.0 ± 0.1 sec−1) resulting in slightly higher enzymatic efficiency when using MTA as a substrate.39
Table 2.
HpMTAN Kinetic parameters.
| SAH | MTA | |||||
|---|---|---|---|---|---|---|
|
KM (µM) |
kcat (sec−1) |
kcat/KM (µM−1 sec−1) |
KM (µM) |
kcat (sec−1) |
kcat/KM (µM−1 sec−1) |
|
| wt | 10 ± 1 | 1.8 ± 0.1 | 0.17 ± 0.02 | 39 ± 5 | 3.8 ± 0.2 | 0.097 ± 0.009 |
| F107A | 19 ± 3 | 0.44 ± 0.02 | 0.023 ± 0.002 | 37 ± 4 | 1.0 ± 0.1 | 0.027 ± 0.002 |
| H109A | 14 ± 4 | 0.74 ± 0.05 | 0.051 ± 0.010 | 33 ± 6 | 1.2 ± 0.1 | 0.037 ± 0.004 |
The reason for the discrepancy in the kinetic parameters is not clear. The kinetics of the S. pnueumoniae MTAN are more like HpMTAN than EcMTAN. However, both the S. pnueumoniae MTAN and EcMTAN possess a tyrosine at the corresponding position of HpMTAN H109.39 Therefore, the difference in kinetics observed in various bacterial MTANs cannot be simply ascribed to an increase in the positive electrostatic potential due to the presence of H109 in the HpMTAN active site. Another factor potentially contributing to the kinetic differences may stem from solvent effects in the different 5’-alkylthio binding pockets. This was previously suggested by Thomas et al. to explain large entropic and enthalpic variation observed when testing a panel of inhibitors to various bacterial MTANs.40 That rationale may apply to data presented here. In other protein-ligand systems, the effects of ligand and active site desolvation have been tested experimentally for both hydrophilic and hydrophobic substrate/ligand binding sites, and the conclusions can be applied to the 5’-alkylthio binding subsite of MTAN.41 Considering that the portion of the HpMTAN 5’-alkylthio binding pocket that interacts with the α-carboxyl and α-amino moieties of SAH is generally hydrophilic, the primary driving force for binding likely stems from favorable entropy due to ligand desolvation and active site desolvation of the hydrophilic portion of the active site. This is displayed clearly in figure 4B, where two ordered water molecules in the MTA complex that exhibit a rather extensive hydrogen bonded network superimpose with portions of the SAH molecule in the SAH complex. Displacement of these ordered waters affords an entropic gain, but enthalpic changes are likely minimal because of maintenance of the specific ionic and hydrogen bonding interactions following SAH binding.
To quantify the effect of these interactions within the HpMTAN 5’-alkylthio binding pocket, we performed kinetic studies using two single mutants, F107A and H109A (Table 2). As predicted from the crystal structure, the KM values of both mutants exhibit a minor increase (1.9-fold for F107A and 1.4-fold for H109A, respectively) when using SAH as a substrate, while the KM remains unchanged when using MTA. This indicates that the cation-π interaction of F107 and the ionic interaction of H109 play a role in recognizing the homocysteine moiety of SAH. Similarly, studies of EcMTAN showed that mutants F105A (F107 in wt-HpMTAN) and Y107F (H109 in wt-HpMTAN) exhibited a four-fold and 1.3-fold increase, respectively, in the KM values for SAH, suggesting these interactions in the 5’-alkylthio binding pocket are common features among different bacterial MTANs.39 The effects of the F107A and H109A mutations on the kcat values are more difficult to interpret. When compared to that of wt-HpMTAN, the kcat values for F107A and H109A using either SAH or MTA as substrate decreased by three to four-fold. We did not anticipate that the catalytic rate when using MTA as a substrate would be affected. A possible rationale for the effect on the turnover rate is that the structure of the loop containing F107 and H109 is altered in the mutants, which could affect the turnover of either SAH or MTA substrates.
Conclusions
Characterizing interactions between enzyme and substrate are at the heart of understanding substrate specificity and enzyme catalysis. For the bacterial MTANs, the interactions within the adenine and ribose subsites are well documented. This study offers the first description of the specific bonding interactions between the 5’-alkylthio binding subsite of any MTAN with a homocysteine-containing ligand by solving the X-ray crystal structure of a binary complex of an inactive HpMTAN mutant with SAH and the structure of a ternary, product complex. These structures not only highlight previously unobserved interactions between the enzyme and either substrate or product, but also show that this portion of the enzyme active site is invariant with respect to its substrate-bound state and substrate identity. Additionally, the wt-HpMTAN/adenine/SRH ternary complex structure shows that the interactions between the homocysteine moiety and the enzyme active site are consistent with those observed for the HpMTAN-D198N/SAH complex. The four-fold lower KM and the two-fold lower kcat observed when using SAH as a substrate versus MTA reflects the additional interactions between the 5’-alkylthio binding subsite and the homocysteine moiety of either the substrate or product. Taken together, these results suggest that the 5’-alkylthio binding subsite is a prime target for the design of new MTAN inhibitors that are highly specific to bacterial MTANs but will selectively kill H. pylori while sparing beneficial commensal bacteria endogenous to the human gastrointestinal tract.
ACKNOWLEDGMENT
The authors thank Chih-chin Huang for helpful comments during manuscript preparation. The authors also thank the LS-CAT for the beam time essential for this study. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
This research was supported by an NIH grant to D.R. Ronning (AI089653).
ABBREVIATIONS
- MTAN
5’-methylthioadenosine/S-adenosylhomocysteine nucleosidase
- SAH
S-adenosylhomocysteine
- MTA
5’methylthioadenosine
- 5’-DOA
5’-deoxyadenosine
- SAM
S-adenosylmethionine
- AI1
autoinducer I
- AI2
autoinducer II
- SRH
S-ribosylhomocysteine
- AHL
N-acylhomoserine lactone
- MTAP
5’-MTA phosphorylase
- HpMTAN
H. pylori MTAN
- EcMTAN
E. coli MTAN
- GABA
γ-aminobutyric-acid
- AChBP
acetylcholine binding protein
Footnotes
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Choi-Rhee EJ, Cronan JE. A nucleosidase required for in vivo function of the S-Adenosyl-L-Methionine radical enzyme, biotin synthase. Chemistry & Biology. 2005;12:589–593. doi: 10.1016/j.chembiol.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 2.Della Ragione F, Porcelli M, Carteni-Farina M, Zappia V, Pegg AE. Escherichia coli S-adenosylhomocysteine/5'-methylthioadenosine nucleosidase. Purification, substrate specificity and mechanism of action. The Biochemical journal. 1985;232:335–341. doi: 10.1042/bj2320335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller CH, Duerre JA. S-ribosylhomocysteine cleavage enzyme from Escherichia coli. The Journal of biological chemistry. 1968;243:92–97. [PubMed] [Google Scholar]
- 4.Sufrin JR, Meshnick SR, Spiess AJ, Garofalo-Hannan J, Pan XQ, Bacchi CJ. Methionine recycling pathways and antimalarial drug design. Antimicrobial agents and chemotherapy. 1995;39:2511–2515. doi: 10.1128/aac.39.11.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borchardt RT. S-Adenosyl-L-methionine-dependent macromolecule methyltransferases: potential targets for the design of chemotherapeutic agents. Journal of medicinal chemistry. 1980;23:347–357. doi: 10.1021/jm00178a001. [DOI] [PubMed] [Google Scholar]
- 6.Pajula RL, Raina A. Methylthioadenosine, a potent inhibitor of spermine synthase from bovine brain. FEBS letters. 1979;99:343–345. doi: 10.1016/0014-5793(79)80988-6. [DOI] [PubMed] [Google Scholar]
- 7.Parveen N, Cornell KA. Methylthioadenosine/S-adenosylhomocysteine nucleosidase, a critical enzyme for bacterial metabolism. Mol Microbiol. 2011;79:7–20. doi: 10.1111/j.1365-2958.2010.07455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fong KP, Chung WSO, Lamont RJ, Demuth DR. Intra- and interspecies regulation of gene expression by Actinobacillus actinomycetemcomitans LuxS. Infect Immun. 2001;69:7625–7634. doi: 10.1128/IAI.69.12.7625-7634.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vendeville A, Winzer K, Heurlier K, Tang CM, Hardie KR. Making ' sense' of metabolism: Autoinducer-2, LuxS and pathogenic bacteria. Nat Rev Microbiol. 2005;3:383–396. doi: 10.1038/nrmicro1146. [DOI] [PubMed] [Google Scholar]
- 10.Marques JC, Lamosa P, Russell C, Ventura R, Maycock C, Semmelhack MF, Miller ST, Xavier KB. Processing the interspecies quorum-sensing signal autoinducer-2 (AI-2): characterization of phospho-(S)-4,5-dihydroxy-2,3-pentanedione isomerization by LsrG protein. The Journal of biological chemistry. 2011;286:18331–18343. doi: 10.1074/jbc.M111.230227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuqua C, Greenberg EP. Listening in on bacteria: Acyl-homoserine lactone signalling. Nature Reviews Molecular Cell Biology. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Apel D, Gaynor EC, Tanner ME. 5 '-Methylthioadenosine Nucleosidase Is Implicated in Playing a Key Role in a Modified Futalosine Pathway for Menaquinone Biosynthesis in Campylobacter jejuni. Journal of Biological Chemistry. 2011;286:19392–19398. doi: 10.1074/jbc.M111.229781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiratsuka T, Furihata K, Ishikawa J, Yamashita H, Itoh N, Seto H, Dairi T. An alternative menaquinone biosynthetic pathway operating in microorganisms. Science. 2008;321:1670–1673. doi: 10.1126/science.1160446. [DOI] [PubMed] [Google Scholar]
- 14.Seto H, Jinnai Y, Hiratsuka T, Fukawa M, Furihata K, Itoh N, Dairi T. Studies on a new biosynthetic pathway for menaquinone. J Am Chem Soc. 2008;130 doi: 10.1021/ja710207s. 5614-+ [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Haapalainen AM, Yan F, Du Q, Tyler PC, Evans GB, Rinaldo-Matthis A, Brown RL, Norris GE, Almo SC, Schramm VL. A Picomolar Transition State Analogue Inhibitor of MTAN as a Specific Antibiotic for Helicobacter pylori. Biochemistry. 2012;51:6892–6894. doi: 10.1021/bi3009664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee JE, Cornell KA, Riscoe MK, Howell PL. Structure of E-coli 5 '-methylthioadenosine/S-adenosylhomocysteine nucleosidase reveals similarity to the purine nucleoside phosphorylases. Structure. 2001;9:941–953. doi: 10.1016/s0969-2126(01)00656-6. [DOI] [PubMed] [Google Scholar]
- 17.Park EY, Oh SI, Nam MJ, Shin JS, Kim KN, Song HK. Crystal structure of 5 '-methylthioadenosine nucleosidase from Arabidopsis thaliana at 1.5-angstrom resolution. Proteins. 2006;65:519–523. doi: 10.1002/prot.21120. [DOI] [PubMed] [Google Scholar]
- 18.Ronning DR, Iacopelli NM, Mishra V. Enzyme-ligand interactions that drive active site rearrangements in the Helicobacter pylori 5 '-methylthioadenosine/S-adenosylhomocysteine nucleosidase. Protein Science. 2010;19:2498–2510. doi: 10.1002/pro.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siu KK, Lee JE, Smith GD, Horvatin-Mrakovcic C, Howell PL. Structure of Staphylococcus aureus 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase. Acta crystallographica. Section F, Structural biology and crystallization communications. 2008;64:343–350. doi: 10.1107/S1744309108009275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allart B, Gatel M, Guillerm D, Guillerm G. The catalytic mechanism of adenosylhomocysteine/methylthioadenosine nucleosidase from Escherichia coli - Chemical evidence for a transition state with a substantial oxocarbenium character. Eur J Biochem. 1998;256:155–162. doi: 10.1046/j.1432-1327.1998.2560155.x. [DOI] [PubMed] [Google Scholar]
- 21.Cornell KA, Swarts WE, Barry RD, Riscoe MK. Characterization of recombinant Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase: Analysis of enzymatic activity and substrate specificity. Biochem Bioph Res Co. 1996;228:724–732. doi: 10.1006/bbrc.1996.1723. [DOI] [PubMed] [Google Scholar]
- 22.Lee JE, Cornell KA, Riscoe MK, Howell PL. Structure of Escherichia coli 5 '-methylthioadenosine/S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis. Journal of Biological Chemistry. 2003;278:8761–8770. doi: 10.1074/jbc.M210836200. [DOI] [PubMed] [Google Scholar]
- 23.Lee JE, Smith GD, Horvatin C, Huang DJT, Cornell KA, Riscoe MK, Howell PL. Structural snapshots of MTA/AdoHcy nucleosidase along the reaction coordinate provide insights into enzyme and nucleoside flexibility during catalysis. J Mol Biol. 2005;352:559–574. doi: 10.1016/j.jmb.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 24.Guan R, Ho MC, Brenowitz M, Tyler PC, Evans GB, Almo SC, Schramm VL. Entropy-Driven Binding of Picomolar Transition State Analogue Inhibitors to Human 5 '-Methylthioadenosine Phosphorylase. Biochemistry. 2011;50:10408–10417. doi: 10.1021/bi201321x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longshaw AI, Adanitsch F, Gutierrez JA, Evans GB, Tyler PC, Schramm VI. Design and Synthesis of Potent "Sulfur-Free" Transition State Analogue Inhibitors of 5 '-Methylthioadenosine Nucleosidase and 5 '-Methylthioadenosine Phosphorylase. Journal of medicinal chemistry. 2010;53:6730–6746. doi: 10.1021/jm100898v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JE, Singh V, Evans GB, Tyler PC, Furneaux RH, Cornell KA, Riscoe MK, Schramm VL, Howell PL. Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5 '-methylthioadenosine/S-adenosylhomocysteine nucleosidase. Journal of Biological Chemistry. 2005;280:18274–18282. doi: 10.1074/jbc.M414471200. [DOI] [PubMed] [Google Scholar]
- 27.Gutierrez JA, Luo M, Singh V, Li L, Brown RL, Norris GE, Evans GB, Furneaux RH, Tyler PC, Painter GF, Lenz DH, Schramm VL. Picomolar inhibitors as transition-state probes of 5 '-methylthioadenosine nucleosidases. Acs Chemical Biology. 2007;2:725–734. doi: 10.1021/cb700166z. [DOI] [PubMed] [Google Scholar]
- 28.Lee JE, Settembre EC, Cornell KA, Riscoe MK, Sufrin JR, Ealick SE, Howell PL. Structural comparison of MTA phosphorylase and MTA/AdoHcy nucleosidase explains substrate preferences and identifies regions exploitable for inhibitor design. Biochemistry. 2004;43:5159–5169. doi: 10.1021/bi035492h. [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 30.Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr D. 1999;55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- 31.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica. Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, Gopal K, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Pai RK, Read RJ, Romo TD, Sacchettin JC, Sauter NK, Storoni LC, Terwilligerf TC. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat. 2004;11:53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lummis SCR, McGonigle I, Ashby JA, Dougherty DA. Two Amino Acid Residues Contribute to a Cation-pi Binding Interaction in the Binding Site of an Insect GABA Receptor. J Neurosci. 2011;31:12371–12376. doi: 10.1523/JNEUROSCI.1610-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- 36.Dougherty DA. Cation-pi interactions in chemistry and biology: A new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–168. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 37.Siu KKW, Asmus K, Zhang AN, Horvatin C, Li S, Liu T, Moffatt B, Woods VL, Jr, Howell PL. Mechanism of substrate specificity in 5 '-methylthioadenosine/S-adenosylhomocysteine nucleosidases. Journal of Structural Biology. 2011;173:86–98. doi: 10.1016/j.jsb.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh V, Shi W, Evans GB, Tyler PC, Furneaux RH, Almo SC, Schramm VL. Picomolar transition state analogue inhibitors of human 5 '-methylthioadeno sine phosphorylase and X-ray structure with MT-immucillin-A. Biochemistry. 2004;43:9–18. doi: 10.1021/bi0358420. [DOI] [PubMed] [Google Scholar]
- 39.Lee JE, Luong W, Huang DJT, Cornell KA, Riscoe MK, Howell PL. Mutational analysis of a nucleosidase involved in quorum-sensing autoinducer-2 biosynthesis. Biochemistry. 2005;44:11049–11057. doi: 10.1021/bi050493q. [DOI] [PubMed] [Google Scholar]
- 40.Thomas K, Haapalainen AM, Burgos ES, Evans GB, Tyler PC, Gulab S, Guan R, Schramm VL. Femtomolar Inhibitors Bind to 5'-Methylthioadenosine Nucleosidases with Favorable Enthalpy and Entropy. Biochemistry. 2012 doi: 10.1021/bi3009938. [DOI] [PubMed] [Google Scholar]
- 41.Syme NR, Dennis C, Bronowska A, Paesen GC, Homans SW. Comparison of entropic contributions to binding in a "hydrophilic" versus "hydrophobic" ligand-protein interaction. J Am Chem Soc. 2010;132:8682–8689. doi: 10.1021/ja101362u. [DOI] [PMC free article] [PubMed] [Google Scholar]