Abstract
Neural progenitor cells have the ability to give rise to neurons and glia in the embryonic, postnatal and adult brain. During development, the program regulating whether these cells divide and self-renew or exit the cell cycle and differentiate is tightly controlled, and imbalances to the normal trajectory of this process can lead to severe functional consequences. However, our understanding of the molecular regulation of these fundamental events remains limited. Moreover, processes underpinning development of the postnatal neurogenic niches within the cortex remain poorly defined. Here, we demonstrate that Nuclear factor one X (NFIX) is expressed by neural progenitor cells within the embryonic hippocampus, and that progenitor cell differentiation is delayed within Nfix−/− mice. Moreover, we reveal that the morphology of the dentate gyrus in postnatal Nfix−/− mice is abnormal, with fewer subgranular zone neural progenitor cells being generated in the absence of this transcription factor. Mechanistically, we demonstrate that the progenitor cell maintenance factor Sry-related HMG box 9 (SOX9) is upregulated in the hippocampus of Nfix−/− mice and demonstrate that NFIX can repress Sox9 promoter-driven transcription. Collectively, our findings demonstrate that NFIX plays a central role in hippocampal morphogenesis, regulating the formation of neuronal and glial populations within this structure.
Keywords: glia, glial fibrillary acidic protein, neural progenitor cell, nuclear factor one X, SOX9
Introduction
During nervous system formation, one of the most important developmental events to occur is the differentiation of neural progenitor cells into neurons and glia. The embryonic forebrain provides a cogent example of this, with neural progenitor cells within the proliferative ventricular zone, region executing a program of proliferation, then differentiation, to generate the postmitotic cells of the cortex and hippocampus (Sauvageot and Stiles 2002). The abnormal proliferation or differentiation of cortical neural progenitor cells during development can lead to severe functional consequences, such as lissencephaly and microcephaly, both of which can cause mental retardation (Manzini and Walsh 2011). As such, understanding the regulatory processes controlling whether neural progenitor cells either divide and self-renew or exit the cell cycle and differentiate is critical to our understanding of both normal and pathological cortical development.
A number of recent studies have begun to elucidate some of the key molecules and signaling pathways that control how neural progenitor cell proliferation and differentiation is coordinated during development. Examples include the Notch (Shimojo et al. 2008; Imayoshi et al. 2010), fibroblast growth factor (Sahara and O'Leary 2009; Rash et al. 2011), and Sonic hedgehog (SHH; Komada et al. 2008) signaling pathways, all of which have been implicated in regulating progenitor cell identity during development of the cortex. Transcription factors of the Sry-related HMG box (SOX) family have also been shown to play a role in the maintenance of progenitor cell identity (Stolt and Wegner 2010). For instance, both SOX2 and SOX3 are expressed by neural progenitor cells within the developing and adult forebrain (Avilion et al. 2003; Wang et al. 2006), and SOX2 has been implicated in maintaining progenitor cell identity within the developing neocortex (Bani-Yaghoub et al. 2006) and the adult hippocampus (Suh et al. 2007). Another suite of molecules known to play a role in regulating the differentiation of neural progenitor cells are the transcription factors of the Nuclear factor one (NFI) family (Piper et al. 2007; Mason et al. 2009), which in vertebrates comprises 4 members; Nfia, Nfib, Nfic, and Nfix (Rupp et al. 1990; Kruse et al. 1991). Mice lacking either Nfia or Nfib display neurological phenotypes including dysgenesis of the corpus callosum (Shu, Butz, et al. 2003; Shu, Puche, et al. 2003; Piper, Moldrich, et al. 2009; Piper, Plachez, et al. 2009), hippocampal malformation (Barry et al. 2008; Piper et al. 2010), and delays in cerebellar development (Steele-Perkins et al. 2005; Wang et al. 2007). Mechanistically, Nfi genes have been implicated in regulating glial development via promoting the expression of astrocyte-specific genes (Gopalan et al. 2006; Brun et al. 2009), and Nfia and Nfib were recently shown to promote progenitor cell differentiation in a complementary fashion within the developing telencephalon through the repression of the Notch signaling pathway (Piper et al. 2010).
Nfix−/− mice also display severe neurological phenotypes (Driller et al. 2007; Campbell et al. 2008), and NFIX has previously been implicated in driving the expression of astrocytic genes during neural development (Gopalan et al. 2006; Piper et al. 2011). However, the mechanism by which NFIX regulates morphogenesis of the nervous system in vivo remains undefined. Here, using the developing hippocampus of Nfix−/− mice as a model, we reveal that NFIX regulates the differentiation of neural progenitor cells through the transcriptional regulation of progenitor-specific pathways. Our data demonstrate that Nfix−/− mice display delayed progenitor cell differentiation, which culminates in deficits in both neuronal and glial formation. Moreover, the formation of progenitor cells within the postnatal dentate gyrus is abnormal in Nfix−/− mice. Finally, we show that SOX9, a central mediator of progenitor cell self-renewal that acts downstream of SHH signaling during corticogenesis (Scott et al. 2010), is a target for transcriptional repression by NFIX. Taken together, these data reveal a central role for NFIX in orchestrating the timely differentiation of neural progenitor cells within the embryonic hippocampus and for regulating the development of neural progenitor cells within the subgranular zone of the postnatal dentate gyrus.
Materials and Methods
Mouse Strains
Nfix+/+ and Nfix−/− littermate mice were used for the majority of this study. These mice were maintained on a C57Bl/6J background. These animals were bred at the University of Queensland under approval from the institutional animal ethics committee. Timed-pregnant females were obtained by placing Nfix+/− male and Nfix+/− female mice together overnight. The following day was designated as embryonic day (E)0 if the female had a vaginal plug. Embryos were genotyped by polymerase chain reaction (PCR; Campbell et al. 2008). For the in utero electroporation experiments, wild-type CD-1 mice were used.
Hematoxylin Staining
Brains from wild-type or Nfix−/− embryos were dissected from the skull, blocked in 3% noble agar (Difco, Sparks, MS), and sectioned coronally at 50 μm on a vibratome (Leica, Nussloch, Germany). Sections were then mounted and stained with Mayer's hematoxylin using standard protocols.
Immunohistochemistry
Embryos and postnatal pups were drop fixed in 4% paraformaldehyde (PFA; E14 and below) or transcardially perfused with 0.9% saline, followed by 4% PFA (E15 to postnatal day [P] P20), and then postfixed in 4% PFA at 4 °C. Brains were removed and sectioned at 50 μm using a vibratome. Immunohistochemistry using the chromogen 3,3′-diaminobenzidine was performed as described previously (Plachez et al. 2008). Biotin-conjugated goat antirabbit IgG (BA-1000, Vector Laboratories, Burlingame, CA, United States of America) and donkey antimouse IgG (715-065-150, Jackson ImmunoResearch Laboratories, West Grove, PA, United States of America) secondary antibodies were used for chromogenic immunohistochemistry at 1/1000. For all immunohistochemical analyses, at least 3 wild-type and Nfix−/− brains were analyzed. Sections from comparable positions along the rostrocaudal axis were imaged using an upright microscope (Zeiss upright Axio-Imager Z1) fitted with an Axio-Cam HRc camera.
Immunohistochemistry on Paraffin Sections
Brains were perfused as above, embedded in paraffin, and sectioned coronally at 6 μm. Hematoxylin staining and immunohistochemistry were performed as described previously (Barry et al. 2008).
Antibody Parameters
Primary antibodies used for immunohistochemistry on floating sections were anti-NFIX (rabbit polyclonal, 1/10 000; Active Motif, Carlsbad, CA, United States of America); anti-TBR2 (rabbit polyclonal, 1/10 000, a gift from Dr Robert Hevner, University of Washington, Seattle, WA, United States of America); anti-GLAST (rabbit polyclonal, 1/50 000, a gift from Dr Niels Danbolt, University of Oslo, Oslo, Norway); anti-glial fibrillary acidic protein (GFAP) (rabbit polyclonal, 1/15 000, Dako, Glostrup, Denmark); anti-TBR1 (rabbit polyclonal, 1/100 000, a gift from Dr Robert Hevner); anti-prospero-related homeobox 1 (PROX1; rabbit polyclonal, 1/25 000, Millipore Bioscience Research Reagents, Billerica, MA, United States of America); anti-calbindin (rabbit polyclonal, 1/50 000, SWANT, Marly, Switzerland); anti-calretinin (rabbit polyclonal, 1/50 000, SWANT); anti-reelin (mouse monoclonal, 1/100 000, a gift from Dr Andre Goffinet, University of Louvain Medical School, Brussels, Belgium); anti-SOX2 (rabbit polyclonal, 1/1000; Cell Signaling Technology, Danvers, MA, United States of America); anti-cleaved caspase-3 (rabbit polyclonal, 1/5000, Cell Signaling Technology); anti-nestin (mouse monoclonal, 1/1500, Developmental Studies Hybridoma Bank); and anti-tenascin C (rabbit polyclonal, 1/5000, Millipore Bioscience Research Reagents). Primary antibodies used for immunohistochemistry on paraffin sections were anti-PAX6 (rabbit polyclonal, 1:1000; Millipore Bioscience Research Reagents), anti-SOX2 (1:1000), anti-SOX9 (rabbit polyclonal, 1:1000; a gift from Dr Peter Koopman, Institute for Molecular Bioscience, University of Queensland, Brisbane, Australia), and anti-phosphohistone H3 (rabbit polyclonal, 1:1000; Millipore Bioscience Research Reagents).
Quantification of Ventricular Zone Width/Hippocampal Cell Counts
To measure the ventricular zone width in E14–18 wild-type and Nfix−/− brains, sections were hematoxylin-stained and imaged with an upright microscope coupled to AxioVision software (Zeiss). The width of the ventricular zone was measured at 3 points along the hippocampus for each section. Data for both wild-type and knockout hippocampi at each age were then pooled for the comparison of ventricular zone width. For phosphohistone H3-, PAX6-, SOX2-, TBR2-, and SOX9-expressing cell counts, the total number of immunopositive cells per 100 μm in the ventricular zone or subventricular zone of each hippocampus was counted. For PROX1-expressing cell counts performed embryonically, the total number of PROX1-positive cells in the emerging dentate gyrus was counted. For postnatal animals, sections were labeled with fluorescent secondary antibodies and imaged with a confocal microscope (Zeiss LSM 510 META) using Zen software (Zeiss). The number of PROX1-positive cells per 100 μm in the upper and lower blades of the dentate gyrus of wild-type or Nfix−/− hippocampi was then counted. For all experiments involving quantification, data represent pooled results from at least 5 wild-type and 5 Nfix−/− brains. For all cell counts, we also measured the size of the nucleus to determine whether there was a difference between genotypes (Guillery 2002). As no size differences were noted, we did not apply the Abercrombie correction factor. Quantification was performed blind to the genotype of the sample, and statistical analyses were performed using a 2-tailed unpaired t-test. Error bars represent the standard error of the mean.
In Situ Hybridization
Embryos were collected and fixed as described above (n= 3 for both wild-type and knockout). In situ hybridization was performed using antisense probes as previously described (Piper, Moldrich, et al. 2009; Piper, Plachez, et al. 2009) with minor modifications. The hybridization temperature was 70 °C. The color reaction solution was BM Purple (Roche). In situ probes were kindly provided by Dr Shubha Tole (Tata Institute of Fundamental Research, Mumbai, India).
Hippocampal Microarrays
Hippocampal samples from E16 Nfix−/− mice (n= 3) and wild-type littermate controls (n= 3) were collected. Total RNA was extracted, and the microarray analysis performed at the Australian Research Council Special Research Centre for Functional and Applied Genomics (The University of Queensland, Australia) as described previously (Piper et al. 2010). Labeled and amplified material (1.5 μg/sample) was hybridized to Illumina's MouseWG-6 v2.0 Expression BeadChip at 55 °C for 18 h according to the Illumina BeadStation 500X™ protocol. Arrays were washed and then stained with 1 μg/mL cyanine3-streptavidin (Amersham Biosciences). The Illumina BeadArray™ reader was used to scan the arrays according to the manufacturer's instructions. Samples were initially evaluated using the BeadStudio™ software from Illumina. Quality control reports were satisfactory for all samples. The raw data were then imported into GeneSpring GX v7.3 (Agilent). Data were initially filtered using GeneSpring normalization algorithms. Quality control data filtering was then performed using the Bead detection score P-value, and with expression values below background, as determined by the cross-gene error model. Differential expression was determined by the 1-way analysis of variance (ANOVA)-Welch's approximate t-test without a multiple testing correction. A cut-off P-value of 0.05 was used for the mean difference between wild-type and Nfix−/− hippocampal tissue. In addition, a 1.5-fold–change filter was imposed on the genes from the ANOVA data set. The full array data set is listed in Supplementary Table 1. Pathway analysis was performed using the DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/) (Huang da et al. 2009). The full data sets from this analysis are listed in Supplementary Tables 2 and 3.
Reverse Transcription and Quantitative Real-Time PCR
For quantitative real-time PCR (qPCR), hippocampi were dissected and samples were then snap frozen. Total RNA was extracted using an RNeasy Micro Kit (Qiagen). Reverse transcription was performed using Superscript III (Invitrogen). Total RNA (0.5 µg) was reverse transcribed with random hexamers. qPCR reactions were carried out in a Rotor-Gene 3000 (Corbett Life Science) using the SYBR Green PCR Master Mix (Invitrogen). All the samples were diluted 1:100 with RNase/DNase-free water and 5 μL of these dilutions were used for each SYBR Green PCR reaction containing 10 μL SYBR Green PCR Master Mix, 10 μM of each primer, and deionized water. The reactions were incubated for 10 min at 95 °C followed by 40 cycles with 15s denaturation at 95 °C, 20 s annealing at 60 °C, and 30 s extension at 72 °C.
Generation of Gene-Specific Quantitative qPCR Standards
The synthesis of these primers was performed by Sigma-Genosys. The following primer sequences were used:
Sox9 forward (CTCACATCTCTCCTAATGCT).
Sox9 reverse (GACCCTGAGATTGCCCAGA).
Hprt forward (GCAGTACAGCCCCAAAATGG).
Hprt reverse (AACAAAGTCTGGCCTGTATCCAA).
qPCR Data Expression and Analysis
After completion of the PCR amplification, the data were analyzed with the Rotor-Gene software. When quantifying the mRNA expression levels, the housekeeping gene HPRT was used as a relative standard. By means of this strategy, we achieved a relative PCR kinetic of standard and sample. For all qPCR analyses, RNA from 3 independent replicates for both wild-type and Nfix−/− mice or control and treated cells were interrogated. All the samples were tested in triplicate, and each experiment was repeated a minimum of 3 times. Statistical analyses were performed using a 2-tailed unpaired t-test. Error bars represent the standard error of the mean.
Electrophoretic Mobility Shift Assay
Total cortex was removed from E18 brains to isolate nuclear extracts. Nuclear extracts were also isolated from COS cells expressing an HA-tagged NFIX expression construct (Nfix pCAGIG IRES GFP). Protease inhibitor tablets (Roche) were added to the extraction buffers as previously described (Smith et al. 1998). Electrophoretic mobility shift assays (EMSAs) were performed using radiolabeled annealed oligonucleotides containing a control NFI consensus site or the putative Sox9 consensus sites, which were designated −675, −183, +415, and +598. EMSA reactions were carried out as described previously using 1 μg of nuclear extract and 1 μg of poly-[dI-dC] as nonspecific competitor per reaction (Smith et al. 1998). Oligonucleotide sequences were: NFI control, 5′-ggTTTTGGATTGAAGCCAATATGATAA-3′ (upper strand), 5′-ggTTATCATATTGGCTTCAATCCAAAA-3′ (lower strand); −675, 5′-ccgggGCAGAAGCTCCAGTCACCACACCAGCTTCGTTGAAc-3′ (upper strand); 5′-ccgggTTCAACGAAGCTGGTGTGGTGACTGGAGCTTCTGCc-3′(lower strand); −183, 5′-ccgggCATCCACCCTCTGGCTGAGCTCCCCTCCCTTCTCCc-3′ (upper strand); 5′-ccgggGGAGAAGGGAGGGGAGCTCAGCCAGAGGGTGGATGc-3′(lower strand); +415, 5′-ccgggGACCGACGAGCAGGAGAAGGGCCTGTCTGGCGCCCc-3′ (upper strand); 5′-ccgggGGGCGCCAGACAGGCCCTTCTCCTGCTCGTCGGTCc-3′(lower strand). +598, 5′-ccgggGTGCATCCGCGAGGCGGTCAGCCAGGTGCTGAAGGc-3′ (upper strand); 5′-ccgggCCTTCAGCACCTGGCTGACCGCCTCGCGGATGCACc-3′ (lower strand). Additional bases used to generate 5′ overhangs for endfill are indicated in lower case.
Luciferase Reporter Assay
The constructs used in the luciferase assay were a full-length Nfix expression construct driven by the chick β-actin promoter (Nfix pCAGIG IRES GFP), and a construct containing the +598 site derived from the mouse Sox9 coding sequence (a gift from Peter Koopman; Kent et al. 1996). This construct was 250 base pairs in length and was generated using the following primers: Forward 5′-CTCGAGTCTCCTGGACCCCTTC-3′; reverse 5′-AAGCTTCAGCACCTGGCTGACC-3′. A construct containing a mutated NFI consensus sequence was generated in parallel, using an alternative reverse primer: 5′-AAGCTTCAGCACTGGTATGACCGC-3′. The resulting construct, termed Sox9ΔNFI, possessed an NFI-binding site that was changed from GAGGCGGTCAGCCAG to GAGGCGGTCATACCA. The amplicons were inserted into the XhoI and HindIII restriction enzyme sites of the pGL4.23 luc2minP vector (Promega, Madison, WI, United States of America). DNA was transfected into NSC-34 (Cashman et al. 1992) cells using FuGene (Invitrogen). Renilla luciferase (pRL SV40; Promega) was added to each transfection as a normalization control. After 24 h, luciferase activity was assessed using a dual-luciferase system (Promega) as per the manufacturer's instructions. Within each experiment, each treatment was replicated 6 times. Each experiment was also independently replicated a minimum of 3 times. Statistical analyses were performed using an ANOVA. Error bars represent the standard error of the mean.
Bioinformatic Promoter Screen
To obtain an NFI binding site motif, data from a recent study identifying NFI-binding sites in vivo using chromatin immunoprecipitation (ChIP) sequencing (Pjanic et al. 2011) were analyzed. NFI peaks were called using ChIP-Peak (Schmid and Bucher 2010) with the following parameters: Server-resident SGA file: mm9/nf1_wt.sga; strand: Any; centering: 75 bp; repeat masker: Checked; window width: 300 bp; vicinity range: 300 bp; peak threshold: 8; count cut-off: 1; refine peak positions: Checked. The NFI motif was created by running MEME (Bailey et al. 2009) on the sequences of 600 of the 708 peak regions declared by ChIP-Peak. The 600 regions were each trimmed to 100 base pairs in width, and chosen randomly from among the 708. MEME was run with parameters: -dna -minw 6 -maxw 30 -revcomp. Potential NFI binding sites were then identified in the promoter region of Sox9 using FIMO (Grant et al. 2011). The Sox9 promoter sequence, which we defined as the region 1000 base pairs either side of the transcription start site (TSS), was downloaded from the UCSC Genome Browser (mm9, July 2007; Fujita et al. 2011). FIMO was run on the Sox9 promoter sequence using a zero-order background generated on all mouse promoter regions, and a pseudocount of 0.1. All potential binding sites with P-value ≤10−3 were reported.
In Utero Electroporation
E13 CD-1 pregnant mice were anesthetized with 1 mg/mL of zylazine and 15 mg/mL ketamine in sterile phosphate-buffered saline. After the induction of anesthesia, the mice were subjected to abdominal incision to expose the uterine horns. The embryos were visualized through the uterus wall, and ∼0.3 μL plasmid mixture containing 1.5 μg/μL plasmid DNA (pCAGIG IRES GFP or Nfix pCAGIG IRES GFP) plus 0.025% fast green, diluted in phosphate-buffered saline, was injected into the lateral ventricle using a fine glass capillary. Using forceps-shaped electrodes, five 30 V electric pulses were applied, each separated by a 1-s interval. The electrodes were placed such that the DNA was targeted for electroporation into the ventricular zone of the neocortex. The uterine horns were repositioned into the abdominal cavity, and the abdominal wall and skin were sutured. The electroporated pups were perfused 3 days later at E16, and the brains were sectioned coronally, then stained with the nuclear marker 4′,6-diamidino-2-phenylindole (DAPI; 1:1000), and visualized under a fluorescence microscope to ensure successful electroporation. The expression of GFAP was then ascertained using the immunohistochemical protocols described above. For the vector only controls (n= 20), no GFAP staining was observed within the neocortex. For those pups successfully electroporated with the Nfix expression construct (n= 16), all exhibited precocious expression of GFAP within the region overexpressing NFIX.
Results
Embryonic Neural Progenitor Cells Within the Hippocampus Express NFIX
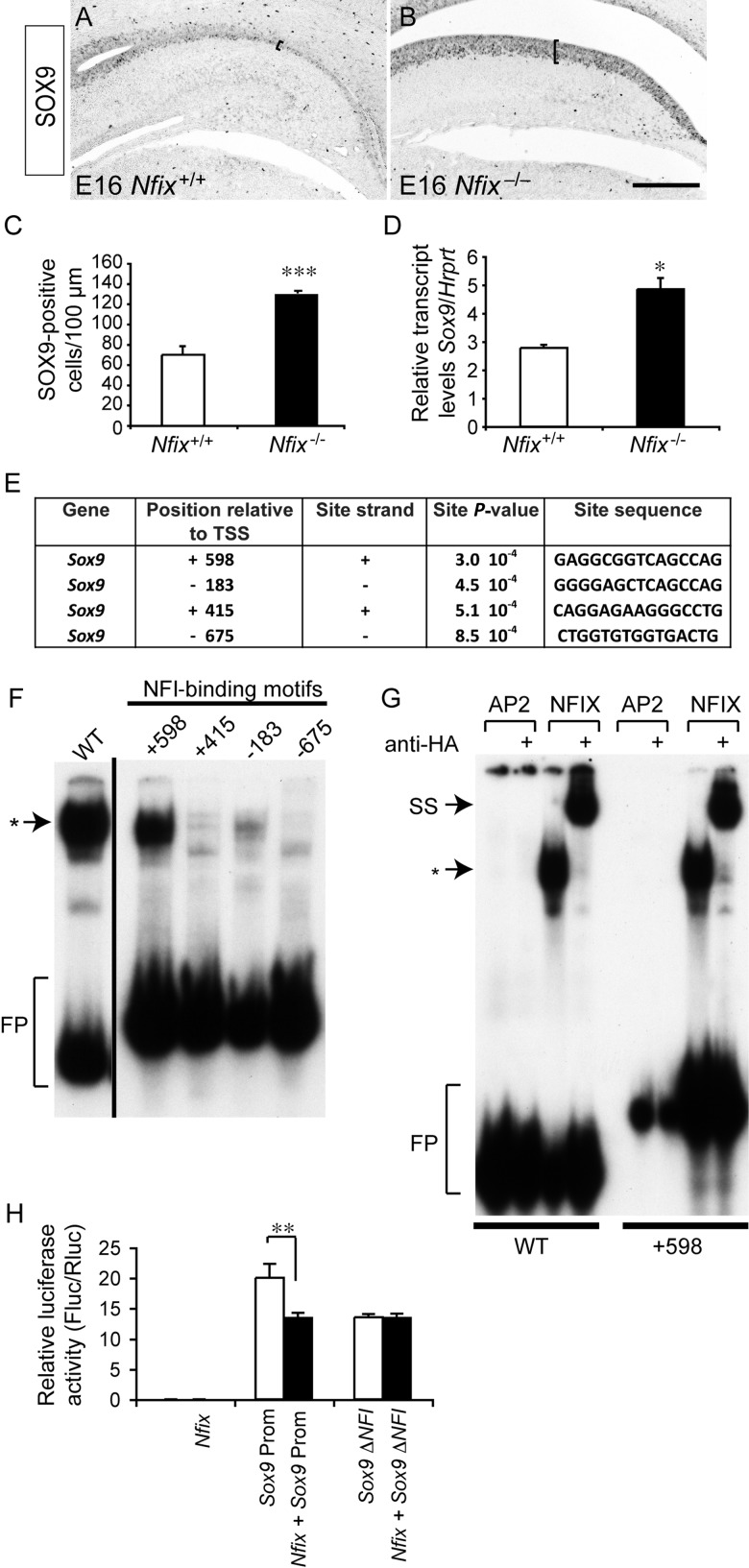
During telencephalic development, NFIX is expressed widely within both the cortex and the hippocampus (Campbell et al. 2008). At E13, NFIX was expressed by neural progenitor cells within the ventricular zone of the hippocampus, but not by cells within the cortical hem region (Fig. 1A,B). This expression pattern was maintained at E14, with progenitors within both the ammonic neuroepithelium and dentate neuroepithelium expressing this transcription factor (Fig. 1C,D). By E17, expression of NFIX within the hippocampus was widespread, encompassing cells within the cornu ammonis (CA) regions and the dentate gyrus, as well neural progenitor cells within the ventricular zone (Fig. 1E,F). NFIX has previously been linked to the regulation of astrocyte-specific genes including Gfap, brain fatty acid-binding protein, and α1-antichymotrypsin (Gopalan et al. 2006; Brun et al. 2009; Piper et al. 2011); however, our findings indicate that NFIX may also regulate transcriptional activity within neural progenitor cells in the developing hippocampus.
Figure 1.
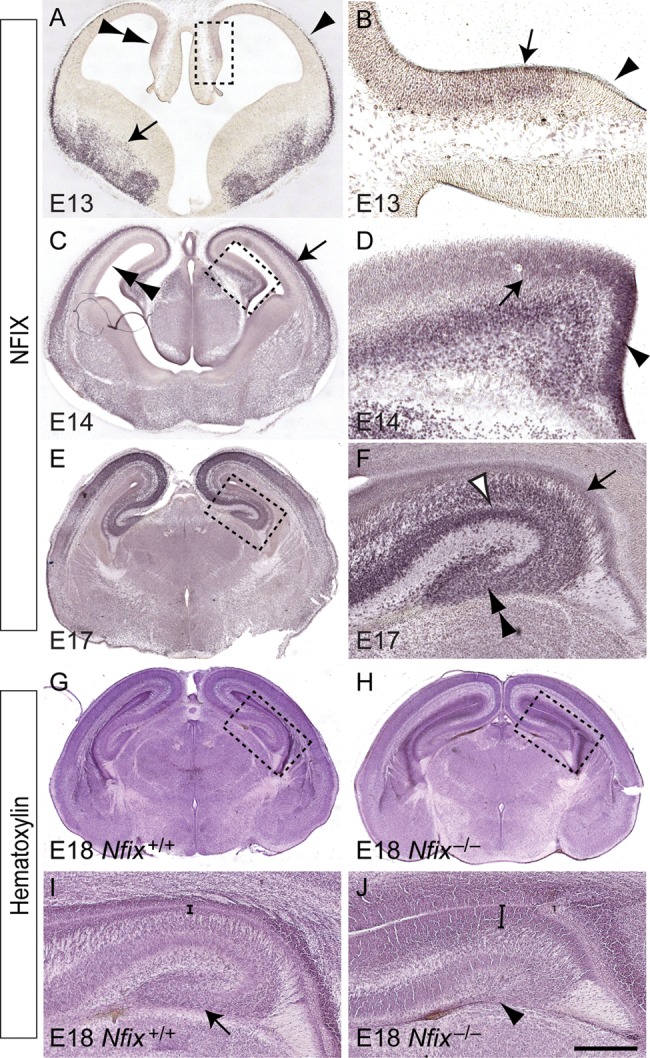
Expression of NFIX in the developing hippocampus. (A–F) Expression of NFIX in coronal sections of the embryonic hippocampus. (A) At E13, NFIX expression was observed within the hippocampus (double arrowhead), the marginal zone of the cortex (arrowhead), and the ventral telencephalon (arrow). (B) Higher magnification view of the boxed region in A. NFIX was expressed within the ammonic neuroepithelium (arrow), but was not expressed within the cortical hem (arrowhead). (C) At E14, NFIX expression was observed within the cortical plate (arrow) and ventricular zone (double arrowhead) of the neocortex. (D) Higher magnification view of the boxed region in C, showing expression of NFIX by progenitor cells within both the ammonic neuroepithelium (arrow) and the dentate neuroepithelium (arrowhead). (E) At E17, NFIX was expressed broadly within the dorsal telencephalon. (F) Higher magnification view of the boxed region in E. Within the E17 hippocampus, NFIX was expressed by ventricular zone progenitor cells (arrow) and by cells within the CA region (open arrowhead) and the dentate gyrus (double arrowhead). (G–J) Hematoxylin-stained coronal sections of E18 wild-type (G) and Nfix−/− (H) brains. (I) Higher magnification view of the boxed region in G, showing the distinctive shape of the dentate gyrus in the wild-type hippocampus (arrow in I). (J) Higher magnification view of the boxed region in H, demonstrating that the hippocampal ventricular zone was markedly wider within Nfix−/− brains (compare brackets in I and J) and that the dentate gyrus was severely reduced in the absence of this transcription factor (arrowhead in J). Scale bar (in J): A, 600 μm; B, 75 μm; C, 750 μm; D, 100 μm; (E, G, H), 1 mm; (F, I, J), 300 μm.
Hippocampal Neural Progenitor Cell Differentiation is Delayed in Nfix−/− Mice
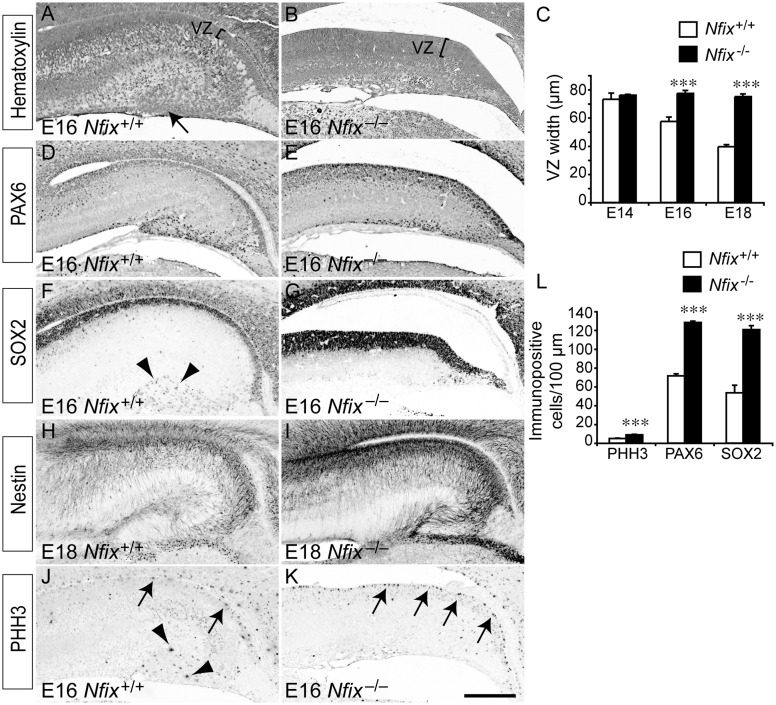
We have previously shown that mice lacking Nfix display gross morphological abnormalities with regard to postnatal neocortical and hippocampal formation (Campbell et al. 2008), although the underlying mechanism by which Nfix regulates neural progenitor cell biology remains undefined. To address this issue, we first analyzed embryonic hippocampal development in Nfix−/− mice. Hematoxylin staining of E18 wild-type and Nfix−/− mice revealed that the dentate gyrus in the mutant was markedly reduced in size, and, moreover, that the size of the hippocampal ventricular zone in the mutant was significantly enlarged in comparison to that of wild-type controls (Figs 1G–J and 2C). These phenotypes were also evident 2 days earlier at E16, the time at which the dentate gyrus is becoming morphologically recognizable in the hippocampus of wild-type mice (Fig. 2A). In Nfix−/− mice at E16, the dentate gyrus had yet to develop at a morphological level (Fig. 2B), and the hippocampal ventricular zone was significantly wider than that in littermate controls (Fig. 2C).
Figure 2.
Delayed differentiation of ventricular zone progenitor cells in the Nfix−/− hippocampus. (A and B) Coronal paraffin sections of E16 wild-type and Nfix−/− brains stained with hematoxylin. The emerging dentate gyrus can be seen clearly in the wild-type (arrow in A), but is absent in the mutant at this age. The brackets delineate the ventricular zone (VZ). (C) The ventricular zone was significantly wider in the mutants than the controls at both E16 and E18. (D–K) Immunostaining of the progenitor cell markers PAX6 (D, E), SOX2 (F, G), and nestin (H, I), and the mitotic marker phosphohistone H3 (PHH3, J, K). There were more PAX6- and SOX2-expressing cells within the ventricular zone of E16 Nfix−/− mice (D–G). There were also markedly higher levels of nestin expression within the hippocampus of Nfix−/− mice at E18 (H, I). There were more mitotically active cells (arrows in J and K) in the ventricular zone of the Nfix−/− hippocampus. However, whereas SOX2-positive and PHH3-positive cells were seen within the dentate gyrus of E16 wild-type mice (arrowheads in F and J), we did not observe such cells within the presumptive dentate gyrus region of Nfix−/− mice. (L) Quantification of the number of immunopositive cells revealed that there were significantly more cells expressing PHH3, PAX6, or SOX2 in the hippocampal ventricular zone of the Nfix−/− mutant than in the wild-type control at E16. ***P< 0.001, Student's t-test. Scale bar (in H): A–G, J, K, 250 μm; H, I, 300 μm.
These findings suggested that the balance between neural progenitor cell self-renewal and differentiation was abnormal in the absence of Nfix, culminating in the maintenance of progenitor cell proliferation for longer than in wild-type mice. To investigate this, we analyzed the expression of the neural progenitor cell-specific marker PAX6 (Gotz et al. 1998). At E14, there was no difference between hippocampal PAX6 expression in wild-type and Nfix−/− mice (data not shown). However, by E16, there were significantly more PAX6-expressing neural progenitor cells within the ventricular zone of Nfix−/− mice (Fig. 2D,E,L). Furthermore, the expression of a second marker for neural progenitor cells, SOX2 (Avilion et al. 2003; Suh et al. 2007), also revealed a significant expansion of the neural progenitor cell pool within Nfix−/− mice (Fig. 2F,G,L).
Neural progenitor cells within the ventricular zone also express the intermediate filament protein nestin (Lendahl et al. 1990; Dahlstrand et al. 1995). Expression of nestin within the hippocampus of late gestation Nfix−/− mice was markedly higher than that within wild-type controls, further emphasizing a delay in the differentiation of neural progenitor cells in the Nfix mutants (Fig. 2H,I). Finally, the expression pattern of phosphohistone H3, a specific marker for cells undergoing mitosis, revealed that there were significantly more mitotically active cells in the hippocampal ventricular zone of Nfix−/− mice at E16 than in their littermate controls (Fig. 2J–L). Importantly, the delay in the differentiation of ventricular zone progenitors was also observed within the neocortical ventricular zone, suggesting that NFIX regulates the differentiation of neural progenitors throughout the pallium (Supplementary Figs 1 and 2). Collectively, these data indicate a shift in the balance of progenitor cell activity toward self-renewal as opposed to differentiation in the absence of Nfix.
Nfix−/− Mice Display Delays in Basal Progenitor Cell Differentiation
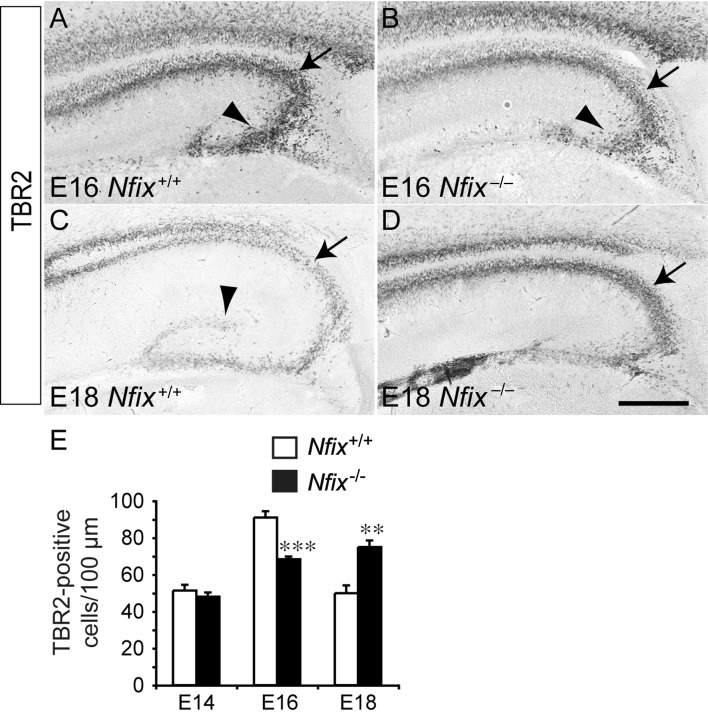
During development, neural progenitor cells within the ventricular zone give rise to a secondary, transient population of progenitors with limited proliferative capacity, known as intermediate progenitor cells (Gotz and Huttner 2005). These progenitors, which are located within the subventricular zone, express specific markers such as TBR2 (Englund et al. 2005). Given the delay in ventricular zone progenitor differentiation, we postulated that delays in intermediate progenitor cell development may also be evident within the subventricular zone of Nfix−/− mice. At E14, there were no significant differences in the number of intermediate progenitor cells between Nfix−/− mice and wild-type controls (Fig. 3E and data not shown). By E16, however, there were more TBR2-positive cells within the hippocampal subventricular zone of wild-type mice than within Nfix mutants (Fig. 3A,B,E), suggestive of a delay in intermediate progenitor cell formation in Nfix−/− mice. At E18 in wild-type mice, there were fewer TBR2-positive intermediate progenitors than at E16, consistent with these cells differentiating to form postmitotic neuronal cells. Interestingly, the decline in intermediate progenitor cell numbers observed between E16 and E18 in wild-type mice was not observed in Nfix−/− mice between these ages. Instead, at E18, there were significantly more TBR2-positive cells within the hippocampal subventricular zone of the mutant mice in comparison to wild-type controls (Fig. 3C–E). These data suggest that intermediate progenitor cell differentiation is delayed in Nfix−/− mice, further demonstrating that this transcription factor plays a central role in regulating the balance of neural progenitor cell proliferation and differentiation during development of the hippocampus.
Figure 3.
Altered trajectory of basal progenitor cell development in the Nfix−/− hippocampus. Expression of TBR2, a basal progenitor cell-specific marker, at E16 (A, B) and E18 (C, D). At E16, strong expression of TBR2 in the wild-type was observed in the subventricular zone of the hippocampus (arrow in A) and within the emerging dentate migratory stream (arrowhead in A). In the mutant, there were fewer TBR2-expressing cells evident within the subventricular zone (arrow in B) and the dentate migratory stream (arrowhead in B) of the hippocampus. In contrast, at E18, expression of TBR2 was more pronounced in the subventricular zone of the mutant hippocampus (arrow in D) than within the wild-type (arrow in C). In the wild-type, but not the Nfix mutant, TBR2-positive cells also demarcated the dentate gyrus at E18 (arrowhead in C). (E) Quantification of the number of TBR2-positive cells within the subventricular zone revealed that there were significantly more basal progenitor cells within the subventricular zone of the wild-type at E16, whereas this situation was reversed at E18, when there were more basal progenitor cells within the Nfix−/− subventricular zone. **P< 0.01, ***P< 0.001, Student's t-test. Scale bar (in D): A, B, 250 μm; C, D, 300 μm.
Glial and Neuronal Development is Delayed in Nfix−/− Mice
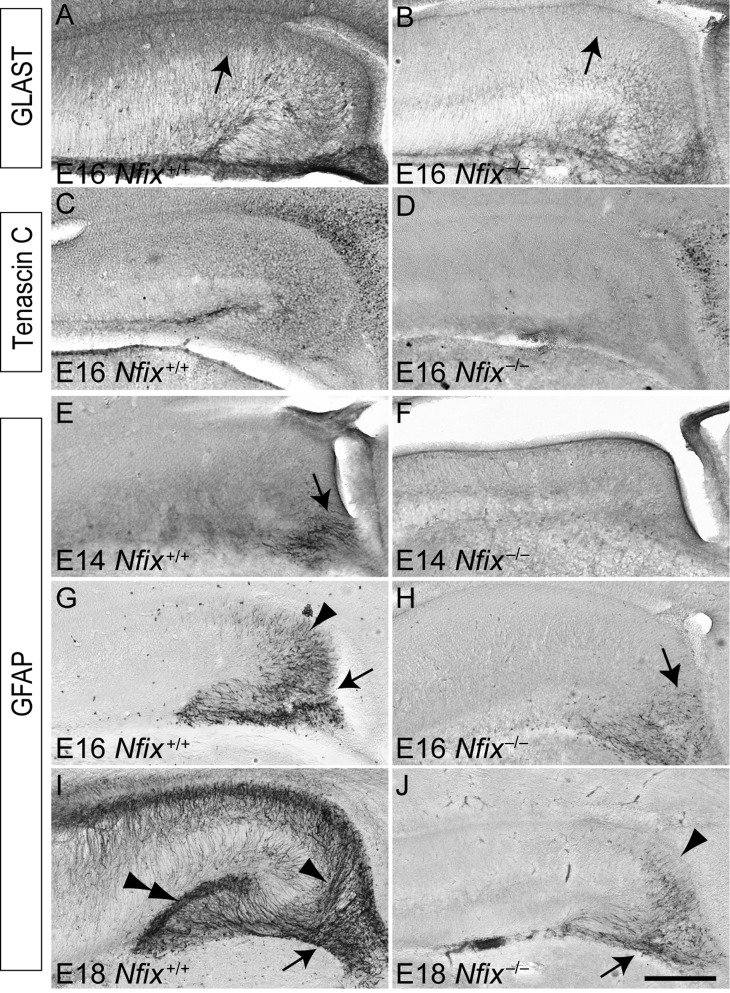
Given the delay in ventricular zone neural progenitor cell differentiation within the hippocampus of Nfix−/− mice, we next sought to analyze the development of postmitotic populations within the hippocampus of these mice. Hippocampal astrocytes are derived from progenitor cells within the ammonic neuroepithelium and the fimbrioglial epithelium of the hippocampal anlage and give rise to the supragranular glial bundle and the fimbrial glial bundle, respectively (Rickmann et al. 1987; Sievers et al. 1992). Analysis of the expression of the astroglial markers astrocyte-specific glutamate transporter GLAST (Shibata et al. 1997) and tenascin C (Gotz et al. 1998) revealed a marked reduction in expression of these proteins within the hippocampal ventricular zone of Nfix−/− mice (Fig. 4A–D), indicating a delay in glial differentiation in the absence of Nfix. Delays in astrocytic formation were even more apparent when expression of GFAP was analyzed. In wild-type mice, expression of GFAP within the fimbrial glia was observed at E14 (Fig. 4E), and by E16 GFAP expression was also observed within glia derived from the ammonic neuroepithelium (Fig. 4G). By E18, GFAP expression within the hippocampus of wild-type mice was extensive, with the supragranular glial bundle and the fimbrial glial bundles clearly delineated, and with GFAP-positive fibers localizing to the hippocampal fissure (Fig. 4I). In contrast, no GFAP expression was present within the hippocampus of E14 Nfix−/− mice (Fig. 4F), and expression was only observed within the fimbrial glia at E16 (Fig. 4H). By E18 in the mutant, GFAP expression was localized to both the supragranular glia and the fimbrial glia, but to a far lesser extent than observed within age-matched controls (Fig. 4J). Instead, GFAP expression in the mutant at E18 was comparable with that in E16 wild-type hippocampi (compare Fig. 4G and J). Similarly, the development of mature glia within the neocortex was delayed in the absence of Nfix (Supplementary Fig. 3). As mature glia are critical for morphogenesis of the dentate gyrus (Barry et al. 2008), these data indicate that the delayed glial differentiation from ventricular zone neural progenitor cells may, in part, underlie the phenotypic abnormalities present within the hippocampus of Nfix−/− mice.
Figure 4.
Delayed development of the fimbrial and supragranular glial bundles in the Nfix−/− hippocampus. Coronal sections of the hippocampus between E14 and E18 in wild-type (A, C, E, G, I) and Nfix−/− (B, D, F, H, J) mice, showing expression of the astroglial markers GLAST (A, B) and tenascin C (C, D), and the mature astrocytic marker, GFAP (E–J). At E16, GLAST was expressed strongly by astroglial cells within the ventricular zone of the wild-type hippocampus (arrow in A), but was expressed at a much lower level in the Nfix−/− hippocampus (arrow in B). Tenascin C was also expressed at a lower level in the hippocampus of Nfix−/− mice (C, D). At E14, expression of GFAP was evident within the fimbrial glioepithelium of the wild-type (arrow in E), but was absent in the mutant (F). At E16, GFAP expression in the wild-type hippocampus was evident within the fimbrial glioepithelium (arrow in G) and the ammonic neuroepithelium (arrowhead in G). In the Nfix−/− hippocampus, GFAP expression was seen within the fimbrial glioepithelium at this age (arrow in H). At E18, GFAP was strongly expressed within the wild-type hippocampus, including within the dentate gyrus (double arrowhead in I), the supragranular glial bundle (arrowhead in I), and the fimbrial glial bundle (arrow in I). In the mutant at E18, however, the expression of GFAP within the ammonic neuroepithelium (arrowhead in J) and the fimbrial glioepithelium (arrow in J) of the hippocampus was markedly reduced. Scale bar (in J): A–D, G, H, 250 μm; E, F, 100 μm; I, J, 300 μm.
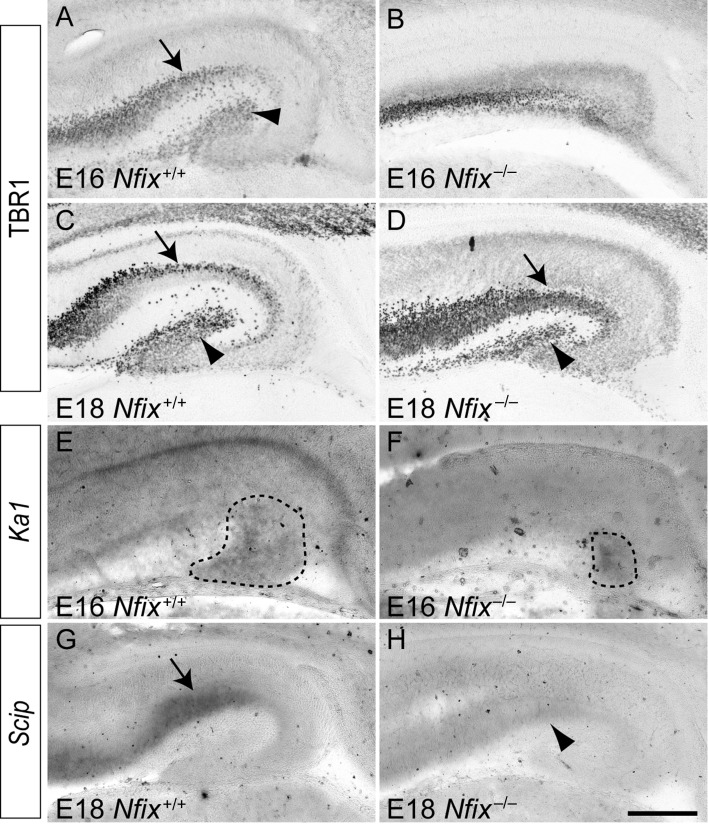
To determine whether the alteration in the balance between neural progenitor cell self-renewal and differentiation within Nfix−/− mice also had consequences for neuronal development, we next investigated neurogenesis within these mice. The transcription factor TBR1 is expressed by both pyramidal neurons within the CA regions of the hippocampus and by dentate granule neurons within the emerging dentate gyrus (Englund et al. 2005). In wild-type sections at E16 and E18, TBR1-expressing neurons were present within these 2 loci of the hippocampus (Fig. 5A,C). In contrast, hippocampal sections from E16 Nfix−/− mice revealed delays in the formation of TBR1-positive cells (Fig. 5B). Indeed, by E18, TBR1 expression within the hippocampus of Nfix−/− mice resembled that within E16 wild-type controls (compare Fig. 5A and D). Delays in neuronal development were also observed within the neocortex of Nfix−/− mice (Supplementary Fig. 4). Furthermore, expression of the hippocampal subfield markers Ka1 (a CA3 marker) at E16 and Scip (a CA1 marker) (Bettler et al. 1990; Frantz et al. 1994) at E18 by in situ hybridization revealed significant developmental delays within Nfix−/− mice (Fig. 5E–H). Taken together, these findings demonstrate that developmental delays are present within both neuronal and glial lineages within the pallium in the absence of Nfix.
Figure 5.
Neuronal development is delayed in the hippocampus of Nfix−/− mice. Expression of the neuronal marker TBR1 (A–D) and the hippocampal subfield markers Ka1 and Scip (E–H) in coronal sections of wild-type (A, C, E, G) and Nfix−/− (B, D, F, H) mice. At E16 (A) and E18 (C) of the wild-type, expression of TBR1 was evident within the pyramidal cell layer of the hippocampus (arrows in A and C) and within the emerging dentate gyrus (arrowheads in A and C). In the mutant at E16, however, the expression of TBR1 within the hippocampus appeared delayed (B). By E18, the expression of TBR1 within the hippocampus of Nfix−/− mice was evident within the pyramidal cell layer of the hippocampus (arrow in D) and within the emerging dentate gyrus (arrowhead in D), in a pattern similar to that observed in the E16 wild-type hippocampus (compare A and D). The expression of the CA3 subfield marker Ka1 revealed a significantly reduced area of expression in the mutant compared with the wild-type control (Ka1 expression is delineated by the dashed lines in E and F). (G) In situ hybridization revealed the expression of the CA1 subfield marker Scip in the wild-type hippocampus (arrow in G). (H) The expression of Scip mRNA was markedly reduced in the mutant at E18 (arrowhead in H). Scale bar (in H): A, B, E, F, 250 μm; C, D, G, H, 300 μm.
Dentate Granule Cell Development is Abnormal in the Absence of Nfix
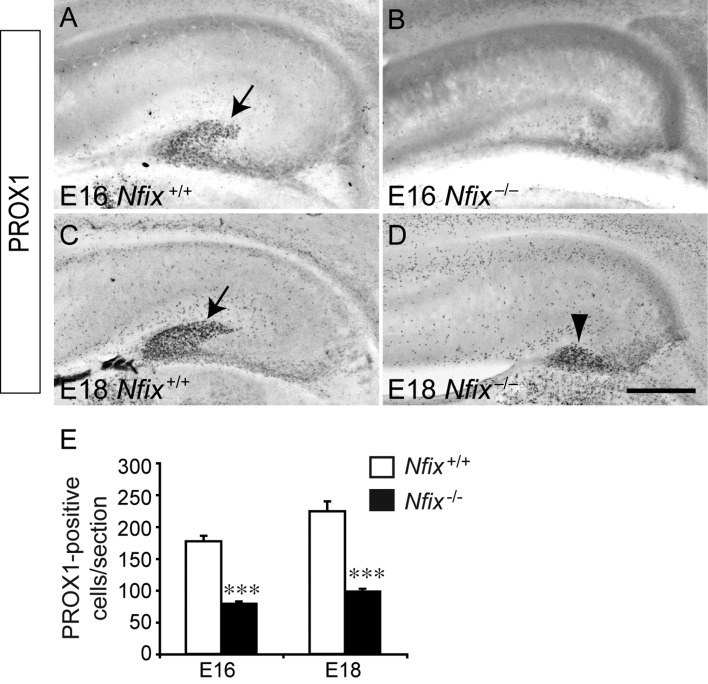
In addition to revealing delays in the formation of pyramidal neurons within the hippocampus of Nfix−/− mice, analysis of TBR1 expression also revealed delays in the development of dentate granule neurons (Fig. 5D). Together with mature glia, dentate granule neurons, are critical for the formation of the dentate gyrus (Zhou et al. 2004). These cells, which can be identified by the expression of the transcription factor PROX1 (Pleasure, Anderson, et al. 2000; Pleasure, Collins, et al. 2000), are derived from neural progenitor cells within the dentate neuroepithelium at approximately E14, and migrate into the granular layer of the emerging dentate gyrus in association with the hippocampal radial glial scaffold (Zhou et al. 2004). The high levels of NFIX expression within the dentate neuroepithelium at E14 (Fig. 1D) hinted at a role for NFIX in dentate granule neuron formation. To assess this we analyzed the expression of PROX1 in wild-type and Nfix−/− mice. In wild-type hippocampi at E16, PROX1-expressing dentate granule neurons were present within the presumptive dentate gyrus, and by E18, the localization of these cells within the dentate gyrus was beginning to resolve into the V-shape formed by the upper and lower blades of the granular zone (Fig. 6A,C). In the mutant, however, there were significantly fewer PROX1-expressing cells within the hippocampus at both E16 and E18 (Fig. 6B,D,E), and the dentate granule neurons had not migrated into the dentate gyrus, perhaps due to the delays in the formation of the supragranular and fimbrial glial bundles (Fig. 4J). Collectively, these data demonstrate that the production of both pyramidal neurons and dentate granule neurons is delayed within the hippocampus of Nfix−/− mice.
Figure 6.
Dentate granule cell development is delayed in the Nfix−/− hippocampus. Expression of the dentate granule cell marker PROX1 in the hippocampus of wild-type (A, C) and Nfix−/− (B, D) mice. At E16, the expression of PROX1 was observed within the developing dentate gyrus of the wild-type (arrow in A), but was significantly reduced within the Nfix mutant at this age (B). By E18, PROX1-expressing cells within the hippocampus of the wild-type clearly demarcated the dentate gyrus (arrow in C). In the Nfix mutant, PROX1-expressing cells were evident by this age (arrowhead in D), although these cells were aberrantly positioned, and did not form the normal chevron shape of the dentate gyrus. (E) Quantification of PROX1-expressing cells revealed significantly fewer dentate granule cells within the hippocampus of Nfix−/− mice at both E16 and E18. ***P< 0.001, Student's t-test. Scale bar (in D): A, B, 250 μm; C, D, 300 μm.
Development of Interneurons and Cajal–Retzius Cells Occurs Normally in Nfix−/− Mice
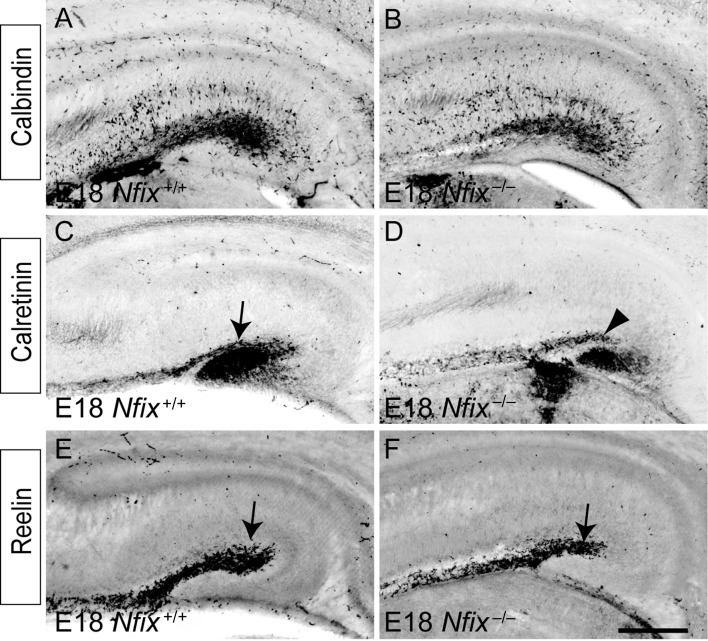
During cortical development, interneurons, which are derived from neural progenitor cells within the ventral pallium, migrate tangentially into all regions of the cortex, including the hippocampus (Pleasure, Anderson, et al. 2000; Pleasure, Collins, et al. 2000). NFIX was not expressed within neural progenitor cells within the ventral pallium at E13 (Fig. 1A), and in line with this, the production of calbindin-positive and calretinin-positive interneurons, and their subsequent migration into the hippocampus, occurred normally within Nfix−/− mice (Fig. 7A–D). Furthermore, development of the choroid plexus, which serves as a source for bone morphogenetic proteins during development (Hebert et al. 2002), was normal within Nfix−/− mice (data not shown). Calretinin expression is also evident within Cajal–Retzius neurons. These specialized cells are derived from a variety of areas within the developing pallium, including the cortical hem (Bielle et al. 2005; Garcia-Moreno et al. 2007). Interestingly, expression of reelin, another marker for Cajal–Retzius cells, was normal within Nfix−/− mice, with the caveat that reelin-expressing cells did not migrate as far dorsally along the incipient hippocampal fissure in the mutant. Importantly, reelin expression during development has been shown to contribute to hippocampal morphogenesis by regulating the formation of the radial glial scaffold (Frotscher et al. 2003). Thus, the lack of NFIX expression within the cortical hem (Fig. 1B), together with the specification and development of Cajal–Retzius cells, suggests that the phenotypic abnormalities within the hippocampus of Nfix−/− mice do not arise as a consequence of impairments to the reelin signaling pathway.
Figure 7.
The migration of hippocampal interneurons and Cajal–Retzius neurons occurs normally the absence of Nfix. Expression of calbindin (A, B), calretinin (C, D), and reelin (E, F) in coronal sections of wild-type (A, C, E) and Nfix−/− (B, D, F) mice at E18. Calbindin-expressing interneurons migrated normally into the hippocampus of Nfix−/− mice (B). Similarly, calretinin-expressing interneurons migrated normally into the hippocampus of the mutant. Although the hippocampal fissure was evident in the wild-type at this age (arrow in C), it was markedly reduced in the mutant (arrowhead in D). Reelin-expressing Cajal–Retzius cells migrated normally into the hippocampus of Nfix−/− mice, although, due to the reduced size of the hippocampal fissure, these cells did not migrate as far dorsally in the mutant as they did in the wild-type (compare arrows in E and F). Scale bar (in F): 300 μm.
Postnatal Nfix−/− Mice Exhibit Abnormal Hippocampal Morphology
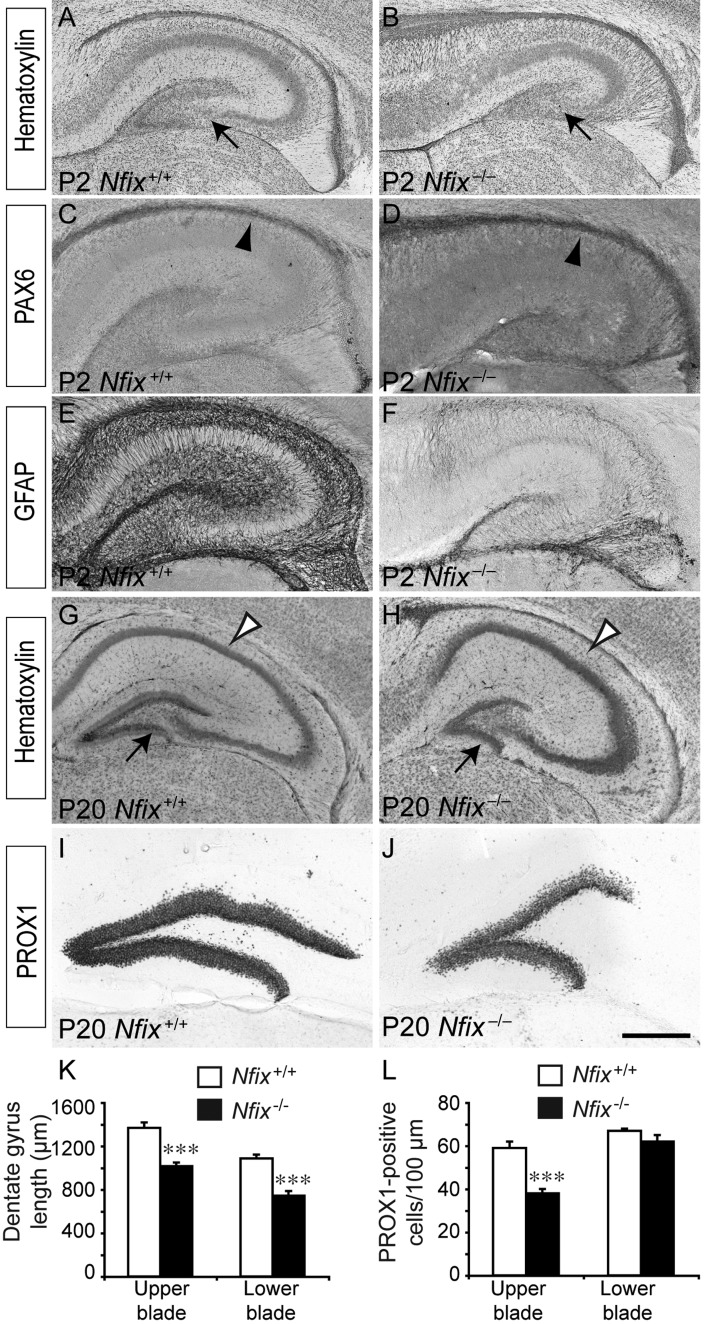
To this point, our data had implicated Nfix in regulating the differentiation of embryonic neural progenitor cells within the emerging hippocampal formation. The hippocampal dentate gyrus is, however, one of the few regions of the postnatal and adult brain in which neural progenitor cells persist, giving rise to new neurons throughout life (Ihrie and Alvarez-Buylla 2008). However, the precise origin of these subgranular zone progenitor cells, and importantly, the molecular mechanisms regulating their development, remain poorly defined. Unlike mice lacking Nfia or Nfib, which die perinatally (Shu, Butz, et al. 2003; Shu, Puche, et al. 2003; Steele-Perkins et al. 2005), Nfix−/− mice survive until approximately P20 on a C57Bl/6J background, enabling the contribution of Nfix to the postnatal development of the hippocampus to be investigated. Analysis of the hippocampus in P2 Nfix−/− mice revealed that the dentate gyrus had indeed developed by this stage, and, furthermore, that very few PAX6-expressing neural progenitor cells were present within the ventricular zone of either wild-type or Nfix−/− mice (Fig. 8A–D). The expression of GFAP, however, was still markedly reduced in the mutant at this age (Fig. 8F). By P20, the morphological consequences of the absence of Nfix during development were manifest, with the CA1 region being dorsally enlarged, and both the upper and lower blades of the dentate gyrus being significantly shortened mediolaterally (Fig. 8G,H,K), akin to the earlier descriptions of this mutant (Campbell et al. 2008). Given the decrease in the size of the dentate gyrus, we quantified the number of PROX1-expressing dentate granule neurons within the blades of the dentate gyrus from P20 wild-type and Nfix−/− mice. Interestingly, there were significantly fewer dentate granule neurons per unit length within the upper blade of the dentate gyrus (Fig. 8L). When considered in light of the decreased length of the dentate gyrus blades within Nfix−/− mice, these data suggest that although morphogenesis of the dentate gyrus does eventually occur in these mice, fewer dentate granule neurons ultimately populate this structure postnatally.
Figure 8.
Abnormal hippocampal morphology in postnatal Nfix−/− mice. Coronal hippocampal sections of P2 (A–F) and P20 (G–J) wild-type and Nfix−/− mice. Hematoxylin staining revealed that, by P2, the dentate gyrus was evident within the hippocampus of both wild-type and Nfix−/− mice (arrows in A and B, respectively). The expression of PAX6 revealed that, in both wild-type (C) and Nfix−/− (D) mice, there were few remaining PAX6-expressing cells within the ventricular zone of the hippocampus (arrowheads in C and D). GFAP expression, however, was still markedly reduced in the hippocampus of Nfix−/− mice (F). (G) Hematoxylin staining of the hippocampus from a P20 wild-type brain. The dentate gyrus (arrow in G) and the CA regions (open arrowhead in G) were clearly evident. (H) In Nfix−/− mice, although the dentate gyrus (arrow in H) and CA (open arrowhead in H) regions were evident, their morphology was abnormal, being shortened along the mediolateral axis, and lengthened along the dorsoventral axis, respectively. (I, J) Expression of PROX1 within the dentate gyrus of wild-type (I) and Nfix−/− (J) mice. (K) The measurement of the lengths of the blades of the dentate gyrus revealed that both the upper and lower blades of the Nfix mutant were significantly shorter than those of their wild-type littermate controls. (L) Furthermore, there were also significantly fewer PROX1-positive dentate granule neurons per unit length within the upper blade of the dentate gyrus of Nfix−/− mice. ***P< 0.001, Student's t-test. Scale bar (in J): A–F, 350 μm; G–J, 650 μm.
Fewer Neural Progenitor Cells are Found Within the Subgranular Zone of Postnatal Nfix−/− Mice
Although it is now well established that neural progenitor cells within the subgranular zone of the dentate gyrus generate new dentate granule neurons within the postnatal and adult mammalian brain (Seri et al. 2001), how these cells arise during development remains unclear. It has been suggested that hippocampal radial glia give rise to subgranular zone progenitors (Eckenhoff and Rakic 1988; Seri et al. 2004), but this is yet to be experimentally verified. Furthermore, the molecular mechanisms regulating this developmental event are poorly defined. A number of lines of evidence suggested a role for Nfix in this process. First, we have previously shown that cells within the subgranular zone express NFIX (Campbell et al. 2008). Secondly, the reduction in the size of the dentate gyrus, and the number of PROX1-expressing cells within this structure (Fig. 8K,L), indicated a deficit in the production of dentate granule neurons. Finally, our analysis of E16 wild-type and Nfix−/− mice had shown a marked reduction in the number of mitotically active (Fig. 2K) and SOX2-positive (Fig. 2G) cells within the emerging dentate gyrus of Nfix knockouts, illustrative of abnormal development of subgranular zone neural progenitors in Nfix mutant mice.
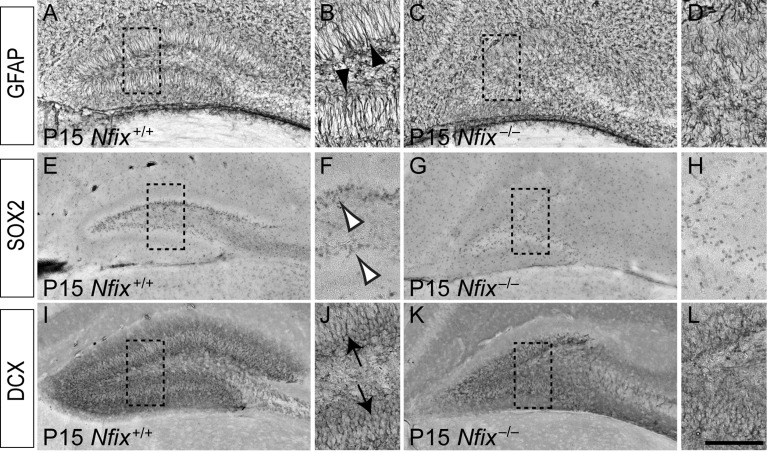
To investigate the role of Nfix in the formation of subgranular zone neural progenitor cells, we analyzed the expression of markers for this population within the dentate gyrus of postnatal Nfix−/− mice. GFAP is expressed by both astrocytes and neural progenitor cells within the dentate gyrus (Seri et al. 2001), but can be used to identify progenitors within the subgranular zone, as they exhibit radially oriented GFAP-positive fibers that extend into the granular cell layer (Seri et al. 2004). This was readily seen within the dentate gyrus of P15 wild-type mice (Fig. 9A,B). In Nfix−/− mice, however, although the expression of GFAP was evident within the dentate gyrus, there were far fewer radially oriented fibers, and the GFAP-expressing cells appeared to be disorganized with regard to their projection into the granular zone (Fig. 9C,D).
Figure 9.
The subgranular zone is abnormal in the dentate gyrus of postnatal Nfix−/− mice. Expression of GFAP (A–D), SOX2 (E–H), and DCX (I–L) in the dentate gyrus of P15 wild-type (A, B, E, F, I, J) and Nfix−/− mice (C, D, G, H, K, L). (A) Expression of GFAP in the dentate gyrus of a P15 wild-type mouse. (B) Higher magnification view of the boxed region in A, showing the radially oriented, GFAP-positive fibers of subgranular zone neural progenitor cells (arrowheads in B). (C) Expression of GFAP in the dentate gyrus of a P15 Nfix−/− mouse. (D) Higher magnification view of the boxed region in C. There were fewer GFAP-expressing cells in the dentate gyrus of Nfix knockout mice that possessed a radially oriented fiber. (E) In the dentate gyrus of a P15 wild-type mouse, SOX2 was expressed by cells in the subgranular zone of the dentate gyrus. (F) Higher magnification view of the boxed region in E, showing SOX2-expressing progenitor cells within the subgranular zone (open arrowheads in F). (G) In Nfix−/− mice, SOX2-expressing progenitor cells did not align within the subgranular zone to the same extent as in wild-type mice. (H) Higher magnification view of the boxed region in G, revealing the disordered localization of progenitor cells within the dentate gyrus of Nfix−/− mice. (I, J) The expression of DCX within the dentate gyrus of wild-type mice revealed that immature neurons exhibited radially arrayed processes that extended into the granular zone (arrows in J). (K, L) In Nfix−/− mice, DCX-expressing cells exhibited a disordered morphology within the dentate gyrus. PanelsJ and L are higher magnification views of the boxed regions in I and K, respectively. Scale bar (in L): A, C, E, G, I, K, 500 μm; B, D, F, H, J, L, 100 μm.
SOX2 expression can also be used to identify self-renewing cells within the subgranular zone (Suh et al. 2007). In wild-type mice, SOX2-positive cells were aligned beneath the granular zone, but in the mutants these cells appeared more scattered throughout the hilus (Fig. 9E–H). This decrease was not due to excessive apoptosis, as we did not observe any increase in cleaved caspase-3–positive cells within the dentate gyrus of Nfix−/− mice either embryonically or postnatally (data not shown). Finally, we analyzed the expression of doublecortin (DCX), a microtubule-associated protein expressed by immature neurons (Gleeson et al. 1999). The expression of DCX within the dentate gyrus of wild-type postnatal brains revealed that DCX-expressing neuroblasts extended radial processes into the granular zone of the dentate gyrus (Fig. 9I,J). In the mutant, however, DCX-expressing cells did not exhibit this ordered array of DCX-expressing fibers, appearing instead disorganized within the dentate gyrus (Fig. 9K,L). These data from the dentate gyrus of postnatal animals suggest that Nfix plays an important role in the development of neural progenitor cells within this neurogenic niche of the adult brain, providing a significant advance in our understanding of the regulatory control of this process.
NFIX Represses Sox9 Expression During Embryonic Hippocampal Development
A salient feature emerging from our study to date was that, during embryogenesis, progenitor cell self-renewal was extended at the expense of differentiation, from which we inferred that Nfix acts, in part, via the repression of progenitor-specific genes. To gain a mechanistic insight into how NFIX acts to regulate neural progenitor cell differentiation, we performed a microarray screen of hippocampal tissue from littermate E16 Nfix+/+ and Nfix−/− mice. This analysis identified over 1000 genes as being differentially expressed within the hippocampus of the mutant mice, using a significance level of P< 0.05 via ANOVA and a fold-change cut-off of 1.5 (Supplementary Table 1). In support of our findings relating to delayed neural progenitor cell differentiation, this analysis identified many neuronal-specific (Prox1, Ka1, and Ncam1) and glial-specific (Gfap and Omg) genes as being significantly downregulated within the hippocampus of Nfix−/− mice. Furthermore, functional annotation of genes downregulated in the hippocampus of the mutant mice identified the processes of neuron development, neuron projection development, and neuron differentiation as being enriched in the mutant (Fig. 10 and Table 1), further emphasizing that progenitor cell differentiation is delayed in the absence of Nfix. The expression of numerous genes was also significantly upregulated in the mutant hippocampus at E16, including a number previously implicated in progenitor cell self-renewal, such as the Notch pathway members Dll1 and Hey2 (Shimojo et al. 2008). Moreover, functional annotation of those genes upregulated in the hippocampus of Nfix−/− mice provided further evidence that the balance between progenitor cell differentiation and self-renewal was shifted toward self-renewal, with many processes involved in proliferation being evident, including cell division, cell cycle, DNA metabolic process, transcription, and mitotic cell cycle (Fig. 10). Taken together, these findings provide further support for the notion that progenitor cell maintenance is prolonged within Nfix−/− mice.
Figure 10.
Microarray and functional classification reveals diverse genes misregulated within the hippocampus of E16 Nfix−/− mice. Microarray analysis was performed on E16 wild-type and Nfix−/− hippocampal tissue. Genes were annotated using the functional annotation tool of DAVID, revealing key biological processes that were downregulated (A) and upregulated (B) in the hippocampus of Nfix−/− mice. Biological pathways involving mRNAs that were downregulated (C) or upregulated (D) in the hippocampus of Nfix−/− mice were also generated within DAVID.
Table 1.
Key examples of transcripts downregulated in the hippocampus of Nfix−/− mice at E16
| Functional classification (DAVID) | Downregulated genes |
|---|---|
| Neuron differentiation | Fezf1; doublecortin; cholecystokinin; MAP2; Slit3; Sema3A; Sema5A |
| Regulation of synaptic transmission | calcium channel, voltage-dependent, P/Q type, alpha 1A subunit; dopamine receptor D1A; glutamate receptor, ionotropic, NMDA2B; glutamate receptor, ionotropic, kainate 1; glutamate receptor, metabotropic 5; alpha synuclein |
| Neurotransmitter transport | Solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator); solute carrier family 6 (neurotransmitter transporter, GABA), member 1; solute carrier family 6 (neurotransmitter transporter, GABA), member 11; synaptotagmin I |
| Integrin-mediated signaling pathway | Integrin alpha 11; integrin alpha 8; integrin alpha V; integrin beta 5; calcium and integrin binding family member 2 |
Interestingly, the analysis of the transcripts upregulated in the hippocampus of Nfix−/− mice at E16 (Table 2) revealed a variety of potential targets for transcriptional regulation by NFIX. Of particular interest was Sox9, a member of the SoxE family of transcription factors, which has recently been implicated in driving the induction and maintenance of cortical neural progenitor cells (Scott et al. 2010). Validation of the array results using qPCR confirmed that there were significantly elevated levels of Sox9 mRNA in the hippocampus of Nfix−/− mice. Furthermore, using a SOX9-specific antibody, immunohistochemical analysis revealed significantly more SOX9-expressing neural progenitor cells within the hippocampal and neocortical ventricular zone of Nfix−/− mice (Fig. 11A–D; Supplementary Fig. 1). We therefore asked whether Sox9 was a direct target for transcriptional control by NFIX. To do this, we first performed an in silico bioinformatic screen of the Sox9 promoter to search for putative NFI consensus DNA-binding sites. The NFI motif DNA-binding site was derived from a recent report that used ChIP sequencing to identify NFI binding sites in vivo (Pjanic et al. 2011). Using this motif, we scanned the region around the TSS of Sox9 (see Materials and methods) and identified multiple putative NFI binding sites within this region, 2 of which were upstream of the TSS, and 2 of which were downstream of the TSS, including 1 within the first exon of the Sox9 gene (+598; Fig. 11E). Although not common, transcription factor-binding sites have been identified in silico within the exonic region of many genes (Gotea et al. 2012). Moreover, the first exon of the elastin gene has been shown to possess a regulatory element that facilitates the expression of this gene (Pierce et al. 2006), while the transcription factor GATA-1 binds to a regulatory region in exon 1 of the C–C chemokine receptor type 3 gene (Zimmermann et al. 2005). In sum, our array data and in silico-binding site predictions provide support for the regulation of Sox9 gene transcription by NFIX.
Table 2.
Key examples of transcripts upregulated in the hippocampus of Nfix−/− mice at E16
| Functional classification (DAVID) | Upregulated genes |
|---|---|
| Transcription | Sox9; Sox5, E2F6; FoxL2; Foxo4; neurogenin 2; TATA bix binding protein; Eya1; Eya3 |
| Cell division | Anillin, actin binding protein; cell division cycle 2 homolog A; cell division cycle associated 7; Wnt3A; polo-like kinase 1; protein regulator of cytokinesis 1 |
| DNA metabolic process | Cyclin E2; cyclin O; exonuclease 1; ligase I, DNA, ATP-dependent; topioisomerase (DNA) I; uracil DNA glycosylase |
| Mitotic cell cycle | Aurora kinase A; aurora kinase B; centromere protein F; cyclin D2; cyclin-dependent kinase 2; Nedd9; checkpoint kinase homolog 1 |
Figure 11.
Upregulation of SOX9 expression in Nfix mutants. (A, B) Coronal sections of E16 wild-type (A) and Nfix−/− (B) hippocampi, showing the expression of SOX9. There were significantly more SOX9-positive cells in the ventricular zone of the Nfix mutant than within the control (C; also compare brackets in A and B). (D) qPCR revealed significantly elevated levels of Sox9 mRNA in the hippocampus of Nfix−/− mice compared with wild-type controls. (E) Potential NFI binding sites reported by FIMO around the Sox9 TSS. We report the position in bases of each potential site relative to the TSS, the strand of the potential site, the P-value of the motif match, and the site sequence. (F) EMSA. E18 mouse brain nuclear extracts were incubated with radiolabeled probes for NFI control (lane 1), +598 (lane 2), +495 (lane 3), −183 (lane 4), and −675 (lane 5) consensus sites. Only the control and the +598 probes exhibited significant binding to the nuclear extract (asterisk). FP, free probe. (G) Supershift assay using nuclear extracts from COS cells expressing an HA-tagged NFIX expression construct. For the control probe (lane 3) and the +598 probe (lane 7), an NFIX complex was produced (asterisk) when the nuclear extract was incubated with either probe. However, the addition of a specific anti-HA antibody to the binding reaction depleted this complex and produced a supershifted complex (SS; lanes 4 and 8). A nonspecific transcription factor, AP2, did not demonstrate any specific binding to either oligonucleotide probe (lanes 1, 2, 5, and 6). (H) Reporter gene assay in NSC-34 cells. Transfection of an Nfix expression vector (Nfix pCAGIG) elicited no luciferase activity, whereas transfection of a luciferase construct under the control of the Sox9 promoter elicited robust induction of the reporter gene. Cotransfection of Nfix with the Sox9 promoter reporter yielded a significantly reduced level of luciferase activity. However, mutation of the putative +598 NFI binding site within the Sox9 promoter (Sox9ΔNFI) abolished NFI-mediated repression of luciferase expression. *P< 0.05, ***P<0.001, Student's t-test; **P<0.01, ANOVA. Scale bar (in B): 250 μm.
To determine which of these predicted sites were functionally relevant, we next performed electrophoretic mobility shift assays using oligonucleotide probes designed to encompass each of the putative NFI binding sites (−695, −183, +415, and +598). First, using E18 mouse cortex nuclear extracts, we showed that only the +598 oligonucleotide probe exhibited significant binding to the nuclear extract derived from the E18 mouse cortex, although the −183 probe also exhibited low levels of binding (Fig. 11F). We next performed supershift assays to determine whether the mobility shift we observed with respect to the +598 probe was due to NFIX. To do this, we transfected an HA-tagged NFIX expression construct into COS cells and isolated nuclear extracts 48h later. Subsequent electrophoretic mobility shift assays with these nuclear extracts revealed supershifting of the +598 probe when an anti-HA antibody was present (Fig. 11G). A nonspecific transcription factor, AP2, did not exhibit any binding to the +598 probe. These data suggest that NFIX can directly interact with the +598 binding site within exon 1 of the Sox9 gene.
To formally address whether NFIX was capable of regulating Sox9 promoter-driven transcriptional activity, we employed a reporter gene assay, whereby the expression of the luciferase gene was under the control of a 250-bp region from the Sox9 gene containing the +598 site. This analysis revealed that NFIX was able to directly repress luciferase expression driven by the Sox9 promoter (Fig. 11H). Moreover, mutagenesis of the +598 site abolished repression of the luciferase gene, indicating the importance of this site for NFIX-mediated repression of Sox9 expression (Fig. 11H). Taken together, these data suggest that NFIX promotes progenitor cell differentiation within the embryonic hippocampus through the direct transcriptional repression of Sox9.
Overexpression of NFIX Drives Precocious Gliogenesis In Vivo
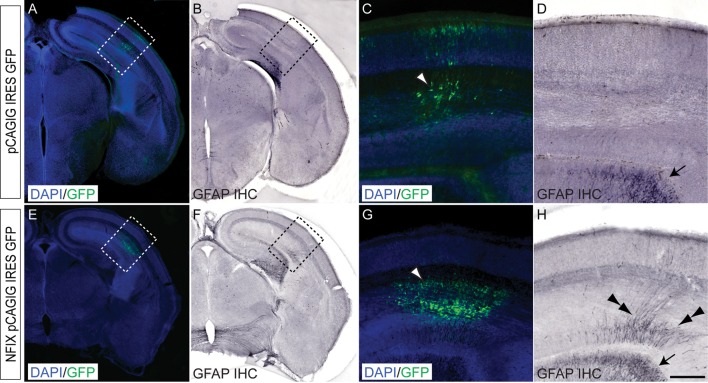
Our data to date suggested that NFIX plays a pivotal role during development of the dorsal telencephalon, driving progenitor cell differentiation in part via direct repression of Sox9 transcription. Given this, we posited that the overexpression of NFIX in vivo would culminate in precocious progenitor cell differentiation. To test this hypothesis, we used in utero electroporation to transfect neural progenitor cells in vivo with an NFIX expression construct (Nfix pCAGIG IRES GFP). As NFIX has been shown to regulate glial development in vitro (Cebolla and Vallejo 2006; Brun et al. 2009; Piper et al. 2011), we used the expression of the mature glial marker GFAP as our read-out of progenitor cell differentiation. Moreover, given that NFIX also regulates neocortical neural progenitor cell differentiation (Supplementary Figs 1–4) and that the expression of GFAP within the neocortex is minimal before E16 (Shu, Butz, et al. 2003; Shu, Puche, et al. 2003), we chose to electroporate neocortical neural progenitor cells at E13 and to analyze GFAP expression 3 days later at E16, reasoning that this would allow the visualization of precocious gliogenesis without the potential confounding presence of endogenous gliogenesis. Electroporation of the vector only control into the neocortical ventricular zone at E13 did not result in precocious gliogenesis within the neocortex at E16 (Fig. 12A–D). However, the overexpression of NFIX at E13 did indeed result in the precocious expression of GFAP within the neocortex at E16 (Fig. 12E–H), demonstrating that NFIX plays a key role in driving the differentiation of neural progenitor cells in vivo.
Figure 12.
Overexpression of NFIX drives gliogenesis in vivo. Overexpression of a vector only control (pCAGIG IRES GFP; A–D) or NFIX (Nfix pCAGIG IRES GFP; E–H) into the neocortex of wild-type CD-1 mice at E13 using in utero electoporation. At E16, brains were fixed and sectioned, then processed for immunofluorescence (A, C, E, G), followed by immunohistochemistry (IHC) using an anti-GFAP antibody (B, D, F, H). Panels A and B show the same section with fluorescence (A) and brightfield (B) microscopy, respectively, as do panels E and F. In the control, expression of GFP can be seen within the neocortex (A, arrowhead in C). However, no GFAP immunoreactivity is present within the neocortex at this time (B, D), although GFAP expression can be seen in the hippocampus (arrow in D). In those embryos electroporated with the NFIX expression construct, expression of GFP can also be seen within the neocortex (E, arrowhead in G). However, as well as GFAP expression being evident within the hippocampus (arrow in H), ectopic expression of this glial marker is also evident within the neocortex (double arrowheads in H). Panels C, D, G and H are higher magnification views of the boxed regions in A, B, E, and F, respectively. Scale bar (in H): A, B, E, F, 600 μm; C, D, G, H, 150 μm.
Discussion
Many factors have been shown to act in concert to regulate the balance between neural progenitor cell self-renewal and differentiation during development. For example, members of the SOX family of transcription factors, such as SOX2, are known to promote the maintenance of progenitor cell identity during development of the embryonic cortex, and within the neurogenic niches of the adult brain (Bani-Yaghoub et al. 2006; Suh et al. 2007). Another member of the SOX family implicated in progenitor cell self-renewal during corticogenesis is SOX9, which has been shown to act downstream of SHH to induce and maintain neural progenitor cell identity (Scott et al. 2010). How SOX9 expression within cortical neural progenitor cells is regulated remains unclear, and indeed, whether or not the regulation of SOX9 by SHH is direct or indirect remains unresolved. Moreover, how SOX9 expression is downregulated to enable progenitor cell differentiation remains unknown. Here, we demonstrate that the transcription factor NFIX plays a key role in this process, promoting the differentiation of hippocampal neural progenitor cells in part via transcriptional repression of Sox9. Using Nfix−/− mice, we demonstrate that ventricular zone progenitor cells in the embryonic hippocampus are retained in the progenitor state for longer in the absence of Nfix, which leads to delays in both neuronal and glial differentiation. Furthermore, we identify multiple potential NFI binding sites in the basal promoter of the Sox9 gene and demonstrate the ability of NFIX to repress Sox9 promoter-driven gene expression. These findings provide a mechanistic insight into how the differentiation of neural progenitor cells within the hippocampus is orchestrated during development and highlight the importance of NFIX in regulating this process.
Although the NFI family were first isolated over 2 decades ago (Rupp et al. 1990; Kruse et al. 1991), the mechanisms by which they drive embryonic development, including that of the nervous system, remain only partially understood. However, work conducted both in vitro and in vivo has begun to reveal the role these transcription factors play during development, and importantly, to identify their downstream transcriptional targets. Expression studies have demonstrated that Nfix mRNA is expressed within the developing mouse telencephalon from approximately E11.5 (Chaudhry et al. 1997). Furthermore, in vitro studies have shown that NFIX drives the expression of astrocyte-specific genes, including Gfap, Sparcl1, brain fatty acid-binding protein, and YKL-40 in various cell lines derived from human glioblastomas (Gopalan et al. 2006; Brun et al. 2009; Singh et al. 2011). Despite this, the role of Nfix during nervous system development remains poorly understood. Gene-specific knockout mice are now providing important insights into the role of Nfix during nervous system development, with knockouts exhibiting a range of neurological abnormalities, including malformation of the hippocampal dentate gyrus, expansion of the cingulate cortex, and increased brain weight (Driller et al. 2007; Campbell et al. 2008). Mechanistically, we have recently shown that NFIX regulates the expression of Gfap during cerebellar development (Piper et al. 2011), but how NFIX acts in neural progenitor cells in vivo was not addressed in this study. Our current findings therefore provide a conceptual advance in our understanding of how Nfix drives neural progenitor cell differentiation during development, revealing an important role for this transcription factor in the repression of the stem cell maintenance gene, Sox9.
The SOX family of transcription factors play a variety of roles during the development of many organs (Stolt and Wegner 2010). Studies have shown that SOX8, SOX9, and SOX10, which comprise a subfamily of the SOX group known as the SOXE family (Bowles et al. 2000), are important determinants of progenitor cell differentiation (Stolt and Wegner 2010). For instance, Sox9 is expressed by ventricular zone progenitor cells within the spinal cord at the time at which gliogenesis is initiated within this structure, and deletion of this gene culminates in deficits in both astrocyte and oligodendrocyte formation (Stolt et al. 2003). Furthermore, SOX10 is also expressed by oligodendrocyte progenitors within the developing spinal cord (Stolt et al. 2002), and compound loss of both Sox9 and Sox10 leads to a more severe phenotype with regard to oligodendrocyte formation than loss of Sox9 alone (Stolt et al. 2003), highlighting the role of these genes in the formation of the oligodendrocyte lineage.
These findings indicate that SOXE family members are key determinants of promoting the gliogenic fate switch within the developing spinal cord (Stolt and Wegner 2010). However, the extent to which they reflect the development of other structures within the nervous system remains unclear. The function of SOX9 provides a pertinent example of this, as this transcription factor appears to play a distinct role during corticogenesis. SOX9 is expressed very early during cortical development, from approximately E10.5, when neural progenitor cells are undergoing the transition to radial glial cells (Scott et al. 2010). This is much earlier than the onset of gliogenesis, which is initiated at approximately E14 within the mouse forebrain (Shu, Butz, et al. 2003; Shu, Puche, et al. 2003). Both loss- and gain-of-function experiments have suggested that SOX9 plays a central role downstream of SHH in the induction and maintenance of cortical neural progenitor cells (Scott et al. 2010). This implies that SOX9 fulfils a different role during cortical development as opposed to spinal cord development, acting to promote progenitor cell self-renewal instead of acting in the switch toward glial fate determination. Interestingly, differences in the activity of the Nfi genes between the developing cortex and spinal cord are also evident. For instance, Nfia, which acts downstream of Notch signaling to induce gliogenesis within the cortex (Namihira et al. 2009), has been shown to downregulate Notch pathway activity within the telencephalon via the repression of the Notch effector Hes1 (Piper et al. 2010), while in the developing spinal cord, Nfia is required for Hes5 expression (Deneen et al. 2006). These findings highlight the important differences between the development of the cortex and spinal cord, and more broadly emphasize that the function of such transcription factors is influenced by the cellular and molecular context in which they act during embryogenesis.
Our data advance our understanding of how the self-renewal gene Sox9 is regulated during hippocampal formation, as, to date, little has been known regarding its transcriptional control during forebrain development. Indeed, much of our understanding of Sox9 regulation has been gleaned from studies within other organ systems. For example, male sex determination is coordinated via the synergistic action of Sry and Sf1 on Sox9 enhancer elements within developing Sertoli cells (Sekido and Lovell-Badge 2008). Lhx2 has also been shown to regulate Sox9 expression within hair follicle stem cells (Mardaryev et al. 2011), whereas Notch1 signaling promotes Sox9 expression during chondrogenesis (Haller et al. 2011). During cortical development, SHH too has been shown to induce Sox9 expression, although whether this effect is direct or indirect remains unclear (Scott et al. 2010). Our data clearly indicate that Nfix plays an important role during the differentiation of embryonic hippocampal neural progenitor cells, repressing Sox9 expression to promote the differentiation of progenitors at the expense of self-renewal. When considered in light of the role of Nfix in driving astrocyte-specific genes (Gopalan et al. 2006; Brun et al. 2009), this reveals that NFIX exerts multifactorial control during neural development. It remains likely, however, that other factors also act to regulate Sox9 expression in addition to SHH and NFIX. LHX2 and Notch1 are likely candidates for contributing to the regulation of Sox9 expression during forebrain development, given their previously reported roles in regulating the development of this structure (Mizutani et al. 2007; Subramanian et al. 2011). Interestingly, a recent report has also linked Sox9 and another Nfi family member, Nfia, in co-operatively regulating astrogliogenesis within the spinal cord. SOX9 was shown to induce Nfia expression during spinal cord development, and these transcription factors were revealed to then form a transcriptional complex that coregulated the expression of Apcdd1 and Mmd2 (Kang et al. 2012). It remains unclear whether SOX9 forms active transcriptional complexes with NFIA, or with other NFI family members, within the developing telencephalon, and, moreover, we did not find evidence for the misregulation of either Apcdd1 or Mmd2 within the hippocampus of Nfix−/− (this study) or Nfia−/− mice (Piper et al. 2010), again indicating context-dependent differences exist in the actions of these transcription factors within the spinal cord and forebrain.
The findings we present here also provide an insight into the development of the subgranular zone of the dentate gyrus. Both the Nfia−/− and Nfib−/− lines die at birth (Shu, Butz, et al. 2003; Shu, Puche, et al. 2003; Steele-Perkins et al. 2005), and as such, the Nfix−/− line provides an opportunity to study Nfi gene function in postnatal hippocampal development. Here we reveal that, although the dentate gyrus does eventually form soon after birth in Nfix mutant mice, the morphology of this structure is aberrant, with neural progenitor cells exhibiting abnormal morphologies, and PROX1-expressing dentate granule neurons being found in significantly smaller numbers. These findings demonstrate that NFIX is important for both the architectural development of the dentate gyrus and the formation of the subgranular zone neurogenic niche. Unfortunately, Nfix mutants on a C57Bl/6J background die at weaning, precluding investigations aimed at determining whether these hippocampal abnormalities culminate in functional deficits with regard to hippocampal function. The development of a conditional floxed Nfix allele may provide one avenue to address this issue.
Another interesting finding arising from these and other recent studies is the phenotypic similarity between mice lacking Nfia, Nfib, or Nfix (Barry et al. 2008; Piper et al. 2010; Heng et al. 2012). In each of these knockout lines, the astroglial development arising from progenitor cells within the ammonic neuroepithelium is delayed, suggesting that Nfi genes act in a common pathway to drive gliogenesis. Our data from postnatal Nfix−/− mice support the idea that NFI family members may act co-operatively in the gliogenic pathway, as gliogenesis within the hippocampus (Fig. 9) and neocortex (Supplementary Fig. 5) does eventually occur in the absence of Nfix. This finding is further supported by studies performed in human glioblastoma cell lines, which indicate that Nfia and Nfib act early during gliogenesis, whereas Nfic and Nfix act later in this process (Wilczynska et al. 2009). However, the extent to which individual NFI proteins act in either common or divergent pathways remains unresolved. Although the data generated both in vitro and from mouse models indicate that Nfi genes contribute to the expression of astrocyte-specific genes, it is unknown whether each family member targets a specific suite of downstream factors during development. However, the structure of the NFI proteins themselves provides insights into this issue. Each NFI has a highly conserved N-terminal DNA-binding domain, and a less well conserved C-terminal domain involved in protein–protein interactions (Mason et al. 2009). The inference from such structural separation between the N- and C-termini is that individual isoforms can potentially act in concert with different binding partners to regulate gene expression. Support for this is provided by the fact that we did not find evidence for the dysregulation of Sox9 in microarrays of hippocampal tissue from Nfia−/− mice (Piper et al. 2010), or for the dysregulation of Hes1, a target for repression by Nfia and Nfib within Nfix−/− mice (Supplementary Table 1). Looking forward, comparative gene expression analyses of different Nfi knockouts in distinct spatial and temporal windows should provide valuable insights into the unique transcriptional signature of each NFI family member during development, thereby clarifying the extent to which these transcription factors act via common or divergent mechanisms throughout development. Such methods will also highlight the similarities and differences in the activities of the NFI proteins within different developmental contexts, such as the developing hippocampus and the developing spinal cord.
Supplementary Material
Supplementary material can be found at: http://www.cercor.oxfordjournals.org/.
Funding
This work was supported by National Health and Medical Research Council (NHMRC) project grants (grant number 1003462 to M.P., grant number 569504 to L.J.R.) and by National Institute of Health and NYSTEM grants (grant numbers HL080624 and C026429 to R.M.G.). The following authors were supported by NHMRC fellowships: M.P. (Biomedical Career Development Fellowship); L.J.R. (Principal Research Fellowship). Y.H.E.H. was supported by a University of Queensland International scholarship.
Supplementary Material
Notes
We thank Rowan Tweedale for critical analysis of the manuscript, and Drs. Peter Koopman, Robert Hevner, Shubha Tole, Andre Goffinet, and Niels Danbolt for kindly providing reagents. Conflict of Interest: None declared.
References
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bani-Yaghoub M, Tremblay RG, Lei JX, Zhang D, Zurakowski B, Sandhu JK, Smith B, Ribecco-Lutkiewicz M, Kennedy J, Walker PR, et al. Role of Sox2 in the development of the mouse neocortex. Dev Biol. 2006;295:52–66. doi: 10.1016/j.ydbio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Barry G, Piper M, Lindwall C, Moldrich R, Mason S, Little E, Sarkar A, Tole S, Gronostajski RM, Richards LJ. Specific glial populations regulate hippocampal morphogenesis. J Neurosci. 2008;28:12328–12340. doi: 10.1523/JNEUROSCI.4000-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettler B, Boulter J, Hermans-Borgmeyer I, O'Shea-Greenfield A, Deneris ES, Moll C, Borgmeyer U, Hollmann M, Heinemann S. Cloning of a novel glutamate receptor subunit, GluR5: expression in the nervous system during development. Neuron. 1990;5:583–595. doi: 10.1016/0896-6273(90)90213-y. [DOI] [PubMed] [Google Scholar]
- Bielle F, Griveau A, Narboux-Neme N, Vigneau S, Sigrist M, Arber S, Wassef M, Pierani A. Multiple origins of Cajal-Retzius cells at the borders of the developing pallium. Nat Neurosci. 2005;8:1002–1012. doi: 10.1038/nn1511. [DOI] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Brun M, Coles JE, Monckton EA, Glubrecht DD, Bisgrove D, Godbout R. Nuclear factor I regulates brain fatty acid-binding protein and glial fibrillary acidic protein gene expression in malignant glioma cell lines. J Mol Biol. 2009;391:282–300. doi: 10.1016/j.jmb.2009.06.041. [DOI] [PubMed] [Google Scholar]
- Campbell CE, Piper M, Plachez C, Yeh YT, Baizer JS, Osinski JM, Litwack ED, Richards LJ, Gronostajski RM. The transcription factor Nfix is essential for normal brain development. BMC Dev Biol. 2008;8:52. doi: 10.1186/1471-213X-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashman NR, Durham HD, Blusztajn JK, Oda K, Tabira T, Shaw IT, Dahrouge S, Antel JP. Neuroblastoma x spinal cord (NSC) hybrid cell lines resemble developing motor neurons. Dev Dyn. 1992;194:209–221. doi: 10.1002/aja.1001940306. [DOI] [PubMed] [Google Scholar]
- Cebolla B, Vallejo M. Nuclear factor-I regulates glial fibrillary acidic protein gene expression in astrocytes differentiated from cortical precursor cells. J Neurochem. 2006;97:1057–1070. doi: 10.1111/j.1471-4159.2006.03804.x. [DOI] [PubMed] [Google Scholar]
- Chaudhry AZ, Lyons GE, Gronostajski RM. Expression patterns of the four nuclear factor I genes during mouse embryogenesis indicate a potential role in development. Dev Dyn. 1997;208:313–325. doi: 10.1002/(SICI)1097-0177(199703)208:3<313::AID-AJA3>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Dahlstrand J, Lardelli M, Lendahl U. Nestin mRNA expression correlates with the central nervous system progenitor cell state in many, but not all, regions of developing central nervous system. Brain Res Dev Brain Res. 1995;84:109–129. doi: 10.1016/0165-3806(94)00162-s. [DOI] [PubMed] [Google Scholar]
- Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron. 2006;52:953–968. doi: 10.1016/j.neuron.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Driller K, Pagenstecher A, Uhl M, Omran H, Berlis A, Grunder A, Sippel AE. Nuclear Factor One X deficiency causes brain malformation and severe skeletal defects. Mol Cell Biol. 2007;27:3855–3867. doi: 10.1128/MCB.02293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckenhoff MF, Rakic P. Nature and fate of proliferative cells in the hippocampal dentate gyrus during the life span of the rhesus monkey. J Neurosci. 1988;8:2729–2747. doi: 10.1523/JNEUROSCI.08-08-02729.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz GD, Bohner AP, Akers RM, McConnell SK. Regulation of the POU domain gene SCIP during cerebral cortical development. J Neurosci. 1994;14:472–485. doi: 10.1523/JNEUROSCI.14-02-00472.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frotscher M, Haas CA, Forster E. Reelin controls granule cell migration in the dentate gyrus by acting on the radial glial scaffold. Cereb Cortex. 2003;13:634–640. doi: 10.1093/cercor/13.6.634. [DOI] [PubMed] [Google Scholar]
- Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno F, Lopez-Mascaraque L, De Carlos JA. Origins and migratory routes of murine Cajal-Retzius cells. J Comp Neurol. 2007;500:419–432. doi: 10.1002/cne.21128. [DOI] [PubMed] [Google Scholar]
- Gleeson JG, Lin PT, Flanagan LA, Walsh CA. Doublecortin is a microtubule-associated protein and is expressed widely by migrating neurons. Neuron. 1999;23:257–271. doi: 10.1016/s0896-6273(00)80778-3. [DOI] [PubMed] [Google Scholar]
- Gopalan SM, Wilczynska KM, Konik BS, Bryan L, Kordula T. Nuclear factor-1-X regulates astrocyte-specific expression of the alpha1-antichymotrypsin and glial fibrillary acidic protein genes. J Biol Chem. 2006;281:13126–13133. doi: 10.1074/jbc.M601194200. [DOI] [PubMed] [Google Scholar]
- Gotea V, Visel A, Westlund JM, Nobrega MA, Pennacchio LA, Ovcharenko I. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2012;20:565–577. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Gotz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillery RW. On counting and counting errors. J Comp Neurol. 2002;447:1–7. doi: 10.1002/cne.10221. [DOI] [PubMed] [Google Scholar]
- Haller R, Schwanbeck R, Martini S, Bernoth K, Kramer J, Just U, Rohwedel J. Notch1 signaling regulates chondrogenic lineage determination through Sox9 activation. Cell Death Differ. 2011;19:461–469. doi: 10.1038/cdd.2011.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert JM, Mishina Y, McConnell SK. BMP signaling is required locally to pattern the dorsal telencephalic midline. Neuron. 2002;35:1029–1041. doi: 10.1016/s0896-6273(02)00900-5. [DOI] [PubMed] [Google Scholar]
- Heng YH, Barry G, Richards LJ, Piper M. Nuclear factor I genes regulate neuronal migration. Neurosignals. 2012;20:159–167. doi: 10.1159/000330651. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ihrie RA, Alvarez-Buylla A. Cells in the astroglial lineage are neural stem cells. Cell Tissue Res. 2008;331:179–191. doi: 10.1007/s00441-007-0461-z. [DOI] [PubMed] [Google Scholar]
- Imayoshi I, Sakamoto M, Yamaguchi M, Mori K, Kageyama R. Essential roles of Notch signaling in maintenance of neural stem cells in developing and adult brains. J Neurosci. 2010;30:3489–3498. doi: 10.1523/JNEUROSCI.4987-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P, Lee HK, Glasgow SM, Finley M, Donti T, Gaber ZB, Graham BH, Foster AE, Novitch BG, Gronostajski RM, et al. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent J, Wheatley SC, Andrews JE, Sinclair AH, Koopman P. A male-specific role for SOX9 in vertebrate sex determination. Development. 1996;122:2813–2822. doi: 10.1242/dev.122.9.2813. [DOI] [PubMed] [Google Scholar]
- Komada M, Saitsu H, Kinboshi M, Miura T, Shiota K, Ishibashi M. Hedgehog signaling is involved in development of the neocortex. Development. 2008;135:2717–2727. doi: 10.1242/dev.015891. [DOI] [PubMed] [Google Scholar]
- Kruse U, Qian F, Sippel AE. Identification of a fourth nuclear factor I gene in chicken by cDNA cloning: NFI-X. Nucleic Acids Res. 1991;19:6641. doi: 10.1093/nar/19.23.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendahl U, Zimmerman LB, McKay RD. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- Manzini MC, Walsh CA. What disorders of cortical development tell us about the cortex: one plus one does not always make two. Curr Opin Genet Dev. 2011;21:333–339. doi: 10.1016/j.gde.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardaryev AN, Meier N, Poterlowicz K, Sharov AA, Sharova TY, Ahmed MI, Rapisarda V, Lewis C, Fessing MY, Ruenger TM, et al. Lhx2 differentially regulates Sox9, Tcf4 and Lgr5 in hair follicle stem cells to promote epidermal regeneration after injury. Development. 2011;138:4843–4852. doi: 10.1242/dev.070284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason S, Piper M, Gronostajski RM, Richards LJ. Nuclear factor one transcription factors in CNS development. Mol Neurobiol. 2009;39:10–23. doi: 10.1007/s12035-008-8048-6. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Namihira M, Kohyama J, Semi K, Sanosaka T, Deneen B, Taga T, Nakashima K. Committed neuronal precursors confer astrocytic potential on residual neural precursor cells. Dev Cell. 2009;16:245–255. doi: 10.1016/j.devcel.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Pierce RA, Moore CH, Arikan MC. Positive transcriptional regulatory element located within exon 1 of elastin gene. Am J Physiol Lung Cell Mol Physiol. 2006;291:L391–L399. doi: 10.1152/ajplung.00441.2004. [DOI] [PubMed] [Google Scholar]
- Piper M, Barry G, Hawkins J, Mason S, Lindwall C, Little E, Sarkar A, Smith AG, Moldrich RX, Boyle GM, et al. NFIA controls telencephalic progenitor cell differentiation through repression of the Notch effector Hes1. J Neurosci. 2010;30:9127–9139. doi: 10.1523/JNEUROSCI.6167-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Dawson AL, Lindwall C, Barry G, Plachez C, Richards LJ. Emx and Nfi genes regulate cortical development and axon guidance in the telencephalon. Novartis Found Symp. 2007;288:230–242. discussion 242–235, 276–281. [PubMed] [Google Scholar]
- Piper M, Harris L, Barry G, Heng YH, Plachez C, Gronostajski RM, Richards LJ. Nuclear factor one X regulates the development of multiple cellular populations in the postnatal cerebellum. J Comp Neurol. 2011;519:3532–3548. doi: 10.1002/cne.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Moldrich RX, Lindwall C, Little E, Barry G, Mason S, Sunn N, Kurniawan ND, Gronostajski RM, Richards LJ. Multiple non-cell-autonomous defects underlie neocortical callosal dysgenesis in Nfib-deficient mice. Neural Dev. 2009;4:43. doi: 10.1186/1749-8104-4-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper M, Plachez C, Zalucki O, Fothergill T, Goudreau G, Erzurumlu R, Gu C, Richards LJ. Neuropilin 1-Sema signaling regulates crossing of cingulate pioneering axons during development of the corpus callosum. Cereb Cortex. 2009;19(Suppl 1):i11–i21. doi: 10.1093/cercor/bhp027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pjanic M, Pjanic P, Schmid C, Ambrosini G, Gaussin A, Plasari G, Mazza C, Bucher P, Mermod N. Nuclear factor I revealed as family of promoter binding transcription activators. BMC Genomics. 2011;12:181. doi: 10.1186/1471-2164-12-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plachez C, Lindwall C, Sunn N, Piper M, Moldrich RX, Campbell CE, Osinski JM, Gronostajski RM, Richards LJ. Nuclear factor I gene expression in the developing forebrain. J Comp Neurol. 2008;508:385–401. doi: 10.1002/cne.21645. [DOI] [PubMed] [Google Scholar]
- Pleasure SJ, Anderson S, Hevner R, Bagri A, Marin O, Lowenstein DH, Rubenstein JL. Cell migration from the ganglionic eminences is required for the development of hippocampal GABAergic interneurons. Neuron. 2000;28:727–740. doi: 10.1016/s0896-6273(00)00149-5. [DOI] [PubMed] [Google Scholar]
- Pleasure SJ, Collins AE, Lowenstein DH. Unique expression patterns of cell fate molecules delineate sequential stages of dentate gyrus development. J Neurosci. 2000;20:6095–6105. doi: 10.1523/JNEUROSCI.20-16-06095.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rash BG, Lim HD, Breunig JJ, Vaccarino FM. FGF signaling expands embryonic cortical surface area by regulating Notch-dependent neurogenesis. J Neurosci. 2011;31:15604–15617. doi: 10.1523/JNEUROSCI.4439-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickmann M, Amaral DG, Cowan WM. Organization of radial glial cells during the development of the rat dentate gyrus. J Comp Neurol. 1987;264:449–479. doi: 10.1002/cne.902640403. [DOI] [PubMed] [Google Scholar]
- Rupp RA, Kruse U, Multhaup G, Gobel U, Beyreuther K, Sippel AE. Chicken NFI/TGGCA proteins are encoded by at least three independent genes: NFI-A, NFI-B and NFI-C with homologues in mammalian genomes. Nucleic Acids Res. 1990;18:2607–2616. doi: 10.1093/nar/18.9.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahara S, O'Leary DD. Fgf10 regulates transition period of cortical stem cell differentiation to radial glia controlling generation of neurons and basal progenitors. Neuron. 2009;63:48–62. doi: 10.1016/j.neuron.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageot CM, Stiles CD. Molecular mechanisms controlling cortical gliogenesis. Curr Opin Neurobiol. 2002;12:244–249. doi: 10.1016/s0959-4388(02)00322-7. [DOI] [PubMed] [Google Scholar]
- Schmid CD, Bucher P. MER41 repeat sequences contain inducible STAT1 binding sites. PLoS One. 2010;5:e11425. doi: 10.1371/journal.pone.0011425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CE, Wynn SL, Sesay A, Cruz C, Cheung M, Gomez Gaviro MV, Booth S, Gao B, Cheah KS, Lovell-Badge R, et al. SOX9 induces and maintains neural stem cells. Nat Neurosci. 2010;13:1181–1189. doi: 10.1038/nn.2646. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- Seri B, Garcia-Verdugo JM, Collado-Morente L, McEwen BS, Alvarez-Buylla A. Cell types, lineage, and architecture of the germinal zone in the adult dentate gyrus. J Comp Neurol. 2004;478:359–378. doi: 10.1002/cne.20288. [DOI] [PubMed] [Google Scholar]
- Seri B, Garcia-Verdugo JM, McEwen BS, Alvarez-Buylla A. Astrocytes give rise to new neurons in the adult mammalian hippocampus. J Neurosci. 2001;21:7153–7160. doi: 10.1523/JNEUROSCI.21-18-07153.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata T, Yamada K, Watanabe M, Ikenaka K, Wada K, Tanaka K, Inoue Y. Glutamate transporter GLAST is expressed in the radial glia-astrocyte lineage of developing mouse spinal cord. J Neurosci. 1997;17:9212–9219. doi: 10.1523/JNEUROSCI.17-23-09212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Shu T, Butz KG, Plachez C, Gronostajski RM, Richards LJ. Abnormal development of forebrain midline glia and commissural projections in Nfia knock-out mice. J Neurosci. 2003;23:203–212. doi: 10.1523/JNEUROSCI.23-01-00203.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Puche AC, Richards LJ. Development of midline glial populations at the corticoseptal boundary. J Neurobiol. 2003;57:81–94. doi: 10.1002/neu.10252. [DOI] [PubMed] [Google Scholar]
- Sievers J, Hartmann D, Pehlemann FW, Berry M. Development of astroglial cells in the proliferative matrices, the granule cell layer, and the hippocampal fissure of the hamster dentate gyrus. J Comp Neurol. 1992;320:1–32. doi: 10.1002/cne.903200102. [DOI] [PubMed] [Google Scholar]
- Singh SK, Bhardwaj R, Wilczynska KM, Dumur CI, Kordula T. A complex of nuclear factor I-X3 and STAT3 regulates astrocyte and glioma migration through the secreted glycoprotein YKL-40. J Biol Chem. 2011;286:39893–39903. doi: 10.1074/jbc.M111.257451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG, Brightwell G, Smit SE, Parsons PG, Sturm RA. Redox regulation of Brn-2/N-Oct-3 POU domain DNA binding activity and proteolytic formation of N-Oct-5 during melanoma cell nuclear extraction. Melanoma Res. 1998;8:2–10. doi: 10.1097/00008390-199802000-00002. [DOI] [PubMed] [Google Scholar]
- Steele-Perkins G, Plachez C, Butz KG, Yang G, Bachurski CJ, Kinsman SL, Litwack ED, Richards LJ, Gronostajski RM. The transcription factor gene Nfib is essential for both lung maturation and brain development. Mol Cell Biol. 2005;25:685–698. doi: 10.1128/MCB.25.2.685-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Rehberg S, Ader M, Lommes P, Riethmacher D, Schachner M, Bartsch U, Wegner M. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Wegner M. SoxE function in vertebrate nervous system development. Int J Biochem Cell Biol. 2010;42:437–440. doi: 10.1016/j.biocel.2009.07.014. [DOI] [PubMed] [Google Scholar]
- Subramanian L, Sarkar A, Shetty AS, Muralidharan B, Padmanabhan H, Piper M, Monuki ES, Bach I, Gronostajski RM, Richards LJ, et al. Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proc Natl Acad Sci USA. 2011;108:E265–E274. doi: 10.1073/pnas.1101109108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H, Consiglio A, Ray J, Sawai T, D'Amour KA, Gage FH. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TW, Stromberg GP, Whitney JT, Brower NW, Klymkowsky MW, Parent JM. Sox3 expression identifies neural progenitors in persistent neonatal and adult mouse forebrain germinative zones. J Comp Neurol. 2006;497:88–100. doi: 10.1002/cne.20984. [DOI] [PubMed] [Google Scholar]
- Wang W, Mullikin-Kilpatrick D, Crandall JE, Gronostajski RM, Litwack ED, Kilpatrick DL. Nuclear factor I coordinates multiple phases of cerebellar granule cell development via regulation of cell adhesion molecules. J Neurosci. 2007;27:6115–6127. doi: 10.1523/JNEUROSCI.0180-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczynska KM, Singh SK, Adams B, Bryan L, Rao RR, Valerie K, Wright S, Griswold-Prenner I, Kordula T. Nuclear factor I isoforms regulate gene expression during the differentiation of human neural progenitors to astrocytes. Stem Cells. 2009;27:1173–1181. doi: 10.1002/stem.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CJ, Zhao C, Pleasure SJ. Wnt signaling mutants have decreased dentate granule cell production and radial glial scaffolding abnormalities. J Neurosci. 2004;24:121–126. doi: 10.1523/JNEUROSCI.4071-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann N, Colyer JL, Koch LE, Rothenberg ME. Analysis of the CCR3 promoter reveals a regulatory region in exon 1 that binds GATA-1. BMC Immunol. 2005;6:7. doi: 10.1186/1471-2172-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.