Abstract
Avian pathogenic Escherichia coli (APEC) causes colibacillosis, which is responsible for morbidity and mortality in chickens. Gene expression patterns have previously been demonstrated to differ between chicken populations that are resistant vs. susceptible to bacterial infection, but little is currently known about gene expression response to APEC. Increased understanding of gene expression patterns associated with resistance will facilitate genetic selection to increase resistance to APEC. Male broiler chicks were vaccinated at 2 weeks of age and challenged with APEC at 4 weeks of age. Peripheral blood leukocytes were collected at 1 and 5 day post-infection. Lesions on the liver, pericardium, and air sacs were used to assign a mild or severe pathology status to non-vaccinated, challenged chicks. Ten treatment groups were therefore generated with a priori factors of vaccination, challenge, day post-infection, and the a posteriori factor of pathology status. Global transcriptomic response was evaluated using the Agilent 44K chicken microarray. APEC infection resulted in more up-regulation than down-regulation of differentially expressed genes. Immune response and metabolic processes were enriched with differentially expressed genes. Although vaccination significantly reduced lesions in challenged bird, there was no detectable effect of vaccination on gene expression. This study investigated the transcriptomic differences in host responses associated with mild vs. severe pathology, in addition to the effects of vaccination and challenge, thus revealing genes and networks associated with response to APEC and providing a foundation for future studies on, and genetic selection for, genetic resistance to APEC.
Abbreviations: APEC, avian pathogenic Escherichia coli; ExPEC, extraintestinal pathogenic Escherichia coli; LB, Luria Bertani; PBL, peripheral blood leukocyte; PBS, phosphate buffered saline
Keywords: Avian pathogenic Escherichia coli, Leukocytes, Microarray, Transcriptome, Immune response, Chicken
1. Introduction
Avian pathogenic Escherichia coli (APEC) are a subpathotye of extraintestinal pathogenic E. coli (ExPEC) that cause extraintestinal diseases in poultry that are collectively known as colibacillosis. Infection can occur through inhalation of contaminated dust or fecal matter, and then develop into airsacculitis. APEC then accesses the bloodstream, resulting in septicemia and potential for development of pericarditis and perihepatitis [18]. APEC may also be a food safety concern, with current research showing strong genetic similarities between APEC and human ExPEC [37,51,60]. These similarities suggest zoonotic potential for APEC and APEC as a source for human ExPEC virulence genes [21,35,60]. Recently, APEC has been shown to cause disease in a rat model for human meningitis [69]. Through horizontal and vertical transmission, APEC can cause disease and death in infected birds, resulting in large monetary losses through condemnation of carcasses and reduced production [4,7,63]. Though there are many serogroups of APEC that cause disease in birds [4,61], O1 has been a focal point for study of pathogen and host response to infection, as it is one of the major serogroups responsible for colibacillosis worldwide [18]. In addition, APEC O1 is one of the most well characterized APEC strains in the literature [37,42,45].
Antimicrobials are commonly used to control colibacillosis in the poultry industry. With rising concern and demonstration of drug resistant bacteria [47], other control methods, such as enhanced host genetics, are a growing area of research. Past experimentation has illustrated the potential for breeding for colibacillosis resistance [2,10], indicating that greater research surrounding the genetic control of mechanisms of resistance is needed as a foundation for more effective breeding programs. Gene expression analysis of immune tissues is commonly used to characterize immune response [13,59] and provides potential candidate genes for disease resistance. Expression levels of immune genes have been shown to be heritable [68], evidence that the tactic of genetic selection for gene expression levels could be successful in a breeding program.
The chicken immune system is equipped with several mechanisms to combat pathogens. Multiple tissues each contribute unique signals and functions to help identify and combat disease. Peripheral blood leukocytes (PBL) are comprised of a dynamic population of cell types that serve in both the innate and adaptive immune responses [20,66]. Heterophils, the avian equivalent of neutrophils, are an innate responder and typically the first cell type to fight infection. Heterophils destroy susceptible bacteria through phagocytosis, oxidative burst and extracellular traps [14,22]. Proper signaling by cytokines and T-helper cells can increase the effectiveness of immune response. The defensive mechanisms of APEC can, however, reduce the effectiveness of the innate immune response and may include resistance to the detrimental effects of phagocytosis and complement, or decreasing the antimicrobial activity of heterophils [28,41,50,54,58]. Enhanced genetics of the host immune response may allow chickens to overcome APEC's defenses. Very few publications have documented differences in gene expression between individuals that are resistant vs. susceptible to APEC. We hypothesize that the global transcriptome of peripheral blood leukocytes will differ depending upon vaccination, challenge, and pathological response to APEC. This study aims to determine these gene expression differences associated with an APEC infection, which will lead to a better understanding of the genetic control of resistance and may serve as biomarkers for genetic selection for improved response to APEC infection.
2. Materials and methods
2.1. Bacteria preparation
APEC O1 strain O1:K1:H7 (NCBI reference sequence: NC_008563.1) was kept in brain heart infusion broth with 10% glycerol at −80 °C. Its genomic sequence is available and has been completely characterized [37]. Two days prior to bacterial challenge, bacteria were removed from the freezer and streaked onto Luria Bertani (LB) agar then incubated overnight at 37 °C. An isolated colony was then placed into 10 ml of LB broth and incubated overnight at 37 °C with shaking. On day of APEC challenge, bacteria were pelleted by centrifugation at 5000×g for 15 minutes. The pellet was then washed in phosphate buffered saline (PBS) 3 times before being enumerated by spectrometric reading at 600 nm. The inoculum was adjusted to the desired bacterial concentration, and counts were confirmed through serial dilution plating onto MacConkey agar overnight.
2.2. Animal experiments
Non-vaccinated, commercial male broiler chicks were purchased at 1 day of age from a local hatchery. Birds were raised on wire-floor cages with ad libitum access to food and water. For each replicate, 120 birds were split by vaccination status, challenge status, and day of necropsy (Fig. 1). Challenged birds were housed separately from non-challenged birds. At 2 weeks of age, 50% of the chicks were intramuscularly vaccinated with 0.5 ml/bird of Iss vaccine [48], containing 2 μg of vaccine and 50 μg of Quil A adjuvant in PBS. Non-vaccinated chicks received 50 μg of Quil A adjuvant in PBS via the same route. Increased serum survival, iss, encodes an outer membrane lipoprotein and is a virulence factor common to most APEC serotypes [36,56,61]. At 4 weeks of age, 80% of the chicks, half vaccinated and half non-vaccinated, were challenged with 0.1 ml containing 108 colony forming units of APEC O1 injected into the left thoracic air sac. Non-challenged chicks received 0.1 ml of PBS via the same route. Birds were sampled and euthanized at 2 time points, 1 and 5 day post-infection, equally splitting birds within each group between the two times. This experimental design was replicated six times, for a total of 720 chickens.
Fig. 1.
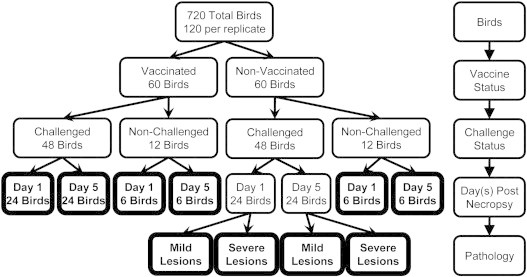
Treatment groups. Chicks were divided into treatment groups at 2 weeks of age by vaccination status, at 4 weeks of age by challenge status, by day of necropsy post-challenge, and by lesion scores assigned at necropsy. Treatment groups are bolded.
Blood samples were collected from the jugular vein into 5 ml vacuum tubes containing EDTA and placed on ice until PBL isolation. Birds were then euthanized and internal lesion scores assigned by a single trained investigator for 3 tissues, air sacs, pericardium and liver, as described by Peighambari et al. [55]. Scores from 0 to 2 were assigned for pericardium and liver; scores from 0 to 3 were assigned for air sacs. A summation of all 3 lesions scores for each individual bird was used to generate a total lesion score. Non-vaccinated, challenged birds were split into two pathology categories based upon total lesion score: mild and severe. Birds with low lesion scores were used to represent mild pathology, with an average lesion score of 0.375, and those with high lesion scores were used to represent severe pathology, with an average lesion score of 6.125. Ten treatment groups were generated from vaccinated (V) or non-vaccinated (NV), challenged (C) or non-challenged (NC), day 1 or day 5 necropsy treatments and 2 pathological categories of mild and severe within the non-vaccinated, challenged groups (Fig. 1).
2.3. PBL isolation
PBL were separated from whole blood samples and red blood cells removed. Phosphate buffered saline was combined with approximately 1–3 ml of blood sample to a total volume of 10 ml. A Histopaque 1077/1119 (Sigma Aldrich, St. Louis, MO) discontinuous gradient was created by placing 10 ml of Histopaque 1119 into a 50 ml tube, overlaying 10 ml of Histopaque 1077, then overlaying 4 ml of blood/PBS mixture. The gradient mixture was centrifuged at 700g for 30 min at room temperature. Cell layers were removed from the plasma/Histopaque 1077 interface (mononuclear cells) and from the 1077/1119 interface (heterophils) with a Pasteur pipette, combined and transferred to a 15 ml tube. Cells were washed by adding 6 ml of PBS and centrifuged at 600g for 10 min at room temperature. Supernatant was discarded and the cell pellet washed a second time with PBS. Supernatant was discarded and PBL resuspended in 1.5 ml of RNAlater (AM7021) (Applied Biosystems, Foster City, CA). Cells were refrigerated in RNAlater for 7 day then excess RNAlater was decanted and cells were stored at −80 °C until RNA isolation.
2.4. RNA isolation
RNA samples were isolated using the Ambion MagMax-96 kit for Microarrays (AM1839) (Applied Biosystems, Foster City, CA). Briefly, PBL were added to 0.6 ml of TRI Reagent Solution (Ambion, Austin, TX). Samples were homogenized and split into two 300 μl aliquots, with 1 aliquot further processed and the other held in reserve. Samples were then processed using the Spin Procedure according to manufacturer's instructions. Total RNA was eluted with 30 μl of Elution Buffer and stored at −80 °C. Quality and quantity of RNA were assessed by Nanodrop (Thermo Scientific, West Palm Beach, FL).
2.5. Microarray experiments
The microarray data for this experiment have been deposited in NCBI's Gene Expression Omnibus (GEO) [5,19] database and are accessible through the series accession GSE31387 〈http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE31387〉.
Gene expression was assessed utilizing the global 2-color chicken 44K Agilent Microarray [43]. A total of 40 samples were hybridized to the microarray, using one individual from each of the ten treatment groups from each of 4 independent experimental replications. Samples were arranged in a reference design, using the NV–NC-day 1 sample as the reference for each experimental replicate, on 36 arrays. Within each replicate, the NV–NC-day 1 sample was hybridized to the other 9 treatment groups. Dye assignments were swapped between replicates. Briefly, 400 ng of total RNA was reverse transcribed into cDNA with a T7 promoter region incorporated, then transcribed back into cRNA labeled with either Cy3 or Cy5 dye. Before hybridization, 825 ng of each labeled sample, Cy3 and Cy5, a blocking agent and fragmentation buffer were mixed together and incubated for 30 minutes at 60 °C. Following incubation, gex hybridization buffer was added and samples were hybridized to the microarray slide for 17 h at 65 °C. After hybridization, slides were washed in Agilent Wash Buffer, Acetonitrile, and Stabilization and Drying Buffer (Agilent Technologies, Santa Clara, CA) then scanned using GenePix 4100A scanner and GenePix Pro software (Molecular Devices Inc., Sunnyvale, CA).
The median backgrounds were subtracted from the median Cy3 and Cy5 foreground intensity reads for each spot and were log 2-transformed. Technical control spots and spots exhibiting an average signal to noise ratio of less than 3 over all 36 arrays were excluded from further analysis. Signal to noise ratios were calculated as (median foreground−median background)/background standard deviation for each dye. Locally Weighted Scatterplot Smoothing (LOWESS) procedure was utilized to correct for intensity dependent dye bias [17]. A linear mixed model approach was used to estimate treatment means by fitting the difference of Cy3 and Cy5 normalized signal intensities with each treatment groups' parameterization for each gene as described by Sandford et al. [64]. Only one random effect, experimental replicate, was included in the model, as likelihood ratio testing determined no effect of slide or array position. P values were obtained for contrasts of interest. False discovery rate was controlled by converting P values to q values using the R package q value [67]. Gene ontology analysis of biological processes for significant genes was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) [32,33].
2.6. Quantitative PCR validation
Quantitative real time PCR (qRT-PCR) was performed as described by [59] to confirm microarray results. The following fifteen genes were selected because of significance in the microarray study: clusters of differentiation (CD) 3ε, CD4, CD5, CD28, toll-like receptors (TLR) 7, TLR15, TLR21, heat shock protein 70 (HSP70), P20K, Rab11a, avian beta-defensins (AvBD) 2, AvBD4, AvBD5, AvBD6, and AvBD7. 28S was utilized as a housekeeping gene to normalize for starting concentration of RNA. Primer sequences for CD4, CD5, TLR7, Rab11a, AvBD2, AvBD4, AvBD5, AvBD6, and AvBD7 were designed using sequences from NCBI and PRIMER3 [62]. Primer sequences for 28S, TLR15, and TLR21 have been previously reported (28S [38]; TLR15 [31]; TLR21 [9]). CD3ε, CD28, and P20K were previously utilized [73] but primer sequences were not published. All unpublished primer sequences can be found in Table 1. Each sample was run in three wells. Cycle threshold (CT) values were recorded for each well and each sample triplicate was averaged. Slopes representing reaction efficiency for each gene were generated through amplification of a serial dilution. CT values were adjusted for RNA concentration and reaction efficiency using the formula: 40−[Sample Mean CT Target Gene+(Median 28S for All Samples−Sample Mean 28S)×(Slope Target Gene/Slope 28S)]. Adjusted CT values were analyzed using the Fit Model procedure in JMP software (SAS Institute Inc., Cary, NC). Validation was carried out utilizing RNA extracted from different birds than those included in the microarray analysis representing the same treatment groups and replicates, allowing for both technical and biological replication.
Table 1.
Primers utilized for qRT-PCR validation experiments.
| Gene | Accession | Forward sequence 5′–3′ | Reverse sequence 5′–3′ |
|---|---|---|---|
| CD3ε | Y08917 | CTGCTGTGTGTGGTTGGTG | CGGATCTGGCTTCCATTTTA |
| CD4 | AY528652 | ATACCGTGGAGGAAGCTCAT | TGCCACCTCATACCAGTGAT |
| CD5 | Y12011 | ACAGGAGGCTGATGAAGAGG | CTCTGCTGCTCCTCCACTCT |
| CD28 | X67915 | CAGTCTTTATAATCTACCGGCAAAA | TTGTTCTTCTGGTGAGGTGGA |
| TLR7 | AJ720504 | CGGAAAATGGTACATCATGC | AAAGTTTTGGGAAACCAACG |
| Rab11a | AJ720402 | GCAAAAGCACCATTGGAGTA | GCACCTCGATAGTACGCTGA |
| P20K | M25784 | CTAGGGAGCGGAACTACACG | GTTTGGGAAGCAGCATTCAT |
| HSP70 | NM_001006685 | ATAGGGTGGGAGCCAAGAAC | GGTTTCGGTCAAGCCAACT |
| AvBD2 | AF033336 | TTTCTCCAGGGTTGTCTTCG | AGCAGCTTCCGACTTTGATT |
| AvBD4 | AY621306 | TTTCATCGTGCTCCTCTTTG | CATAGCCCCAGGTAAGCATT |
| AvBD5 | AY621307 | CCACAAGTCATGTCCTCCAG | CATGGAGATGAACGTGAAGG |
| AvBD6 | AY621308 | TGCAGGTCAGCCCTACTTTT | GTCCACTGCCACATGATCC |
| AvBD7 | AY621309 | CTGCTTTCCAGGGATCTGTC | GCCAGAGAAGCCATTTGGTA |
3. Results
3.1. Transcriptome analysis of PBL
Forty individual samples were analyzed by microarray; 1 bird from each of 10 treatment groups, with 4 replications. After removal of spots with a signal to noise ratio less than 3, a total of 24,387 genes were included in the statistical analysis. Effects of treatment were analyzed through contrasts of treatment groups. Contrasts measured tested effect of time for each treatment group, comparisons of each group to control, effect of vaccination, and effect of pathology. Large numbers (>100) of significantly differentially expressed genes (q value<0.05) were detected in 4 contrasts of interest, all involving the NV–C severe groups. The largest number of differentially expressed genes (1914 genes) was found in the contrast of NV–C severe day 5 vs. NV–C mild day 5. The second largest number of differentially expressed genes (1097 genes) was found in the contrast of NV–C severe group vs. NV–NC control group on day 1. This number was reduced to 506 differentially expressed genes for the same contrast on day 5. The number of differentially expressed genes in the contrast of NV–C severe day 1 vs. NV–C severe 5 was 107 genes.
Numbers of shared and unique differentially expressed genes between 3 contrasts analyzing differences in treatment group on the same day are displayed in Fig. 2. A total of 417 differentially expressed genes found in the contrast of NV–C severe day 5 vs. NV–NC control day 5 were also differentially expressed in the NV–C severe day 5 vs. NV–C mild day 5 contrast; 280 genes had increased expressed in the severe group in both contrasts, 137 genes had decreased expressed in the severe group in both contrasts. In all of the shared genes between the contrasts represented in Fig. 2, 99% of genes were expressed in the same direction relative to the severe group; either all more highly expressed in the severe group across all contrasts or more lowly expressed among all contrasts.
Fig. 2.
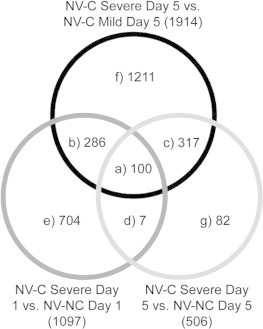
Shared significant genes. Comparison of shared significant genes within the 3 contrasts of NV–C mild day 5 vs. NV–C severe day 5, NV–C severe day 1 vs. NV–NC group day 1, and NV–C severe day 5 vs. NV–NC group day 5. (a) Represents shared genes over all 3 contrasts, (b–d) represent shared genes between 2 contrasts, and (e–g) represent genes unique to a contrast. The numbers in parenthesis represent the total number of significant genes within each contrast.
The differences between internal lesion scores of vaccinated and non-vaccinated, challenged birds were tested through two-sample t-tests for each day. There was a significant (P value<0.001) reduction in internal lesions among vaccinated, challenged birds compared to those non-vaccinated, challenged birds on both days, however, no differentially expressed genes due to vaccination effect were detected. On day 1, the mean±standard deviation of lesion scores for vaccinated birds was 1.50±1.27 (N=76) and unvaccinated birds 3.06±1.68 (N=87). On day 5, the mean±standard deviation of lesion scores for vaccinated birds was 2.94±2.08 (N=80) and unvaccinated birds 4.28±1.94 (N=85). Vaccination effect on gene expression was tested both through the V–NC group vs. NV–NC contrast on both days and through a contrast utilizing a combination of all treatment groups, non-challenged and challenged, on both days.
Samples from individuals with severe pathology or collected 5 day post-infection exhibited more gene induction than repression. Of differentially expressed genes with a minimum fold change of 1.5, 60–80% of genes exhibited induction in the 4 contrasts that include: NV–C severe day 5 vs. NV–C mild day 5, NV–C severe day 1 vs. NV–NC day 1, NV–C severe day 5 vs. NV–NC day 5, NV–C severe day 1 vs. NV–C severe day 5. Many differentially expressed genes showed large fold changes. In the 4 contrasts described, 25–31% of differentially expressed genes had a fold change of 3 or greater.
Heatmaps were generated to characterize patterns of gene expression between similar contrasts by including genes with a minimum q value of 0.05 in any contrast (Figs. 3 and 4). The NV–C mild group on day 5 showed more similarities to the V–NC groups than to the challenged groups (Fig. 3). The remaining challenged groups, both vaccinated and non-vaccinated, exhibited similar expression patterns. The only group with notable expression changes over time was the NV–C severe group (Fig. 4).
Fig. 3.
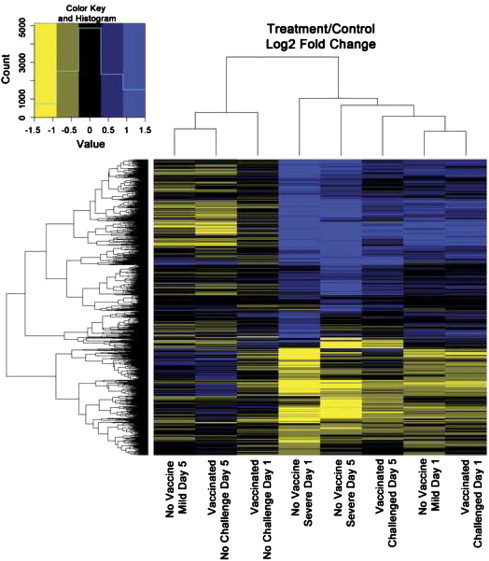
Treatments compared to controls. Heatmap comparison of fold change for each treatment group compared to the day appropriate NV–NC control. Fold change was calculated as log 2 (treatment)−log 2 (control).
Fig. 4.
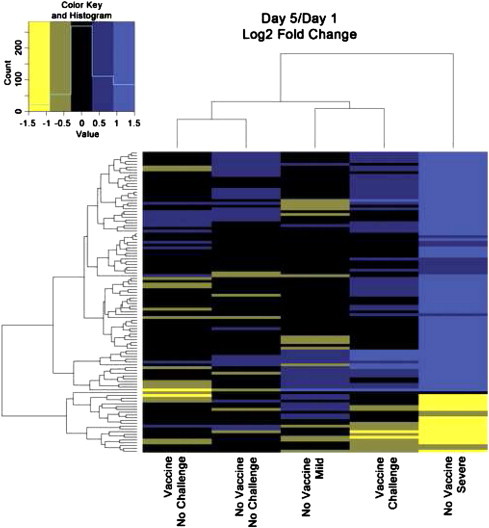
Day 5 compared to day 1. Heatmap comparison of fold change for each day 5 treatment group compared the day 1 treatment group. Fold change was calculated as log 2 (day 5)−log 2 (day 1).
3.2. Gene ontology analysis of significant genes
Gene ontology analysis focused on biological process terms among significant genes. Larger numbers of significantly enriched GO terms were found in contrasts with a higher number of genes with differential expression. Three GO terms related to response (response to stimulus, response to stress, and defense response) were discovered among significantly differentially expressed genes in the contrast of NV–C severe day 1 vs. NV–C severe 5. Among the other 3 contrasts described (NV–C severe day 5 vs. NV–C mild day 5, NV–C severe day 1 vs. NV–NC control day 1, NV–C severe day 5 vs. NV–NC control day 5), a variety of metabolic and biosynthetic processes were common. Prominent within NV–C severe day 5 vs. NV–C mild day 5, were GO terms for signal transduction, immune system processes, ion homeostasis and, surprisingly, several GO terms centered on reproduction. Within NV–C severe vs. NV–NC control, response terms were prominent on day 1, and ion homeostasis and DNA structural terms were prominent on day 5.
GO analysis of unique and shared differentially expressed genes for 3 contrasts (Fig. 2) was carried out; NV–C severe day 5 vs. NV–C mild day 5, NV–C severe day 1 vs. NV–NC control day 1, NV–C severe day 5 vs. NV–NC control day 5. Many of the genes shared among contrasts were related to immune response. These included (a) CD4, tumor necrosis factor receptor, and Rab11a shared by all 3 contrasts; (b) ATPase, CD5, interferon gamma receptor, and toll-like receptor 15 shared by NV–C severe day 1 vs. NV–NC control day 1 and NV–C severe day 5 vs. NV–NC control day 5; (c) ATPase, CD3ε, CD200R1, toll-like receptor 7 shared by NV–C severe day 5 vs. NV–NC control day 5 and NV–C severe day 5 vs. NV–NC control day 5. Avian beta-defensins, CD74 and interleukin-8 were unique to NV–C severe vs. NV–NC control on day 1 (e). Unique to NV–C severe vs. NV–C mild on day 5 (f) were genes related to ion transport and energy (ATPases and ATP synthases), immune response (CD28, CD79b, interleukin 4 receptor, interleukin 10 receptor beta, toll-like receptor 21), and reproduction.
Only two contrasts, NV–C severe day 5 vs. NV–C mild day 5 and NV–C severe day 1 vs. NV–NC control day 1, had significantly enriched KEGG pathways detected by DAVID (P value<0.10, Tables 2 and 3). Many of the pathways exhibited relatedness to defense mechanisms, including lysosome pathway, signaling pathways, apoptosis, and NK cell mediated toxicity.
Table 2.
Effect of severity of pathology at day 5 in non-vaccinated, APEC challenged birds on KEGG pathway enrichment (P value<0.10).
Table 3.
Effect of severe pathology at day 1 compared to non-vaccinated, non-challenged birds on KEGG pathway enrichment (P value<0.10).
| KEGG pathway | Gene count | Accessions |
|---|---|---|
| MAPK signaling pathway gga04010 | 12 | AJ851529, AY033635, CR387146, AJ720776, AJ851546, X68073, BX933236, AJ719540, AF296875, AF167296, U10329, AJ851639 |
| VEGF signaling pathway gga04370 | 6 | AJ851529, AY033635, AJ720776, AJ719540, M64990, U10329 |
| Natural killer cell mediated cytotoxicity gga04650 | 7 | AJ851529, AY033635, AJ720191, AJ719540, AF296875, AY957508, AJ002317 |
| NOD-like receptor signaling pathway gga04621 | 5 | AY033635, AJ720776, BX935364, AY057939, M16199 |
| Apoptosis gga04210 | 6 | AJ720191, BX931997, U26645, AJ719540, AF296875, AY057939 |
| Arachidonic acid metabolism gga00590 | 3 | AJ006405, M64990, U10329 |
| DNA replication gga03030 | 4 | AJ720172, AJ719992, AJ719763, AJ719578 |
The majority of pathways had higher expression in the severe group compared to either mild or control groups, though some, like the cell adhesion molecules pathway and the regulation of actin cytoskeleton, had less expression among severe groups.
3.3. Microarray validation by quantitative real time PCR
Fifteen significant genes from the microarray study were validated through qRT-PCR analysis: CD 3ε, CD4, CD5, CD28, TLR7, TLR15, TLR21, HSP70, P20K, Rab11a, AvBD2, AvBD4, AvBD5, AvBD6, and AvBD7. These genes had significant differential expression in microarray analysis (q value<0.05). Validation was performed on two contrasts of interest, NV–C severe day 5 vs. NV–C mild day 5 for CD 3ε, CD4, CD5, CD28, TLR7, TLR15, TLR21, HSP70, P20K, and Rab11a, and NV–C severe day 1 vs. NV–NC day 1 for AvBD2, AvBD4, AvBD5, AvBD6, and AvBD7. Results show similar trends in direction of fold change for 12 of the 15 genes analyzed; TLR7, TLR15, and Rab11a were expressed in the opposite direction from the microarray, although the results were non-significant.
4. Discussion
Gene expression differences associated with challenge, pathology, or vaccination status, can provide valuable insights into the host response. The largest amount of gene expression differences occurred between pathology categories classified as mild and severe in the NV–C day 5 group. Several prominent receptor types and clusters of differentiation important to immune response and signaling were differentially expressed between the mild and severe groups. The toll-like receptors (TLR) 7, 15, and 21 all exhibited higher expression in the severe group than the mild group. Surprisingly, two of these TLRs showed fold change in the opposite direction in the qRT-PCR results. There are several differences in the technical and statistical approaches between microarray and qRT-PCR. These conflicting results, however, do still indicate an importance of the TLR family with regards to response to infection and illustrate the need for further experimentation. TLRs have shown expression changes due to pathogen challenge in multiple tissues [1,46,59]. In vitro Salmonella stimulation of heterophils from a Salmonella-resistant population of birds revealed higher expression of TLR15 than heterophils from a Salmonella susceptible population [53], consistent with the results of the current qRT-PCR validation. Cultured macrophages have also shown up-regulation of TLR15 when stimulated with E. coli- or Salmonella-derived LPS [11]. It is surprising that in the current study, TLR4 and TLR5 were not among those genes differentially expressed, because they are considered to be the main TLRs for recognizing features of Gram-negative bacteria such as APEC [8]. TLR4 expression differences have been reported in heterophils due to bacterial challenge and genetic background [59]. TLR4 and TLR5 had differential expression in the cecum, TLR4 in the spleen and TLR5 within males in the spleen of Salmonella infected chicks [1].
Interleukin receptors 4 and 10, and interferon gamma (IFNγ) receptor 2 all had higher expression in the severe group than the mild group. Changes in interleukin (IL) receptor genes have been noted in response to pathogen challenge [15], although expression changes in IL4R and IL10R have not been extensively studied. IFNγR2 in macrophages exposed to Salmonella endotoxin in vitro was up-regulated 4 h post-stimulation [12]. Tregs, which control the expression of IL-4 and IFNγ to prevent autoimmunity, have been shown to fail under high antigen dose in vitro [24]. Higher expression of receptors for these pro-inflammatory cytokines may allow for greater downstream signaling triggered by IL-4 and IFNγ, promoting the severe pathology through autoimmunity. Cytokines are commonly produced by PBL after pathogen challenge and have shown differential expression due to bacterial challenge and genetic background in heterophils in vitro [59]. Although several receptors were up-regulated, no cytokine genes were significantly differentially expressed. Leukocytes with increased cytokine receptor levels may more readily receive and process signals, resulting in a variety of pleiotropic effects, even if the cytokine levels are unaltered in the animal.
Clusters of differentiation (CD) are cell surface molecules common to leukocytes that have roles in the immune response. Differential expression of CD molecules in response to Salmonella and Campylobacter infections has been observed in multiple chicken tissues: heterophils [13] and jejunum [65] with Salmonella and ceca with Campylobacter [44]. In the current study, many CD molecules had higher expression among the mild vs. severe pathology group, including CD3ε, CD4, CD5, CD28, CD79b, and CD200R1. CD81 was also differentially expressed, but showed higher expression in the severe pathology (susceptible) group. Previous expression studies have reported higher levels of CD4 among heterophils from Salmonella resistant chickens compared to susceptible lines [13]. This is particularly noteworthy as this difference was noted in non-challenged birds, presenting CD4 expression level as a potential pre-challenge assessment of susceptibility. The current study utilized bacterial challenge to assess pathology and found higher levels of CD4 among birds showing mild pathology (resistant) than in birds demonstrating severe pathology (susceptible). Many CD molecules are associated with or have higher prevalence of specific PBL types (CD3ε among T-cells [26], CD4 among T-cells [40], ggCD200R-B1 among macrophages [71]), suggestive of differences in PBL population composition between mildly and severely affected birds that influence downstream pathology. Higher CD4 expression may be indicative of a higher T-helper population within birds with mild pathology. In humans, CD200R1 acts as a regulator of myeloid cell activation and pro-inflammatory response [52]. Avian CD200R, specifically ggCD200R-B1, has high homology to mammalian CD200R [71]. Lower expression among the severe pathology group would allow an unchecked pro-inflammatory response, leading to greater host damage.
The Ras superfamily can be divided into five major family groups: Ras, Rab, Rho, Ran, and Arf. The superfamily has many roles related to immune response, Ras genes can cause regulatory changes in cell proliferation, differentiation and survival, Rho are Ras homologous proteins with roles in the cell cycle, Rab proteins have roles in vesicle formation and transport and Arf also has roles in vesicle transport [72]. Ten members of the Ras superfamily, Ras, Rab, Rho, and Arf groups, were differentially expressed between pathology groups on day 5, introducing a new family of genes to be explored in greater depth in the role of immune response. Seven had higher expression among the severe group and 3 among the mild group. Ras p21 protein activator 3, Rasa3, which is involved in a signaling pathway for B-cells to avoid pro-apoptotic signals [49], was higher amongst the mild group. Rab11a has been shown to have roles in TLR4 trafficking to phagosome and control interferon regulatory factor-3 in human monocytes [34]. The conflicting result of the qRT-PCR validation in Rab11a, along with the lack of literature on the Ras family in chickens, illustrates the need for more attention to this gene group in the investigation of immune response.
Differences between the NV–C severe group and the NV–NC group were observed on both days. The number of differentially expressed genes in this contrast decreased over time from 1097 genes on day 1 to 506 on day 5. This may be due to the rapid response of PBL to infection. Unique to the day 1 comparison of the NV–C severe and NV–NC group for PBL, genes encoding avian beta-defensins (AvBD) and interleukin 8 were up-regulated. The genes for AvBD2, 4, 5, 6, and 7 were all rapidly up-regulated by APEC infection. The antimicrobial properties of beta-defensins have been well described [70]. AvBD2, 5, 6, and 7 have been found to be expressed in leukocytes [13,70]. TLR agonists, such as LPS, increase AvBD2 in heterophils [39]. Additionally, structural variants in AvBD genes have been associated with response to Salmonella in chickens [30,29], indicating the feasibility of their use in marker-assisted selection to enhance the anti-bacterial response on a population level. The in vitro response of macrophages to Salmonella endotoxin is typified by a significant induction of IL8 at 1, 2, 4, and 8 h post-stimulation [12]. The chemotactic ability of IL8 for other peripheral blood mononuclear cells and heterophils [3] is consistent with a role in early response to infection, as seen here, with IL8 only significantly induced in the day 1 response.
Differential expression was seen among clusters of differentiation genes and in the Rab and Rho family groups on day 1 between the NV–C severe and NV–NC group, similar to the differential expression between mild and severe pathologies at day 5. All differentially expressed CD genes were down-regulated following APEC challenge: CD4, CD5, CD74, CD82, CD83, and CD247. A strain of APEC (APEC17) was previously shown to activate caspase 3/7 in macrophages, inducing apoptosis [6]. APEC O1 in the current study may result in APEC-induced PBL death, shifting the PBL population structure compared to basal (non-challenged) levels. CD247, also known as the T-cell receptor (TCR) ζ-chain, is well conserved between chickens and mammals [25], and is responsible for aiding in assembly of the TCR complex and receptor signaling. In vitro studies of the human ζ-chain have shown degradation by activated caspases [23], indicating a possible mechanism by which APEC could reduce the abilities of T-cells and of the cell-mediated response, resulting in more severe pathology. Among the Rab and Rho genes that were differently expressed, only RhoB was down-regulated in the severe pathology group. Under stress, RhoB inhibits apoptosis and activates NF-κB in rats [42,45], such that decrease of expression in severe pathology would allow greater apoptosis and limit NF-κB activation. Rab11a was again higher in the severe pathology group in this contrast, along with Rab18, 32, and 35.
Fewer significantly differentially expressed genes limited GO analysis and interpretation of the NV–C severe day 5 and NV–NC day 5 group comparison. Similar to other contrasts, three CD groups were significantly differentially expressed. CD3ε, CD4, and CD200R1 showed less expression in the NV–C severe group, suggestive of continued reduction in CD4+ leukocytes, such as T-cells, and in regulators of pro-inflammatory response. Expression patterns within prominent GO groupings for ion homeostasis and cellular developmental processes were inconsistent, with no clear trend of greater expression in one treatment group compared to the other.
Many genes were significantly expressed in more than one contrast (Fig. 2), which is reinforced by the common patterns seen within the treatment/control heatmap (Fig. 3). Similarities between NV–C severe and NV–C mild on day 5 and NV–C severe vs. NV–NC group on day 1 suggest similarities between mild pathology on day 5 and the control groups. This could be the result of a return to homeostasis after a successful defense against APEC. The changes between the severe pathology group and the control non-challenged group over time appear to be driven by the NV–C severe group, as this was the only group to exhibit large changes between day 1 and day 5 (Fig. 4).
Only two contrasts, NV–C severe day 5 vs. NV–C mild day 5 and NV–C severe day 1 vs. NV–NC control day 1, had significantly enriched KEGG pathways, as detected by DAVID (P value<0.10, Tables 2 and 3). Between pathology states on day 5, genes that enriched metabolic pathways were more highly expressed in the severe group, potentially mobilizing more energy to fight infection. The effect of severe status compared to the non-challenged control on day 1 illustrates the importance of signaling pathways during early response to infection.
The lack of a detectible vaccination effect, given the large impact on total lesion scores and the tissue analyzed, is surprising. Vaccination against Newcastle disease virus increases serum antibody titers and can impact T cell populations in the 9 weeks following vaccination [16]. Significant changes in IFNα and IFNγ mRNA expression have been reported in peripheral blood at 1 and 7 day post-vaccination to Marek's disease vaccine in 6–8 week old chickens [57]. These chickens were then sampled only 4 h after challenge to observe differential expression patterns due to vaccination and challenge [57]. Several cytokines showed significant expression changes 1 day post-vaccination with complete Freund's adjuvant [27]. Time of vaccination, time of challenge and time of tissue sampling all impact the observed mRNA expression patterns. Our sampling at 15 and 19 day post-vaccination may have been too late to observe expression changes in PBL due to vaccination alone, and sampling at 1 and 5 day post-infection may have been too late to observe rapid, vaccine-induced effects in response to infection.
Great insights about how specific cell types react to a foreign agent can be gained through targeted in vitro experimentation. A limitation of in vitro experimentation, however, is in knowing how well the information gained from a reduced system will translate to response at the organismal level. The in vivo holistic approach of this experiment allowed the assessment of each animal's gene expression response, not simply an individual cell type. By utilizing samples taken from whole blood, the current study simultaneously detected expression differences due to up-(or down)regulation in specific cell types and the differences caused by changes in the proportion of circulating cell types; that is, the holistic response of the animal. Understanding the systemic nature of complex infections, such as those caused by APEC, requires study of a whole organism's response and multiple tissues, which is better accomplished through an in vivo experiment. This is further highlighted through the differences in information gained from the current experiment on PBL and prior examination of spleen transcriptomes from these same individuals [64].
Peripheral blood leukocytes are comprised of cells with roles in innate/adaptive response and cell/humoral-mediated responses. Transcriptome interrogation of this system reveals which gene expression patterns play an important role in immune response. Additionally, these cells can be collected from live birds without the need to harvest the breeding animal to assay the cellular response. Through this greater understanding of host response, candidate genes to improve genetic resistance to APEC infection can be identified. Applying selection for beneficial genotypes in breeding populations will generate enhanced host responses against APEC infection.
Acknowledgments
This work was supported by National Research Initiative Competitive Grant no. 2008-35604-18805 from the USDA National Institute of Food and Agriculture Microbial Genome Program. ES support provided by USDA National Needs Graduate Fellowship Competitive Grant no. 2007-38420-17767 from the National Institute of Food and Agriculture. MS support National Science Foundation Research Experience for Undergraduates DBI-1062211.
The authors acknowledge the group of researchers (Darrell Trampel, Luke Baldwin, Thomas Denagamage, Christine Fanelli, Ashraf Hussein, Kalinda Kaluarachchi, Ganwu Li, Catherine Logue, Paul Mangiamele, Kelly Tivendale, and Yvonne Wannemuehler) involved in conducting the animal experiments and collecting numerous tissues, in particular Michael Kaiser for peripheral blood collection and Jennifer Cheeseman, Ceren Ciraci and Behnam Abasht for PBL isolation.
References
- 1.Abasht B., Kaiser M.G., Lamont S.J. Toll-like receptor gene expression in cecum and spleen of advanced intercross line chicks infected with Salmonella enterica serovar Enteritidis. Veterinary Immunology and Immunopathology. 2008;123:314–323. doi: 10.1016/j.vetimm.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 2.Ask B., van der Waaij E.H., Stegeman J.A., van Arendonk J.A. Genetic variation among broiler genotypes in susceptibility to colibacillosis. Poultry Science. 2006;85:415–421. doi: 10.1093/ps/85.3.415. [DOI] [PubMed] [Google Scholar]
- 3.Barker K.A., Hampe A., Stoeckle M.Y., Hanafusa H. Transformation-associated cytokine 9E3/CEF4 is chemotactic for chicken peripheral blood mononuclear cells. Journal of Virology. 1993;67:3528–3533. doi: 10.1128/jvi.67.6.3528-3533.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnes H.J., Nolan L.K., Vaillancourt J.P. Colibacillosis. In: Saif Y.M., editor. Diseases of Poultry. twelfth ed. Blackwell Publishing; Ames, Iowa: 2008. pp. 691–732. [Google Scholar]
- 5.Barrett T., Troup D.B., Wilhite S.E., Ledoux R., Rudnev D., Evangelista C. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Research. 2009;37:D5–D15. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bastiani M., Vidotto M.C., Horn F. An avian pathogenic Escherichia coli isolate induces caspase 3/7 activation in J774 macrophages. FEMS Microbiology Letters. 2005;253:133–140. doi: 10.1016/j.femsle.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 7.Bisaillon J.R., Meek A.H., Feltmate T.E. An assessment of condemnations of broiler chicken carcasses. Canadian Journal of Veterinary Research. 1988;52:269–276. [PMC free article] [PubMed] [Google Scholar]
- 8.Brownlie R., Allan B. Avian toll-like receptors. Cell and Tissue Research. 2011;343:121–130. doi: 10.1007/s00441-010-1026-0. [DOI] [PubMed] [Google Scholar]
- 9.Brownlie R., Zhu J., Allan B., Mutwiri G.K., Babiuk L.A., Potter A. Chicken TLR21 acts as a functional homologue to mammalian TLR9 in the recognition of CpG oligodeoxynucleotides. Molecular Immunology. 2009;46:3163–3170. doi: 10.1016/j.molimm.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 10.Cavero D., Schmutz M., Philipp H.C., Preisinger R. Breeding to reduce susceptibility to Escherichia coli in layers. Poultry Science. 2009;88:2063–2068. doi: 10.3382/ps.2009-00168. [DOI] [PubMed] [Google Scholar]
- 11.Ciraci C., Lamont S.J. Avian-specific TLRs and downstream effector responses to CpG-induction in chicken macrophages. Developmental and Comparative Immunology. 2011;35:392–398. doi: 10.1016/j.dci.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 12.Ciraci C., Tuggle C.K., Wannemuehler M.J., Nettleton D., Lamont S.J. Unique genome-wide transcriptome profiles of chicken macrophages exposed to Salmonella-derived endotoxin. BMC Genomics. 2010;11:545. doi: 10.1186/1471-2164-11-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang H.I., Swaggerty C.L., Kogut M.H., Dowd S.E., Li X., Pevzner I.Y. Gene expression profiling in chicken heterophils with Salmonella enteritidis stimulation using a chicken 44K Agilent microarray. BMC Genomics. 2008;9:526. doi: 10.1186/1471-2164-9-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chuammitri P., Ostojić J., Andreasen C.B., Redmond S.B., Lamont S.J., Palić D. Chicken heterophil extracellular traps (HETs): novel defense mechanism of chicken heterophils. Veterinary Immunology and Immunopathology. 2009;129:126–131. doi: 10.1016/j.vetimm.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Crowley T.M., Haring V.R., Moore R. Chicken anemia virus: an understanding of the in-vitro host response over time. Viral Immunology. 2011;24:3–9. doi: 10.1089/vim.2010.0064. [DOI] [PubMed] [Google Scholar]
- 16.Dalgaard T.S., Norup L.R., Pedersen A.R., Handberg K.J., Jørgensen P.H., Juul-Madsen H.R. Flow cytometric assessment of chicken T cell-mediated immune responses after Newcastle disease virus vaccination and challenge. Vaccine. 2010;28:4506–4514. doi: 10.1016/j.vaccine.2010.04.044. [DOI] [PubMed] [Google Scholar]
- 17.Dudoit S., Yang Y.H., Callow M.J., Speed T.P. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Statistica Sinica. 2002;12:111–139. [Google Scholar]
- 18.Dziva F., Stevens M.P. Colibacillosis in poultry: unravelling the molecular basis of virulence of avian pathogenic Escherichia coli in their natural hosts. Avian Pathology. 2008;37:355–366. doi: 10.1080/03079450802216652. [DOI] [PubMed] [Google Scholar]
- 19.Edgar R., Domrachey M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erf G.F. Cell-mediated immunity in poultry. Poultry Science. 2004;83:580–590. doi: 10.1093/ps/83.4.580. [DOI] [PubMed] [Google Scholar]
- 21.Ewers C., Li G., Wilking H., Kieβling S., Alt K., Antáo E.M. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? International Journal of Medical Microbiology. 2007;297:163–176. doi: 10.1016/j.ijmm.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Farnell M.B., Donoghue A.M., de Los Santos F.S., Blore P.J., Hargis B.M., Tellez G. Upregulation of oxidative burst and degranulation in chicken heterophils stimulated with probiotic bacteria. Poultry Science. 2006;85:1900–1906. doi: 10.1093/ps/85.11.1900. [DOI] [PubMed] [Google Scholar]
- 23.Gastman B.R., Johnson D.E., Whiteside T.L., Rabinowich H. Caspase-mediated degradation of T-cell receptor ζ-chain. Cancer Research. 1999;59:1422–1427. [PubMed] [Google Scholar]
- 24.George T.C., Bilsborough J., Viney J.L., Norment A.M. High antigen dose and activated dendritic cells enable Th cells to escape regulatory T cell-mediated suppression in vitro. European Journal of Immunology. 2003;33:502–511. doi: 10.1002/immu.200310026. [DOI] [PubMed] [Google Scholar]
- 25.Göbel T.W., Bolliger L. The chicken TCR ζ-chain restores the function of a mouse T cell hybridoma. Journal of Immunology. 1998;160:1552–1554. [PubMed] [Google Scholar]
- 26.Göbel T.W., Fluri M. Identification and analysis of the chicken CD3ε gene. European Journal of Immunology. 1997;27:194–198. doi: 10.1002/eji.1830270129. [DOI] [PubMed] [Google Scholar]
- 27.Hangalapura B.N., Kaiser M.G., Poel J.J., Parmentier H.K., Lamont S.J. Cold stress equally enhances in vivo pro-inflammatory cytokine gene expression in chicken lines divergently selected for antibody responses. Developmental and Comparative Immunology. 2006;30:503–511. doi: 10.1016/j.dci.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Harmon B.G. Avian heterophils in inflammation and disease resistance. Poultry Science. 1998;77:972–977. doi: 10.1093/ps/77.7.972. [DOI] [PubMed] [Google Scholar]
- 29.Hasenstein J.R., Lamont S.J. Chicken gallinacin gene cluster associated with Salmonella response in advanced intercross line. Avian Diseases. 2007;51:561–567. doi: 10.1637/0005-2086(2007)51[561:CGGCAW]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 30.Hasenstein J.R., Zhang G., Lamont S.J. Analysis of five gallinacin genes and the Salmonella enterica serovar Enteritidis response in poultry. Infection and Immunity. 2006;74:3375–3380. doi: 10.1128/IAI.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgs R., Cormican P., Cahalane S., Allan B., Lloyd A.T., Meade K. Induction of a novel chicken toll-like receptor following Salmonella enterica serovar Typhimurium infection. Infection and Immunity. 2006;74:1692–1698. doi: 10.1128/IAI.74.3.1692-1698.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths towards the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang D.W., Sherman B.T., Lempicki R.A. Systemic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Husebye H., Aune M.H., Stenvik J., Samstad E., Skeldal F., Halaas O. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory facor-3 on phagosomes. Immunity. 2010;33:583–596. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakobsen L., Spangholm D.J., Pedersen K., Jensen L.B., Emborg H.D., Agersø Y. Broiler chickens, broiler chicken meat, pigs and pork as sources of ExPEC related virulence genes and resistance in Escherichia coli isolates from community-dwelling humans and UTI patients. International Journal of Food Microbiology. 2010;142:264–272. doi: 10.1016/j.ijfoodmicro.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 36.Johnson T.J., Siek K.E., Johnson S.J., Nolan L.K. DNA sequence of a ColV plasmid and prevalence of selected plasmid-encoded virulence genes among avian Escherichia coli strains. Journal of Bacteriology. 2006;188:745–758. doi: 10.1128/JB.188.2.745-758.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson T.J., Kariyawasam S., Wannamuehler Y., Mangiamele P., Johnson S.J., Doetkott C. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. Journal of Bacteriology. 2007;189:3228–3236. doi: 10.1128/JB.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaiser P., Rothwell L., Galyov E.E., Barrow P.A., Burnside J., Wigley P. Differential cytokine expression in avian cells in response to invasion by Salmonella typhimurium, Salmonella enteritidis and Salmonella gallinarum. Microbiology. 2000;12:3217–3226. doi: 10.1099/00221287-146-12-3217. [DOI] [PubMed] [Google Scholar]
- 39.Kannan L., Liyanage R., Lay J.O., Jr., Rath N.C. Evaluation of beta defensin 2 production by chicken heterophils using direct MALDI mass spectrometry. Molecular Immunology. 2009;46:3151–3156. doi: 10.1016/j.molimm.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 40.Koskinen R., Salomonsen J., Tregaskes C.A., Young J.R., Goodchild M., Bumstead N. The chicken CD4 gene has remained conserved in evolution. Immunogenetics. 2002;54:520–525. doi: 10.1007/s00251-002-0490-4. [DOI] [PubMed] [Google Scholar]
- 41.Kottom T.J., Nolan L.K., Robinson M., Brown J., Gustad T., Horne S.M. Further characterization of a complement-sensitive mutant of a virulent avian Escherichia coli isolate. Avian Diseases. 1997;41:817–823. [PubMed] [Google Scholar]
- 42.Li G., Tivendale K.A., Liu P., Feng Y., Wannemuehler Y., Cai W. Transcriptome analysis of avian pathogenic Escherichia coli O1 in chicken serum reveals adaptive responses to systemic infection. Infection and Immunity. 2011;79:1951–1960. doi: 10.1128/IAI.01230-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li X., Chiang H.I., Zhu J., Dowd S.E., Zhou H. Characterization of a newly developed chicken 44K Agilent microarray. BMC Genomics. 2008;9:60. doi: 10.1186/1471-2164-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X., Swaggerty C.L., Kogut M.H., Chiang H.I., Wang Y., Genovese K.J. Gene expression profiling of the local cecal response of genetic chicken lines that differ in their susceptibility to Campylobacter jejuni colonization. PLoS One. 2010;5:e11827. doi: 10.1371/journal.pone.0011827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Y.D., Liu Y.P., Cao D.M., Yan Y.M., Hou Y.N., Zhao J.Y. Induction of small G protein RhoB by non-genotoxic stress inhibits apoptosis and activates NF-κB. Journal of Cellular Physiology. 2011;226:729–738. doi: 10.1002/jcp.22394. [DOI] [PubMed] [Google Scholar]
- 46.Lu Y., Sarson A.J., Gong J., Zhou H., Zhu W., Kang Z. Expression profiles of genes in Toll-like receptor-mediated signaling of broiler infected with Clostridium perfringens. Clinical and Vaccine Immunology. 2009;16:1639–1647. doi: 10.1128/CVI.00254-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lutful Kabir S.M. Avian colibacillosis and salmonellosis: a closer look at epideminology, pathogenesis, diagnosis, control and public health concerns. International Journal of Environmental Research and Public Health. 2010;7:89–114. doi: 10.3390/ijerph7010089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lynne A.M., Foley S.L., Nolan L.K. Immune response to recombinant Escherichia coli Iss protein in poultry. Avian Diseases. 2006;50:273–276. doi: 10.1637/7441-092105R.1. [DOI] [PubMed] [Google Scholar]
- 49.Maréchal Y., Pesesse X., Jia Y., Pouillon V., Pérez-Morga D., Daniel J. Inositol 1,3,4,5-tetrakisphosphate controls proapoptotic Bim gene expression and survival in B cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13978–13983. doi: 10.1073/pnas.0704312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mellata M., Dho-Moulin M., Dozois C.M., Curtiss R., III, Lehoux B., Fairbrother J.M. Role of avian pathogenic Escherichia coli virulence factors in bacterial interaction with chicken heterophils and macrophages. Infection and Immunity. 2003;71:494–503. doi: 10.1128/IAI.71.1.494-503.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moulin-Schouleur M., Schouler C., Tailliez P., Kao M.R., Bree A., Germon P. Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. Journal of Clinical Microbiology. 2006;44:3484–3492. doi: 10.1128/JCM.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mukhopadhyay S., Plüddemann A., Hoe J.C., Williams K.J., Varin A., Makepeace K. Immune inhibitory ligand CD200 induction by TLRs and NLRs limits macrophage activation to protect the host from menigococcal septicemia. Cell Host and Microbe. 2010;8:236–247. doi: 10.1016/j.chom.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Nerren J.R., Swaggerty C.L., MacKinnon K.M., Genovese K.J., He H., Pevzner I. Differential mRNA expression of the avian-specific toll-like receptor 15 between heterophils from Salmonella-susceptible and -resistant chickens. Immunogenetics. 2009;61:71–77. doi: 10.1007/s00251-008-0340-0. [DOI] [PubMed] [Google Scholar]
- 54.Nolan L.K., Horne S.M., Giddings C.W., Foley S.L., Johnson T.J., Lynne A.M. Resistance to serum complement, iss, and virulence of avian Escherichia coli. Veterinary Research Communications. 2003;27:101–110. doi: 10.1023/a:1022854902700. [DOI] [PubMed] [Google Scholar]
- 55.Peighambari S.M., Julian R.J., Gyles C.L. Experimental Escherichia coli respiratory infection in broilers. Avian Diseases. 2000;44:759–769. [PubMed] [Google Scholar]
- 56.Pfaff-McDonough S.J., Horne S.M., Giddings C.W., Doetkott J.O., Smith M.H., Nolan L.K. Complement resistance-related traits among Escherichia coli isolates from apparently healthy birds and birds with colibacillosis. Avian Diseases. 2000;44:23–33. [PubMed] [Google Scholar]
- 57.Quéré P., Rivas C., Ester K., Novak R., Ragland W.L. Abundance of IFN-α and IFN-γ mRNA in blood of resistant and susceptible chickens infected with Marek's disease virus (MDV) or vaccinated with turkey herpesvirus; and MDV inhibition of subsequent induction of IFN gene transcription. Archives of Virology. 2005;150:507–519. doi: 10.1007/s00705-004-0435-3. [DOI] [PubMed] [Google Scholar]
- 58.Qureshi M.A. Avian macrophage and immune response: an overview. Poultry Science. 2003;82:691–698. doi: 10.1093/ps/82.5.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Redmond S.B., Chuammitri P., Andreasen C.B., Palić D., Lamont S.J. Chicken heterophils from commercially selected and non-selected genetic lines express cytokines differently after in vitro exposure to Salmonella enteritidis. Veterinary Immunology and Immunopathology. 2009;1132:129–134. doi: 10.1016/j.vetimm.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 60.Rodrigues-Siek K.E., Gidding C.W., Doetkott C., Johnson T.J., Fakhr M.K., Nolan L.K. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology. 2005;151:2097–2110. doi: 10.1099/mic.0.27499-0. [DOI] [PubMed] [Google Scholar]
- 61.Rodrigues-Siek K.E., Gidding C.W., Doetkott C., Johnson T.J., Nolan L.K. Characterizing the APEC pathotype. Veterinary Research. 2005;36:241–256. doi: 10.1051/vetres:2004057. [DOI] [PubMed] [Google Scholar]
- 62.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biology programmers. Methods in Molecular Biology. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 63.Russell S.M. The effect of airsacculitis on bird weights, uniformity, fecal contamination, processing errors, and populations of Campylobacter spp. and Escherichia coli. Poultry Science. 2003;82:1326–1331. doi: 10.1093/ps/82.8.1326. [DOI] [PubMed] [Google Scholar]
- 64.Sandford E.E., Orr M., Balfanz E., Bowerman N., Li X., Zhou H. Spleen transcriptome response to infection with avian pathogenic Escherichia coli in broiler chickens. BMC Genomics. 2011;12:469. doi: 10.1186/1471-2164-12-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schokker D., Smits M.A., Hoekman A.J., Parmentier H.K., Rebel J.M. Effects of Salmonella on spatial–temporal processes of jejunal development in chickens. Developmental and Comparative Immunology. 2010;34:1090–1100. doi: 10.1016/j.dci.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 66.Sharma J.M. Host factors for disease resistance. In: Saif Y.M., editor. Diseases of Poultry. twelfth ed. Blackwell Publishing; Ames, Iowa: 2008. pp. 47–58. [Google Scholar]
- 67.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Swaggerty C.L., Pevzner I.Y., Kaiser P., Kogut M.H. Profiling pro-inflammatory cytokine and chemokine mRNA expression levels as a novel method for selection of increased innate immune responsiveness. Veterinary Immunology and Immunopathology. 2008;126:35–42. doi: 10.1016/j.vetimm.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 69.Tivendale K.A., Logue C.M., Kariyawasam S., Jordan D., Hussein A., Li G. Avian-pathogenic Escherichia coli strains are similar to neonatal meningitis E. coli strains and are able to cause meningitis in the rat model of human disease. Infection and Immunity. 2010;78:3412–3419. doi: 10.1128/IAI.00347-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.van Dijk A., Veldhuizen E.J., Haagsman H.P. Avian defensins. Veterinary Immunology and Immunopathology. 2008;124:1–18. doi: 10.1016/j.vetimm.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Viertlboeck B.C., Hanczaruk M.A., Schmitt F.C., Schmitt R., Göbel T.W. Characterization of the chicken CD200 receptor family. Molecular Immunology. 2008;45:2097–2105. doi: 10.1016/j.molimm.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 72.Wennerberg K., Rossman K.L., Der C.J. The Ras superfamily at a glance. Journal of Cell Science. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 73.Zhou H., Lamont S.J. Global gene expression profile after Salmonella enterica Serovar enteritidis challenge in two F8 advanced intercross chicken lines. Cytogenetic and Genome Research. 2007;117:131–138. doi: 10.1159/000103173. [DOI] [PubMed] [Google Scholar]


