Graphical abstract
Highlights
► Description of the RNA editing process in trypanosomatids. ► Editosome: the multiprotein complex that carries RNA editing. ► Virtual screening for inhibitors against editosome proteins. ► High-throughput screening for inhibitors of editosome activity. ► Current inhibitors of trypanosomatid RNA editing.
Keywords: Trypanosomatids, RNA editing inhibitor, Virtual screening, High-throughput screening
Abstract
The related trypanosomatid pathogens, Trypanosoma brucei spp., Trypanosoma cruzi and Leishmania spp. cause devastating diseases in humans and animals and continue to pose a major challenge in drug development. Mitochondrial RNA editing, catalyzed by multi-protein complexes known as editosomes, has provided an opportunity for development of efficient and specific chemotherapeutic targets against trypanosomatid pathogens. This review will discuss both methods for discovery of RNA editing inhibitors, as well as inhibitors against the T. brucei editosome that were recently discovered through creative virtual and high throughput screening methods. In addition, the use of these inhibitors as agents that can block or perturb one or more steps of the RNA editing process will be discussed. These inhibitors can potentially be used to study the dynamic processing and assembly of the editosome proteins. A thorough understanding of the mechanisms and specificities of these new inhibitors is needed in order to contribute to both the functional studies of an essential gene expression mechanism and to the possibility of future drug development against the trypanosomatid pathogens.
1. Introduction
The three major trypanosomatid pathogens, Trypanosoma brucei, Trypanosoma cruzi and Leishmania major are related parasitic protozoa of kinetoplastids, and cause different diseases, including Human African trypanosomiasis, Chagas disease, and Leishmaniasis, respectively. Leishmaniasis reportedly has a mortality rate of 50,000 individuals per year and annual loss of 2.1 million disability-adjusted life years (DALYs), followed by sleeping sickness with 48,000 deaths per year (1.5 million DALYs per year) (WHO, 2002). Chagas disease also causes 15,000 deaths and loss of 700,000 DALYs annually. The available drugs for these diseases are not ideal, since they are toxic, costly, and have invasive routes of administration (Croft et al., 2005; Delespaux and de Koning, 2007; Stuart et al., 2008; Teixeira et al., 2006). Also, resistance against many of these drugs has already emerged; hence there is an urgent need for development of new drugs (Buckner et al., 1998; den Boer and Davidson, 2006; Olliaro et al., 2005; Wilkinson et al., 2008).
Since Rob Benne discovered RNA editing in 1986 (Benne et al., 1986), we have come to learn many details about the mechanisms and major players involved in this remarkable post-transcriptional RNA maturation process, including its potential as an effective anti-trypanosomal drug target. Most mitochondrial mRNAs in kinetoplastids undergo RNA editing to produce mature and functional mRNAs that are translated into multiple essential components of the mitochondrial oxidative phosphorylation system (Hannaert et al., 2003; Madison-Antenucci et al., 2002). The insect stage, procyclic form (PF) T. brucei has a highly active mitochondrion (Vickerman, 1965) and generates ATP by oxidative phosphorylation, and hence RNA editing is crucial for its energy metabolism. Although oxidative phosphorylation is repressed in bloodstream form (BF), RNA-editing is nevertheless required to maintain an active mitochondrion, which is needed for other essential metabolic pathways such as calcium homeostasis and fatty acid metabolism, (Hashimi et al., 2010; Schnaufer et al., 2005). Encouragingly, several proteins involved in editing process were found to be essential for the growth and survival of the BF (Baldassarre et al., 2003; Carnes et al., 2005; Deng et al., 2005; Drozdz et al., 2002; Guo et al., 2008; Law et al., 2008; O’Hearn et al., 2003; Schnaufer et al., 2001; Trotter et al., 2005). Therefore, editing is an essential process in both life stages. This observation suggests the important possibility that the RNA editing process may provide similar and efficient chemotherapeutic targets throughout the medically important trypanosomatid parasites. Furthermore, this type of RNA editing is unique to these parasites; hence, targeting this process should have a few or no side effects in the human host.
Here we will review the recent advances in development of new methods to discover potential inhibitors of RNA editing in T. brucei. We will also highlight how recently reported inhibitors of RNA editing may contribute to both the functional studies of this essential RNA processing mechanism and to the exciting possibility of future drug development against the three related trypanosomatid pathogens.
2. RNA editing process
In vitro studies using mitochondrial extract from T. brucei (Seiwert et al., 1996) indicate that RNA editing is mediated by a series of coordinated catalytic steps of multi-protein complexes known as editosomes, which insert and delete uridylates (Us) as specified by guide RNAs (gRNAs) (Aphasizhev and Aphasizheva, 2011; Blum et al., 1990; Osato et al., 2009; Simpson et al., 2003, 2000; Sturm and Simpson, 1990). The initial event in the editing reaction is hybridization of the 5′ end of the gRNA to the precursor mRNA (pre-edited mRNA) just downstream of the first editing site, to form an anchor duplex (Blum et al., 1990). The catalytic cascade of RNA editing occurs by endonucleolytic cleavage of the precursor mRNA at the editing site, U addition by 3′-terminal uridylyl transferase (TUTase) activity or removal by 3′ exouridylylase activity (exoUase) both at the 3′ end of the 5′ cleavage fragment, and subsequent ligation of the RNA fragments by RNA ligase activity, reviewed in (Hajduk and Ochsenreiter, 2010; Stuart et al., 1997) (Fig. 1). In addition, the editosome contains RNA editing-associated RNA helicase activity, which may affect gRNA/mRNA interactions and/or displace gRNA after its use (Missel et al., 1997).
Fig. 1.
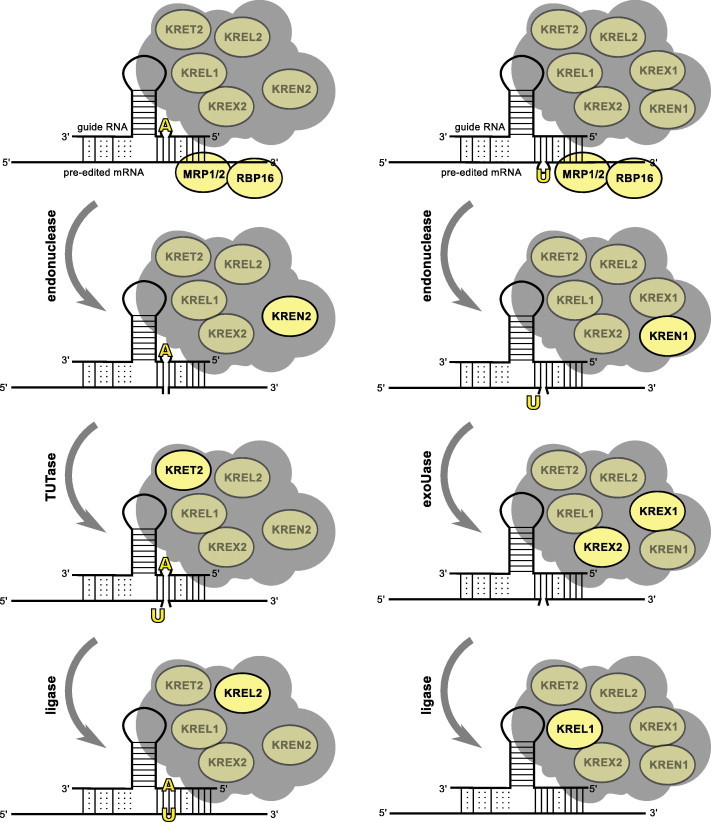
Schematic presentation of RNA editing reactions. RNA editing entails maturation of the mitochondrial mRNAs by insertion (left panel) and deletion (right panel) of uridylates (Us) as specified by small guide RNAs (gRNAs). The specific proteins involved in each catalytic reaction are highlighted.
RNA editing appears to involve dynamic interactions of multiple proteins and complexes that are not directly part of the core editosome, but indirectly regulate the efficiency of editing and editosome stability. For example, the MRP complex, which consists of MRP1 and MRP2 proteins, has a matchmaking type of RNA annealing activity (Muller and Goringer, 2002; Schumacher et al., 2006; Zikova et al., 2008). RBP16 also plays a role in gRNA/pre-mRNA interaction and has been shown to have an overlapping function with MRP1 and MRP2 proteins (Ammerman et al., 2008; Fisk et al., 2009). The mitochondrial RNA binding complex 1 (MRB1) or the guide RNA binding complex (GRBC), which share a number of common protein components and likely represent the same particle, have recently been described to play a role in diverse aspects of mitochondrial RNA metabolism such as gRNA utilization and expression and/or stability of gRNAs as well as edited and pre-edited mRNAs (Acestor et al., 2009; Ammerman et al., 2008, 2011, 2010; Hashimi et al., 2009, 2008; Hernandez et al., 2010; Panigrahi et al., 2008; Weng et al., 2008).
3. The editosome composition
The functional editosome proteins were identified by several methods including mass spectrometric analysis of editosomes that were purified by serial ion exchange and gel permeation chromatography, immunoaffinity chromatography specific to editosome proteins, or tandem affinity purification (TAP) based on tagged RNA editing proteins (Aphasizhev et al., 2003a; Panigrahi et al., 2006, 2001a,b, 2003; Rusche et al., 1997). Several nomenclatures have been proposed in literature to describe the editosome proteins (Panigrahi et al., 2001a; Peris et al., 1997; Rusche et al., 1997; Simpson et al., 2010; Stuart et al., 2005). Here we will use the nomenclature proposed by Stuart et al., with Kinetoplastid RNA editing (KRE) as prefix followed by abbreviation for its function if experimentally known (Stuart et al., 2005). The letter K will be replaced with species’ abbreviation when specifically discussed throughout this article. Proteins that catalyze exoUase (KREX1 and KREX2) (Ernst et al., 2009; Kang et al., 2005; Rogers et al., 2007), TUTase (KRET2) (Aphasizhev et al., 2003c; Ernst et al., 2003), ligase (KREL1 and KREL2) (McManus et al., 2001; Rusche et al., 2001; Schnaufer et al., 2001), endonuclease (KREN1, KREN2, and KREN3) (Carnes et al., 2005, 2008; Trotter et al., 2005), and helicase (KREH1) (Li et al., 2009; Missel et al., 1997) activities of editing have been identified and functionally characterized (Fig. 1). The number of proteins in the fully functional editosome is not known; however, the most recent studies have identified around 20 proteins (Table 1) as highly dynamic components of three complexes, all with a common set of core proteins, but each associated with a different endonuclease (KREN1–3) (Carnes et al., 2011, 2008; Panigrahi et al., 2006; Trotter et al., 2005). These complexes sediment at ∼ 20 Svedberg (20S) on glycerol gradients and hence are known as ‘20S editosome’ (Carnes et al., 2008; Panigrahi et al., 2006). These editosomes may represent a “catalytic core complex” and additional proteins may contribute to RNA editing in vivo. Multi-subunit complexes comprised of separate insertion and deletion activities each sedimenting at ∼5–10S have also been proposed which may contribute to the assembly of the entire system (Schnaufer et al., 2003). Whether editosomes contain catalytic or structural RNA is unknown.
Table 1.
Biological importance and biochemical functions of editosome proteins – alternative nomenclatures are also mentioned (e.g. TbMP63, T. brucei mitochondrial protein of 63 kDa).
| Protein | Proposed function | Essentiality | Technique used for inhibition | Effects on RNA editing activities and the editosome complex | References |
|---|---|---|---|---|---|
| KREPA1/TbMP81 | RNA/protein interaction | BF and PF | RNAi-mediated repression |
|
Drozdz et al. (2002) O’Hearn et al. (2003) |
| KREPA2/TbMP63 | RNA/protein interaction | PF | RNAi-mediated repression |
|
Huang et al. (2002) |
| KREPA3/TbMP42 | RNA/protein interaction | BF and PF | Regulatable knockout in BF RNAi-mediated repression in PF |
|
Brecht et al. (2005), Law et al. (2008), Guo et al. (2008), Guo et al. (2010) |
| KREPA4/TbMP24 | RNA/protein interaction | PF | RNAi-mediated repression |
|
Kala and Salavati (2010), Salavati et al. (2006) |
| KREPA5/TbMP19 | RNA/protein interaction | N.D. | N.D. | Carnes et al. (2011), Panigrahi et al. (2006) | |
| KREPA6/TbMP18 | RNA/protein interaction | PF | RNAi-mediated repression |
|
Tarun et al. (2008) |
| KREN1/TbMP90 | U deletion-specific endonuclease | BF and PF | RNAi-mediated repression in PF Regulatable knockout in BF |
|
Kang et al. (2006), Trotter et al. (2005) |
| KREN3/TbMP67 | Endonuclease specific for Cytochrome oxidase II pre-edited mRNA | BF | Regulatable knockout |
|
Carnes et al. (2008) |
| KREN2/TbMP61 | Insertion-specific endonuclease activity | BF and PF | RNAi-mediated repression in PF Regulatable knockout in BF |
|
Carnes et al. (2005) |
| KREPB4/TbMP46 | RNA/protein interaction | PF | RNAi-mediated repression |
|
Babbarwal et al. (2007) |
| KREPB5/TbMP44 | RNA/protein interaction | BF | Regulatable knockout |
|
Baldassarre et al. (2003) |
| KREPB6/TbMP49 | RNA/protein interaction | N.D. | N.D. | Panigrahi et al. (2006), Carnes et al. (2011) | |
| KREPB7/TbMP47 | RNA/protein interaction | N.D. | N.D. | Panigrahi et al. (2006), Carnes et al. (2011) | |
| KREPB8/TbMP41 | RNA/protein interaction | N.D. | N.D. | Panigrahi et al. (2006), Carnes et al. (2011) | |
| KREL1/TbMP52 | Deletion-specific Ligase | BF | Regulatable knockout |
|
Schnaufer et al. (2001), Cruz-Reyes et al. (2002) |
| KREL2/TbMP48 | Insertion-specific Ligase | Not essential in BF and PF | RNAi-mediated repression |
|
Drozdz et al. (2002), Gao and Simpson (2003), Cruz-Reyes et al. (2002) |
| KREX1/TbMP100 | 3′–5′ exonuclease responsible for U-deletion | PF | RNAi-mediated repression |
|
Kang et al. (2005), Rogers et al. (2007), Ernst et al. (2009) |
| KREX2/TbMP99 | 3′–5′ exonuclease activity | PF (Minor effect on cell growth) | RNAi-mediated repression |
|
Rogers et al. (2007), Ernst et al. (2009) |
| KRET1 | Guide RNA-specific TUTase | PF | RNAi-mediated repression |
|
Aphasizhev et al. (2002), Ernst et al. (2003) |
| KRET2/TbMP57 | U insertion-specific TUTase | PF | RNAi-mediated repression |
|
Aphasizhev et al. (2003c), Ernst et al. (2003) |
| KREH1/mHel61p | Helicase activity | PF | Knockout |
|
Missel et al. (1997), Li et al. (2011) |
N.D.: not determined.
The editosome proteins are related in pairs or sets by sequence and motif similarities which reflect their functions with predicted catalytic and/or RNA interaction motifs (Worthey et al., 2003). For instance, the two editosome RNA ligases are similar (41% identity) and several lines of evidence have suggested that KREL1 is involved in U-deletion editing and KREL2 is involved in U-insertion editing, leading to the proposed structural and functional division of the editosome model into insertion and deletion sub-complexes (Schnaufer et al., 2003). KREPA1–6 are six additional editosome proteins which share a C-terminal motif associated with an oligonucleotide-binding (OB) fold with varying degrees of relatedness (Worthey et al., 2003). The largest three of these five proteins also have C2H2 zinc finger motifs, suggesting possible RNA/protein-binding functions. The specific functions of these six proteins are not yet known; however, recent data propose extensive protein–protein interactions that are mediated by OB-fold proteins, coordinating the editosome function (Schnaufer et al., 2010).
4. Editosome as drug target
Gene inactivation studies of RNA editing ligase 1 in T. brucei (TbREL1) showed for the first time that RNA editing is essential in BF (Schnaufer et al., 2001). Subsequent RNAi mediated repression and gene knockout studies of several functionally characterized editosome proteins has shown that RNA editing is essential in both life cycle stages of the parasite (Table 1). It should be mentioned that there are “dyskinetoplastic” trypanosomes that have partially lost their kinetoplast (Brun et al., 1998), in which the RNA editing process has been suggested to be dispensable. For example, it has been shown that in the dyskinetoplastic Trypanosoma evansi, although editosome proteins are expressed and even imported into the mitochondrion, their function is not essential (Paris et al., 2011). This loss of essentiality has been hypothesized to be the result of a compensatory mutation in a nuclear-encoded subunit of the mitochondrial ATP synthase (Lai et al., 2008; Schnaufer et al., 2005, 2002). Therefore, drugs targeting RNA editing machinery will likely not work against dyskinetoplastic parasites of livestock, such as T. evansi and T. equipedrum. However, as mentioned before, RNA editing is essential in all known trypanosomatid parasites of human, including the closely related T. brucei, T. cruzi and Leishmania spp. Therefore, RNA editing is a viable drug target, indicating the need for development of methods for discovery of new chemical compounds that can inhibit this process. In the next sections, we will discuss such methods and the compounds that have been discovered.
5. High-throughput screening (HTS) methods: virtual and chemical
In this review, we have chosen to focus our attention on the discovery and development of innovative therapeutic agents that target RNA editing. Target-driven approaches can identify chemicals that specifically affect a certain biological process with minimal off-targets effects (Ilag et al., 2002; Westwell and Stevens, 2004). Identification of validated and druggable biological targets with essential roles in the cell is the first step towards target-driven drug discovery. The target can be a specific protein with an essential function, such as TbREL1. Alternatively, an essential protein complex or biological process can be considered as the target, such as the whole editosome. In this case, the drug discovery process is unbiased towards a specific protein, and the chances of identifying an effective compound are higher. The second step consists of a “molecule hunt” which involves HTS of libraries of small-molecule compounds in order to identify a hit. Modern approaches such as combinatorial chemistry, structural biology and computational analysis can be leveraged to increase the chances of success in this step and minimize the required time. The next step is known as hit-to-lead development, with the aim of identifying a lead compound with drug-like properties that is active in the cellular context. Lastly, the lead development step will test and optimize the pharmacokinetic and pharmacodynamic properties of the lead compounds in vivo, towards the identification of a clinical candidate. In this section, we will focus on recent developments on virtual and chemical screening of libraries towards identification of chemical compounds that target the editosome.
5.1. Virtual screening methods
Virtual screening emerged as an important tool for the discovery of drug-like compounds against a specific target (Kuntz, 1992; Maryanoff, 2004; Shoichet, 2004; Shoichet et al., 2002; Varney et al., 1992; von Itzstein et al., 1993). Currently, various protocols are available for virtual screening of chemical compound databases. Overall, virtual screening contains the following two steps: (i) rapid docking algorithms are used in order to identify the most likely position and orientation of the available candidate compounds within the active site of protein of interest; (ii) the compound activity is ranked by analyzing the steric and electrostatic properties of the predicted protein–ligand complex (Reddy et al., 2007). This approach has been used recently (Amaro et al., 2008; Durrant et al., 2010; Moshiri et al., 2011) in order to find potential drug-like compounds against TbREL1, one of the essential enzymes of the editosome whose N-terminal high-resolution crystal structure is available (Deng et al., 2004). Moreover, there are no close human homologs; hence, TbREL1 can be used as an ideal chemotherapeutic target for designing selective inhibitors that block the essential RNA ligase function.
The ligation mechanism of TbREL1 is similar to that of DNA ligases, although their structures are different. The ligation reaction follows three steps (Fig. 2): (i) the adenylylation step, in which the conserved catalytic lysine attacks α-phosphate of ATP and releases pyrophosphate. At this step, an enzyme-AMP intermediate is formed through the phosphoamide linkage; (ii) The deadenylylation step, in which TbREL1-AMP can recognize the double-stranded nicked mRNA/gRNA and transfer its bound AMP to the 5′-phosphate of the RNA molecule, forming an adenylylated RNA with a 5′,5′-phosphoanhydride bond; (iii) The ligation step, in which the free 3′-hydroxyl of the 5′ fragment attacks the phosphoanhydride bond of the adenylylated 3′ RNA fragment at the nick site, leading to the formation of phosphodiester bond and the release of AMP.
Fig. 2.
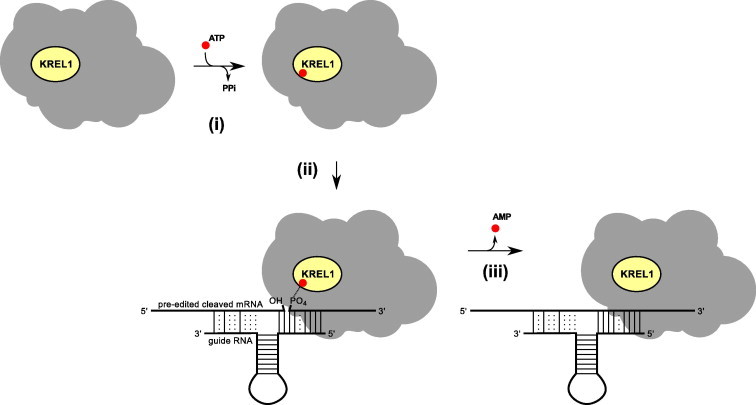
The schematic presentation of the three-step ligation mechanism of RNA editing ligase 1.
Recent studies have performed virtual screening of various compounds against high-resolution structure of TbREL1 in order to identify compounds that can inhibit the adenylylation step by competing with ATP for the active site. In the first study, Amaro et al. (2008) initially used structure-based virtual screening of a library of compounds from National Cancer Institute (NCI) to identify the top compounds with the highest binding affinity. This resulted in a set of drug-like candidate compounds, named V1 to V8. Since these compounds were predicted to bind to the ATP-binding pocket of TbREL1, their effect on TbREL1 adenylylation was tested experimentally; V1 and V4 showed the highest inhibition of the adenylylation step among the candidates. Amaro et al. then used a ligand-based virtual screening, in which they searched for compounds with the highest similarity to V1. They came up with six more compounds that were predicted to have very high affinity for TbREL1 ATP-binding pocket, named S1 to S6, among which S5 was shown experimentally to have the highest inhibitory effect on TbREL1 (Table 2). These drugs were reported to have an in vitro effect on the adenylylation step, although they did not show any phenotypic effect on the cultured T. brucei cells, most probably because the cells were not permeable to these compounds. Furthermore, their inhibitory effect on TbREL1 and the closely related T4 RNA ligase 2 in comparison to human DNA ligase III suggests selectivity for RNA ligases. Later, in another study, Durrant et al. performed virtual screening against TbREL1 using a library of compounds that were similar to the core naphthalene scaffold of previously found TbREL1 inhibitors, resulting in four novel compounds named V1 to V4 (Durrant et al., 2010). Among these compounds, V2 and V4 were shown to inhibit the adenylylation step most efficiently, and V4 was able to kill the parasite as well. Simultaneously, Moshiri et al., performed structure-based virtual screening against TbREL1 (Moshiri et al., 2011), resulting in 12 potential inhibitors of TbREL1 adenylylation, of which one was shown experimentally to inhibit full-round RNA editing reaction. This compound, named C35, turned out to be the same as the compound V2 found by Durrant et al. while Moshiri et al. performed the docking on a single protein conformation based on the available crystal structure of TbREL1, Durrant et al. used the “relaxed complex scheme”, in which the compounds were docked on an ensemble of 30 protein conformations obtained from molecular dynamic simulations.
Table 2.
The most potent inhibitors of RNA editing and their possible mode of action.
| ID/source | Structure | IC50 (μM) based on adenylylation activity | IC50 (μM) based on full-round RNA editing activity | Potential mode of inhibition | References |
|---|---|---|---|---|---|
| V4, Sigma Mordant Black 25 | 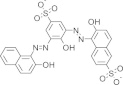 |
1.59 ± 1.10 | N.D. | Adenylylation | Durrant et al. (2010) |
| C35/V2, NSC162535 | 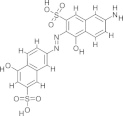 |
1.53 ± 1.17 | N.D. | Deadenylylation/adenylylation/RNA–protein interaction | Durrant et al. (2010), Moshiri et al. (2011) |
| V1, NSC45609 | 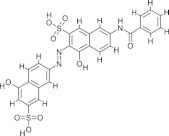 |
2.16 ± 1.20 | N.D. | Adenylylation | Durrant et al. (2010) |
| V3, NSC1698 | 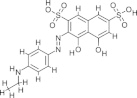 |
8.36 ± 1.71 | N.D. | Adenylylation | Durrant et al. (2010) |
| S5, NSC16209 | 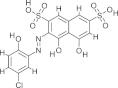 |
1.01 ± 0.16 | 2.1 ± 0.4 | Deadenylylation/adenylylation/RNA–protein interaction | Amaro et al. (2008), Moshiri et al. (2011), Moshiri and Salavati (2010) |
| S1, NSC100234 | 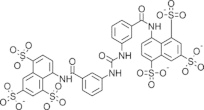 |
1.95 ± 0.61 | N.D. | Adenylylation | Amaro et al. (2008) |
| NF023 | 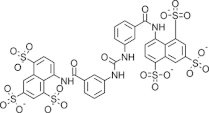 |
N.D. | 2.9 ± 0.7 | RNA editing before or at the endonuclease cleavage step/global effect on editing complex | Liang and Connell (2010) |
| Mitoxantrone | 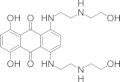 |
N.D. | 2.6 ± 0.5 | RNA editing before or at the endonuclease cleavage step/global effect on editing complex | Liang and Connell (2010) |
| D-Sphingosine | N.D. | 2.0 ± 0.1 | RNA editing after the endonuclease cleavage step/global effect on editing complex | Liang and Connell (2010) | |
| GW5074 | 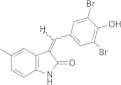 |
N.D. | 2.9 ± 0.4 | RNA editing before or at the endonuclease cleavage step | Liang and Connell (2010) |
| Protoporphyrin IX | 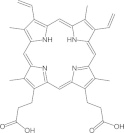 |
N.D. | 0.9 ± 0.1 | RNA editing before or at the endonuclease cleavage step | Liang and Connell (2010) |
N.D.: not determined.
According to these studies, the predicted binding mode of C35 (V2) (Durrant et al., 2010; Moshiri et al., 2011) is as follow: theoretically, the interaction of this compound with TbREL1 mimics certain aspects of ATP binding. Similar to the adenine moiety of ATP, one of the naphthalene rings of C35 forms a π–π stacking interaction with Phe209. Also, similar to the triphosphate tail of ATP, a sulfonate group forms an ion pair with the guanidine group of Arg111. Aminonaphthyl group forms a water-mediated interaction with Arg288 similar to the N1 atom of the adenine moiety in ATP. While this inhibitor is predicted to bind in the deep binding pocket of the active site, the study by Durrant et al. revealed a novel predicted drug binding site on the periphery of the TbREL1 active site, which can potentially be harnessed to design specific inhibitors, given the low similarity of this region with its counterpart in human DNA ligase.
Drug discovery relies on experimental validation for identification of compounds with real inhibitory effect on the target. In the next section, we will discuss methods for validation of computationally identified chemicals, as well as for direct discovery of inhibitory compounds from chemical compound libraries.
5.2. Chemical HTS methods
In order to design a successful high-throughput screening assay, several factors should be taken into consideration. For example, sensitivity, specificity and reproducibility of the assay [collectively measured by an important statistical parameter called the Z-factor (Zhang et al., 1999)], manageability of the compound library, and suitability of reagents for large-scale screening are among the most important factors in order to develop a successful HTS assay. Two types of HTS assays can be developed; homogeneous assays and heterogeneous assay. Homogeneous assays are simpler and include straightforward steps such as addition of reagents, incubation, and reading, whereas heterogeneous assays are more cumbersome due to their requirement for extra steps such as several washing steps, filtration, and centrifugation. Hence, an ideal HTS assay should be homogenous, inexpensive, with minimal requirement for complex instrumentation.
Recently, two HTS assays were designed and developed for screening of different chemical libraries against the editosome (Fig. 3) (Liang and Connell, 2009; Moshiri and Salavati, 2010). Liang et al. developed a heterogeneous RNA aptamer-based assay in which RNA editing reaction can be monitored using electrochemiluminescent signal. In this study, upon successful U-insertion RNA editing, the RNA aptamer undergoes a conformational change that activates its streptavidin-binding region, resulting in a measurable electrochemiluminescent signal. In another study by Moshiri et al., a simple ‘mix and measure’ homogeneous HTS assay based on fluorescence resonance energy transfer (FRET) has been developed. This assay uses hammerhead ribozyme as an RNA editing substrate. Upon successful U-deletion RNA editing, the inactive ribozyme becomes active and is able to cleave a 16-nucleotide FRET substrate that carries a fluorescent reporter (FAM) on the 5′ end and a quencher (TAMRA) on the 3′ end, resulting in a detectable signal. While these two methods monitor the activity of the whole editosome in the context of U-deletion and U-insertion RNA editing, novel methods are being developed that can specifically monitor the ligase activity of TbREL1 (Hall and Schnaufer, Kinetoplastid Molecular Cell Biology meeting, 2011). The availability of these different methods provides the opportunity to find inhibitors against a wide range of RNA editing-related activities. In the next section, we will describe the recently found inhibitors of RNA editing based on these assays as well as virtual screening.
Fig. 3.
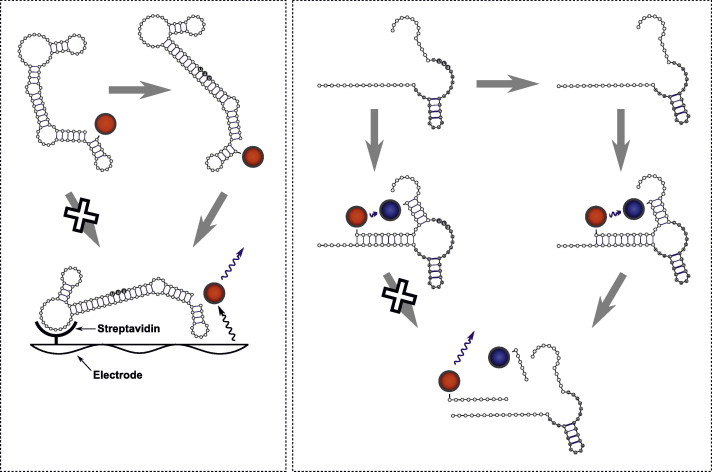
RNA editing assays that are developed for high-throughput screening – Left panel illustrates the RNA aptamer-based assay. In this assay, an aptamer that is labeled with ruthenium (red ball) is used, which upon successful addition of three Us changes conformation and, becomes activated, and thus binding to the streptavidin that coats the microtiter plate. Upon electrical stimulation, the ruthenium complex generates a measurable ECL signal. Right panel illustrates the FRET-based assay, in which the 16nt-long reporter RNA is labeled with a fluorescent reporter (FAM, red ball) and a fluorescent quencher (TAMRA, blue ball). In this assay, upon successful deletion of three Us from the catalytic core of HHR, the inactive HHR becomes active and cleaves the reporter RNA and generates a detectable fluorescent signal. (For interpretation of reference to colors in this figure legend, the reader is referred to the web version of this article.)
6. Editosome inhibitors
Liang et al. used their aptamer-based assay to screen a library of pharmacologically active compounds (LOPAC) against RNA editing machinery, and found 28 hits with strong inhibitory effect on RNA editing (Liang and Connell, 2010). By performing secondary assays, two of the compounds were found to perturb endonuclease activity at the initial steps of RNA editing process (Table 2). While Liang et al. used the whole editosome as the initial target for drug discovery, the Amaro group as well as the Salavati group used the essential TbREL1 as a specific target to screen several chemical compounds for inhibitors of RNA editing. The Amaro group identified several naphthalene-based compounds through virtual screening, and reported inhibitory effect of these compounds on the adenylylation step of ligation in vitro using recombinant TbREL1 (Table 2). Two of these compounds, namely S5 and C35 (V2), were later tested by the Salavati group on whole editosome, and showed strong inhibition of U-deletion RNA editing (Moshiri et al., 2011). Secondary assays confirmed that this compound could inhibit the adenylylation of TbREL1 at very low editosome concentrations, whereas at higher editosome concentration, which is required for obtaining full-round RNA editing, the deadenylylation of TbREL1 was inhibited, but not adenylylation. Moshiri et al. thus suggested that although these compounds are able to inhibit adenylylation of TbREL1, this might not be the mechanism through which the drug exerts its effect in the functional biological context (Moshiri et al., 2011). Based on the observation that these compounds inhibited all editing steps that required RNA–editosome interaction, Moshiri et al. suggested that C35 and S5 inhibit the interaction of the editosome with its substrate RNA, and supported their claim by directly examining the effect of these compounds on RNA–editosome interaction. Consistent with these observations, C35 addition could also alter the sedimentation profile of the 20S editosomes which most likely results from direct perturbation of RNA–protein interactions and the editosome assembly and/or integrity. These observations highlight the potential use of such compounds and their importance in the better understanding of TbREL1-inhibitor interaction and editosome assembly. Taken together, these naphthalene-based compounds appear to find additional targets other than the anticipated ATP-binding pocket of TbREL1 in the context of partially purified functional editosomes. These studies raise a few essential questions with respect to naphthalene-based inhibitors-TbREL1 interaction; do the inhibitors bind similar or distinct targets? Do the interactions of inhibitors affect TbREL1 sub-complex assembly and/or stability, or do they block recruitment of the editing substrates to the editosome? Studying the mechanistic details of inhibition of TbREL1 activity by these compounds will not only provide valuable insights to the editosome assembly, but will be instrumental in determining the mode of action and designing more effective inhibitors of the editosome.
7. Conclusion and prospects
Although recent studies have provided much insight on the structure of editosome (Golas et al., 2009; Li et al., 2009), the mechanism and sequence of editosome assembly is largely unknown. Acting as an editosome subunit, a gRNA–protein complex may bring gRNA to the editosome; however, this has yet to be confirmed (McManus et al., 2000). In several studies, it has been shown that treatment of editosome complex with nucleases results in disassembly of complex and loss of editing activities, suggesting a role for RNA to maintain the editosome complex integrity (Aphasizhev et al., 2003b; Salavati et al., 2002). On the other hand, it has been reported that mutants that lack mitochondrial DNA and, hence, lack mitochondrial mRNA and gRNA contain catalytically active editosomes (Domingo et al., 2003). These data suggest mitochondrial encoded RNA may not be required for assembly of functional editosomes. Therefore, it is not known whether RNA editing is catalyzed by an editosome that is formed by the association of multi-subunit complexes upon RNA binding, similar to the ribosome, or it is rather with the RNAs in a stepwise manner and with discrete intermediates, similar to the spliceosome (Hoskins et al., 2011). Asides from their use as chemotherapeutic agents, small compounds have provided a wealth of information regarding the conformational dynamics of ribosome. For instance, paromomycin, puromycin, erythromycin, and viomycin, have been quite useful in helping to understand ribosomal function and assembly, its recognition of tRNAs, and translation by trapping the ribosome in an intermediate state (Douthwaite et al., 1985; Ermolenko et al., 2007; Moazed and Noller, 1989; Yoshizawa et al., 1999). Similarly, compounds identified against the spliceosome have allowed a deeper understanding of the assembly of the functional spliceosome by blocking various stages of assembly with some possessing antitumor activities (Albert et al., 2007; Kaida et al., 2007; Kotake et al., 2007; O’Brien et al., 2008). We anticipate that the chemical compounds generated from the high throughput screening against the editosome will block or otherwise affect one or more steps in the editing cycle. These steps include potentially various stages of the editosome assembly, leading to the initiation of the catalytic steps, editing of multiple sites with the use of multiple gRNAs, and a termination step. Due to multi-subunit nature of the editosome which may involve assembly of such units, we may find the chemical genetics approach more advantageous compared to RNAi or gene replacement strategies, interfering with the editosome function and assembly by direct editosome binding. The characteristics of the alterations will indicate likely functions for specific components of the editosome proteins and their order of assembly. Thus, development and characterization of editosome inhibitors will not only provide opportunities for possible therapeutics, but also tools for studying the details of editosome assembly and function.
References
- Acestor N., Panigrahi A.K., Carnes J., Zikova A., Stuart K.D. The MRB1 complex functions in kinetoplastid RNA processing. RNA. 2009;15:277–286. doi: 10.1261/rna.1353209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert B.J., Sivaramakrishnan A., Naka T., Czaicki N.L., Koide K. Total syntheses, fragmentation studies, and antitumor/antiproliferative activities of FR901464 and its low picomolar analogue. J. Am. Chem. Soc. 2007;129:2648–2659. doi: 10.1021/ja067870m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro R.E., Schnaufer A., Interthal H., Hol W., Stuart K.D., McCammon J.A. Discovery of drug-like inhibitors of an essential RNA-editing ligase in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 2008;105:17278–17283. doi: 10.1073/pnas.0805820105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman M.L., Fisk J.C., Read L.K. GRNA/pre-mRNA annealing and RNA chaperone activities of RBP16. RNA. 2008;14:1069–1080. doi: 10.1261/rna.982908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman M.L., Hashimi H., Novotna L., Cicova Z., McEvoy S.M., Lukes J., Read L.K. MRB3010 is a core component of the MRB1 complex that facilitates an early step of the kinetoplastid RNA editing process. RNA. 2011;17:865–877. doi: 10.1261/rna.2446311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman M.L., Presnyak V., Fisk J.C., Foda B.M., Read L.K. TbRGG2 facilitates kinetoplastid RNA editing initiation and progression past intrinsic pause sites. RNA. 2010;16:2239–2251. doi: 10.1261/rna.2285510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R., Aphasizheva I. Uridine insertion/deletion editing in trypanosomes: a playground for RNA-guided information transfer. Wiley Interdiscip. Rev. RNA. 2011;2:669–685. doi: 10.1002/wrna.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R., Aphasizheva I., Nelson R.E., Gao G., Simpson A.M., Kang X., Falick A.M., Sbicego S., Simpson L. Isolation of a U-insertion/deletion editing complex from Leishmania tarentolae mitochondria. EMBO J. 2003;22:913–924. doi: 10.1093/emboj/cdg083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R., Aphasizheva I., Nelson R.E., Simpson L. A 100-kDa complex of two RNA-binding proteins from mitochondria of Leishmania tarentolae catalyzes RNA annealing and interacts with several RNA editing components. RNA. 2003;9:62–76. doi: 10.1261/rna.2134303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R., Aphasizheva I., Simpson L. A tale of two TUTases. Proc. Natl. Acad. Sci. USA. 2003;100:10617–10622. doi: 10.1073/pnas.1833120100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R., Sbicego S., Peris M., Jang S.H., Aphasizheva I., Simpson A.M., Rivlin A., Simpson L. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–648. doi: 10.1016/s0092-8674(02)00647-5. [DOI] [PubMed] [Google Scholar]
- Babbarwal V.K., Fleck M., Ernst N.L., Schnaufer A., Stuart K. An essential role of KREPB4 in RNA editing and structural integrity of the editosome in Trypanosoma brucei. RNA. 2007;13:737–744. doi: 10.1261/rna.327707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassarre H., Keefer C., Wang B., Lazaris A., Karatzas C.N. Nuclear transfer in goats using in vitro matured oocytes recovered by laparoscopic ovum pick-up. Clon. Stem Cells. 2003;5:279–285. doi: 10.1089/153623003772032781. [DOI] [PubMed] [Google Scholar]
- Benne R., Van den Burg J., Brakenhoff J.P., Sloof P., Van Boom J.H., Tromp M.C. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Blum B., Bakalara N., Simpson L. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell. 1990;60:189–198. doi: 10.1016/0092-8674(90)90735-w. [DOI] [PubMed] [Google Scholar]
- Brecht M., Niemann M., Schluter E., Muller U.F., Stuart K., Goringer H.U. TbMP42, a protein component of the RNA editing complex in African trypanosomes, has endo-exoribonuclease activity. Mol. Cell. 2005;17:621–630. doi: 10.1016/j.molcel.2005.01.018. [DOI] [PubMed] [Google Scholar]
- Brun R., Hecker H., Lun Z.R. Trypanosoma evansi and T. equiperdum: distribution, biology, treatment and phylogenetic relationship (a review) Vet. Parasitol. 1998;79:95–107. doi: 10.1016/s0304-4017(98)00146-0. [DOI] [PubMed] [Google Scholar]
- Buckner F.S., Wilson A.J., White T.C., Van Voorhis W.C. Induction of resistance to azole drugs in Trypanosoma cruzi. Antimicrob. Agents Chemother. 1998;42:3245–3250. doi: 10.1128/aac.42.12.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J., Soares C.Z., Wickham C., Stuart K. Endonuclease associations with three distinct editosomes in Trypanosoma brucei. J. Biol. Chem. 2011;286:19320–19330. doi: 10.1074/jbc.M111.228965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J., Trotter J.R., Ernst N.L., Steinberg A., Stuart K. An essential RNase III insertion editing endonuclease in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA. 2005;102:16614–16619. doi: 10.1073/pnas.0506133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnes J., Trotter J.R., Peltan A., Fleck M., Stuart K. RNA editing in Trypanosoma brucei requires three different editosomes. Mol. Cell. Biol. 2008;28:122–130. doi: 10.1128/MCB.01374-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft S.L., Barrett M.P., Urbina J.A. Chemotherapy of trypanosomiases and leishmaniasis. Trends Parasitol. 2005;21:508–512. doi: 10.1016/j.pt.2005.08.026. [DOI] [PubMed] [Google Scholar]
- Cruz-Reyes J., Zhelonkina A.G., Huang C.E., Sollner-Webb B. Distinct functions of two RNA ligases in active Trypanosoma brucei RNA editing complexes. Mol. Cell. Biol. 2002;22:4652–4660. doi: 10.1128/MCB.22.13.4652-4660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delespaux V., de Koning H.P. Drugs and drug resistance in African trypanosomiasis. Drug Resist. Updat. 2007;10:30–50. doi: 10.1016/j.drup.2007.02.004. [DOI] [PubMed] [Google Scholar]
- den Boer M., Davidson R.N. Treatment options for visceral leishmaniasis. Expert Rev. Anti. Infect. Ther. 2006;4:187–197. doi: 10.1586/14787210.4.2.187. [DOI] [PubMed] [Google Scholar]
- Deng J., Ernst N.L., Turley S., Stuart K.D., Hol W.G. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J., Schnaufer A., Salavati R., Stuart K.D., Hol W.G. High resolution crystal structure of a key editosome enzyme from Trypanosoma brucei: RNA editing ligase 1. J. Mol. Biol. 2004;343:601–613. doi: 10.1016/j.jmb.2004.08.041. [DOI] [PubMed] [Google Scholar]
- Domingo G.J., Palazzo S.S., Wang B., Pannicucci B., Salavati R., Stuart K.D. Dyskinetoplastic Trypanosoma brucei contains functional editing complexes. Eukaryot. Cell. 2003;2:569–577. doi: 10.1128/EC.2.3.569-577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douthwaite S., Prince J.B., Noller H.F. Evidence for functional interaction between domains II and V of 23S ribosomal RNA from an erythromycin-resistant mutant. Proc. Natl. Acad. Sci. USA. 1985;82:8330–8334. doi: 10.1073/pnas.82.24.8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdz M., Palazzo S.S., Salavati R., O’Rear J., Clayton C., Stuart K. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 2002;21:1791–1799. doi: 10.1093/emboj/21.7.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant J.D., Hall L., Swift R.V., Landon M., Schnaufer A., Amaro R.E. Novel naphthalene-based inhibitors of Trypanosoma brucei RNA editing ligase 1. PLoS Negl. Trop. Dis. 2010;4:e803. doi: 10.1371/journal.pntd.0000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolenko D.N., Spiegel P.C., Majumdar Z.K., Hickerson R.P., Clegg R.M., Noller H.F. The antibiotic viomycin traps the ribosome in an intermediate state of translocation. Nat. Struct. Mol. Biol. 2007;14:493–497. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- Ernst N.L., Panicucci B., Carnes J., Stuart K. Differential functions of two editosome exoUases in Trypanosoma brucei. RNA. 2009;15:947–957. doi: 10.1261/rna.1373009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst N.L., Panicucci B., Igo R.P., Jr., Panigrahi A.K., Salavati R., Stuart K. TbMP57 is a 3′ terminal uridylyl transferase (TUTase) of the Trypanosoma brucei editosome. Mol. Cell. 2003;11:1525–1536. doi: 10.1016/s1097-2765(03)00185-0. [DOI] [PubMed] [Google Scholar]
- Fisk J.C., Presnyak V., Ammerman M.L., Read L.K. Distinct and overlapping functions of MRP1/2 and RBP16 in mitochondrial RNA metabolism. Mol. Cell. Biol. 2009;29:5214–5225. doi: 10.1128/MCB.00520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao G., Simpson L. Is the Trypanosoma brucei REL1 RNA ligase specific for U-deletion RNA editing, and is the REL2 RNA ligase specific for U-insertion editing? J. Biol. Chem. 2003;278:27570–27574. doi: 10.1074/jbc.M303317200. [DOI] [PubMed] [Google Scholar]
- Golas M.M., Bohm C., Sander B., Effenberger K., Brecht M., Stark H., Goringer H.U. Snapshots of the RNA editing machine in trypanosomes captured at different assembly stages in vivo. EMBO J. 2009;28:766–778. doi: 10.1038/emboj.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Ernst N.L., Carnes J., Stuart K.D. The zinc-fingers of KREPA3 are essential for the complete editing of mitochondrial mRNAs in Trypanosoma brucei. PLoS One. 2010;5:e8913. doi: 10.1371/journal.pone.0008913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Ernst N.L., Stuart K.D. The KREPA3 zinc finger motifs and OB-fold domain are essential for RNA editing and survival of Trypanosoma brucei. Mol. Cell. Biol. 2008;28:6939–6953. doi: 10.1128/MCB.01115-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajduk S., Ochsenreiter T. RNA editing in kinetoplastids. RNA Biol. 2010;7:229–236. doi: 10.4161/rna.7.2.11393. [DOI] [PubMed] [Google Scholar]
- Hannaert, V., Bringaud, F., Opperdoes, F.R., Michels, P.A., 2003. Evolution of energy metabolism and its compartmentation in Kinetoplastida. Kinetoplastid biology and disease 2, 11. [DOI] [PMC free article] [PubMed]
- Hashimi H., Benkovicova V., Cermakova P., Lai D.H., Horvath A., Lukes J. The assembly of F(1)F(O)-ATP synthase is disrupted upon interference of RNA editing in Trypanosoma brucei. Int. J. Parasitol. 2010;40:45–54. doi: 10.1016/j.ijpara.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Hashimi H., Cicova Z., Novotna L., Wen Y.Z., Lukes J. Kinetoplastid guide RNA biogenesis is dependent on subunits of the mitochondrial RNA binding complex 1 and mitochondrial RNA polymerase. RNA. 2009;15:588–599. doi: 10.1261/rna.1411809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimi H., Zikova A., Panigrahi A.K., Stuart K.D., Lukes J. TbRGG1, an essential protein involved in kinetoplastid RNA metabolism that is associated with a novel multiprotein complex. RNA. 2008;14:970–980. doi: 10.1261/rna.888808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez A., Madina B.R., Ro K., Wohlschlegel J.A., Willard B., Kinter M.T., Cruz-Reyes J. REH2 RNA helicase in kinetoplastid mitochondria: ribonucleoprotein complexes and essential motifs for unwinding and guide RNA (gRNA) binding. J. Biol. Chem. 2010;285:1220–1228. doi: 10.1074/jbc.M109.051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins A.A., Friedman L.J., Gallagher S.S., Crawford D.J., Anderson E.G., Wombacher R., Ramirez N., Cornish V.W., Gelles J., Moore M.J. Ordered and dynamic assembly of single spliceosomes. Science. 2011;331:1289–1295. doi: 10.1126/science.1198830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.E., O’Hearn S.F., Sollner-Webb B. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 2002;22:3194–3203. doi: 10.1128/MCB.22.9.3194-3203.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilag L.L., Ng J.H., Beste G., Henning S.W. Emerging high-throughput drug target validation technologies. Drug Discov. Today. 2002;7:S136–142. doi: 10.1016/s1359-6446(02)02429-7. [DOI] [PubMed] [Google Scholar]
- Kaida D., Motoyoshi H., Tashiro E., Nojima T., Hagiwara M., Ishigami K., Watanabe H., Kitahara T., Yoshida T., Nakajima H., Tani T., Horinouchi S., Yoshida M. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 2007;3:576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- Kala S., Salavati R. OB-fold domain of KREPA4 mediates high-affinity interaction with guide RNA and possesses annealing activity. RNA. 2010;16:1951–1967. doi: 10.1261/rna.2124610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Gao G., Rogers K., Falick A.M., Zhou S., Simpson L. Reconstitution of full-round uridine-deletion RNA editing with three recombinant proteins. Proc. Natl. Acad. Sci. USA. 2006;103:13944–13949. doi: 10.1073/pnas.0604476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X., Rogers K., Gao G., Falick A.M., Zhou S., Simpson L. Reconstitution of uridine-deletion precleaved RNA editing with two recombinant enzymes. Proc. Natl. Acad. Sci. USA. 2005;102:1017–1022. doi: 10.1073/pnas.0409275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotake Y., Sagane K., Owa T., Mimori-Kiyosue Y., Shimizu H., Uesugi M., Ishihama Y., Iwata M., Mizui Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
- Kuntz I.D. Structure-based strategies for drug design and discovery. Science. 1992;257:1078–1082. doi: 10.1126/science.257.5073.1078. [DOI] [PubMed] [Google Scholar]
- Lai D.H., Hashimi H., Lun Z.R., Ayala F.J., Lukes J. Adaptations of Trypanosoma brucei to gradual loss of kinetoplast DNA: Trypanosoma equiperdum and Trypanosoma evansi are petite mutants of T. brucei. Proc. Natl. Acad. Sci. USA. 2008;105:1999–2004. doi: 10.1073/pnas.0711799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., O’Hearn S.F., Sollner-Webb B. Trypanosoma brucei RNA editing protein TbMP42 (band VI) is crucial for the endonucleolytic cleavages but not the subsequent steps of U-deletion and U-insertion. RNA. 2008;14:1187–1200. doi: 10.1261/rna.899508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Ge P., Hui W.H., Atanasov I., Rogers K., Guo Q., Osato D., Falick A.M., Zhou Z.H., Simpson L. Structure of the core editing complex (L-complex) involved in uridine insertion/deletion RNA editing in trypanosomatid mitochondria. Proc. Natl. Acad. Sci. USA. 2009;106:12306–12310. doi: 10.1073/pnas.0901754106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Herrera J., Zhou S., Maslov D.A., Simpson L. Trypanosome REH1 is an RNA helicase involved with the 3′–5′ polarity of multiple gRNA-guided uridine insertion/deletion RNA editing. Proc. Natl. Acad. Sci. USA. 2011;108:3542–3547. doi: 10.1073/pnas.1014152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S., Connell G.J. An electrochemiluminescent aptamer switch for a high-throughput assay of an RNA editing reaction. RNA. 2009;15:1929–1938. doi: 10.1261/rna.1720209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S., Connell G.J. Identification of specific inhibitors for a trypanosomatid RNA editing reaction. RNA. 2010;16:2435–2441. doi: 10.1261/rna.2347310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison-Antenucci S., Grams J., Hajduk S.L. Editing machines: the complexities of trypanosome RNA editing. Cell. 2002;108:435–438. doi: 10.1016/s0092-8674(02)00653-0. [DOI] [PubMed] [Google Scholar]
- Maryanoff B.E. Inhibitors of serine proteases as potential therapeutic agents: the road from thrombin to tryptase to cathepsin G. J. Med. Chem. 2004;47:769–787. doi: 10.1021/jm030493t. [DOI] [PubMed] [Google Scholar]
- McManus M.T., Adler B.K., Pollard V.W., Hajduk S.L. Trypanosoma brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Mol. Cell. Biol. 2000;20:883–891. doi: 10.1128/mcb.20.3.883-891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus M.T., Shimamura M., Grams J., Hajduk S.L. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA. 2001;7:167–175. doi: 10.1017/s1355838201002072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missel A., Souza A.E., Norskau G., Goringer H.U. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 1997;17:4895–4903. doi: 10.1128/mcb.17.9.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Noller H.F. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–148. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- Moshiri H., Acoca S., Kala S., Najafabadi H.S., Hogues H., Purisima E., Salavati R. Naphthalene-based RNA editing inhibitor blocks RNA editing activities and editosome assembly in Trypanosoma brucei. J. Biol. Chem. 2011;286:14178–14189. doi: 10.1074/jbc.M110.199646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moshiri H., Salavati R. A fluorescence-based reporter substrate for monitoring RNA editing in trypanosomatid pathogens. Nucleic Acids Res. 2010;38:e138. doi: 10.1093/nar/gkq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U.F., Goringer H.U. Mechanism of the gBP21-mediated RNA/RNA annealing reaction: matchmaking and charge reduction. Nucleic Acids Res. 2002;30:447–455. doi: 10.1093/nar/30.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K., Matlin A.J., Lowell A.M., Moore M.J. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J. Biol. Chem. 2008;283:33147–33154. doi: 10.1074/jbc.M805556200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hearn S.F., Huang C.E., Hemann M., Zhelonkina A., Sollner-Webb B. Trypanosoma brucei RNA editing complex: band II is structurally critical and maintains band V ligase, which is nonessential. Mol. Cell. Biol. 2003;23:7909–7919. doi: 10.1128/MCB.23.21.7909-7919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olliaro P.L., Guerin P.J., Gerstl S., Haaskjold A.A., Rottingen J.A., Sundar S. Treatment options for visceral leishmaniasis: a systematic review of clinical studies done in India, 1980–2004. Lancet Infect. Dis. 2005;5:763–774. doi: 10.1016/S1473-3099(05)70296-6. [DOI] [PubMed] [Google Scholar]
- Osato D., Rogers K., Guo Q., Li F., Richmond G., Klug F., Simpson L. Uridine insertion/deletion RNA editing in trypanosomatid mitochondria: in search of the editosome. RNA. 2009;15:1338–1344. doi: 10.1261/rna.1642809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Ernst N.L., Domingo G.J., Fleck M., Salavati R., Stuart K.D. Compositionally and functionally distinct editosomes in Trypanosoma brucei. RNA. 2006;12:1038–1049. doi: 10.1261/rna.45506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Gygi S.P., Ernst N.L., Igo R.P., Jr., Palazzo S.S., Schnaufer A., Weston D.S., Carmean N., Salavati R., Aebersold R., Stuart K.D. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001;21:380–389. doi: 10.1128/MCB.21.2.380-389.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Schnaufer A., Carmean N., Igo R.P., Jr., Gygi S.P., Ernst N.L., Palazzo S.S., Weston D.S., Aebersold R., Salavati R., Stuart K.D. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 2001;21:6833–6840. doi: 10.1128/MCB.21.20.6833-6840.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Schnaufer A., Ernst N.L., Wang B., Carmean N., Salavati R., Stuart K. Identification of novel components of Trypanosoma brucei editosomes. RNA. 2003;9:484–492. doi: 10.1261/rna.2194603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi A.K., Zikova A., Dalley R.A., Acestor N., Ogata Y., Anupama A., Myler P.J., Stuart K.D. Mitochondrial complexes in Trypanosoma brucei: a novel complex and a unique oxidoreductase complex. Mol. Cell. Proteomics. 2008;7:534–545. doi: 10.1074/mcp.M700430-MCP200. [DOI] [PubMed] [Google Scholar]
- Paris Z., Hashimi H., Lun S., Alfonzo J.D., Lukes J. Futile import of tRNAs and proteins into the mitochondrion of Trypanosoma brucei evansi. Mol. Biochem. Parasitol. 2011;176:116–120. doi: 10.1016/j.molbiopara.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peris M., Simpson A.M., Grunstein J., Liliental J.E., Frech G.C., Simpson L. Native gel analysis of ribonucleoprotein complexes from a Leishmania tarentolae mitochondrial extract. Mol. Biochem. Parasitol. 1997;85:9–24. doi: 10.1016/s0166-6851(96)02795-8. [DOI] [PubMed] [Google Scholar]
- Reddy A.S., Pati S.P., Kumar P.P., Pradeep H.N., Sastry G.N. Virtual screening in drug discovery – a computational perspective. Curr. Protein Pept. Sci. 2007;8:329–351. doi: 10.2174/138920307781369427. [DOI] [PubMed] [Google Scholar]
- Rogers K., Gao G., Simpson L. Uridylate-specific 3′–5′-exoribonucleases involved in uridylate-deletion RNA editing in trypanosomatid mitochondria. J. Biol. Chem. 2007;282:29073–29080. doi: 10.1074/jbc.M704551200. [DOI] [PubMed] [Google Scholar]
- Rusche L.N., Cruz-Reyes J., Piller K.J., Sollner-Webb B. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 1997;16:4069–4081. doi: 10.1093/emboj/16.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche L.N., Huang C.E., Piller K.J., Hemann M., Wirtz E., Sollner-Webb B. The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol. 2001;21:979–989. doi: 10.1128/MCB.21.4.979-989.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salavati R., Ernst N.L., O’Rear J., Gilliam T., Tarun S., Jr., Stuart K. KREPA4, an RNA binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2006;12:819–831. doi: 10.1261/rna.2244106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salavati R., Panigrahi A.K., Morach B.A., Palazzo S.S., Igo R.P., Stuart K. Endoribonuclease activities of Trypanosoma brucei mitochondria. Mol. Biochem. Parasitol. 2002;120:23–31. doi: 10.1016/s0166-6851(01)00431-5. [DOI] [PubMed] [Google Scholar]
- Schnaufer A., Clark-Walker G.D., Steinberg A.G., Stuart K. The F1-ATP synthase complex in bloodstream stage trypanosomes has an unusual and essential function. EMBO J. 2005;24:4029–4040. doi: 10.1038/sj.emboj.7600862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaufer A., Domingo G.J., Stuart K. Natural and induced dyskinetoplastic trypanosomatids: how to live without mitochondrial DNA. Int. J. Parasitol. 2002;32:1071–1084. doi: 10.1016/s0020-7519(02)00020-6. [DOI] [PubMed] [Google Scholar]
- Schnaufer A., Ernst N.L., Palazzo S.S., O’Rear J., Salavati R., Stuart K. Separate insertion and deletion subcomplexes of the Trypanosoma brucei RNA editing complex. Mol. Cell. 2003;12:307–319. doi: 10.1016/s1097-2765(03)00286-7. [DOI] [PubMed] [Google Scholar]
- Schnaufer A., Panigrahi A.K., Panicucci B., Igo R.P., Jr., Wirtz E., Salavati R., Stuart K. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science. 2001;291:2159–2162. doi: 10.1126/science.1058955. [DOI] [PubMed] [Google Scholar]
- Schnaufer A., Wu M., Park Y.J., Nakai T., Deng J., Proff R., Hol W.G., Stuart K.D. A protein-protein interaction map of trypanosome 20S editosomes. J. Biol. Chem. 2010;285:5282–5295. doi: 10.1074/jbc.M109.059378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher M.A., Karamooz E., Zikova A., Trantirek L., Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–711. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- Seiwert S.D., Heidmann S., Stuart K. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell. 1996;84:831–841. doi: 10.1016/s0092-8674(00)81062-4. [DOI] [PubMed] [Google Scholar]
- Shoichet B.K. Virtual screening of chemical libraries. Nature. 2004;432:862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoichet B.K., McGovern S.L., Wei B., Irwin J.J. Lead discovery using molecular docking. Curr. Opin. Chem. Biol. 2002;6:439–446. doi: 10.1016/s1367-5931(02)00339-3. [DOI] [PubMed] [Google Scholar]
- Simpson L., Aphasizhev R., Lukes J., Cruz-Reyes J. Guide to the nomenclature of kinetoplastid RNA editing: a proposal. Protist. 2010;161:2–6. doi: 10.1016/j.protis.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L., Sbicego S., Aphasizhev R. Uridine insertion/deletion RNA editing in trypanosome mitochondria: a complex business. RNA. 2003;9:265–276. doi: 10.1261/rna.2178403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson L., Thiemann O.H., Savill N.J., Alfonzo J.D., Maslov D.A. Evolution of RNA editing in trypanosome mitochondria. Proc. Natl. Acad. Sci. USA. 2000;97:6986–6993. doi: 10.1073/pnas.97.13.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K., Allen T.E., Heidmann S., Seiwert S.D. RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev. 1997;61:105–120. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K., Brun R., Croft S., Fairlamb A., Gurtler R.E., McKerrow J., Reed S., Tarleton R. Kinetoplastids: related protozoan pathogens, different diseases. J. Clin. Invest. 2008;118:1301–1310. doi: 10.1172/JCI33945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart K.D., Schnaufer A., Ernst N.L., Panigrahi A.K. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Sturm N.R., Simpson L. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell. 1990;61:879–884. doi: 10.1016/0092-8674(90)90198-n. [DOI] [PubMed] [Google Scholar]
- Tarun S.Z., Jr., Schnaufer A., Ernst N.L., Proff R., Deng J., Hol W., Stuart K. KREPA6 is an RNA-binding protein essential for editosome integrity and survival of Trypanosoma brucei. RNA. 2008;14:347–358. doi: 10.1261/rna.763308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira A.R., Nitz N., Guimaro M.C., Gomes C., Santos-Buch C.A. Chagas disease. Postgrad. Med. J. 2006;82:788–798. doi: 10.1136/pgmj.2006.047357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter J.R., Ernst N.L., Carnes J., Panicucci B., Stuart K. A deletion site editing endonuclease in Trypanosoma brucei. Mol. Cell. 2005;20:403–412. doi: 10.1016/j.molcel.2005.09.016. [DOI] [PubMed] [Google Scholar]
- Varney M.D., Marzoni G.P., Palmer C.L., Deal J.G., Webber S., Welsh K.M., Bacquet R.J., Bartlett C.A., Morse C.A., Booth C.L. Crystal-structure-based design and synthesis of benz[cd]indole-containing inhibitors of thymidylate synthase. J. Med. Chem. 1992;35:663–676. doi: 10.1021/jm00082a006. [DOI] [PubMed] [Google Scholar]
- Vickerman K. Polymorphism and mitochondrial activity in sleeping sickness trypanosomes. Nature. 1965;208:762–766. doi: 10.1038/208762a0. [DOI] [PubMed] [Google Scholar]
- von Itzstein M., Wu W.Y., Kok G.B., Pegg M.S., Dyason J.C., Jin B., Van Phan T., Smythe M.L., White H.F., Oliver S.W. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature. 1993;363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- Weng J., Aphasizheva I., Etheridge R.D., Huang L., Wang X., Falick A.M., Aphasizhev R. Guide RNA-binding complex from mitochondria of trypanosomatids. Mol. Cell. 2008;32:198–209. doi: 10.1016/j.molcel.2008.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwell A.D., Stevens M.F. Hitting the chemotherapy jackpot: strategy, productivity and chemistry. Drug. Discov. Today. 2004;9:625–627. doi: 10.1016/S1359-6446(04)03162-9. [DOI] [PubMed] [Google Scholar]
- WHO, 2002. WHO – Global Burden of Disease Estimates.
- Wilkinson S.R., Taylor M.C., Horn D., Kelly J.M., Cheeseman I. A mechanism for cross-resistance to nifurtimox and benznidazole in trypanosomes. Proc. Natl. Acad. Sci. USA. 2008;105:5022–5027. doi: 10.1073/pnas.0711014105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthey E.A., Schnaufer A., Mian I.S., Stuart K., Salavati R. Comparative analysis of editosome proteins in trypanosomatids. Nucleic Acids Res. 2003;31:6392–6408. doi: 10.1093/nar/gkg870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa S., Fourmy D., Puglisi J.D. Recognition of the codon–anticodon helix by ribosomal RNA. Science. 1999;285:1722–1725. doi: 10.1126/science.285.5434.1722. [DOI] [PubMed] [Google Scholar]
- Zhang J.H., Chung T.D., Oldenburg K.R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- Zikova A., Kopecna J., Schumacher M.A., Stuart K., Trantirek L., Lukes J. Structure and function of the native and recombinant mitochondrial MRP1/MRP2 complex from Trypanosoma brucei. Int. J. Parasitol. 2008;38:901–912. doi: 10.1016/j.ijpara.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]


