Graphical abstract
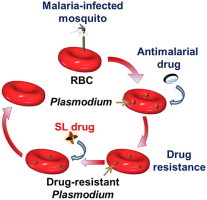
Abbreviations: SL, synthetic lethal; P., Plasmodium; ADR, antimalarial drug resistance; SW, Smith–Waterman; GO, gene ontology; GOA, gene ontology annotation; PDB, Protein Data Bank
Keywords: Drug-resistant malaria, Synthetic lethality, Drug targeting, Antimalarial therapy
Highlights
-
•
Potential antimalarial drug targets were suggested using by homology analysis of yeast–human–Plasmodium.
-
•
A antimalarial drug candidates were inferred by searching drugs that cause a fitness defect in yeast SL genes.
-
•
Information on new usage for already-described drugs are provided.
Abstract
Many antimalarial drugs kill malaria parasites, but antimalarial drug resistance (ADR) and toxicity to normal cells limit their usefulness. To solve this problem, we suggest a new therapy for drug-resistant malaria. The approach consists of data integration and inference through homology analysis of yeast–human–Plasmodium. If one gene of a Plasmodium synthetic lethal (SL) gene pair has a mutation that causes ADR, a drug targeting the other gene of the SL pair might be used as an effective treatment for drug-resistant strains of malaria. A simple computational tool to analyze the inferred SL genes of Plasmodium species (malaria parasites Plasmodium falciparum and Plasmodium vivax for human malarial therapy, and rodent parasite Plasmodium berghei for in vivo studies of human malarias) was established to identify SL genes that can be used as drug targets. Information on SL gene pairs with ADR genes and their first neighbors was inferred from yeast SL genes to search for pertinent antimalarial drug targets. We not only suggest drug target gene candidates for further experimental validation, but also provide information on new usage for already-described drugs. The proposed specific antimalarial drug candidates can be inferred by searching drugs that cause a fitness defect in yeast SL genes.
1. Introduction
Malaria is one of the most serious causes of morbidity and mortality in tropical regions. The World Health Organization recently estimated that malaria causes about 240 million bouts of illness and around 0.86 million deaths annually (http://www.who.int/mediacentre/factsheets/fs094/en/print.html, world health organization malaria fact sheet No. 94, 2010). Several Plasmodium species (P. falciparum, P. vivax, P. ovale, P. malariae, and P. knowlesi) have been identified to be responsible for a significant number of human malaria infections in Southeast Asia. Among these species, P. falciparum is the most severe and responsible for about 90% of malaria deaths (Cox-Singh et al., 2008), predominantly in Africa (Eisenstein, 2012).
P. vivax has a considerably lower mortality rate but a far greater geographical distribution. It causes widespread malaria outside Africa, mainly afflicting Asia and the Americas (Price et al., 2007). The human malaria parasite P. vivax is responsible for 25–40% of the annual bouts of malaria worldwide. Although infection by the parasite is seldom fatal, such infection induces severe, debilitating clinical symptoms and often causes relapses months after a primary infection is cleared. Despite being a major human pathogen, studies on P. vivax are very limited compared with P. falciparum because P. vivax is not contagiously propagated in the laboratory and in vitro culture is not amenable (Carlton et al., 2008).
The major problem with the currently available antimalarial drugs is that some malaria parasites have continuously developed resistance to these drugs. Several special treatments for specific malarial strains have been introduced. However, the parasites have become increasingly resistant to conventional antimalarial drugs, thereby increasing morbidity and mortality. Numerous malarial drugs kill malaria parasites, but antimalarial drug resistance (ADR) and toxicity to normal cells limit their usefulness. Such emerging drug resistance and a lack of effective vaccines require urgent discovery of new antimalarial drugs and vaccines against malarial parasites (Anderson, 2009; Huthmacher et al., 2010). Most malaria researchers are eager to identify and act on drug target genes. However, ideal drug and vaccine targets are not easy to determine. Currently, the systematic disruption of every gene in Plasmodium is technically difficult. A computational approach to prioritizing potential drug targets was recently established (Yeh et al., 2004). As massive screening methods, bioinformatics and computational approaches play predominant roles in drug design. Nevertheless, a novel strategy is required to overcome the technological limitations encountered by the current single-drug approach for effective treatments (Fernández et al., 2009).
We therefore investigated targeting synthetic lethal (SL) gene pairs as an alternative antimalarial drug therapy. When the mutation of either component of a gene pair is not lethal but the mutation of both leads to death or a significant decrease in the fitness of the organism, the two genes are called synthetic lethal (Conde-Pueyo et al., 2009). Fig. 1 represents the rationale for SL applications in antimalarial therapy. Given that malarial parasites exhibit various types of drug resistance, the identification of proper SL gene partners may be helpful in finding specific antimalarial drug targets. Such identification provides new insights into the design of a selective antimalarial therapy by exploiting SL partners that have ADR genes. By filtering out the SL targets that have analogues of the human proteome, minor damage is expected in the human cell, as shown in the conceptual schematics of selective antimalarial therapy (Fig. 1). Therefore, this method would be helpful in dramatically overcoming the limitations in the design of a suitable malarial drug. However, large-scale screenings for SL gene pairs have been performed mostly in yeast (Forsburg, 2001; Pan et al., 2004; Ooi et al., 2006; Boone et al., 2007), and to a lesser extent in Caenorhabitis elegans (Jorgensen and Mango, 2002; Baugh et al., 2005; Lehner et al., 2006) and other organisms. Screening research on Plasmodium SL gene pairs has yet to be accomplished.
Fig. 1.
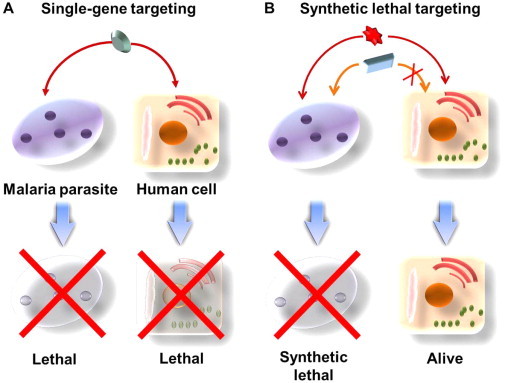
Rationale for SL applications in antimalarial therapy. Rationale for synthetic lethality applied in the design of novel antimalarial therapies. Two genes are synthetic lethal (SL) if the mutation of either gene alone is compatible with viability; the mutation of both leads to death. (A) Single gene targeting is more likely to express toxicity in both the malaria parasite and the human cell. (B) If one gene of the SL pair is mutated and the other SL partner is not mutated, the human cells would not die. However, if both SL partners are inactivated in the malaria parasite, the parasite is selectively damaged.
Consequently, we investigated Plasmodium SL gene pair candidates in the present study using inference methods. Fig. 2 shows the basic concept of searching for drug targets for antimalarial therapy. SL screening in yeast is often used to identify the genes involved in cell polarity, secretion, DNA repair, and the cell cycle (Tong et al., 2001, 2004). Considering the high conservation of genome integrity and cell cycle-related genes from yeast to higher organisms (Yuen et al., 2007), a massive screening for yeast SL interactions may provide information on using SL inference to search for novel malarial therapies. The present study aimed not to discuss the general inference method for the SL network from one organism to another, but to present the rationale for drug design by supplying suitable Plasmodium SL gene pairs that can be used as a potential basis for future pharmacologic tests. Another objective was to provide an excellent framework for handling very large genetic systems. In particular, studies on genetic diseases (Goh et al., 2007; Platzer et al., 2007), gene targets (Yildirim et al., 2007), and SL networks (Boone et al., 2007; Paladugu et al., 2008; Chipman and Singh, 2009) have contributed to the understanding of these systems as a whole. In the present study, network framework data from several databases were integrated to increase the reliability of the supplied candidates.
Fig. 2.
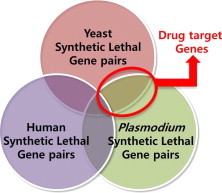
Information diagram showing the basic concept of searching for drug targets for use in malarial therapy and a new SL therapy for the elimination of antimalarial drug-resistant Plasmodium. Information diagram of the basic concept of a drug target search for malarial therapy. In this study, the phylogenetic inference of synthetic lethal (SL) genes extracted from yeast and malaria parasites was utilized for pharmacologic purposes because most existing SL data are restricted to yeast screenings. SL pairs with human orthologs were not considered.
2. Materials and methods
Potential antimalarial drug target genes were identified by integrating whole biological information from many databases. Fig. 3 illustrates the methodology used for data collection and the selection of potential antimalarial drug target genes. The phylogenetically inferred Plasmodium SL gene network was reconstructed by using phylogenetic analysis and database manipulation to search for potential antimalarial drug targets. The databases used include BioGRID (Biological General Repository for Interaction Datasets) (http://thebiogrid.org) for yeast SL genes, KEGG SSDB (Kyoto Encyclopedia of Genes and Genomes Sequence Similarity DataBase) (http://www.genome.jp/kegg), KEGG OC (Ortholog Cluster) (http://www.genome.jp/tools/oc) (Nakaya et al., 2013), PlasmoDB Version 9.1 (http://plasmodb.org), and Gene DB (http://www.genedb.org) for Plasmodium genes. The inferred Plasmodium SL networks were constructed using Cytoscape (version 2.8.1) (Shannon et al., 2003).
Fig. 3.
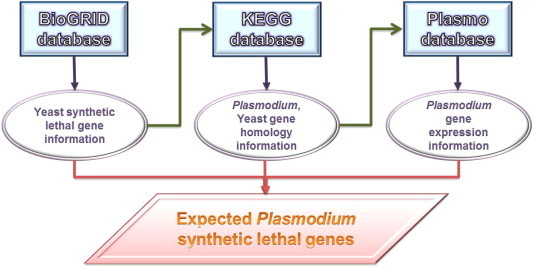
Schematic diagram of the synthetic lethal (SL) gene identification methodology. Biological information from different databases including BioGRID, KEGG, and PlasmoDB were integrated. To search for potential antimalarial drug targets, the inferred Plasmodium SL gene network was reconstructed by phylogenetic analysis and database manipulation. The databases employed in this study were BioGRID for yeast SL genes, as well as KEGG and PlasmoDB for Plasmodium genes.
2.1. Inference of Plasmodium SL genes from yeast SL genes
The list of yeast SL gene pairs was obtained from the ‘BIOGRID-ALL-3.1.72.tab2’ data source provided by the BioGRID database. The collected data on yeast genetic screens included synthetic lethality. The yeast SL network was constructed using the yeast SL pairs. Then, three Plasmodium species (P. falciparum, P. vivax, and P. berghei) SL gene networks were inferred from the yeast SL gene data. Plasmodium–yeast gene ortholog data were obtained from the ortholog information in the KEGG SSDB. Among the yeast genes (14,548 genes in total), approximately 29% were SL genes (4,284 genes). Plasmodium gene information, such as gene expression levels and stages, were obtained from the PlasmoDB version 9.1 (PlasmoDB: http://plasmodb.org/plasmo/webcite, Aug. 31, 2012).
2.2. Extraction of ADR genes from inferred Plasmodium SL genes
We identified eight P. falciparum ADR genes (PfABC, PfCRT, PfDHFR-TS, PfDHPS, PfMT, PfMDR1, PfMDR2, and PfNHE1) through a systematic literature survey (Wilson et al., 1989; Foote et al., 1990; Wellems et al., 1990; Cowman et al., 1991, 1994; Peel et al., 1993, 1994; Zalis et al., 1993; Rubio and Cowman, 1996; Reed et al., 2000; Mutabingwa et al., 2001; Sibley et al., 2001; Giancarlo et al., 2009; Briolant et al., 2012). By phylogenetic inference, four ADR genes (PfDHFR-TS, PfMDR1, PfMDR2, and PfNHE1) were discovered to contain the inferred SL genes. PfDHFR-TS exhibits a homologous relation with YBR205W and YOR074C. Pfmdr1 has a homologous relation with YKL209C, YGR281W, YKL188C, and YLL048C. PfMDR2 has a homologous relation with YMR301C. PfNHE1 has a homologous relation with YHR081W and YDR456W. Detailed information is included in Supplementary file 1.
2.3. Search for Plasmodium genes without similarity to human genes
We searched for information about protein similarities in the Protein Data Bank (PDB) provided by PlasmoDB. Plasmodium genes which do not have protein similarities to human genes were selected. The protein structure data in the PDB have been used actively in the studies of protein function, evolution, and structure prediction (Bernstein et al., 1977; Noguchi and Akiyama, 2003). Information on the Plasmodium genes which do not have human orthologs was obtained from the KEGG OC database. Detailed information is included in Supplementary file 2.
2.4. Search for drug candidates
Fitness data from the Yeast Fitness DB (http://fitdb.stanford.edu, 2012) (Hillenmeyer et al., 2008, 2010) on the yeast homologous genes of the selected Plasmodium genes were searched to find new drugs for the clinical treatment of malaria. A list of drug candidates was obtained through the top three experiments that gave rise to a fitness defect in yeast genes by heterozygous or homozygous deletion. To utilize the yeast drugs provided by the FitDB, we conjectured the possibility of evolutionary conservation between the yeast genes and Plasmodium homologs by identifying protein similarities in the PDB. From the list of drug candidates, we selected the final drug candidates by filtering out the compounds which do not exist in the DrugBank 3.0 database (http://www.drugbank.ca) (Knox et al., 2011). A detailed report is available in Supplementary file 3.
2.5. Statistical analysis of gene function
The networks associated with gene ontology (GO) annotation (GOA) terms were statistically analyzed using BINGO 2.44 Cytoscape plugging (Maere et al., 2005). BINGO is a useful tool for determining statistically over-represented GO categories in a set of genes or subgraphs of a biological network. P. falciparum GOA data were obtained using BINGO. The GO biological process, molecular functions, and cellular components were selected from the ontology list, whereas P. falciparum was selected from the organism list. Finally, the over-represented GO categories were analyzed using Cytoscape. The gene functions of the inferred P. falciparum SL genes, as well as the ADR genes and their first neighbors, were analyzed from the inferred P. falciparum gene networks using BINGO.
The significance of their cellular components, molecular functions, and biological processes was evaluated using a hypergeometric test, in which the Benjamini and Hochberg false discovery rate with a P value less than 0.05 was used as the minimum cutoff threshold. Using this method, only 608 genes out of a total of 1,423 inferred P. falciparum SL genes were found to be associated with GOA terms. Only 23 genes out of 50 ADR genes and their first neighbors extracted from the inferred P. falciparum gene network were associated with GOA terms. The extracted list of genes related to specific GOA terms is available in Supplementary file 4. In addition, we analyzed the gene functions of the genes selected ADR genes and their first neighbors from the inferred Plasmodium gene network using the Plasmodium genomics resource in the PlasmoDB. A detailed report including the list of genes related to specific GOA terms is available in Supplementary files 2 and 3.
3. Results
3.1. Inference of Plasmodium SL genes from yeast SL genes
We inferred human and Plasmodium SL gene pairs from yeast SL gene pairs. A total of 4284 yeast SL genes were obtained from the BioGRID database (version 3.1.72, http://thebiogrid.org). We then inferred the Plasmodium (P. falciparum, P. vivax, and P. berghei) SL gene pairs from the yeast SL gene pairs using the KEGG database. The inferred P. falciparum, P. vivax, and P. berghei SL networks are composed of 1536 nodes and 7600 links, 1449 nodes and 6458 links, as well as 1299 nodes and 7608 links, respectively.
3.2. Extraction of ADR genes from inferred Plasmodium SL genes
Due to mutations in ADR genes, some antimalarial are not effective in certain patients. Through a literature survey, we identified eight ADR genes: P. falciparum ABC Transporter (PfABC), P. falciparum putative metabolite/drug transporter (PfMT), P. falciparum dihydropteroate synthase (PfDHPS), P. falciparum multidrug resistance 1 (PfMDR1), P. falciparum multidrug resistance 2 (PfMDR2), P. falciparum sodium/hydrogen exchanger 1 (PfNHE1), P. falciparum dihydrofolate reductase-thymidylate synthase (PfDHFR-TS), and P. falciparum chloroquine-resistance transporter gene (PfCRT) (Wilson et al., 1989; Foote et al., 1990; Wellems et al., 1990; Cowman et al., 1991; Peel et al., 1993, 1994; Zalis et al., 1993; Cowman et al., 1994; Rubio and Cowman, 1996; Reed et al., 2000; Mutabingwa et al., 2001; Sibley et al., 2001; Giancarlo et al., 2009; Briolant et al., 2012). Four genes (PfMDR1, PfMDR2, PfDHFR-TS, and PfNHE1) among the eight P. falciparum ADR genes are inferred as Plasmodium SL genes.
3.3. Extraction of of ADR genes’ SL partners
The SL partners of the ADR genes were extracted from the yeast SL gene pairs. Fig. 4A and B show typical combinations of Plasmodium SL pairs inferred from the yeast SL pairs. Each entry in the homology data file contains the gene’s evolutionary history, which corresponds to a gene tree diverging from a common ancestor (Conde-Pueyo et al., 2009). In this study, the gene conservation relations in yeast and Plasmodium are categorized into three types. The simplest is the one-to-one relation between yeast and Plasmodium genes (orthologous relation). However, evolutionary duplication events lead to two alternative types; one yeast gene has more than one Plasmodium homolog (one-to-m relation), and vice versa (n-to-one relation). The n-to-m relation corresponds to the case where n Plasmodium genes are homologs of m yeast genes (Sonnhammer and Koonin, 2002). The Plasmodium genes selected in this study have one-to-one and one-to-n (Plasmodium to yeast) relations. These relations include auto-loop genes (marked x in Fig. 4) as SL partners; when an SL pair was composed of the same single Plasmodium gene, we defined it as an ‘auto-loop gene’. We discovered four auto-loop genes in P. falciparum, one in P. vivax, and two in P. berghei from the inferred SL networks composed of ADR genes and their SL partners. The auto-loop nodes are represented as yellow background genes in Supplementary file 1. In this study, the SL pairs containing auto-loop genes were excluded from the list of drug target candidates.
Fig. 4.
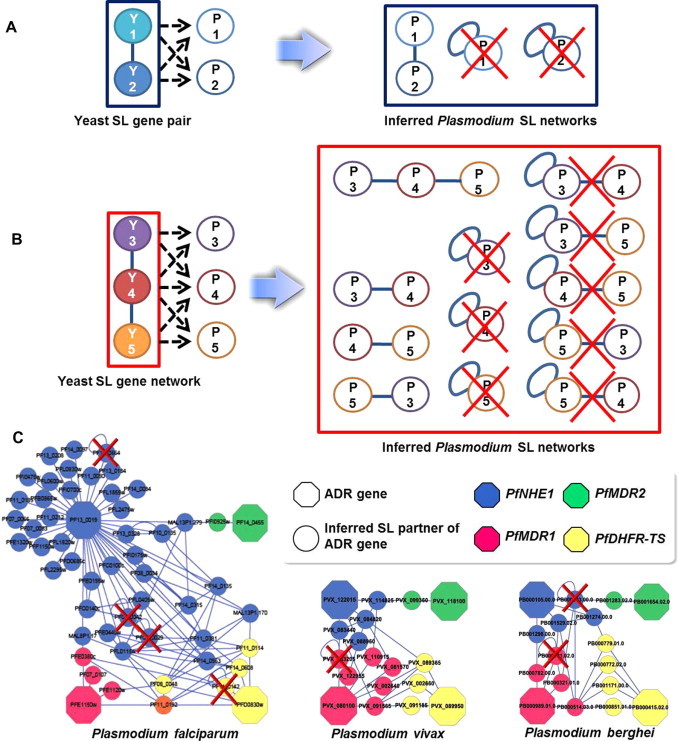
Networks of ADR genes and their SL partners extracted from the inferred Plasmodium SL genes. Plasmodium SL gene pairs were inferred from yeast SL pairs. In this network, nodes represent genes and the link between them indicates an SL interaction. When both are simultaneously mutated, an SL condition is satisfied. Gene conservation between Plasmodium and yeast can be categorized into two types of relations. The simplest case is a one-to-one relation between Plasmodium and yeast genes (orthologous relation). However, duplication events during evolution cause a one-to-n relation in which two or more yeast genes are homologous to one Plasmodium gene. (A) For example, the one-to-one case corresponds to one yeast gene having one Plasmodium homolog; the SL pairs Y1 and Y2 correspond to SL pair P1 and P2. In the one-to-n case, several yeast genes have the same Plasmodium homolog; SL pairs Y1 and Y2 can correspond to only one gene between P1 and P2. In this case, an autolink (auto-loop) is formed in the network. (B) For yeast SL networks represented by Y3, Y4, and Y5, various relations such as one-to-one, n-to-one, auto-loop, and SL pair-containing auto-loop genes can be combined. (C) Networks of the four inferred ADR genes in the Plasmodium species. When two SL yeast partners are phylogenetically related to a single Plasmodium gene, an auto-link appears in the SL network. Single gene targeting is possibly more dangerous than double gene targeting. Therefore, we eliminated the auto-loop nodes from the drug target candidates and denoted them as red X’s. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.4. Networks consisting of ADR genes and their SL partners
Fig. 4C shows the networks composed of four ADR genes and their inferred SL partners as inferred from the yeast SL gene networks. Detailed information is given in Supplementary files 1. The inferred SL networks composed of ADR genes and their SL partners have 50 nodes and 86 links in P. falciparum, 17 nodes and 27 links in P. vivax, as well as 15 nodes and 19 links in P. berghei. Octagons denote ADR-related nodes and circles indicate their SL partners. The links connecting the ADR genes are depicted as blue lines. PfNHE1, PfMDR1, PfMDR2 and PfDHFR-TS are four representative ADR genes inferred as SL genes. Red indicates the PfMDR1 gene and its inferred SL partners, yellow represents the PfDHFR-TS gene and its inferred SL partners, green indicates the PfMDR2 gene and its inferred SL partners, and blue indicates the PfNHE1 gene and its inferred SL partners. PF11_0192 (orange circle) is the inferred SL partner of the PfMDR1 (red octagon) and PfDHFR-TS (yellow octagon) genes. Four ADR genes and their 46 SL partners were extracted from the inferred P. falciparum SL gene network. Meanwhile, 4 ADR genes and 13 SL partners were selected from the inferred P. vivax network, whereas four ADR genes and 11 partners were extracted from the inferred P. berghei network. Detailed functional information on the drug target candidates selected from the PlasmoDB are summarized and provided in Table 1 and Supplementary file 2. In this study, we selected genes which do not have protein similarities between Plasmodium genes and their human orthologs. Interestingly, almost all of the selected genes are connected to interrelated biological functions, such as ATPase activity, ATP binding, and ion transportation.
Table 1.
Selected antimalarial drug target genes and their GO terms.
| Inferred Plasmodium SL genes | Plasmodium GO terms | Yeast homolog genes of Plasmodium | Yeast GO terms |
|---|---|---|---|
| PB000415.02.0 (ADR gene: PfDHFR-TS) | Biological Process: dTMP biosynthetic process, glycine biosynthetic process, one-carbon metabolic process, nucleotide biosynthetic process, oxidation-reduction process; Molecular Function: dihydrofolate reductase activity, thymidylate synthase activity | YOR074C | Biological Process: dTMP biosynthetic process; Molecular Function: thymidylate synthase activity; Cellular Component: nucleus |
| PB000779.01.0 (PB000415.02.0’s neighbor gene) | Molecular Function: nucleic acid binding, DNA binding, helicase activity, ATP binding | YDR334W | Biological Process: chromatin remodeling, histone exchange; Molecular Function: structural molecule activity; Cellular Component: Swr1 complex, nucleus |
| PFD0830w (ADR gene: PfDHFR-TS) | Biological Process: dTMP biosynthetic process, glycine biosynthetic process, one-carbon metabolic process, nucleotide biosynthetic process, oxidation-reduction process; Molecular Function: dihydrofolate reductase activity, thymidylate synthase activity | YOR074C | Biological Process: dTMP biosynthetic process; Molecular Function: thymidylate synthase activity; Cellular Component: nucleus |
| PF08_0048 (PFD0830w’s neighbor gene) | Molecular Function: nucleic acid binding, DNA binding, helicase activity, ATP binding, ATP-dependent helicase activity | YDR334W | Biological Process: chromatin remodeling, histone exchange; Molecular Function: structural molecule activity; Cellular Component: Swr1 complex, nucleus |
| PVX_089950 (ADR gene: PfDHFR-TS) | Biological Process: dTMP biosynthetic process, glycine biosynthetic process, one-carbon metabolic process, nucleotide biosynthetic process, oxidation–reduction process; Molecular Function: dihydrofolate reductase activity, thymidylate synthase activity | YOR074C | Biological Process: dTMP biosynthetic process; Molecular Function: thymidylate synthase activity; Cellular Component: nucleus |
| PVX_089365 (PVX_089950’s neighbor gene) | Molecular Function: nucleic acid binding, DNA binding, helicase activity, ATP binding | YLR085C | Biological Process: chromatin remodeling, histone exchange; Molecular Function: nucleosome binding; Cellular Component: Swr1 complex, cytoplasm |
| PVX_118100 (ADR gene: PfMDR2) | Biological Process: transport, transmembrane transport; Molecular Function: ATP binding, ATPase activity, nucleoside-triphosphatase activity; Cellular Component: integral to membrane | YMR301C | Biological Process: cellular iron ion homeostasis, transmembrane transport; Molecular Function: ATPase activity, coupled to transmembrane movement of substances; Cellular Component: integral to membrane, mitochondrial inner membrane, mitochondrion |
| PVX_099360 (PVX_118100’s neighbor gene) | Biological Process: glutathione biosynthetic process; Molecular Function: glutamate–cysteine ligase activity | YJL101C | Biological Process: glutathione biosynthetic process, response to cadmium ion, response to hydrogen peroxide; Molecular Function: glutamate–cysteine ligase activity; Cellular Component, cytoplasm, intracellular |
In this table, we can see detailed functional information on the drug target candidates. We selected genes which do not have protein similarities between Plasmodium genes and their human orthologs. Interestingly, almost all of the selected genes are connected to interrelated biological functions, such as ATPase activity, ATP binding, and ion transportation. In this table, we compare the GO terms of yeast SL genes with that of Plasmodium SL genes inferred from yeast SL gene pairs. In addition, through PlasmoDB, we investigated similarities to Protein Data Bank chains between the yeast gene and its Plasmodium homolog gene. Although these are examples of similarity in the biological process, molecular function, or cellular component of Plasmodium evolutionary conserved from yeast, their synthetic lethality might conserve in Plasmodium from yeast in the same vein.
In Table 1, we compare the GO terms of yeast SL genes with that of the Plasmodium SL genes inferred from yeast SL gene pairs. The GO terms of PFD0830w, PVX_089950, and PB000415.02.0 (Plasmodium homologous genes of ADR gene- PfDHFR-TS) match the GO terms of YOR074C (yeast homolog gene of PFD0830w, PVX_089950, and PB000415.02.0) well. Their biological process is the dTMP biosynthetic process and their molecular function is thymidylate synthase activity. The GO terms of the biological process, molecular function, and cellular component of PVX_118100 (P. berghei homologous genes of ADR gene: PfMDR2) and YMR301C (yeast homologous gene of PVX_118100) are particularly well matched. Their biological process is transmembrane transport and their molecular function is ATPase activity. In addition, the cellular component is an integral factor to the membrane, and the GO terms of the biological process (glutathione biosynthetic process) and the molecular function (glutamate–cysteine ligase activity) of PVX_099360 (PVX_118100’s neighbor gene) and YJL101C (yeast homologous gene of PVX_099360) match each other. In Table 2, PVX_099360, which is the neighbor gene of PVX_118100 (ADR gene: PfMDR2), is a putative gamma-glutamylcysteine synthetase. YJL101C is a yeast homolog gene of PVX_099360. In addition, using the PlasmoDB, we investigated the similarities of the protein chains between the yeast and its plasmodium. YJL101C and PVX_099360 were seen to exhibit protein similarities to glutamate–cysteine ligase with an identity of 32% and a P value of 7.6 × 10−64.
Table 2.
Suggested antimalarial drug target genes and possibility of evolutionary conservation of yeast synthetic lethal genes.
| Plasmodium | Plasmodium | Saccharomyces cerevisiae |
|---|---|---|
| ADR gene | ADR gene’s inferred SL partner | Similarities to Protein Data Bank Chains (ADR gene’s inferred SL partner) |
| PB000415.02.0 (ADR gene: PfDHFR-TS)/bifunctional dihydrofolate reductase-thymidylate synthase, putative | PB000779.01.0 (PB000415.02.0’s neighbor gene)/Snf2-related CBP activator, putative | Chromo domain-containing protein 1 (% of PlasmoDB protein Covered: 42;% Identity: 32; P-value: 2.5 × 10−78) |
| PFD0830w (ADR gene: PfDHFR-TS)/bifunctional dihydrofolate reductase-thymidylate synthase | PF08_0048 (PFD0830w’s neighbor gene)/Snf2-related CBP activator, putative | Chromo domain-containing protein 1 (% of PlasmoDB protein Covered: 34;% Identity: 33; P-value: 1.5 × 10−75) |
| PF13_0019 (ADR gene: PfNHE1)/sodium/hydrogen exchanger, Na, H antiporter | MAL13P1.170 (PF13_0019’s neighbor gene)/nucleotidyltransferase, putative | Poly(A) RNA polymerase protein 2 (% of PlasmoDB protein Covered: 32; % Identity: 31; P-value: 5.2 × 10−29) |
| PVX_089950 (ADR gene: PfDHFR-TS)/bifunctional dihydrofolate reductase-thymidylate synthase 1, putative | PVX_089365 (PVX_089950’s neighbor gene)/helicase, putative | Chromo domain-containing protein 1 (% of PlasmoDB protein Covered: 32,% Identity: 35; P-value: 3.5 × 10−75) |
| PVX_118100 (ADR gene: PfMDR2)/multidrug resistance protein 2, putative | PVX_099360 (PVX_118100’s neighbor gene)/gamma-glutamylcysteine synthetase, putative | Glutamate–cysteine ligase (% of PlasmoDB protein Covered: 63;% Identity: 32; P-value: 7.6 × 10−64) |
The list of selected SL gene pairs is shown in this table. Potential antimalarial drug targets (inferred SL partners for ADR genes) are suggested for future experimental validation. All SL genes represented in this table do not have any human homologous gene. For application to yeast drugs provided by the FitDB, we conjectured the possibility of evolutionary conservation between the yeast genes and Plasmodium homologs by identifying similarities in protein chains in the Protein Data Bank. The five drug target genes are also shown in Fig. 5.
3.5. Prediction of antimalarial drug targets
To narrow down the list of drug target candidates, we examined the developmental stages in which Plasmodium genes were expressed (Supplementary file 1). In addition, we searched for the protein similarities within the protein databank (PDB) provided by the PlasmoDB. These ADR genes and their SL partners extracted from the inferred Plasmodium SL gene networks can serve as alternative antimalarial drug targets for malaria patients with ADR genes. In addition, we conjectured the possibility of evolutionary conservation between the yeast genes and Plasmodium homologs using the same identification process involving protein similarities. We obtained the five inferred SL partners (marked by red loops in Fig. 5) having the inferred Plasmodium SL gene networks which have protein similarities among yeast orthologs and Plasmodium orthologs. In the three networks, the filtered Plasmodium SL pairs (ADR genes and their inferred SL partners) do not include human homologous genes. The filtered Plasmodium SL pairs also do not have human orthologs in the KEGG OC database.
Fig. 5.
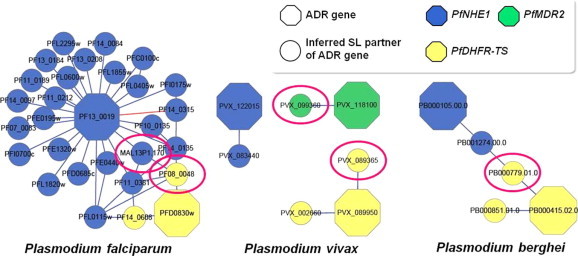
Inferred Plasmodium SL gene networks to select antimalarial drug targets. To narrow down the list of drug target candidates, potential antimalarial drug target genes are suggested for future experimental validation. The resulting network was filtered by identifying protein similarities in the PDB provided by the PlasmoDB. In three networks, all the filtered inferred Plasmodium SL pairs (ADR genes and their inferred SL partners) do not include human orthologs except for some ADR genes such as PFD0830w, PVX_089950, and PB000415.02.0. These ADR genes and their SL partners extracted from the inferred Plasmodium SL gene network can serve as alternative antimalarial drug targets for malaria patients with ADR genes. In addition, we conjectured the possibility of evolutionary conservation between the yeast genes and Plasmodium homologs through the identification of protein similarities. We then obtained the five inferred SL partners (marked by red loops) having the inferred Plasmodium SL gene networks, which exhibit protein similarities among the yeast orthologs and Plasmodium orthologs. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.6. Analysis of biological function of selected drug targets
We carried out a statistical analysis on the biological function of the selected drug targets using the BINGO 2.44 program (Maere et al., 2005). BINGO has been used to determine which GO categories are statistically over-represented in a set of genes or a subset of biological networks. We obtained only P. falciparum GO data because the program provides information on the GO of P. falciparum genes among the Plasmodium species. Fig. 6 shows the relationship between the biological processes of the PFE0195W and PF13_0019 genes as a typical example of GO term analysis. These genes are the ADR genes and their first neighbors extracted from the inferred P. falciparum SL networks. Information on the biological functions of other species (P. vivax and P. berghei) can be obtained from the PlasmoDB. We analyzed the biological processes, molecular functions, and cellular components of the inferred P. falciparum SL genes, the inferred ADR genes, and their SL partners. Detailed information, including the GO terms, is available in Supplementary files 2–4. Table 1 summarizes the biological functions of the selected drug target genes analyzed using the PlasmoDB.
Fig. 6.
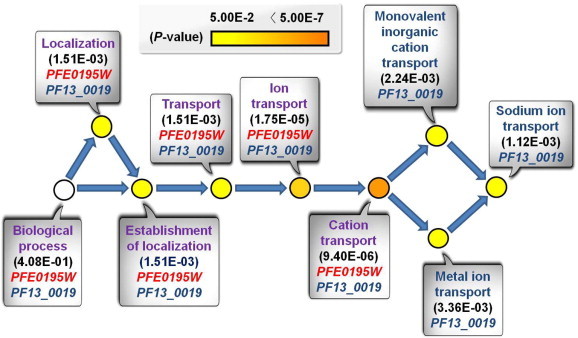
Relationship between the biological processes of the PFE0195W and PF13_0019 genes. We conducted statistical GO analysis on the biological function of the selected antimalarial drug target genes using BINGO 2.44. Using this GO term network (BINGO graph), the biological process of the PFE0195W and PF13_0019 genes were compared. The BINGO graph visualizes the GO categories that are statistically over-represented in the context of the GO hierarchy. The PFE0195W and PF13_0019 genes are the ADR genes and their first-neighbor genes inferred from yeast SL pairs. The gene pair is one of the inferred SL gene pairs that contain the most potential malaria drug target genes. For each circle, the GO term related to the biological process and the P value on the two corresponding genes are specified. The P value is a good indicator for the prominence of a given functional category; white, yellow, and orange denote the different P value scales. The uncolored nodes are not over-represented; they are the parents of over-represented categories at downstream (biological process in white box). Yellow nodes represent GO categories that are significantly over-represented. For more significant P values, the saturation of the node color becomes denser (color legend panel). Blue arrows denote the stage and direction of biological processes. The GO term and P value are shown in each white box. Purple represents the biological processes that are related only to the two genes. Monovalent inorganic cation transport, metal ion transport, and sodium ion transport are biological processes that are related only to the PF13_0019 gene. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.7. New malarial drug candidates
The five selected drug target gene pairs are listed in Table 2 and depicted in Fig. 5. According to the information obtained through the identification of protein similarities, the selected SL genes do not have human homologous genes. The drug candidates for the selected drug target genes were inferred from the Yeast Fitness DB and the results are summarized in Table 3. We searched the fitness data for the yeast homologs of the selected Plasmodium SL genes to find new drugs for clinical malaria treatment.
Table 3.
New malarial drug candidates.
| Inferred Plasmodium SL genes (Description) |
Yeast homologs of Plasmodium | Heterozygous knockout drug (Description) |
Homozygous knockout drug (Description) |
|---|---|---|---|
|
PB000415.02.0 (ADR gene: PfDHFR-TS) |
YOR074C | 5-Fluorouridine (antineoplastic), floxuridine (antiviral) | x |
|
PB000779.01.0 (PB000415.02.0’s neighbor gene) |
YDR334W | Caspofungin (antifungal) | Mechlorethamine (alkylating) |
|
PFD0830w (ADR gene: PfDHFR-TS) |
YOR074C | Methotrexate (antineoplastic antimetabolite), 5-fluorouridine (antineoplastic), floxuridine (antiviral) |
x |
|
PF08_0048 (PFD0830w’s neighbor gene) |
YDR334W | Caspofungin (antifungal) | Mechlorethamine (alkylating) |
|
PF13_0019 (ADR gene: PfNHE1) |
YHR081W | Miconazole (antifungal), fluconazole (antifungal), fenpropimorph (antifungal) | x |
|
MAL13P1.170 (PF13_0019’s neighbor gene) |
YOL115W | 5-fluorouracil (antineoplastic antimetabolite), 5-fluorouridine (antineoplastic) | x |
|
PVX_089950 (ADR gene: PfDHFR-TS) |
YOR074C | Methotrexate (antineoplastic antimetabolite), 5-fluorouridine (antineoplastic), floxuridine (antiviral) |
x |
|
PVX_089365 (PVX_089950’s neighbor gene) |
YLR085C | Alverine citrate (psychoactive) | Mechlorethamine (alkylating), busulfan (alkylating) |
|
PVX_118100 (ADR gene: PfMDR2) |
YMR301C | Fluconazole (antifungal) | x |
|
PVX_099360 (PVX_118100’s neighbor gene) |
YJL101C | Itraconazole (antifungal), methotrexate (antineoplastic antimetabolite) | x |
In future antimalarial drug research, a couple of the best candidates must be selected from the list, and they should be experimentally demonstrated to have the desired effect on the parasite. Drug target candidates were finally selected from ADR genes and their SL partners containing inferred Plasmodium (P. falciparum, P. vivax, and P. berghei) SL genes. We analyzed the fitness data for the yeast homologs of the selected Plasmodium genes to find new drugs for clinical malaria treatment. The right column of this table shows which drugs obtained from the top three experiments can be searched in the Drug Bank database.
All Plasmodium genes summarized in Tables 1–3 are not human orthologs except for ADR genes such as PFD0830w, PVX_089950 and PB000415.02.0. Hence, the selective mutation of the SL genes might have strong therapeutic potential in malaria elimination.
4. Discussion
In the present study, a methodology based on the SL relation of genes is proposed to select drug targets and drug candidates for human antimalarial therapy and for experimental validation. A simple computational tool was established to identify SL genes as proper drug targets by analyzing the inferred SL genes of Plasmodium species (P. falciparum and P. vivax for human malarial therapy, and P. berghei for an in vivo experimental model). The inferred Plasmodium SL genes can be used to search for drug targets for effective antimalarial therapy through in vitro or in vivo experiments. However, in vitro experiments on P. vivax are not easy to perform. P. vivax can only be cultured in vitro for short periods (at least partially due to this parasite’s preference for infecting short-lived reticulocytes) and drug studies must therefore be performed at sites where P. vivax is readily available. In addition, genetic manipulation of this parasite is virtually impossible (Eisenstein, 2012). The lack of good experimental models is another limiting factor. Alternatively, the inferred P. berghei target genes can be used for in vivo tests. Commercial drug candidates can eliminate malaria parasites, but they are toxic for people with mutating human genes.
The incomplete effectiveness of a specific antimalarial drug in a certain patient results from the mutation of one of the ADR genes. To ensure successful therapy for these patients, the first neighbors of the mutated ADR gene should be the target of antimalarial therapy such as chemical therapy with SL gene-specific drugs. If the mutation of the ADR genes coincides with the mutation of their SL partners, the malaria parasites would die. However, definitive results are impossible to obtain because of the long evolutionary history and different architectures of yeast and Plasmodium genomes.
The evolutionary conservation of SL pairs is controversial (Tarailo et al., 2007; Yuen et al., 2007; Dixon et al., 2008; Tischler et al., 2008), although a significant conservation of SL interactions between eukaryotes has been studied (Dixon et al., 2008). Not all yeast SL gene pairs are conserved in distantly related organisms. However, previous studies have reported SL genes conserved from yeast. For instance, the evolutionary conserved gene MHO1 is synthetic lethal with PLC1 (Schlatter et al., 2012).
P-type ATPase 3 seems to have no merit as a drug target because the gene has been proposed as a target by other researchers (Rozmajzl et al., 2001). Therefore, we regarded the PfATP3 (PFE0195w) gene as a drug target under special conditions, such as when its SL partner mutates.
The drug may have an adverse effect on Plasmodium but not on human cells. To avoid such unwanted side effects on human hosts, parasitic enzymes should be targeted instead of homologous human enzymes (Yeh et al., 2004). If the homologs of drug target genes exist in humans, the enzyme would neither be essential to the human host nor have different inhibitory effects because of their different protein structure and regulation.
Potential drug target genes have to be expressed in some or all of the developmental stages of parasites. We do not know the developmental stage at which the effect of the drug therapy would appear. However, we conjectured that the therapeutic effect would be better when the expressed SL genes in some of the developmental stages of the parasites are simultaneously targeted. Provisional drug targets can be examined computationally and experimentally to extract these features. Experimental analyses can be performed under in vitro and in vivo conditions for Plasmodium, but such experimental studies are costly and laborious. Comparisons between the present predictions with such experimental data would be particularly interesting.
Homology data provide important information that can be used to predict the functions of unknown proteins of closely related species. A large number of gene or protein sequences are stored in many databases, but the functions of the majority of these proteins are unknown. An experimental analysis for all of them is almost impossible (Itoh et al., 2004). Thus, computational methods to predict protein functions are commonly adopted, and homology searching is the most efficient and prevalent means (Itoh et al., 2004). Interestingly, all of the selected genes express protein products with interrelated biological functions, such as ATPase and ion transportation. Thus, ADR genes and their partners with these functions can reasonably be expected to give SL interactions. In this study, we conjectured that the key metabolic pathways, housekeeping functions, and repertoire of predicted membrane transporters were highly conserved between the P. vivax and P. falciparum proteomes (Gardner et al., 2002). Consequently, the two species had similar metabolic potentials (Carlton et al., 2008). We surveyed the fitness data for the homolog yeast genes of the selected genes to find new drugs for clinical malaria treatment. In this survey, the yeast chemogenomic assays were used to predict its therapeutic potential in malaria (Hillenmeyer et al., 2010). The proposed drug candidates were searched in the DrugBank 3.0 database, which contains comprehensive information on the target diseases, proteins, genes, and organisms on which these drugs act. Researchers can obtain helpful information from the DrugBank database to verify a specific antimalarial therapy for the proposed antimalarial drug candidates. Some of the drug candidates in Table 3, such as methotrexate, fluoxuridine, mechlorethamine, fluorouracil, alverine, busulfan, and miconazole, have influence on humans. If these drugs are to be used as antimalarial drugs, the severity of their side effects should be checked. A drug can bind to a human protein even if it is not a close analogue of the parasite target, as long as the two have a strong similarity in their ligand-binding domains. Among the list of drug candidates in Table 3, caspofungin and itraconazole are worthwhile for drug screening because there is no report on toxicity or effect on the human organism for these two drugs. No significant lethality was observed when itraconazole was administered orally to mice and rats. Therefore, based on previous studies, we suggest a new methodology for antimalarial therapy. This methodology provides not only new potential gene candidates for further experimental tests and for drug design by supplying suitable Plasmodium SL gene pairs that can be used as a potential basis for future pharmacologic tests, but also information on new usage for existing drugs.
5. Conclusions
Taking a specific antimalarial drug is not always effective for some patients because of the mutation of ADR genes in malaria parasites. To solve this problem, we proposed a novel approach for searching for pertinent antimalarial drug targets based on the identification of SL pairs of proteins with ADR genes. We suggested a new concept of antimalarial therapy using data integration and inference by homology analysis of yeast–human–Plasmodium. Potential antimalarial drug targets and drug candidates were suggested for future experimental validation. ADR genes and their first neighbors extracted from the inferred Plasmodium SL gene networks can be used as alternative antimalarial drug targets for a malaria parasite with ADR genes. The inference of SL gene pairs in Plasmodium and humans was described based on yeast SL gene data. Yeast SL interaction data as well as homologous information on Plasmodium, human, yeast, and ADR genes were integrated to provide a list of candidate genes for malarial therapy. The proposed candidates can be used as drug targets in antimalarial therapy for future experimental validation. If one gene of a Plasmodium SL gene pair has a mutation that causes ADR, the drug targeted at another gene can be used for effective therapy with largely reduced injury to human cells.
The Plasmodium cells can be killed, while keeping the human cells intact by mutating or blocking the SL partner of the mutated gene that causes ADR. SL gene pairs have been used to kill malaria parasites selectively and leave human cells unharmed. The present SL screening analysis and the use of highly curated databases are very helpful for discovering proper drugs and identifying correct gene targets in antimalarial research. In conclusion, the present computational approach is inexpensive and very simple to adopt. The approach also has strong potential in the preparation of chemical therapy with SL gene-specific chemical drugs or gene therapy and can help identify suitable drug candidates with apparent antimalarial function for clinical malaria treatments.
Acknowledgements
We thank Sunghyun Kim, Jae Hong Lim, Sungsook Ahn, Vivek Gupta, Jeongeun Ryu, and Youngran Ha for their thoughtful discussions and support during this project. This research was supported by the World Class University program of the National Research Foundation (NRF) of Korea, and funded by the Creative Research Initiatives (Center for Biofluid and Biomimic Research) of the MSIP and NRF of Korea (No. 2008-0061991).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Appendix A. Supplementary data
References
- Anderson T. Mapping the spread of malaria drug resistance. PLoS Med. 2009;6(4):e1000054. doi: 10.1371/journal.pmed.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh L.R., Wen J.C., Hill A.A., Slonim D.K., Brown E.L., Hunter C.P. Synthetic lethal analysis of Caenorhabditis elegans posterior embryonic patterning genes identifies conserved genetic interactions. Genome Biol. 2005;6(5):r45. doi: 10.1186/gb-2005-6-5-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F.C., Koetzle T.F., Williams G.J., Meyer E.F., Jr., Brice M.D., Rodgers J.R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Boone C., Bussey H., Andrews B.J. Exploring genetic interactions and networks with yeast. Nat. Rev. Genet. 2007;8(6):437–449. doi: 10.1038/nrg2085. [DOI] [PubMed] [Google Scholar]
- Briolant S., Bogreau H., Gil M., Bouchiba H., Baret E., Amalvict R., Rogier C., Pradines B. The F423Y mutation in the pfmdr2 gene and mutations N51I, C59R, and S108N in the pfdhfr gene are independently associated with pyrimethamine resistance in Plasmodium falciparum isolates. Antimicrob. Agents Chemother. 2012;56:2750–2752. doi: 10.1128/AAC.05618-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J.M., Adams J.H., Silva J.C., Bidwell S.L., Lorenzi H., Caler E., Crabtree J., Angiuoli S.V., Merino E.F., Amedeo P., Cheng Q., Coulson R.M., Crabb B.S., Del Portillo H.A., Essien K., Feldblyum T.V., Fernandez-Becerra C., Gilson P.R., Gueye A.H., Guo X., Kang’a S., Kooij T.W., Korsinczky M., Meyer E.V., Nene V., Paulsen I., White O., Ralph S.A., Ren Q., Sargeant T.J., Salzberg S.L., Stoeckert C.J., Sullivan S.A., Yamamoto M.M., Hoffman S.L., Wortman J.R., Gardner M.J., Galinski M.R., Barnwell J.W., Fraser-Liggett C.M. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008;455(7214):757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipman K., Singh A. Predicting genetic interactions with random walks on biological networks. BMC Bioinformatics. 2009;10:17. doi: 10.1186/1471-2105-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde-Pueyo N., Munteanu A., Solé R.V., Rodríguez-Caso C. Human synthetic lethal inference as potential anti-cancer target gene detection. BMC Syst. Biol. 2009;3:116. doi: 10.1186/1752-0509-3-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman A.F., Karcz S., Galatis D., Culvenor J.G. A P-glycoprotein homologue of Plasmodium falciparum is localized on the digestive vacuole. J. Cell Biol. 1991;113:1033–1042. doi: 10.1083/jcb.113.5.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman A.F., Galatis D., Thompson J.K. Selection for mefloquine resistance in Plasmodium falciparum is linked to amplification of the pfmdr1 gene and cross-resistance to halofantrine and quinine. Proc. Natl. Acad. Sci. USA. 1994;91:1143–1147. doi: 10.1073/pnas.91.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox-Singh J., Davis T.M., Lee K.S., Shamsul S.S., Matusop A., Ratnam S., Rahman H.A., Conway D.J., Singh B. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin. Infect. Dis. 2008;46:165–171. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S.J., Fedyshyn Y., Koh J.L.Y., Prasad T.S.K., Chahwan C., Chua G., Toufighi K., Baryshnikova A., Hayles J., Hoe K.L., Kim D.U., Park H.O., Myers C.L., Pandey A., Durocher D., Andrews B.J., Boone C. Significant conservation of synthetic lethal genetic interaction networks between distantly related eukaryotes. Proc. Natl. Acad. Sci. USA. 2008;105(43):16653–16658. doi: 10.1073/pnas.0806261105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenstein M. Drug development: holding out for reinforcements. Nature. 2012;484:s16–s18. doi: 10.1038/484S16a. [DOI] [PubMed] [Google Scholar]
- Fernández A., Crespo A., Tiwari A. Is there a case for selectively promiscuous anticancer drugs? Drug Discov. Today. 2009;14(1–2):1–5. doi: 10.1016/j.drudis.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Foote S.J., Kyle D.E., Martin R.K., Oduola A.M., Forsyth K., Kemp D.J., Cowman A.F. Several alleles of the multidrug-resistance gene are closely linked to chloroquine resistance in Plasmodium falciparum. Nature. 1990;345:255–258. doi: 10.1038/345255a0. [DOI] [PubMed] [Google Scholar]
- Forsburg S.L. The art and design of genetic screens: yeast. Nat. Rev. Genet. 2001;2(9):659–668. doi: 10.1038/35088500. [DOI] [PubMed] [Google Scholar]
- Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., Paulsen I.T., James K., Eisen J.A., Rutherford K., Salzberg S.L., Craig A., Kyes S., Chan M.S., Nene V., Shallom S.J., Suh B., Peterson J., Angiuoli S., Pertea M., Allen J., Selengut J., Haft D., Mather M.W., Vaidya A.B., Martin D.M., Fairlamb A.H., Fraunholz M.J., Roos D.S., Ralph S.A., McFadden G.I., Cummings L.M., Subramanian G.M., Mungall C., Venter J.C., Carucci D.J., Hoffman S.L., Newbold C., Davis R.W., Fraser C.M., Barrell B. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giancarlo A.B., Patrick G.B., Stephen A.W.D.L. Mechanisms of antimalarial drug resistance. In: Mayers D.L., editor. Antimicrobial Drug Resistance. Humana Press; New York: 2009. pp. 561–574. [Google Scholar]
- Goh K.I., Cusick M.E., Valle D., Childs B., Vidal M., Barabási A.L. The human disease network. Proc. Natl. Acad. Sci. USA. 2007;104(21):8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M.E., Fung E., Wildenhain J., Pierce S.E., Hoon S., Lee W., Proctor M., St Onge R.P., Tyers M., Koller D., Altman R.B., Davis R.W., Nislow C., Giaever G. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320(5874):362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillenmeyer M.E., Ericson E., Davis R.W., Nislow C., Koller D., Giaever G. Systematic analysis of genome-wide fitness data in yeast reveals novel gene function and drug action. Genome Biol. 2010;11(3):r30. doi: 10.1186/gb-2010-11-3-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthmacher C., Hoppe A., Bulik S., Holzhütter H.G. Antimalarial drug targets in Plasmodium falciparum predicted by stage-specific metabolic network analysis. BMC Syst. Biol. 2010;4:120. doi: 10.1186/1752-0509-4-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M., Akutsu T., Kanehisa M. Clustering of database sequences for fast homology search using upper bounds on alignment score. Genome Inform. 2004;15(1):93–104. [PubMed] [Google Scholar]
- Jorgensen E.M., Mango S.E. The art and design of genetic screens: Caenorhabditis elegans. Nat. Rev. Genet. 2002;3(5):356–369. doi: 10.1038/nrg794. [DOI] [PubMed] [Google Scholar]
- Knox C., Law V., Jewison T., Liu P., Ly S., Frolkis A., Pon A., Banco K., Mak C., Neveu V., Djoumbou Y., Eisner R., Guo A.C., Wishart D.S. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B., Crombie C., Tischler J., Fortunato A., Fraser A.G. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat. Genet. 2006;38(8):896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- Maere S., Heymans K., Kuiper M. Bingo: a cytoscape plugging to assess over-representation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Mutabingwa T., Nzila A., Mberu E., Nduati E., Winstanley P., Hills E., Watkins W. Chlorproguanil-dapsone for treatment of drug- resistant falciparum malaria in Tanzania. Lancet. 2001;358:1218–1223. doi: 10.1016/S0140-6736(01)06344-9. [DOI] [PubMed] [Google Scholar]
- Nakaya A., Katayama T., Itoh M., Hiranuka K., Kawashima S., Moriya Y., Okuda S., Tanaka M., Tokimatsu T., Yamanishi Y., Yoshizawa A.C., Kanehisa M., Goto S. KEGG OC: a large-scale automatic construction of taxonomy-based ortholog clusters. Nucleic Acids Res. 2013;41:D353–D357. doi: 10.1093/nar/gks1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi T., Akiyama Y. PDB-REPRDB: a database of representative protein chains from the Protein Data Bank (PDB) in 2003. Nucleic Acids Res. 2003;31(1):492–493. doi: 10.1093/nar/gkg022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S.L., Pan X., Peyser B.D., Ye P., Meluh P.B., Yuan D.S., Irizarry R.A., Bader J.S., Spencer F.A., Boeke J.D. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 2006;22:56–63. doi: 10.1016/j.tig.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Paladugu S.R., Zhao S., Ray A., Raval A. Mining protein networks for synthetic genetic interactions. BMC Bioinformatics. 2008;9:426. doi: 10.1186/1471-2105-9-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Yuan D.S., Xiang D., Wang X., Sookhai-Mahadeo S., Bader J.S., Hieter P., Spencer F., Boeke J.D. A robust toolkit for functional profiling of the yeast genome. Mol. Cell. 2004;16(3):487–496. doi: 10.1016/j.molcel.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Peel S.A., Merritt S.C., Handy J., Baric R.S. Derivation of highly mefloquine-resistant lines from Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 1993;48:385–397. doi: 10.4269/ajtmh.1993.48.385. [DOI] [PubMed] [Google Scholar]
- Peel S.A., Bright P., Yount B., Handy J., Baric R.S. A strong association between mefloquine and halofantrine resistance and amplification, overexpression, and mutation in the P-glycoprotein gene homolog (pfmdr) of Plasmodium falciparum in vitro. Am. J. Trop. Med. Hyg. 1994;51:648–658. doi: 10.4269/ajtmh.1994.51.648. [DOI] [PubMed] [Google Scholar]
- Platzer A., Perco P., Lukas A., Mayer B. Characterization of protein-interaction networks in tumors. BMC Bioinformatics. 2007;8:224. doi: 10.1186/1471-2105-8-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R.N., Tjitra E., Guerra C.A., Yeung S., White N.J., Anstey N.M. Vivax malaria: neglected and not benign. Am. J. Trop. Med. Hyg. 2007;77:79–87. [PMC free article] [PubMed] [Google Scholar]
- Reed M.B., Saliba K.J., Caruana S.R., Kirk K., Cowman A.F. Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature. 2000;403:906–909. doi: 10.1038/35002615. [DOI] [PubMed] [Google Scholar]
- Rozmajzl P.J., Kimura M., Woodrow C.J., Krishna S., Meade J.C. Characterization of P-type ATPase 3 in Plasmodium falciparum. Mol. Biochem. Parasitol. 2001;116(2):117–126. doi: 10.1016/s0166-6851(01)00319-x. [DOI] [PubMed] [Google Scholar]
- Rubio J.P., Cowman A.F. The ATP-binding cassette (ABC) gene family of Plasmodium falciparum. Parasitol. Today. 1996;12:135–140. doi: 10.1016/0169-4758(96)10003-x. [DOI] [PubMed] [Google Scholar]
- Schlatter I.D., Meira M., Ueberschlag V., Hoepfner D., Movva R., Hynes N.E. MHO1, an evolutionarily conserved gene, is synthetic lethal with PLC1; Mho1p has a role in invasive growth. PLoS One. 2012;7(3):e32501. doi: 10.1371/journal.pone.0032501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibley C.H., Hyde J.E., Sims P.F., Plowe C.V., Kublin J.G., Mberu E.K., Cowman A.F., Winstanley P.A., Watkins W.M., Nzila A.M. Pyrimethamine–sulfadoxine resistance in Plasmodium falciparum: what next? Trends Parasitol. 2001;17:582–588. doi: 10.1016/s1471-4922(01)02085-2. [DOI] [PubMed] [Google Scholar]
- Sonnhammer E.L.L., Koonin E.V. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18(12):619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- Tarailo M., Tarailo S., Rose A.M. Synthetic lethal interactions identify phenotypic “interologs” of the spindle assembly checkpoint components. Genetics. 2007;177(4):2525–2530. doi: 10.1534/genetics.107.080408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischler J., Lehner B., Fraser A.G. Evolutionary plasticity of genetic interaction networks. Nat. Genet. 2008;40(4):390–391. doi: 10.1038/ng.114. [DOI] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Pagé N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., Andrews B., Tyers M., Boone C. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294(5550):2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Tong A.H.Y., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., Chen Y., Cheng X., Chua G., Friesen H., Goldberg D.S., Haynes J., Humphries C., He G., Hussein S., Ke L., Krogan N., Li Z., Levinson J.N., Lu H., Ménard P., Munyana C., Parsons A.B., Ryan O., Tonikian R., Roberts T., Sdicu A.M., Shapiro J., Sheikh B., Suter B., Wong S.L., Zhang L.V., Zhu H., Burd C.G., Munro S., Sander C., Rine J., Greenblatt J., Peter M., Bretscher A., Bell G., Roth F.P., Brown G.W., Andrews B., Bussey H., Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303(5659):808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- Wellems T.E., Panton L.J., Gluzman I.Y., Rosario V.E., Gwadz R.W., Walker-Jonah A., Krogstad D.J. Chloroquine resistance not linked to mdr-like genes in a Plasmodium falciparum cross. Nature. 1990;345:253–255. doi: 10.1038/345253a0. [DOI] [PubMed] [Google Scholar]
- Wilson C.M., Serrano A.E., Wasley A., Bogenschutz M.P., Shankar A.H., Wirth D.F. Amplification of a gene related to mammalian mdr genes in drug-resistant Plasmodium falciparum. Science. 1989;244:1184–1186. doi: 10.1126/science.2658061. [DOI] [PubMed] [Google Scholar]
- Yeh I., Hanekamp T., Tsoka S., Karp P.D., Altman R.B. Computational analysis of Plasmodium falciparum metabolism: organizing genomic information to facilitate drug discovery. Genome Res. 2004;14:917–924. doi: 10.1101/gr.2050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim M.A., Goh K.I., Cusick M.E., Barabási A.L., Vidal M. Drug-target network. Nat. Biotechnol. 2007;25:1119–1126. doi: 10.1038/nbt1338. [DOI] [PubMed] [Google Scholar]
- Yuen K.W.Y., Warren C.D., Chen O., Kwok T., Hieter P., Spencer F.A. Systematic genome instability screens in yeast and their potential relevance to cancer. Proc. Natl. Acad. Sci. USA. 2007;104(10):3925–3930. doi: 10.1073/pnas.0610642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalis M.G., Wilson C.M., Zhang Y., Wirth D.F. Characterization of the pfmdr2 gene for Plasmodium falciparum. Mol. Biochem. Parasitol. 1993;62:83–92. doi: 10.1016/0166-6851(93)90180-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


