Graphical abstract
Highlights
► We examined the relative importance of the 198 and 200 beta-tubulin SNPs in a benzimidazole resistant H. contortus isolate. ► The most resistant worms in the population showed high resistance allele frequencies at the 198 position. ► A degree of mutual exclusivity was apparent for the 198 and 200 resistance SNPs. ► Interactions between resistance mechanisms need to be understood in order to interpret molecular resistance tests.
Keywords: Haemonchus contortus, Benzimidazole, Resistance, Beta tubulin
Abstract
While the F200Y SNP in the beta-tubulin gene is most commonly associated with benzimidazole resistance in trichostrongylid nematodes, other SNPs as well as drug efflux pathways have been implicated in the resistance. The relative contributions of all these mechanisms are not understood sufficiently to allow expected drug efficacy to be inferred from molecular data. As a component of developing better means to interpret molecular resistance tests, the present study utilised a drug resistant Haemonchus contortus isolate which possesses two of the principal benzimidazole resistance SNPs (E198A and F200Y) in order to assess the relative degree of resistance conferred by the two SNPs. We exposed larvae to a range of thiabendazole concentrations in in vitro development assays, and collected the surviving L3 larvae at each drug concentration to establish sub-populations showing increasing levels of resistance. We then sequenced the isotype 1 beta-tubulin gene in pooled larval samples, and measured allele frequencies at the two SNP positions. The frequency of the resistance allele at the 198 position increased as the thiabendazole concentration increased, while the frequency of the resistance allele at the 200 position decreased. Genotyping of individual larvae showed that the highest drug concentration was associated with the removal of all genotypes except for homozygous resistance at the 198 position alongside homozygous susceptible at the 200 position. This indicates that, at least for larval life stages, the E198A SNP is able to confer higher levels of resistance to benzimidazole drugs than the F200Y SNP, and that the homozygosity at 198 in the highly resistant individuals is mutually exclusive with heterozygosity or resistant homozygosity at the 200 position. This study illustrates the need to understand the relative contributions of different resistance mechanisms in order to maximise the degree to which molecular tests are able to inform on drug resistance phenotype.
1. Introduction
As increasing anthelmintic resistance threatens the sustainability of livestock industries worldwide (Kaplan, 2004; Sutherland and Leathwick, 2011) there is a need to monitor resistance levels so that appropriate drug-use decisions can be made on grazing properties. The only readily available means of determining the resistance status of ruminant nematodes remains the faecal egg count reduction test (FECRT), which is laborious and lacks sensitivity. In vitro tests on the free-living life stages of some important helminth parasites have been developed, however they have not been widely applied in the field as they generally also lack sensitivity, and in some cases apply to only a limited number of drug/species combinations (Coles et al., 2006). Molecular-based tests offer much greater sensitivity, however they are not available for use at present due largely to an incomplete knowledge of the molecular basis of anthelmintic resistances, and, hence, uncertainty as to the nature of the relationship between genotype and resistance phenotype. The FECRT therefore remains the test of choice for graziers.
Among the commonly used anthelmintic drug groups, the molecular basis of resistance has been most studied, and is considered to be most understood, for the benzimidazole group (Wolstenholme et al., 2004; von Samson-Himmelstjerna, 2006; von Samson-Himmelstjerna et al., 2007). Haemonchus contortus has been well studied in this regard, and may be the best characterized of the economically-important species. However an examination of current knowledge on the basis of benzimidazole resistance in H. contortus reveals a degree of uncertainty in the ability to interpret the results of molecular-based resistance tests. Benzimidazole resistance in this species is considered to be primarily due to a F200Y SNP in the beta-tubulin gene (Kwa et al., 1994, 1995). However, a F167Y SNP has also been found to be associated with resistance (Prichard, 2001), and more recently, another SNP, E198A, has also been implicated in resistance in some isolates (Ghisi et al., 2007; de Lourdes Mottier and Prichard, 2008; Rufener et al., 2009). In addition, P-glycoprotein may play a role in resistance to these drugs (Blackhall et al., 2008).
The primary role of the F200Y SNP in benzimidazole resistance in trichostrongylid nematodes is well established (Kwa et al., 1994; Beech et al., 1994; Elard et al., 1996; Elard and Humbert, 1999; von Samson-Himmelstjerna et al., 2009). However, the existence of the other mechanisms described above, and a lack of knowledge as to the relative effects of multiple mechanisms in cases where they may occur simultaneously, illustrates the need to develop an approach to molecular testing based on a broader understanding of all contributing mechanisms, and the degree to which each component will impact on the observed resistance phenotype. This will allow for a more informed interpretation of molecular data. The presence of both the E198A and F200Y SNPs in an isolate of H. contortus from Australia (previously described by de Lourdes Mottier and Prichard, 2008) provided an opportunity to evaluate the relative contributions of the two SNPs to the observed level of resistance. The present study aimed to examine genotypes in this Wallangra isolate of H. contortus by dividing L3 stage larvae into sub-populations based on their ability to survive increasing thiabendazole concentrations in a larval development assay, that is, on their relative resistance status. This approach offered significant advantages over the previous comparisons of genotypes between populations showing different levels of resistance (often multiple drug resistances) but also differing significantly in genetic background, hence making it difficult to associate genotype with resistance to specific drugs. In contrast, the present study aimed to avoid these issues by subdividing a single population of worms on the basis of resistance of larval life stages to a single drug (thiabendazole) in order to be able to examine correlations between genotype and resistance to just this single compound. Resistance allele frequencies at both the 198 and 200 SNP positions (GCA and TAC, respectively) were measured for each of the sub populations recovered from larval development assays at different concentrations of thiabendazole. The original and most highly drug-pressured populations were compared further by genotyping individual larvae at the two SNP positions.
2. Materials and methods
2.1. Parasites
The H. contortus used for this study were from the drug susceptible Kirby 1986 (Albers and Burgess, 1988) isolate and the multi-drug resistant isolate Wallangra 2003 (Love et al., 2003). Prior to being used in the present study these isolates were kept as frozen stock as described by Hunt et al. (2008). Infections were maintained in sheep at the CSIRO Chiswick Laboratory in Armidale, and faeces was sent regularly by courier to the CSIRO laboratory in Brisbane for either recovery of eggs, or to establish cultures to provide L3 stage larvae for molecular analyses.
2.2. Larval development assay
Larval development assays were conducted using an agar-based 96-well format modified from Gill et al. (1995). The thiabendazole concentration gradient ranged from 0.005 to 10.4 μg/ml. Nematode eggs were recovered from sheep faeces by filtration and sucrose density centrifugation, and placed onto assay plates. The next day, each well of the plate received 10 μl of a nutrient solution containing a fresh culture of Escherichia coli diluted in growth medium (Hubert and Kerboeuf, 1984) as described by Kotze et al. (2009). For initial assessment of IC50 values (defined as the concentration of drug required to inhibit the development of larvae to the infective L3 stage to 50% of that observed in control wells), the assays were terminated after 6 days by the addition of Lugol’s iodine and the numbers of L3 larvae were counted and compared to control (no drug) wells.
Subsequent experiments involved the exposure of larvae to thiabendazole in the agar-based format described above, but at the end of the incubation period the surviving larvae were collected rather than killed with iodine. Whole 96-well plates were set-up with a single concentration of thiabendazole in each well (concentrations: 0.33, 0.65, 1.3, 2.6 μg/ml), and eggs were added to each well on day 0. The larvae were fed on day 1 as described above. On day 6, the larvae were flushed out of the wells using water and placed into the upper chamber of a filter apparatus. They were allowed to migrate through a 20 μm mesh filter over several hours into the lower chamber. Larvae were recovered from the lower chamber and several aliquots were counted in order to determine the total number of individuals. Larvae were frozen at −20 °C in groups of approximately 3000–8000 for later molecular analysis (= pooled larval samples). For genotyping of individuals, the larvae were collected as described above and then exsheathed by incubation in 0.9% saline at 42 °C with a gas mixture consisting of 40% CO2 and 60% N2 bubbling through the solution. Larvae were examined under a microscope to confirm exsheathment, and individuals were picked into separate wells of 96-well plates in approximately 2 μl of water, and frozen at −20 °C for later molecular analysis.
2.3. Preparation of genomic DNA, PCR, and sequencing
DNA was prepared from four sample types:
-
(1)
Pooled Kirby or Wallangra L3 larvae that had migrated from faecal cultures (10,000 per sample).
-
(2)
Pooled Wallangra L3 larvae recovered from LDA plates containing thiabendazole at one of four concentrations (3000–8000 per sample).
-
(3)
Individual Wallangra L3 larvae that had migrated from faecal cultures.
-
(4)
Individual Wallangra L3 larvae recovered from LDA plates containing thiabendazole at 2.6 μg/ml.
Pooled larval samples were homogenised by shaking in a 2 ml tube containing three 3 mm glass beads and approximately 0.3 g of 1 mm ceramic zirconia silica beads, and 160 μl lysis solution (Qiagen QIAamp DNA miniprep kit) using a Savant FP 120 BIO121 bead beater. DNA was then recovered following the QIAamp DNA miniprep kit instructions.
DNA was recovered from individual L3 stage larvae using the technique described by von Samson-Himmelstjerna et al. (2009). Larvae were digested overnight in 10 μl of a digestion buffer (10 mM Tris–HCl, pH 8.3, containing 50 mM KCl, 2.5 mM MgCl2, 0.45% Nonident p-40, 0.45% Tween-20, and 100 μg/ml proteinase K), followed by a 20 min incubation at 75 °C.
PCR primers were designed to differentiate between H. contortus beta-tubulin isotype 1 and 2 genes (Table 1). PCR conditions for pooled larvae DNA samples were as follows: 98 °C for 30 s, followed by 35 cycles of 98 °C for 10 s, 60 °C for 30 s and 72 °C for 25 s, followed by 72 °C for 10 min, using Finnzymes Phusion Hot Start Polymerase II. PCR on single larva DNA was as follows: 98 °C for 30 s, followed by 50 cycles of 98 °C for 10 s, 57 °C for 15 s and 72 °C for 45 s, followed by 72 °C for 7 min, using BioRad iProof high-fidelity DNA polymerase. PCR products were examined on 1.5% agarose gels.
Table 1.
Primer sequences for H. contortus beta-tubulin isotype 1 and 2 PCRs, and sequencing across the 198 and 200 SNP positions of isotype 1 in the forward and reverse directions.
| Purpose | Sequence 5′–3′ | Expected product size |
|---|---|---|
| PCR | ||
| Pooled larvae | ||
| Isotype 1 forward | TTCTGGACCGTATGGACAG | 895 |
| Isotype 1 reverse | GTAAGCTCAGCAACTGTCGAA | |
| Isotype 2 forward | GCTCCGGACCTTTTGGTGCT | 2176 |
| Isotype 2 reverse | CGTGAGCTCAGAAACAGTTAGT | |
| Single larvae | ||
| Isotype 1 forward | CTAGTTGATAACGTATTAGACGTTGTC | 791 |
| Isotype 1 reverse | GTAAGCTCAGCAACTGTCGAA | |
| Sequencing | ||
| Isotype 1 forward | GAGGCACTGGATCTGGAATG | – |
| Isotype 1 reverse | TCTCCATAGGTTGGATTTGTGAG | – |
Sequencing reaction methods differed for the pooled and single larvae preparations. For pooled larvae, the PCR products were purified using Qiagen QIAquick spin columns as per the manufacturers’ instructions. For individual larvae, PCR products were prepared for sequencing by incubation in Exonuclease I (New England Biolabs) (6000 U/μl), calf intestinal alkaline phosphatase (Finnzymes) (6.3 U/μl) and sequencing buffer for 97 °C for 20 min, followed by 80 °C for 20 min. Sequencing reactions were then performed for pooled and individual DNA with the sequencing primers listed in Table 1 using BigDye Terminator 3.1 (BDT) (Applied Biosystems). BDT reaction products were prepared for sequencing by either ethanol precipitation (pooled larval DNA) or using the Agencourt CleanSEQ cleanup protocol (individual larval DNA). All samples were sequenced using an ABI Prism 3100 Genetic Analyser.
2.4. Determination of allele frequencies
Sequence traces were examined using ChromasPro version 1.5 software, focusing only on the F167Y (TTC/TAC), E198A (GAA/GCA), and F200Y (TTC/TAC) SNPs. For pooled larvae, when only one peak was present, the sample was recorded as being 100% of the resistant or susceptible allele at that position; where two peaks were present, the height of the nucleotide known to be associated with resistance was expressed as a percentage of the combined height of the two peaks. For individual larvae, the presence of a single peak was recorded as indicating that the individual was homozygous (either susceptible or resistant) at that position, while the presence of two peaks indicated that the individual was heterozygous at that position. The threshold value of heterozygote detection was set at 50%, that is, a secondary peak was only considered indicative of heterozygosity if its height was at least 50% of the dominant peak. An online calculator tool (available at: http://www.tufts.edu/~mcourt01/Documents/Court%20lab%20-%20HW%20calculator.xls) was used to determine if the observed genotype frequencies were consistent with the Hardy–Weinberg equilibrium.
3. Results
Dose responses of the Wallangra and Kirby isolates in larval development assays with thiabendazole are shown in Fig. 1. The Wallangra isolate showed a significant level of resistance to the drug: IC50 (95% Confidence Interval) Kirby 0.030 μg/ml (0.025–0.036), Wallangra 0.397 μg/ml (0.326–0.482), resistance ratio 13. The arrows on Fig. 1 indicate the drug concentration levels subsequently used to select for sub-populations of the Wallangra isolate that were able to survive increasing concentrations of the drug.
Fig. 1.
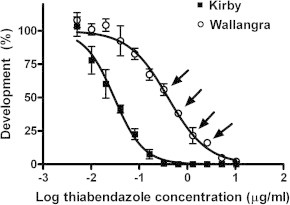
Response of Kirby and Wallangra isolates of H. contortus to thiabendazole in larval development assays. Each data point represents mean ± SE, n = 3 separate assay wells. Where SEs are not visible, they are smaller than the dimensions of the data symbols. Arrows indicate the drug concentrations that were used subsequently to partition the Wallangra isolate into sub-populations with different levels of drug resistance.
While beta-tubulin isotype 2 PCRs with Kirby routinely produced an amplicon of expected size, only rarely was a product generated with DNA prepared from Wallangra worms. Hence, the genotyping aspects of this study were only performed on PCR amplicons generated using primers specific for isotype 1.
Genotyping was initially performed on pooled larval samples by simple measurement of relative sequence-trace peak heights, and expression of the height of the peak for the resistance-associated nucleotide at each SNP position as a % of the combined heights of both nucleotides at each position (Table 2). The Kirby isolate primarily showed the susceptible genotypes at each position of interest, however it did show a low level of the resistance allele at the 200 position (TAC), as well a trace of the resistance allele at the 167 position (TAC), however the latter was only present in one out of the three samples examined. The Wallangra isolate showed significant levels of both the E198A and F200Y SNPs. In all pairs of forward and reverse sequence data, the resistance allele frequencies at either position were higher when the PCR products were sequenced in the reverse direction than in the forward direction.
Table 2.
Resistance allele frequencies for the F167Y, E198A and F200Y SNPs in isotype 1 beta-tubulin from pooled larval samples of Kirby and Wallangra isolates of H. contortus.
| Isolate | SNP position | Sequence direction | % Resistant allele (mean ± SE)a |
|---|---|---|---|
| Kirby | 167 | For | 0 |
| Rev | 4 ± 4 | ||
| 198 | For | 0 | |
| Rev | 0 | ||
| 200 | For | 7 + 3 | |
| Rev | 16 ± 3 | ||
| Wallangra | 167 | For | 0 |
| Rev | 2 ± 1 | ||
| 198 | For | 40 ± 2b | |
| Rev | 58 ± 2 | ||
| 200 | For | 32 ± 2b | |
| Rev | 38 ± 1 | ||
n = 3 separate pooled samples of larvae.
Denotes that within an isolate, and within a SNP position, the forward and reverse sequencing data were significantly different (P < 0.05) (data were not analysed where means of one or both sequencing directions were zero).
The results of genotyping at the 198 and 200 positions for pooled samples of Wallangra larvae (not treated with thiabendazole) as well as the survivors of the various drug concentrations (from Fig. 1) are shown in Fig. 2. It is clear that as the concentration of thiabendazole in the assay increased, the resistance allele frequency at position 198 increased, alongside a decrease at the 200 position. This relationship was apparent for both forward and reverse-sequenced samples, although, as described above, the resistance allele frequencies were consistently higher for the reverse sequences.
Fig. 2.
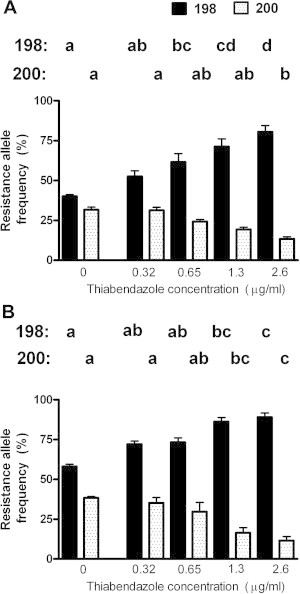
Resistance allele frequencies at the 198 and 200 SNP positions in isotype 1 beta-tubulin from pooled samples of Wallangra larvae not exposed to thiabendazole, or populations of Wallangra larvae that had survived various levels of the drug in larval development assays; data was derived from sequencing reactions in the forward (A) or reverse (B) directions. Each column represents mean ± SE, n = 3 separate pooled larval samples. Within a SNP position, columns labelled with different letters are significantly different (P < 0.05).
Table 3 shows individual larval genotypes for Wallangra worms not treated with thiabendazole as well as the survivors of 2.6 μg/ml. The Wallangra isolate was primarily composed of larvae which were heterozygous at both positions (45.5%) or homozygous resistant at one position and homozygous susceptible at the other (21.8% homozygous resistant at 198, and 23.7% homozygous resistant at 200). The genotype frequencies at both positions were consistent with the Hardy–Weinberg equilibrium (P = 0.69 at 198, P = 0.73 at 200) in the population that was not treated with thiabendazole. No individuals were homozygous resistant at one position alongside heterozygosity at the other, and none were homozygous resistant at both positions. All larvae which were able to survive 2.6 μg/ml thiabendazole were homozygous resistant at the 198 SNP, alongside homozygous susceptible at the 200 position. The allele frequency for 198A was 0.50 in the absence of selection and 1.0 in the thiabendazole-selected individuals, while the allele frequency of 200Y was 0.45 in the absence of selection and 0.0 in the thiabendazole-selected individuals; both observed differences in allele frequency were highly significant (P < 0.01).
Table 3.
Individual worm genotypes at the 198 and 200 SNP positions in isotype 1 beta-tubulin for Wallangra worms not exposed to thiabendazole and the survivors of exposure to the drug at 2.6 μg/ml in larval development assays.
| Genotypea | Wallangrab |
Wallangra 2.6 μg/mlc |
||
|---|---|---|---|---|
| Number of larvae | % of total | Number of larvae | % of total | |
| Hs-198, Hs-200 | 0 | 0 | 0 | 0 |
| Het-198, Hs-200 | 4 | 7.2 | 0 | 0 |
| Hs-198, Het-200 | 1 | 1.8 | 0 | 0 |
| Het-198, Het-200 | 25 | 45.5 | 0 | 0 |
| Hs-198, HR-200 | 12 | 21.8 | 0 | 0 |
| HR-198, Hs-200 | 13 | 23.7 | 62 | 100 |
| HR-198, Het-200 | 0 | 0 | 0 | 0 |
| Het-198, HR-200 | 0 | 0 | 0 | 0 |
| HR-198, HR-200 | 0 | 0 | 0 | 0 |
Hs = homozygous susceptible; Het = heterozygote; HR = homozygous resistant.
Total of 55 larvae genotyped from the group not exposed to drug.
Total of 62 larvae genotyped from the group which survived 2.6 μg/ml thiabendazole.
4. Discussion
The present study has for the first time examined the relative degree of resistance conferred by different beta-tubulin SNPs in a parasitic nematode. Many studies have compared genotypes in different populations with quite different genetic backgrounds and often with only one relevant SNP present (F200Y), and found that resistance allele frequencies are related to some degree with the observed levels of resistance. In contrast, this study has examined a single population which possesses two relevant SNPs, and then subdivided it on the basis of relative resistance status in order to highlight the resistance mechanism utilised by the most resistant individuals in the population. The study has indicated that the E198A SNP confers a higher level of resistance than does the F200Y SNP in the larvae of a field isolate of H. contortus. It is clear that while the Wallangra isolate contains a significant number of larvae showing heterozygosity at both the 198 and 200 positions, or homozygous resistant at one position only, it is the individuals which are homozygous resistant at the 198 position that show the highest levels of resistance.
An important limitation of the present study is that we have examined the relationship between beta-tubulin SNPs and benzimidazole sensitivity in larval stages of H. contortus, whereas the relationship in adult life stages will be more important in terms of the impact of the beta-tubulin SNPs on the ability to control worms with benzimidazole drugs in the field. The relationship between SNP levels and the ability of adult worms to survive increasing concentrations of benzimidazole drugs remains to be determined. However, the response of H. contortus in larval development assays is known to be indicative of the relative sensitivity of adult stages to benzimidazole drugs (Coles et al., 2006). Hence, it is likely that our association of the relative impact of the E198A and F200Y SNPs on drug sensitivity in the larval stages is also indicative of adult worm relationships.
We found that larvae that were homozygous resistant at one position were always homozygous susceptible at the other SNP. While individuals could be heterozygous at both positions, no heterozygosity was seen in larvae homozygous resistant at the other position. This suggests a degree of fitness disadvantage associated with these combinations that may otherwise be expected to confer high levels of resistance. Similarly, de Lourdes Mottier and Prichard (2008) reported an absence of double homozygous resistant individuals among a Wallangra population. Ghisi et al. (2007) found that E198A was not observed in sequences from resistant H. contortus isolates that had F200Y, suggesting that the two SNPs were mutually exclusive. This is also suggested by the data in Fig. 2. Increasing larval development assay drug concentrations led to an increasing resistance allele frequency at the 198 SNP position, with a concomitant decrease in the resistance allele frequency at the 200 position.
While PCRs of isotype 2 beta tubulin routinely produced amplicons with DNA from pooled samples of Kirby larvae, such PCRs often failed to produce a product visible on agarose gels for Wallangra. This is consistent with the reported loss of isotype 2 beta-tubulin in resistant isolates of H. contortus (Kwa et al., 1993a,b).
The measurement of genotype frequencies for pooled DNA samples using sequence trace peak heights (from von Samson-Himmelstjerna et al., 2007) was useful for the present study in describing the relationship between SNP frequencies and in vitro resistance status, however its general use as a worm genotyping tool is questionable. This method at best provides only an estimate of genotype frequencies. We found that when resistant genotypes were present at only low levels the method was inconsistent between separate sequencing reactions, which resulted in large standard errors when mean values were low (from Table 2). In addition, the frequency values were consistently higher for the reverse direction. This may be due to different nucleotide incorporation efficiencies for the various nucleotides under our experimental conditions. Different nucleotide incorporation efficiencies are known to effect relative peak heights within a sequence trace, as well as relative peak heights between sequencing reactions primed in the forward or reverse directions (for example, Parker et al., 1995, 1996; Hancock et al., 2005). More appropriate methods for routine molecular resistance testing have been reported, most notably by von Samson-Himmelstjerna et al. (2009) who described pyrosequencing and real time PCR assays useful for benzimidazole resistance testing at each of the candidate beta-tubulin SNP positions.
In order for molecular tests to adequately describe the resistance status of worm populations, a better understanding of the relationship between molecular data, either in the form of genotype frequencies (specifically, beta-tubulin resistance allele frequencies for benzimidazole resistance) or gene expression levels (for example, P-glycoprotein genes), and phenotype is clearly needed. The present difficulty of interpreting molecular-based resistance data was illustrated by von Samson-Himmelstjerna et al. (2009) who described pyrosequencing and real time PCR assays for the detection of benzimidazole resistance in H. contortus. The interpretation of test results based on the use of a threshold frequency of the susceptible allele (TTC) at the 200 position of 90%, was shown by the authors to be an effective means to identify the presence of drug resistance in the isolates they examined. However, the TTC allele frequency was not a quantitative measure of resistance. For example, two isolates with allele frequencies of 70–74% showed very similar egg hatch assay EC50 values (range 0.08–0.14 μg/ml) as two isolates with TTC allele frequencies of approximately 2% and 7%. In addition, three isolates showing similarly low TTC allele frequencies (range 0–7%) showed much higher EC50 values (range 0.4–0.66 μg/ml). Both comparisons suggest that other mechanisms may be contributing to the observed resistance phenotypes. Only a low level of the F167Y SNP was detected in one isolate in this study, and no E198A was detected, indicating that these SNPs were not contributing to the resistance in the isolates examined. In addition, the difference in drug sensitivity between two isolates which showed significantly different egg hatch EC50 values alongside similar TTC frequencies was not explained by the measurement of individual genotype frequencies, with the numbers of heterozygous and homozygous resistant individuals similar in both isolates. This illustrates the need to understand the contribution of different mechanisms to the overall resistance phenotype if the molecular data are to be able to predict drug efficacy.
The present study is an initial step in understanding interactions of different resistance mechanisms in a nematode population. The results indicate that the presence of the E198A SNP in H. contortus larvae is associated with a higher level of thiabendazole resistance in larval life stages than the F200Y SNP. Importantly, while this represents an advance in our understanding of the interaction of the various beta-tubulin SNPs in benzimidazole sensitivity, this specific relationship occurs in a minority of studied cases as most published analyses of resistance to this drug group show the presence of the F200Y SNP and not the E198A SNP (for example, de Lourdes Mottier and Prichard, 2008; von Samson-Himmelstjerna et al., 2009). Nevertheless, the present study highlights a general need to more clearly define the relationships between benzimidazole drug efficacy and resistance mechanisms (SNP frequencies at the 167, 198 and 200 positions, as well as P-glycoprotein expression levels) in order to develop molecular tests which are able to more accurately predict drug efficacy.
References
- Albers G.A., Burgess S.K. Serial passage of Haemonchus contortus in resistant and susceptible sheep. Vet. Parasitol. 1988;28:303–306. doi: 10.1016/0304-4017(88)90077-5. [DOI] [PubMed] [Google Scholar]
- Beech R.N., Prichard R.K., Scott M.E. Genetic variability of the beta-tubulin genes in benzimidazole-susceptible and -resistant strains of Haemonchus contortus. Genetics. 1994;138:103–110. doi: 10.1093/genetics/138.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackhall W.J., Prichard R.K., Beech R.N. P-glycoprotein selection in strains of Haemonchus contortus resistant to benzimidazoles. Vet. Parasitol. 2008;152:101–107. doi: 10.1016/j.vetpar.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Coles G.C., Jackson F., Pomroy W.E., Prichard R.K., von Samson-Himmelstjerna G., Silvestre A., Taylor M.A., Vercruysse J. The detection of anthelmintic resistance in nematodes of veterinary importance. Vet. Parasitol. 2006;136:167–185. doi: 10.1016/j.vetpar.2005.11.019. [DOI] [PubMed] [Google Scholar]
- Elard L., Comes A.M., Humbert J.F. Sequences of beta-tubulin cDNA from benzimidazole-susceptible and -resistant strains of Teladorsagia circumcincta, a nematode parasite of small ruminants. Mol. Biochem. Parasitol. 1996;79:249–253. doi: 10.1016/0166-6851(96)02664-3. [DOI] [PubMed] [Google Scholar]
- Elard L., Humbert J.F. Importance of the mutation of amino acid 200 of the isotype 1 beta-tubulin gene in the benzimidazole resistance of the small-ruminant parasite Teladorsagia circumcincta. Parasitol. Res. 1999;85:452–456. doi: 10.1007/s004360050577. [DOI] [PubMed] [Google Scholar]
- Ghisi M., Kaminsky R., Mäser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet. Parasitol. 2007;144:313–320. doi: 10.1016/j.vetpar.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Gill J.H., Redwin J.M., van Wyk J.A., Lacey E. Avermectin inhibition of larval development in Haemonchus contortus – effects of ivermectin resistance. Int. J. Parasitol. 1995;25:463–470. doi: 10.1016/0020-7519(94)00087-5. [DOI] [PubMed] [Google Scholar]
- Hancock D.K., Tully L.A., Levin B.C. A Standard Reference Material to determine the sensitivity of techniques for detecting low-frequency mutations, SNPs, and heteroplasmies in mitochondrial DNA. Genomics. 2005;86:446–461. doi: 10.1016/j.ygeno.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Hubert J., Kerboeuf D. A new method for culture of larvae used in diagnosis of ruminant gastrointestinal strongylosis: comparison with fecal cultures. Can. J. Comp. Med. 1984;48:63–71. [PMC free article] [PubMed] [Google Scholar]
- Hunt P.W., Knox M.R., Le Jambre L.F., McNally J., Anderson L.J. Genetic and phenotypic differences between isolates of Haemonchus contortus in Australia. Int. J. Parasitol. 2008;38:885–900. doi: 10.1016/j.ijpara.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Kaplan R.M. Drug resistance in nematodes of veterinary importance: a status report. Trends Parasitol. 2004;20:477–481. doi: 10.1016/j.pt.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Kotze A.C., O’Grady J., Emms J., Toovey A.F., Hughes S., Jessop P., Bennell M., Vercoe P.E., Revell D.K. Exploring the anthelmintic properties of Australian native shrubs with respect to their potential role in livestock grazing systems. Parasitology. 2009;136:1065–1080. doi: 10.1017/S0031182009006386. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Kooyman F.N., Boersema J.H., Roos M.H. Effect of selection for benzimidazole resistance in Haemonchus contortus on beta-tubulin isotype 1 and isotype 2 genes. Biochem. Biophys. Res. Commun. 1993;191:413–419. doi: 10.1006/bbrc.1993.1233. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Roos M.H. Molecular characterisation of beta-tubulin genes present in benzimidazole-resistant populations of Haemonchus contortus. Mol. Biochem. Parasitol. 1993;60:133–143. doi: 10.1016/0166-6851(93)90036-w. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., Roos M.H. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Mol. Biochem. Parasitol. 1994;63:299–303. doi: 10.1016/0166-6851(94)90066-3. [DOI] [PubMed] [Google Scholar]
- Kwa M.S., Veenstra J.G., van Dijk M., Roos M.H. Beta-tubulin genes from the parasitic nematode Haemonchus contortus modulate drug resistance in Caenorhabditis elegans. J. Mol. Biol. 1995;246:500–510. doi: 10.1006/jmbi.1994.0102. [DOI] [PubMed] [Google Scholar]
- de Lourdes Mottier M., Prichard R.K. Genetic analysis of a relationship between macrocyclic lactone and benzimidazole anthelmintic selection on Haemonchus contortus. Pharmacogenet. Genomics. 2008;18:129–140. doi: 10.1097/FPC.0b013e3282f4711d. [DOI] [PubMed] [Google Scholar]
- Love S.C.J., Neilson F.J.A., Biddle A.J., McKinnon R. Moxidectin-resistant Haemonchus contortus in sheep in northern New South Wales. Aus. Vet. J. 2003;81:359–360. doi: 10.1111/j.1751-0813.2003.tb11514.x. [DOI] [PubMed] [Google Scholar]
- Parker L.T., Deng Q., Zakeri H., Carlson C., Nickerson D.A., Kwok P.Y. Peak height variations in automated sequencing of PCR products using Taq dye-terminator chemistry. Biotechniques. 1995;19:116–121. [PubMed] [Google Scholar]
- Parker L.T., Zakeri H., Deng Q., Spurgeon S., Kwok P.Y., Nickerson D.A. AmpliTaq DNA polymerase, FS dye-terminator sequencing: analysis of peak height patterns. Biotechniques. 1996;21:694–699. doi: 10.2144/96214rr02. [DOI] [PubMed] [Google Scholar]
- Prichard R.K. Genetic variability following selection of Haemonchus contortus with anthelmintics. Trends Parasitol. 2001;17:445–453. doi: 10.1016/s1471-4922(01)01983-3. [DOI] [PubMed] [Google Scholar]
- Rufener L., Kaminsky R., Mäser P. In vitro selection of Haemonchus contortus for benzimidazole resistance reveals a mutation at amino acid 198 of beta-tubulin. Mol. Biochem. Parasitol. 2009;168:120–122. doi: 10.1016/j.molbiopara.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Sutherland I.A., Leathwick D.M. Anthelmintic resistance in nematode parasites of cattle: a global issue? Trends Parasitol. 2011;27:176–181. doi: 10.1016/j.pt.2010.11.008. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G. Molecular diagnosis of anthelmintic resistance. Vet. Parasitol. 2006;136:99–107. doi: 10.1016/j.vetpar.2005.12.005. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Blackhall W.J., McCarthy J.S., Skuce P.J. Single nucleotide polymorphism (SNP) markers for benzimidazole resistance in veterinary nematodes. Parasitology. 2007;134:1077–1086. doi: 10.1017/S0031182007000054. [DOI] [PubMed] [Google Scholar]
- von Samson-Himmelstjerna G., Walsh T.K., Donnan A.A., Carrière S., Jackson F., Skuce P.J., Rohn K., Wolstenholme A.J. Molecular detection of benzimidazole resistance in Haemonchus contortus using real-time PCR and pyrosequencing. Parasitology. 2009;136:349–358. doi: 10.1017/S003118200800543X. [DOI] [PubMed] [Google Scholar]
- Wolstenholme A.J., Fairweather I., Prichard R., von Samson-Himmelstjerna G., Sangster N.C. Drug resistance in veterinary helminths. Trends Parasitol. 2004;20:469–476. doi: 10.1016/j.pt.2004.07.010. [DOI] [PubMed] [Google Scholar]


