Graphical abstract

Highlights
► We compile 1508 ms4760 alleles of the pfnhe-1 gene from various geographical regions. ► New numbering of ms4760 allele and classification grouping alleles are proposed. ► Pfnhe-1 allele polymorphism as a marker of quinine resistance appears restricted to South Asia.
Keywords: Malaria, Plasmodium falciparum, Na+/H+ exchanger, Quinine resistance, Genetic polymorphism
Abstract
The aim of this study was to provide a comprehensive analysis of the worldwide genetic polymorphism of ms4760 alleles of the pfnhe-1 gene and to discuss their usefulness as molecular marker of quinine resistance (QNR). A new numbering of ms4760 allele, classification grouping ms4760 alleles according to the number of DNNND and DDNHNDNHNND repeat motifs in blocks II and V was also proposed.
A total of 1508 ms4760 sequences from isolates, culture-adapted parasites or reference strains from various geographical regions were retrieved from GenBank (last update on 15th June 2012) or from publications and were used for genetic analyses. The association of different alleles of pfnhe-1 with resistance to quinoline antimalarial drugs showed marked geographic disparities.
The validity and reliability of candidate polymorphisms in pfnhe-1 gene as molecular markers of QNR appeared restricted to endemic areas from South Asia or possibly East African countries and needs to be confirmed.
1. Introduction
Quinine (QN), a natural compound found in Cinchona bark, has been used for centuries in malaria endemic regions (Baird, 2005). It is currently recommended for treating severe malaria cases, malaria in pregnant women or as second-line therapy in combination with antibiotic for uncomplicated malaria (World Health Organization, 2010a). Though clinical failures have been reported in Asia and South America in the 1960s and later on, although more rarely in Africa, resistance to QN (QNR) remains particularly punctual and rare (Chongsuphajaisiddhi et al., 1983; Pukrittayakamee et al., 1994, 2000; de Vries et al., 2000; McGready et al., 2000, 2005; Rahman et al., 2001; Adam et al., 2005; Adegnika et al., 2005; Achan et al., 2009; World Health Organization, 2010a,b).
QN, a quinoline derivative, is a monoprotic weak base that accumulates within the low pH environment of the parasite digestive vacuole of Plasmodium falciparum. QN presumably acts by interference with the detoxification of heme produced during hemoglobin degradation by P. falciparum asexual blood stages, leading to toxic degradation by-products (Hawley et al., 1998). However, the mechanism of QNR is not well known. Several reports have documented associations between in vitro susceptibility to QN with other structurally related drugs such as amino-4-quinolines (chloroquine, amodiaquine) or aryl-amino-alcohol (mefloquine, halofantrine), suggesting that a common genetic determinant may affect the parasite response to these antimalarials (Simon et al., 1986; Warsame et al., 1991; Basco and Le Bras, 1992; Brasseur et al., 1992). Particularly, QNR has been associated with mutations in the P. falciparum multidrug resistance 1 gene (pfmdr-1) and the P. falciparum chloroquine resistance transporter gene (pfcrt) (Wongsrichanalai et al., 2002; Valderramos and Fidock, 2006). More recently, other genetic polymorphisms, such as mutations in the P. falciparum multi-resistance protein 1 gene (pfmrp-1) have been suggested (Mu et al., 2003), but not confirmed (Anderson et al., 2005). However, PfMRP knock-out parasite lines displayed increase susceptibly to several antimalarial drugs, including chloroquine, QN and artemisinin derivatives (Raj et al., 2009). The degree of implication or linkage of the three genes in QNR remains uncertain, probably because additional genes are involved. In 2004, by using quantitative trait loci (QTL) analysis on the genetic cross of the HB3 and Dd2 clones, Ferdig et al. (2004) identified genes associated with QN reduced susceptibility (Ferdig et al., 2004), namely pfmdr-1 on chromosome 5, pfcrt on chromosome 7 and pfnhe-1 (P. falciparum Na+/H+ exchanger-1) on chromosome 13. To test for an association of QN response with this latter gene, pfnhe-1 was resequenced from the HB3 and Dd2 parents and the identified coding frame polymorphisms were surveyed in 71 P. falciparum culture-adapted isolates and reference lines from South-East Asia, Africa and Central and South America. Sequences of pfnhe-1 showed multiple and complex variations. Three point polymorphisms at three separate codons (790 gtc/ttc, 894 aat/aaa, 950 ggg/gtg) and microsatellite variations in three different repeat sequences (msR1, ms3580 and ms4760) were observed (Fig. 1). Moreover, there was a significant association between variations in ms4760 and in vitro QN response. One of the eight ms4760 profiles, ms4760-1, was relatively frequent in lines with reduced susceptibility to QN (i.e. higher IC90), but it was also present in fully susceptible parasites. More interestingly, the authors reported that presence of more than 2 DNNND repeat motifs in block II was associated with higher in vitro IC90 for QN compared with presence of only one repeat (Ferdig et al., 2004).
Fig. 1.
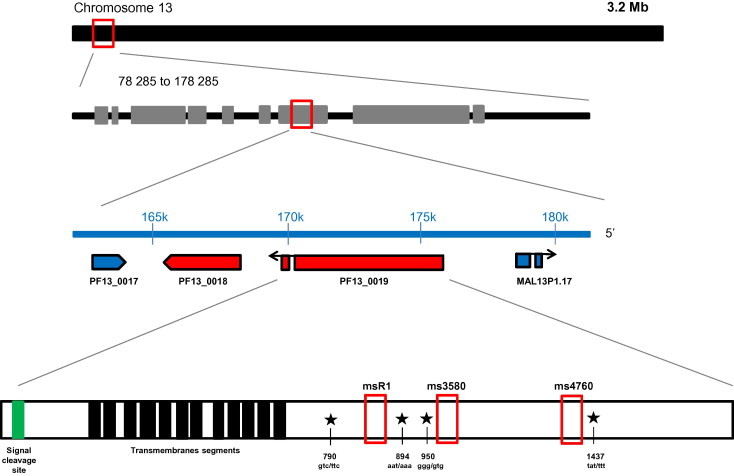
Schematic representation of pfnhe-1 gene (PF13_0019) on chromosome 13 and positions of codons polymorphisms (790, 894, 950 and 1437) and microsatellite variations (msR1, ms3580 and ms4760).
The physiological role of PfNHE-1 is still debated. In all living organisms, the fundamental homeostatic mechanisms are ubiquitous and vital. These physiological processes which regulate cellular pH, volume, and ion composition are supported by transmembrane exchange of cations implying several transporters like the family of Na+/H+ exchangers (NHEs) (Pouyssegur et al., 1984; Putney and Barber, 2003). Investigations performed in 1993 by the group of Ginsburg have shown that the major role of P. falciparum Na+/H+ exchanger was to increase the cytosolic pH (pHcyt) and to compensate acidosis caused by anaerobic glycolysis (Bosia et al., 1993). PfNHE-1, a 226 kDa protein with 12 predicted trans-membrane segments (Gardner et al., 2002; Ferdig et al., 2004), is supposed by some authors to reside in the parasite’s plasma membrane (Bosia et al., 1993; Bennett et al., 2007) but others underlined that the subcellular localization of this protein is not established (Nkrumah et al., 2009). Saliba and Kirk (1999) demonstrated that P. falciparum maintains its pHcyt by using mainly a V-type H1-ATPase, which serves as the major route for the efflux of H+ ions (Saliba and Kirk, 1999). Later, in 2007, Bennett et al. (2007) showed that high level of QNR was correlated to an increased PfNHE-1 activity which determines pHcyt. They also demonstrated that antimalarial drug resistances were related to modifications of ion transport across plasma (pHcyt) and digestive vacuole (pHDV) membranes and concluded that pairwise interactions of genetic determinants located on chromosome 13 and chromosome 9 affecting pHcyt and PfNHE-1 were involved in QNR (Bennett et al., 2007). However, using the protocols of Bennett et al., Spillman et al. (2008) showed that the Na+-dependent efflux of H+ from parasites acidified using nigericin/BSA was attributable to Na+/H+ exchange via residual nigericin remaining in the parasite plasma membrane, rather than to endogenous transporter activity (Spillman et al., 2008). Likewise, Nkrumah et al. (2009) were unable to reproduce the Na+/H+-exchanger activity observed by Bennett et al., (Nkrumah et al., 2009) but they provided evidences that PfNHE-1 expression levels influenced QN sensitivity in concert with additional parasite genetic factors such as PfCRT, PfMDR1 and possibly additional yet unidentified parasite proteins. However, variations in PfNHE-1 expression levels did not impact on pHcyt.
Since the seminal work by Ferdig et al. (2004), several studies have been conducted in different countries to evaluate the pfnhe-1 polymorphisms and its association with in vitro QN susceptibility (Vinayak et al., 2007; Henry et al., 2009; Andriantsoanirina et al., 2010, 2012; Baliraine et al., 2010; Briolant et al., 2010, 2011; Meng et al., 2010; Okombo et al., 2010; Pelleau et al., 2011; Sinou et al., 2011). Conflicting data have been reported, likely due to the different geographical origin of parasites (implying different genetic backgrounds), the type of parasites used (fresh isolates, culture-adapted strains and reference lines) and the method used to assess in vitro QN susceptibility (Okombo et al., 2011; Pelleau et al., 2011). Thus, the implication of PfNHE-1 polymorphisms in QNR remains to be studied in detail.
The aim of this study was to provide a comprehensive analysis of the worldwide genetic polymorphism of ms4760 alleles of the pfnhe-1 gene and to discuss their usefulness as molecular marker of quinine resistance.
2. Plasmodium falciparum Na+/H+ exchanger (Pfnhe-1) allele polymorphism
Following the initial work of Ferdig et al. (2004) on genetic polymorphism of ms4760 within the pfnhe-1 gene, a total of 1508 ms4760 sequences from isolates, culture-adapted parasites or reference strains from various geographical regions were retrieved from GenBank (last update on 15th June 2012) or from publications (Table 1) and were used for genetic analyses. A new numbering of ms4760 allele according to the chronological order of the data of the publication was conducted. Classification grouping ms4760 alleles according to the number of DNNND and DDNHNDNHNND repeat motifs in blocks II and V was also performed.
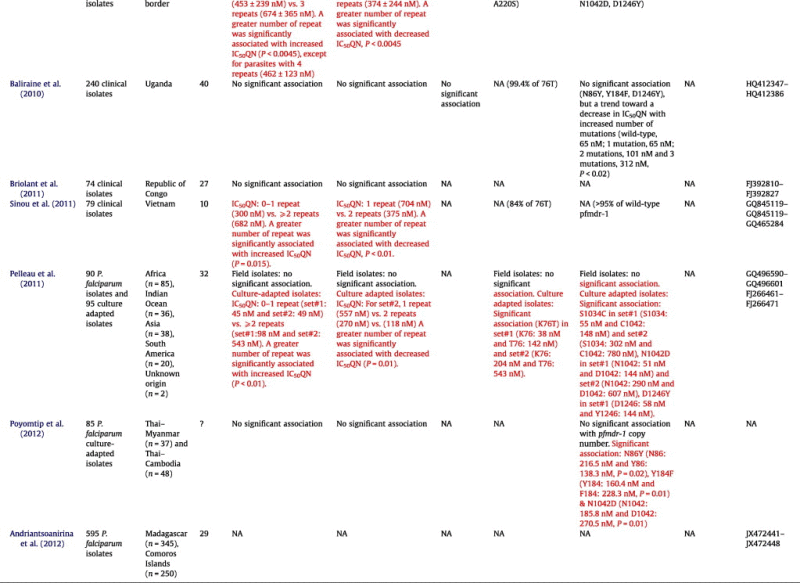
Table 1.
Summarized findings of the studies describing relationships between polymorphisms in pfnhe-1, pfcrt, pfmdr-1 and pfmrp genes and in vitro susceptibility to quinine.
|
NA, Not available; QN, Quinine; IC50, 50% inhibitory concentration; IC90, 90% inhibitory concentration; Significant associations are shown in red.
Ms4760 sequences were aligned and compared using the Clustal W multiple alignment algorithm in BioEdit Sequence Alignment editor (Hall, 1999). Genetic diversity was assessed by Nei’s unbiased expected heterozygosity (He) from haploid data and calculated as He = [n/(n − 1)][1 − pi] (n = the number of isolates sampled; pi = the frequency of the ith allele) (Nei, 1978). Population genetic differentiation was measured using Wright’s F statistics (Wright, 1965); population genetic parameters were computed with FSTAT software, v2.9.4 (Goudet, 1995).
The Mann-Whitney U test or Kruskal–Wallis method were used for non-parametric comparisons, and Student’s t test or one-way analysis of variance for parametric comparisons. For categorical variables, Chi-squared or Fisher’s exact tests were used to assess significant differences in proportions.
All reported P-values are two-sided and were considered statistically significant if less than 0.05.
2.1. Global genetic polymorphism of ms4760 in pfnhe-1 gene
Amongst the 1508 studied sequences, 101 different ms4760 alleles were observed. Ms4760 alleles were renumbered according to the chronology of the publication of the studies (ranging from ms4760-1 to ms4760-101) and are presented in Table 2. Alignment of sequences of blocks I–VI in ms4760 are displayed in Fig. 2.
Table 2.
Classification of pfnhe-1 sequences in 101 different alleles according to the geographical location of the isolates.
 |
Countries are designed by the 3-letters codes used by the United Nations: THA, Thailand; KHM, Cambodia; VNM, Vietnam; MYS, Malaysia; CHN, China; MMR, Myanmar; IND, India; MRT, Mauritania; SEN, Senegal; GMB, Gambia; MLI, Mali; GNB, Guinea-Bissau; SLE, Sierra Leone; LBR, Liberia; CIV, Côte d’Ivoire; GHA, Ghana; TGO, Togo; BEN, Benin; BFA, Burkina Faso, NER, Niger; SDN, Soudan; KEN, Kenya; UGA, Uganda; DJI, Djibouti; CAF, Central African Republic; TCD, Chad; COG, Congo; COD, Congo, the Republic Democratic of the; ZAF, South Africa; MOZ, Mozambique; COM, Comoros; MDG, Madagascar; HND, Honduras; COL, Colombia; ECU, Ecuador; PER, Peru; BRA, Brazil; PNG, Papua New Guinea; HTI, Haiti.
Grey box means “ms-4760 haplotype never detected” and black box means “ms-4760 haplotype detected at least once”.
Fig. 2.

Alignment of the 101 ms4760 protein sequences found in Genbank and in publications.
2.1.1. Geographical distribution of ms4760 alleles: between continents
According to the location of the sample collection, 39 ms4760 alleles were observed in Asia (n = 398), 74 in Africa (n = 1070), 5 in South America (n = 17), 2 in Papua New Guinea (n = 5) and one in Haiti (n = 1). Five alleles were globally distributed (ms4760-1, ms4760-3 ms4760-5, ms4760-6, ms4760-7) while others were exclusively found in Asia and in Africa (n = 24, ms4760-2, ms4760-8, ms4760-9, ms4760-12, ms4760-14, ms4760-15 and ms4760-18 to ms4760-35) or only in Asia (n = 10, ms4760-4, ms4760-10, ms4760-11, ms4760-13, ms4760-16, ms4760-17 and ms4760-98 to ms4760-101) or only in Africa (n = 45, from ms4760-36 to ms4760-64, from ms4760-66 to ms4760-173 & ms4760-90 to ms4760-97). Seventeen alleles have unknown origins (ms4760-65 & ms4760-74 to ms4760-89). Data are presented in Table 2 and Fig. 3.
Fig. 3.
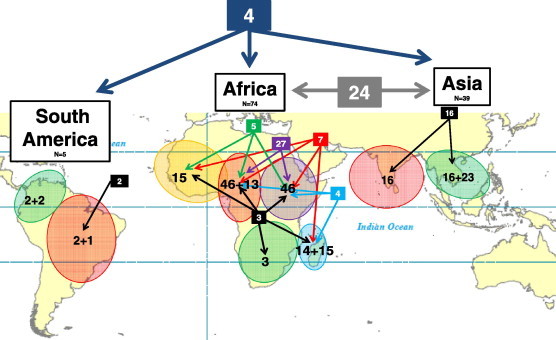
Geographical distribution of the pfnhe-1 ms4760 profile between and with continents. Numbers in boxes indicate shared ms4760 profiles between continents (blue and grey boxes) and regions (black, green, red, purple and light blue boxes). For each region, numbers of ms4760 profiles are split into shared profiles (first number) and local profiles (second number) (i.e., Madagascar: 14 shared profiles and 15 local profiles). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.1.2. Geographical distribution of ms4760 alleles: within continents and between regions
Amongst the 39 alleles found in Asia, 16 (41%) were shared between South East Asia (Thailand, Cambodia, Vietnam, Malaysia and Myanmar/China border) and Central Asia (India), whereas 23 were specific to South East Asia. In Africa, the 74 observed alleles were distributed as follows: three were shared by all regions (West Africa, East Africa, Central Africa, South Africa, Indian Ocean Islands), seven were present in all regions except South Africa, five alleles were shared by West Africa, East Africa and Central Africa, four by East Africa, Central Africa and Indian Ocean Islands and 27 by East Africa and Central Africa. Thirteen alleles were found only in Central Africa and fifteen in Indian Ocean Islands only. Among the five alleles described in South America, only two were shared by Western countries (Honduras, Colombia, Ecuador and Peru) and Eastern country (Brazil). Two were specific from Western countries and one from Eastern countries (Table 2 and Fig. 3).
2.2. ms4760 profiles: geographical distribution and prevalence
According to the number of repeats in block II (DNNND) and block V (DDNHNDNHNND) which have been associated with modulation of in vitro QNR (Henry et al., 2009; Andriantsoanirina et al., 2010), ms4760 alleles were grouped in 15 allele profiles (from ms4760-A to ms4760-O) presented in Table 3. The number of repeats in block II (DNNND) varied from 0 (ms-4760-A) to 6 (ms4760-O) while the number of repeats in block V (DDNHNDNHNND) from 1 (ms4760-B, ms4760-E, ms4760-I, ms4760-L, ms4760-O) to 4 (ms4760-H). Sixty-six percent of ms-4760 alleles were grouped in 4 profiles: ms-4760-C [(DNNND)1; (DDNHNDNHNND)2] (23%), ms-4760-F [(DNNND)2; (DDNHNDNHNND)2] (15%), ms-4760-E [(DNNND)2; (DDNHNDNHNND)1] (14%) and ms-4760-I [(DNNND)3; (DDNHNDNHNND)1] (14%). Four profiles were globally distributed (ms-4760-C, ms-4760-E, ms-4760-F and ms-4760-L), five were observed in both Asia and Africa (ms-4760-A, ms-4760-B, ms-4760-D, ms-4760-H and ms-4760-J), two were only found in Asia (ms-4760-I, ms-4760-N) and four only in Africa (ms-4760-G, ms-4760-K, ms-4760-M and ms-4760-O) (Fig. 3).
Table 3.
Phne-1 ms4760 profile groups according to the number of repeat in block II (DNNND) and block V (DDNHNDNHNND) and the geographical location of the isolates.
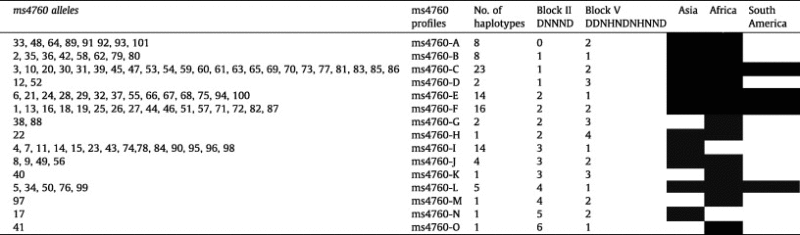 |
For each profile (A to O) and continent (Asia, Africa and South America), white box means “ms-4760 profile never detected” and black box means “ms-4760 profile detected at least once”.
The mean number of DNNND repeats was significantly higher in Asia (2.30, SD = 0.78) compared to Africa (2.06, SD = 0.90, P < 0.001). Inversely, the number of DDNHNDNHNND repeats was significantly lower in Asia (1.34, SD = 0.51) compared to Africa (1.72, SD = 0.54, P < 0.001). Consequently, the mean ratio of DNNND/DDNHNDNHNND repeats was significantly higher in Asia (2.03 ± 0.98 vs. 1.46 ± 1.05, P < 0.001).
The prevalence of the pfnhe-1 ms4760 profiles according to the geographical location of the isolates (continent & country) significantly differed between continents (P < 0.0001, Table 4 and Fig. 4). In both continents (Asia & Africa), 11 profiles had a low prevalence (<10%). Three profiles were predominant in Africa (ms-4760-F, 32.9%; ms-4760-C, 21.3% and ms-4760-I, 15.5%), and four in Asia (ms-4760-I, 37.1%; ms-4760-E, 28.1%; ms-4760-C, 14.3% and ms-4760-F, 10.9%). Interestingly, the prevalence of the ms-4760-C (1 DNNND repeat) decreased from Central Africa (38%) to East Africa (21%), Indian Ocean (19%), Central Asia (19%) and South East Asia (6%) whereas the prevalence of the ms-4760-I (3 DNNND repeats) increased along the same west to east axis (7%, 14%, 17%, 23% and 61%).
Table 4.
Prevalence and expected heterozygosity of the pfnhe-1 ms4760 groups according to the geographical location of the isolates (continent & country)
| ms4760 profiles (%) | Prevalence by Continent/Country | Total | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Africa | Asia | ||||||||
| Congo | Uganda | Kenya | Madagascar | ComorosIslands | India | China/Myanmar | Vietnam | ||
| n = 74 | n = 172 | n = 29 | n = 386 | n = 251 | n = 244 | n = 60 | n = 79 | ||
| ms4760-A | 1.3 | 0.0 | 0.0 | 2.0 | 1.5 | 0.0 | 0.0 | 1.3 | 1.1 |
| ms4760-B | 4.1 | 1.7 | 7.1 | 4.4 | 1.1 | 0.0 | 1.6 | 0.0 | 2.2 |
| ms4760-C | 38.3 | 22.6 | 14.2 | 24.1 | 11.9 | 19.2 | 4.9 | 6.3 | 19.2 |
| ms4760-D | 5.4 | 1.1 | 0.0 | 2.3 | 3.5 | 2.8 | 0.0 | 0.0 | 2.3 |
| ms4760-E | 9.5 | 6.3 | 3.5 | 9.8 | 6.3 | 36.0 | 19.7 | 10.1 | 13.9 |
| ms4760-F | 26.0 | 33.7 | 53.5 | 31.1 | 34.2 | 10.2 | 14.7 | 10.1 | 26.3 |
| ms4760-G | 0.0 | 1.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 |
| ms4760-H | 0.0 | 0.0 | 0.0 | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 |
| ms4760-I | 6.8 | 13.9 | 14.2 | 14.0 | 21.5 | 23.2 | 50.8 | 69.6 | 21.9 |
| ms4760-J | 8.2 | 11.0 | 7.1 | 8.5 | 5.1 | 6.5 | 3.3 | 0.0 | 7.0 |
| ms4760-K | 0.0 | 5.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.7 |
| ms4760-L | 0.0 | 2.3 | 0.0 | 2.3 | 14.3 | 1.6 | 4.9 | 2.6 | 4.5 |
| ms4760-M | 0.0 | 0.0 | 0.0 | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 |
| ms4760-N | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.4 | 0.0 | 0.0 | 0.1 |
| ms4760-O | 0.0 | 0.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 |
| He | 0.765 | 0.797 | 0.658 | 0.805 | 0.794 | 0.766 | 0.680 | 0.493 | |
He: expected heterozygosity.
Fig. 4.
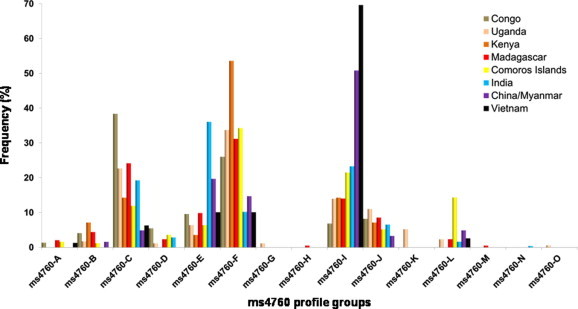
Prevalence of the pfnhe-1 ms4760 profile groups according to the geographical location of the isolates (continent and country). Phne-1 ms4760 profile groups are described in Table 3 (according to the number of repeat in block II, DNNND and block V, DDNHNDNHNND).
2.3. Genetic diversity and genetic differentiation between parasite populations
Genetic diversity, assessed by Nei’s unbiased expected heterozygosity (He) was significantly higher in Africa (Congo = 0.7649, Uganda = 0.7975, Kenya = 0.6582), Indian Ocean (Madagascar = 0.8053, Comoros Islands = 0.7946) or India (0.6807) compared to China/Myanmar (0.6807, P = 0.04) or Vietnam (0.4981, P < 0.0001) (Table 4).
The degree of genetic differentiation of the ms4760 profiles within parasite populations, estimated by Fst values, indicated a large divergence between Asian populations and African populations (Table 5). The highest differences were observed between populations from Vietnam or China/Myanmar and populations from Kenya (Fst = 0.319 and 0.183), Congo (Fst = 0.291 and 0.176), Uganda (Fst = 0.219 and 0.121), Madagascar (Fst = 0.202 and 0.111), Comoros Islands (Fst = 0.171 and 0.083) and India (Fst = 0.171 and 0.069). On the other hand, populations from Africa (Congo, Uganda, Kenya and Madagascar) showed very low divergence or were similar (Fst from 0.0001 to 0.076) and population from India was intermediate (Fst from 0.070 to 0.163).
Table 5.
Degree of genetic differentiation of the ms4760 profile groups within parasite populations (African and Asian countries), estimated by pairwise population genetic distances (FST).
| Uganda | Kenya | Madagascar | Comoros Islands | India | China/Myanmar | Vietnam | |
|---|---|---|---|---|---|---|---|
| Congo | 0.01739 | 0.07610 | 0.01025 | 0.06382 | 0.08763 | 0.17689 | 0.29186 |
| Uganda | 0.01959 | 0.00011 | 0.01911 | 0.08776 | 0.12179 | 0.21948 | |
| Kenya | 0.02770 | 0.02876 | 0.16355 | 0.18367 | 0.31962 | ||
| Madagascar | 0.02089 | 0.07075 | 0.11195 | 0.20280 | |||
| Comoros Islands | 0.09344 | 0.08370 | 0.17157 | ||||
| India | 0.06970 | 0.17199 | |||||
| China/Myanmar | 0.02694 |
3. Discussion
The biostatistical analyses performed in this study showed a large global genetic polymorphism of ms4760 in pfnhe-1 gene. The African continent displayed the highest number of different alleles, followed by Asia and South America. While a few alleles were shared by three continents, others appeared restricted to Asia and/or Africa. Similarly, within continents, some alleles were shared between various regions while others appeared restricted to specific areas as Central Africa or Indian Ocean. The geographical distribution and prevalence of ms4760 profiles, defined by variations in the number of repeats in block II and V, differed also significantly between continents. Interestingly, the mean ratio of DNNND/DDNHNDNHNND repeats, proposed by some authors as associated with in vitro QNR, was significantly higher in Asia than in Africa. The predominant ms4760 profiles in Africa were the same that in Asia, except for the ms-4760-E which was predominant only in Asia. Some data suggested a geographical diffusion of some alleles. The prevalence of a particular profile, the ms-4760-C (1 DNNND repeat), decreased from Central Africa to East Africa, Indian Ocean, Central Asia and South East Asia while the prevalence of another “opposite” profile, the ms-4760-I (3 DNNND repeats), increased along the same west to east axis. Genetic analysis using expected heterozygosity He showed a higher genetic diversity in parasites from Africa (including Indian Ocean Islands) than in those from Asia. In addition, Fst values indicated a large divergence between Asian and African populations. These data showed that the Asian and African populations were clearly differentiated.
Several studies have described the relationships between polymorphisms in pfnhe-1 and in vitro susceptibility to quinine (Table 1). Some of these studies have also considered the association of in vitro susceptibility to quinine with polymorphisms in pfcrt, pfmdr-1 and pfmrp genes, with conflicting results for pfcrt and pfmrp, and no association (except in one study) with SNPs polymorphisms of pfmdr-1 (Table 1).
Following the inaugural study of Ferdig et al. (2004), Henry et al. (2009) investigated a series of 23 culture-adapted isolates or reference strains. The relationship between the number of DNNND repeats and the inhibitory concentration 50% values (IC50) to QN was confirmed and an increased number of the DDNHNDNHNND repeat motif was associated with decreased IC50s to QN. A limitation of these studies was that the in vitro QN susceptibility and polymorphisms determinations were performed on culture-adapted cloned isolates or reference strains, which could lead to biased results due to accumulated mutations selected by in vitro conditions or to selection of specific alleles during the culture. Indeed, a recent study showed an association between pfnhe-1 polymorphism and in vitro QN response on cultured adapted isolates but not in field isolates (Pelleau et al., 2011).
An increased number of DNNND repeats was positively associated with in vitro QNR in six studies (Ferdig et al., 2004; Henry et al., 2009; Meng et al., 2010; Okombo et al., 2010; Pelleau et al., 2011; Sinou et al., 2011) (Table 1). All these studies used culture-adapted parasites, except study from Sinou et al. (2011). It is worth noting that Ferdig et al. (2004) used IC90s rather than IC50s in the other studies (Ferdig et al., 2004). Parasites having 2 or more repeats had higher IC90s than parasites having 1 repeat (P < 0.05 for Asia and South America lines). Okombo et al. (2010) observed this association only for parasites from Kenya having 2 repeats compared to 1 repeat (P < 0.05) (Okombo et al., 2010). Meng et al. (2010) reported a strong positive association in a series of 60 adapted isolates from the China–Myanmar border (Meng et al., 2010). Sinou et al. (2011) also reported a positive association in a series of 51 clinical fresh isolates from Vietnam: isolates with two or more DNNND motifs were less susceptible to QN than those harbouring zero or one DNNND repeats (Sinou et al., 2011). Discordant results were reported in a Thai study by Poyomtip et al. (2012) who did not observe an association between the number of DNNND repeats and in vitro QNR in a series of 81 culture-adapted isolates obtained from the Thai–Myanmar border and the Thai–Cambodia border (Poyomtip et al., 2012).
Pradines and colleagues (2009–2011) published 3 studies of pfnhe-1 polymorphisms (Henry et al., 2009; Briolant et al., 2010, 2011). In the first one, Henry et al., 2009 including 23 reference strains or culture-adapted isolates of various geographic origin, found a positive association between the number of DNNND repeats and IC50s (Henry et al., 2009). In another study Briolant et al. (2010, 2011) including 23 reference strains or culture adapted isolates of similar geographic origin did not found any association (Briolant et al., 2010). Lastly, in a series of 74 clinical isolates from Republic of Congo, Briolant et al. (2011) did not found an association either (Briolant et al., 2011). Two other studies including respectively 83 and 172 clinical isolates from African countries did not find any association between an increased number of repeats in DNNND and in vitro QNR (Andriantsoanirina et al., 2010; Baliraine et al., 2010). The studies conducted in Asian areas reported an overrepresentation of the ms4760-7 allele, harboured by 49.2% and 68.3% of isolates in China–Myanmar border and Vietnam, respectively (Meng et al., 2010; Sinou et al., 2011). In the study by Meng et al. (2010), ms4760-7 isolates were among those having the lowest in vitro susceptibility to QN but other ms4760-7 isolates of that series displayed perfect susceptibility (Meng et al., 2010). The ms4760-7 allele was also overrepresented in the study by Henry et al. (2009) including 4 Asian isolates with high IC50s, ranging from 599 to 1310 nM (Henry et al., 2009). The presence of the ms4760-7 allele was not rare in other areas, in particular in African countries (Ferdig et al., 2004; Andriantsoanirina et al., 2010, 2012; Okombo et al., 2010; Briolant et al., 2011) but without obvious association with QNR, many isolates harbouring this allele showing full in vitro susceptibility. Thus, further studies are needed to confirm whether the ms4760-7 allele is necessary for the emergence QN resistance and can be used in monitoring the QNR spread in South East Asia.
The number of DDNHNDNHNND repeat motif was associated with reduced in vitro susceptibility to QN (P < 0.01) in one study of 83 clinical isolates obtained in African countries (Andriantsoanirina et al., 2010). Conversely, an increased number of DDNHNDNHNND repeats was associated with higher in vitro susceptibility to QN in studies of isolates from the China–Myanmar border (Meng et al., 2010) or Vietnam (Sinou et al., 2011). The same association was observed in one study (Henry et al., 2009) but not confirmed in 2 subsequent studies by the same team (Briolant et al., 2010, 2011). Three other studies did not find this association either (Baliraine et al., 2010; Okombo et al., 2010; Poyomtip et al., 2012).
The usefulness of PfNHE-1 polymorphisms as marker of in vitro QNR may be inferred from some publications. In parasites from Asian areas, the number of DNNND repeats has been positively associated with in vitro QNR in culture-adapted isolates from the China–Myanmar border (Meng et al., 2010) and in isolates from Vietnam (Sinou et al., 2011) but not in culture-adapted isolates from the Thai–Myanmar border and the Thai–Cambodia border (Poyomtip et al., 2012). In parasites from Africa, the existing data show no evidence of association of the number of DNNND repeats and QN susceptibility, excluding its use as a molecular marker of QNR (Andriantsoanirina et al., 2010; Baliraine et al., 2010; Briolant et al., 2010, 2011). This may change in the future and the situation could be particular in Kenya (Okombo et al., 2010) as resistance genotypes originating from the South-East Asia may have reached this country as it was the case in the past for chloroquine and antifolates resistant P. falciparum.
The number of DDNHNDNHNND repeats does not seem correlated with in vitro QNR or contributing to QNR in Asian areas, so it does not appear as an interesting marker. Choudhary and Sharma, 2009 studied the polymorphisms in flanking microsatellites of the pfnhe-1 gene in 108 Indian isolates (Choudhary and Sharma, 2009). They observed an expected heterozygosity of 10 flanking microsatellites in the vicinity of ±40 kb of pfnhe-1 gene comparable to any other neutral loci. Thus, no selective sweep or valley of reduced variation around ±40 kb of this gene was observed, indicating that there was no strong selection pressure on the pfnhe-1 gene. In addition, these authors did not find an association between DNNND repeat polymorphisms and microsatellite alleles.
The association of PfNHE-1 polymorphism and clinical resistance remains to be evaluated. Currently, only 2 cases of clinical failures have been reported. Pradines and colleagues (2011) studied a QN treatment failure in a traveller from Senegal, and observed the association of two repeats of DNNND with a reduced in vitro susceptibility (IC50 = 829 nM) (Pradines et al., 2011). The second case, a QN treatment failure in a traveller from French Guiana, did not show this association as the ms4760 microsatellite showed 1 repeat of DNNND and 2 repeats of DDNHNDNHNND, though the isolate had a reduced susceptibility to QN (IC50 = 1019 nM) (Bertaux et al., 2011).
The finding of association of polymorphisms in putative genes with clinical failures and/or in vitro susceptibility constitutes a pivotal step in the development of tools for the surveillance of emergence and spreading of P. falciparum resistant strains. Such associations must be verified on numerous isolates originating from various geographical areas and supported by molecular studies to specifically assess the involvement of the candidate genes in drug resistance. Recent genetic and physiological studies reinforced the conclusion that QNR is a complex trait requiring multiple actors (Nkrumah et al., 2009). Several transporters have been identified as determinants of resistance to quinoline antimalarial drugs. The available data on molecular surveys of potential contributors to QN resistance do not allow to propose a simple molecular typing methodology of global application. It is possibly a consequence of the multigene nature of the QNR trait, which involves multiple gene interactions. Such gene interactions depend on the alleles at play in each genetic background and likely show substantial geographic variations. In particular pfcrt and pfmdr1 known to contribute to QN susceptibility have different alleles in different geographic settings (Wellems et al., 2009). For example, CQR P. falciparum strains have originated from at least six different geographic locations spread across Southeast Asia, Latin America and the Pacific region (Wootton et al., 2002; Wellems et al., 2009). African CQR strains have their origins in a single foundation event, a strain apparently imported from Southeast Asia. In the case of QN (and of most other antimalarial drugs), the drug pressure that selected for resistance varied considerably with respect to intensity and time in the different geographic areas. As a result, the association of different alleles of transporters with resistance to quinoline antimalarial drugs may show geographic disparities. Likewise, the amplification of pfmdr1, associated with in vitro resistance to QN, mefloquine, and halofantrine is frequent in Asia (Price et al., 2004) but rare in the African continent. Analogous processes may have occurred for pfnhe-1. However, the absence of selective sweep in 108 Indian P. falciparum isolates and the lack of association of microsatellite markers with DNNND repeats, possibly indicates that there is no strong selection pressure on the pfnhe-1 gene (Choudhary and Sharma, 2009). Studies summarized in this paper do not exclude a potential role for PfNHE-1 in QNR in a strain-dependent manner.
In this context, the validity and reliability of candidate polymorphisms in pfnhe-1 gene as molecular markers of QNR appears restricted to endemic areas from South Asia or possibly East African countries and needs to be confirmed.
Funding
This work was supported by Grants from Natixis Banques and the Genomics Platform, Pasteur Génopôle, Pasteur Institute, France. Sample collection was funded in Comoros Islands by the French Foreign Ministry (FSP-RAI project) and in Madagascar by the Global Fund project round 3 (Grant MDG-304-G05-M). Didier Ménard is supported by the French Ministry of Foreign Affairs, Benoit Witkowski by a post-doctoral fellowship from the Division International – Institut Pasteur (2011–2013) and Christophe Benedet by a grant from the Fondation Pierre Ledoux – Jeunesse Internationale (2012).
Conflict of interest
None declared.
Acknowledgements
We thank the patients and healthcare workers involved in the studies performed in Madagascar and Comoros Islands. We are grateful to Christiane Bouchier and Magali Tichit for performing sequencing reactions (Genomics Platform, Pasteur Génopôle, Pasteur Institute, France) and Carol H. Sibley for her advices.
Contributor Information
Didier Ménard, Email: dmenard@pasteur-kh.org.
Rémy Durand, Email: remy.durand@avc.aph.fr.
References
- Achan J., Tibenderana J.K., Kyabayinze D., Wabwire Mangen F., Kamya M.R., Dorsey G., D’Alessandro U., Rosenthal P.J., Talisuna A.O. Effectiveness of quinine versus artemether–lumefantrine for treating uncomplicated falciparum malaria in Ugandan children: randomised trial. BMJ. 2009;339:b2763. doi: 10.1136/bmj.b2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam I., Ali D.M., Noureldien W., Elbashir M.I. Quinine for the treatment of chloroquine-resistant Plasmodium falciparum malaria in pregnant and non-pregnant Sudanese women. Ann. Trop. Med. Parasitol. 2005;99:427–429. doi: 10.1179/136485905X36217. [DOI] [PubMed] [Google Scholar]
- Adegnika A.A., Breitling L.P., Agnandji S.T., Chai S.K., Schutte D., Oyakhirome S., Schwarz N.G., Grobusch M.P., Missinou M.A., Ramharter M., Issifou S., Kremsner P.G. Effectiveness of quinine monotherapy for the treatment of Plasmodium falciparum infection in pregnant women in Lambarene, Gabon. Am. J. Trop. Med. Hyg. 2005;73:263–266. [PubMed] [Google Scholar]
- Anderson T.J., Nair S., Qin H., Singlam S., Brockman A., Paiphun L., Nosten F. Are transporter genes other than the chloroquine resistance locus (pfcrt) and multidrug resistance gene (pfmdr) associated with antimalarial drug resistance? Antimicrob. Agents Chemother. 2005;49:2180–2188. doi: 10.1128/AAC.49.6.2180-2188.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriantsoanirina V., Menard D., Rabearimanana S., Hubert V., Bouchier C., Tichit M., Bras J.L., Durand R. Association of microsatellite variations of Plasmodium falciparum Na+/H+ exchanger (Pfnhe-1) gene with reduced in vitro susceptibility to quinine: lack of confirmation in clinical isolates from Africa. Am. J. Trop. Med. Hyg. 2010;82:782–787. doi: 10.4269/ajtmh.2010.09-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriantsoanirina, V., Khim, N., Ratsimbasoa, A., Witkowski, B., Benedet, C., Canier, L., Bouchier, C., Tichit, M., Durand, R., Ménard, D., 2012. Short report: Plasmodium falciparum Na+/H+ exchanger (Pfnhe-1) genetic polymorphism in Indian Ocean malaria endemic areas. Am. J. Trop. Med. Hyg. <http://www.ajtmh.org/cgi/doi/10.4269/ajtmh.2012.12-0359>. [DOI] [PMC free article] [PubMed]
- Baird J.K. Effectiveness of antimalarial drugs. New Engl. J. Med. 2005;352:1565–1577. doi: 10.1056/NEJMra043207. [DOI] [PubMed] [Google Scholar]
- Baliraine F.N., Nsobya S.L., Achan J., Tibenderana J.K., Talisuna A.O., Greenhouse B., Rosenthal P.J. Limited ability of Plasmodium falciparum pfcrt, pfmdr1, and pfnhe1 polymorphisms to predict quinine in vitro sensitivity or clinical effectiveness in Uganda. Antimicrob. Agents Chemother. 2010;55:615–622. doi: 10.1128/AAC.00954-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basco L.K., Le Bras J. In vitro activity of halofantrine and its relationship to other standard antimalarial drugs against African isolates and clones of Plasmodium falciparum. Am. J. Trop. Med. Hyg. 1992;47:521–527. doi: 10.4269/ajtmh.1992.47.521. [DOI] [PubMed] [Google Scholar]
- Bennett T.N., Patel J., Ferdig M.T., Roepe P.D. Plasmodium falciparum Na+/H+ exchanger activity and quinine resistance. Mol. Biochem. Parasitol. 2007;153:48–58. doi: 10.1016/j.molbiopara.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertaux L., Kraemer P., Taudon N., Trignol A., Martelloni M., Saidi R., Parzy D., Pradines B., Simon F. Quinine-resistant malaria in traveler returning from French Guiana, 2010. Emerg. Infect. Dis. 2011;17:943–945. doi: 10.3201/eid1705.101424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosia A., Ghigo D., Turrini F., Nissani E., Pescarmona G.P., Ginsburg H. Kinetic characterization of Na+/H+ antiport of Plasmodium falciparum membrane. J. Cell Physiol. 1993;154:527–534. doi: 10.1002/jcp.1041540311. [DOI] [PubMed] [Google Scholar]
- Brasseur P., Kouamouo J., Moyou-Somo R., Druilhe P. Multi-drug resistant falciparum malaria in Cameroon in 1987–1988. I. Stable figures of prevalence of chloroquine- and quinine-resistant isolates in the original foci. Am. J. Trop. Med. Hyg. 1992;46:1–7. doi: 10.4269/ajtmh.1992.46.1. [DOI] [PubMed] [Google Scholar]
- Briolant S., Henry M., Oeuvray C., Amalvict R., Baret E., Didillon E., Rogier C., Pradines B. Absence of association between piperaquine in vitro responses and polymorphisms in the pfcrt, pfmdr1, pfmrp, and pfnhe genes in Plasmodium falciparum. Antimicrob. Agents Chemother. 2010;54:3537–3544. doi: 10.1128/AAC.00183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briolant S., Pelleau S., Bogreau H., Hovette P., Zettor A., Castello J., Baret E., Amalvict R., Rogier C., Pradines B. In vitro susceptibility to quinine and microsatellite variations of the Plasmodium falciparum Na+/H+ exchanger (Pfnhe-1) gene: the absence of association in clinical isolates from the Republic of Congo. Malar. J. 2011;10:37. doi: 10.1186/1475-2875-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chongsuphajaisiddhi T., Sabchareon A., Attanath P. Treatment of quinine resistant falciparum malaria in Thai children. Southeast Asian J. Trop. Med. Public Health. 1983;14:357–362. [PubMed] [Google Scholar]
- Choudhary V., Sharma Y.D. Extensive heterozygosity in flanking microsatellites of Plasmodium falciparum Na+/H+ exchanger (pfnhe-1) gene among Indian isolates. Acta Trop. 2009;109:241–244. doi: 10.1016/j.actatropica.2008.11.004. [DOI] [PubMed] [Google Scholar]
- de Vries P.J., Bich N.N., Van Thien H., Hung L.N., Anh T.K., Kager P.A., Heisterkamp S.H. Combinations of artemisinin and quinine for uncomplicated falciparum malaria: efficacy and pharmacodynamics. Antimicrob. Agents Chemother. 2000;44:1302–1308. doi: 10.1128/aac.44.5.1302-1308.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdig M.T., Cooper R.A., Mu J., Deng B., Joy D.A., Su X.Z., Wellems T.E. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol. Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- Gardner M.J., Shallom S.J., Carlton J.M., Salzberg S.L., Nene V., Shoaibi A., Ciecko A., Lynn J., Rizzo M., Weaver B., Jarrahi B., Brenner M., Parvizi B., Tallon L., Moazzez A., Granger D., Fujii C., Hansen C., Pederson J., Feldblyum T., Peterson J., Suh B., Angiuoli S., Pertea M., Allen J., Selengut J., White O., Cummings L.M., Smith H.O., Adams M.D., Venter J.C., Carucci D.J., Hoffman S.L., Fraser C.M. Sequence of Plasmodium falciparum chromosomes 2, 10, 11 and 14. Nature. 2002;419:531–534. doi: 10.1038/nature01094. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT (Version 1.2): a computer program to calculate F-statistics. J. Hered. 1995:485–486. [Google Scholar]
- Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hawley S.R., Bray P.G., Mungthin M., Atkinson J.D., O’Neill P.M., Ward S.A. Relationship between antimalarial drug activity, accumulation, and inhibition of heme polymerization in Plasmodium falciparum in vitro. Antimicrob. Agents Chemother. 1998;42:682–686. doi: 10.1128/aac.42.3.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M., Briolant S., Zettor A., Pelleau S., Baragatti M., Baret E., Mosnier J., Amalvict R., Fusai T., Rogier C., Pradines B. Plasmodium falciparum Na+/H+ exchanger 1 transporter is involved in reduced susceptibility to quinine. Antimicrob. Agents Chemother. 2009;53:1926–1930. doi: 10.1128/AAC.01243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGready R., Brockman A., Cho T., Cho D., van Vugt M., Luxemburger C., Chongsuphajaisiddhi T., White N.J., Nosten F. Randomized comparison of mefloquine–artesunate versus quinine in the treatment of multidrug-resistant falciparum malaria in pregnancy. Trans. R. Soc. Trop. Med. Hyg. 2000;94:689–693. doi: 10.1016/s0035-9203(00)90235-9. [DOI] [PubMed] [Google Scholar]
- McGready R., Ashley E.A., Moo E., Cho T., Barends M., Hutagalung R., Looareesuwan S., White N.J., Nosten F. A randomized comparison of artesunate–atovaquone–proguanil versus quinine in treatment for uncomplicated falciparum malaria during pregnancy. J. Infect Dis. 2005;192:846–853. doi: 10.1086/432551. [DOI] [PubMed] [Google Scholar]
- Meng H., Zhang R., Yang H., Fan Q., Su X., Miao J., Cui L., Yang Z. In vitro sensitivity of Plasmodium falciparum clinical isolates from the China–Myanmar border area to quinine and association with polymorphism in the Na+/H+ exchanger. Antimicrob. Agents Chemother. 2010;54:4306–4313. doi: 10.1128/AAC.00321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J., Ferdig M.T., Feng X., Joy D.A., Duan J., Furuya T., Subramanian G., Aravind L., Cooper R.A., Wootton J.C., Xiong M., Su X.Z. Multiple transporters associated with malaria parasite responses to chloroquine and quinine. Mol. Microbiol. 2003;49:977–989. doi: 10.1046/j.1365-2958.2003.03627.x. [DOI] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkrumah L.J., Riegelhaupt P.M., Moura P., Johnson D.J., Patel J., Hayton K., Ferdig M.T., Wellems T.E., Akabas M.H., Fidock D.A. Probing the multifactorial basis of Plasmodium falciparum quinine resistance: evidence for a strain-specific contribution of the sodium–proton exchanger PfNHE. Mol. Biochem. Parasitol. 2009;165:122–131. doi: 10.1016/j.molbiopara.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okombo J., Kiara S.M., Rono J., Mwai L., Pole L., Ohuma E., Borrmann S., Ochola L.I., Nzila A. In vitro activities of quinine and other antimalarials and pfnhe polymorphisms in Plasmodium isolates from Kenya. Antimicrob. Agents Chemother. 2010;54:3302–3307. doi: 10.1128/AAC.00325-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okombo J., Ohuma E., Picot S., Nzila A. Update on genetic markers of quinine resistance in Plasmodium falciparum. Mol. Biochem. Parasitol. 2011;177:77–82. doi: 10.1016/j.molbiopara.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Pelleau S., Bertaux L., Briolant S., Ferdig M.T., Sinou V., Pradines B., Parzy D., Jambou R. Differential association of Plasmodium falciparum Na+/H+ exchanger polymorphism and quinine responses in field- and culture-adapted isolates of Plasmodium falciparum. Antimicrob. Agents Chemother. 2011;55:5834–5841. doi: 10.1128/AAC.00477-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyssegur J., Sardet C., Franchi A., L’Allemain G., Paris S. A specific mutation abolishing Na+/H+ antiport activity in hamster fibroblasts precludes growth at neutral and acidic pH. Proc. Natl. Acad. Sci. USA. 1984;81:4833–4837. doi: 10.1073/pnas.81.15.4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyomtip T., Suwandittakul N., Sitthichot N., Khositnithikul R., Tan-ariya P., Mungthin M. Polymorphisms of the pfmdr1 but not the pfnhe-1 gene is associated with in vitro quinine sensitivity in Thai isolates of Plasmodium falciparum. Malar. J. 2012;11:7. doi: 10.1186/1475-2875-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradines B., Pistone T., Ezzedine K., Briolant S., Bertaux L., Receveur M.C., Parzy D., Millet P., Rogier C., Malvy D. Quinine-resistant malaria in traveler returning from Senegal, 2007. Emerg. Infect. Dis. 2011;16:546–548. doi: 10.3201/eid1603.091669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R.N., Uhlemann A.C., Brockman A., McGready R., Ashley E., Phaipun L., Patel R., Laing K., Looareesuwan S., White N.J., Nosten F., Krishna S. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukrittayakamee S., Supanaranond W., Looareesuwan S., Vanijanonta S., White N.J. Quinine in severe falciparum malaria: evidence of declining efficacy in Thailand. Trans. R. Soc. Trop. Med. Hyg. 1994;88:324–327. doi: 10.1016/0035-9203(94)90102-3. [DOI] [PubMed] [Google Scholar]
- Pukrittayakamee S., Chantra A., Vanijanonta S., Clemens R., Looareesuwan S., White N.J. Therapeutic responses to quinine and clindamycin in multidrug-resistant falciparum malaria. Antimicrob. Agents Chemother. 2000;44:2395–2398. doi: 10.1128/aac.44.9.2395-2398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney L.K., Barber D.L. Na–H exchange-dependent increase in intracellular pH times G2/M entry and transition. J. Biol. Chem. 2003;278:44645–44649. doi: 10.1074/jbc.M308099200. [DOI] [PubMed] [Google Scholar]
- Rahman M.R., Paul D.C., Rashid M., Ghosh A., Bangali A.M., Jalil M.A., Faiz M.A. A randomized controlled trial on the efficacy of alternative treatment regimens for uncomplicated falciparum malaria in a multidrug-resistant falciparum area of Bangladesh – narrowing the options for the National Malaria Control Programme? Trans. R. Soc. Trop. Med. Hyg. 2001;95:661–667. doi: 10.1016/s0035-9203(01)90108-7. [DOI] [PubMed] [Google Scholar]
- Raj D.K., Mu J., Jiang H., Kabat J., Singh S., Sullivan M., Fay M.P., McCutchan T.F., Su X.Z. Disruption of a Plasmodium falciparum multidrug resistance-associated protein (PfMRP) alters its fitness and transport of antimalarial drugs and glutathione. J. Biol. Chem. 2009;284:7687–7696. doi: 10.1074/jbc.M806944200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba K.J., Kirk K. pH regulation in the intracellular malaria parasite, Plasmodium falciparum. H(+) extrusion via a V-type h(+)-ATPase. J. Biol. Chem. 1999;274:33213–33219. doi: 10.1074/jbc.274.47.33213. [DOI] [PubMed] [Google Scholar]
- Simon F., Le Bras J., Charmot G., Girard P.M., Faucher C., Pichon F., Clair B. Severe chloroquine-resistant falciparum malaria in Gabon with decreased sensitivity to quinine. Trans. R. Soc. Trop. Med. Hyg. 1986;80:996–997. doi: 10.1016/0035-9203(86)90290-7. [DOI] [PubMed] [Google Scholar]
- Sinou V., Quang le H., Pelleau S., Huong V.N., Huong N.T., Tai le M., Bertaux L., Desbordes M., Latour C., Long L.Q., Thanh N.X., Parzy D. Polymorphism of Plasmodium falciparum Na(+)/H(+) exchanger is indicative of a low in vitro quinine susceptibility in isolates from Vietnam. Malar. J. 2011;10:164. doi: 10.1186/1475-2875-10-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spillman N.J., Allen R.J., Kirk K. Acid extrusion from the intraerythrocytic malaria parasite is not via a Na(+)/H(+) exchanger. Mol. Biochem. Parasitol. 2008;162:96–99. doi: 10.1016/j.molbiopara.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Valderramos S.G., Fidock D.A. Transporters involved in resistance to antimalarial drugs. Trends Pharmacol. Sci. 2006;27:594–601. doi: 10.1016/j.tips.2006.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak S., Alam M.T., Upadhyay M., Das M.K., Dev V., Singh N., Dash A.P., Sharma Y.D. Extensive genetic diversity in the Plasmodium falciparum Na+/H+ exchanger 1 transporter protein implicated in quinine resistance. Antimicrob. Agents Chemother. 2007;51:4508–4511. doi: 10.1128/AAC.00317-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warsame M., Wernsdorfer W.H., Willcox M., Kulane A.A., Bjorkman A. The changing pattern of Plasmodium falciparum susceptibility to chloroquine but not to mefloquine in a mesoendemic area of Somalia. Trans. R. Soc. Trop. Med. Hyg. 1991;85:200–203. doi: 10.1016/0035-9203(91)90018-t. [DOI] [PubMed] [Google Scholar]
- Wellems T.E., Hayton K., Fairhurst R.M. The impact of malaria parasitism: from corpuscles to communities. J. Clin. Invest. 2009;119:2496–2505. doi: 10.1172/JCI38307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsrichanalai C., Pickard A.L., Wernsdorfer W.H., Meshnick S.R. Epidemiology of drug-resistant malaria. Lancet Infect. Dis. 2002;2:209–218. doi: 10.1016/s1473-3099(02)00239-6. [DOI] [PubMed] [Google Scholar]
- Wootton J.C., Feng X., Ferdig M.T., Cooper R.A., Mu J., Baruch D.I., Magill A.J., Su X.Z. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- World Health Organization, 2010. Global report on antimalarial efficacy and drug resistance: 2000–2010. <http://whqlibdoc.who.int/publications/2010/9789241500470_eng.pdf> (accessed 15.06.12).
- World Health Organization, 2010. Guidelines for the treatment of malaria 2011. <http://whqlibdoc.who.int/publications/2010/9789241547925_eng.pdf> (accessed 15.06.12).
- Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19:395–420. [Google Scholar]


